Pseudomonas aeruginosa is an opportunistic pathogen that can cause infections in animals, humans, and plants. The formation of biofilms by P. aeruginosa is the central mode of action to persist in hosts and evade immune and antibiotic attacks. Cyclic-di-GMP (c-di-GMP) is an important second messenger involved in the regulation of biofilm formation. In P. aeruginosa PAO1 strain, there are around 40 genes that encode enzymes for making and breaking this dinucleotide. A major missing piece of information in this field is the phylogeny and expression profile of those genes. Here, we took a systemic approach to investigate this mystery. We found that among 40 c-di-GMP metabolizing genes, 5 have well-conserved phylogenetic distribution and invariable expression profiles, suggesting that there are enzymes required for the basal level of c-di-GMP in P. aeruginosa. This study thus provides putative therapeutic targets against P. aeruginosa infections.
KEYWORDS: diguanylate cyclases, phosphodiesterases, phylogeny, Pseudomonas aeruginosa
ABSTRACT
Cyclic diguanosine monophosphate (c-di-GMP) is an important second messenger involved in bacterial switching from motile to sessile lifestyles. In the opportunistic pathogen Pseudomonas aeruginosa, at least 40 genes are predicted to encode proteins for the making and breaking of this signal molecule. However, there is still paucity of information concerning the systemic expression pattern of these genes and the functions of uncharacterized genes. In this study, we analyzed the phylogenetic distribution of genes from P. aeruginosa that were predicted to have a GGDEF domain and found five genes (PA5487, PA0285, PA0290, PA4367, and PA5017) with highly conserved distribution across 52 public complete pseudomonad genomes. PA5487 was further characterized as a typical diguanylate cyclase (DGC) and was named dgcH. A systemic analysis of the gene expression data revealed that the expression of dgcH is highly invariable and that dgcH probably functions as a conserved gene to maintain the basal level of c-di-GMP, as reinforced by gene expression analyses. The other four conserved genes also had an expression pattern similar to that of dgcH. The functional analysis suggested that PA0290 encoded a DGC, while the others functioned as phosphodiesterases (PDEs). Our data revealed that there are five DGC and PDE genes that maintain the basal level of c-di-GMP in P. aeruginosa.
IMPORTANCE Pseudomonas aeruginosa is an opportunistic pathogen that can cause infections in animals, humans, and plants. The formation of biofilms by P. aeruginosa is the central mode of action to persist in hosts and evade immune and antibiotic attacks. Cyclic-di-GMP (c-di-GMP) is an important second messenger involved in the regulation of biofilm formation. In P. aeruginosa PAO1 strain, there are around 40 genes that encode enzymes for making and breaking this dinucleotide. A major missing piece of information in this field is the phylogeny and expression profile of those genes. Here, we took a systemic approach to investigate this mystery. We found that among 40 c-di-GMP metabolizing genes, 5 have well-conserved phylogenetic distribution and invariable expression profiles, suggesting that there are enzymes required for the basal level of c-di-GMP in P. aeruginosa. This study thus provides putative therapeutic targets against P. aeruginosa infections.
INTRODUCTION
Biofilms are communities of microorganisms embedded in extracellular polymeric substances (1). The biofilm protects microorganisms from hostile conditions and is the main feature of chronic and persistent infections (2). This biofilm mode of living is physiologically distinct from free-living planktonic status and can lead to high resistance to antibiotics, thus making infections refractory to medical treatments (3). The second messenger bis-(3′,5′)-cyclic dimeric GMP (c-di-GMP) plays an important role in biofilm formation (4). Generally, a high level of c-di-GMP stimulates the biosynthesis of adhesins and exopolysaccharides to enhance biofilm formation, and a low level of c-di-GMP is associated with an increase in motility and virulence (5). c-di-GMP is synthesized from two GTP molecules by diguanylate cyclases (DGCs) that harbor a conserved GGDEF domain and is hydrolyzed by c-di-GMP-specific phosphodiesterases with conserved EAL or HD-GYP domains, which degrade c-di-GMP to pGpG (5), followed by subsequently splitting into two GMP molecules via oligoribonuclease Orn (6, 7). To exert its function, c-di-GMP has to bind to downstream effectors and allosterically alter their structure and the output function (5, 8). In some bacteria, such as Pseudomonas aeruginosa, there are multiple genes that are predicted to have a conserved GGDEF domain (9), and the role and phylogenetic status of these genes remain largely unclear.
P. aeruginosa is a Gram-negative gammaproteobacterium that can grow in diverse habitats and acts as an opportunistic pathogen over a wide range of hosts, including humans, animals, and plants (10). This bacterium is notorious for its ability to cause chronic pulmonary infections and mortality in cystic fibrosis patients (2). The success of P. aeruginosa infections relies on both the production of acute virulence factors and the ability to form biofilms (11). At least three exopolysaccharides (Psl, Pel, and alginate) have been identified in P. aeruginosa biofilms, and they play distinct roles in structure maintenance and antibiotic resistance (1, 12). Recent evidence indicated that in addition to its physical role, the Psl exopolysaccharide could act as a signaling molecule to stimulate the biofilm formation by elevating the c-di-GMP level through two DGCs, SiaD and SadC (13). With the assistance of genomic analysis, 41 genes were predicted to be involved in the synthesis and degradation of c-di-GMP, encoding 17 different proteins with a GGDEF domain, 5 with an EAL or HD-GYP domain, and 16 with both types of domains (9). Functional roles have been investigated for several of these genes, including DGC-encoding genes such as PA0169 (siaD), PA1120 (yfiN), PA3702 (wspR), PA4332 (sadC), and PA4843 (adcA) and phosphodiesterase (PDE)-encoding genes such as PA2133, PA3947 (rocR), PA4367 (bifA), and PA5017 (pch) (14–21).
However, a few studies have focused on the comprehensive and systemic features of these c-di-GMP-related genes or proteins in bacteria (9, 22, 23). The first comprehensive studies of c-di-GMP signaling were performed in P. aeruginosa and showed that the c-di-GMP-related DGC and PDE proteins function in diverse patterns and demonstrate distinct phenotypes when mutated individually (9). The authors of that study further hypothesized that the formation and degradation of c-di-GMP are highly localized and linked to certain targets of c-di-GMP action. This was also confirmed by another study, which showed that the phenotypes controlled by two different DGC enzymes have discrete outputs despite the same total level of c-di-GMP, supporting the notion that localized c-di-GMP signaling likely contributes to coordination of the action of multiple proteins involved in the synthesis, degradation, and/or binding of this critical signal (22). In addition, systemic analyses of c-di-GMP signaling in Vibrio cholerae pointed out that c-di-GMP functions via a high-specificity signaling pathway characterized by individual DGCs or PDEs that are specifically associated with downstream c-di-GMP-mediated responses rather than via a simple, low-specificity signaling pathway (23).
In this study, we analyzed the phylogenetic distribution of all predicted DGC-encoding genes in P. aeruginosa PAO1 and found that PA5487 (named dgcH) showed a highly conserved distribution across the entire Pseudomonas genus. We systematically analyzed the expression data from more than 160 conditions deposited in the GEO database (https://www.ncbi.nlm.nih.gov/geo) and found that the expression of dgcH is invariable under all conditions and that dgcH probably functions as a conserved gene to generate c-di-GMP constantly. The typical functions of DgcH as a DGC were further investigated by phenotypic analyses. In addition, we show that DgcH is also implicated in virulence during plant infection. Finally, our results also reveal four other conserved c-di-GMP-related genes in pseudomonads.
RESULTS
Phylogenetic analysis suggested five conserved genes encoding proteins with the GGDEF domain in pseudomonads.
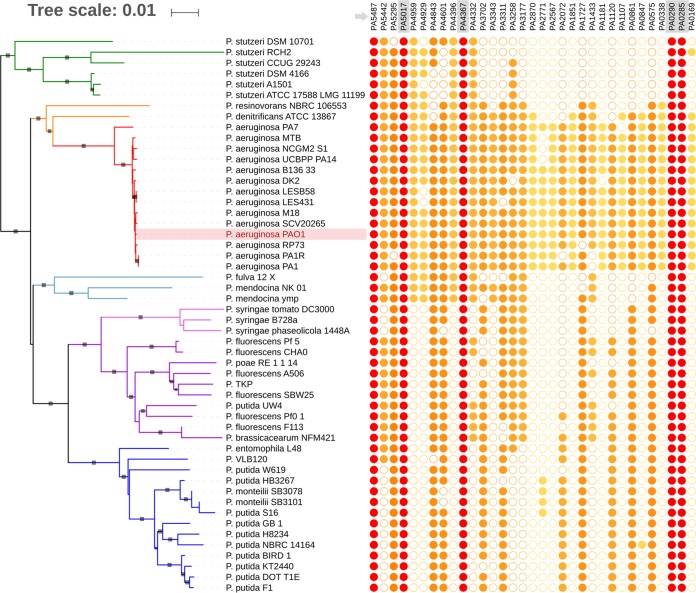
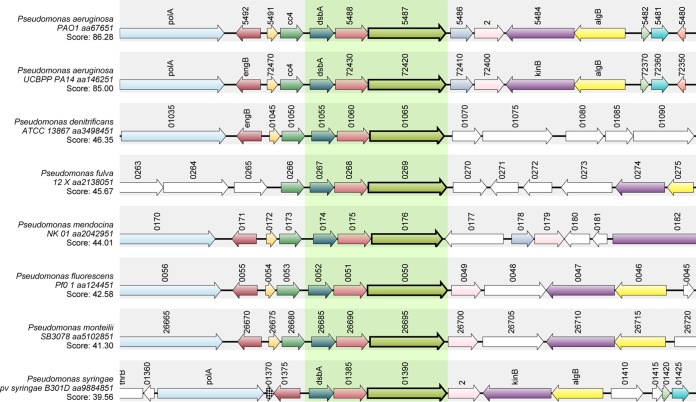
In P. aeruginosa PAO1, 33 genes were predicted to have a conserved GGDEF domain (9). The high redundancy of predicted DGCs in P. aeruginosa led us to investigate the phylogenetic distribution of those genes using 52 public complete pseudomonad genomes at the time of the study (24). We searched all 33 genes, encoding proteins with either a single GGDEF domain or hybrid with other domains such as EAL, PAS-PEC, and REC, against the complete pseudomonad genomes and found that several genes showed highly conserved distribution across tested Pseudomonas species. Among them, PA5487, which encoded a protein with a single GGDEF domain, was detected in the 52 pseudomonad genomes (Fig. 1, indicated by an arrow). In addition, another four genes—PA0285, PA0290, PA4367 (bifA), and PA5017 (pch) (Fig. 1, boxed)—were all conserved in tested pseudomonad genomes; two of them (PA4367 and PA5017) have been characterized previously (14, 21). In contrast, we noticed that several genes, such as PA2870, PA2771, and PA2587, are mainly present in P. aeruginosa, indicating that the phylogenetic distribution of DGC-encoding genes is highly variable in pseudomonads. To confirm the conserved status of PA5487, we compared its genomic context among all sequenced pseudomonad genomes in GenBank using SyntTax explorer (25) and found that across all typical selected Pseudomonas species, the genomic context of PA5487 was well conserved, particularly the PA5487-PA5488-dsbA gene cluster (Fig. 2, shown in the shaded region).
FIG 1.
Phylogenetic distribution of P. aeruginosa PAO1 DGC genes among pseudomonads. The unrooted pseudomonad phylogenetic tree was reconstructed using the neighbor-joining method from the concatenated alignment of seven housekeeping genes. The bootstrap support resulting from 100 replicates is indicated by a gray box when ≥90%; larger boxes represent higher bootstrap support (most being at 100%). The presence of an ortholog for each P. aeruginosa PAO1 DGC gene among pseudomonad genomes is indicated by a filled circle, and their absence is indicated by an open circle. Circle color is related to the distribution of the gene, from light yellow (specific to P. aeruginosa genomes) to red (found in all genomes).
FIG 2.
Synteny analysis of PA5487. Complex syntenies were obtained using the P. aeruginosa PA5487 protein sequence as a query sequence to search against the indicated pseudomonad species. Consistent color coding permitted the correct identification of both orthologs and paralogs. Genes corresponding to the query proteins are indicated in boldface and boxed with light green. The gene maps were produced by SyntTax in “.pdf” format and were edited for clarity.
PA5487-encoded protein functions as a bona fide DGC.
The presence of the GGDEF domain at the C terminus of PA5487 suggested that this gene might encode a functional DGC. To determine that PA5487 is an active and bona fide DGC in P. aeruginosa PAO1, we generated an in-frame deletion mutant of PA5487 and examined its cellular c-di-GMP level. The fluorescence-based reporter pCdrA::gfpC was used to detect the c-di-GMP concentration in both wild-type PAO1 and the PA5487 mutant. The relative green fluorescent protein (GFP) intensity was greatly reduced in the PA5487 mutant compared to that of wild-type PAO1 (Fig. 3A), indicating that PA5487 is indeed involved in the generation of c-di-GMP in P. aeruginosa. Furthermore, we extracted c-di-GMP from both PAO1 and PA5487 mutant strains and measured the c-di-GMP quantity using a high-pressure liquid chromatography–tandem mass spectrometry (HPLC-MS/MS) analysis pipeline. We found that the mutant produced much less c-di-GMP than the wild-type PAO1, suggesting that the PA5487-encoded protein is a functional DGC in P. aeruginosa PAO1 (Fig. 3B). Due to the highly conserved pattern of PA5487, we investigated its expression dynamics during the entire growth phase. The transcription of PA5487 increased, along with the growth curve, and demonstrated a precise cooccurrence with bacterial growth (Fig. 3C). This result showed that the expression of PA5487 was highly correlated to its growth phase, which is a common feature of conserved genes. Therefore, we proposed to name PA5487 as dgcH.
FIG 3.
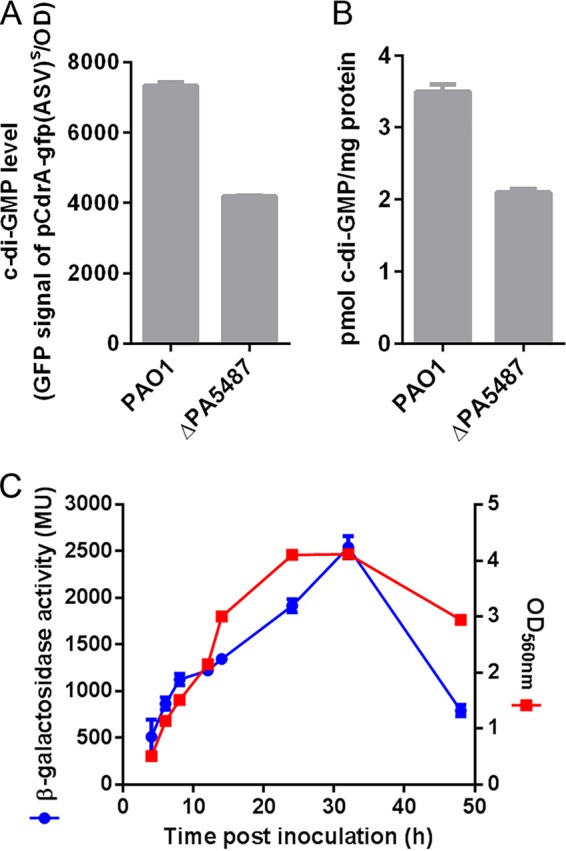
Expression of PA5487 and its effect on intracellular c-di-GMP. (A) c-di-GMP concentration measurement by fluorescence determination. The fluorescence from P. aeruginosa PAO1 and PA5487 mutant strains was measured, and the relative fluorescence unit values are arbitrary fluorescence intensity units corrected for cell density. The results are averages of triplicate measurements on test tube cultures in stationary growth phase (12 h). (B) c-di-GMP concentration measurement by HPLC-MS/MS determination. Bacterial cultures of PAO1 and the PA5487 mutant were collected from the stationary growth phase (12 h), and the c-di-GMP concentration was detected using the HPLC-MS/MS method as described in the text. Results are averages of triplicate measurements on test tube cultures. (C) The expression of PA5487 is consistent with its growth stage. PAO1::mini-CTX-dgcH-lacZ was used to detect the expression of PA5487, and control strain PAO1::mini-CTX-lacZ was used to calibrate the background expression. A standard β-galactosidase assay was performed in LB broth for 24 h. We also tested the expression in Jensen’s medium, which showed the same result as that using obtained LB medium (data not shown).
dgcH affects bacterial motilities, biofilm formation, and the production of the Psl exopolysaccharide.
The cellular c-di-GMP level impacts bacterial motility, biofilm formation, and exopolysaccharide production. We therefore sought to determine the effect of dgcH on these phenotypes. We first tested the swarming and swimming motility. The swarming zone of the dgcH mutant was significantly larger than that of wild-type strain PAO1, whereas the expression of dgcH on a high-copy-number plasmid totally inhibited bacterial swarming (Fig. 4A, upper panel). Consistently, the dgcH mutant formed a larger swimming zone than PAO1, and overexpression of dgcH inhibited the swimming motility. In contrast to what was observed for swarming motility, the swimming motility was not completely inhibited when dgcH was expressed in a plasmid (Fig. 4A, lower panel).
FIG 4.
dgcH affects bacterial motilities, biofilm formation, and the production of Psl exopolysaccharide. (A) Swarming and swimming motility analyses of dgcH and PAO1 strains containing either empty vector or wild-type dgcH. The diameters of both swarming and swimming strains were measured, and the standard errors are shown. (B) Biofilm formation analysis of the dgcH mutant and its complemented strain. Biomasses were detected for P. aeruginosa PAO1, the dgcH mutant, and its complemented strain with the wild-type dgcH gene in the dgcH mutant. CV staining was used, and three independent experiments were performed to calculate the final biomass formation. (C) Confocal laser scanning microscopy (CLSM) observation of biofilm formation of dgcH at 12 and 24 h. Pellicle biofilms after 1 day of growth for PAO1 and the dgcH mutant were observed and imaged by CLSM. Representative images are shown. The P. aeruginosa biofilms were stained with the green fluorescent probe SYTO9. (D) COMSTAT analysis of biofilm biomass and maximum thickness for both PAO1 and the dgcH mutant. Data from two different time points were chosen for analyses. (E) Psl dot blotting analysis of the dgcH mutant and its derivatives. A representative immune dot blotting result is shown above each corresponding column. The relative Psl production was calculated based on the intensity of blotted Psl dots.
We next examined the effect of dgcH on biofilm formation (Fig. 4B). The dgcH mutant significantly reduced the biofilm formation in a microtiter dish assay, and overexpression of dgcH could restore the biofilm formation of the ΔdgcH mutant to a higher level than that of PAO1 (Fig. 4B). Deletion of dgcH also decreased the pellicle formation (Fig. 4C), indicated by biofilm biomass and thickness (Fig. 4D). The pellicle formation of the dgcH mutant displayed a delayed and reduced level at different time points (12 h versus 24 h) compared to that of its parental wild-type strain PAO1 (Fig. 4C).
Psl exopolysaccharide is a key matrix component of P. aeruginosa PAO1 biofilms. We reasoned that the reduction in biofilm formation would be associated with the decreased Psl polysaccharide synthesis. Therefore, we examined the contribution of dgcH to the synthesis of Psl polysaccharide. A custom-made anti-Psl serum was generated to detect the intensity of Psl polysaccharide (Fig. 4E). The deletion of dgcH reduced the Psl polysaccharide production, and complementation of wild-type dgcH could greatly promote Psl exopolysaccharide synthesis (Fig. 4E), in agreement with the biofilm phenotype. Altogether, we showed that dgcH deletion enhanced motilities and reduced biofilm formation and exopolysaccharide synthesis in P. aeruginosa, which was consistent with the function of a DGC.
The dgcH mutant has elevated virulence.
We further evaluated the effect of dgcH on virulence by using Chinese cabbage leaves as an infection model (Fig. 5). The wild-type P. aeruginosa PAO1 strain inoculated into the middle rib of the Chinese cabbage was able to cause typical necrosis by 3 days after inoculation. However, the dgcH mutant could induce more severe necrosis after inoculation. Complementation with dgcH in trans partially restored the virulence of the dgcH mutant toward Chinese cabbage (Fig. 5). Interestingly, overexpression of dgcH in the wild-type P. aeruginosa PAO1 strain led to a complete loss of necrosis 3 days after inoculation. In addition, enumeration of P. aeruginosa cells from infected plants indicated that the numbers of bacterial cells in plants were proportional to the necrosis area (R2 = 0.97). Previous reports showed that a reduction of cellular c-di-GMP level enhanced bacterial virulence (26). The increased virulence of the ΔdgcH mutant was also in agreement with its impact on cellular c-di-GMP level (Fig. 3A and B).
FIG 5.
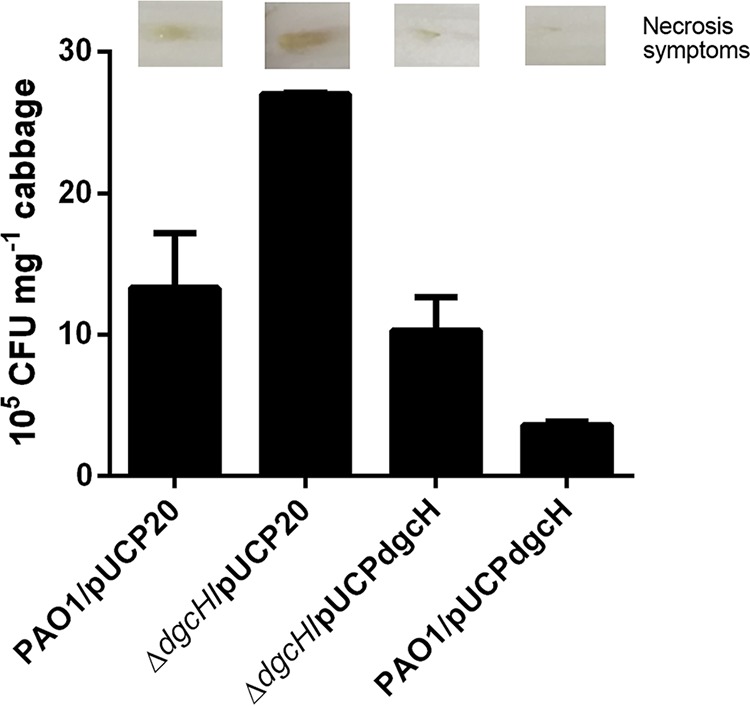
The dgcH mutant has elevated virulence. The virulence of the wild type, the dgcH mutant, and the dgcH mutant complemented with wild-type dgcH in a Chinese cabbage leaf virulence assay was assessed. The numbers of bacterial cells (indicated as CFU) present in 1 mg of cabbage midrib at 3 days postinjection are shown. Error bars were calculated from three independent experiments. A representative picture of infected midribs is also shown for each strain.
Meta-analyses of P. aeruginosa transcriptomes further suggest that DgcH is an invariable DGC.
Given the conserved distribution of dgcH in pseudomonad species, we hypothesized that DgcH functions as a conserved DGC to maintain a cellular c-di-GMP pool in P. aeruginosa. To test this hypothesis, we performed a meta-analysis of the DGC-encoding gene transcription from the GEO database, which provides a large amount of gene expression data related to P. aeruginosa studies. After quality control and normalization of the published data sets, we were able to identify 85 Affymetrix data sets, which contain 162 different conditions for P. aeruginosa studies (see Table S1 in the supplemental material), including different temperatures, growth media, antibiotic treatments, and oxidative stresses. Analysis of these data sets showed that the transcription level of dgcH showed almost no significant changes under all experimental conditions, except under one condition (Fig. 6A, black line), where the fold change of dgcH is −1.3 (log2 scale, Table S1; Fig. 6A, indicated by an arrow). However, as a control, the other well-studied DGC-encoding gene, mucR, exhibited a highly variable transcriptional pattern under all tested conditions (27) (Fig. 6A, gray line).
FIG 6.
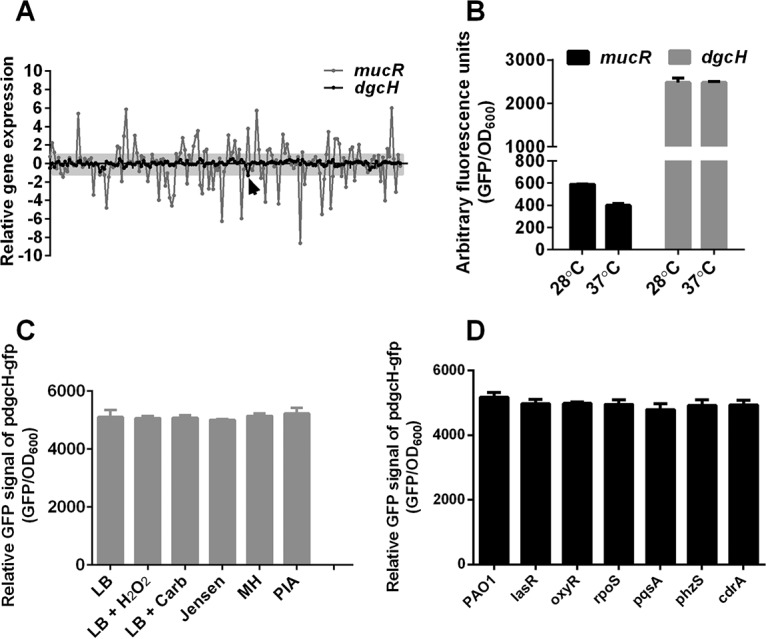
Meta-analyses of P. aeruginosa transcriptomes suggest that DgcH is an invariable DGC. (A) Relative expression profiling patterns of dgcH in P. aeruginosa meta-array analyses. In total, 162 conditions were tested for expression analyses of dgcH, and we found that there was no significant difference within these conditions except one. The x axis represents 162 different conditions, and the y axis indicates the expression values calculated from a meta-analysis of the GEO data sets. The expression value is on a log2 scale. (B) mucR and dgcH expression in different temperatures. Temperatures of 28 and 37°C were used to detect the expression of mucR and dgcH promoter fusion, with GFP as the readout. (C) dgcH expression under different environmental conditions. Freshly inoculated P. aeruginosa cells were treated with a final concentration of 10 mM H2O2 and 300 μg/ml carbenicillin, grown at 28°C, and incubated with different nutritional media, namely, Jensen’s medium, MH medium (BD), and PIA medium (BD). (D) dgcH expression in different genetic backgrounds. The GFP reporter strain pProbe-dgcH was conjugated into multiple mutant strains of P. aeruginosa and tested for promoter activity of dgcH in these strains. (B to D) Arbitrary fluorescence units were defined by the GFP intensity calibrated using the OD600.
To verify the transcriptional pattern of dgcH, we constructed a reporter plasmid carrying the transcriptional fusion of the dgcH promoter driving the expression of gfp (Table 1, pdgcH-gfp). We compared first the impact of growth temperature on the transcription of mucR and dgcH. The transcription of dgcH was much higher (4- to 6-fold) than that of mucR and yet underwent little change at two temperature (28 and 37°C), whereas mucR exhibited a distinct gene expression pattern (Fig. 6B). We next examined the transcription of dgcH under different physiological conditions, such as oxidative stress treatment (10 mM H2O2), antibiotic treatment (carbenicillin), and growth media (Luria-Bertani [LB], Jensen, Mueller-Hinton [MH], and Pseudomonas isolation agar [PIA] medium) (Fig. 6C). No significant changes were observed under these conditions compared to the untreated control, indicating that dgcH transcription is not influenced by these physiological conditions. In addition, we also randomly selected several gene deletion mutants to examine whether the dgcH transcription is changed in these mutants. These genes encoded important transcriptional regulators and signaling factors in P. aeruginosa, including the quorum-sensing (QS) regulator LasR, the oxidative stress response regulator OxyR, the stress response-related σ factor RpoS, the PQS signal synthesis protein PqsA, monooxygenase PhzS, and c-di-GMP-responsive adhesin CdrA. Our results showed that the expression levels of dgcH were similar in all of these deletion mutant strains (Fig. 6D). These data are consistent with our meta-analysis of the Affymetrix data sets. Taken together, the transcriptional pattern of dgcH further suggested that DgcH is an invariable DGC.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, primer, or plasmid | Description or sequencea | Source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Prototroph | Lab collection |
| ΔPA5487 | PAO1 with in-frame deletion of PA5487 gene; ΔdgcH | This study |
| ΔPA0285 | PAO1 with in-frame deletion of PA0285 gene | This study |
| ΔPA0290 | PAO1 with in-frame deletion of PA0290 gene | This study |
| ΔPA4367 | PAO1 with in-frame deletion of PA4367 gene | This study |
| ΔPA5217 | PAO1 with in-frame deletion of PA5217 gene | This study |
| PAO1/pUCP20 | PAO1 containing the empty vector pUCP20 (Cbr) | This study |
| PAO1/pUCPdgcH | PAO1 containing the recombinant vector pUCPdgcH (Cbr) | This study |
| ΔdgcH/pUCP20 | ΔdgcH containing the empty vector pUCP20 (Cbr) | This study |
| ΔdgcH/pUCPdgcH | ΔdgcH containing the recombinant vector pUCPdgcH (Cbr) | This study |
| PAO1/pProbe-AT’ | PAO1 containing the promoterless gfp reporter plasmid pProbe-AT′ (Cbr) | This study |
| PAO1/pdgcH-gfp | PAO1 containing dgcH-gfp reporter plasmid pProbe-dgcH (Cbr) | This study |
| PAO1/pmucR-gfp | PAO1 containing mucR-gfp reporter plasmid pProbe-mucR (Cbr) | This study |
| phzS/pdgcH-gfp | phzS containing dgcH-gfp reporter plasmid pProbe-dgcH (Cbr) | This study |
| oxyR/pdgcH-gfp | oxyR containing dgcH-gfp reporter plasmid pProbe-dgcH (Cbr) | This study |
| rpoS/pdgcH-gfp | rpoS containing dgcH-gfp reporter plasmid pProbe-dgcH (Cbr) | This study |
| cdrA/pdgcH-gfp | cdrA containing dgcH-gfp reporter plasmid pProbe-dgcH (Cbr) | This study |
| pqsA/pdgcH-gfp | pqsA containing dgcH-gfp reporter plasmid pProbe-dgcH (Cbr) | This study |
| lasR/pdgcH-gfp | lasR containing dgcH-gfp reporter plasmid pProbe-dgcH (Cbr) | This study |
| PAO1::mini-CTX-lacZ | PAO1 with mini-CTX-lacZ integrated in the chromosome | This study |
| PAO1::mini-CTX-dgcH-lacZ | PAO1 with mini-CTX-dgcH-lacZ integrated in the chromosome | This study |
| E. coli | ||
| S17-1 λpir | thi pro hsdR hsdM+ recA RP4-2-Tc::Mu-Km::Tn7 λpir | Lab collection |
| DH5α | recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 (lacZYA-argF)U169 [ϕ80dlacZΔM15] | Lab collection |
| Plasmids | ||
| pUCP20 | E. coli-P. aeruginosa shuttle plasmid (Apr) | Lab collection |
| pProbe-AT′ | Promoterless gfp reporter plasmid (Apr) | Lab collection |
| pUCPdgcH | pUCP20 containing coding sequence of dgcH gene (Apr) | This study |
| pProbe-dgcH | Promoter sequence of dgcH inserted into gfp reporter plasmid pProbe-AT′ (Apr) | This study |
| pProbe-mucR | Promoter sequence of mucR inserted into gfp reporter plasmid pProbe-AT′ (Apr) | This study |
| Mini-CTX lacZ | Self-proficient integration vector with tet, Ω-FRT-attP-MCS, ori, int, and oriT (Tcr) | Lab collection |
| Mini-CTX-dgcH-lacZ | Promoter of dgcH cloned in mini-CTX-lacZ (Tcr) | This study |
| pFLP2 | FLP recombinase expressing plasmid (Apr) | Lab collection |
| pEX18Ap | Gene replacement vector for P. aeruginosa (Apr) | Lab collection |
| pEX18Ap-dgcH | Upstream and downstream of dgcH cloned into pEX18Ap (Apr) | This study |
| pCdrA::gfp(ASV)s | c-di-GMP reporter plasmid | Lab collection |
| Primers | ||
| PA5487-UPFw | GCTCTAGAGCTGGAAGATCATCCGTTGC | |
| PA5487-UPRv | GAAATGGAACGGACACTCCTGCTGCTCGATGTTC | |
| PA5487-DwFw | ATCGAGCAGCAGGAGTGTCCGTTCCATTTCAAGGG | |
| PA5487-DwRv | CCCAAGCTTTGAGGAAGAGCAGCAGCAG | |
| PA5487-Comp-UP | GGAATTCATGAGTCGCGACGACGTCCAG | |
| PA5487-Comp-Dw | CGGGATCCCCTTCAGGCCACTTCCAGG | |
| PA5487-proFw | GGAATTCGTCCAGCATGTGACCAACG | |
| PA5487-proRv | CGGGATCCCAGCATGGCGGTGAGAATC | |
| PA0285-UPFw | GCTCTAGACTGTGGTCAACCACAACCAG | |
| PA0285-UPRv | GTACCAGTAGCCTTGGATGGCGATTGCGAGATAAGC | |
| PA0285-DwFw | CTCGCAATCGCCATCCAAGGCTACTGGTACGGAC | |
| PA0285-DwRv | CCCAAGCTTGATGGGGTATCGAACGGATG | |
| PA0290-UPFw | GCTCTAGACATCGCTACCCTGGTCTTC | |
| PA0290-UPRv | GATGCAGTCCTTGCCGGTTAGGTCGTCCATGGATC | |
| PA0290-DwFw | ATGGACGACCTAACCGGCAAGGACTGCATCGTCGT | |
| PA0290-DwRv | CCCAAGCTTGAACGGAAGCGTGTGCATG | |
| PA4367-UPFw | GCTCTAGAGCTGGCTGGTGAAGAAGG | |
| PA4367-UPRv | GCTTGCTGTACAGGTAGCCTTGAGCGAGAGGCTGTG | |
| PA4367-DwFw | CACAGCCTCTCGCTCAAGGCTACCTGTACAGCAAGC | |
| PA4367-DwRv | CCCAAGCTTCGATCGATACCCTGCGAC | |
| PA5017-UPFw | GCTCTAGA CAGCGAATGCCACAGGAC | |
| PA5017-UPRv | GACTACCTTGAGCTTGAGGGGTAGCTGCGTGACAACC | |
| PA5017-DwFw | GGTTGTCACGCAGCTACCCCTCAAGCTCAAGGTAGTC | |
| PA5017-DwRv | CCCAAGCTTCTTGAGCAGCTGTTCGAAGC |
Apr, ampicillin resistance; Tcr, tetracycline resistance; Cbr, carbenicillin resistance.
Characterization of other housekeeping DGCs and PDEs in P. aeruginosa.
In addition to dgcH, PA0285, PA0290, PA4367 (bifA), and PA5017 (pch) were all conserved in pseudomonads (Fig. 1, boxed). Among these four genes, PA0290 has only one GGDEF conserved domain, while other the three genes contain a conserved EAL domain together with the GGDEF domain (Fig. 7A). We have also unraveled the expression pattern of the four genes, which all displayed invariable expression under 162 conditions (Fig. 7A). We constructed in-frame deletion mutants in PAO1 for all of them and measured the biofilm-related phenotypes, including motilities, Psl production, and biofilm formation. As shown in Fig. 7B and C, the deletion of PA0285, PA4367 (bifA), and PA5017 (pch) led to increased biofilm formation and Psl production, typical features of EAL genes in P. aeruginosa, which was consistent with the previous reports on bifA (20) and pch (21) in P. aeruginosa strain PA14. In addition, we also determined that PA0290 is a bona fide DGC since it positively regulates biofilm formation and Psl production but negatively controls the motilities of P. aeruginosa (data not shown). Altogether, our results suggested that there were five DGC and PDE genes that maintained the basal level of c-di-GMP in P. aeruginosa.
FIG 7.

Characterization of other conserved genes in P. aeruginosa. (A) Relative expression profiling patterns of PA0285, PA0290, PA4367, and PA5017 in P. aeruginosa meta-array analyses. In total, 162 conditions were tested for expression analyses of all four genes, and we found that there was no significant difference within these conditions. The x axis represents 162 different conditions, and the y axis indicates the expression values calculated from a meta-analysis of the GEO data sets. The expression value is shown on a log2 scale. (B) Relative biofilm analysis of four DGC gene mutants. The biomasses were detected for P. aeruginosa PAO1 and DGC mutants; CV staining was used and three independent experiments were performed to calculate the final biomass formation. The results are presented as the biofilm formation relative to wild-type PAO1. (C) Relative Psl production of four DGC gene mutants. The diameters of swimming bacteria were measured, and relative swarming was calculated by evaluating the wild-type PAO1. The standard errors are shown.
DISCUSSION
Bacterial second messenger c-di-GMP has received extensive attention during the last half century and been referred to as Sleeping Beauty in bacterial signal transduction and regulation (4, 5, 11). In the opportunistic pathogen P. aeruginosa, 41 genes are predicted to be responsible for the synthesis and degradation of c-di-GMP (9). Extensive studies have been undertaken to study their functions by individually deleting these genes and dissecting the biological functions in respect to biofilm formation and motilities (9, 15, 16, 21, 28–30).
In this study, we investigated the phylogenetic distribution and the expression pattern of these c-di-GMP-metabolizing genes and found that several of them are highly conserved in all tested pseudomonad genomes. Furthermore, we also noticed that some genes are relatively less conserved or even specifically found only in certain species, e.g., P. aeruginosa. These results raised the possibility that some c-di-GMP-metabolizing genes are generally acting as conserved genes, while other genes that were specifically distributed may function in certain fixed pathways. Here, we focused on dgcH, which is found in the investigated Pseudomonas species, at a conserved genomic position. In addition, when we detected the expression pattern of the c-di-GMP-metabolizing genes using GEO data sets, we found another striking feature for dgcH: among 160 conditions, no significant changes in gene transcription were observed. Both traits indicated that dgcH may function as a basic c-di-GMP generating gene that stably produces a general c-di-GMP pool in P. aeruginosa. Our study presents the phylogenetic distribution and expression landscape of c-di-GMP-metabolizing genes in P. aeruginosa and sheds light on the biological functions of dgcH. Since the overwhelming number of these c-di-GMP-metabolizing genes are found in Gram-negative bacteria, it is vital to apply phylogenetic tools or use omics data sets to assist the functional analysis of these genes.
To further prove the DGC activity, we detected the c-di-GMP using different methods. Interestingly, more genes could have contributed to the c-di-GMP pool since when we measured the c-di-GMP level, it was not totally abolished in the dgcH mutant. The biological functions of dgcH were also studied, and we found that dgcH can regulate the switch from a motile to a sessile lifestyle. This is reminiscent of pioneering work on sadC in P. aeruginosa PA14 or PAK strains (13, 19, 22, 31, 32). In these studies, the function of sadC was thoroughly studied and revealed that SadC is a membrane-associated DGC that could control the switch between biofilm formation and motility by regulating Pel polysaccharide synthesis (31). In addition, SadC was demonstrated to be involved in the GacS/GacA/RsmA-mediated biofilm formation pathway, and this involvement is very specific. DgcH was chosen in this study as one of the negative controls, and overexpression of the small RNA-binding protein RsmA exhibited no effect on its expression; this further confirmed our conclusion that DgcH acts as one of the conserved enzymes in P. aeruginosa (33).
It was also interesting that there is a correlation between biofilm formation and Psl polysaccharide synthesis in the P. aeruginosa PAO1 strain. In PA14, the increased biofilm formation was correlated with the enhanced production of Pel polysaccharide that was caused by the overproduction of SadC (22). It was further proved that PelD is the c-di-GMP receptor that mediates c-di-GMP regulation of Pel polysaccharide biosynthesis, explaining the regulatory mechanism of c-di-GMP in promoting Pel synthesis and biofilm formation (34). However, in our study, we found that the overexpression of dgcH could promote the Psl polysaccharide synthesis and biofilm formation. It is still a mystery, however, whether a c-di-GMP receptor is located in the psl operon or in another genomic locus in P. aeruginosa.
Furthermore, we also tested the virulence phenotype of the dgcH mutant in a plant model. We found that the dgcH mutant, which has reduced c-di-GMP levels, showed enhanced necrosis after injection into the ribs of Chinese cabbage. In contrast, the overexpression of dgcH, which has increased c-di-GMP levels, led to attenuated virulence in a plant model. This confirmed the conclusion that high c-di-GMP levels repress virulence (5). In support of this conclusion, our results further confirmed that dgcH is really important for virulence, as evidenced by the inoculation of a dgcH homologue mutant in a murine model of acute infection (9). In that study, the researchers used PA14 as a type strain and tested several DGC mutants; they found a lack of correlation between cytotoxicity and virulence in mice and suggested that some other DGCs may have roles not involved in cytotoxicity in P. aeruginosa.
It is also interesting that the dgcH mutant has a lower level of c-di-GMP, which is linked with lower biofilm formation seen for the mutant than for wild-type PAO1. However, in the virulence experiment, the dgcH mutant was more virulent than the wild-type PAO1. Indeed, a lower level of c-di-GMP was shown to correlate with more virulence in acute infection models (9). Further evidence showed that low c-di-GMP levels could increase the expression of both rhl and pqs QS system-regulated genes (26). In addition to upregulated QS-regulated genes, the increased swimming and swarming motilities of dgcH would also contribute to more virulence in plant and animal models. Although the biofilm formation is positively correlated with virulence, the stronger biofilm is believed to be connected with chronic infection but not with acute infection. In the present experiment, we only used an acute infection model to examine the virulence of the dgcH mutant. It would be intriguing to evaluate the virulence phenotype in a chronic infection model.
The meta-analyses of the expression pattern of DGC genes provided us with a tremendous amount of interesting information. In regard to the dgcH gene, we found that its expression pattern is highly invariable and that only under one condition did it show a relative 1.3-fold reduction in expression level. When we verified the expression of dgcH in randomly selected mutants, we further proved its stable expression pattern, suggesting that dgcH is an invariable gene and could function as a c-di-GMP generator under all circumstances. However, in our study, we focused primarily on the deposited microarray data sets in the GEO database. With the introduction of transcriptome sequencing (RNA-seq) technology, there are many more interesting data concerning the expression of P. aeruginosa genes. Therefore, it would be intriguing to know whether our data are consistent with those upcoming data sets, and analysis of increasing data sets would definitely enhance our conclusions and reveal more important findings.
Finally, other than dgcH, we also showed that there are several other genes with a conserved distribution across the entire Pseudomonas genus, suggesting that there are also conserved enzymes controlling the basic c-di-GMP pool in P. aeruginosa. Surprisingly, when we deleted these four genes, we found only one bona fide DGC gene with a DGC activity, whereas the rest of them exhibited PDE activity. This is perhaps due to the existence of additional EAL domains in these three genes, leading to a PDE activity masking the DGC activity. Another possibility would be that their DGC activity is silenced by other, as-yet-unknown genetic factors. Further investigations will focus on the functions of these genes in the maintenance of the basic c-di-GMP pool in P. aeruginosa.
All in all, we have undertaken comprehensive phylogenetic and expression analyses of c-di-GMP-metabolizing genes in P. aeruginosa. The conclusions from this study will provide insight into the development of therapeutic agents against bacterial infections, especially those with dozens of c-di-GMP-metabolizing genes.
MATERIALS AND METHODS
Strains and growth conditions.
Strains, plasmids, and primers used in this study are listed in Table 1 . P. aeruginosa strain PAO1 was used in the study. Unless otherwise stated, plasmid-harboring strains were selected and maintained on Jensen’s (a chemically defined medium) plates or LB medium containing appropriate antibiotics (35). Biofilms of P. aeruginosa were cultured in Jensen’s medium at 30°C.
Phylogenetic distribution analysis of DGC genes.
The 52 complete pseudomonad genomes available in GenBank in April 2014 were downloaded, and the DNA sequences of the seven housekeeping genes gyrB, rpoB, rpoD, ftsZ, groEL, gmk, and tpiA were extracted from each genome. An alignment was produced for each gene independently using MAFFT v7.271 (36), and all resulting alignments were concatenated as a superalignment. Genes absent in some genomes were replaced by stretches of gaps in the length of the gene. A neighbor-joining unrooted phylogenetic tree was then estimated using MEGA v.6.6 (37) with default parameters, except a 75% partial deletion threshold. Orthologs of the 33 DGC proteins defined for P. aeruginosa PAO1 in Kulasekara et al. (9) were searched within each of the 52 pseudomonad genomes using TBLASTN, with a minimum identity of 55% on at least 66% of the sequence. Using these parameters, all genes matched at most once within each genome; these matches were therefore considered orthologs. Lowering the minimum identity threshold to 50% led to the erroneous identification of additional DGC sequences other than those of the orthologs. The phylogenetic tree and the distribution of DGC orthologs among genomes were generated using iTOL v.3.4.3 (38). The synteny of dgcH was determined according to the protocols described in the SyntTax documentation (25).
Meta-analysis of GEO data sets.
The expression profiling of 41 c-di-GMP-metabolizing genes was performed using custom-made R scripts. Briefly, the protocols were as follows. (i) First, we extracted data from the publicly available expression profiling data sets on P. aeruginosa in the GEO database within the NCBI database. Since the start of this project, most of the data sets deposited in GEO are generated from the Affymetrix Pseudomonas aeruginosa array (GLP84 chip); only these types of data sets were chosen for further analysis. (ii) Second, we filtered these data sets and obtained the raw data for 116 GSE data sets. (iii) We instituted quality control (QC) and normalization processes for these Affymetrix data sets. After QC and removal of poor-quality samples, MAS5 normalization implemented in the “Affy” package was used to carry out the normalization to obtain expression values for all 5,900 probe sets on the GLP84 chips, and all of the chips were scaled to a trimmed mean value of 100. (iv) Meta-analysis of the obtained expression profiles was performed, in particular for the 41 genes of interest, which included 33 DGC genes.
Molecular manipulation.
Isolation of plasmid and genomic DNA from Escherichia coli and restriction enzyme digestion were performed according to the manufacturer’s instructions (Qiagen). DNA cloning, transformations, and agarose gel electrophoresis were performed as previously described (39). An in-frame and unmarked dgcH deletion mutant in P. aeruginosa PAO1 was constructed using the suicide vector pEX18Ap. This vector contains the lethal sacB gene encoding levansucrase, and recombination of the vector onto the chromosome confers ampicillin resistance to the host strain. Approximately 500 bp upstream and downstream sequences flanking the gene of interest were amplified by PCR and fused with overlapping PCR. The PCR product was excised from the agarose gel and cloned into the pEX18Ap vector, resulting in the plasmid pEX18Ap-dgcH. The pEX18Ap-based deletion allele was mobilized to P. aeruginosa and integrated into the chromosome by a single crossover using E. coli S17-1 λpir as the delivery strain. Double-crossover events were subsequently selected by growth in the presence of 5% sucrose. Deletion mutants were confirmed by both PCR and sequencing. Complementation analysis was accomplished by cloning the intact dgcH gene into the shuttle vector pUCP20 to generate pUCPdgcH and introducing it into the deletion mutants.
Promoter activity assay.
The GFP reporter plasmid pProbe-AT′ was used to construct transcriptional fusions with the dgcH promoter (40). The promoter region was amplified by PCR. The DNA fragment was digested and cloned into digested pProbe-AT′. The resulting plasmids were mobilized into P. aeruginosa by conjugation using E. coli S17-1 as the donor strain. The promoter activity was evaluated by measuring GFP intensity via recording the optical density at 600 nm (OD600) values, and the corresponding GFP fluorescence was captured with a Synergy H4 hybrid reader (Ex/Em 488/520; BioTek).
c-di-GMP detection.
The c-di-GMP levels of different strains were determined according to a previously described method using pCdrA::gfp as a reporter (41). The growth curve and green fluorescent signal of PAO1/pCdrA::gfp and ΔdgcH/pCdrA::gfp were measured by a Synergy H4 hybrid reader (BioTek) via recording the OD600 values, and the corresponding GFP fluorescence captured (Ex/Em 488/520). The promoter activity was calibrated with the OD600 and is expressed as the relative fluorescence divided by the OD600.
Determination of c-di-GMP was further conducted as previously described (17). Strains were incubated in Jensen’s medium for 24 h. A portion (1 ml) of bacterial culture was used for protein quantification by a BCA assay (Thermo), and 10 ml of medium was harvested and lysed with 0.3 ml of 0.6 M HClO4 on ice for 30 min. Cell debris was removed by centrifugation, and the supernatants were neutralized to pH 6.0 by the addition of 2.5 M K2CO3. The precipitated KClO4 was removed by centrifugation, and the supernatant was used for relative quantification of the c-di-GMP. Samples were analyzed by using an HPLC-MS/MS system. Separation was achieved with an HPLC apparatus (Agilent 1200 series) using a C18 column (4.6 by 250 mm; Epic). Temperature was uncontrolled, the flow rate was set up to 1 ml/min, and the cycle time was 22 min. The c-di-GMP level was detected by MS/MS multiple reaction monitoring using an Agilent 6460 Triple-Quad mass spectrometer in positive electrospray ionization. The m/z 691 > 152 transition was used for quantification; 691 > 248 and 691 > 540 were monitored as confirmatory signals. The collision energies were 30, 24, and 24 eV, respectively; the fragmentor was set to 225 V in all cases.
dgcH transcriptional fusion strain construction.
A 956-bp DNA fragment containing the promoter of the dgcH operon from the PAO1 chromosome was obtained by PCR using the primers PA5487-proFw (5′-GGAATTCGTCCAGCATGTGACCAACG-3′) and PA5487-proRv (5′-CGGGATCCCAGCATGGCGGTGAGAATC-3′) (underlining denotes the restriction enzyme sites). The PCR product and mini-CTX-lacZ vector were doubly digested with EcoRI and BamHI and ligated to generate fusion plasmid. The recombinant plasmids were conjugated into the chromosomal attB sites of P. aeruginosa strains used in this study, as described previously (42).
β-Galactosidase assays.
The β-galactosidase activity was quantitatively assayed as described elsewhere, with some modifications (43). P. aeruginosa strains were grown in nutrient broth liquid medium with or without arabinose at 37°C with shaking at 250 rpm for 2 days. One-milliliter culture aliquots were resuspended in 200 μl of Z-buffer (16.1 g/liter Na2HPO4·7H2O, 5.5 g/liter NaH2PO4·7H2O, 0.75 g/liter KCl, 0.24 g/liter MgSO4·7H2O, pH 7.0) and frozen/thawed three times to lyse the bacteria. Cell lysates were assayed for both the β-galactosidase activity and the total protein content using a BCA assay kit (Pierce). All β-galactosidase activity units were normalized based on the total protein per ml in aliquots. All samples were tested in triplicate.
Biofilm formation detection.
Microtiter dish biofilm assay was performed as previously described (44). Each well of a polyvinyl chloride plate contained 100 μl of Jensen’s medium with different concentrations of arabinose. Strains grown for 24 h in Jensen’s culture were incubated in wells to a final OD600 of 0.01. The plates were incubated at 30°C for different times, followed by staining with 0.1% crystal violet (CV) at 30°C for 30 min. CV was dissolved with 30% acetic acid and quantified by measuring the OD560.
Confocal laser scanning microscopy observation of biofilms.
To assess the air-liquid interface biofilms, pellicles were grown in glass chambers with individual chamber dimensions of 1 × 1 × 4 cm (chambered no. 1.5 German cover glass system; Nunc, Inc.). To observe the structures of the pellicles, SYTO9 (Molecular Probes, Invitrogen) was used to stain the pellicles, and then buffers were removed from glass chambers to allow the pellicles to drop onto the coverslips. Fluorescent and differential interference contrast images were acquired using a FV1000 confocal laser scanning microscope (Olympus, Japan). The biofilm biomass and thickness were quantified by using COMSTAT software (45).
Motility assays.
A swimming assay was performed as previously described (46). A single colony was stabbed into a Jensen’s 0.3% agar plate. After incubation at 37°C overnight, the swimming zone was measured. Swarming motility was assessed according to previously described methods (46). Twitching motility was assayed by stabbing strains through thin Jensen’s 1% agar medium (47).
Psl blotting.
Strains were incubated in Jensen’s liquid culture for 24 h. Crude bacterial surface-bound polysaccharide extracts were extracted from cultures with an OD600 of 10, resuspended in 100 μl of 0.5 M EDTA, and boiled at 100°C for 5 min. The protein concentration was measured by a BCA protein assay kit (Thermo) to ensure the same amount of cell lysate was used in each experiment. After centrifugation at 13,000 × g for 10 min, the supernatant fraction was transferred and treated with 0.5 mg/ml proteinase K at 60°C for 1 h and then at 80°C for 30 min to inactivate the proteinase K. Immunoblotting was performed as previously described (48). ImageJ software was used to quantify the immunoblot data (49).
Virulence assays.
A virulence test was performed as described previously with modifications (50). P. aeruginosa strains were grown overnight at 37°C in LB medium until they reached the stationary phase. After centrifugation, the bacterial cells were washed in 10 mM MgSO4 and diluted to 108 CFU ml−1, and then 10 μl of this suspension was injected with a syringe into the midrib of Chinese cabbage (Brassica rapa subsp. pekinensis) leaves, previously washed with 5% bleach. The leaves were placed on dishes containing a Whatman filter impregnated with 10 mM MgSO4. The plates were kept in a growth chamber at 37°C, and rot symptoms were monitored for 3 days. The experiments were repeated at least three times on independent days.
Statistical analysis.
The data for the c-di-GMP level, biofilm formation, biomass and minimum thickness, relative Psl production, virulence test, and expression verification were analyzed by one-way analysis of variance (ANOVA). A Student’s t test was used when one-way ANOVA revealed significant differences (P < 0.05). All statistical analyses were performed using GraphPad Prism statistical software (GraphPad Software, La Jolla, CA) with the help of Excel (Microsoft).
Supplementary Material
ACKNOWLEDGMENTS
We thank Di Wang at the Institute of Microbiology, Chinese Academy of Sciences, for constructive suggestions to the study.
This study was supported by the National Natural Science Foundation of China (grants 31570126 to L.Z.M., 31770152 to S.W., and 31300066 to Q.W.) and by the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (Q.W.).
We declare that we have no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01194-19.
REFERENCES
- 1.Wei Q, Ma LZ. 2013. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int J Mol Sci 14:20983–21005. doi: 10.3390/ijms141020983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15:194–222. doi: 10.1128/cmr.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drenkard E, Ausubel FM. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740–743. doi: 10.1038/416740a. [DOI] [PubMed] [Google Scholar]
- 4.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 6.Cohen D, Mechold U, Nevenzal H, Yarmiyhu Y, Randall TE, Bay DC, Rich JD, Parsek MR, Kaever V, Harrison JJ, Banin E. 2015. Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:11359–11364. doi: 10.1073/pnas.1421450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orr MW, Donaldson GP, Severin GB, Wang J, Sintim HO, Waters CM, Lee VT. 2015. Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc Natl Acad Sci U S A 112:E5048–E5057. doi: 10.1073/pnas.1507245112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alm RA, Bodero AJ, Free PD, Mattick JS. 1996. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol 178:46–53. doi: 10.1128/jb.178.1.46-53.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulasekara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A 103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2:1051–1060. doi: 10.1016/S1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 11.Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. 2012. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev 76:46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryder C, Byrd M, Wozniak DJ. 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol 10:644–648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irie Y, Borlee BR, O’Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. 2012. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 109:20632–20636. doi: 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klebensberger J, Birkenmaier A, Geffers R, Kjelleberg S, Philipp B. 2009. SiaA and SiaD are essential for inducing autoaggregation as a specific response to detergent stress in Pseudomonas aeruginosa. Environ Microbiol 11:3073–3086. doi: 10.1111/j.1462-2920.2009.02012.x. [DOI] [PubMed] [Google Scholar]
- 15.Malone JG, Jaeger T, Spangler C, Ritz D, Spang A, Arrieumerlou C, Kaever V, Landmann R, Jenal U. 2010. YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog 6:e1000804. doi: 10.1371/journal.ppat.1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu B, Liu C, Liu S, Cong H, Chen Y, Gu L, Ma LZ. 2016. Membrane association of SadC enhances its diguanylate cyclase activity to control exopolysaccharides synthesis and biofilm formation in Pseudomonas aeruginosa. Environ Microbiol 18:3440–3452. doi: 10.1111/1462-2920.13263. [DOI] [PubMed] [Google Scholar]
- 18.Jones C, Newsom D, Kelly B, Rie Y, Jennings L, Xu B, Limoli D, Harrison JJ, Parsek MR, White P, Wozniak D. 2014. ChIP-seq and RNA-seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog 10:e1003984. doi: 10.1371/journal.ppat.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol 55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- 20.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O’Toole GA. 2007. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulasekara BR, Kamischke C, Kulasekara HD, Christen M, Wiggins PA, Miller SI. 2013. c-di-GMP heterogeneity is generated by the chemotaxis machinery to regulate flagellar motility. Elife 2:e01402. doi: 10.7554/eLife.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merritt JH, Ha DG, Cowles KN, Lu W, Morales DK, Rabinowitz J, Gitai Z, O’Toole GA. 2010. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio 1:e00183-10. doi: 10.1128/mBio.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massie JP, Reynolds EL, Koestler BJ, Cong JP, Agostoni M, Waters CM. 2012. Quantification of high-specificity cyclic diguanylate signaling. Proc Natl Acad Sci U S A 109:12746–12751. doi: 10.1073/pnas.1115663109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39:D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oberto J. 2013. SyntTax: a web server linking synteny to prokaryotic taxonomy. BMC Bioinformatics 14:4. doi: 10.1186/1471-2105-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Chua S, Liu Y, Li Y, Jun Ting H, Kohli GS, Cai Z, Suwanchaikasem P, Kau Kit Goh K, Pin Ng S, Tolker-Nielsen T, Yang L, Givskov M. 2017. Reduced intracellular c-di-GMP content increases expression of quorum sensing-regulated genes in Pseudomonas aeruginosa. Front Cell Infect Microbiol 7:451. doi: 10.3389/fcimb.2017.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hay ID, Remminghorst U, Rehm BH. 2009. MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl Environ Microbiol 75:1110–1120. doi: 10.1128/AEM.02416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kazmierczak BI, Lebron MB, Murray TS. 2006. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol Microbiol 60:1026–1043. doi: 10.1111/j.1365-2958.2006.05156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryan RP, Lucey J, O’Donovan K, McCarthy Y, Yang L, Tolker-Nielsen T, Dow JM. 2009. HD-GYP domain proteins regulate biofilm formation and virulence in Pseudomonas aeruginosa. Environ Microbiol 11:1126–1136. doi: 10.1111/j.1462-2920.2008.01842.x. [DOI] [PubMed] [Google Scholar]
- 30.Roy AB, Petrova OE, Sauer K. 2012. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol 194:2904–2915. doi: 10.1128/JB.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merritt JH, Brothers KM, Kuchma SL, O’Toole GA. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J Bacteriol 189:8154–8164. doi: 10.1128/JB.00585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De N, Pirruccello M, Krasteva PV, Bae N, Raghavan RV, Sondermann H. 2008. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol 6:e67. doi: 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moscoso JA, Jaeger T, Valentini M, Hui K, Jenal U, Filloux A. 2014. The diguanylate cyclase SadC is a central player in Gac/Rsm-mediated biofilm formation in Pseudomonas aeruginosa. J Bacteriol 196:4081–4088. doi: 10.1128/JB.01850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol 65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen SE, Fecycz IT, Campbell JN. 1980. Nutritional factors controlling exocellular protease production by Pseudomonas aeruginosa. J Bacteriol 144:844–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarighi S, Wei Q, Camara M, Williams P, Fletcher MP, Kajander T, Cornelis P. 2008. The PA4204 gene encodes a periplasmic gluconolactonase (PpgL) which is important for fitness of Pseudomonas aeruginosa. Microbiology 154:2979–2990. doi: 10.1099/mic.0.2008/018465-0. [DOI] [PubMed] [Google Scholar]
- 40.Miller WG, Leveau JH, Lindow SE. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol Plant Microbe Interact 13:1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- 41.Rybtke MT, Borlee BR, Murakami K, Irie Y, Hentzer M, Nielsen TE, Givskov M, Parsek MR, Tolker-Nielsen T. 2012. Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl Environ Microbiol 78:5060–5069. doi: 10.1128/AEM.00414-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 43.Hassett DJ, Woodruff WA, Wozniak DJ, Vasil ML, Cohen MS, Ohman DE. 1993. Cloning and characterization of the Pseudomonas aeruginosa sodA and sodB genes encoding manganese- and iron-cofactored superoxide dismutase: demonstration of increased manganese superoxide dismutase activity in alginate-producing bacteria. J Bacteriol 175:7658–7665. doi: 10.1128/jb.175.23.7658-7665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 45.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 46.Wang S, Parsek MR, Wozniak DJ, Ma LZ. 2013. A spider web strategy of type IV pilus-mediated migration to build a fibre-like Psl polysaccharide matrix in Pseudomonas aeruginosa biofilms. Environ Microbiol 15:2238–2253. doi: 10.1111/1462-2920.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 48.Byrd MS, Sadovskaya I, Vinogradov E, Lu HP, Sprinkle AB, Richardson SH, Ma LY, Ralston B, Parsek MR, Anderson EM, Lam JS, Wozniak DJ. 2009. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol 73:622–638. doi: 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. 2015. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol Reprod Dev 82:518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fito-Boncompte L, Chapalain A, Bouffartigues E, Chaker H, Lesouhaitier O, Gicquel G, Bazire A, Madi A, Connil N, Veron W, Taupin L, Toussaint B, Cornelis P, Wei Q, Shioya K, Deziel E, Feuilloley MG, Orange N, Dufour A, Chevalier S. 2011. Full virulence of Pseudomonas aeruginosa requires OprF. Infect Immun 79:1176–1186. doi: 10.1128/IAI.00850-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.