As researchers try to understand and combat the development of antibiotic resistance in pathogens, there is a growing need to thoroughly understand the physiology and metabolism of the microbes. Staphylococcus aureus is a threatening pathogen with increased antibiotic resistance and well-studied virulence mechanisms. However, the adaptive mechanisms used by this pathogen to survive environmental stresses remain unclear, mostly due to the lack of information about its metabolic requirements. Defining the minimal metabolic requirements for S. aureus growth is a first step toward unraveling the mechanisms by which it adapts to metabolic stresses. Here, we present the development of a chemically defined minimal medium supporting growth of three S. aureus strains, and we reveal key genetic mutations contributing to improved growth in minimal medium.
KEYWORDS: adaptive laboratory evolution, Staphylococcus aureus, systems biology
ABSTRACT
Staphylococcus aureus is a Gram-positive pathogenic bacterium that colonizes an estimated one-third of the human population and can cause a wide spectrum of disease, ranging from superficial skin infections to life-threatening sepsis. The adaptive mechanisms that contribute to the success of this pathogen remain obscure partially due to a lack of knowledge of its metabolic requirements. Systems biology approaches can be extremely useful in predicting and interpreting metabolic phenotypes; however, such approaches rely on a chemically defined minimal medium as a basis to investigate the requirements of the cell. In this study, a chemically defined minimal medium formulation, termed synthetic minimal medium (SMM), was investigated and validated to support growth of three S. aureus strains: LAC and TCH1516 (USA300 lineage), as well as D592 (USA100 lineage). The formulated SMM was used in an adaptive laboratory evolution experiment to probe the various mutational trajectories of all three strains leading to optimized growth capabilities. The evolved strains were phenotypically characterized for their growth rate and antimicrobial susceptibility. Strains were also resequenced to examine the genetic basis for observed changes in phenotype and to design follow-up metabolite supplementation assays. Our results reveal evolutionary trajectories that arose from strain-specific metabolic requirements. SMM and the evolved strains can also serve as important tools to study antibiotic resistance phenotypes of S. aureus.
IMPORTANCE As researchers try to understand and combat the development of antibiotic resistance in pathogens, there is a growing need to thoroughly understand the physiology and metabolism of the microbes. Staphylococcus aureus is a threatening pathogen with increased antibiotic resistance and well-studied virulence mechanisms. However, the adaptive mechanisms used by this pathogen to survive environmental stresses remain unclear, mostly due to the lack of information about its metabolic requirements. Defining the minimal metabolic requirements for S. aureus growth is a first step toward unraveling the mechanisms by which it adapts to metabolic stresses. Here, we present the development of a chemically defined minimal medium supporting growth of three S. aureus strains, and we reveal key genetic mutations contributing to improved growth in minimal medium.
INTRODUCTION
Staphylococcus aureus is a Gram-positive pathogenic bacterium that has colonized an estimated one-third of the human population (1). Due to its high virulence potential and widespread reports of antibiotic resistance, deep-seated S. aureus infections are difficult to treat (2) and are associated with a high mortality rate (1). Methicillin-resistant S. aureus (MRSA) strains are especially concerning due to their prevalence in hospital settings and the paucity of reliable treatment options (3). The adaptive mechanisms of MRSA remain obscure, partially due to a lack of knowledge of the bacterium’s fundamental metabolic requirements. Systems biology approaches can be used to predict and interpret metabolic phenotypes through genome-scale modeling and bioinformatics approaches (4, 5). However, a validated chemically defined minimal medium is critical to further investigate the constraints of the cell. Such constraints and requirements can provide information on functional states of the cell and unveil the underlying mechanisms for growth and pathogenicity.
Antibiotic susceptibility testing is a cornerstone in determining the proper measures to treat, control, and prevent S. aureus infections (6). Susceptibility testing in the clinical laboratory to determine the MIC of an antibiotic is typically performed in rich bacteriological media such as Mueller-Hinton broth (MHB). However, several recent reports have illustrated large medium-dependent alterations in antibiotic activity when testing is performed in more physiological medium types such as tissue culture media for mammalian cell culture (7–9). These findings call into question the in vivo relevance of in vitro susceptibility testing in standard enriched bacteriological media. Medium-dependent variations in S. aureus antibiotic susceptibility highlight the need for a defined minimal medium to provide a more consistent nutritional environment (8). Understanding the relationship between the nutritional environment and antibiotic susceptibility may guide future approaches to enhance in vitro prediction of clinical drug activity.
Several efforts have been undertaken to reveal the nutritional requirements for growth of S. aureus. Prior studies by Gladstone (10) demonstrated the essentiality of ammonia as the main source of nitrogen and suggested that prior cultivation conditions affect future nutritional requirements among S. aureus strains (10). In addition, prior work revealed the importance of members of the vitamin B complex (11) and carbohydrates (12), as well as the essentiality of various amino acids (13, 14), in promoting S. aureus growth. AAM, a chemically defined formulation originally implemented for the isolation of amino acid auxotrophs (15), was recently analyzed using a metabolic model (16). As a result, a reduced formulation, AAM–, was created based on a metabolomic essentiality analysis that removed five amino acids and one vitamin from the original recipe (16). Although this work laid the foundation for identifying the metabolic requirements of S. aureus, a further reduced defined medium is critical to fully understand the metabolic network. Here, a chemically defined minimal medium formulation, termed synthetic minimal medium (SMM), further adapted from AAM–, was investigated, modified, and validated to enable growth of three S. aureus strains: LAC, TCH1516, and D592 (all MRSA). The formulated SMM was utilized in an adaptive laboratory evolution (ALE) experiment to further probe the optimal capabilities of the targeted strains in this nutritional environment and uncover mechanisms underlying the optimized states. The resulting SMM and the evolved strains can serve as important tools for fully resolving the metabolic network and identifying essential metabolic functions of pathogenic S. aureus, enabling improved approaches to drug design against this foremost human pathogen.
RESULTS
Development of a synthetic minimal medium for S. aureus.
A previously reported minimal defined medium demonstrated to support growth of S. aureus (AMM–) was used as the starting point for development of the SMM and subsequently modified to support reproducible growth (Table 1). Three strains were selected: two USA300 strains (TCH1516 and LAC), as well as a clinical isolate of a USA100 strain isolated from a male in his 50s who had experienced prolonged and persistent MRSA bacteremia (17). Initially, AMM– alone was insufficient to support growth of USA300 S. aureus strains (TCH1516 and LAC) beyond the first flask. Valid growth was defined as the capability of a medium to support growth for at least three consecutive passages under the same condition, with approximately three generations per flask, and reaching an appreciable final cell density (i.e., the optical density at 600 nm [OD600]) between 0.20 and 0.25 (see Materials and Methods). The passaging criterion is essential as maladapted strains will often display positive growth in the first flask due to nutrient carryover from the starter cultures. Turning to previous work to augment the starting minimal media, studies have shown arginine and proline are needed for S. aureus protein synthesis and growth in the presence of glucose, which led us to explore adding l-arginine to AMM– (18). Supplementation of AAM– with l-arginine, supported the growth of the three strains. Additional screening of the two USA300 strains (LAC and TCH1516) in AAM– plus l-arginine (termed AAM++) was performed to validate growth. All independent replicate cultures (n = 5) of both USA300 strains remained viable after three transfers, confirming the essentiality of l-arginine.
TABLE 1.
Composition of synthetic minimal medium (SMM) and other minimal media reported for Staphylococcus
| Component | Composition (mg)a
|
||
|---|---|---|---|
| AAM (15) | AAM– (16) | SMM (this study) | |
| Salts | |||
| NaCl (g) | 9.5 | 9.5 | 9.5 |
| KCl (g) | 3 | 3 | 3 |
| MgSO4⋅7H2O (g) | 1.3 | 1.3 | 1.3 |
| (NH4)2SO4 (g) | 4 | 4 | 4 |
| CaCl2⋅2H2O | 22 | 22 | 22 |
| KH2PO4 | 140 | 140 | 140 |
| FeSO4⋅7H2O | 6 | 6 | 6 |
| MnSO4⋅4H2O | 10 | 10 | 10 |
| Citric acid | 6 | 6 | |
| Tris (g) | 12.1 | 12.1 | 12.1 |
| Carbon source (g) | |||
| Glucose | 5 | 5 | 5 |
| Amino acids | |||
| l-Arginine | 125 | 125 | |
| l-Cysteine | 80 | 80 | 80 |
| l-Leucine | 150 | 150 | |
| l-Glutamic acid | 250 | ||
| l-Proline | 200 | 200 | 200 |
| l-Threonine | 150 | ||
| l-Valine | 150 | ||
| l-Phenylalanine | 150 | ||
| Vitamins | |||
| Nicotinic acid | 2 | 2 | 2 |
| Thiamine | 2 | 2 | 2 |
| Calcium pantothenate | 2 | 2 | 2 |
| Biotin | 0.1 | ||
| pH | 7.9 | 7.4 | 7.4 |
Values are expressed in milligrams (per 1 liter of medium) unless noted otherwise in column 1; all values are per liter.
The essentiality of the components of the modified AAM++ formulation was investigated experimentally to determine whether the medium could be further reduced. Citric acid, l-glutamic acid, l-leucine, l-cysteine, thiamine, nicotinic acid, and calcium pantothenate were selected for further essentiality analysis. Each component was removed individually or in combination from the base AAM++ media. Media that excluded citric acid, l-glutamic acid, and l-leucine, individually or in combination, demonstrated the capacity to support growth based on the aforementioned selection criteria for validity. Therefore, citric acid, l-glutamic acid, and l-leucine were determined to be nonessential and removed. The medium without the nonessential compounds (termed SMM; Table 1) was further validated and utilized to characterize the growth of USA300 MRSA strain TCH1516 in triplicate.
Although strains replicated in SMM, growth events were stochastic, and the growth rates were low (TCH1516, 0.189 ± 0.084 h−1) compared to rich media (1.18 ± 0.09 h−1). Therefore, in an attempt to optimize S. aureus strains (LAC, THC1516, and D592) for growth in SMM, adaptive laboratory evolution was utilized.
Adaptive laboratory evolution.
Adaptive laboratory evolution (ALE) uses inherited selective advantages in a given growth environment to uncover underlying growth-promoting molecular mechanisms. Owing to the natural capability of cells to adapt to a defined growth environment, robust strains can be produced for use in a number of application areas (19–22). Thus, an automated cell culture platform was used (23) to evolve S. aureus on SMM to generate strains with advanced growth capabilities (i.e., faster growth and increased reproducibility) and to explore growth-promoting mutations that could point to additional nutritional requirements. Briefly, in this approach, cultures were consecutively passaged in mid-exponential phase for more than 30 days, an average of 1.3 × 1012 ± 1.88 × 1011 cumulative number of cell divisions (see Materials and Methods for more details).
At the beginning of the ALE experiments, there was a surprisingly high degree of variability in the ability of identical replicates to grow on SMM for the three different S. aureus strains tested. Multiple replicates were used for each strain, since the utility of ALE increases when identically evolved replicates can be compared on a genetic level to find commonly mutated genes. Specifically, when 10, 10, and 15 replicates were started on SMM for LAC, TCH1516, and D592, only 7, 8, and 3 of the strains grew reproducibly after initial passage attempts. This surprising degree of stochasticity was observed but not further pursued. The replicates that did grow reproducibility were evolved to select for mutants with increased growth rates.
The ALE experiments resulted in observed fitness increases for evolved S. aureus strains that grew reproducibly in the SMM culturing conditions. The overall fitness trajectories for the approximately month-long experiment for three replicates of each strain are shown in Fig. 1 (see Fig. S1 in the supplemental material for all experiments). The average initial growth rates for USA300 evolved replicates (i.e., initial 10 flasks) were 0.189 ± 0.084 h−1 (TCH1516) and 0.194 ± 0.071 (LAC). At the end of the evolutions, the average growth rates for the evolved populations were 0.449 ± 0.079 (TCH1516) and 0.477 ± 0.077 (LAC), corresponding to increases of 137 and 145%, respectively. Strain D592 (USA100) had only three successfully evolved replicates, with inconsistent initial growth rates that could not be accurately determined (see Materials and Methods), but as the evolution proceeded, accurate growth rates could be collected and an increase in growth rate was evident. Overall, there was a significant increase in growth rate for each of the evolved populations (P < 0.0001, Student t test), and it appears that most replicates underwent one or two major jumps in fitness. Clones were isolated from each of the evolved populations from the final flask, i.e., an endpoint clone, for whole-genome sequencing and further phenotypic testing (see Table S1 in the supplemental material).
FIG 1.
Fitness trajectories of representative replicates for the ALE experiments on SMM. A3, A4, and A7 are three biological replicates of S. aureus USA300 strain LAC. A14, A19, and A20 are three biological replicates of USA300 strain TCH1516. A25, A30, and A35 are three biological replicates of USA100 strain D592. All strains demonstrated a growth rate increase over the course of the experiment. The endpoint clones were used for physiological characterizations, and both clones and populations were sequenced to understand the genetic basis for increased growth rate. The displayed growth rates were calculated for flasks that had ≥3 OD measurements in the exponential growth phase. Some of the cultures, especially those corresponding to strain D592, did not have accurate growth rate calls for earlier flasks in the evolution.
Genetic analysis of the mutations present in the evolved strains.
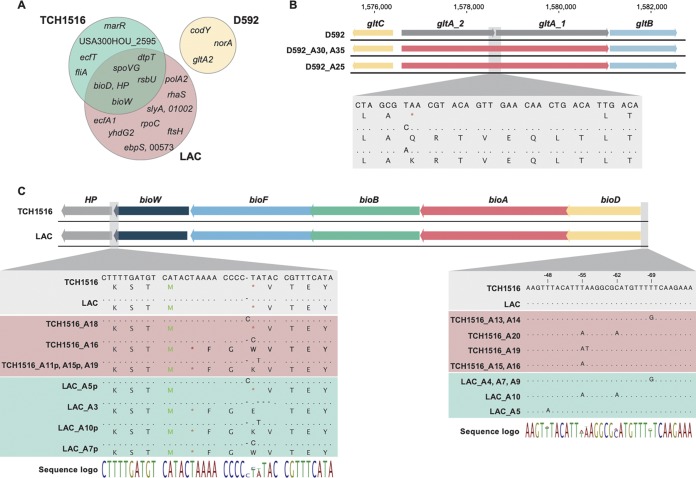
Whole-genome sequencing of the evolved S. aureus strains revealed a high level of parallel evolution between identical starting-strain replicates and between the two different evolved USA300 strains. Overall, sequencing revealed a total of 237 observed mutations across all strains (see Tables S2 to S5 in the supplemental material). Strain D592 (USA100), with only three replicates, had the lowest number of mutations identified (n = 19), although strains TCH1516 and LAC (USA300) had 77 and 144 identified mutations, respectively. One isolated clone, LAC A10, harbored a very high number of the observed mutations (n = 75), although no obvious hypermutator mutations could be identified. All other clones had a range of 5 to 14 mutations per strain. The USA300 strains presented a similar overall number of unique mutations across all samples (77 and 69 [LAC A10 excluded]), and a high degree of parallelism could be observed in the evolutions of these two different strains (Fig. 2A).
FIG 2.
Key mutations identified in the adapted strains. (A) Venn diagram of key mutated (i.e., occurring in parallel) genes identified. (B) Two different point mutations in D592 evolved strains result in the loss of the stop codon in the glutamate synthase gene, leading to possible gene repair. (C) Alignment of the biotin biosynthetic cluster showing mutations in the large majority of USA300 evolved strains, both TCH1516 and LAC. HP, hypothetical protein; p, denotes mutations identified only in population samples (i.e., not in the isolated and sequenced clone); M (in green), methionine start codon; * (in red), stop codon. Dots indicate the conservation of a base pair, while letters represent mutations.
Instances of parallel evolution were explored further to understand the genetic basis for adaptation on SMM. This analysis was led by looking at key mutations that occurred in multiple replicates or at genetic regions with two or more unique mutations (23). Of the 20 key mutated regions identified (Fig. 2A), 5 occurred in both USA300 strains, while 15 were strain specific. The majority of the identified key mutations were related to transport (norA, ecfT, dtpT, ecfA1, and yhdG2) and regulation (codY, rsbU, marR, rhaS, slyA, and spoVG). Two key mutations occurred in cofactor biosynthetic pathways (bioD and bioW), and only one occurred in a gene involved in energy metabolism (gltA2).
There were no identical mutated genes or regions that appeared in evolutions from all three strains or any between the two USA300 (LAC and TCH1516) and USA100 (D592) starting strains (Fig. 2A). However, there was a high degree of parallelism across all three replicates of D592 with mutations affecting the three genes norA, codY, and gltA2.
Interestingly, the D592 wild-type strain carried an early stop codon in gltA2, resulting in a truncated open reading frame and a corresponding protein sequence containing 674 amino acids instead of the 1,499 found in the USA300 strains (i.e., the sequence length was reduced to 45% of its original length). Mutations in gltA2 appeared to lead to the repair of gltA gene (which was originally annotated as gltA1 and gltA2) (Fig. 2B) through the loss of a stop codon in all three strains. Single nucleotide polymorphisms (SNPs) in the first base pair of the stop codon occurred three times and in two versions, replacing the stop codon with a glutamine (Q) or a lysine (K), leading ultimately to the repair of glutamate synthase (gltA) by fusion of open reading frames gltA1 and gltA2 (Fig. 2B). In USA300 strains the gltA gene is a single open reading frame similar to the one in USA100 evolved clones. Nevertheless, the disruption of this gene could be identified in other S. aureus strains, with at least three different patterns of gene disruption (Fig. S5). The other key mutations found in the USA100 strain were in codY, which codes for a global metabolic regulator considered to be a regulatory link between metabolism and virulence, with >200 genes being part of its regulon (24–27), and in norA, which encodes a multidrug efflux MFS transporter, thus pointing to a possible change in transport activities. Mutations in norA were deletions of 60, 129, and 1 bp in A25, A30, and A35, respectively, most likely resulting in loss of function. NorA has been shown to be involved in the transport of a wide range of structurally dissimilar drugs and possibly in siderophore export, playing a role in iron homeostasis (28). Therefore, deletion of this gene might abolish the low-level multidrug resistance of these USA100 strains, while also affecting iron homeostasis. In fact, USA100 strains have been shown to be more sensitive to norA iron regulation than USA300 (28), possibly explaining why this gene was mutated in USA100 but not in USA300 strains. Regarding codY, two of the mutations were amino acid substitutions (A25 and A35), while one was the deletion of 60 bp, most likely resulting in loss of function (A30) (Table S4). Although it is difficult to predict the outcome of single amino acid substitutions in CodY activity, the consequences of its deletion have been previously reported (25, 27). CodY is a direct repressor of several genes coding for enzymes of amino acid biosynthesis, amino acid and peptide transporter, and other nutrient transporters (25).
For the USA300 strains, as previously mentioned, the majority of key mutations were in genetic regions related with transport and regulation. Specifically, genes related to the transport of cobalt (ecfT and ecfA1) and dipeptides and tripeptides (dtpT) presented mutations that might change their affinities and/or efficiencies, pointing to a high pressure for efficient transport in this minimal medium. Mutations in ecfT and ecfA1 were present in two replicates each, but only one of each resulted in a nonsynonymous SNP, leading to an amino acid substitution. Furthermore, the ecfA1 mutations were only identified at the population level (∼50%), but no endpoint sequenced isolates presented these mutations (Table S5). It is not clear what the result of these mutations might be, since no cobalt was added to the media; nevertheless, it is possible that the Ecf proteins are responsible for the transport of other nutrients in the media. Mutations in genes coding for regulatory proteins were the second most common group, with marR, rsbU, rhaS, slyA, and spoVG affected. Prediction of the effect of nondeleterious changes in regulatory genes is more complicated, since these conceivably impact the entire regulon. Although no key mutations occurred in energy metabolism genes, the cofactor biosynthetic pathway responsible for biotin biosynthesis was mutated in several instances. Analysis revealed mutations in the biotin biosynthetic cluster, within the promoter region upstream of bioD (ATP-dependent dethiobiotin synthetase), and/or in the end of the bioW gene (6-carboxyhexonate-coenzyme A ligase) (Fig. 2C). Mutations in the promoter region of the bioD gene, the first gene of the biotin biosynthetic operon, likely contribute to the enhancement of transcription and/or translation of the operon, possibly increasing the biotin production levels. Full lists of key mutations are given in Tables S2 to S4 in the supplemental material, and a full list of mutations is presented in Table S5.
Phenotypic characterization of the ALE-adapted strains.
Representative isolates from the endpoint flasks were selected for further phenotypic characterization (Table S1). This process included determining the growth rate, the glucose uptake rate (GUR), the acetate secretion rate (ASR), and the lactate secretion rate (LSR). Three independently evolved isolates of S. aureus strains LAC and TCH1516 were grown and characterized alongside the parental strains, except for uptake and secretion rate determinations, where only evolved strains were analyzed. The averaged growth rates were 0.20 ± 0.01, 0.28 ± 0.04, and 0.28 ± 0.03 h−1 for the LAC strains (A3, A4, and A7, respectively) compared to 0.13 ± 0.01 h−1 for the LAC wild type. For the TCH1516 strains, the averaged growth rates were 0.24 ± 0.02, 0.29 ± 0.01, and 0.28 ± 0.01 h−1 (A14, A19, and A20, respectively) compared to 0.14 ± 0.02 h−1 for the TCH1516 wild type (Fig. 3A). These isolated clone growth rate values differ from those from the ALE experiments since they were cultured in different formats (plates versus culture vessels), most likely due to aeration, but the values correlate well between experiments (Fig. S2). In addition to an increase in growth rate, a shortening of the lag phase was also observed for evolved clones (Fig. S3). For the LAC evolved strains, the averaged GUR, ASR, and LSR were 5.99 ± 0.38, 5.42 ± 0.25, and 0.53 ± 0.23 mmol g (dry weight) (gDW)−1 h−1, respectively. Similarly, for the TCH1516 evolved strains, the averaged GUR, ASR and LSR were 4.89 ± 0.60, 4.84 ± 0.61, and 0.36 ± 0.10 mmol gDW−1 h−1, respectively (Fig. S4).
FIG 3.
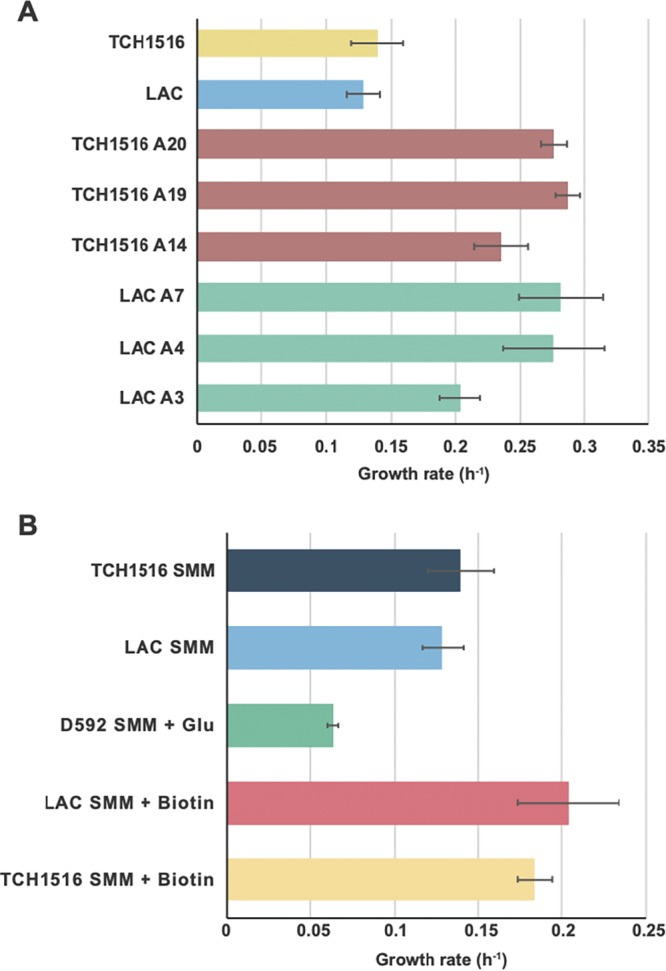
(A) Growth rates of wild-type and evolved USA300 strains. (B) Growth rates of wild-type strains in SMM supplemented with glutamic acid (SMM + Glu; 250 mg/liter) or biotin (SMM + biotin; 0.1 mg/liter). The growth rate of the D592 wild-type strain was not included since no clear exponential growth was observed (Fig. S3).
In an effort to better understand the causality of mutations found during the ALE experiments on SMM, supplementation experiments were carried out, and physiological measurements were taken for unevolved (wild-type) strains. We focused specifically on mutations related to energy metabolism and cofactor biosynthesis, since this can be directly correlated to a fitness increase. The mutations related to biotin biosynthesis and glutamine metabolism were explored by supplementing biotin and l-glutamic acid to SMM for USA300 and USA100 strains, respectively. Wild-type strains cultured in SMM supplemented with each metabolite had a higher growth rate (a 30 to 50% increase) in supplemented media compared to unsupplemented SMM (Fig. 3B). Further validation of the essentiality of bioD, bioW, and gltA genes mutated in the evolution experiments was performed using the Nebraska transposon mutant library (29). Strains with deletions in these genes were not able to grow in SMM, while supplementation with biotin or glutamate restored growth (Table S6). This finding confirms the importance of these metabolites for growth of the studied S. aureus strains in SMM and supports the likely beneficial role of biotin biosynthesis (bioD and bioW) and glutamate synthase (gltA2) mutations for growth in SMM.
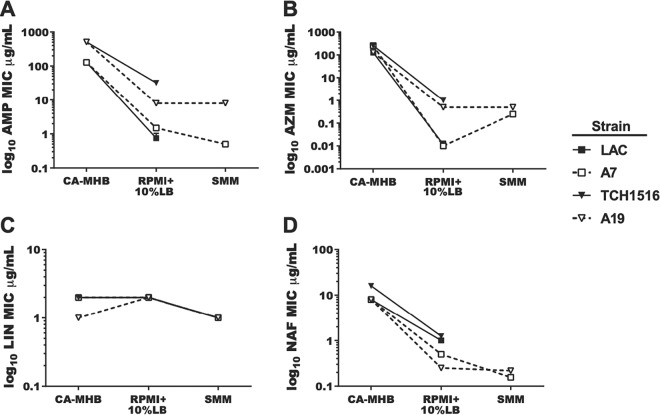
In order to understand medium-specific antibiotic resistance mechanisms, an evolved clone from each of the USA300 strains (LAC A7 and TCH1516 A19) was chosen for MIC testing using four antibiotics in both SMM and RPMI plus 10% Luria-Bertani (LB) media (RPMI+10%LB) (7). RPMI+10%LB has been proposed to represent a more relevant physiological medium due to it more closely mimicking the nutritional environment of the human host (30). Comparison of this medium to standard MHB testing medium confirms that a medium-dependent susceptibility exists (7, 8). Therefore, four antibiotics were selected to test strain sensitivity: ampicillin (AMP), azithromycin (AZM), linezolid (LNZ), and nafcillin (NAF). Overall, there were no significant differences in antibiotic susceptibility when we compared wild-type strains to the SMM-adapted strains in RPMI+10%LB (Fig. 4). This finding implies that the mutations present in the evolved strains do not impact antibiotic susceptibility in the more physiological RPMI+10%LB medium conditions. However, when cultivated in SMM, a shift in the susceptibility to AZM for the evolved strains was seen, with an ∼32-fold MIC increase compared to RPMI+10%LB for both evolved clones (LAC A7 and TCH1516 A19). No significant change was observed for any other antibiotics. The change in susceptibility to AZM for both USA300 strains in SMM confirms an SMM effect on antibiotic susceptibility and adds another observed case to medium-specific susceptibility.
FIG 4.
Adapted strains maintain relevant antibiotic sensitivities in synthetic minimal media. Strains LAC, A7 (adapted from LAC), TCH1516, and A19 (adapted from TCH1516) were analyzed for their sensitivities to antibiotics, as measured by the determination of the MIC of drug required to inhibit >90% of the growth from the untreated controls (MIC90), in CA-MHB, RPMI+10%LB, and SMM. All experiments were performed in triplicate. Error bars are present but only visible in panel A. (A) AMP; (B) AZM; (C) LIN; (D) NAF.
DISCUSSION
In this study, a chemically defined minimal medium (SMM) was explored, redefined, and verified for the cultivation of the multidrug-resistant USA300 S. aureus strains LAC, TCH1516, and USA100 strain D592. Although low growth rates and stochasticity in growth were observed for all strains in SMM, after an approximate month-long ALE experiment, the resulting evolved strains presented improved growth rates (Fig. 3A) and adaptabilities in SMM, such as a shorter lag phase (Fig. S3). Next-generation sequencing was used to reveal the genotypic changes occurring during the adaptation process and key mutations were identified, highlighted, and compared across strains.
Starting with 10 and 15 replicates for USA300 strains and USA100 strain, respectively, 8 TCH1516 strains (USA300), 7 LAC strains (USA300), and 3 D592 strains (USA100) were able to grow consistently throughout the cultivation period. This surprisingly stochastic growth of S. aureus, with inconsistent results observed among all strains examined, hints at a complex process inherent to S. aureus in adaptation to a new environment, requiring S. aureus to mutate early or undergo major regulatory rearrangements in order to proliferate in the growth experiments. Further, the finding that more replicates of the USA300 strains were able to grow compared to the USA100 strains and the overlap of mutations observed (Fig. 2A) suggests that there exists a lineage-specific (i.e., USA300 versus USA100) behavior which can provide insights on this stochasticity.
The mutational analysis provides mechanisms required for enhanced growth of these strains in a minimal nutritional environment, since the same gene mutating more than once across independent ALE replicates indicates a high probability of the mutation providing an adaptive benefit (31). The only genetic mutation directly related to metabolism identified in the USA100 evolved strains was a glutamate synthase pseudogene repair (Fig. 2B), which is required for growth in SMM (Table S6), since glutamic acid was removed from this formulation. Even though glutamic acid supplementation of SMM enhanced growth of the USA100 wild type, it did not fully restore the growth phenotype of evolved strains, which may be due to the impact of incorporating glutamic acid in the media versus efficient production intracellularly. This explains the low number of replicates that were successful in growing in SMM during adaptive laboratory evolution (3 of 15), since a very specific mutation was needed for successful growth of this strain in SMM. In the USA300 strains, this gene is fully functional, but we identified disruption of the same gene in several other S. aureus strains (Fig. S5). Although pseudogene repair driven by selection pressure in bacteria has been reported previously (32), this study reports such pseudogene repair specifically in the genus Staphylococcus. The ability of S. aureus to repair open reading frames upon necessity might explain the versatility of strains in this species, such as the previously observed reversion of auxotrophies (33).
For USA300 strains, some of the mutations observed were focused on the biotin biosynthetic cluster (Fig. 2C), both upstream of bioD and within the terminal region of the bioW gene. These mutations might be enhancing the transcription and/or translation of the operon, since upon supplementation of SMM with biotin the wild-type strains presented an improved growth rate (Fig. 3B). These mutations happened multiple times not only across replicates of the same strain but also across strains of the same lineage, with both LAC and TCH1516 acquiring the same point mutations in several instances (Fig. 2C). Biotin is a critical nutrient, since it is involved in a number of metabolic bioprocesses such as carboxylation and, more importantly, it is highly demanded by the bacteria during infection (34, 35). Interestingly, these metabolic requirements seem to be strain specific, and SMM formulation might have to be adjusted to support growth of different S. aureus strains. For example, in the case of the USA300 strains in this study this would be the addition of biotin to SMM, while for strain D592 (USA100) it would be glutamic acid supplementation.
Most of the other mutated genes are involved in energy transport, nutrient acquisition, and regulation (Fig. 2A), with some being related to bacterial pathogenesis. For example, the phosphatase-encoding gene rsbU is known for its role in the activation of SigB, an important transcription factor (36, 37). The SigB transcription factor is related to the expression of high-level methicillin resistance and regulated in response to growth phase and environmental stress (38, 39). Moreover, spoVG, a SigB-controlled protein, is a critical factor that impacts cell wall synthesis, and it is likewise involved in antibiotic resistance and virulence (40, 41).
Even though some of these mutations occurred in genes involved in antibiotic resistance, no significant antibiotic susceptibility differences were observed between wild type and evolved strains, but the same was not true for the different media tested. MIC testing of the evolved USA300 strains provided an interesting differential medium-specific behavior (Fig. 4). The lower susceptibilities of the evolved strains to azithromycin in SMM suggests that cellular responses activated during cultivation in SMM impaired the efficacy of the antibiotic or that SMM components interact with the antibiotic, making it less available and therefore less effective. It has been shown that ribosome-targeting antibiotics which bind reversibly to the 50S ribosomal subunit, such as azithromycin, are less effective at low growth rates (42, 43), which is the case of growth in SMM. On the other hand, other medium-specific antibiotic susceptibilities have been reported (7, 17), and the development of SMM comes as an opportunity to look at more specific relationships between medium additives and bacteria to further elucidate the mechanisms behind such phenomena.
In summary, a chemically defined minimal medium has been designed and validated. ALE was used to produce robust and faster-growing S. aureus strains for phenotypic profiling and MIC testing and to understand the mechanisms at work to enable optimal growth. Further investigation of the strains and specific results presented here are warranted, but the strains and medium compositions provide valuable tools for the study of this important pathogen.
MATERIALS AND METHODS
Preparation of SMM.
The components of SMM were prepared in bulk as stock solutions and were split into three categories, i.e., base, salts, and supplements. These were dissolved in Milli-Q water to the final concentrations listed in Table 1. The salt mixture includes FeSO4⋅7H2O and MnSO4⋅H2O, CaCl2⋅2H2O and KH2PO4, and was sterilized by vacuum filtration. The base contains KCl, NaCl, MgSO4·7H2O, (NH4)2SO4, and Tris and was autoclaved after adjusting its pH to 7.4 with 4 M HCl. The remaining components are the supplements and were prepared and vacuum filtered individually except for l-arginine and l-proline, which were autoclaved separately. All the sterilized solutions were mixed and vacuum filtered again to form SMM.
Cell culture and ALE.
The parental S. aureus strains (TCH1516, LAC, and D592) were grown on LB agar plates and individual colonies were isolated and inoculated into RPMI+10%LB. The precultures were cultivated overnight, and 1 ml of each overnight culture was transferred into flasks containing 15 ml of SMM at 37°C. These 15-ml cultures were constantly stirred at high speed (1,000 rpm) for sufficient aeration and serially transferred once the Optical Density reached between 0.20 and 0.25 (Tecan Sunrise plate reader, equivalent to an OD600 between 0.67 and 0.83 with 1-cm light path length) when the cultures were proliferating exponentially. Each passage was given a flask number, representing the passage number. Replicates that did not grow after passaging, reaching ODs of ∼0.20, were not passed further. The average duration of the ALE experiments was approximately 34 days, and frozen stocks of the endpoint populations were created from the last available flask. One clone was isolated from each of the evolved populations from the final flask, i.e., an endpoint clone, for whole-genome sequencing and further phenotypic testing (Table S1). The population in the final flask was also sequenced to ensure the endpoint clone was representative of the population.
Physiological characterizations.
The parental strains and endpoint strains, LAC (A3, A4, and A7), TCH1516 (A14, A19, and A20), and D592 (A25, A30, and A35) were first plated on LB agar plates. Individual colonies were selected and cultured overnight in LB medium in triplicate for physiological characterizations. The overnight cultures washed in the respective media to be used (SMM or any of the supplemented versions), diluted to an OD600 of 0.05, and the optical densities were monitored over time using BioScreen C (Labsystems, Finland). The growth rates (h−1) were calculated using two time points within the exponential growth phase, using the following formula:
At a few sampling points, small portions of the growing cultures were collected and filtered for determinations of metabolites, including glucose, acetate, and lactate, and their production/secretion rates were evaluated using high-performance liquid chromatography (HPLC). The filtrates were injected through HPLC column (Aminex HPX-87H column 125-0140) and analyzed for the concentrations of the metabolites mentioned above by comparison to the concentrations of the standards. The production rates and the secretion rates were then computed by linear regression and converted into yield (mmol gDW−1 h−1).
Transposon mutants from the Nebraska transposon library were streaked onto LB agar plates supplemented with erythromycin (5 μg/ml). Cultures for growth in SMM were prepared as described above for the ALE strains. Strains were considered able to grow if OD600 values of ≥0.3 were registered.
Susceptibility testing.
Azithromycin (Fresenius Kabi) and Linezolid (Pfizer) were purchased from a clinical pharmacy. Ampicillin and nafcillin was purchased from Sigma-Aldrich. All drugs were resuspended in 1× Dulbecco modified phosphate-buffered saline (DPBS; Corning). The bacterial strains used in antibiotic susceptibility testing were first streaked onto Luria agar plates from stocks stored at –80°C (in 20% glycerol–80% MHB) and grown at 37°C overnight. Isolated colonies were picked from the plate and inoculated into 5 ml of either CA-MHB (MHB [Difco] amended with 20 mg/liter Ca2+ and 10 mg/liter Mg2+) or RPMI+ (phenol-free RPMI [Gibco 1640] amended with 10% LB [Criterion]) medium in a 14-ml Falcon polypropylene round-bottom snap-cap tube (Corning, catalog no. 352059) and grown with shaking at 100 rpm at 37°C overnight. The following day, the overnight cultures were subcultured 1:50 in the desired medium in 14-ml snap cap tubes and grown with shaking at 100 rpm at 37°C until they reached the mid-logarithmic phase (OD600 of ∼0.4). Unless otherwise noted, the experiments were conducted in Costar flat-bottom 96-well plates (Corning, catalog no. 3370) with a final volume of 200 μl/well. For the MIC experiments, the bacteria were cultured in the same medium throughout (CA-MHB or RPMI+) prior to the addition of antibiotics. The mid-logarithmic-phase cultures were diluted to approximately 5 × 105 CFU (OD600 of ∼0.002), and 180 μl was added to each experimental well of the 96-well flat-bottom plate (Costar, catalog no. 3370). Either 20 μl of 1× DPBS or 20 μl of the desired 10× drug stock was added to each culture-containing well. The plates were then incubated with shaking at 100 rpm at 37°C overnight. Bacterial growth, as determined by measuring the OD600 of each well, was determined by utilizing an Enspire Alpha multimode plate reader (Perkin-Elmer). To determine the MIC90, defined as the amount of drug required to inhibit ≥90% of the growth of the untreated controls, the density of each drug-treated well was compared to the untreated control.
Next-generation DNA sequencing and mutational analysis.
Genomic DNA was isolated using the Zymo Quick-DNA fungal/bacterial miniprep kit. DNA was quantified and quality controlled using a Qubit dsDNA high-sensitivity assay. Paired-end sequencing libraries were prepared by using a KAPA HyperPlus kit and sequenced using Illumina NextSeq 500. The sequencing reads are available at the NCBI SRA database (www.ncbi.nlm.nih.gov/sra) under BioProject number PRJNA513389. Reads were quality controlled, and mutations were identified by a bioinformatics pipeline (44). Reads generated from the USA300 TCH1516 were aligned to a reference TCH1516 genome (NCBI accession no. NC_010079), while the other two strains (USA300 LAC and USA100 D592) were aligned to genomes generated de novo (see below). These genomes are available under NCBI accession numbers CP035369 and CP035370 (LAC) and CP035791 and CP035792 (D592). The output mutations across all three strains were first filtered by a mutation frequency threshold set at 40%, followed by manual inspections and filtrations of the mismapped results. All samples were confirmed for an averaged mapped coverage of at least 31×.
De novo hybrid assembly using ONT MinIon.
The Oxford Nanopore Technologies (ONT) MinIon was used to obtain long sequencing reads and combined with the Illumina reads described above to assemble high-quality, closed reference genomes for mutational analysis. The long reads were obtained from genomic DNA that was isolated using a Zymo Quick-DNA fungal/bacterial kit (catalog no. D6007) and then sheared to an ∼8 kb average size using a Covaris g-Tube (catalog no. 520079). Libraries were prepared according to standard library preparation using ONT’s ligation sequencing kit 1D and then sequenced using the ONT MinION sequencer. The genomes were assembled using Unicycler (45) v0.4.2 and annotated with PROKKA (46) v1.1.2. Both the LAC and the D592 sequences were resolved to a single genomic and plasmid sequence with N50 scores of 2,875,046 and 2,820,177, respectively.
Accession number(s).
Newly determined sequence data were deposited in the NCBI database under accession numbers SRX5223031 to SRX5223065.
Supplementary Material
ACKNOWLEDGMENTS
H.M., L.L.W., N.D., Y.S., M.H., V.N., B.O.P., and A.M.F. were funded through an NIH NIAID grant (U01-AI124316).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01773-19.
REFERENCES
- 1.Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LaPlante KL, Rybak MJ. 2004. Impact of high-inoculum Staphylococcus aureus on the activities of nafcillin, vancomycin, linezolid, and daptomycin, alone and in combination with gentamicin, in an in vitro pharmacodynamic model. Antimicrob Agents Chemother 48:4665–4672. doi: 10.1128/AAC.48.12.4665-4672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verhoeven PO, Gagnaire J, Botelho-Nevers E, Grattard F, Carricajo A, Lucht F, Pozzetto B, Berthelot P. 2014. Detection and clinical relevance of Staphylococcus aureus nasal carriage: an update. Expert Rev Anti-Infect Ther 12:75–89. doi: 10.1586/14787210.2014.859985. [DOI] [PubMed] [Google Scholar]
- 4.Monk JM, Charusanti P, Aziz RK, Lerman JA, Premyodhin N, Orth JD, Feist AM, Palsson BØ. 2013. Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments. Proc Natl Acad Sci U S A 110:20338–20343. doi: 10.1073/pnas.1307797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feist AM, Palsson BØ. 2008. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotechnol 26:659–667. doi: 10.1038/nbt1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown DFJ, Edwards DI, Hawkey PM, Morrison D, Ridgway GL, Towner KJ, Wren M. 2005. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA). J Antimicrob Chemother 56:1000–1018. doi: 10.1093/jac/dki372. [DOI] [PubMed] [Google Scholar]
- 7.Lin L, Nonejuie P, Munguia J, Hollands A, Olson J, Dam Q, Kumaraswamy M, Rivera H Jr, Corriden R, Rohde M, Hensler ME, Burkart MD, Pogliano J, Sakoulas G, Nizet V. 2015. Azithromycin synergizes with cationic antimicrobial peptides to exert bactericidal and therapeutic activity against highly multidrug-resistant gram-negative bacterial pathogens. EBioMedicine 2:690–698. doi: 10.1016/j.ebiom.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ersoy SC, Heithoff DM, Barnes L V, Tripp GK, House JK, Marth JD, Smith JW, Mahan MJ. 2017. Correcting a fundamental flaw in the paradigm for antimicrobial susceptibility testing. EBioMedicine 20:173–181. doi: 10.1016/j.ebiom.2017.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farha MA, French S, Stokes JM, Brown ED. 2018. Bicarbonate alters bacterial susceptibility to antibiotics by targeting the proton motive force. ACS Infect Dis 4:382–390. doi: 10.1021/acsinfecdis.7b00194. [DOI] [PubMed] [Google Scholar]
- 10.Gladstone GP. 1937. The nutrition of Staphylococcus aureus: nitrogen requirements. Br J Exp Pathol 18:322–333. [Google Scholar]
- 11.Knight B. 1937. The nutrition of Staphylococcus aureus: nicotinic acid and vitamin B1. Biochem J 31:731–737. doi: 10.1042/bj0310731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hussain M, Hastings JG, White PJ. 1991. A chemically defined medium for slime production by coagulase-negative staphylococci. J Med Microbiol 34:143–147. doi: 10.1099/00222615-34-3-143. [DOI] [PubMed] [Google Scholar]
- 13.Mah RA, Fung DY, Morse SA. 1967. Nutritional requirements of Staphylococcus aureus S-6. Appl Microbiol 15:866–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller RD, Fung DY. 1973. Amino acid requirements for the production of enterotoxin B by Staphylococcus aureus S-6 in a chemically defined medium. Appl Microbiol 25:800–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudin L, Sjöström JE, Lindberg M, Philipson L. 1974. Factors affecting competence for transformation in Staphylococcus aureus. J Bacteriol 118:155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee D-S, Burd H, Liu J, Almaas E, Wiest O, Barabási A-L, Oltvai ZN, Kapatral V. 2009. Comparative genome-scale metabolic reconstruction and flux balance analysis of multiple Staphylococcus aureus genomes identify novel antimicrobial drug targets. J Bacteriol 191:4015–4024. doi: 10.1128/JB.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakoulas G, Okumura CY, Thienphrapa W, Olson J, Nonejuie P, Dam Q, Dhand A, Pogliano J, Yeaman MR, Hensler ME, Bayer AS, Nizet V. 2014. Nafcillin enhances innate immune-mediated killing of methicillin-resistant Staphylococcus aureus. J Mol Med 92:139–149. doi: 10.1007/s00109-013-1100-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halsey CR, Lei S, Wax JK, Lehman MK, Nuxoll AS, Steinke L, Sadykov M, Powers R, Fey PD. 2017. Amino acid catabolism in Staphylococcus aureus and the function of carbon catabolite repression. mBio 8:e01434-16. doi: 10.1128/mBio.01434-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SY, Kim HU. 2015. Systems strategies for developing industrial microbial strains. Nat Biotechnol 33:1061–1072. doi: 10.1038/nbt.3365. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Khushk I, Xiao Y, Gao Q, Bao J. 2018. Tolerance improvement of Corynebacterium glutamicum on lignocellulose derived inhibitors by adaptive evolution. Appl Microbiol Biotechnol 102:377–388. doi: 10.1007/s00253-017-8627-4. [DOI] [PubMed] [Google Scholar]
- 21.Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 22.Barrick JE, Lenski RE. 2013. Genome dynamics during experimental evolution. Nat Rev Genet 14:827–839. doi: 10.1038/nrg3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaCroix RA, Sandberg TE, O’Brien EJ, Utrilla J, Ebrahim A, Guzman GI, Szubin R, Palsson BO, Feist AM. 2015. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol 81:17–30. doi: 10.1128/AEM.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balasubramanian D, Harper L, Shopsin B, Torres VJ. 2017. Staphylococcus aureus pathogenesis in diverse host environments. Pathog Dis 75:ftx005. doi: 10.1093/femspd/ftx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, Bodi K, Sonenshein AL. 2010. Direct targets of CodY in Staphylococcus aureus. J Bacteriol 192:2861–2877. doi: 10.1128/JB.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, Sonenshein AL. 2008. Staphylococcus aureus CodY negatively regulates virulence gene expression. J Bacteriol 190:2257–2265. doi: 10.1128/JB.01545-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pohl K, Francois P, Stenz L, Schlink F, Geiger T, Herbert S, Goerke C, Schrenzel J, Wolz C. 2009. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J Bacteriol 191:2953–2963. doi: 10.1128/JB.01492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng X, Sun F, Ji Q, Liang H, Missiakas D, Lan L, He C. 2012. Expression of multidrug resistance efflux pump gene norA is iron responsive in Staphylococcus aureus. J Bacteriol 194:1753–1762. doi: 10.1128/JB.06582-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velegraki A, Alexopoulos EC, Kritikou S, Gaitanis G. 2004. Use of fatty acid RPMI 1640 media for testing susceptibilities of eight Malassezia species to the new triazole posaconazole and to six established antifungal agents by a modified NCCLS M27-A2 microdilution method and Etest. J Clin Microbiol 42:3589–3593. doi: 10.1128/JCM.42.8.3589-3593.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bailey SF, Rodrigue N, Kassen R. 2015. The effect of selection environment on the probability of parallel evolution. Mol Biol Evol 32:1436–1448. doi: 10.1093/molbev/msv033. [DOI] [PubMed] [Google Scholar]
- 32.Anand A, Olson CA, Yang L, Sastry AV, Catoiu E, Choudhary KS, Phaneuf PV, Sandberg TE, Xu S, Hefner Y, Szubin R, Feist AM, Palsson BO. 2019. Pseudogene repair driven by selection pressure applied in experimental evolution. Nat Microbiol 4:386. doi: 10.1038/s41564-018-0340-2. [DOI] [PubMed] [Google Scholar]
- 33.Emmett M, Kloos WE. 1975. Amino acid requirements of staphylococci isolated from human skin. Can J Microbiol 21:729–733. doi: 10.1139/m75-107. [DOI] [PubMed] [Google Scholar]
- 34.Peters-Wendisch P, Götker S, Heider SAE, Komati Reddy G, Nguyen AQ, Stansen KC, Wendisch VF. 2014. Engineering biotin prototrophic Corynebacterium glutamicum strains for amino acid, diamine and carotenoid production. J Biotechnol 192(Pt B):346–354. doi: 10.1016/j.jbiotec.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 35.Pendini NR, Yap MY, Traore DAK, Polyak SW, Cowieson NP, Abell A, Booker GW, Wallace JC, Wilce JA, Wilce M. 2013. Structural characterization of Staphylococcus aureus biotin protein ligase and interaction partners: an antibiotic target. Protein Sci 22:762–773. doi: 10.1002/pro.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pané-Farré J, Jonas B, Hardwick SW, Gronau K, Lewis RJ, Hecker M, Engelmann S. 2009. Role of RsbU in controlling SigB activity in Staphylococcus aureus following alkaline stress. J Bacteriol 191:2561–2573. doi: 10.1128/JB.01514-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giachino P, Engelmann S, Bischoff M. 2001. B activity depends on RsbU in Staphylococcus aureus. J Bacteriol 183:1843–1852. doi: 10.1128/JB.183.6.1843-1852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kullik I, Giachino P. 1997. The alternative sigma factor σB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch Microbiol 167:151–159. doi: 10.1007/s002030050428. [DOI] [PubMed] [Google Scholar]
- 39.Rachid S, Ohlsen K, Wallner U, Hacker J, Hecker M, Ziebuhr W. 2000. Alternative transcription factor sigma B is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J Bacteriol 182:6824–6826. doi: 10.1128/JB.182.23.6824-6826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X, Zhang S, Sun B. 2016. SpoVG regulates cell wall metabolism and oxacillin resistance in methicillin-resistant Staphylococcus aureus strain N315. Antimicrob Agents Chemother 60:3455–3461. doi: 10.1128/AAC.00026-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulthess B, Meier S, Homerova D, Goerke C, Wolz C, Kormanec J, Berger-Bächi B, Bischoff M. 2009. Functional characterization of the σB-dependent yabJ-spoVG operon in Staphylococcus aureus: role in methicillin and glycopeptide resistance. Antimicrob Agents Chemother 53:1832–1839. doi: 10.1128/AAC.01255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levin BR, McCall IC, Perrot V, Weiss H, Ovesepian A, Baquero F. 2017. A numbers game: ribosome densities, bacterial growth, and antibiotic-mediated stasis and death. mBio 8:e02253-16. doi: 10.1128/mBio.02253-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greulich P, Scott M, Evans MR, Allen RJ. 2015. Growth-dependent bacterial susceptibility to ribosome-targeting antibiotics. Mol Syst Biol 11:796. doi: 10.15252/msb.20145949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pabinger S, Dander A, Fischer M, Snajder R, Sperk M, Efremova M, Krabichler B, Speicher MR, Zschocke J, Trajanoski Z. 2014. A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform 15:256–278. doi: 10.1093/bib/bbs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.