Coxiella burnetii is an intracellular bacterium that causes query, or Q fever, a disease that typically manifests as a severe flu-like illness. The initial target of C. burnetii is the alveolar macrophage. Here, it regulates vesicle trafficking pathways and fusion events to establish a large replication vacuole called the Coxiella-containing vacuole (CCV). Similar to a phagolysosome, the CCV has an acidic pH and contains lysosomal hydrolases obtained via fusion with late endocytic vesicles.
KEYWORDS: Coxiella, Coxiella-containing vacuole, GNPTAB knockout cells, LC3, Q fever, autophagy, hydrolases, lysosome, mannose-6-phosphate receptor, vesicular trafficking
ABSTRACT
Coxiella burnetii is an intracellular bacterium that causes query, or Q fever, a disease that typically manifests as a severe flu-like illness. The initial target of C. burnetii is the alveolar macrophage. Here, it regulates vesicle trafficking pathways and fusion events to establish a large replication vacuole called the Coxiella-containing vacuole (CCV). Similar to a phagolysosome, the CCV has an acidic pH and contains lysosomal hydrolases obtained via fusion with late endocytic vesicles. Lysosomal hydrolases break down various lipids, carbohydrates, and proteins; thus, it is assumed C. burnetii derives nutrients for growth from these degradation products. To investigate this possibility, we utilized a GNPTAB−/− HeLa cell line that lacks lysosomal hydrolases in endocytic compartments. Unexpectedly, examination of C. burnetii growth in GNPTAB−/− HeLa cells revealed replication and viability are not impaired, indicating C. burnetii does not require by-products of hydrolase degradation to survive and grow in the CCV. However, although bacterial growth was normal, CCVs were abnormal, appearing dark and condensed rather than clear and spacious. Lack of degradation within CCVs allowed waste products to accumulate, including intraluminal vesicles, autophagy protein LC3, and cholesterol. The build-up of waste products coincided with an altered CCV membrane, where LAMP1 was decreased and CD63 and LAMP1 redistributed from a punctate to uniform localization. This disruption of CCV membrane organization may account for the decreased CCV size due to impaired fusion with late endocytic vesicles. Collectively, these results demonstrate lysosomal hydrolases are not required for C. burnetii survival and growth but are needed for normal CCV development. These data provide insight into mechanisms of CCV biogenesis while raising the important question of how C. burnetii obtains essential nutrients from its host.
INTRODUCTION
Coxiella burnetii is a bacterium that causes Q fever, a flu-like disease primarily transmitted through inhalation of dust contaminated by infected cattle, sheep, and goats. In rare cases, chronic illness develops, leading to severe complications, including endocarditis and vascular disease (1, 2). C. burnetii is an intracellular bacterium that replicates within a large vacuole termed the Coxiella-containing vacuole (CCV). The CCV is similar to a phagolysosome, having an acidic pH and containing lysosomal hydrolases acquired via prolific fusion with late endocytic vesicles (3–5).
The mechanisms by which intracellular pathogens that reside in a host-derived vacuole obtain nutrients are poorly understood. Intravacuolar pathogens have the advantage of evading host immune defenses, but this then poses the challenge of transporting nutrients through a vacuole membrane barrier. Legionella pneumophila is phylogenetically related to C. burnetii and exploits the host’s neutral amino acid transporter, solute carrier (SLC) 1A5, to obtain amino acids from the cytoplasm (6–8). Unlike other vacuolar bacteria, C. burnetii resides within a harsh phagolysosome-like compartment that is thought to be a rich source of nutrients delivered by fusion with endocytic and autophagic vesicles (9, 10). Lysosomal hydrolases break down various cellular macromolecules into their constituent parts, generating nutrients in the form of amino acids and carbohydrates that are predicted to support C. burnetii growth (10–12).
More than sixty lysosomal hydrolases that break down proteins, lipids, nucleic acids, and carbohydrates have been identified (11, 12). Hydrolases are synthesized in the endoplasmic reticulum (ER) and trafficked through the Golgi network to late endosomes. The vast majority of soluble acid hydrolases are trafficked from the Golgi network to endosomes via a mannose-6-phosphate receptor (M6PR)-mediated pathway (13, 14). Few known hydrolases are transported solely by M6PR-independent means, including being transported as integral membrane proteins (lysosomal acid phosphatase) or via the receptors sortilin (cathepsin H) and LIMP2 (β-glucocerebrosidase) (15). In the Golgi network, N-acetylglucosamine-1-phosphotransferase (GNPT) attaches phosphates to mannose residues of newly synthesized hydrolases to generate mannose-6-phosphate (M6P) tags. M6P-tagged enzymes are bound by M6PRs and are packaged into vesicles for delivery to late endosomes (13, 16). Upon fusion with late endosomes, vesicle acidity causes the lysosomal hydrolases to dissociate from M6PRs. The ligand-free M6PRs return to the trans-Golgi network via retrograde trafficking while hydrolases deposited in the endosomal lumen undergo cleavage to generate active enzymes (17).
Inhibition of M6PR-mediated transport of lysosomal hydrolases results in production of inclusions that are evident in micrographs as enlarged, dark endocytic vesicles packed with cellular waste products. In humans, mutations in the GNPTAB gene disrupt the α and β subunits of GNPT, resulting in the severe lysosomal storage disease mucolipidosis type II (MLII), which is also called inclusion-cell (I-cell) disease. Mutant GNPTAB genes direct the synthesis of GNPTs that fail to phosphorylate hydrolases, thereby resulting in enzymes that are trafficked to the plasma membrane and exocytosed (18). van Meel et al. (19) developed a GNPT α and β knockout (GNPTAB−/−) HeLa cell line with defects similar to those of MLII patient fibroblasts, including enlarged late endocytic vesicles and drastically reduced acid hydrolase activity.
It is unknown whether C. burnetii requires lysosomal hydrolases for nutrient acquisition. Here, we investigated this possibility by analyzing infected GNPTAB−/− HeLa cells. We report that defects in CCV formation occurred in HeLa cells lacking GNPT activity but that C. burnetii replication and viability were normal. CCV accumulation of intraluminal vesicles, autophagy protein LC3, and cholesterol in GNPTAB−/− cells demonstrated a severe lack of hydrolase activity. These results demonstrate C. burnetii survival and growth do not require host lysosomal hydrolases but normal CCV formation does.
RESULTS
CCVs in GNPTAB−/− HeLa cells lack hydrolase activity.
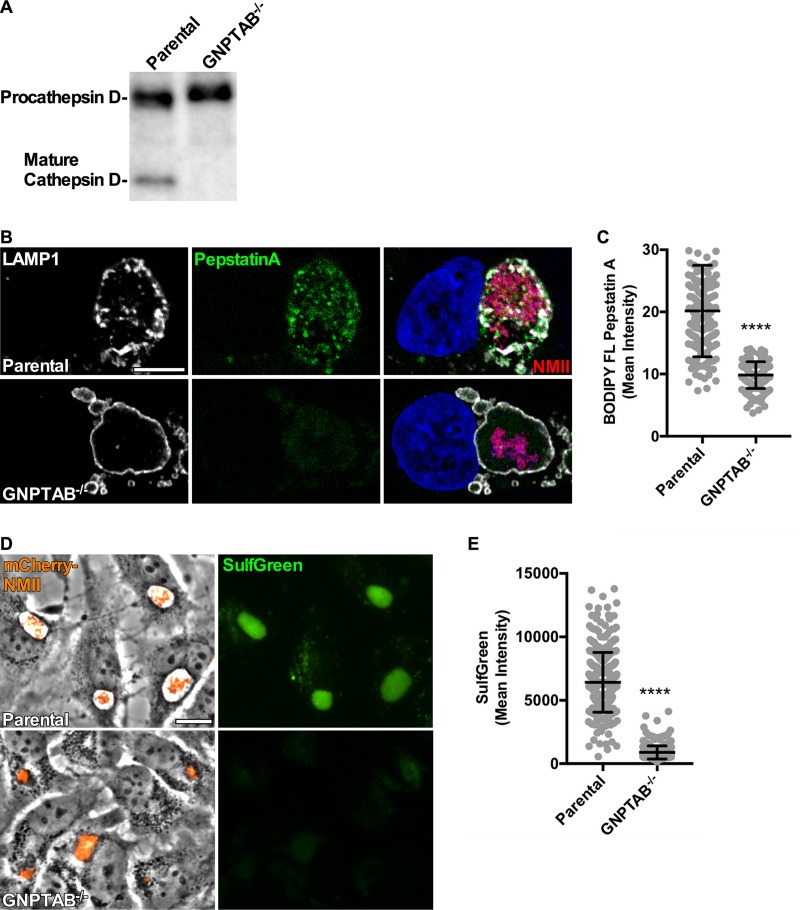
A prevailing hypothesis predicts that C. burnetii requires hydrolytic breakdown of host macromolecules by lysosomal hydrolases to acquire essential nutrients. To investigate this prediction, we used a GNPTAB−/− HeLa cell line that is deficient in active lysosomal hydrolases. Previously, van Meel et al. (19) demonstrated GNPTAB−/− HeLa cells lack enzymatic activity for a panel of known hydrolases. To confirm hydrolases remained inactive in GNPTAB−/− cells infected with C. burnetii, we examined the hydrolytic activity of cathepsin D and sulfatases. Cathepsin D activation requires proteolytic cleavage of procathepsin D within the late endosome (20). Immunoblots of lysates from parental and GNPTAB−/− cells 3 days postinfection (dpi) revealed an absence of mature cathepsin D in the knockout samples (Fig. 1A). Fluorescent BODIPY FL-conjugated pepstatin A is an inhibitor that binds the active site of cathepsin D (21). CCVs of GNPTAB−/− cells lacked BODIPY FL staining, indicating an absence of cathepsin D (Fig. 1B and C). Sulfatase activity within the CCV was examined using SulfGreen substrate, which fluoresces when hydrolyzed. CCVs in GNPTAB−/− cells showed a severe defect in sulfatase activity (Fig. 1D and E). Together, these results show CCVs of GNPTAB−/− HeLa cells lack hydrolase activity.
FIG 1.
Infected GNPTAB−/− HeLa cells lack active lysosomal hydrolases. (A) The active form of cathepsin D is absent from infected GNPTAB−/− cells. Immunoblot of procathepsin and mature forms of cathepsin D in cells at 3 dpi. (B and C) CCVs in GNPTAB−/− cells are deficient in cathepsin D. Cells were infected for 3 days with mCherry-NMII, fixed with PFA, and fluorescently stained for cathepsin D using BODIPY FL pepstatin A and the CCV membrane using anti-LAMP1 antibody. Images were analyzed for intensity of BODIPY FL pepstatin A per CCV. (D and E) CCVs in GNPTAB−/− cells lack sulfatase activity. Cells were infected for 3 days with mCherry-NMII, incubated for 18 h with SulfGreen, and imaged live. Images were analyzed for intensity of SulfGreen per CCV. Graphs represent the means ± standard deviations (SD) of ≥150 cells from 3 independent experiments. Statistical significance was determined by Student's t test (****, P < 0.0001). Nuclei were stained with Hoechst stain. Scale bar, 10 μm.
GNPTAB−/− HeLa cells support C. burnetii growth, but CCVs are abnormal.
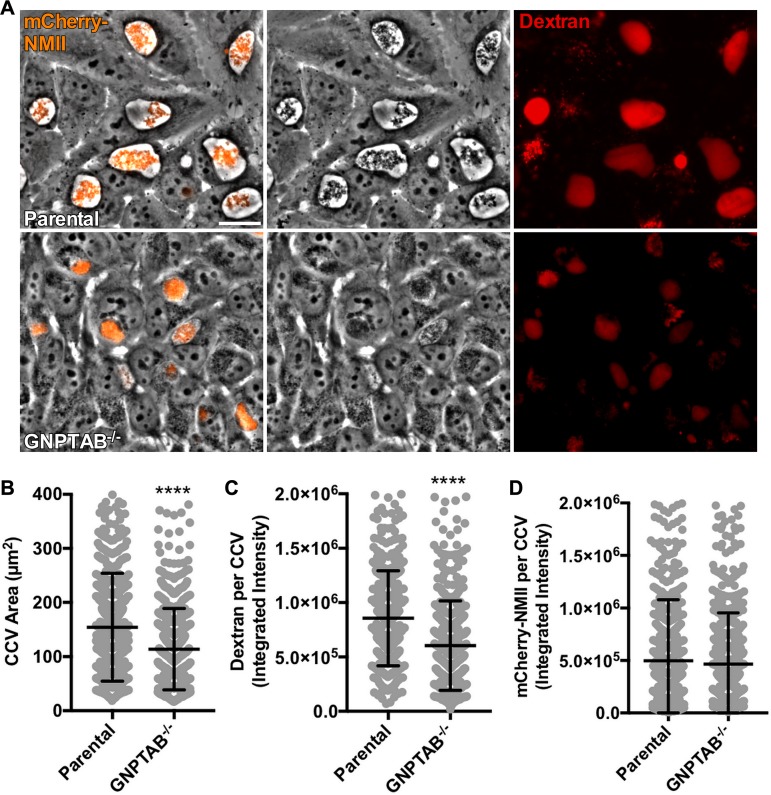
To determine whether C. burnetii requires lysosomal hydrolases for survival within a host cell, bacterial replication and CCV formation were examined in GNPTAB−/− cells. Cells were infected with an mCherry-expressing Nine Mile phase II strain (mCherry-NMII) of C. burnetii for 3 days and examined using live microscopy. Phase contrast images revealed infected GNPTAB−/− cells contain abnormal CCVs with reduced size and dark appearance (Fig. 2A and B). Decreased CCV area suggested reduced fusion with endosomes (22, 23). To test this idea, HeLa cells were incubated with fluorescent dextran for 18 h, and then fluorescence intensity of CCVs was measured (Fig. 2A and C). CCVs in GNPTAB−/− cells had decreased dextran intensity compared to that of parental cells. Although CCVs within GNPTAB−/− cells were irregular, analysis of fluorescence intensity of mCherry-NMII per CCV was similar to that of parental cells (Fig. 2A and D), suggesting comparable C. burnetii growth.
FIG 2.
GNPTAB−/− HeLa cells support C. burnetii growth, but CCVs are abnormal. (A to C) C. burnetii growing in GNPTAB−/− cells generates dark, condensed CCVs with decreased dextran fluorescence. Cells were infected with mCherry-NMII for 3 days, incubated for 18 h with Alexa Fluor 647 dextran, and imaged live. Images were analyzed for CCV area and dextran fluorescence intensity. (D) The growth of C. burnetii per vacuole is normal in GNPTAB−/− cells. Images from panel A were analyzed for mCherry-NMII fluorescence intensity per CCV. Graphs represent the means ± SD of ≥150 cells from 3 independent experiments. Statistical significance was determined by Student's t test (****, P < 0.0001). Scale bar, 10 μm.
C. burnetii replication and viability do not require lysosomal hydrolases.
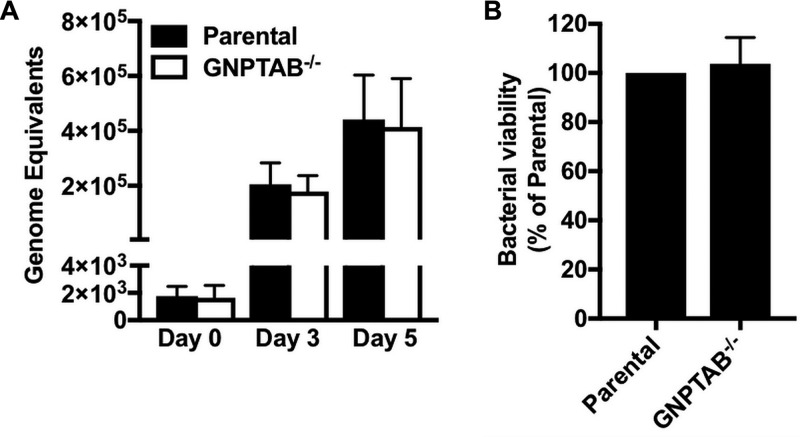
C. burnetii growth was further assessed by measuring bacterial genome equivalents using quantitative PCR (qPCR). Consistent with mCherry-NMII fluorescence results (Fig. 2A and D), no defect in replication was detected in GNPTAB−/− cells relative to levels in the parental cells (Fig. 3A). Furthermore, a fluorescent infectious focus-forming unit (FFU) assay demonstrated bacterial viability was similar for C. burnetii grown in GNPTAB−/− cells and parental cells (Fig. 3B). These data show lysosomal hydrolases are not required for C. burnetii growth or viability.
FIG 3.
C. burnetii replication and viability are normal in GNPTAB−/− HeLa cells. (A) The growth of C. burnetii in GNPTAB−/− cells is similar to that in parental cells. Genome equivalents were determined using qPCR of dotA. (B) The viability of C. burnetii grown in GNPTAB−/− cells is normal. C. burnetii from HeLa cells at 3 dpi was used to infect monolayers of Vero cells, and CCVs were enumerated 5 dpi by a fluorescent infectious focus-forming unit (FFU) assay. Graphs represent the means ± SD from 3 independent experiments.
CCVs in GNPTAB−/− HeLa cells accumulate material.
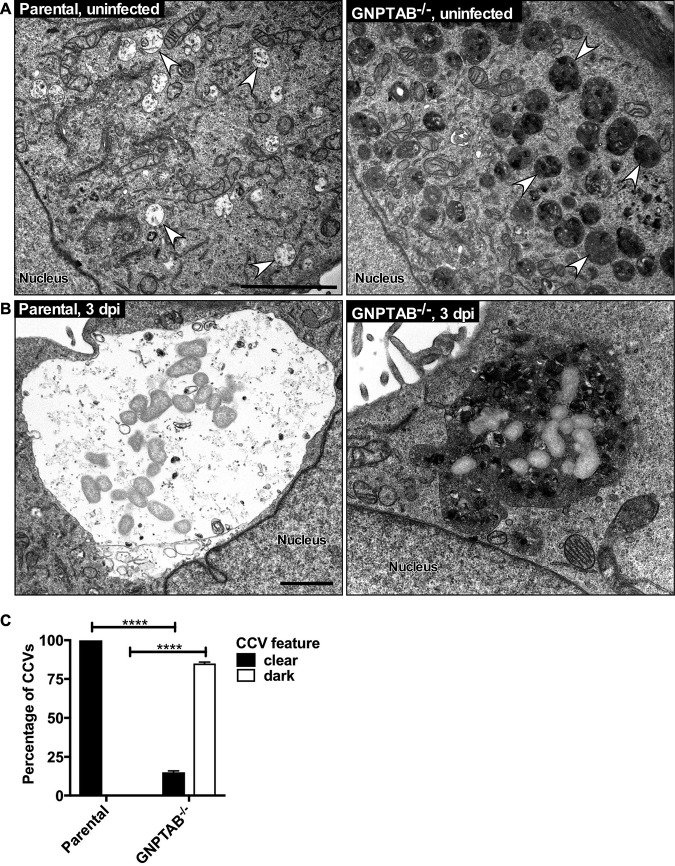
CCV biogenesis occurs primarily via prolific fusion with late endocytic vesicles. The abnormal nature of CCVs in GNPTAB−/− cells was further investigated by transmission electron microscopy (TEM). Uninfected GNPTAB−/− cells contained numerous electron-dense inclusions, similar to MLII fibroblasts (Fig. 4A). CCVs in GNPTAB−/− cells resembled gigantic inclusions, with the majority of CCVs appearing dark and dense rather than clear and spacious (Fig. 4B and C). Interestingly, vesicular structures, possibly intraluminal vesicles, were contained within CCVs, suggesting deficiency in degradation of host waste products. Overall, these results show that waste materials accumulate in CCVs lacking lysosomal hydrolases.
FIG 4.
CCVs in GNPTAB−/− HeLa cells accumulate electron-dense material. (A) By TEM, uninfected GNPTAB−/− cells contain abundant inclusions, observed as enlarged, dark endocytic vesicles. Arrows point to representative late endocytic vesicles. (B and C) CCVs in parental cells are clear and spacious, whereas the majority of CCVs in GNPTAB−/− cells are dense and filled with material. TEM images are of cells at 3 dpi. Images were analyzed for percentage of clear versus dark vacuoles. Magnification of TEM images was at 11,000×. Graphs represent the means ± SD of ≥60 cells from 2 independent experiments. Statistical significance determined by Student's t test (****, P < 0.0001). Scale bar, 1 μm.
Lysosomal hydrolases eliminate intraluminal vesicles from inside the CCV.
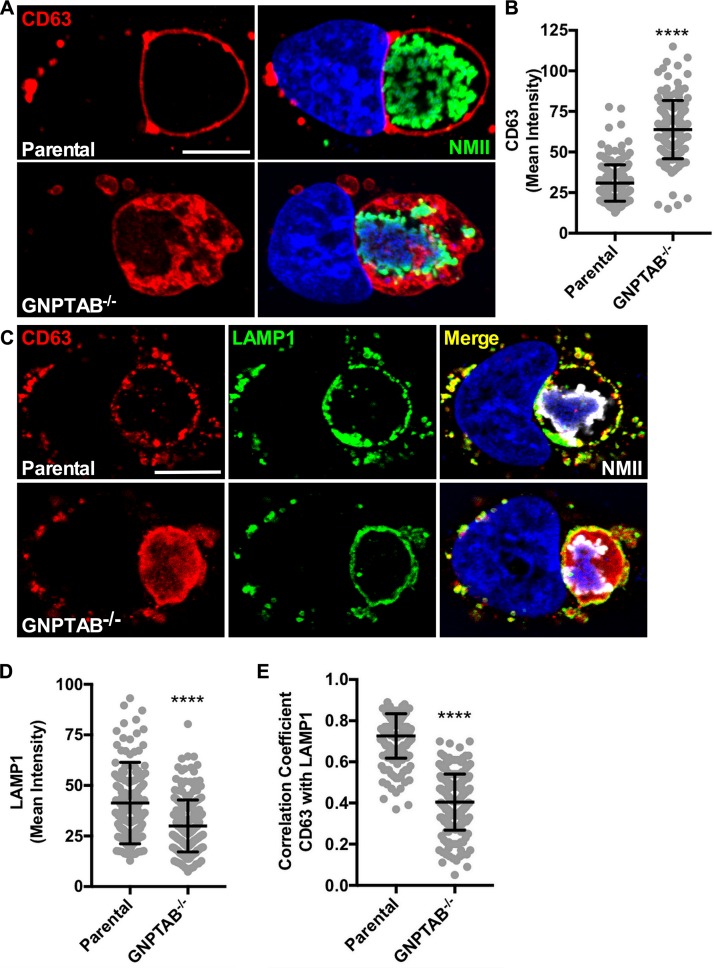
TEM imaging revealed a buildup of vesicles within CCVs in GNPTAB−/− cells. To determine if these were intraluminal vesicles, cells were stained for CD63, a transmembrane protein localized on intraluminal vesicles and late endosomal membranes (24). Methanol fixation followed by fluorescence staining of CD63 revealed that, unlike 3 dpi parental cells that contained CD63 primarily on the CCV membrane, CCVs in GNPTAB−/− cells accumulated CD63+ vesicles within the vacuole lumen (Fig. 5A and B). This indicated intraluminal vesicles are not being degraded by hydrolases or removed by back-fusion, whereby intraluminal vesicles fuse with the limiting membrane of the vacuole (25). These results demonstrate that lysosomal hydrolases are active in the CCV and are required for elimination of CD63+ intraluminal vesicles.
FIG 5.
CCVs in GNPTAB−/− HeLa cells accumulate intraluminal vesicles and have redistributed CCV membrane proteins. (A and B) CCVs in GNPTAB−/− cells have increased amounts of intraluminal CD63+ vesicles. Cells at 3 dpi were fixed with methanol and fluorescently stained for CD63. Images were analyzed for CD63 intensity per CCV. (C to E) LAMP1 on CCV membranes of GNPTAB−/− cells is redistributed, with decreased levels and colocalization with CD63 relative to those of parental cells. Cells at 3 dpi were fixed with 4% PFA and fluorescently stained for CD63 and LAMP1. Images were analyzed for LAMP1 intensity per CCV and colocalization of CD63 with LAMP1. Graphs represent the means ± SD of ≥150 cells from 3 independent experiments. Colocalization analysis of CCVs was determined using Pearson’s correlation coefficient. Statistical significance was determined by Student's t test (****, P < 0.0001). Nuclei were stained with Hoechst stain and C. burnetii with NMII-specific antibody. Scale bar, 10 μm.
Absence of lysosomal hydrolases redistributes CCV membrane proteins.
The CCV membrane regulates fusion with vesicles, which is impaired in GNPTAB−/− cells. In addition to CD63, the CCV membrane contains the lysosomal membrane protein LAMP1 (3, 26). Fluorescence imaging was used to examine infected GNPTAB−/− cells to determine whether the lack of hydrolases disrupts the organization of LAMP1 and CD63 domains on the CCV membrane. For fluorescence staining, cells were fixed with paraformaldehyde instead of methanol to better preserve the natural state of membrane structures. In both parental and knockout cells, CCVs at 3 dpi labeled with LAMP1 and CD63, but the distribution of the proteins on the limiting membrane differed (Fig. 5C). CCVs in parental cells exhibited punctate localization of LAMP1 and CD63, but the knockout cells exhibited uniform localization. Aberrant distribution of LAMP1 of CCVs in GNPTAB−/− cells correlated with reduced protein levels (Fig. 5C and D). Moreover, colocalization between CD63 and LAMP1 on CCVs in GNPTAB−/− cells was reduced, with the majority of CD63 localized to the lumen of the CCV (Fig. 5C and E). These results show lysosomal hydrolase activity is needed for normal CCV membrane organization.
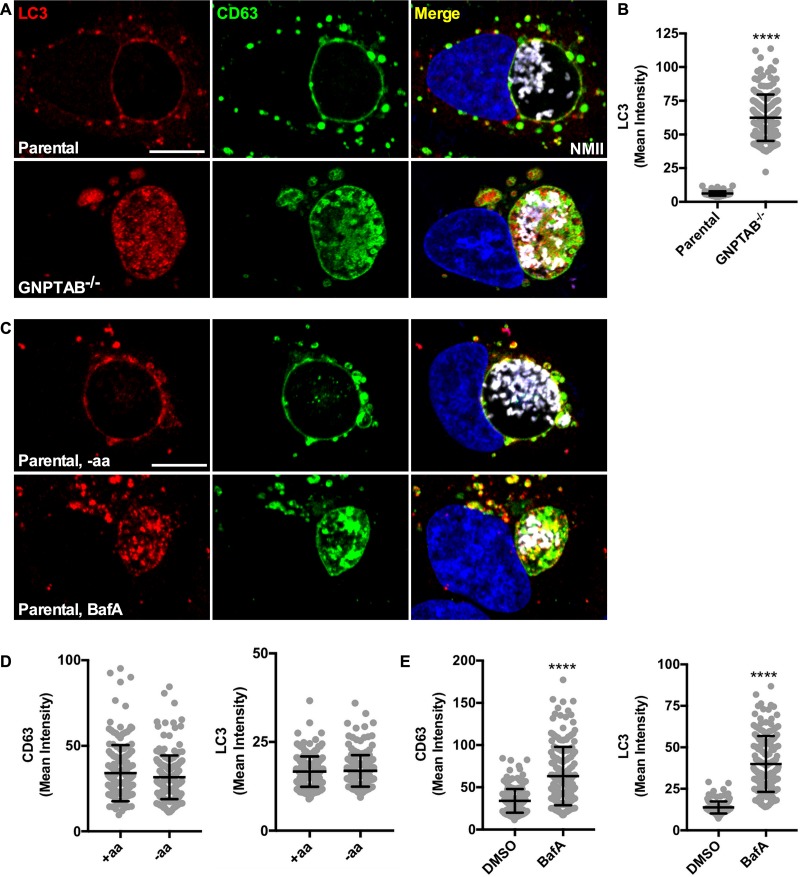
LC3 accumulates in CCVs without lysosomal hydrolases.
The CCV fuses not only with late endocytic vesicles but also with autophagosomes (22, 27); thus, we speculated that GNPTAB−/− cells would accumulate autophagic components. To examine this possibility, cells at 3 dpi were methanol fixed and stained for LC3, an autophagy protein that labels autophagic vesicles and is degraded upon fusion with late endosomes and lysosomes (28, 29). In parental cells, LC3 localized to the CCV membrane. Conversely, in GNPTAB−/− cells, large amounts of LC3 accumulated within the lumen of the CCV (Fig. 6A and B). To rule out the possibility that LC3 accumulation resulted from increased autophagic flux, LC3 was examined when autophagy was induced by amino acid starvation. Induction of autophagy for 4 h did not increase LC3 within CCVs of parental cells. However, a 4-h treatment with bafilomycin A1, an inhibitor of vacuolar (H+)-ATPase that blocks vacuole acidification and hydrolase activity (30), caused LC3 and CD63 accumulation (Fig. 6C to E). Thus, active hydrolases efficiently degrade CCV LC3 even when autophagic flux is increased, but lack of hydrolase activity results in accumulation of LC3.
FIG 6.
Autophagy protein LC3 accumulates within CCVs in GNPTAB−/− HeLa cells. (A and B) LC3 levels are elevated within CCVs in GNPTAB−/− cells. Cells at 3 dpi were fixed with methanol and fluorescently stained for LC3 and CD63. Images were analyzed for LC3 intensity per CCV. (C to E) Bafilomycin treatment, but not induction of autophagy, accumulates LC3 and CD63 within CCVs of parental cells. Parental cells at 3 dpi were amino acid starved (−aa) or treated with bafilomycin A1 (BafA) for 4 h and then fixed with methanol and stained for LC3 and CD63. Images were analyzed for CD63 and LC3 intensity per CCV. Graphs represent the means ± SD of ≥150 cells from 3 independent experiments. Statistical significance was determined by Student's t test (****, P < 0.0001). Nuclei were stained with Hoechst and C. burnetii with NMII-specific antibody. Scale bar, 10 μm.
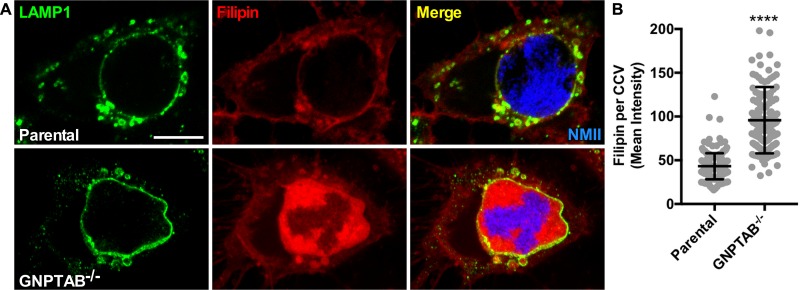
CCVs in GNPTAB−/− cells accumulate cholesterol.
Neimann-Pick C2 (NPC2) is an M6PR pathway-dependent lysosomal protein that transports cholesterol out of endosomes. GNPTAB−/− cells lack NPC2 in endosomes, resulting in accumulation of cholesterol within late endocytic compartments (31). Cells were stained with filipin to investigate whether cholesterol levels are elevated in CCVs of GNPTAB−/− cells. Cholesterol primarily localized to the CCV membrane of infected parental cells, which was in contrast to localization in the CCV lumen of GNPTAB−/− cells (Fig. 7A and B). These results demonstrate that without M6PR pathway-dependent lysosomal proteins, cholesterol accumulates within the CCV.
FIG 7.
CCVs in GNPTAB−/− HeLa cells accumulate cholesterol. (A and B) Cholesterol builds up inside CCVs in GNPTAB−/− cells. At 3 dpi with mCherry-NMII, cells were fixed with 4% PFA and stained for cholesterol using filipin and the CCV membrane using anti-LAMP1 antibody. Images were analyzed for filipin intensity per CCV. Graphs represent the means ± SD of ≥150 cells from 2 independent experiments. Statistical significance was determined by Student's t test (****, P < 0.0001). Scale bar, 10 μm.
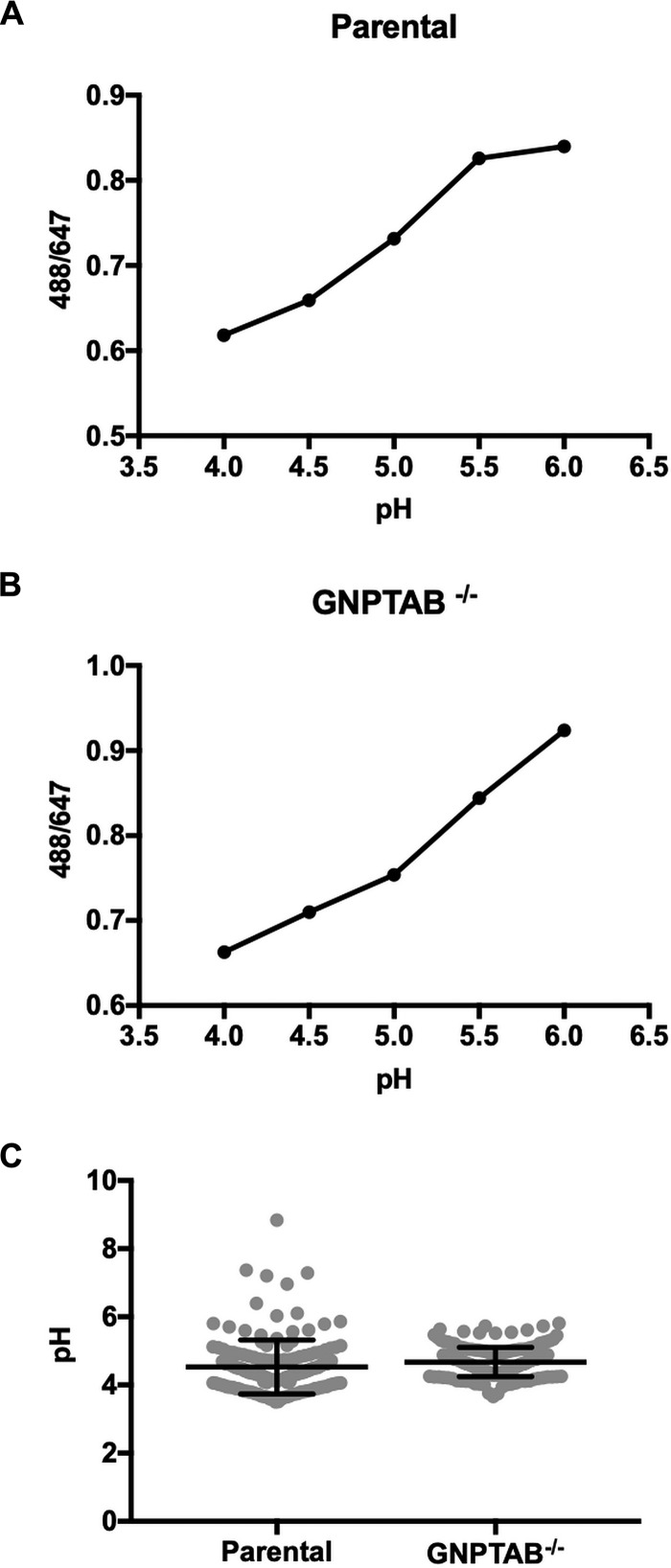
The pH of CCVs in GNPTAB−/− and parental cells is similar.
Mulye et al. (32) demonstrated that elevation of CCV cholesterol by pharmacological treatment results in C. burnetii lysis. CCV accumulation of cholesterol is associated with decreased CCV pH relative to that of untreated cells, a factor proposed to increase hydrolase activity and consequently bacterial death. We observed no significant difference in the pH of CCVs in GNPTAB−/− (pH 4.67) and parental (pH 4.53) cells (Fig. 8), despite accumulation of cholesterol by knockout cells.
FIG 8.
pH of CCVs in parental and GNPTAB−/− HeLa cells is similar. (A and B) The standard curves for CCVs of parental and knockout cells were generated by graphing fluorescent intensity ratios of Oregon Green 488/Alexa Fluor 647 with pH. Cells at 3 dpi with mCherry-NMII were incubated for 18 h with Oregon Green 488-dextran and Alexa Fluor 647-dextran. Standard curve samples were made by incubating cells with buffers of different pHs containing nigericin and monensin. (C) CCVs in GNPTAB−/− and parental cells have a similar pH. The pH of CCVs was determined using the standard curves. CCVs in GNPTAB−/− and parental cells have a pH of 4.67 and 4.53, respectively. Graphs represent the means ± SD of ≥150 cells from 2 independent experiments.
DISCUSSION
In this study, we show lysosomal hydrolases are required for normal CCV development but not for C. burnetii survival and growth. C. burnetii replication and viability are normal in hydrolase-deficient GNPTAB−/− HeLa cells, but CCVs are smaller and densely packed with material, resembling electron-dense inclusions of cells from MLII patients. This phenotype results from failed degradation of cellular waste products. Similarly, we show CCVs accumulate intraluminal vesicles, autophagy protein LC3, and cholesterol. Additionally, the CCV membrane is altered, with a decrease in LAMP1 levels and a redistribution of CD63 and LAMP1. These results suggest lysosomal hydrolases have a functional role in CCV biogenesis.
The reduced size of CCVs in GNPTAB−/− cells correlates with decreased sequestration of dextran, a fluid phase marker. This behavior likely reflects defects in fusion between the CCV and endocytic vesicles that manifest due to changes on the CCV membrane that are induced by accumulated waste products. CD63 and LAMP1 are uniformly distributed on CCVs in GNPTAB−/− cells as opposed to a typical punctate localization (33). The uniform distribution of CCV membrane molecules could result from deficient lipid raft formation. Lipid rafts are membrane microdomains of lipids and proteins that are important in fusion events (34). Furthermore, fusion may also be affected by decreased LAMP1 in CCV membranes of GNPTAB−/− cells. LAMP1 promotes fusion of lysosomes with phagosomes (35). Indeed, Schulze-Luehrmann et al. (36) showed decreased CCV size in LAMP1/LAMP2 double knockout cells, indicating a role for LAMPs in CCV expansion.
The expansive growth of the CCV is supported by enhanced fusion with late endosomes, lysosomes, and autophagosomes, all of which provide membrane for the CCV but also deliver waste products. In the absence of degradative lysosomal hydrolases, waste products, including cholesterol, build up in the CCV. This material does not adversely affect growth or viability of C. burnetii. Interestingly, a previous report showed that the CCV becomes more acidic, and consequently bactericidal, when cholesterol accumulates (32). Increased hydrolase activity is one mechanism proposed for C. burnetii killing. We demonstrate that CCVs of both GNPTAB−/− and parental cells have a typical lysosomal pH. It should be noted that because CCVs in GNPTAB−/− cells lack hydrolases, conditions that may potentiate hydrolase activity are irrelevant with respect to C. burnetii viability.
Given the degradative, phagolysosome-like character of the CCV, it was presumed that C. burnetii scavenges nutrients generated by hydrolytic breakdown of host waste material. Because C. burnetii survival and replication do not require lysosomal hydrolases, the question remains of how C. burnetii obtains nutrients from the host. One possibility is that C. burnetii breaks down host macromolecules using its own secreted enzymes. This scenario is unlikely, since CCVs in GNPTAB−/− cells accumulate material, suggesting enzymes are not degrading host waste products. An emerging concept in bacterial pathogenesis is nutritional virulence, whereby intracellular bacteria are specifically adapted to obtain essential nutrients by exploiting host metabolic processes (37). Several bacteria, including Anaplasma phagocytophilum and Francisella tularensis, utilize host autophagy for nutrient acquisition (6). Investigators have speculated that C. burnetii can also obtain nutrients from the host via CCV fusion with autophagosomes (6, 9). However, induction of autophagy by amino acid starvation or Torin-1 treatment does not benefit C. burnetii growth (38). Furthermore, C. burnetii infection does not increase autophagic flux. C. burnetii and L. pneumophila both use amino acids as main carbon and energy sources. They share auxotrophies for several amino acids, which consequently must be obtained from the host (6, 39, 40). Like L. pneumophila, intravacuolar Chlamydia trachomatis and Salmonella enterica use SLC transporters to derive critical nutrients, including glucose (SLC35D2) and arginine (SLC7A1), respectively (6–8). Lysosomes contain several SLC transporters, including those that pump amino acids (SLC7A14 and SLC36A1) and glucose (SLC2A8) into the vesicle lumen (41). Given the lysosomal character of the CCV, the vacuole likely contains these and other transport systems that provide C. burnetii with basic nutrients. Indeed, studies show that both cytosolic and lysosomal concentrations of essential amino acids largely exceed levels needed to support robust growth of C. burnetii in a defined medium (42–44). C. burnetii grows to tremendous numbers in host cells without obvious negative impact on cell physiology and division (45). The precise mechanisms by which host cell amino acid homeostasis is maintained to support such a parasitic burden is an intriguing area of future investigation.
MATERIALS AND METHODS
Cell culture and infection.
Parental (wild-type) and GNPTAB−/− HeLa cells were kindly provided by Stuart Kornfeld of the Washington University School of Medicine, St. Louis, Missouri. Generation of the knockout cell line by CRISPR/Cas9 genome editing is described in van Meel et al. (19). HeLa cell lines were cultured in Dulbecco’s modified Eagle medium (DMEM) (Life Technologies) supplemented with 10% fetal bovine serum (FBS) and 1 mM sodium pyruvate (Life Technologies) and incubated at 37°C with 5% CO2. Vero (ATCC CCL-81; African green monkey epithelial) cells were cultured in RPMI 1640 medium (Life Technologies) supplemented with 10% FBS and incubated at 37°C with 5% CO2.
Coxiella burnetii Nine Mile RSA439, phase II, clone 4 (NMII) was grown in Vero cells and purified as previously described (46). NMII expressing mCherry (mCherry-NMII) was propagated in second-generation acidified citrate cysteine medium (ACCM-2) at 37°C with 5% CO2 and 2.5% O2 (47). For infections, HeLa cells were seeded in 24-well plates at 6 × 104 cells per well and infected at a multiplicity of infection (MOI) of 10 by centrifuging bacteria onto monolayers at 500 × g for 30 min at room temperature.
CCV fusion assay.
HeLa cells seeded on glass-bottom 24-well SensoPlates (662892; Greiner Bio-One) were infected with mCherry-NMII. At 2 dpi, Alexa Fluor 647-dextran of 10,000 molecular weight (D22914; ThermoFisher) was added to the cells at 1 mg/ml in complete DMEM, and cells were incubated for 18 h. Cells were then washed with fresh medium and imaged using a Nikon eclipse Ti2 epifluorescence microscope. Analysis was performed using Fiji (Image J) (NIH), whereby mCherry-positive CCVs were selected for measurements of area and fluorescence intensities of dextran and mCherry-NMII.
C. burnetii growth in HeLa cells.
Following infection, HeLa cells were washed to remove free bacteria. Samples were collected by trypsinization of cells at day 0 (immediately after infection) and days 3 and 5. To release bacterial DNA from cells, samples were bead beaten with 0.1-mm zirconia/silica beads (BioSpec) in a homogenizer (FastPrep FP120; ThermoElectron), followed by 10 min of boiling. To determine genome equivalents (GE), TaqMan qPCR using a StepOnePlus real-time PCR system (Applied Biosystems) was performed with primers specific for dotA (48).
C. burnetii viability assay.
At 3 dpi, HeLa cells were lysed with 250 μl of water and lysates were diluted 1:5 with RPMI containing 2% FBS. Confluent monolayers of Vero cells in 24-well plates were infected with serial dilutions of samples for 4 h and then washed to remove free bacteria. At 5 dpi, cells were fixed with methanol for 30 min and stained using anti-NMII rabbit antibody followed by Alexa Fluor 488-goat anti-rabbit antibody (Life Technologies). To enumerate CCVs, a fluorescent infectious focus-forming unit (FFU) assay was conducted, with 10 fields of view per well counted on a Zeiss Axiovert 25 fluorescence microscope at ×32 magnification.
Western blot for procathepsin and mature cathepsin D.
Cell lysates of infected HeLa cells were run on an SDS-PAGE gel and transferred onto nitrocellulose membrane. After blocking with 5% skim milk, the membrane was probed with anti-cathepsin D monoclonal rabbit antibody (clone EPR3057Y; ab75852; Abcam), followed by horseradish peroxidase-goat anti-rabbit IgG antibody (A16104; ThermoFisher). Immunodetection of cathepsin D was performed using enhanced chemiluminescence substrate (ThermoFisher) and the blot imaged with an Azure C600 imager (Azure Biosystems).
Cathepsin D staining.
At 3 dpi with mCherry-NMII, HeLa cells were fixed with 4% paraformaldehyde (PFA). Cells were blocked and permeabilized with 0.05% saponin in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA). To stain for cathepsin D, cells were incubated for 1 h with 2 μg/ml of BODIPY FL pepstatin A (P12271; ThermoFisher). For staining the CCV membrane, cells were incubated for 1 h with anti-LAMP1 antibody (ab24170; Abcam), followed by Alexa Fluor 488 goat anti-rabbit antibody for 30 min. All antibody incubations were in saponin permeabilization/blocking buffer. Nuclei were stained with Hoechst 33342 (ThermoFisher) in PBS. Coverslips were mounted using Prolong Gold antifade mountant (ThermoFisher). Cells were imaged using a Zeiss LSM-710 confocal fluorescence microscope. Fluorescence intensities of BODIPY FL pepstatin A in CCVs were measured using Fiji.
Sulfatase activity assay.
HeLa cells seeded on glass-bottom 24-well SensoPlates were infected with mCherry-NMII. A MarkerGene LysoLive lysosomal sulfatase assay kit was used to determine sulfatase activity by fluorescence imaging. At 2 dpi, cells were incubated for 18 h with 200 μM SulfGreen and 1 mg/ml of Alexa Fluor 647-dextran of 10,000 MW. Cells were washed with fresh medium and imaged using a Nikon eclipse Ti2 epifluorescence microscope. Analysis was performed using Fiji, whereby mCherry-positive CCVs, marked by dextran, were selected for analysis of fluorescence intensities of SulfGreen.
Transmission electron microscopy.
For TEM, HeLa cells were seeded on Thermanox plastic coverslips (Ted Pella, Inc.) in 24-well plates and infected with C. burnetii. Cells at 3 dpi were fixed overnight at 4°C in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer. After 3 washes with 0.1 M sodium cacodylate buffer, the cells were postfixed for 1 h with 0.5% osmium tetroxide and 0.8% potassium ferricyanide in 0.1 M sodium cacodylate buffer. Following 3 washes with 0.1 M sodium cacodylate buffer, the cells were stained for 1 h with 1% tannic acid in distilled water (dH2O), rinsed 3 times with dH2O, and stained for 1 h at 4°C with 2% samarium acetate in dH2O. The cells were dehydrated in a graded ethanol series, followed by three exchanges in 100% ethanol. The cells were infiltrated and embedded with Spurr’s resin and cured overnight at 68°C. The samples were sectioned at 80 nm using a UC6 ultramicrotome (Leica Microsystems) and viewed on a Tecnai BioTwin Spirit (Thermo Fisher Scientific) at 120 kV. Images were acquired with a Hamamatsu ORCA-HR digital camera system (Advanced Microscopy Techniques).
Immunofluorescence staining of intraluminal vesicles, LC3, and the CCV membrane.
For staining intraluminal vesicles and LC3, cells at 3 dpi were fixed with cold methanol for 30 min. Cells were blocked using 0.1% Triton X-100 in PBS containing 1% BSA and then stained with anti-CD63 monoclonal mouse antibody (clone H5C6; 556019; BD Pharmingen) or anti-LC3B monoclonal rabbit antibody (clone D11; 3868S; Cell Signaling Technology). Antibody incubations were for 30 to 60 min in 0.1% Triton X-100 in PBS containing 1% BSA.
For CCV membrane staining, cells at 3 dpi were fixed with 4% paraformaldehyde in PBS. Cells were permeabilized and blocked for 30 min with 0.05% saponin in PBS containing 1% BSA and then stained with anti-CD63 monoclonal mouse antibody or anti-LAMP1 rabbit antibody. Antibody incubations were in saponin permeabilization/blocking buffer for 30 to 60 min.
NMII was stained with anti-C. burnetii guinea pig antibody and nuclei with Hoechst 33342 (ThermoFisher). Alexa Fluor 647-, 568-, and 488-conjugated secondary antibodies were from Life Technologies. Coverslips were mounted using Prolong Gold antifade mountant. A Zeiss LSM-710 confocal fluorescence microscope was used for imaging. Fiji was used for analysis, where fluorescence intensities of CD63, LC3, and LAMP1 were determined for CCVs. Correlation coefficients (Pearson’s correlation coefficient) were determined using Coloc 2.
Bafilomycin A1 treatment and amino acid starvation.
HeLa cells were treated with bafilomycin A1 (B-1183; A.G. Scientific, Inc.) at 400 nM in culture medium for 4 h at 37°C with 5% CO2. For amino acid starvation, cells were incubated for 4 h at 37°C with 5% CO2 in RPMI 1640 medium modified without amino acids, plus glucose (MyBioSource).
Cholesterol staining.
At 3 dpi with mCherry-NMII, HeLa cells were fixed with 4% PFA for 30 min. Cells were stained for cholesterol by incubating for 1 h with 100 μg/ml of filipin (70440; Cayman Chemical) in PBS. The CCV membrane was stained by incubating cells for 30 min with anti-LAMP1 antibody in PBS followed by incubation with Alexa Fluor 488 goat anti-rabbit antibody in PBS for 30 min. Coverslips were then mounted using Prolong Gold antifade mountant. Cells were imaged using a Zeiss LSM-710 confocal fluorescence microscope (Carl Zeiss). Fluorescence intensity of filipin per CCV was measured using Fiji.
CCV pH determination.
The pH of CCVs was determined as previously described (49). Briefly, HeLa cells seeded on glass-bottom 24-well SensoPlates were infected with mCherry-NMII for 3 days. The cells were incubated for 18 h with 1 mg/ml of both Oregon Green 488-dextran and Alexa Fluor 647-dextran of 10,000 MW (ThermoFisher). Standard samples were made by incubating cells with buffers of different pHs containing 10 μM both nigericin and monensin (Sigma). Cells were imaged at room temperature using a Nikon eclipse Ti2 epifluorescence microscope. Fluorescence intensity analysis of dextrans was performed using Fiji. A standard curve was generated for CCVs of parental and knockout cells by graphing fluorescent intensity ratios of Oregon Green 488/Alexa Fluor 647 with pH. The pHs of the experimental CCVs were then determined using the standard curve.
Graphing and statistics.
GraphPad Prism (GraphPad Software) was used for all graphing and statistics. Unpaired Student's t test was used to determine statistical significance.
ACKNOWLEDGMENTS
We are grateful to Stuart Kornfeld of the Washington University School of Medicine for GNPT knockout cells. We appreciate the advice and support contributed by Lin Liu of the Washington University School of Medicine. We are thankful to Charlie Larson for critical review of the manuscript.
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Eldin C, Melenotte C, Mediannikov O, Ghigo E, Million M, Edouard S, Mege JL, Maurin M, Raoult D. 2017. From Q fever to Coxiella burnetii infection: a paradigm change. Clin Microbiol Rev 30:115–190. doi: 10.1128/CMR.00045-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Schaik EJ, Chen C, Mertens K, Weber MM, Samuel JE. 2013. Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat Rev Microbiol 11:561–573. doi: 10.1038/nrmicro3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinzen RA, Scidmore MA, Rockey DD, Hackstadt T. 1996. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun 64:796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veras PS, de Chastellier C, Moreau MF, Villiers V, Thibon M, Mattei D, Rabinovitch M. 1994. Fusion between large phagocytic vesicles: targeting of yeast and other particulates to phagolysosomes that shelter the bacterium Coxiella burnetii or the protozoan Leishmania amazonensis in Chinese hamster ovary cells. J Cell Sci 107:3065–3076. [DOI] [PubMed] [Google Scholar]
- 5.Howe D, Shannon JG, Winfree S, Dorward DW, Heinzen RA. 2010. Coxiella burnetii phase I and II variants replicate with similar kinetics in degradative phagolysosome-like compartments of human macrophages. Infect Immun 78:3465–3474. doi: 10.1128/IAI.00406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Best A, Abu Kwaik Y. 2019. Nutrition and bipartite metabolism of intracellular pathogens. Trends Microbiol 27:550–561. doi: 10.1016/j.tim.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manske C, Hilbi H. 2014. Metabolism of the vacuolar pathogen Legionella and implications for virulence. Front Cell Infect Microbiol 4:125. doi: 10.3389/fcimb.2014.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wieland H, Ullrich S, Lang F, Neumeister B. 2005. Intracellular multiplication of Legionella pneumophila depends on host cell amino acid transporter SLC1A5. Mol Microbiol 55:1528–1537. doi: 10.1111/j.1365-2958.2005.04490.x. [DOI] [PubMed] [Google Scholar]
- 9.Gutierrez MG, Colombo MI. 2005. Autophagosomes: a fast-food joint for unexpected guests. Autophagy 1:179–181. doi: 10.4161/auto.1.3.2063. [DOI] [PubMed] [Google Scholar]
- 10.Heinzen RA, Hackstadt T, Samuel JE. 1999. Developmental biology of Coxiella burnettii. Trends Microbiol 7:149–154. doi: 10.1016/S0966-842X(99)01475-4. [DOI] [PubMed] [Google Scholar]
- 11.Xu H, Ren D. 2015. Lysosomal physiology. Annu Rev Physiol 77:57–80. doi: 10.1146/annurev-physiol-021014-071649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Appelqvist H, Waster P, Kagedal K, Ollinger K. 2013. The lysosome: from waste bag to potential therapeutic target. J Mol Cell Biol 5:214–226. doi: 10.1093/jmcb/mjt022. [DOI] [PubMed] [Google Scholar]
- 13.Coutinho MF, Prata MJ, Alves S. 2012. Mannose-6-phosphate pathway: a review on its role in lysosomal function and dysfunction. Mol Genet Metab 105:542–550. doi: 10.1016/j.ymgme.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Reitman ML, Varki A, Kornfeld S. 1981. Fibroblasts from patients with I-cell disease and pseudo-Hurler polydystrophy are deficient in uridine 5’-diphosphate-N-acetylglucosamine: glycoprotein N-acetylglucosaminylphosphotransferase activity. J Clin Investig 67:1574–1579. doi: 10.1172/jci110189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coutinho MF, Prata MJ, Alves S. 2012. A shortcut to the lysosome: the mannose-6-phosphate-independent pathway. Mol Genet Metab 107:257–266. doi: 10.1016/j.ymgme.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 16.Hasanagic M, Waheed A, Eissenberg JC. 2015. Different pathways to the lysosome: sorting out alternatives. Int Rev Cell Mol Biol 320:75–101. doi: 10.1016/bs.ircmb.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Braulke T, Bonifacino JS. 2009. Sorting of lysosomal proteins. Biochim Biophys Acta 1793:605–614. doi: 10.1016/j.bbamcr.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 18.Braulke T, Raas-Rothschild A, Kornfeld S. 2014. I-cell disease and pseudo-Hurler polydystrophy: disorders of lysosomal enzyme phosphorylation and localization In Beaudet AL, Vogelstein B, Kinzler KW, Antonarakis SE, Ballabio A, Gibson KM, Mitchell G (ed), The online metabolic and molecular bases of inherited disease. The McGraw-Hill Companies, Inc, New York, NY. [Google Scholar]
- 19.van Meel E, Lee WS, Liu L, Qian Y, Doray B, Kornfeld S. 2016. Multiple domains of GlcNAc-1-phosphotransferase mediate recognition of lysosomal enzymes. J Biol Chem 291:8295–8307. doi: 10.1074/jbc.M116.714568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaidi N, Maurer A, Nieke S, Kalbacher H. 2008. Cathepsin D: a cellular roadmap. Biochem Biophys Res Commun 376:5–9. doi: 10.1016/j.bbrc.2008.08.099. [DOI] [PubMed] [Google Scholar]
- 21.Chen CS, Chen WN, Zhou M, Arttamangkul S, Haugland RP. 2000. Probing the cathepsin D using a BODIPY FL-pepstatin A: applications in fluorescence polarization and microscopy. J Biochem Biophys Methods 42:137–151. doi: 10.1016/S0165-022X(00)00048-8. [DOI] [PubMed] [Google Scholar]
- 22.Beron W, Gutierrez MG, Rabinovitch M, Colombo MI. 2002. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect Immun 70:5816–5821. doi: 10.1128/IAI.70.10.5816-5821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campoy EM, Mansilla ME, Colombo MI. 2013. Endocytic SNAREs are involved in optimal Coxiella burnetii vacuole development. Cell Microbiol 15:922–941. doi: 10.1111/cmi.12087. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi T, Vischer UM, Rosnoblet C, Lebrand C, Lindsay M, Parton RG, Kruithof EK, Gruenberg J. 2000. The tetraspanin CD63/lamp3 cycles between endocytic and secretory compartments in human endothelial cells. Mol Biol Cell 11:1829–1843. doi: 10.1091/mbc.11.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huotari J, Helenius A. 2011. Endosome maturation. EMBO J 30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghigo E, Capo C, Tung CH, Raoult D, Gorvel JP, Mege JL. 2002. Coxiella burnetii survival in THP-1 monocytes involves the impairment of phagosome maturation: IFN-gamma mediates its restoration and bacterial killing. J Immunol 169:4488–4495. doi: 10.4049/jimmunol.169.8.4488. [DOI] [PubMed] [Google Scholar]
- 27.Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, Rabinovitch M, Colombo MI. 2005. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol 7:981–993. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 28.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. 2005. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 29.Tanida I, Ueno T, Kominami E. 2008. LC3 and autophagy. Methods Mol Biol 445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- 30.Mauvezin C, Neufeld TP. 2015. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy 11:1437–1438. doi: 10.1080/15548627.2015.1066957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willenborg M, Schmidt CK, Braun P, Landgrebe J, von Figura K, Saftig P, Eskelinen EL. 2005. Mannose 6-phosphate receptors, Niemann-Pick C2 protein, and lysosomal cholesterol accumulation. J Lipid Res 46:2559–2569. doi: 10.1194/jlr.M500131-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Mulye M, Samanta D, Winfree S, Heinzen RA, Gilk SD. 2017. Elevated cholesterol in the Coxiella burnetii intracellular niche is bacteriolytic. mBio 8:e02313-16. doi: 10.1128/mBio.02313-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller HE, Larson CL, Heinzen RA. 2018. Actin polymerization in the endosomal pathway, but not on the Coxiella-containing vacuole, is essential for pathogen growth. PLoS Pathog 14:e1007005. doi: 10.1371/journal.ppat.1007005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanzal-Bayer MF, Hancock JF. 2007. Lipid rafts and membrane traffic. FEBS Lett 581:2098–2104. doi: 10.1016/j.febslet.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 35.Huynh KK, Eskelinen EL, Scott CC, Malevanets A, Saftig P, Grinstein S. 2007. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J 26:313–324. doi: 10.1038/sj.emboj.7601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schulze-Luehrmann J, Eckart RA, Ölke M, Saftig P, Liebler-Tenorio E, Lührmann A. 2016. LAMP proteins account for the maturation delay during the establishment of the Coxiella burnetii-containing vacuole. Cell Microbiol 18:181–194. doi: 10.1111/cmi.12494. [DOI] [PubMed] [Google Scholar]
- 37.Abu Kwaik Y, Bumann D. 2013. Microbial quest for food in vivo: “nutritional virulence” as an emerging paradigm. Cell Microbiol 15:882–890. doi: 10.1111/cmi.12138. [DOI] [PubMed] [Google Scholar]
- 38.Larson CL, Sandoz KM, Cockrell DC, Heinzen RA. 2019. Noncanonical inhibition of mTORC1 by Coxiella burnetii promotes replication within a phagolysosome-like vacuole. mBio 10:e02816-18. doi: 10.1128/mBio.02816-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallejo Esquerra E, Yang H, Sanchez SE, Omsland A. 2017. Physicochemical and nutritional requirements for axenic replication suggest physiological basis for Coxiella burnetii niche restriction. Front Cell Infect Microbiol 7:190. doi: 10.3389/fcimb.2017.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Häuslein I, Cantet F, Reschke S, Chen F, Bonazzi M, Eisenreich W. 2017. Multiple substrate usage of Coxiella burnetii to feed a bipartite-type metabolic network. Front Cell Infect Microbiol 7:285. doi: 10.3389/fcimb.2017.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bissa B, Beedle AM, Govindarajan R. 2016. Lysosomal solute carrier transporters gain momentum in research. Clin Pharmacol Ther 100:431–436. doi: 10.1002/cpt.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abu-Remaileh M, Wyant GA, Kim C, Laqtom NN, Abbasi M, Chan SH, Freinkman E, Sabatini DM. 2017. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358:807–813. doi: 10.1126/science.aan6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baydoun AR, Emery PW, Pearson JD, Mann GE. 1990. Substrate-dependent regulation of intracellular amino acid concentrations in cultured bovine aortic endothelial cells. Biochem Biophys Res Commun 173:940–948. doi: 10.1016/s0006-291x(05)80876-9. [DOI] [PubMed] [Google Scholar]
- 44.Sandoz KM, Beare PA, Cockrell DC, Heinzen RA. 2016. Complementation of arginine auxotrophy for genetic transformation of Coxiella burnetii by use of a defined axenic medium. Appl Environ Microbiol 82:3042–3051. doi: 10.1128/AEM.00261-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baca OG, Scott TO, Akporiaye ET, DeBlassie R, Crissman HA. 1985. Cell cycle distribution patterns and generation times of L929 fibroblast cells persistently infected with Coxiella burnetii. Infect Immun 47:366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cockrell DC, Beare PA, Fischer ER, Howe D, Heinzen RA. 2008. A method for purifying obligate intracellular Coxiella burnetii that employs digitonin lysis of host cells. J Microbiol Methods 72:321–325. doi: 10.1016/j.mimet.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Omsland A, Beare PA, Hill J, Cockrell DC, Howe D, Hansen B, Samuel JE, Heinzen RA. 2011. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl Environ Microbiol 77:3720–3725. doi: 10.1128/AEM.02826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coleman SA, Fischer ER, Howe D, Mead DJ, Heinzen RA. 2004. Temporal analysis of Coxiella burnetii morphological differentiation. J Bacteriol 186:7344–7352. doi: 10.1128/JB.186.21.7344-7352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samanta D, Gilk SD. 2017. Measuring pH of the Coxiella burnetii parasitophorous vacuole. Curr Protoc Microbiol 47:6c.3.1–6c.3.11. doi: 10.1002/cpmc.38. [DOI] [PMC free article] [PubMed] [Google Scholar]