Salmonella infection can cause gastroenteritis in healthy individuals or a serious, systemic infection in immunocompromised patients and has a global impact. CD4 Th1 cells represent the main lymphocyte population that participates in bacterial clearance during both primary and secondary infections in mice of the H-2b haplotype.
KEYWORDS: CD4 T cells, CD8 T cells, MHC, Salmonella
ABSTRACT
Salmonella infection can cause gastroenteritis in healthy individuals or a serious, systemic infection in immunocompromised patients and has a global impact. CD4 Th1 cells represent the main lymphocyte population that participates in bacterial clearance during both primary and secondary infections in mice of the H-2b haplotype. Previous studies have used congenic mice to examine the function of major histocompatibility complex (MHC) molecules in elimination of this pathogen from the host. In this study, we further characterized the ability of H-2b, H-2k, and H-2u molecules to influence adaptive immunity to Salmonella in MHC congenic mice. By depleting different cell populations during infection, we unexpectedly found that CD8 T cells, in addition to CD4 T cells, play a major role in accelerated clearance of bacteria from H-2k congenic hosts. Our data suggest that CD8 T cells accelerate clearance in some MHC congenic mouse strains and could therefore represent an unexpected contributor to the protective efficacy of Salmonella vaccines outside the typical studies in C57BL/6 mice.
INTRODUCTION
Enteric fever, caused by Salmonella enterica serovar Typhi, continues to be a major health concern in the developing world, infecting over 26 million people annually (1). Salmonella serovars can also cause gastroenteritis and invasive nontyphoidal salmonellosis (NTS), a systemic disease prevalent in sub-Saharan Africa (1–3). Although there are vaccines available for treatment of infections by Salmonella Typhi, none are currently available for other Salmonella serovars, including Salmonella Typhimurium (4, 5). Since Salmonella Typhi replicates only in a human host, it has been difficult to model this disease in vivo, and Salmonella Typhimurium infection of inbred mice is widely used as a model of systemic typhoidal and nontyphoidal disease (6, 7).
Mouse models have uncovered several mechanisms by which Salmonella spp. are able to invade and disseminate within the infected host. The bacteria initially exploit intestinal epithelial M cells to gain entry into Peyer’s patches, where they subsequently infect dendritic cells and macrophages (8, 9), before migrating to the mesenteric lymph node and blood via the lymphatic system (10). Under some circumstances, Salmonella spp. also infect lamina propria phagocytes that directly sample intestinal contents (11–13) or breach the epithelial barrier by disrupting tight junctions (14). Once infection is initiated in the intestine, it rapidly spreads to systemic tissues, where Salmonella replicates in the liver, spleen, and bone marrow (10).
Host innate and adaptive immune responses are initiated rapidly after Salmonella infection (15, 16). The major mechanism of bacterial killing during systemic salmonellosis is via the activation of macrophages by Th1 cell-secreted gamma interferon (IFN-γ) (17–19). Mice lacking CD4 T cells demonstrated delayed bacterial clearance and had higher bacterial burdens after a month of infection (14, 20). Data from human studies support a strong association between individual resistance to enteric fever and allelic variation within the HLA class II HLA-DRB1 gene (21). On the basis of these observations in both mice and humans, the relationships among major histocompatibility complex (MHC) class II gene variation, CD4 T cell activation, and mouse resistance to Salmonella infection deserve further investigation.
There are several different models for studying Salmonella infection in mice. Some laboratories choose to infect resistant mouse strains, while others predominantly use susceptible mouse strains that lack the protective SLC11A1 gene (22). Infection of susceptible C57BL/6 mice with an attenuated strain of Salmonella Typhimurium elicits robust CD4 T cell responses that contribute to bacterial clearance (20, 23, 24). In contrast, infecting resistant mouse strains with virulent Salmonella typically elicits strong antibody-mediated protection (25, 26). Despite robust expansion of CD4 T cells during Salmonella infection, depleting CD4 T cells increases bacterial replication only modestly (by around 1 to 2 log) (20), suggesting that other protective mechanisms are important. Previous work has shown that different mouse strains eliminate Salmonella Typhiumurium at vastly different rates, with C57BL/6 mice among the slowest to eradicate bacteria (27). MHC alleles themselves are influential in determining how quickly congenic mice can eradicate Salmonella infection (27). On the basis of these historical data, we hypothesized that the I-Ab molecule was particularly poor at initiating protective CD4 T cell responses and that stronger protective CD4 T cell responses would develop in C57BL/6 mice expressing other MHC haplotypes. The present study therefore examined whether H-2 congenic mouse strains with enhanced resistance to Salmonella infection elicited superior CD4 T cell-dependent protective responses. Surprisingly, our results show that, although CD4 T cells contribute to anti-Salmonella immunity in different MHC congenic strains, CD8 T cells are essential to the enhanced protection evident in comparisons between strains.
RESULTS
Congenic mice expressing H-2k and H-2u molecules demonstrated rapid clearance of Salmonella Typhimurium.
We initially examined whether MHC congenic mice displayed different rates of Salmonella clearance, as had been previously reported (27). Mice possessing variant H-2 molecules at the class I and class II alleles, as well as congenic control strains, were infected intravenously with 5 × 105 CFU of Salmonella Typhimurium, and bacterial burdens were assessed over the course of 28 days. Mice were infected intravenously because NTS is a systemic disease that is not typically associated with high bacterial burdens in the gut lamina propria (10). No significant differences were observed in the rates of bacterial clearance from the spleen or liver of two control strains [C57BL/6ByJ (H-2b) and BALB/cJ (H-2d)], or from those of their MHC congenic counterparts B6.C-H-2d/bByJ and C.B10-H2b/LilMcdJ, at any time point over the 4 weeks of infection (Fig. 1A and B). Thus, allelic substitutions of b and d haplotypes at the MHC locus had no discernible effect on Salmonella infection in C57BL/6ByJ or BALB/cJ mice.
FIG 1.
Congenic mice expressing H-2k and H-2u molecules demonstrate rapid clearance of Salmonella Typhimurium. Levels of clearance of Salmonella Typhimurium from the spleen (top panels) and liver (bottom panels) were determined over 4 weeks. All mice were intravenously infected with 5 × 105 CFU of S. Typhimurium strain BRD509. Data represent results of analyses of the kinetics of eradication of S. Typhimurium from C57BL/6ByJ (H-2b) mice and the B6.C-H-2d/bByJ congenic strain (A), BALB/cJ (H-2d) mice and the C.B10-H2b/LilMcdJ congenic strain (B), C57BL/10SnJ mice (H-2b) and the B10.BR-H2k2H2-Tl8a/SgSnJJrep strain (C), and the B10.PL-H-2uH2-Tl8a/(73NS)SnJ congenic strain (D). Significance values were determined using Holm-Sidak t tests (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). n = 3 to 4 mice per group per time point, and data are representative of results from two experiments. p.i., postinfection.
Next, we compared the clearance rates of Salmonella in C57BL/10SnJ mice possessing H-2b MHC haplotype with the clearance rates in congenic strains expressing H-2k and H-2u haplotypes [strains B10.BR-H2k2H2-Tl8a/SgSnJJre and B10.PL-H-2uH2-Tl8a/(73NS)SnJ], respectively. C57BL/10SnJ mice with the H-2k haplotype or the H-2u haplotype cleared S. Typhimurium from the spleen and liver at a higher rate than the control strain expressing H-2b (Fig. 1C and D). Bacterial burdens began to significantly diverge as early as 2 weeks after infection, and there was an approximately 100-fold difference in tissue bacterial loads between the H-2k-expressing or H-2u-expressing MHC congenic mice and the H-2b-expressing control mice by 28 days after infection. This finding is broadly in agreement with a previous study by Nauciel et al., which reported that MHC alleles influence the ability of mice to resolve infection with Salmonella Typhimurium (27).
To test whether the enhanced protection observed in MHC congenic strains was specific to Salmonella, we next examined host immunity to another intracellular bacterium, Chlamydia muridarum. We chose this intracellular bacterium because protection against this pathogen in C57BL/6 mice is mediated by CD4 T cells, which disseminate to the spleen (28). Congenic and control strains were infected vaginally with Chlamydia, and bacterial burdens were assessed regularly by intermittent swabbing of the vaginal vault. The rate of clearance of C. muridarum from the reproductive tracts of MHC congenic strains expressing H-2k or H-2u molecules was not statistically different from that seen with the control strains (see Fig. S1A and B in the supplemental material). Thus, the protective effect of these H-2 alleles is specific to Salmonella infection and not does not represent a general form of resistance to intracellular bacterial infections.
Protection in MHC congenic mice is not due to an increase in the percentage of T cells.
To initially determine which leukocyte subsets responded to S. Typhimurium infection in the MHC congenic strains, a general screen of splenic leukocyte populations was performed at 2 weeks postinfection (a time when both MHC congenic strains started to display faster clearance). This analysis showed significant increases in the percentages of Gr-1+ and CD11b+ cells, but not in the T cell populations, which we suspect may be responsible for the greater rate of clearance in the MHC congenic mice (Fig. S2). Although increased T cell percentages did not correlate with enhanced protection in MHC congenic mice, it was possible that the levels of T cell effector functions were elevated and contributed to Salmonella clearance. Therefore, we focused subsequent analysis on comparisons of T cell effector properties in these MHC congenic mice.
Accelerated Salmonella clearance in MHC congenic mice correlates with enhanced IFN-γ production from CD8 T cells, but not CD4 T cells.
IFN-γ production by Th1 cells is essential for the successful resolution of Salmonella infection (19). Given that H-2k and H-2u C57BL/10 mice displayed faster clearance of Salmonella than H-2b mice, we examined the functional capacity of T-bet-expressing Th1 cells during infection. At 2 weeks after infection with Salmonella Typhimurium, mice were stimulated with lipopolysaccharide (LPS) to activate CD4 and CD8 T cells via noncognate stimulation (29). Antigen-experienced Th1 cells were identified by gating on CD44 and T-bet-expressing CD4 T cells, while effector CD8 T cells were identified using CD44 surface staining (Fig. 2A and B). Consistent with previous studies (20), around 60% of the Tbet+ CD4+ T cells from H-2b mice produced IFN-γ (Fig. 2B and D), indicative of a strong Th1 response to Salmonella infection. Despite the accelerated bacterial clearance evident in H-2k-expressing and H-2u-expressing mice, both strains displayed lower percentages of IFN-γ-positive Th1 cells than control mice (Fig. 2B and D). As expected, the level of CD8 T cell production of IFN-γ in all mouse strains was considerably lower than that seen with CD4 Th1 cells (Fig. 2A and C). While H-2b mice and H-2u mice had similar frequencies of IFN-γ-producing CD8 T cells, this population was significantly increased in H-2k mice, correlating with a greater role of CD8 T cell-mediated control of Salmonella infection (Fig. 3B and C). These data suggest that greater protection against Salmonella infection in these MHC congenic mouse strains is likely to be independent of Th1 IFN-γ production.
FIG 2.
Accelerated Salmonella clearance in congenic mice correlates with enhanced IFN-γ production from CD8 T cells but not from CD4 T cells. Mice were infected intravenously (i.v.) with 5 × 105 CFU S. Typhimurium, restimulated after 2 weeks i.v. with 10 μg of LPS, and euthanized after 4 h. (A) Gating strategy for identification of activated CD4 T and CD8 T cells. (B) IFN-γ production in CD44+Tbet+ CD4+ and CD44+CD8+ T cells determined by flow cytometry. (C and D) Summary of the percentages of IFN-γ+ CD44hi CD8+ T cells (C) and IFN-γ+ Tbet+ CD44hi CD4+ T cells (D) as a proportion of the total population of Tbet+ CD4+ T cells. Significant differences were calculated using Holm-Sidak t tests (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; n = 1 to 4 mice per group). Data are representative of results from two separate experiments. Error bars represent standard deviations.
FIG 3.
CD4 and CD8 T cells control Salmonella burden in H-2k congenic mice. Mice were infected i.v. with 5 × 105 CFU of S. Typhimurium and treated with anti-CD4 antibody alone, with anti-CD4 and anti-CD8 antibody, or with an isotype control (Isotope ctrl) every 3 to 4 days from day 7 to day 30. (A) Gating strategy for analysis of CD4 and CD8 T cells. (B) Log CFU of S. Typhimurium in the spleen (top panel) and liver (bottom panel) of B10 strains H-2b, H-2k, and H-2u on day 30 after infection. n = 3 to 6 mice per group; data represent results from 3 experimental replicates. Significant differences between strains were calculated using Mann-Whitney t tests (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). Error bars represent standard deviations.
CD4 and CD8 T cells control Salmonella burden in H-2k congenic mice.
Given the relatively minor differences in levels of T cell IFN-γ production between MHC congenic strains, it was important to directly test whether accelerated clearance is dependent on CD4 and/or CD8 T cells. From this point on, we narrowed our studies to focus on the H-2k congenic mice. As previously reported (20), antibody depletion of CD4 T cells in control H-2b-expressing mice resulted in bacterial burdens in the spleen and liver that were 1 to 2 log higher (Fig. 3), demonstrating the essential role of CD4 T cells in bacterial clearance. The effects of depletion of both CD4 and CD8 T cells in H-2b mice did not differ markedly from the effects of depletion of CD4 T cells alone with respect to affecting bacterial clearance from the spleen or liver (Fig. 3B). However, depletion of CD8 T cells alone resulted in significantly increased bacterial burdens in the liver, although the effect was modest overall (Fig. 3B). Thus, CD8 T cells made a small contribution to primary clearance in H-2b mice, as previously reported (16). Surprisingly, the depletion of both CD4 and CD8 T cells from H-2k mice resulted in significantly greater bacterial growth than CD4 depletion alone (Fig. 3). In the spleens of the H-2k mice, CD8 T cells apparently provided more protection against Salmonella than CD4 T cells, since individual depletion of CD8 T cells had a stronger effect. There seemed to be a redundant protective effect of CD4 and CD8 T cells in the liver of H-2k mice, since depleting either of the cell types alone did not result in any loss of protection. These data suggest that CD8 T cells play an important role in the accelerated bacterial clearance observed in H-2k-expressing MHC congenic mouse strains.
CD8 T cells display enhanced effector phenotypes during S. Typhimurium infection in H-2k mice.
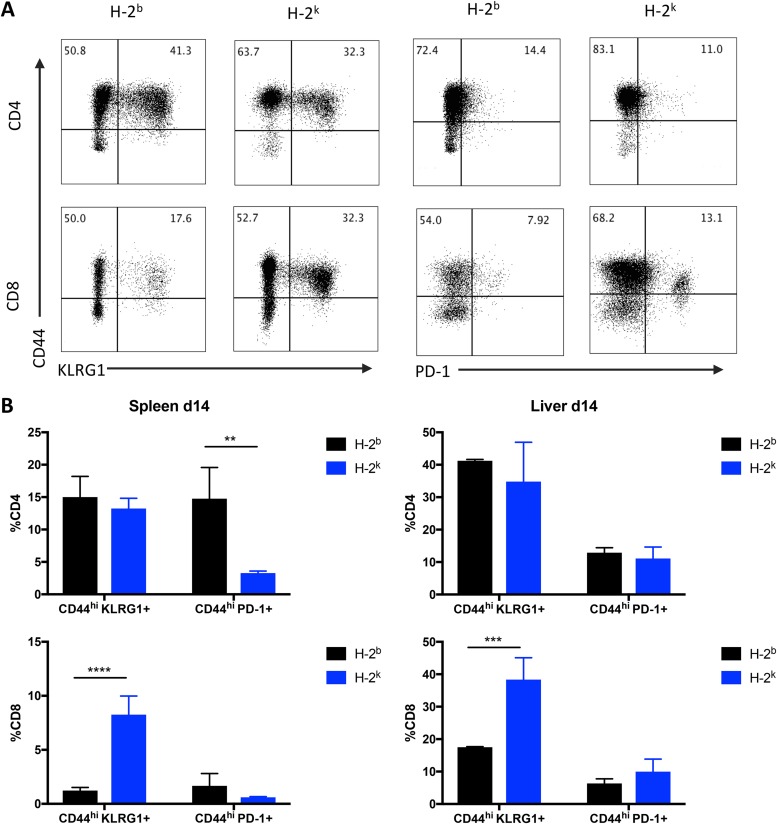
Because of the marked role of CD8 T cells in controlling Salmonella infection in the H-2k congenic strain, we examined functional markers of CD8 T cell activation and effector differentiation that might explain their protective capacity. At 14 days after infection, we observed a significantly greater proportion of CD44hi KLRG1+ CD8 T cells in both the spleen and liver of H-2k mice (Fig. 4). Additionally, there were significantly fewer CD44hi PD-1+ CD4 T cells in the spleen of the H-2k congenic strain (Fig. 4). The increase in the level of CD8 T cells expressing the cytotoxic marker KLRG1 and the reduction in the level of CD4 T cells expressing the inhibitory marker PD-1 suggest an overall enhanced proinflammatory state in H-2k mice that was likely responsible for the accelerated clearance of Salmonella Typhimurium in this congenic strain.
FIG 4.
CD8 T cells displayed an enhanced effector phenotype during S. Typhimurium infection in H-2k mice. Mice were infected i.v. with 5 × 105 CFU S. Typhimurium and analyzed after 2 weeks. (A) Flow plots of CD4 and CD8 T cell marker expression in the liver. Data represent a summary of the percentages of CD44hi KLRG1+ and CD44hi PD-1+ CD4 and CD8 T cells in the spleens and livers of H-2b and H-2k congenic mice. (B) Log CFU of S. Typhimurium in the spleen (left panels) and liver (right panels) of B10 strains H-2b and H-2k on day 14 (d14) after infection. Significant differences were calculated using Holm-Sidak t tests (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; n = 3 mice per group). Data are representative of results from one experiment.
Congenic mice expressing H-2k are more resistant to challenge with virulent Salmonella.
Given that virulent Salmonella challenge is quickly lethal in C57BL/10 mice, we wondered whether the congenic H-2k allele would show enhanced survival in response to lethal Salmonella challenge. Congenic mice were challenged intravenously with Salmonella Typhimurium SL1344, analyzed for bacterial burdens after 4 days, and monitored for overall survival. H-2k congenic mice did not control initial bacterial growth any better than the H-2b controls at 4 days after challenge (Fig. 5A). However, the H-2k mice did survive significantly longer than the H-2b mice (Fig. 5B). Together, these observations indicate that mice with the congenic H-2k allele were able to tolerate the infection slightly longer than the H-2b mice.
FIG 5.
Mice expressing H-2k are more resistant to challenge with virulent Salmonella. Mice were infected i.v. with 5 × 105 CFU S. Typhimurium SL1344 and analyzed 4 days later for bacterial burden and separately for overall survival. (A) Comparison of Salmonella bacterial burdens 4 days after infection in the spleen and liver using multiple t tests (n = 4 mice per group). (B) Comparison of survival rates of congenic strains using log rank Mantel-Cox test (n = 3 to 4 mice per group). Data represent results from one experiment.
DISCUSSION
The objective of this study was to examine whether CD4 T cells restricted to H-2b molecules were particularly poor at Salmonella clearance compared to CD4 T cells restricted to other MHC molecules. Such a conclusion could be inferred from previous data showing that C57BL/6 mice show considerably slower clearance than almost all other MHC congenic mouse strains (27). Furthermore, a recent epitope discovery has allowed better analysis of Salmonella-specific CD4 T cells within H-2b-expressing C57BL/6 mice and these data paint a somewhat confusing picture of multiple low-frequency I-Ab-restricted clones, some of which lack the ability to survive and propagate into the memory pool (22). This is an important issue to resolve since almost every study of CD4 T cells in the mouse model of salmonellosis has focused narrowly on I-Ab-restricted CD4 T cells.
The results of analysis of bacterial clearance in MHC congenic mouse strains in this study support the conclusions of previous studies which suggested that differences in MHC haplotypes drastically affect primary clearance of Salmonella (27, 30). Examination of Chlamydia infection indicates that this is a particular feature of Salmonella clearance in these inbred mice, since there was little effect on Chlamydia shedding using these same mouse strains. At face value, these data might indicate that the overall frequency or functional activity of Salmonella-specific CD4 T cells is amplified in certain MHC congenic mouse strains or, alternatively, that those other mouse strains lack a major suppressive activity with respect to CD4 T cells that is evident in C57BL/6 mice. However, a basic characterization of splenocyte populations in Salmonella-infected MHC congenic strains demonstrated that there are no obvious differences in CD4 and CD8 T cell percentages during active bacterial clearance. When the requirement of these cells was examined directly by antibody depletion of CD4 and CD8 T cells in each of the congenic lines, we observed a similar requirement of CD4 T cells for protection in these congenic lines. However, CD8 T cells make a larger contribution to protection in the H-2k congenic mice. Thus, while it is true that some MHC congenic mice display faster clearance of Salmonella, it is unrelated to the number and/or functional activity of CD4 T cells in these mice.
Depletion of CD8 T cells alone or in conjunction with CD4 T cells had an exaggerated impact on bacterial burdens in congenic mice displaying accelerated Salmonella clearance. This is surprising since the impact of MHC class-I-restricted T cells is relatively modest in C57BL/6 mice (16). However, CD8 T cells are activated in humans and mice during Salmonella infection (23, 31–33) and our data would suggest that they could play a more important role in these mouse strains. Given that all of these mouse strains are congenic for the MHC class I allele, it is not surprising that they might have an altered CD8 response to the typical H-2b strain. The protection mediated by the CD8 response implies that different or unique Salmonella antigens are displayed on MHC class I molecules, resulting in an amplification of CD8-dependent protective immunity. It might be instructive to examine some of these epitopes in further studies.
The accelerated Salmonella clearance rate seen in some congenic mice correlated with a very modestly heightened IFN-γ response by CD8 T cells which occurred in concert with lower levels of IFN-γ production from CD4 T cells. The lower Th1 cell activity in these mice could be explained by reduced antigen availability, which would be expected to reduce effector T cell activity (10). Since CD8 T cells did display enhanced IFN-γ production in H-2k mouse strains, this cytokine may be responsible for some of the accelerated bacterial clearance observed in these mice. However, no change in IFN-γ production was observed in H-2u mice compared to H-2b mice. It seems more likely that enhanced CD8 T cell responses rely on a perforin-dependent cell killing mechanism or on the ability of CD8 T cells to rapidly recruit innate cells to infected tissues. Indeed, we observed that the H-2k mice had a significantly higher proportion of CD44hiKLRG1+ CD8 T cells in the spleen and liver than the H-2b controls. These congenic mice also had fewer PD-1-expressing CD4 T cells in the spleen, indicating less inhibition of activated lymphocytes. KLRG1 expression is an indicator of CD8 T cell cytotoxicity and is modulated by MHC class I molecules (34). It is possible that this enhanced killing capacity is beneficial for killing Salmonella-infected phagocytes. Indeed, data from previous studies support the idea that macrophages can execute cross-presentation of vacuolar antigen and prime cytotoxic T lymphocyte (CTL) responses in mice and humans (35–37). It seems reasonable to hypothesize, then, that these CTLs kill infected macrophages through recognition of Salmonella antigens expressed on MHC class I molecules. Further studies will be required to better characterize the precise mechanism of CD8 killing in MHC congenic mouse strains.
In conclusion, this study confirmed previous observations showing that MHC congenic mouse lines display marked differences in the speed of Salmonella clearance. However, our data show that these differences are less dependent on CD4 responses than would be expected and thus do not support the idea that particular murine MHC alleles can elicit more-robust CD4 T cell responses. Instead, our data are consistent with the idea of a larger role for CD8 T cells in some MHC congenic inbred mouse strains and may indicate a greater role for cytotoxic T cell activity in clearing Salmonella infection.
MATERIALS AND METHODS
Mice.
C57BL/10SnJ (Jax 000666), B10.BR-H2k2H2-Tl8a/SgSnJJrep (Jax 004804), B10.PL-H-2uH2-Tl8a/(73NS)SnJ (Jax 000458), BALB/cJ (Jax 000651), C.B10-H2b/LilMcdJ (Jax 001952), C57BL/6ByJ (Jax 001139), and B6.C-H-2d/bByJ (Jax 000359) mice were all purchased from the Jackson Laboratories at 6 to 8 weeks of age. Mice were bred and housed at the Center for Comparative Medicine, University of California, Davis (UC Davis). Male and female mice (6 to 12 weeks old) were used for all experimental studies. Congenic mice were genotyped by PCR according to the recommended protocol provided by Jackson Laboratories. All experiments in this study followed protocols approved by the Animal Care and Use Committee.
Bacterial strains and infections.
Salmonella infection experiments were performed using the BRD509 ΔaroA or SL1344 strains of Salmonella enterica serovar Typhimurium. While BRD509 is typically considered to be an ΔaroA ΔaroD mutant, genomic sequencing has revealed a functional aroD gene (38). Mice were infected intravenously with 5 × 105 CFU of BRD509 that had been grown overnight in LB broth at 37°C and resuspended in phosphate-buffered saline (PBS). Mice were challenged either intravenously with SL1344 at 1 × 103 CFU or orally by water bottle administration at 1 × 109 CFU/ml (39). Bacterial concentrations were determined by spectrophotometry at an optical density at 600 nm (OD600) and adjusted to the correct challenge inoculum. For measurements of vaginal shedding of Chlamydia muridarum, mice were injected with 2.5 mg of medroxyprogesterone acetate (Depo-Provera) subcutaneously and then infected a week later by pipetting of 105 IFU of Chlamydia into the vaginal vault (40).
Determination of bacterial loads.
Livers and spleens of infected mice were collected in PBS on ice and homogenized, and the final volume of the homogenate was recorded. After vortex mixing was performed, serial dilutions of organ homogenate were plated onto MacConkey agar plates. Plates were incubated overnight at 37°C for 16 h, and their contents were enumerated to determine the number of CFU present. The total CFU count/organ was determined by back-calculation from the plate counts, sample dilution, and homogenate volume. For measuring Chlamydia bacterial burdens, vaginal swabs were collected every 3 to 4 days over a 28-day period. Supernatants from swabs were pipetted on a HeLa cell monolayer, fixed, and stained using anti-Chlamydia antibody (40). Chlamydia inclusions were enumerated under a fluorescence microscope.
In vivo LPS stimulation.
A 10-μg volume of ultrapure lipopolysaccharide (LPS) from Escherichia coli strain EH100Rα (Alexis strain; Toll-like receptor [TLR] grade) was diluted in PBS and injected intravenously into infected mice, 4 h before harvesting of organs and examination of T cell responses. This methodology has previously been shown to elicit noncognate T cell activation and to allow analysis of cytokine production by determination of endogenous polyclonal T cell responses (20).
Flow cytometry.
Single-cell suspensions of splenocytes were subjected to lysis using ACK lysis buffer (0.15 M NH4Cl, 1 mM KHCO3, 0.1 mM EDTA) for 4 min and washed with fluorescence-activated cell sorter (FACS) buffer. A range of 2 × 106 cells and 5 × 106 cells were Fc blocked for 15 min and stained with the following antibodies: CD8 PercpEF710, CD11b AF488, CD45R PECy7, NK1.1 phycoerythrin (PE), CD11c AF700, CD4 EF660, Gr-1 EF450, fluorescein isothiocyanate (FITC) (CD11b, CD11c, F4/80, B220), CD4 PECy7, CD8 PE, CD44 EF450, IFN-γ allophycocyanin (APC), T-bet Percp Cy5.5, CD4 BV650, IFN-γ BV785, T-bet PE, granzyme B EF450, KLRG1 APC, PD-1 PeCy7, and CD44 AF700. Cells were fixed and permeabilized overnight using a Foxp3 staining kit (BD) before addition of intracellular stains. Cells were analyzed on a BD Fortessa cell analyzer using appropriate compensation controls, and the resulting data were analyzed using FlowJo analysis software.
Antibody depletion of CD4 and CD8 cells.
CD4 and CD8 cells were depleted from infected mice by administration of anti-CD4 clone GK1.5 and anti-CD8a clone 2.43 monoclonal antibodies (purchased from Bio X Cell). Isotype control antibody clone LTF2 was used in nondepleted mice. Seven days after intravenous infection with Salmonella, monoclonal antibodies were diluted in PBS and administered to mice intraperitoneally every 3 to 4 days for up to 30 days postinfection. Mice received 200 μg of each antibody at the first two time points and 300 μg for the remainder of the time points.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (AI139047 and AI139410 to S.J.M.). J.C.L. was supported by an NIH T32 training grant (AI60555). O.H.P. was supported by a Vietnam Education Foundation Fellowship.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00588-19.
REFERENCES
- 1.Wain J, Hendriksen RS, Mikoleit ML, Keddy KH, Ochiai RL. 2015. Typhoid fever. Lancet 385:1136–1145. doi: 10.1016/S0140-6736(13)62708-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States–major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilchrist JJ, MacLennan CA, Hill A. 2015. Genetic susceptibility to invasive Salmonella disease. Nat Rev Immunol 15:452–463. doi: 10.1038/nri3858. [DOI] [PubMed] [Google Scholar]
- 4.Tennant SM, Levine MM. 2015. Live attenuated vaccines for invasive Salmonella infections. Vaccine 33(Suppl 3):C36–C41. doi: 10.1016/j.vaccine.2015.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacLennan CA, Martin LB, Micoli F. 2014. Vaccines against invasive Salmonella disease: current status and future directions. Hum Vaccin Immunother 10:1478–1493. doi: 10.4161/hv.29054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dougan G, John V, Palmer S, Mastroeni P. 2011. Immunity to salmonellosis. Immunol Rev 240:196–210. doi: 10.1111/j.1600-065X.2010.00999.x. [DOI] [PubMed] [Google Scholar]
- 7.Hormaeche CE. 1979. Natural resistance to Salmonella typhimurium in different inbred mouse strains. Immunology 37:311–318. [PMC free article] [PubMed] [Google Scholar]
- 8.Jones BD, Ghori N, Falkow S. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J Exp Med 180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrovska L, Aspinall RJ, Barber L, Clare S, Simmons CP, Stratford R, Khan SA, Lemoine NR, Frankel G, Holden DW, Dougan G. 2004. Salmonella enterica serovar Typhimurium interaction with dendritic cells: impact of the sifA gene. Cell Microbiol 6:1071–1084. doi: 10.1111/j.1462-5822.2004.00419.x. [DOI] [PubMed] [Google Scholar]
- 10.Griffin AJ, McSorley SJ. 2011. Development of protective immunity to Salmonella, a mucosal pathogen with a systemic agenda. Mucosal Immunol 4:371–382. doi: 10.1038/mi.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rescigno M, Rotta G, Valzasina B, Ricciardi-Castagnoli P. 2001. Dendritic cells shuttle microbes across gut epithelial monolayers. Immunobiology 204:572–581. doi: 10.1078/0171-2985-00094. [DOI] [PubMed] [Google Scholar]
- 12.Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. 2001. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol 2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 13.Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, Littman DR, Reinecker HC. 2005. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 14.Hapfelmeier S, Stecher B, Barthel M, Kremer M, Muller AJ, Heikenwalder M, Stallmach T, Hensel M, Pfeffer K, Akira S, Hardt WD. 2005. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol 174:1675–1685. doi: 10.4049/jimmunol.174.3.1675. [DOI] [PubMed] [Google Scholar]
- 15.Kirby AC, Yrlid U, Wick MJ. 2002. The innate immune response differs in primary and secondary Salmonella infection. J Immunol 169:4450–4459. doi: 10.4049/jimmunol.169.8.4450. [DOI] [PubMed] [Google Scholar]
- 16.Lee S-J, Dunmire S, McSorley SJ. 2012. MHC class-I-restricted CD8 T cells play a protective role during primary Salmonella infection. Immunol Lett 148:138–143. doi: 10.1016/j.imlet.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hess J, Ladel C, Miko D, Kaufmann SH. 1996. Salmonella typhimurium aroA− infection in gene-targeted immunodeficient mice: major role of CD4+ TCR-alpha beta cells and IFN-gamma in bacterial clearance independent of intracellular location. J Immunol 156:3321–3326. [PubMed] [Google Scholar]
- 18.VanCott JL, Chatfield SN, Roberts M, Hone DM, Hohmann EL, Pascual DW, Yamamoto M, Kiyono H, McGhee JR. 1998. Regulation of host immune responses by modification of Salmonella virulence genes. Nat Med 4:1247–1252. doi: 10.1038/3227. [DOI] [PubMed] [Google Scholar]
- 19.Ravindran R, Foley J, Stoklasek T, Glimcher LH, McSorley SJ. 2005. Expression of T-bet by CD4 T cells is essential for resistance to Salmonella infection. J Immunol 175:4603–4610. doi: 10.4049/jimmunol.175.7.4603. [DOI] [PubMed] [Google Scholar]
- 20.O'Donnell H, Pham OH, Li L-X, Atif SM, Lee S-J, Ravesloot MM, Stolfi JL, Nuccio S-P, Broz P, Monack DM, Baumler AJ, McSorley SJ. 2014. Toll-like receptor and inflammasome signals converge to amplify the innate bactericidal capacity of T helper 1 cells. Immunity 40:213–224. doi: 10.1016/j.immuni.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunstan SJ, Hue NT, Han B, Li Z, Tram TTB, Sim KS, Parry CM, Chinh NT, Vinh H, Lan NPH, Thieu NTV, Vinh PV, Koirala S, Dongol S, Arjyal A, Karkey A, Shilpakar O, Dolecek C, Foo JN, Phuong LT, Lanh MN, Do T, Aung T, Hon DN, Teo YY, Hibberd ML, Anders KL, Okada Y, Raychaudhuri S, Simmons CP, Baker S, de Bakker PIW, Basnyat B, Hien TT, Farrar JJ, Khor CC. 2014. Variation at HLA-DRB1 is associated with resistance to enteric fever. Nat Genet 46:1333–1336. doi: 10.1038/ng.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McSorley SJ. 2014. Immunity to intestinal pathogens: lessons learned from Salmonella. Immunol Rev 260:168–182. doi: 10.1111/imr.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mittrucker HW, Kohler A, Kaufmann SH. 2002. Characterization of the murine T-lymphocyte response to Salmonella enterica serovar Typhimurium infection. Infect Immun 70:199–203. doi: 10.1128/iai.70.1.199-203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srinivasan A, Foley J, McSorley SJ. 2004. Massive number of antigen-specific CD4 T cells during vaccination with live attenuated Salmonella causes interclonal competition. J Immunol 172:6884–6893. doi: 10.4049/jimmunol.172.11.6884. [DOI] [PubMed] [Google Scholar]
- 25.Eisenstein TK, Killar LM, Sultzer BM. 1984. Immunity to infection with Salmonella typhimurium: mouse-strain differences in vaccine- and serum-mediated protection. J Infect Dis 150:425–435. doi: 10.1093/infdis/150.3.425. [DOI] [PubMed] [Google Scholar]
- 26.Johanns TM, Law CY, Kalekar LA, O’Donnell H, Ertelt JM, Rowe JH, Way SS. 2011. Early eradication of persistent Salmonella infection primes antibody-mediated protective immunity to recurrent infection. Microbes Infect 13:322–330. doi: 10.1016/j.micinf.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nauciel C, Ronco E, Guenet JL, Pla M. 1988. Role of H-2 and non-H-2 genes in control of bacterial clearance from the spleen in Salmonella typhimurium-infected mice. Infect Immun 56:2407–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L-X, McSorley SJ. 2013. B cells enhance antigen-specific CD4 T cell priming and prevent bacteria dissemination following Chlamydia muridarum genital tract infection. PLoS Pathog 9:e1003707. doi: 10.1371/journal.ppat.1003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pham OH, O’Donnell H, Al-Shamkhani A, Kerrinnes T, Tsolis RM, McSorley SJ. 2017. T cell expression of IL-18R and DR3 is essential for non-cognate stimulation of Th1 cells and optimal clearance of intracellular bacteria. PLoS Pathog 13:e1006566. doi: 10.1371/journal.ppat.1006566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nauciel C, Ronco E, Pla M. 1990. Influence of different regions of the H-2 complex on the rate of clearance of Salmonella typhimurium. Infect Immun 58:573–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salerno-Goncalves R, Pasetti MF, Sztein MB. 2002. Characterization of CD8(+) effector T cell responses in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J Immunol 169:2196–2203. doi: 10.4049/jimmunol.169.4.2196. [DOI] [PubMed] [Google Scholar]
- 32.Salerno-Gonçalves R, Wyant TL, Pasetti MF, Fernandez-Viña M, Tacket CO, Levine MM, Sztein MB. 2003. Concomitant induction of CD4+ and CD8+ T cell responses in volunteers immunized with Salmonella enterica serovar typhi strain CVD 908-htrA. J Immunol 170:2734–2741. doi: 10.4049/jimmunol.170.5.2734. [DOI] [PubMed] [Google Scholar]
- 33.Luu RA, Gurnani K, Dudani R, Kammara R, van Faassen H, Sirard JC, Krishnan L, Sad S. 2006. Delayed expansion and contraction of CD8+ T cell response during infection with virulent Salmonella typhimurium. J Immunol 177:1516–1525. doi: 10.4049/jimmunol.177.3.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corral L, Hanke T, Vance RE, Cado D, Raulet DH. 2000. NK cell expression of the killer cell lectin-like receptor G1 (KLRG1), the mouse homolog of MAFA, is modulated by MHC class I molecules. Eur J Immunol 30:920–930. doi:. [DOI] [PubMed] [Google Scholar]
- 35.Tang-Huau TL, Gueguen P, Goudot C, Durand M, Bohec M, Baulande S, Pasquier B, Amigorena S, Segura E. 2018. Human in vivo-generated monocyte-derived dendritic cells and macrophages cross-present antigens through a vacuolar pathway. Nat Commun 9:2570. doi: 10.1038/s41467-018-04985-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dinter J, Duong E, Lai NY, Berberich MJ, Kourjian G, Bracho-Sanchez E, Chu D, Su H, Zhang SC, Le Gall S. 2015. Variable processing and cross-presentation of HIV by dendritic cells and macrophages shapes CTL immunodominance and immune escape. PLoS Pathog 11:e1004725. doi: 10.1371/journal.ppat.1004725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernhard CA, Ried C, Kochanek S, Brocker T. 2015. CD169+ macrophages are sufficient for priming of CTLs with specificities left out by cross-priming dendritic cells. Proc Natl Acad Sci U S A 112:5461–5466. doi: 10.1073/pnas.1423356112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benoun JM, Peres NG, Wang N, Pham OH, Rudisill VL, Fogassy ZN, Whitney PG, Fernandez-Ruiz D, Gebhardt T, Pham Q-M, Puddington L, Bedoui S, Strugnell RA, McSorley SJ. 2018. Optimal protection against Salmonella infection requires noncirculating memory. Proc Natl Acad Sci U S A 115:10416–10421. doi: 10.1073/pnas.1808339115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Donnell H, Pham OH, Benoun JM, Ravesloot-Chávez MM, McSorley SJ. 2015. Contaminated water delivery as a simple and effective method of experimental Salmonella infection. Future Microbiol 10:1615–1627. doi: 10.2217/fmb.15.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L-X, Labuda JC, Imai DM, Griffey SM, McSorley SJ. 2017. CCR7 deficiency allows accelerated clearance of Chlamydia from the female reproductive tract. J Immunol 199:2547–2554. doi: 10.4049/jimmunol.1601314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.