Streptococcus pneumoniae (pneumococcus) causes multiple infectious diseases. The pneumococcal competence system facilitates genetic transformation, spreads antibiotic resistance, and contributes to virulence. DNA-processing protein A (DprA) regulates the exit of pneumococcus from the competent state. Previously, we have shown that DprA is important in both bacteremia and pneumonia infections. Here, we examined the mechanisms of virulence attenuation in a ΔdprA mutant.
KEYWORDS: Streptococcus pneumoniae, competence system, DprA, allolytic factors, ComM, bacteremia, competence system, pneumonia
ABSTRACT
Streptococcus pneumoniae (pneumococcus) causes multiple infectious diseases. The pneumococcal competence system facilitates genetic transformation, spreads antibiotic resistance, and contributes to virulence. DNA-processing protein A (DprA) regulates the exit of pneumococcus from the competent state. Previously, we have shown that DprA is important in both bacteremia and pneumonia infections. Here, we examined the mechanisms of virulence attenuation in a ΔdprA mutant. Compared to the parental wild-type D39, the ΔdprA mutant enters the competent state when exposed to lower concentrations of the competence-stimulating peptide CSP1. The ΔdprA mutant overexpresses ComM, which delays cell separation after division. Additionally, the ΔdprA mutant overexpresses allolytic factors LytA, CbpD, and CibAB and is more susceptible to detergent-triggered lysis. Disabling of the competent-state-specific induction of ComM and allolytic factors compensated for the virulence loss in the ΔdprA mutant, suggesting that overexpression of these factors contributes to virulence attenuation. Finally, the ΔdprA mutant fails to downregulate the expression of multiple competence-regulated genes, leading to the excessive energy consumption. Collectively, these results indicate that an inability to properly exit the competent state disrupts multiple cellular processes that cause virulence attenuation in the ΔdprA mutant.
INTRODUCTION
Streptococcus pneumoniae (pneumococcus) is a respiratory tract commensal that causes otitis media, community-acquired pneumonia, pneumonia-derived sepsis, and multiorgan dysfunction, meningitis, and other diseases. The pneumococcal competence system for genetic transformation contributes to the dissemination of antibiotic resistance and its high genomic plasticity (1–3). Under appropriate in vitro conditions, pneumococci have the ability to enter the competent state spontaneously. The competence-stimulating peptide (CSP), encoded by the comC gene, is exported by the ABC transporter ComAB and serves as a signal that activates competence regulon (4–8). Previously, it was thought that when the secreted CSP reaches a threshold concentration in a quorum-sensing (QS) manner, it will bind and activate the cognate receptor ComD, leading to the phosphorylation of the response regulator ComE to drive the whole pneumococcal population into the competent state. However, a recent study suggests that in some pneumococcal strains, including the virulent serotype 2 strain D39, CSP is not diffused but instead is retained by its cognate receptor ComD and activates the competence of neighboring pneumococcus by cell-cell contact (9). Activated ComD-ComE (10, 11) induces the expression of approximately 24 “early” genes, including comAB, comCDE, and comX (12). ComX is an alternate sigma factor (13–15) that activates expression of approximately 80 “late” genes, of which 16 genes are directly involved in DNA uptake and recombination. Among the factors encoded by the “late” genes which are dispensable for genetic transformation are LytA, CbpD, and CibAB, which mediate the release of DNA from neighboring pneumococcus through a process called allolytic fratricide (16–20). Especially, CbpD, LytA, and CibAB target noncompetent cells which do not express the competence-induced allolysis immunity proteins ComM (21) and CibC (18). Finally, the regulation of competence-activated “delayed” genes, most of which are involved in stress response or metabolism, is still not well characterized.
The competence system can be artificially induced by providing CSP to the pneumococcal culture. The competent state peaks at 15 min postexposure, after which it declines rapidly, and competent pneumococci become resistant to a second CSP challenge (12). Recently, it was discovered that DNA-processing protein A (DprA), encoded by one of the ComX-regulated “late” genes, is responsible for competence shutoff (22–24) by dissociating and inactivating the phosphorylated ComE dimers (22). DprA remained highly expressed for an extended period of time, accounting for the resistance to a new wave of induction by fresh CSP.
Intriguingly, we have found that a ΔdprA mutant is attenuated in mouse models of acute pneumonia and bacteremia (25). We chose to focus our investigation on the link between DprA and the competence state because the difference in virulence between dprA+ and ΔdprA mutant strains was no longer evident in a ΔcomB background (e.g., ΔcomB versus ΔdprA ΔcomB), suggesting that the loss of competence function accounted for virulence attenuation in the ΔdprA mutant (25). High levels of DprA ensure the fitness of pneumococcal transformation by mediating competence shutoff (24) as well as by protecting the incoming single-stranded DNA (ssDNA) from being degraded by DNase (26). However, ssDNA protection by DprA may not be important for virulence because deletion of other key genes in DNA recombination, including coiA and ssbB, did not alter pneumococcal virulence (25). Rather, virulence attenuation in the ΔdprA mutant is likely caused by the inability of pneumococcus to exit the competent state. In this study, we examined the underlying mechanisms that contribute to virulence attenuation in the ΔdprA mutant.
RESULTS
DprA deletion negatively affects pneumococcal virulence and causes an exponential-phase-growth delay under CSP-induced competence.
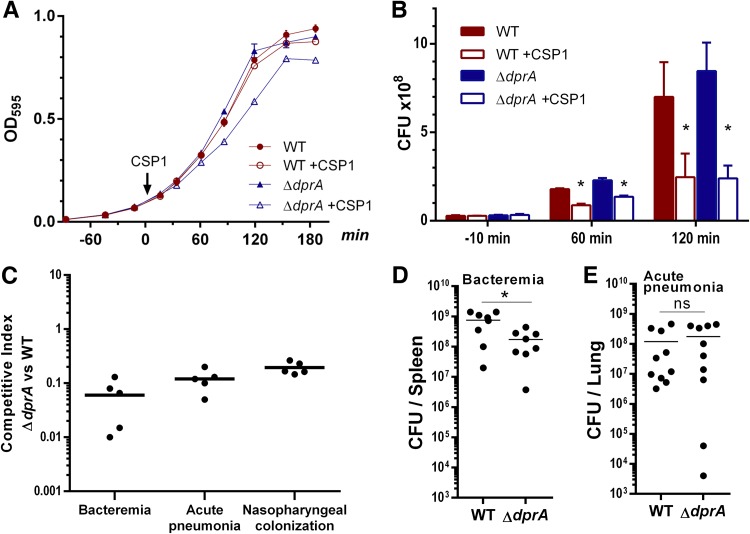
Compared to its parental wild-type (WT) strain D39, the ΔdprA mutant exhibited normal growth in Todd-Hewitt broth (THB) supplemented with 0.5% yeast extracts (THY) (Fig. 1A). When the competence system was activated by 100 ng ml−1 CSP1, ΔdprA cells showed a growth delay in the exponential phase, similar to that seen in previously published studies albeit not as severe (22, 23). Competence induction decreased ΔdprA CFU, but interestingly, also reduced WT CFU (Fig. 1B). Similar results were observed with 10 ng ml−1 CSP1 (see Fig. S1A and B in the supplemental material). The mechanism of pneumococcal CFU reduction was further examined as described in another section (see Fig. 4). To verify the aforementioned results, we examined the growth patterns of the CP1250 strain during competence induction. CP1250 is a nonencapsulated hypercompetent strain derived from the Rx strain, which itself is a nonencapsulated derivative of D39 (9, 10, 27). We first attempted to use THY medium, but interestingly, CP1250 could easily enter the competent state spontaneously without the provision of exogenous CSP1 (see Fig. S3 in the supplemental material). To overcome this problem, we compared the growth of CP1250 and its ΔdprA derivative in the competence-optimized casein-tryptone (CAT) medium supplemented with CSP1 as previously described (22–24). We did not observe a significant growth delay in the CP1250-derived ΔdprA mutant (see Fig. S4A and B in the supplemental material).
FIG 1.
The ΔdprA mutant displays a growth delay in the exponential phase during CSP1 exposure and is attenuated in virulence. (A) The ΔdprA mutant showed a growth delay after competence induction. Pneumococcal growth was assayed in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY) at 37°C in 5% CO2. CSP1 (100 ng ml−1) was added at an OD595 of ∼0.1. (B) Both the D39 WT and ΔdprA strains showed reduced CFU upon CSP1 induction. CFU were measured by serial-dilution plating on THY agar in parallel to the growth studies at the designated time points shown in panel A. Two-tailed unpaired Student t tests were used to determine the significance of CFU differences (P < 0.05). (C) The ΔdprA mutant is attenuated in the mouse models of competitive bacteremia and acute-pneumonia infection and in nasopharyngeal colonization. For bacteremia, CD-1 mice (n = 5) were intraperitoneally inoculated with a 1:1 ratio (1 × 104 CFU per strain) of the WT and ΔdprA strains. Bacteria in spleens were enumerated at 24 h postinfection. For acute pneumonia, CD-1 mice (n = 5) were intranasally inoculated with a 1:1 ratio (1 × 106 CFU per strain) of the WT and ΔdprA strains. Bacteria in lungs were enumerated at 48 h postinfection. For nasopharyngeal colonization, CD-1 mice (n = 5) were intranasally inoculated with a 1:1 ratio (1 × 106 CFU per strain) of the WT and ΔdprA strains. Bacteria in nasal washes were enumerated at 48 h postinoculation. (D) The ΔdprA mutant is attenuated in a mouse model of single-bacteremia infection. CD-1 mice (n = 8) were intraperitoneally inoculated with 1 × 104 CFU of the WT or ΔdprA strain. The bacterial burden in spleens was enumerated at 24 h postinfection. The two-tailed unpaired Student t test indicates a significant difference (P = 0.0117). (E) The ΔdprA mutant is not attenuated in a mouse model of single acute-pneumonia infection. CD-1 mice (n = 10) were intranasally infected with 1 × 106 CFU of the WT or ΔdprA strain. The bacterial burden in lungs was enumerated at 48 h postinfection. The two-tailed unpaired Student t test indicates no significant difference (ns) (P = 0.4985).
As previously shown by us and others, the ΔdprA mutant is attenuated in mouse models of infection (25, 28). When competed against the WT, the ΔdprA mutant was less competitive in bacteremia, acute-pneumonia infections, and nasopharyngeal colonization (Fig. 1C). Additionally, compared with the WT, the ΔdprA mutant was found to be attenuated in single-bacteremia (Fig. 1D) but not in acute-pneumonia (Fig. 1E) infections. These results indicate that DprA confers competitive advantages during host infection and colonization by pneumococcus.
DprA does not cause additional allolytic cell death under CSP1-induced competence in vitro.
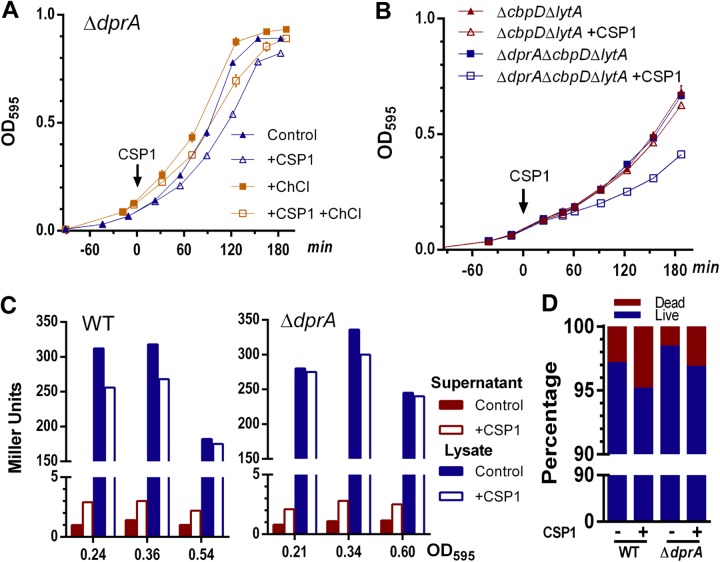
As discussed above, competent pneumococci are capable of releasing allolytic proteins CbpD, LytA, and CibAB, which target noncompetent cells that do not express allolysis immunity proteins ComM (21) and CibC (18). Deletion of three major allolytic genes lytA, lytC, and cbpD greatly reduces pneumococcal allolysis (19, 20, 25, 29). We investigated whether the exponential-phase-growth defect (Fig. 1A) and decreased CFU (Fig. 1B) in competent-state ΔdprA cells were caused by excessive allolysis of the noncompetent cells. In the presence of an allolysis inhibitor (2% choline chloride [ChCl]) (30), CSP1-treated ΔdprA cells still grew slower during the exponential phase than unexposed ΔdprA cells (Fig. 2A). Consistent with the ChCl inhibition results, ΔdprA ΔcbpD ΔlytA cells still exhibited a growth delay upon competence induction by CSP1 (Fig. 2B). These results suggest that in the CSP1-induced competent state in vitro, the exponential-phase-growth delay in the ΔdprA mutant is independent of allolytic killing of noncompetent cells.
FIG 2.
Allolysis does not contribute to exponential-phase-growth delay in the ΔdprA mutant during competence induction in vitro. (A) Inhibition of allolysis does not relieve growth delay in the ΔdprA mutant during CSP1-induced competence. Growth curves of the ΔdprA mutant in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY) in the presence or absence of the allolysis inhibitor 2% choline chloride (ChCl) are shown. CSP1 (100 ng ml−1) was added to the bacterial cultures at an OD595 of ∼0.1. (B) Double deletion of the major allolytic genes lytA and cbpD does not relieve the growth delay in the ΔdprA mutant during competence. CSP1 (100 ng ml−1) was added to the ΔlytA ΔcbpD mutant cultured in THY at an OD595 of ∼0.1. (C) The ΔdprA mutant does not exhibit increased cell lysis after competence induction. CSP1 (100 ng ml−1) was added to WT (D39) and ΔdprA strains constitutively expressing a lacZ gene reporter, and β-galactosidase activities in supernatants and lysates were measured. (D) The ΔdprA mutant does not exhibit increased cell death after competence induction. CSP1 (100 ng ml−1) was added to the bacterial cultures at an OD595 of ∼0.1. After 1 h of incubation at 37°C, cells were washed and stained with live/dead staining reagents. Percentages of dead and live pneumococcal cells were quantified under a fluorescence microscope. Bacteria in 10 representative fields (n = 300 bacteria per field) were counted.
Previously, It was reported that in the capsule-deficient hypercompetent strain Rx, a derivative of D39, up to 20% of pneumococcal cells underwent allolysis when exposed to 250 ng ml−1 CSP1 in the casein-tryptone (CAT) medium (16). We adopted a similar strategy by transcriptionally fusing the lacZ gene to the promoter of the constitutively expressed rplL gene. Pneumococcal cells which undergo allolysis will release β-galactosidase into the culture supernatants, which could be examined by a colorimetric assay (16). Across three different growth stages in THY medium, there was a slight increase of β-galactosidase in the supernatant of CSP1-induced cultures for both the WT and ΔdprA strains (Fig. 2C). However, the ΔdprA mutant did not show higher lysis than the WT, suggesting that both strains did not experience the high levels of CSP1-mediated cell lysis observed in the Rx strain (16). Both D39 and CP1250 were also tested in CAT medium, and no significant cell lysis was found (Fig. S4C). By live/dead fluorescence staining, we confirmed that CSP1 did not trigger significant differences in death between the ΔdprA and WT strains (Fig. 2D; see Fig. S2A in the supplemental material).
Most recently, it was shown that ΔdprA cells undergo frequent cell lysis after 110 min of incubation with CSP1 (24) in C+Y medium (31, 32), which is optimized for competence development. However, we observed no difference in cell lysis of the WT and ΔdprA strains after 1 h (data not shown) and 3 h (Fig. S2B) of CSP1 induction in C+Y medium.
Genetic evidence that allolysis contributes to ΔdprA attenuation in vivo.
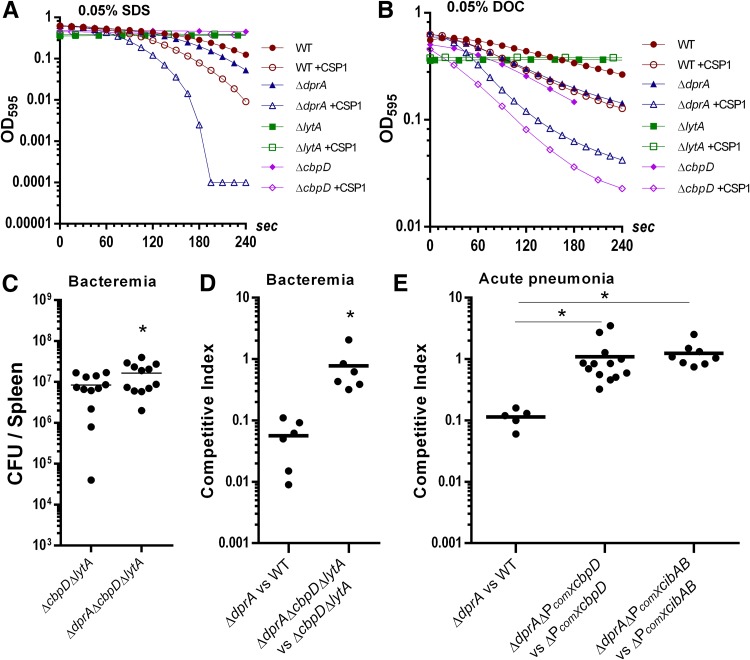
The inability of the ΔdprA mutant to turn off the competent state is expected to cause unwanted overexpression of allolytic factors. We tested the hypothesis that elevated levels of CbpD and LytA are held intracellularly and that perturbation of pneumococcal cell membrane by detergents (e.g., sodium dodecyl sulfate [SDS] and deoxycholate [DOC]) would release these allolytic factors to enhance pneumococcal lysis. Both ΔlytA and ΔcbpD cells were resistant to SDS, indicating that both allolytic factors were responsible for SDS-mediated cell lysis (Fig. 3A). Importantly, the ΔdprA mutant lysed more rapidly than the WT when treated with 0.5% SDS (Fig. 3A) and DOC (Fig. 3B). The difference in the kinetics of detergent-mediated lysis between the WT and ΔdprA strains were more pronounced in the presence of CSP1 (Fig. 3A and B).
FIG 3.
CbpD- and CibAB-mediated allolysis contributes to ΔdprA attenuation in vivo. (A and B) CSP1-induced ΔdprA bacteria are more susceptible to SDS- and deoxycholate (DOC)-mediated lysis. Competence was induced in the D39 WT and ΔdprA strains by adding CSP1 (100 ng ml−1) at an OD595 of 0.1. When both cultures reached an OD595 of 0.5, SDS (0.05%) (A) or DOC (0.05%) (B) was added. Cell lysis was monitored temporally using a spectrophotometer. Noninduced and CSP1-induced ΔlytA and ΔcbpD strains were used as controls. (C) Deletion of allolytic genes cbpD and lytA ameliorates virulence attenuation of the ΔdprA mutant in a single-bacteremia infection. CD-1 mice (n = 12) were intraperitoneally infected with 1 × 104 CFU of the ΔcbpD ΔlytA or ΔdprA ΔcbpD ΔlytA mutant. Mouse spleens were harvested at 24 h postinfection for bacterial enumeration. The two-tailed unpaired Student t test indicates a significant difference (P = 0.0480). (D) Deletion of the cbpD and lytA genes ameliorates virulence attenuation of the ΔdprA mutant in a competitive bacteremia infection. CD-1 mice (n = 6) were infected with a 1:1 ratio (1 × 104 CFU) of the indicated strains. Mouse spleens were harvested at 24 h postinfection for bacterial enumeration. The two-tailed unpaired Student t test indicates a significant difference (P = 0.0226). (E) Deletion of the ComX binding site (combox) on the promoters of cbpD and cibAB ameliorates virulence attenuation of the ΔdprA mutant in a competitive acute-pneumonia infection. CD-1 mice (n = 5 to 13) were intranasally infected with a 1:1 ratio (1 × 106 CFU) of the indicated strains. Mouse lungs were harvested at 48 h postinfection for bacterial enumeration. The two-tailed unpaired Student t test indicates a significant difference between the indicated groups (P < 0.05).
DOC is a bile salt that not only is capable of disrupting the cell membrane but also directly activates LytA and causes rapid pneumococcal lysis (33). Both WT and ΔcbpD cells were lysed by DOC, while the ΔlytA mutant was resistant to DOC, indicating that LytA was responsible for DOC-mediated cell lysis (Fig. 3A and B). The ΔcbpD mutant lysed much faster than the WT, suggesting that it might have structural deficiencies in the cell wall that rendered it more sensitive to DOC-LytA-mediated lysis (Fig. 3B). Increased susceptibility to SDS and DOC was correlated with elevated expression of LytA (see Fig. S5A and B in the supplemental material) and CbpD (see Fig. S6A and B in the supplemental material), respectively, in competent ΔdprA cells.
We further investigated whether overexpression of allolytic factors contributed to the virulence attenuation in the ΔdprA mutant. In a mouse model of single-bacteremia infection, the difference in virulence between dprA+ (ΔcbpD ΔlytA) and ΔdprA mutant (ΔdprA ΔcbpD ΔlytA) bacteria was abolished (Fig. 3C). Similarly, in the competitive bacteremia infection between ΔcbpD ΔlytA and ΔdprA ΔcbpD ΔlytA bacteria, the competitive index (CI) increased to 0.78, compared to 0.06 for infection between the ΔdprA mutant and the WT (Fig. 3D). These results suggest that deletion of the dprA gene in the allolysis-deficient ΔcbpD ΔlytA strain did not confer additional virulence attenuation (Fig. 3C and D) compared to deletion of the dprA gene in the WT background (Fig. 1C and D), supporting the idea that virulence attenuation in the ΔdprA mutant is dependent on CbpD- and LytA-mediated allolysis in vivo.
The in vivo virulence-associated genetic studies (Fig. 3C and D), which indicate that excessive allolysis causes attenuation in the ΔdprA mutant, are inconsistent with the in vitro β-galactosidase release and live/dead assays (Fig. 2C and D). Therefore, a more detailed examination of the role of allolysis in virulence attenuation of the ΔdprA mutant was needed. CbpD is a murein hydrolase which targets the cell division zone of pneumococcus (20). The expression of the cbpD gene is very low during normal growth but is greatly elevated in the competent state (17). CbpD is directly involved in allolysis in liquid medium, whereas the competence-induced two-peptide bacteriocin CibAB is important for allolysis on agar plates (18). Because the expression levels of CbpD and CibAB during in vivo infection are unknown, we exploited a more direct approach by deleting the ComX binding site (combox) in the promoters of both the cbpD and cibAB genes to derive the ΔPcomX cbpD and ΔPcomX cibAB strains, respectively. We did not generate a ΔPcomX lytA mutant because the basal constitutive expression of the lytA gene is high, and CSP1 induction increased the protein levels only temporarily and moderately (Fig. S3B) (20). Additionally, lytA is the fourth gene downstream of its comX binding site, with at least two additional promoters that modulate its basal expression (34).
To delete the combox without causing polarity effects, we used the Sweet Janus cassette (SJC) to generate scarless mutants (35), with some modifications (see the supplemental material). Successful construction of ΔPcomX cbpD or ΔPcomX cibAB mutants was verified by PCR and DNA sequencing (see Fig. S7A in the supplemental material). The expression of cbpD in the ΔPcomX cbpD strain was examined by transcriptionally fusing to a firefly luciferase reporter, and no cbpD expression was detected under CSP1 stimulation (data not shown). CbpD protein levels were also examined by attaching a 3×FLAG tag to the C terminus of the cbpD gene product in the ΔPcomX cbpD strain, and no CbpD protein was detected during competence induction with CSP1 (Fig. S7B and C). Importantly, in an acute-pneumonia model of competitive infection between the ΔdprA ΔPcomX cbpD and ΔPcomX cbpD strains, the CI was 1.1, compared to 0.1 in mice infected by a combination of the ΔdprA and WT strains (Fig. 3E). Similarly, a CI of 1.4 was obtained during competitive infection between the ΔdprA ΔPcomX cibAB and ΔPcomX cibAB strains (Fig. 3E). Collectively, these results suggest that dysregulation of competence-induced allolysis mediated by CbpD and CibAB contributes to the virulence attenuation in the ΔdprA mutant.
Because competence-induced ΔdprA cells lysed faster when exposed to SDS and DOC, we used transmission electron microscopy (TEM) to examine whether cell walls of CSP1-exposed ΔdprA bacteria were compromised. No discernible cell wall defects were found (see Fig. S8 in the supplemental material).
Competence-induced CFU reduction in the ΔdprA mutant is caused by cell division delay mediated by ComM.
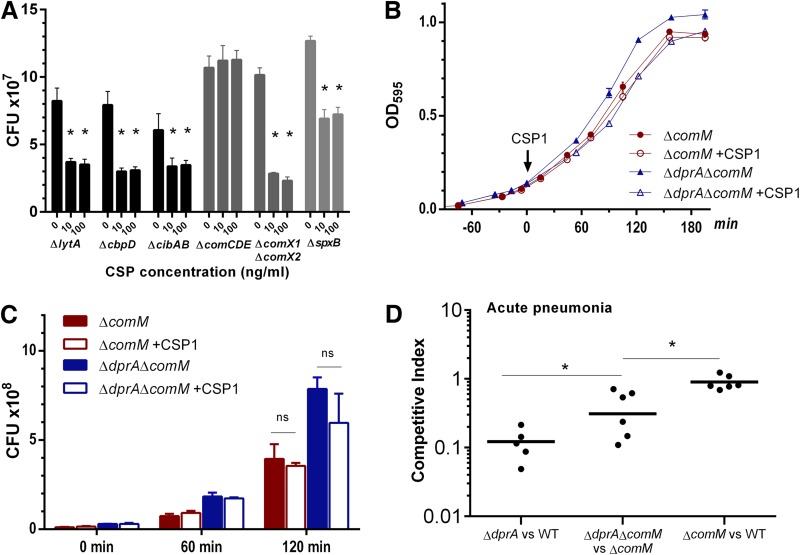
Because competence induction caused exponential-phase-growth delay in the ΔdprA mutant, the reduction of CFU was not unexpected. However, the CFU reduction in competent WT culture was surprising (Fig. 1; see Fig. S1 in the supplemental material). One plausible explanation for CFU reduction was the increase in chain length of WT cells in liquid culture, which could reduce the CFU but not dramatically affect measurement of optical density at 595 nm (OD595). We attempted to identify the gene(s) responsible for the CFU reduction. Under in vitro experimental conditions, allolysis was not required for CFU reduction (Fig. 4A). The “late” competence genes did not appear to play a role, because the ΔcomX1 ΔcomX2 mutant still showed significant CFU reduction upon exposure to CSP1. Also, we ruled out a contribution from oxidative stress, as deletion of the spxB gene, encoding pyruvate kinase for H2O2 production, still led to CFU reduction. Significantly, CFU reduction was abolished in the ΔcomCDE mutant exposed to CSP1 (Fig. 4A).
FIG 4.
CSP1-mediated CFU reduction and virulence attenuation are partially caused by ComM. (A) CSP1-mediated CFU reduction is caused by “early” genes in the competence regulon. Competence was induced in designated mutant cultures by CSP1 (100 ng ml−1) at an OD595 of ∼0.1. After 1.5 h of incubation at 37°C, CFU were measured by serial-dilution plating. Two-tailed unpaired Student t tests were used to determine the significance of CFU differences (P < 0.05). (B) Growth rates of the ΔcomM and ΔdprA ΔcomM strains during exposure to CSP1. Pneumococcal growth was compared in Todd-Hewitt broth supplied with 0.5% yeast extract at 37°C. CSP1 (100 ng ml−1) was added at an OD595 of ∼0.1. (C) CFU reduction is attenuated in both the ΔcomM and ΔdprA ΔcomM strains during exposure to CSP1. CFU were determined by serial-dilution plating in parallel to the growth studies at designated time points as described in panel B. Two-tailed unpaired Student t tests were used to determine the significance of CFU differences (P < 0.05). (D) ComM contributes virulence reduction in the ΔdprA mutant. CD-1 mice were intranasally infected with a 1:1 ratio (1 × 106 CFU per strain) of the indicated strains. Mouse lungs were harvested at 48 h postinfection for bacterial enumeration. The two-tailed unpaired Student t test indicates significant differences between the indicated groups (P < 0.05).
Based on the aforementioned results, overexpression of an “early” gene was likely the cause of the CFU reduction. Recently, it was found that in the competent state, overexpression of the fratricide immunity protein gene comM delayed cell division (36, 37). A ΔcomM strain was created by replacing the gene with an erythromycin resistance gene and was confirmed with PCR and sequencing (see Fig. S9 in the supplemental material). The growth rate of the ΔcomM strain was similar to that of the WT (see Fig. S10A in the supplemental material) in the presence or absence of CSP1, but the CFU reduction caused by CSP1 treatment was eliminated (Fig. S10B). The transformation rate of the ΔcomM strain was reduced (Fig. S10C).
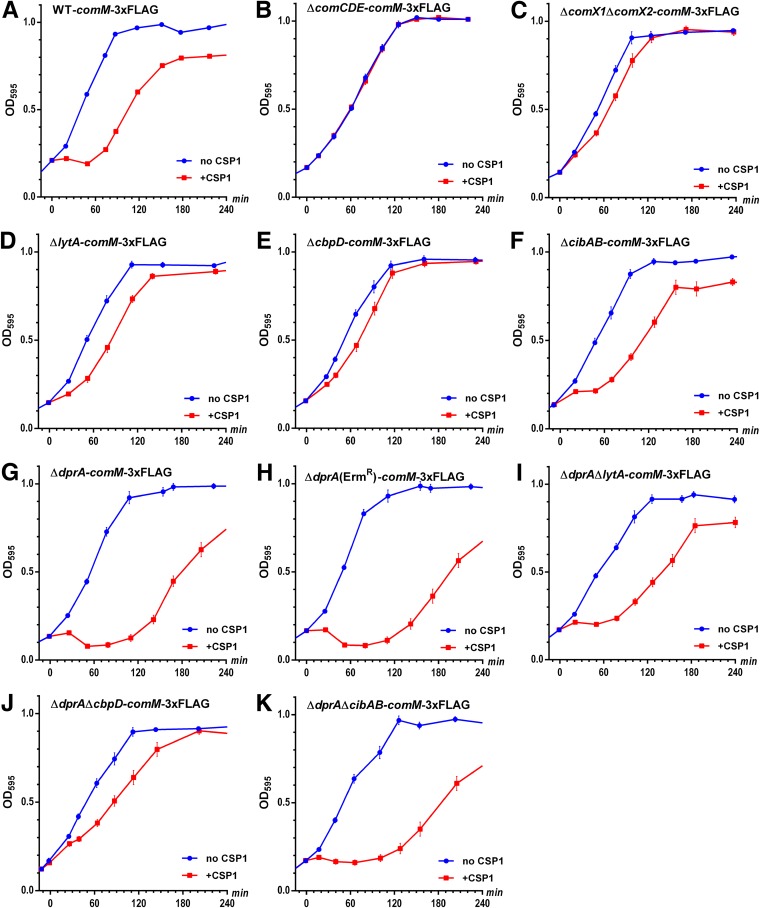
Next, we compared the growth rate and CFU reduction in the ΔdprA ΔcomM strain in the presence or absence of CSP1. The exponential-phase-growth delay was still present (Fig. 4B), but the CFU reduction was eliminated (Fig. 4C). By using a firefly luciferase reporter and 3×FLAG tag, the ΔdprA mutant was found to express higher levels of the comM gene (see Fig. S11 in the supplemental material) as well as ComM protein (see Fig. S12 in the supplemental material) for a prolonged time period. An unexpected observation was that the WT-comM-3×FLAG strain showed basal expression of ComM-3×FLAG even without CSP1 treatment (Fig. S12), which contradicts a previous report (36) and our transcriptional data (Fig. S11), indicating that the degradation of ComM may be impaired. Furthermore, both the WT-comM-3×FLAG and ΔdprA-comM-3×FLAG strains suffered severe growth arrest after competence induction as shown by decreasing amounts of total proteins in the loading controls (Fig. S12) and stalled or decreased OD595 cell density (Fig. 5A and G), although the severity was much more pronounced in the latter.
FIG 5.
Deletion of cbpD ameliorates the competence-induced growth arrest in comM-3×FLAG strains. (A to K) Pneumococcal strains were cultured in THY medium and treated with CSP1 (100 ng ml−1) at time zero, and bacterial growth was monitored for 4 h. Fusion of a 3×FLAG tag to the C terminus of the comM gene product caused growth arrest of both the WT and ΔdprA strains after competence induction by CSP1. The ΔdprA strain had a more severe growth arrest which lasted for more than 1.5 h. This growth arrest is competence dependent and is caused largely by the expression of the competence late genes encoding CbpD and LytA, as shown by the alleviation of growth arrest in the ΔcomX1ΔcomX2-comM-3×FLAG, ΔcbpD-comM-3×FLAG, and ΔlytA-comM-3×FLAG strains.
To determine whether competence-regulated proteins were involved in the CSP1-induced growth arrest of the WT-comM-3×FLAG and ΔdprA-comM-3×FLAG strains, we deleted the comCDE and comX1 comX2 genes in the WT-comM-3×FLAG strains (Fig. 5B and C). Growth arrest mediated by CSP1 was eliminated in the ΔcomCDE-comM-3×FLAG strain and largely abolished in the ΔcomX1ΔcomX2-comM-3×FLAG strain, suggesting that one or more ComX-regulated “late” competence genes might play a role in the growth arrest. We deleted the ComX-regulated lytA, cbpD, and cibAB genes individually in the WT-comM-3×FLAG background and found that deletion of cbpD, and to a lesser extent lytA, largely rescued the growth arrest (Fig. 5D to F). When we deleted lytA, cbpD, and cibAB in the ΔdprA-comM-3×FLAG strain, again deletion of cbpD, and to a lesser extent lytA, largely rescued the growth arrest (Fig. 5I to K). Because these strains (Fig. 5I to K) were constructed with a different dprA knockout strategy (replacement by an erythromycin resistance gene rather than by a kanamycin resistance gene in Fig. 5G and other dprA knockout strains), we verified that the new ΔdprA (Ermr) comM-3×FLAG strain had a similar stalled growth that lasted >1.5 h (Fig. 5H). These results indicate that competence-specific induction of CbpD and LytA is likely the cause of competence-mediated growth arrest in the comM-3×FLAG strain.
Next, we determined whether the inability to shut off ComM expression accounted for the virulence attenuation in the ΔdprA mutant. In a mouse model of competitive acute pneumonia between the ΔcomM and ΔdprA ΔcomM strains, a CI of 0.39 was achieved (Fig. 4D), which is higher than the CI value of 0.1 in the competition between the ΔdprA and WT strains (Fig. 4D), indicating that the absence of ComM improved the competitiveness of the ΔdprA mutant. These results suggest that overexpression of ComM protein partially mediates CFU reduction in vitro and contributes to the virulence attenuation of the ΔdprA mutant. In contrast, the ΔcomM strain was as competitive against the WT (Fig. 4D), suggesting that ComM does not play an important role in pneumococcal virulence.
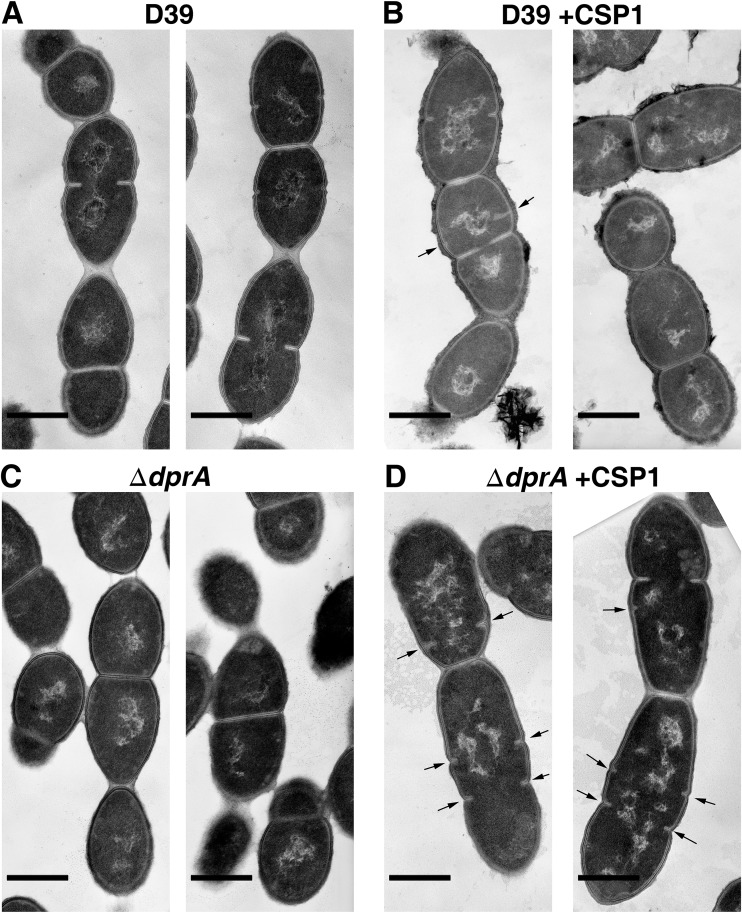
We also compared the cellular morphology of the ΔdprA mutant to that of the WT in the presence and absence or CSP1 by TEM. The morphology of the untreated ΔdprA mutant was similar to that of the WT with or without CSP1 treatment (compare Fig. 6A and B to Fig. 6C). In contrast, in the competent state, the ΔdprA mutant exhibited partially synthesized septa while cells were still at the previous round of division compared to the WT (compare Fig. 6B to Fig. 6D). The cellular morphology of competent ΔdprA cells is similar to that of the previously reported ComM overexpression strain (36). Collectively, these results suggest that overexpression of ComM in the ΔdprA mutant leads to growth delay and virulence attenuation.
FIG 6.
The ΔdprA mutant exhibits cell division delay during CSP1-induced competence. (A to D) Transmission electron microscopy images show the cellular morphologies of the WT and ΔdprA strains in the presence or absence of 100 ng ml−1 CSP1 for 60 min. Arrows point to partially synthesized septa in dividing pneumococcal cells. Scale bars, 0.5 μm.
The ΔdprA mutant is more responsive than the WT to competence induction at low concentrations of CSP1.
As we have shown above, CFU reduction was observed in both the WT and ΔdprA strains when exposed to CSP1 at concentrations of 10 ng ml−1 (Fig. S1) or 100 ng ml−1 (Fig. 1B). These concentrations are relatively low compared to those commonly used for competence studies in vitro (16, 19). The physiologically relevant concentrations of CSP1 for genetic transformation have not been firmly established. We examined the lower limits of CSP concentrations needed to confer CFU reduction (see Fig. S13 in the supplemental material). Both the WT and ΔdprA strains were susceptible to CFU reduction by CSP1 at concentrations of 100 ng ml−1 to 0.5 ng ml−1. However, at 0.25 ng ml−1, the ΔdprA mutant was still susceptible to CSP1, while the WT showed no response (Fig. S13A). These experiments were performed by adding concentrated CSP1 stock solution to culture. At lower final concentrations below 1 ng ml−1, it is not feasible to reduce the pipetting volume of CSP1 solution. Therefore, we increased the volume of freshly aliquoted culture (from 10 ml to 80 ml) to achieve a lower final concentration while maintaining the pipetting volume at 10 μl (Fig. S13A).
To verify the results in Fig. S13A, we adopted a different approach. At final concentrations of CSP1 lower than 1 ng ml−1, the CSP1 stock solution was diluted from 1 ng μl−1 to 0.125 ng μl−1 (Fig. S13B) while maintaining the culture volume at 10 ml and the CSP1 stock volume at 10 μl. Interestingly, in this experimental setting, the CFU reduction by CSP1 in the ΔdprA mutant observed at 0.25 ng ml−1 (Fig. S13A) was abolished (Fig. S13B). Although both approaches achieve the same CSP1 final concentration of 0.25 ng ml−1, the main difference between them is the concentration of CSP1 stock used (1 ng μl−1 versus 0.25 ng μl−1). Since both experiments were performed by adding a relatively small volume of CSP1 to large-volume cultures and then mixing by swirling, there was subset of cells that were exposed to the initial higher concentration of CSP1 stock before mixture. When the subset of ΔdprA cells were exposed to the higher-concentration CSP1 stock (1 ng μl−1 compared to 0.25 ng μl−1), they were more likely to synthesize and secrete more CSP1 into the medium to increase the CSP1 concentration from 0.25 ng ml−1 to achieve the threshold concentration required to induce competence in the whole population. In contrast, when WT bacteria were exposed to higher CSP1 (1 ng μl−1), they could not enter competence (Fig. S13A), suggesting that ΔdprA bacteria were more easily induced to enter competence. Additionally, the different results from the two approaches suggest that both the “local” and “final” CSP1 concentrations could affect the outcome of competence induction. This may be important because during in vivo infection, pneumococcus cells may be restrained by different barriers and therefore be highly heterogeneous. When a subset of ΔdprA cells enters the competence state, they may be able to spread competence more easily than the WT.
To further examine the proposed model of ΔdprA versus WT response to low concentrations of CSP1, we used a firefly luciferase reporter strain to map the expression profile of the “late” gene ssbB, which encodes single-stranded DNA binding protein B, at the CSP1 concentrations and under the experimental conditions described in Fig. S13A. Higher concentrations of CSP1 consistently induced longer and higher expression of the ssbB gene in the ΔdprA mutant than in the WT (Fig. S13C and D). The WT was not responsive to 0.5 ng ml−1 of CSP1 and did not express detectable ssbB (Fig. S13C, arrow). In contrast, even though the ΔdprA mutant exposed to 0.5 ng ml−1 of CSP1 was not able to induce ssbB immediately, once initiated at 40 min postexposure, the induction peaked at 70 min and continued to 120 min postexposure (Fig. S13C and D). Collectively, these results suggest that the concentration of CSP1 in the initial ΔdprA culture was inadequate to boost ssbB expression until additional CSP1 was synthesized by the cells. Additionally, the ΔdprA mutant appears to be more responsive than the WT to low levels of CSP1 that drive the self-amplifying loop for additional synthesis of the competence peptide that resulted in the ComM-mediated CFU reduction (Fig. 4).
Inability to turn off the competence system leads to excessive energy consumption in the ΔdprA mutant.
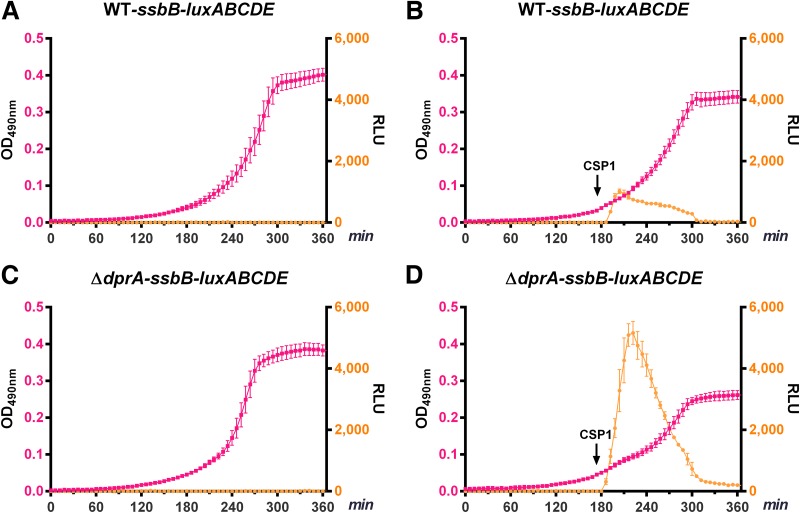
As discussed above, DprA is required to shut off the competent state and the expression of ∼100 “early” and “late” competence genes (22, 23). We have shown that the ΔdprA mutant overexpresses the “early” gene comM, as well as ComX-regulated “late” genes, including lytA, cbpD, and cibAB. Higher expression levels of these competence genes in the ΔdprA mutant are likely to cause unwanted cellular stress and excessive energy consumption. We transcriptionally fused the bacterial bioluminescent luxABCDE genes (38) to ssbB and used the reporter as a surrogate to measure excessive energy consumption in the ΔdprA mutant. The light production catalyzed by luciferase encoded by luxA and luxB involves the oxidization of long-chain fatty aldehyde by oxygen. In order to maintain continuous light output, a key substrate, fatty aldehyde, must be regenerated by a fatty acid reductase complex encoded by luxCDE. Fatty acid reduction consumes ATP (39). When “late” competence genes, including ssbB, are overexpressed by the reporter strain, excessive energy is consumed by the bioluminescent reaction instead of being used for cell growth. In the absence of competence induction, both the WT ssbB-luxABCDE and ΔdprA ssbB-luxABCDE strains did not emit light, and they exhibited similar growth rates (Fig. 7A and C). In contrast, CSP1 induced a more prolonged and heightened expression of ssbB-luxABCDE in the ΔdprA mutant than in the WT, resulting in a significantly lower growth rate (Fig. 7B and D). To further validate the aforementioned results, we also constructed a constitutively expressed rplL-luxABCDE reporter. After competence induction, the WT rplL-luxABCDE and ΔdprA rplL-luxABCDE strains both produced less light, which is attributable to reduced ATP availability and/or reduced transcription and translation of luxABCDE (see Fig. S14 in the supplemental material). The reduction in light output was more severe in the ΔdprA rplL-luxABCDE mutant, indicating the negative effect imposed by competence induction on the normal cellular activities.
FIG 7.
Overexpression of the “late” competence gene ssbB leads to excessive energy consumption and growth delay in the ΔdprA mutant during competence induction. Energy expenditure was tracked indirectly by transcriptionally fusing the bacterial bioluminescent reporter luxABCDE genes to the promoter of the “late” competence gene ssbB. CSP1 (100 ng ml−1) was added at an OD590 of ∼0.05 (180 min). The expression of ssbB-luxABCDE in the WT ssbB-luxABCDE (A and B) and ΔdprA ssbB-luxABCDE (C, D) strains was compared.
DISCUSSION
The role of DprA in competence shutoff was recently reported (22, 23). Independently, during a comprehensive genetic deletion screen of “late” genes regulated by ComX, we have shown that DprA is important for host infection, and this process is competence dependent (25). However, the mechanism of virulence attenuation in the ΔdprA mutant was preliminarily attributed to slight exponential-phase-growth delay in vitro, but this was not mechanistically interrogated. In this study, we found that the ΔdprA mutant is more sensitive to induction by lower concentrations of CSP1 and enters a prolonged competent state, leading to deleterious overexpression of allolytic factors LytA, CbpD, and CibAB, as well as the fratricide immunity protein ComM. Removal of the competent-state-specific expression of allolytic factors and ComM rescued the virulence loss, suggesting dysregulation of these proteins to virulence attenuation in the ΔdprA mutant. Additionally, overexpression of ∼100 competence genes drives excessive energy consumption, leading to competitive disadvantages in the ΔdprA mutant.
Contrary to the previously reported severe growth arrest and allolysis in ΔdprA mutants derived from the noncapsulated, hypercompetent strains Rx, R800, and CP1250 (22, 23), we found that competent-state ΔdprA derived from the encapsulated WT strain D39 showed only a slight growth delay during the exponential phase, with no evidence of increased allolysis. Similarly, we did not find that CP1250 exhibited severe growth arrest and increased cell lysis, suggesting that these phenotypes might be caused by specific culture conditions and/or experimental handlings that we were not able to reproduce.
It has been reported that high levels of intracellular DprA ensure the fitness of genetic transformants by a complete competence shutdown, which also prevents severe growth delay and lysis (24). Our previous in vivo studies have shown that ComX, DprA, LytA, CbpD, and CibAB are competence state-specific virulence factors that are important for bacteremia and acute-pneumonia infections in mice (25). However, our current study suggests that excessive expression of allolytic factors, as happens in the ΔdprA mutant, actually conveys a competitive disadvantage during host infection. The exact mechanisms by which LytA, CbpD, and CibAB contribute to virulence inside the host environment and their regulation by the competence system require further investigation.
The importance of DprA in maintaining homeostatic levels of ComM appears to be crucial for rapid recovery of pneumococcus from the stress imposed by the competent state. ComM induction temporarily pauses cell division, as indicated by reduced CFU in the competent state (Fig. 4), until DNA recombination is completed (37). Overexpression and the extended presence of ComM lead to morphological changes in pneumococcal cells (36). Our data show that ΔdprA cells overexpress ComM and display delayed cell division and separation, which likely contributed to growth delay during the exponential phase (Fig. 6; see Fig. S11 and S12 in the supplemental material). These results are consistent with the notion that DprA tightly regulates ComM levels to prevent prolonged cell division arrest while genomic DNA is replicating.
ComM is expressed at very low basal levels and can be degraded by membrane proteases (21, 36). Previously, it was reported that insertion-duplication into the C terminus of the comM gene product in the R800 strain caused a CbpD-dependent severe cell lysis, and therefore, ComM was considered an immunity protein (21). We adopted a similar strategy by using a pEVP3-derived plasmid to attach a 3×FLAG tag to the C terminus of the comM gene product via insertion-duplication. We found that the 3×FLAG-tagged ComM could be detected in noncompetent cells, and CSP1 treatment increased ComM expression, resulting in stalled growth and lysis. These phenotypes are more severe in the ΔdprA mutant but are rescued by CbpD and LytA deficiency. Our data suggest that 3×FLAG tagging at the C terminus of the comM gene product interfered with membrane protease-mediated degradation and therefore led to prolonged stability and accumulation and pneumococcal lysis. Therefore, it is possible that Havarstein et al. generated a ComM-stabilized strain by disrupting the C terminus of ComM (21), similar to our ComM-3×FLAG strain. Interestingly, when we replaced the comM gene with an antibiotic marker, the ΔcomM mutant did not exhibit significant cell lysis. Our ΔcomM results are in agreement with the phenotypes of another previously published ΔcomM mutant, which did not show significant cell death after competence induction (37). Collectively, these results suggest that ComM is not the immunity protein against CbpD. Rather, it is the increased stability of 3×FLAG-tagged ComM and the C terminus insertion-duplication reported by Havarstein et al. (21) that cause pneumococcal lysis. The exact effect of 3×FLAG tagging on ComM function requires further investigation.
Because the deleterious effect of excessive ComM on cell growth can be mitigated by the loss of CbpD and LytA, it is likely that ComM is not an immunity protein but rather is a contributor to CbpD- and LytA-mediated fratricide. However, because pneumococcus has high basal expression of LytA but CbpD expression is competence dependent, CbpD is the likely contributor to competence-triggered cell lysis in the ComM-3×FLAG strain. Additionally, ComM expression causes cell separation delay which is manifested as CFU reduction. However, delay in cell separation is independent of any “late” competence genes, suggesting that this particular physiological function of ComM is not dependent on CbpD. Further investigations are needed to unveil the interactions between ComM and CbpD.
The finding that the ΔdprA mutant is more responsive to low concentrations of CSP1 is interesting. The majority of competence studies in pneumococcus, including those involving ΔdprA in both hypercompetent Rx and R800 strains, were performed using high concentration of CSP1 (≥100 ng ml−1) (22, 23), which may not be not physiologically meaningful. Recently a non-QS-based model of competence induction has been proposed (9, 40) and challenged (41). In this model, competent cells of certain pneumococcus strains, including D39, retain CSP on their ComD receptors, and the competence signal is propagated by transferring the receptor-bound CSP to ComD receptors on the recipient cells via direct cell-cell contact. This model proposes that CSP peptides are secreted and freely diffusible in a traditionally described QS manner (9). Our studies show that, regardless of the models, once competence is induced in a small subpopulation of pneumococcus, the ΔdprA mutant is likely to express more CSP1 and therefore is more likely to spread competence to the whole population (see Fig. S13 in the supplemental material). For example, the ΔdprA ssbB-luc strain exposed to 0.5 ng ml−1 CSP1 (Fig. S13D) eventually caught up with the pace of competence induction, and the lag time most likely suggested a period of “CSP1 accumulation in the medium” or “cell-cell contact signal propagation.” Future studies will examine the differences in competence induction between the ΔdprA mutant and D39 in vivo. Unlike the near homogeneity in the test tubes, pneumococcus encounters different niches and physical barriers inside mouse lungs and other host tissues.
In summary, detailed characterization indicates that dysregulated expression of allolytic factors, excessive energy consumption as a result of an inability to shut off the transcription of ∼100 competence genes, and deleterious effects associated with ComM-mediated cell division delay and death contribute to competence-specific virulence attenuation in the ΔdprA mutant. Increased understanding of the DprA-regulated competence-specific virulence mechanism may lead to better understanding of pneumococcal pathogenesis and disease control.
MATERIALS AND METHODS
Chemicals.
Unless stated otherwise, chemicals were purchased from Sigma-Aldrich.
Bacterial strains and growth conditions.
S. pneumoniae D39 and derivatives (see Table S1 in the supplemental material) were cultured overnight on Todd-Hewitt broth (THB) (Thermo Fisher Scientific) agar plates containing 5% defibrinated sheep blood at 37°C with 5% CO2. Single colonies were picked and cultured in fresh THY liquid medium to a desirable density. CAT (23) and C+Y (32) media were also used in designated in vitro experiments. Antibiotics were used at the following concentrations: kanamycin for Janus cassette, 200 μg ml−1; kanamycin for Sweet Janus cassette, 500 μg ml−1; streptomycin, 100 μg ml−1; and chloramphenicol, 4 μg ml−1. For sucrose selection, 10% sucrose was added to THB agar plates.
Construction of gene replacement mutants.
To generate kanamycin-resistant pneumococcal strains (Table S1), the kanamycin resistance gene was cloned from the Janus cassette. Flanking sequences from target genes were amplified by PCR by using the Q5 high-fidelity DNA polymerase (New England Biolabs) and spliced with the kanamycin resistance gene using the NEBuilder HiFi DNA assembly master mix (New England Biolabs). Spliced genes were used as templates for PCR amplification. PCR products were purified with a gel purification kit (ZYMO Research), and used to transform recipient pneumococcal strains. For chloramphenicol resistance, the target gene to be deleted was PCR amplified and cloned into plasmid pEVP3 (see Table S2 in the supplemental material), and the resultant plasmid was used to transform the recipient strains. To generate scarless mutants, the Sweet Janus cassette was used as described in detail in the supplemental material. To construct reporter plasmids (Table S2), we performed gene assembly using the NEBuilder HiFi DNA assembly master mix. Detail procedures for mutant and plasmid constructions are provided in the supplemental material.
Ethics statement.
Animal studies were performed in strict accordance with the U.S. National Research Council’s Guide for the Care and Use of Laboratory Animals and the U.S. Public Health Service’s Policy on Humane Care and Use of Laboratory Animals. The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Illinois at Urbana-Champaign (UIUC).
Mouse models.
Six-week-old male and female CD-1 mice (Charles River, Boston, MA) were housed in positively ventilated microisolator cages with automatic recirculating water located in a room with laminar, high-efficiency particle accumulation-filtered air. The animals received autoclaved food, water, and bedding and were acclimated for 5 to 7 days before infection. Briefly, for the acute-pneumonia model, mice were anesthetized by exposing them to inhaled isoflurane by placing them in a vaporizer with a scavenger system, followed by intranasal inoculation with 106 CFU of pneumococcal cells in 50 μl saline. After 48 h, mice were euthanized, and the bacterial burden in the homogenized lungs was enumerated by serial-dilution plating. For the bacteremia model, isoflurane-anesthetized CD-1 mice were intraperitoneally inoculated with 104 CFU pneumococcal cells in 100 μl saline. Bacterial burdens in the spleens were enumerated at 24 h after infection.
For nasopharyngeal colonization, unanesthetized CD-1 mice were intranasally inoculated with 106 CFU pneumococcal cells in 20 μl saline. After 48 h, mice were euthanized, and nasal cavities were dissected and homogenized. The bacterial burden was enumerated after serial-dilution plating on THB agar. Because both male and female mice responded similarly in all three mouse models of infection, data were collated and presented together.
In vivo competitions were performed as previously described (41). Briefly, CD-1 mice were inoculated with a pneumococcal suspension containing a 1:1 mixture of the parental strain and its derivative, using the concentration of each strain described above for the single infections. At designated times, mouse lungs or spleens were harvested, homogenized, and serially diluted onto THB agar plates with and without antibiotics. The CI was defined as the output ratio of derivative to parental bacteria divided by the input ratio of derivative to parental bacteria. A CI value of 0.7 or lower was defined as attenuated (41).
β-Galactosidase assays.
Pneumococcus strains constitutively expressing lacZ reporter gene were grown to the indicated OD595 and treated with 100 ng ml−1 CSP1 for 30 min. β-Galactosidase assays were performed with culture supernatant and whole culture lysates as previously reported (23).
Live/dead staining.
Pneumococcal strains were grown to an OD595 of 0.1 and treated with CSP1 at 100 ng ml−1 for 1 h or 3 h. Bacteria were gently washed with sterile saline at 4,000 × g for 3 min and stained with the LIVE/DEAD BacLight bacterial viability kit (Thermo Fisher Scientific). Stained bacteria were examined using a fluorescence microscope. The number of live and dead bacteria (n = 300 per field) were counted in 10 representative fields.
TEM.
Pneumococcal strains were grown to an OD595 of 0.1 and treated with CSP1 (100 ng ml−1) for 1 h. Bacteria were gently washed with saline, pelleted by centrifugation at 4,000 × g for 3 min, and fixed using Karnovsky’s fixative. Fixed bacteria were washed with the Sorensen’s phosphate buffer and incubated in osmium tetroxide, followed by a wash in sterile water. Bacteria were then dehydrated in ethanol followed by acetonitrile and then immersed sequentially in 1:1 epoxy-acetonitrile, 3:1 epoxy-acetonitrile, and 100% epoxy and finally embedded in Lx112 resin (LADD Research Industries). The pellet was then hardened in a microcentrifuge tube at 80°C overnight, removed, and reembedded on an epoxy stub. Embedded bacteria were sectioned to 90-nm thickness by using a diamond blade on a Reichert ultramicrotome and stained with uranyl acetate and lead citrate. Images were captured by using a Hitachi H600 transmission electron microscope (TEM) at 75 kV using plate film at the UIUC Material Research Laboratory.
Western blot analysis.
Pneumococcal culture (1 ml) was centrifuged at 4,000 × g for 3 min at 4°C. Pneumococcal reporter strains harboring lytA-FLX3 and cbpD-FLX3 were lysed by Bullet Blender (Next Advance). Pneumococcal strains expressing 3×FLAG-tagged ComM were lysed with 0.1% Triton X-100 and 0.01% SDS. Ten microliters of total cell lysates was separated by 10% SDS-PAGE, transferred onto nitrocellulose membranes, subjected to Western blot analysis by probing with primary antibody against FLAG tag (Agilent number 200474, 1:1000 dilution), and visualized with a secondary goat anti-rat IgG–horseradish peroxidase (HRP) (Cell Signaling Technology number 7077, 1:2,000 dilution) by using the ECL substrate (Bio-Rad number 170-5060). Relative abundances of proteins were quantified by densitometry using the ImageJ software (NIH).
In vitro luciferase assays.
Pneumococcal strains harboring bacterial luxABCDE or firefly luciferase (luc) were cultured in a 96-well plate at 37°C in a Wallac Victor 2 Multilabel Counter (PerkinElmer). d-Luciferin potassium (GoldBio) was added to cultures of luc-bearing strains to a final concentration of 0.65 mM. Pneumococcal growth (OD495) and reporter gene expression (luminescence) were measured automatically. CSP1 was added to induce competence to a final concentration of 100 ng ml−1.
Statistical analyses.
Quantitative data were expressed as the mean ± standard deviation. Statistical significance was determined using the GraphPad Prism statistical software package. A significant difference was considered to be indicated by a P value of <0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following individuals for sharing various reagents: David Briles (University of Alabama at Birmingham) for the pneumococcal strain D39, Donald Morrison (University of Illinois at Chicago) for both the Janus cassette and pEVP3 plasmid, Marc Lipsitch (Harvard School of Public Health) for the Sweet Janus cassette, and Malcolm Winkler (Indiana University) for the ΔspxB mutant.
This work was supported in part by an American Lung Association DeSouza Research Award (DS-192835-N), the NIH (HL090699 and HL142626A1), and a University of Illinois Research Board award (RB16002) to G. W. Lau.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The authors have no competing financial interests and are solely responsible for the experimental designs and data analysis.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00349-19.
REFERENCES
- 1.Hiller NL, Ahmed A, Powell E, Martin DP, Eutsey R, Earl J, Janto B, Boissy RJ, Hogg J, Barbadora K, Sampath R, Lonergan S, Post JC, Hu FZ, Ehrlich GD. 2010. Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog 6:e1001108. doi: 10.1371/journal.ppat.1001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chancey ST, Agrawal S, Schroeder MR, Farley MM, Tettelin H, Stephens DS. 2015. Composite mobile genetic elements disseminating macrolide resistance in Streptococcus pneumoniae. Front Microbiol 6:26. doi: 10.3389/fmicb.2015.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hui FM, Zhou L, Morrison DA. 1995. Competence for genetic transformation in Streptococcus pneumoniae: organization of a regulatory locus with homology to two lactococcin A secretion genes. Gene 153:25–31. doi: 10.1016/0378-1119(94)00841-f. [DOI] [PubMed] [Google Scholar]
- 5.Havarstein LS, Coomaraswamy G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claverys JP, Prudhomme M, Martin B. 2006. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol 60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- 7.Johnston C, Campo N, Berge MJ, Polard P, Claverys JP. 2014. Streptococcus pneumoniae, le transformiste. Trends Microbiol 22:113–119. doi: 10.1016/j.tim.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Johnston C, Martin B, Fichant G, Polard P, Claverys JP. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 9.Prudhomme M, Berge M, Martin B, Polard P. 2016. Pneumococcal competence coordination relies on a cell-contact sensing mechanism. PLoS Genet 12:e1006113. doi: 10.1371/journal.pgen.1006113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pestova EV, Havarstein LS, Morrison DA. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol 21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 11.Havarstein LS, Gaustad P, Nes IF, Morrison DA. 1996. Identification of the streptococcal competence-pheromone receptor. Mol Microbiol 21:863–869. doi: 10.1046/j.1365-2958.1996.521416.x. [DOI] [PubMed] [Google Scholar]
- 12.Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, Walling J, Li H, Mintz M, Tsegaye G, Burr PC, Do Y, Ahn S, Gilbert J, Fleischmann RD, Morrison DA. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol 51:1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- 13.Lee MS, Morrison DA. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol 181:5004–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo P, Morrison DA. 2003. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J Bacteriol 185:349–358. doi: 10.1128/jb.185.1.349-358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo P, Li H, Morrison DA. 2003. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol Microbiol 50:623–633. doi: 10.1046/j.1365-2958.2003.03714.x. [DOI] [PubMed] [Google Scholar]
- 16.Steinmoen H, Knutsen E, Havarstein LS. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc Natl Acad Sci U S A 99:7681–7686. doi: 10.1073/pnas.112464599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kausmally L, Johnsborg O, Lunde M, Knutsen E, Havarstein LS. 2005. Choline-binding protein D (CbpD) in Streptococcus pneumoniae is essential for competence-induced cell lysis. J Bacteriol 187:4338–4345. doi: 10.1128/JB.187.13.4338-4345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guiral S, Mitchell TJ, Martin B, Claverys JP. 2005. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci U S A 102:8710–8715. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eldholm V, Johnsborg O, Haugen K, Ohnstad HS, Havarstein LS. 2009. Fratricide in Streptococcus pneumoniae: contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiology 155:2223–2234. doi: 10.1099/mic.0.026328-0. [DOI] [PubMed] [Google Scholar]
- 20.Eldholm V, Johnsborg O, Straume D, Ohnstad HS, Berg KH, Hermoso JA, Havarstein LS. 2010. Pneumococcal CbpD is a murein hydrolase that requires a dual cell envelope binding specificity to kill target cells during fratricide. Mol Microbiol 76:905–917. doi: 10.1111/j.1365-2958.2010.07143.x. [DOI] [PubMed] [Google Scholar]
- 21.Havarstein LS, Martin B, Johnsborg O, Granadel C, Claverys JP. 2006. New insights into the pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol Microbiol 59:1297–1307. doi: 10.1111/j.1365-2958.2005.05021.x. [DOI] [PubMed] [Google Scholar]
- 22.Mirouze N, Berge MA, Soulet AL, Mortier-Barriere I, Quentin Y, Fichant G, Granadel C, Noirot-Gros MF, Noirot P, Polard P, Martin B, Claverys JP. 2013. Direct involvement of DprA, the transformation-dedicated RecA loader, in the shut-off of pneumococcal competence. Proc Natl Acad Sci U S A 110:E1035–E1044. doi: 10.1073/pnas.1219868110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weng L, Piotrowski A, Morrison DA. 2013. Exit from competence for genetic transformation in Streptococcus pneumoniae is regulated at multiple levels. PLoS One 8:e64197. doi: 10.1371/journal.pone.0064197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston C, Mortier-Barriere I, Khemici V, Polard P. 2018. Fine-tuning cellular levels of DprA ensures transformant fitness in the human pathogen Streptococcus pneumoniae. Mol Microbiol 109:663–675. doi: 10.1111/mmi.14068. [DOI] [PubMed] [Google Scholar]
- 25.Zhu L, Lin J, Kuang Z, Vidal JE, Lau GW. 2015. Deletion analysis of Streptococcus pneumoniae late competence genes distinguishes virulence determinants that are dependent or independent of competence induction. Mol Microbiol 97:151–165. doi: 10.1111/mmi.13016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortier-Barriere I, Velten M, Dupaigne P, Mirouze N, Pietrement O, McGovern S, Fichant G, Martin B, Noirot P, Le Cam E, Polard P, Claverys JP. 2007. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell 130:824–836. doi: 10.1016/j.cell.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 27.Ravin AW. 1959. Reciprocal capsular transformations of pneumococci. J Bacteriol 77:296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu Y, Chang D, Xu H, Zhang X, Pan L, Xu C, Huang B, Zhou H, Li J, Guo J, Liu C. 2017. The virulence of Streptococcus pneumoniae partially depends on dprA. Braz J Microbiol 48:225–231. doi: 10.1016/j.bjm.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moscoso M, Claverys JP. 2004. Release of DNA into the medium by competent Streptococcus pneumoniae: kinetics, mechanism and stability of the liberated DNA. Mol Microbiol 54:783–794. doi: 10.1111/j.1365-2958.2004.04305.x. [DOI] [PubMed] [Google Scholar]
- 30.Briese T, Hakenbeck R. 1985. Interaction of the pneumococcal amidase with lipoteichoic acid and choline. Eur J Biochem 146:417–427. doi: 10.1111/j.1432-1033.1985.tb08668.x. [DOI] [PubMed] [Google Scholar]
- 31.Tomasz A, Hotchkiss RD. 1964. Regulation of the transformability of pheumococcal cultures by macromolecular cell products. Proc Natl Acad Sci U S A 51:480–487. doi: 10.1073/pnas.51.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens KE, Chang D, Zwack EE, Sebert ME. 2011. Competence in Streptococcus pneumoniae is regulated by the rate of ribosomal decoding errors. mBio 2:e00071-11. doi: 10.1128/mBio.00071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mellroth P, Daniels R, Eberhardt A, Ronnlund D, Blom H, Widengren J, Normark S, Henriques-Normark B. 2012. LytA, major autolysin of Streptococcus pneumoniae, requires access to nascent peptidoglycan. J Biol Chem 287:11018–11029. doi: 10.1074/jbc.M111.318584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortier-Barriere I, de Saizieu A, Claverys JP, Martin B. 1998. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol Microbiol 27:159–170. doi: 10.1046/j.1365-2958.1998.00668.x. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Thompson CM, Lipsitch M. 2014. A modified Janus cassette (Sweet Janus) to improve allelic replacement efficiency by high-stringency negative selection in Streptococcus pneumoniae. PLoS One 9:e100510. doi: 10.1371/journal.pone.0100510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Straume D, Stamsas GA, Salehian Z, Havarstein LS. 2017. Overexpression of the fratricide immunity protein ComM leads to growth inhibition and morphological abnormalities in Streptococcus pneumoniae. Microbiology 163:9–21. doi: 10.1099/mic.0.000402. [DOI] [PubMed] [Google Scholar]
- 37.Berge MJ, Mercy C, Mortier-Barriere I, VanNieuwenhze MS, Brun YV, Grangeasse C, Polard P, Campo N. 2017. A programmed cell division delay preserves genome integrity during natural genetic transformation in Streptococcus pneumoniae. Nat Commun 8:1621. doi: 10.1038/s41467-017-01716-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Francis KP, Joh D, Bellinger-Kawahara C, Hawkinson MJ, Purchio TF, Contag PR. 2000. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect Immun 68:3594–3600. doi: 10.1128/iai.68.6.3594-3600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meighen EA. 1993. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J 7:1016–1022. doi: 10.1096/fasebj.7.11.8370470. [DOI] [PubMed] [Google Scholar]
- 40.Weyder M, Prudhomme M, Berge M, Polard P, Fichant G. 2018. Dynamic modeling of Streptococcus pneumoniae competence provides regulatory mechanistic insights into its tight temporal regulation. Front Microbiol 9:1637. doi: 10.3389/fmicb.2018.01637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreno-Gamez S, Sorg RA, Domenech A, Kjos M, Weissing FJ, van Doorn GS, Veening JW. 2017. Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat Commun 8:854. doi: 10.1038/s41467-017-00903-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.