ABSTRACT
Histone 3 lysine 9 trimethylation (H3K9me3) is a conserved histone modification that is best known for its role in constitutive heterochromatin formation and the repression of repetitive DNA elements. More recently, it has become evident that H3K9me3 is also deposited at certain loci in a tissue-specific manner and plays important roles in regulating cell identity. Notably, H3K9me3 can repress genes encoding silencing factors, pointing to a fundamental principle of repressive chromatin auto-regulation. Interestingly, recent studies have shown that H3K9me3 deposition requires protein SUMOylation in different contexts, suggesting that the SUMO pathway functions as an important module in gene silencing and heterochromatin formation. In this Review, we discuss the role of H3K9me3 in gene regulation in various systems and the molecular mechanisms that guide the silencing machinery to target loci.
KEY WORDS: Chromatin, Heterochromatin, Epigenetics, Gene regulation, Transcriptional repression, Transposons, Germline, Cell fate maintenance
Summary: This Review covers how the H3K9me3 mark is established and maintained, and the modes by which it functions to regulate gene expression and cell identity during development.
Introduction
In eukaryotic nuclei, DNA associates with proteins to form a higher-order complex known as chromatin. Early observations distinguished differently stained regions of interphase chromatin, termed ‘euchromatin’ and ‘heterochromatin’, reflecting regions that decondense or remain condensed during interphase (Heitz, 1928). Although euchromatin is mostly associated with active transcription, heterochromatin is usually (but not always) associated with gene silencing. The repressive properties of heterochromatin have been attributed to the denser packing of DNA that might make it less accessible to transcription factors and the transcriptional machinery. There are two broad types of heterochromatin. Constitutive heterochromatin is present in all cell types and is typically composed of repeat-rich and gene-poor regions around centromeres and telomeres. In contrast, facultative heterochromatin is established in a cell type-specific manner on genomic regions that generally have normal gene density (Elgin and Reuter, 2013). In this context, the establishment of facultative heterochromatin on specific genomic regions usually correlates with transcriptional repression.
The basic unit of chromatin is the chromatosome, which consists of DNA wrapped around octamers containing two copies each of the histone proteins H2A, H2B, H3 and H4, and a linker histone H1. Specific residues on histones can be post-translationally modified via the covalent addition of chemical groups, such as acetyl, methyl and phosphoryl, as well as by small protein modifiers such as ubiquitin and SUMO (Small Ubiquitin-like Modifier) (Bannister and Kouzarides, 2011). The activity of the enzymatic ‘writers’ that carry out these modifications is counterbalanced by ‘eraser’ enzymes that can remove the modifications. Histone modifications regulate the accessibility of DNA to the transcriptional machinery and can serve as marks to recruit effector proteins with diverse functional outcomes. Trimethylation of histone 3 lysine 9 (H3K9me3) and lysine 27 (H3K27me3) are the best-known histone modifications associated with gene repression and heterochromatin. These marks are established and recognized by distinct writer and reader complexes, and are typically localized to different genomic regions, suggesting that they associate with distinct types of chromatin. H3K27me3 is found on many regions silenced in a cell-specific manner, such as the X-chromosome and the developmentally regulated homeotic (HOX) genes (Beuchle et al., 2001; Plath et al., 2003; Ringrose and Paro, 2004; Schuettengruber et al., 2017). The role of H3K27me3 in controlling expression of developmentally regulated genes has been extensively studied (reviewed by Aloia et al., 2013; Schuettengruber et al., 2007; Schuettengruber et al., 2017). Although H3K27me3-marked domains are often referred to as ‘facultative heterochromatin’, not all cell-specific heterochromatic domains are marked by H3K27me3 (see below) and some H3K27me3-marked regions are not condensed (e.g. Becker et al., 2017).
In contrast, H3K9me3 is enriched in constitutive heterochromatin such as centromeric and telomeric repeats from yeast to human (Richards and Elgin, 2002). H3K9me3 is also associated with stable repression of transposable elements (TEs), abundant nuclear parasites that can propagate within host genomes causing DNA damage and mutations in both Drosophila and vertebrate systems (Karimi et al., 2011; Klenov et al., 2011; LeThomas et al., 2013; Matsui et al., 2010; Mikkelsen et al., 2007; Pezic et al., 2014; Riddle et al., 2011; Rowe et al., 2010; Rozhkov et al., 2013; Sienski et al., 2012; Wang and Elgin, 2011). TE insertions scattered throughout the genome are often marked by local H3K9me3 peaks in otherwise euchromatic regions. Owing to this concentration at TEs, repetitive regions and chromosomal ends, H3K9me3-marked constitutive heterochromatin is best known for its role in chromosome architecture and genome stability, as it is required for proper chromosome segregation and to prevent unequal recombination between repeats (Janssen et al., 2018). However, genome-wide profiling of H3K9me3 in mammals and Drosophila have revealed that this mark is also present outside of repeat-rich and gene-poor regions, suggesting that it plays an important role in host gene regulation, including the repression of developmentally restricted genomic regions, thereby acting as a key regulator of cell fate.
In this Review, we discuss how the H3K9me3 mark is established and maintained and review the modes by which it functions to regulate gene expression and cell identity in development, with an emphasis on the murine and Drosophila systems. We also highlight recent findings that have identified a conserved role for the SUMO pathway in H3K9me3 establishment. Finally, we discuss findings suggesting that heterochromatin effectors are themselves regulated by the H3K9me3 silencing mark they deposit, indicating a homeostatic mechanism for heterochromatin maintenance.
Mechanisms of H3K9me3 establishment and maintenance
Readers, writers and erasers of H3K9 methylation
Factors involved in heterochromatin formation first emerged from genetic screens for factors affecting position effect variegation (PEV) in Drosophila – the phenomenon by which the relocation of protein-coding genes that normally reside in euchromatin next to heterochromatin leads to their variegated repression (reviewed by Elgin and Reuter, 2013). These screens identified over 100 loci encoding putative chromatin regulators, referred to as suppressors of PEV [Su(var)] and enhancers of PEV [En(var)]. Many Su(var) genes encode factors that are necessary for heterochromatin-induced gene repression. Their molecular characterization identified proteins that establish and maintain heterochromatin structure, including writers and readers of the H3K9me3 mark (Fig. 1). The Su(var)3-9 gene family was found to be the first known H3K9-specific histone methyltransferases (H3K9-HMTs) in human (Suv39h), Drosophila [Su(var)3-9] and yeast (clr4) (Czermin et al., 2001; Nakayama et al., 2001; Rea et al., 2000). Studies in metazoan systems identified two other conserved families of H3K9-specific HMTs, including SETDB1/ESET (dSetDB1/eggless in Drosophila) (Schultz et al., 2002) and G9a/GLP (Tachibana et al., 2001). Despite similar activities in vitro, H3K9-HMTs differ in vivo in their tissue specificity, their genomic regions of activity, their bias towards a specific methyltransferase activity (mono-, di- or tri-methylation) and their dispensability. For example, the SUV39-family proteins are typically associated with di- and tri-methylation of H3K9 at centromeric and telomeric regions, whereas SETDB1 was first identified as a HMT that primarily acts on euchromatic regions such as TEs scattered throughout the genome (Aagaard et al., 1999; Karimi et al., 2011; Peters et al., 2003; Rice et al., 2003; Schotta et al., 2002; Schultz et al., 2002). In addition, SetDB1 acts at telomeric heterochromatin in mouse embryonic stem cells (ESCs) (Gauchier et al., 2019). In mammals, G9a-GLP predominantly regulates H3K9 mono- and di-methylation, and is essential for embryogenesis (Tachibana et al., 2001, 2002). Conversely, among the three H3K9-specific HMTs in Drosophila – G9a, Su(var)3-9 and dSetDB1/Eggless – only the last is essential (Brower-Toland et al., 2009). However, a comprehensive picture of the specificity and functional redundancy of HMTs in different organisms has not yet been established. The activity of H3K9-HMTs is counterbalanced by erasers from the jumonji (JmjC) domain-containing demethylase families, with the JMJD2/KDM4 family displaying activity towards H3K9me2/me3 residues (as well as methylated H3K36), and JMJD1/KDM3 proteins displaying activity towards H3K9me2/1 (reviewed by Cloos et al., 2008; Nottke et al., 2009).
Fig. 1.
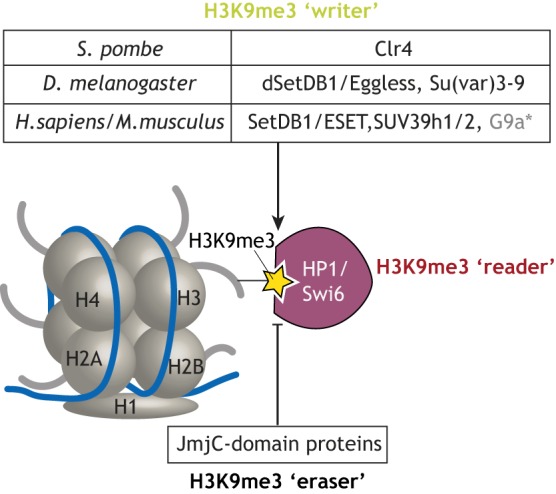
Schematic of enzymatic machineries involved in H3K9me3 regulation. H3K9me3 (yellow star) is deposited by ‘writers’ and is recognized by the chromodomain present in numerous ‘reader’ proteins, which include the HP1 family of proteins. It is removed by ‘erasers’ from the JmjC-domain family of demethylases. The table depicts species-specific H3K9me3 writers. *G9a primarily acts as a H3K9 mono- and dimethylase, but can catalyze H3K9 trimethylation in vitro with slow kinetics (Collins et al., 2005).
Proteins from the highly conserved heterochromatin protein 1 (HP1/CBX/Swi6) family are H3K9me3 readers and central effectors of heterochromatin formation from yeast to human. HP1 proteins consist of a N-terminal chromodomain (CD), which is required for their specific interaction with methylated H3K9 (Bannister et al., 2001; Jacobs et al., 2001; Lachner et al., 2001), a hinge region and a C-terminal chromoshadow domain (CSD). A current model suggests that CSD domain-mediated dimerization of two HP1 proteins bound to H3K9me3 residues on adjacent nucleosomes brings these nucleosomes in closer proximity, thereby causing chromatin condensation (Canzio et al., 2011; Hiragami-Hamada et al., 2016; Machida et al., 2018). The CSD also mediates HP1 interaction with additional proteins (Platero et al., 2004), and may recruit other chromatin remodeling and modifying complexes. Drosophila and mammalian genomes encode several paralogs of the H3K9me3 reader HP1 family that exhibit distinct localization patterns and post-translational modifications (Lomberk et al., 2006a,b).
H3K9me3 reader and writer activities can be coupled. For example, members of the SUV39/Clr4 family of H3K9-HMTs have a conserved chromodomain that can mediate binding to H3K9me3, implying direct binding of the mark by its own writer. The S. pombe Clr4 CD binds H3K9me in vitro and is required for maintaining a repressed state in vivo (Ragunathan et al., 2015; Zhang et al., 2008). In Drosophila, Su(var)3-9 interacts directly with the reader HP1, and the CD of Su(var)3-9 is required for its proper localization to chromatin (Schotta et al., 2002; Zhang et al., 2008). These and other examples of interactions or inter-dependencies between histone mark writer and reader complexes are thought to confer a feed-forward loop that ensures H3K9me3 maintenance and propagation (reviewed by Allshire and Madhani, 2018; Vermaak and Malik, 2009).
Recruitment of H3K9-modifying complexes to target genomic regions
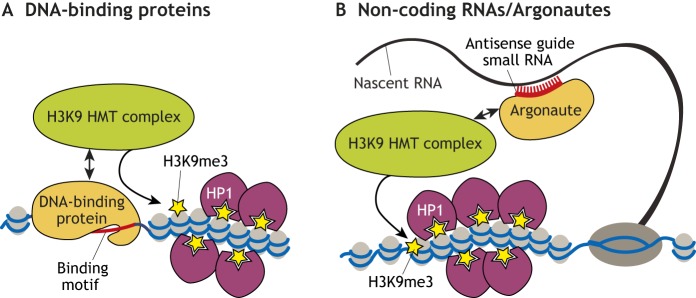
In order to ensure H3K9me3 deposition at appropriate targets and avoid ectopic silencing, the recruitment of silencing effectors to specific regions of the genome must be precisely regulated. H3K9-HMTs and HP1 proteins cannot recognize specific sequences and bind DNA directly, and therefore require additional factors for their recruitment. Studies have identified two major modes of H3K9-HMT recruitment to chromatin: through sequence-specific DNA-binding proteins, and through small RNA guides complementary to nascent transcripts (Fig. 2).
Fig. 2.
Modes of recruiting the H3K9me3 machinery to specific genomic targets. Target specificity for recruitment of HMTs is either provided by: (A) DNA-binding proteins with sequence-specific DNA binding motifs; or (B) by small non-coding RNAs associated with Argonaute proteins.
Most of our current knowledge of DNA-binding proteins involved in silencing complex recruitment comes from mammalian systems. Perhaps the best-characterized factors that act in H3K9-HMT recruitment are members of the large vertebrate-specific family of Krüppel-associated box (KRAB)-containing zinc-finger proteins (KRAB-ZFPs). The majority of KRAB-ZFPs target sites are located within TEs, and specific KRAB-ZFPs have been shown to induce SETDB1/H3K9me3-dependent silencing at endogenous retrovirus (ERV) targets (Ecco et al., 2016; Imbeault et al., 2017; Lupo et al., 2013; Wolf et al., 2015a,b). Biochemical studies have demonstrated that the KRAB domain interacts with the universal co-repressor KAP1/Trim28, which in turn recruits SetDB1 (Friedman et al., 1996; Peng et al., 2000; Schultz et al., 2002). The retinoblastoma (Rb) protein and MAX have also been implicated in SUV39H1- and SetDB1-mediated silencing of specific genes in mammals, respectively (Maeda et al., 2013; Nielsen et al., 2001; Tatsumi et al., 2018). Studies in murine cell lines have also identified several transcription factors that localize to H3K9me3-rich pericentric satellites and are required for heterochromatin integrity (Bulut-Karslioglu et al., 2012; Vassen et al., 2006; Yamashita et al., 2007); however, a mechanistic link with H3K9-HMT recruitment is not well established. Of note, H3K9-related factors can interact with and share some targets with other chromatin modifiers, such as DNA methylase1 and NuRD, in various contexts, indicating complex context-dependent interactions between different silencing pathways (e.g. Ivanov et al., 2007; Lehnertz et al., 2003; Robertson et al., 2000; Schultz et al., 2001; Tatsumi et al., 2018; Uchimura et al., 2006). In Drosophila, DNA-binding proteins that recruit H3K9-HMTs have not been identified to date, although several lines of evidence (discussed below) point to their existence.
Small RNA-based targeting mechanisms for H3K9me3-induced transcriptional silencing also occur and seem to function in organisms from yeast to mammals. RNA interference (RNAi) is a conserved mechanism of gene regulation in which short RNAs (microRNAs, siRNAs or piRNAs) are loaded into proteins from the Argonaute (Ago) family and guide them to complementary regions at RNA targets with different regulatory outcomes. In the nucleus, small RNA-associated Argonautes recognize complementary regions in nascent RNAs and guide silencing effectors to induce co-transcriptional repression and heterochromatin formation (reviewed by Holoch and Moazed, 2015). The role of RNAi in heterochromatin formation was first demonstrated in fission yeast, where complexes containing Argonaute-bound small RNA guides are required for H3K9me3 deposition at centromeric regions (Hall, 2002; Volpe et al., 2002). RNAi-directed mechanisms also have a well-established role in heritable epigenetic silencing in C. elegans, and analogous pathways have been described in plants and ciliates (Ashe et al., 2012; Chalker et al., 2013; Zilberman et al., 2003). In metazoans, a dedicated RNAi-based silencing pathway – the piRNA pathway, which consists of proteins from the Piwi clade of Argonautes associated with short RNAs (piRNAs) – guides H3K9me3 deposition at TE targets in the germline (LeThomas et al., 2013; Pezic et al., 2014; Rozhkov et al., 2010; Sienski et al., 2012).
It is important to note that de novo deposition and maintenance of H3K9me3 can be achieved by different pathways. Given reader/writer coupling, once established, H3K9me3 could be maintained independently of the initial inducer. In S. pombe, H3K9me is not completely lost upon Argonaute deletion; furthermore, H3K9me at a reporter locus can be maintained in the absence of the initial inducer if demethylation is inhibited (Ragunathan et al., 2015). Similarly, it has been proposed that, in Drosophila, embryonic piRNAs may contribute to the initial establishment of H3K9me3 profiles in both somatic and germ cells, and that these H3K9me3 patterns are later maintained in a piRNA-independent manner (Akkouche et al., 2017; Gu and Elgin, 2013).
The role of SUMO in recruiting the silencing complex
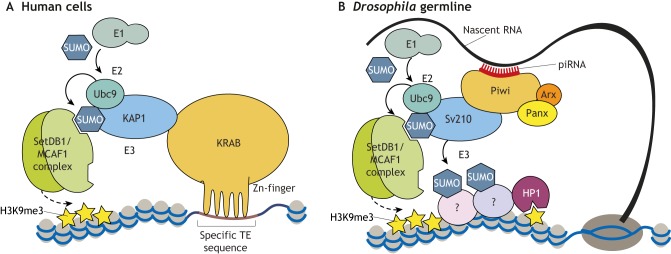
Post-translational modification by SUMO (see Box 1) has been recognized as a mechanism that functions in different silencing pathways and model systems. For example, SUMO (smt3) and Swi6 SUMOylation have been shown to be required for heterochromatin stability at the silent mating type loci in fission yeast (Shin et al., 2005). SUMOylation is also implicated in several aspects of heterochromatin formation in different genomic contexts in mammals. Studies in murine cell lines have shown that SUMOylation of HP1α regulates its de novo localization to pericentric chromatin, with Suv39h1 acting as a SUMO E3 ligase (Maison et al., 2011, 2016). SUMOylation of the core histone H4 induces HP1γ recruitment and local silencing in human cell lines (Shiio and Eisenman, 2003). Notably, studies in both vertebrate and invertebrate systems demonstrate that the SUMO moiety can act as a docking site for the recruitment of silencing effectors containing SUMO-interacting motifs (SIMs). For example, SUMOylation of the transcription factor Sp3 induces the recruitment of SetDB1 and HP1 proteins, resulting in H3K9me3 deposition and silencing of reporter genes in human cells (Stielow et al., 2008). SUMO is also involved in KAP1/SetDB1-mediated silencing (Fig. 3A). SetDB1 contains SIMs, and autocatalytic SUMOylation of KAP1 mediates the recruitment of SetDB1 to chromatin and enhances its methyltransferase activity in human cells (Ivanov et al., 2007). The conserved SetDB1 co-factor MCAF1/ATF7IP also localizes to chromatin targets in a SUMO-dependent manner (Uchimura et al., 2006). In line with their involvement in SetDB1 recruitment, SUMO and SUMO-conjugating enzymes have emerged as factors required for ERV repression in embryonic carcinomas and ESCs (Fukuda et al., 2018; Yang et al., 2015). Finally, SUMO was found to be enriched at H3K9me3-marked regions, including ERVs in ESCs, and its depletion leads to global reduction of H3K9 methylation and ERV de-repression (Cossec et al., 2018). Together, these studies indicate that SUMOylation is an important modification for KRAB-KAP1-SetDB1-mediated ERV silencing.
Box 1. The SUMO pathway.
SUMO (Small Ubiquitin-like Modifier) is a ∼10 kDa protein that acts as a reversible covalent post-translational modifier. The SUMO conjugation mechanism is highly conserved and resembles that of ubiquitin conjugation. In brief, the SUMO E1 activation complex Aos1/Uba2 passes SUMO to the SUMO E2-conjugating enzyme Ubc9. Ubc9 then catalyzes the transfer of SUMO to its substrate by forming an isopeptide bond between the C-terminal glycine of SUMO and a lysine in the substrate, typically within the consensus motif ΨKxD/E. SUMO E3 ligases are thought to facilitate this transfer and provide substrate selectivity but, overall, this class of SUMO enzymes remains poorly understood.
SUMO modification may lead to changes in protein stability or conformation with different regulatory outcomes. SUMOylation is often implicated in large protein assemblies, as the SUMO moiety provides a binding surface for partners containing SUMO-interacting motifs (SIMs) and can thereby promote protein-protein interactions. Interestingly, while post-translational modifications typically involve a highly specific interaction between a substrate and a modifying enzyme, with a single event triggering a particular regulatory outcome, SUMOylation appears to function in a different manner. Indeed, studies in yeast indicate that collective SUMOylation of co-localized proteins, rather than individual factors is functionally important to promote the assembly, stability and/or activity of large protein complexes (Psakhye and Jentsch, 2012). Such is the case of the homologous recombination pathway in yeast, where multiple factors are collectively SUMOylated by the chromatin-bound SUMO E3 ligase Siz2 upon recruitment to DNA (Psakhye and Jentsch, 2012). As many of these proteins also have SIMs, it has been proposed that ubiquitous SUMOylation of co-localized proteins results in multiple SUMO-SIM interactions that may stabilize protein complexes. Together, the loose subtract selectivity of the SUMO-conjugation machinery, and the high frequency of SUMOylation and SIMs, lead to a model wherein a SUMO ligase induces a local ‘SUMO spray’ that promotes the formation of large protein assemblies (Psakhye and Jentsch, 2012). Whether such collective SUMOylation is involved in heterochromatin formation remains to be determined.
Fig. 3.
The SUMO pathway is involved in SetDB1-dependent H3K9me3 deposition downstream of different silencing pathways. (A) Model of KRAB-ZFP/KAP1-mediated TE silencing in human cells. KRAB-ZFPs recognize specific TE sequences in DNA. The KRAB-ZFP co-repressor KAP1 is a SUMO E3 ligase that undergoes autocatalytic SUMOylation (involving the SUMO E1 and E2 ligases). It then recruits the H3K9-specific HMT SetDB1 (via the interaction between SUMO and the SUMO-interacting motif present in SetDB1), which in turn induces methylation of H3K9. Adapted from Ivanov et al. (2007). (B) Model of piRNA-mediated TE silencing in the Drosophila female germline. piRNAs guide the Piwi complex to complementary nascent TE transcripts. The Piwi complex interacts with the SUMO E3 ligase Su(var)2-10 (Sv210), which is auto-SUMOylated and may also induce the SUMOylation of additional chromatin factors. The SUMO moiety then recruits the SetDB1/Wde (MCAF1 homolog) complex, which induces methylation of H3K9. Adapted from Ninova et al. (2019a preprint).
Components of the SUMO pathway also emerged as factors required for TE repression in a genome-wide screen in Drosophila (Muerdter et al., 2013). Subsequent studies showed that the SUMO E3 ligase Su(var)2-10 acts downstream of piRNA-Piwi complexes to recruit the SetDB1 silencing complex, which deposits H3K9me3 at TE targets in the female germline (Fig. 3B, Ninova et al., 2019a preprint). Furthermore, Su(var)2-10 is required for H3K9me3 deposition and for transcriptional repression at loci that are not targeted by piRNAs, indicating that it can act downstream of different guides (Ninova et al., 2019a preprint,b).
Collectively, these observations suggest that the SUMO pathway is a highly conserved module acting in the recruitment of silencing complexes in different genomic contexts and pathways (Fig. 3). However, although the role of SUMO in silencing is now well documented, with thousands of proteins being identified as SUMOylation targets (Cubeñas-Potts and Matunis, 2013), only a few specific substrates (such as KAP1 in mammals) are known in the context of heterochromatin formation.
Diverse modes of gene regulation by H3K9me3
The establishment and maintenance of constitutive heterochromatin is essential for genome stability (reviewed by Janssen et al., 2018). Although H3K9 methylation is often studied in the context of repetitive, gene-poor heterochromatic regions such as centromeres, recent data from multiple model systems demonstrate that it also affects the transcription of host genes. Genetic and biochemical studies have identified several major modes by which H3K9me3 can affect the host transcriptome.
Heterochromatin and H3K9 methylation are typically associated with gene silencing, as first evidenced by the phenomenon of PEV (Elgin and Reuter, 2013). Loss of H3K9me3 following depletion of silencing guides or effectors, such as H3K9-HMTs, KRAB-ZFP or piRNA pathway components, is associated with transcriptional upregulation of TE targets but also, in some cases, of host genes that are normally decorated by this mark (e.g. Karimi et al., 2011; Klenov et al., 2011; LeThomas et al., 2013; Matsui et al., 2010; Rozhkov et al., 2013; Sienski et al., 2012; Smolko et al., 2018; Wolf et al., 2015b). Furthermore, artificial recruitment of H3K9-HMTs, such as SetDB1, or of other silencing factors to reporter loci in euchromatin results in transcriptional silencing of the reporter gene in both somatic and germ cells (Ayyanathan et al., 2003; Ivanov et al., 2007; Li et al., 2003; Ninova et al., 2019a preprint; Schultz et al., 2002; Sienski et al., 2015; Yu et al., 2015). However, H3K9me3 is not always associated with silencing, especially if it is excluded from the transcription start site (TSS). For example, heterochromatin-residing yet active genes in Drosophila are usually enriched in H3K9me3 across their bodies but not around the TSS (Ninova et al., 2019b; Riddle et al., 2011). Studies in flies have shown that some active genes that reside in constitutive heterochromatin require this environment for their proper expression, as they become silent if translocated to euchromatin (Wakimoto et al., 1990; Yasuhara and Wakimoto, 2006). Loss of H3K9me3 results in transcriptional downregulation of a significant fraction of genes residing in constitutive heterochromatin and the H3K9me3-rich chromosome 4 in various tissues, as well as of genomic loci encoding piRNA precursors in the germline (Ninova et al., 2019b; Rangan et al., 2011; Tzeng et al., 2007). These findings suggest that, despite its common silencing function, H3K9me3 enrichment over gene bodies outside of the TSS is not only compatible with transcription but is even required for proper expression of heterochromatic genes. In the case of piRNA precursor loci in Drosophila germ cells, H3K9me3 acts as a docking site for the so-called RDC complex and associated factors necessary for piRNA biogenesis (Andersen et al., 2017; Mohn et al., 2014). Why many protein-coding genes residing in heterochromatin require H3K9me3 for their expression is not understood.
H3K9me3 installment on host genes can occur either via direct recruitment of the chromatin modifier to the gene (as discussed earlier) or by indirect mechanisms. The best-studied indirect mechanisms of HMT recruitment to host genes involves the targeting of TEs in the gene proximity for H3K9me3-associated silencing. It is well established that the H3K9me3 mark is deposited at TE sequences both at heterochromatic loci and at TE insertions in euchromatin. However, as H3K9me3 can spread in cis several kilobases (Karimi et al., 2011; Mikkelsen et al., 2007; Pezic et al., 2014; Rebollo et al., 2011), TE insertions in the vicinity of host genes can induce their epigenetic silencing. Moreover, although TEs are often viewed as ‘junk DNA’ or harmful elements, they are also an important source of regulatory sequences and ncRNAs (Chuong et al., 2017; Kapusta et al., 2013; Slotkin and Martienssen, 2007). Indeed, depending on the tissue, 6-30% of transcripts in mouse and human cells initiate within TEs, and this phenomenon is most widespread in embryonic tissues (Faulkner et al., 2009). TEs, particularly the ERV1 family, contain many of the predicted binding motifs for the core pluripotency factors OCT4/POU5F1 and Nanog in mammals (Kunarso et al., 2010). In addition, many TE-KRAB-ZFP pairs are conserved despite the TEs being long inactive, suggesting that some binding sites were co-opted as regulatory elements of host genomes (Imbeault et al., 2017). Although not all computationally predicted TE-derived regulatory regions are functional, several elements were found to act as enhancers in ESCs (Todd et al., 2019). Thus, the targeting of H3K9me3 to TE sequences can also modulate the activity of some associated promoter and/or enhancer elements with diverse regulatory outcomes on proximal genes. The role of TE-related gene regulation has mostly been investigated in mammals; however, evidence from other systems suggests that TE co-option in developmental processes is a common phenomenon (Chuong et al., 2017).
Finally, recent advances in chromosome conformation capture methods have revealed that, at least in mammals, H3K9me3 plays a role in gene expression by maintaining the three-dimensional organization of the genome by suppressing cryptic CTCF binding sites (Jiang et al., 2017).
Gene regulation by H3K9me3 in development
The role of H3K9me3 in gene regulation in somatic tissues
In metazoans, gametogenesis and early embryogenesis are accompanied by extensive epigenetic reprogramming during which most chromatin marks, including H3K9me3, are erased to grant totipotency to the zygote and are later re-established. H3K9me3 re-establishment in somatic tissues is essential for normal developmental progression in Drosophila and mammals. Interestingly, loss of silencing effectors that primarily control H3K9me3 deposition at constitutive heterochromatin leads to less severe phenotypes than disruption of H3K9me3 deposition outside constitutive heterochromatin. For example, mutant mice double null for the two SUV39 paralogs, which primarily localize to centromeric regions, display chromosomal instability and multiple defects, yet some animals survive to adulthood (Peters et al., 2001). Conversely, upon loss of SetDB1/ESET, which is primarily involved in H3K9me3 deposition outside constitutive heterochromatin, the inner cell mass of the pre-implantation embryo fails to form properly, leading to pre-implantation lethality (Dodge et al., 2004). Likewise, in Drosophila, Su(var)3-9 null mutants are viable and fertile (Tschiersch et al., 1994), while SetDB1 loss-of-function mutations are homozygous lethal (Seum et al., 2007). Loss of several members of the HP1 family of H3K9me3 readers, including the Drosophila Su(var)2-5/HP1a and the mouse Cbx1/HP1β, also result in developmentally lethal phenotypes (Aucott et al., 2008; Eissenberg et al., 1990; Eissenberg et al., 1992; Kellum and Alberts, 1995), highlighting the essential role of H3K9me3 in early development.
H3K9me3 in the early embryo and ESCs
Most current knowledge about the role of H3K9me3 in early somatic development comes from studies in murine systems, which have shown that gene silencing by H3K9me3 is particularly important during pre-implantation embryonic development, ESC self-renewal, cell differentiation and cell lineage commitment.
It is well established that there is a complex interplay between H3K9me and DNA methylation in mammalian embryos (Allis and Jenuwein, 2016; Cedar and Bergman, 2009). DNA methylation provides a stable and mitotically heritable mode of silencing, which is temporarily erased during gametogenesis and upon fertilization, with re-methylation occurring at the time of implantation (Reik et al., 2001; Smith et al., 2012; Wu et al., 2016). During this time, H3K9me3 mediates gene and transposon repression, and guides the re-establishment of DNA methylation later on (Allis and Jenuwein, 2016; Cedar and Bergman, 2009). As development progresses, pluripotency-associated genes are silenced, while genes involved in alternative cell fates become activated; these processes also involve H3K9me3. High-resolution mapping of H3K9me3 in the mouse embryo by ChIP-seq has revealed a finely regulated timing of H3K9me3 establishment at different genomic elements (Wang et al., 2018). H3K9me3 is present at some developmental genes and some long terminal repeats (LTRs) in oocytes and zygotes, but is lost at the two-cell stage. However, globally the two-cell stage is characterized by a stark increase in H3K9me3, which initially accumulates predominantly on LTRs (Wang et al., 2018). Depletion of SetDB1, KAP1/Trim28, Sumo2 and the histone chaperone Chaf1a leads to H3K9me3 loss and upregulation of several LTRs. In addition, many embryos arrest at the blastocyst stage upon knockdown of these factors, highlighting their importance for proper early development (Wang et al., 2018). As development continues into the implantation stage, H3K9me3 also begins to appear at host genes where different lineages acquire distinct H3K9me3 signatures. Typically, in a specific cell type H3K9me3 is deposited at genes that are characteristic of alternative cell fates (Wang et al., 2018). Thus, the H3K9me3 mark appears to suppress lineage-inappropriate gene expression.
The role of SetDB1-mediated TE, and gene repression and cell fate control is also apparent from studies in mouse ESCs. As in early embryos, LTRs in ESCs are marked by H3K9me3. Depletion of KAP1/Trim28, SetDB1/ESET and its co-factor MCAF1/ATF7IP, the KRAB-ZFP Zfp809, Morc2a, Chaf1a/b, Sumo2 and SUMO pathway enzymes leads to pervasive upregulation of multiple (partially overlapping) ERV targets (Cossec et al., 2018; Fukuda et al., 2018; Karimi et al., 2011; Martens et al., 2005; Matsui et al., 2010; Mikkelsen et al., 2007; Rowe et al., 2010; Wolf and Goff, 2007; Yang et al., 2015). In addition, depletion of the H3K9me3 effectors SetDB1, KAP1 and several KRAB-ZFPs from ESCs leads to de-repression of a subset of protein-coding genes (Ecco et al., 2016; Karimi et al., 2011; Rowe et al., 2013; Wolf et al., 2015b). Notably, a significant fraction of genes activated upon SetDB1 or Trim28 depletion reside in proximity to TEs (mostly ERV and LINEs), and many become transcribed from alternative TSSs residing in concomitantly upregulated ERV regions, thereby forming chimeric transcripts (Karimi et al., 2011; Rowe et al., 2013). Furthermore, SetDB1 and H3K9me3 have been reported to occupy promoter regions in ESCs and early embryos, suggesting that they directly target specific host genes (Bilodeau et al., 2009; Karimi et al., 2011; Wang et al., 2018; Yuan et al., 2009). Consistent with a role for H3K9me3-dependent silencing in maintaining cell fate, depletion of silencing factors from ESCs is generally associated with loss of cell identity. For example, depletion of KAP1/Trim28, Chaf1a, SUMO2/3 or the SUMO E2 ligase Ubc9 leads to conversion of the transcription profile of ESCs to a state resembling that of the two-cell embryo, i.e. a two-cell (2C)-like state, suggesting that these factors maintain the ESC state by repressing 2C-specific genes, including the master regulator of the 2C state Dux (Cossec et al., 2018; Ishiuchi et al., 2015; Macfarlan et al., 2012). Elimination of SetDB1 from ESCs also alters their fate, with cells reported to either die or shift to trophoblast-like fate (Bilodeau et al., 2009; Yeap et al., 2009; Yuan et al., 2009). A large fraction of genes repressed by SetDB1/H3K9me3 are developmental regulators (Bilodeau et al., 2009; Karimi et al., 2011; Yuan et al., 2009). Notably, a subset of genes repressed by SetDB1/H3K9me3 in ESCs also encode factors normally expressed in testis and oocytes (Karimi et al., 2011). A recent study suggested that SetDB1 can be directly guided to at least some germline-specific genes in ESCs by the transcription factor MAX (Tatsumi et al., 2018). Moreover, many genes associated with the germline transcriptional program, such as P-granule components and meiosis genes, are also occupied by SUMO in ESCs (Cossec et al., 2018). Together, these data suggest that, in ESCs, H3K9me3-mediated repression involving SetDB1 and SUMO also plays an important role in maintaining cell identity by suppressing alternative fates. Interestingly, some targets of SetDB1/H3K9me3 in ESCs are also marked by H3K27me3 and DNA methylation, indicating several layers of repression for certain genomic targets (Bilodeau et al., 2009; Karimi et al., 2011).
H3K9me3 functions in lineage commitment and cell differentiation
Gene repression through SetDB1-dependent H3K9 methylation is not restricted to ESCs and pre-gastrulation embryos. Despite a prevailing model that TEs in adult somatic tissues of mammals are silenced by DNA methylation, KRAB-ZFP/KAP1/SetDB1-dependent transcriptional repression was reported to control several cell type-specific subsets of ERVs in a range of adult mouse cell types, including embryonic fibroblasts (MEFs), pre-adipocytes, hepatocytes and B-lymphocytes (Collins et al., 2015; Ecco et al., 2016; Fasching et al., 2015; Kato et al., 2018; Wolf et al., 2015b).
Multiple examples highlight a role for H3K9me3 in cell type-specific gene regulation. High resolution analysis of heterochromatin formation in murine cells from different germ layers, and from hepatic and pancreatic lineages revealed that the number of H3K9me3-marked regions in different lineages increases from early developmental stages until gastrulation, although H3K9me3 is subsequently removed as cells progress into specific lineages (Nicetto et al., 2019). Transient deployment of H3K9me3 in germ layer cells is required to repress genes associated with mature cell function, and failure to properly establish this mark leads to expression of lineage-inappropriate genes later on (Nicetto et al., 2019). Silencing by SUV39H1-dependent H3K9me3 and HP1α deposition was shown to be involved in lineage commitment of Th2 lymphocytes by repressing Th1-specific loci (Allan et al., 2012), and in adipogenesis by restricting the expression of master regulatory genes until differentiation is required (Matsumura et al., 2015). H3K9me3-mediated regulation of host genes and TEs by SetDB1 has also been implicated in transcriptome regulation and normal cell fate switches during murine neurogenesis and oligodendrocyte differentiation (Jiang et al., 2017; Liu et al., 2015; Tan et al., 2012). Notably, SetDB1-repressed genes in neuronal tissue are enriched in factors characteristic for other lineages, and particularly in germline-specific genes (Tan et al., 2012). Germline genes are also targets of SetDB1- and SUMO-mediated repression in ESCs (as mentioned above), pointing to a ubiquitous role of this pathway in suppressing germ cell fate. Finally, Hi-C analysis of SetDB1-depleted postnatal mouse forebrain neurons revealed alterations in chromosomal conformation resulting from CTCF binding to cryptic sites normally occupied by H3K9 and DNA methylation (Jiang et al., 2017).
The H3K9me3 mark has also been found to impede cell re-programming (Becker et al., 2016). Studies in human embryonic fibroblasts, for example, identified over 200 H3K9me3-enriched genomic regions, with an average size of 2.2 Mb, that are refractory to binding of the pioneer transcription factors Oct4, Sox2, Klf4 and Myc (OKSM), thereby impeding re-programming to pluripotency (Soufi et al., 2012). Knockdown of the HMTs SUV39H1 and SetDB1, the histone chaperone CAF1 subunits Chaf1a and Chaf1b, Cbx3/HP1γ, Sumo2 and SUMO pathway components, or overexpression of the Jmjd2c demethylase, improve OKSM binding and reprogramming of fibroblasts to induced pluripotent cells in human or murine systems (Borkent et al., 2016; Cheloufi et al., 2015; Chen et al., 2013; Cossec et al., 2018; Onder et al., 2012; Soufi et al., 2012; Sridharan et al., 2013). Similar ‘reprogramming-resistant regions’ (RRRs) marked by H3K9me3 impede epigenetic reprogramming upon somatic cell nuclear transfer, with overexpression of the H3K9 demethylase Kdm4d or simultaneous depletion of the two SUV39 paralogs partially releasing this impediment (Matoba et al., 2014). Notably, a detailed study of the role of SUMO in the reprogramming of MEFs to pluripotency revealed that, in this context, SUMO is required to maintain the activity of fibroblast-specific enhancers (Cossec et al., 2018). This function is in stark contrast to the role of SUMO in ESCs, where it suppresses RRRs and the 2C-like transcriptome, highlighting a context-dependent function of protein SUMOylation (Cossec et al., 2018).
The role of H3K9me3 in somatic tissues in fruit flies is less well understood. dSetDB1/Eggless is the only essential HMT in D. melanogaster. dSetDB1 appears to be responsible for initial deposition of H3K9me3 and HP1 at many regions in the early embryo (Seller et al., 2019), and dSetDB1 mutations are associated with a wide variety of developmental defects and lethality (Brower-Toland et al., 2009; Stabell et al., 2006; Tzeng et al., 2007). As SetDB1-dependent H3K9me3 is present at multiple genomic loci, including nearly the entire chromosome 4 (which contains 79 genes and is enriched in repeats), the severe phenotypes of SetDB1 loss-of-function mutations are likely due to pleiotropic effects. Factors that recruit SetDB1 to its genomic targets in fly somatic tissues have yet to be established. In Drosophila, mutations in RNAi factors, including Piwi, affect PEV and H3K9me3 in somatic tissues not known to have an active piRNA pathway (Gu and Elgin, 2013; Pal-Bhadra et al., 2004). As piRNA/Piwi are maternally loaded into the egg (but not zygotically expressed outside of the gonads), an attractive model is that maternal piRNA/Piwi complexes guide initial heterochromatin establishment in the early embryo, which is later maintained piRNA independently. There are also some H3K9me3-marked genes in regions that do not have local TEs and cannot be targeted by piRNA, pointing to the existence of piRNA-independent targeting mechanisms (Ninova et al., 2019b). DNA-binding proteins that recruit SetDB1 analogous to the vertebrate-specific KRAB-ZFP family have not been identified.
H3K9me3 and gene silencing in germ cells
Germline specification, gonad development and gametogenesis are highly orchestrated processes associated with extensive epigenetic re-programming. As germ cells carry the genetic material to be transmitted to offspring, they must also be well protected from damaging TE activity. Chromatin modification by H3K9me3 plays an essential role in germ cell development and fertility in both vertebrate and invertebrate animals. Most current understanding of transcriptional repression by H3K9me3 in germ cells comes from studies in the male germline of mice, and in the female germline of Drosophila.
In mice, a population of epiblast cells in the post-implantation embryo forms primordial germ cells (PGCs): the precursors of oocytes and spermatozoa. SetDB1 depletion at early stages of development (prior to E6.5 by Sox2Cre cKO and at E9.5 by TnapCre cKO) was shown to repress PGC formation and lead to gonadal hypotrophy in adults (Liu et al., 2014; Mochizuki et al., 2018). During PGC-like cell induction, SetDB1 was suggested to directly repress several transcription factors involved in mesoderm cell fate, thereby maintaining proper cell identity (Mochizuki et al., 2018). In E13.5 PGCs, SetDB1 was shown to control H3K9me3 levels and repress a subset of retrotransposons from the ERV and LINE1 classes, as well as a number of host genes (Liu et al., 2014). As in other systems, many genes deregulated upon SetDB1 loss are not directly marked by H3K9me3 but reside in the proximity of or initiate their transcription from within TEs (Liu et al., 2014). The factors that guide H3K9 methylation by SetDB1 in early PGCs are not known. Metazoan germline cells typically possess an active piRNA pathway. However, Miwi2, the only nuclear Piwi protein in mice, is not expressed until E14.5-15.5 (Aravin et al., 2008), thus H3K9me3 deposition before this stage is likely piRNA independent. It is possible that, as in other tissues, TEs in early germ cells are repressed by KRAB-ZFPs, but this hypothesis needs to be addressed.
In addition to functioning in PGCs and testis, SetDB1 has been shown to regulate the expression of host genes, several TEs and associated chimeric transcripts in mouse oocytes (Eymery et al., 2016; Kim et al., 2016). While mammalian oocytes express Piwi proteins and piRNAs, the mechanisms of H3K9me3 establishment at different genomic targets in this system has not been comprehensively characterized.
In Drosophila, H3K9me3-mediated silencing is best understood in the ovary. Of the two main H3K9 HMTs that induce trimethylation in Drosophila, SetDB1/Eggless is required throughout the entire course of oogenesis, from germ cell differentiation to egg maturation, as well as for the somatic follicular cells that support the ovary, while Su(var)3-9 is not essential for fertility (Clough et al., 2007; Clough et al., 2014). Functionally, SetDB1/Eggless acts at multiple levels, including the control of TE expression by the piRNA pathway and repression of lineage-specific genes. Unlike Su(var)3-9 (Sienski et al., 2015), SetDB1 is involved in piRNA-dependent TE repression not only by being part of the piRNA-mediated transcriptional silencing pathways but also by regulating piRNA production. In D. melanogaster, primary piRNAs are generated from discrete genomic loci termed piRNA clusters (Brennecke et al., 2007). Most piRNA clusters in germ cells are characterized by a unique epigenetic landscape consisting of H3K9me3, the germline-specific HP1 variant Rhino/HP1d (a Drosophila-specific HP1 homolog) and several other factors that are required for their transcription and piRNA production (Andersen et al., 2017; Chen et al., 2016; Mohn et al., 2014; Rangan et al., 2011). In the nucleus, piRNA-loaded Piwi proteins recognize nascent transcripts of active TEs and induce local H3K9 trimethylation and co-transactional silencing (Klenov et al., 2011; LeThomas et al., 2013; Rozhkov et al., 2013; Sienski et al., 2012). SetDB1/Eggless depletion leads to H3K9me3 loss from TE targets and loss of piRNAs (Rangan et al., 2011). While loss of H3K9me3 at TE targets is probably partly due to loss of piRNA guides, several lines of evidence show that SetDB1 is also directly involved in H3K9me3 deposition downstream of the piRNA/Piwi complex. Piwi is not known to interact with any HMTs. However, two of Piwi's interacting partners, Panoramix (Panx)/Silencio and the SUMO E3 ligase Su(var)2-10, induce H3K9me3 deposition when recruited to chromatin in a process that is dependent on SetDB1 and its conserved co-factor Wde (ATF7IP/MCAF1 in mammals) (Ninova et al., 2019a preprint; Sienski et al., 2015; Yu et al., 2015). Furthermore, SetDB1/Wde recruitment requires SUMO and the SUMO E3 ligase activity of Su(var)2-10 (Ninova et al., 2019a preprint). Collectively, these findings lead to a model in which Su(var)2-10 interacts with Piwi/Arx/Panx and acts to induce SUMO-dependent recruitment of Wde/SetDB1, which in turn deposits H3K9me3 at piRNA targets (Ninova et al., 2019a preprint) (Fig. 3B).
As in mammalian systems, epigenetic silencing of TEs affects the host transcriptome of Drosophila germ cells. For example, H3K9me3 loss is associated with activation of cryptic promoters within TE sequences, the appearance of chimeric or truncated transcripts and mis-regulation of canonical gene isoforms (Ninova et al., 2019b). Finally, even though the piRNA pathway is the only known mode of H3K9me3 deposition in Drosophila ovaries, a recent ChIP-seq study revealed a number of discrete H3K9me3 peaks at euchromatic genes that are conserved, show no evidence of TE insertions or targeting by piRNAs, and do not lose H3K9me3 upon Piwi depletion, i.e. are likely piRNA-independent (Ninova et al., 2019b). About 20% of these H3K9me3-marked genes become upregulated upon knockdown of the SUMO ligase Su(var)2-10, suggesting that they are regulated in a SUMO-dependent manner and possibly through SetDB1. Notably, this set primarily includes genes characteristic of other tissues such as the testis or the central nervous system (Ninova et al., 2019b). It was recently demonstrated that the H3K9me3 effectors SetDB1, Wde and HP1a are required to confer transcriptional repression of male germline fate in the ovary (Smolko et al., 2018). Among other targets, SetDB1/Wde-dependent H3K9me3 suppresses the male-specific isoform of the master regulator of sex identity phf7 (Smolko et al., 2018). Thus, in addition to its role in constitutive heterochromatin and TE repression, epigenetic regulation by H3K9me3 in the female germline appears to grant tissue-specific gene repression to secure female germ cell identity (Ninova et al., 2019b; Smolko et al., 2018). The presence of discrete and TE-independent H3K9me3 peaks in otherwise euchromatic regions in female germ cells suggests the existence of a piRNA-independent mode of SetDB1 recruitment, and a regulatory mechanism that restricts H3K9me3 spreading in this genomic context.
Interestingly, a recent study in Drosophila showed a role for the conserved factor L(3)mbt in lineage-inappropriate gene repression in the female germline and soma (Coux et al., 2018). The mammalian L3MBTL2 homolog (involved in PRC1.6) is also required for the repression of germline-specific genes in mouse ESCs (Maeda et al., 2013; Stielow et al., 2018; Tatsumi et al., 2018). In the future, it would be worthwhile comparing targets of SetDB1/H3K9me3 and other silencing complexes, and investigating any potential cooperation between them.
Negative feedback regulation of H3K9me3 and heterochromatin effectors
In addition to developmental genes, several factors involved in heterochromatin formation and maintenance are themselves marked by H3K9me3. For example, genes encoding KRAB-ZFPs in mouse and human (often organized in tandem arrays) are enriched in H3K9me3, KAP1 and SetDB1 (Fig. 4A) (Frietze et al., 2010; O'Geen et al., 2007), as well as SUV39H1 and Cbx1/HP1β in various cell lines (Vogel et al., 2006). Moreover, at least two KRAB-ZFPs (ZNF274 and ZNF75D) are enriched at genomic ZFP clusters in human cells (Frietze et al., 2010; Imbeault et al., 2017). It has been proposed that KRAB-ZFPs auto-regulate through KAP1/SetDB1 (O'Geen et al., 2007). However, as KRAB-ZFPs are typically not silenced and H3K9me3 accumulates at the 3′UTR, it is unclear whether this mark causes transcriptional regulation or serves another function, e.g. preventing deletion due to unequal recombination between repetitive ZFP sequences (Vogel et al., 2006). Why KRAB-ZFPs deposit H3K9me3 at their own genes remains to be determined.
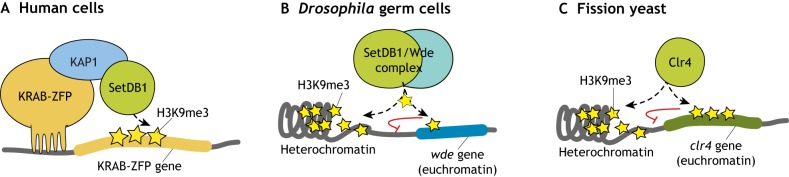
Fig. 4.
Auto-regulation of factors involved in H3K9me3 deposition in different systems. (A) Model of KRAB-ZFP gene autoregulation in human cells. KRAB-ZFP genes themselves are marked by H3K9me3 deposited by KRAB-ZFP-mediated recruitment of the H3K9-HMT SetDB1. Adapted from O'Geen et al. (2007). (B) Model of auto-regulation of the SetDB1 co-factor Wde in Drosophila germ cells. The wde gene is repressed through H3K9me3 accumulation that is dependent on the SetDB1/Wde complex, which thereby limits its own expression. Adapted from Ninova et al. (2019b). (C) Model of the negative feedback of Clr4 regulation and heterochromatin assembly in fission yeast. Genome-wide H3K9me3 is controlled by modulating the expression of the HMT Clr4 via accumulation of the repressive H3K9me3 mark on the clr4 gene region. Adapted from Wang et al. (2015).
A case of negative-feedback transcriptional regulation of heterochromatin effectors through H3K9me3 was recently found in the female germline of D. melanogaster. In the fly ovary, the gene encoding the SetDB1 co-factor Wde has a prominent H3K9me3 peak near its TSS, and H3K9me3 deposition and wde expression are regulated by SetDB1, Su(var)2-10, SUMO and its own protein product (Fig. 4B) (Ninova et al., 2019b). Similar H3K9me3 peaks are present at several other known or predicted components of heterochromatin formation in D. melanogaster, and are conserved in the distantly related D. virilis. These findings indicate that cellular levels of H3K9 methylation may be regulated through a homeostatic mechanism that involves Wde and perhaps additional factors acting as sensors for H3K9me3 levels. Strikingly, the human Wde homolog ATF7IP is also enriched in H3K9me3 in several human cell lines (ENCODE data). A negative-feedback mechanism regulating H3K9me3 levels was also identified in fission yeast; in this context, H3K9me3 accumulates at the locus encoding the single H3K9-HMT clr4, when cells experience excessive heterochromatin spreading (Fig. 4C) (Wang et al., 2015). H3K9me3 accumulation at clr4 leads to its repression and reduces Clr4 protein levels, thereby restricting heterochromatin spreading (Wang et al., 2015). Negative feedback is a common mechanism for providing homeostatic regulation in biological circuits, and it appears that this system has been used in the regulation of heterochromatin effectors and H3K9 methylation in yeast, flies and potentially in mammals. In the future, it will be important to address the precise architecture of gene regulatory networks involved in heterochromatin formation.
Conclusions
The role of H3K9me3 in genome regulation and silencing of selfish genetic elements is well established. However, despite its central role in genome stability, neither the molecular mechanism of H3K9-HMT targeting and recruitment, nor the breadth of its targets and functional roles are fully understood.
Although different general modes of H3K9-HMT recruitment to genomic targets – through sequence-specific DNA-binding proteins and small RNAs – have been identified, our understanding of cell- and genomic context-dependent targeting mechanisms is far from complete. We still lack a comprehensive picture of the factors that guide H3K9-HMTs to targets such as satellite repeats or protein-coding genes that lack proximal TEs. In fact, in Drosophila, the only known mechanism of H3K9me3 targeting is piRNA-dependent SetDB1 recruitment to TEs in the germline. What guides H3K9me3 to TE-independent loci, including to numerous developmentally regulated genes, both in the germline and in somatic cells that lack a functional piRNA pathway, requires further study.
How H3K9-HMTs are recruited upon target recognition is also not well understood. Recent findings point towards protein SUMOylation as a conserved mode of recruiting the SetDB1 silencing complex to chromatin, which seems to be used by different upstream target recognition pathways, such as piRNAs and KRAB-ZFP. However, SUMO is reported to occupy distinct chromatin types in different cell lines and is also implicated in gene activation (e.g. Cossec et al., 2018). Thus, it is likely that the role of SUMO modification is highly context dependent, and future work is required to dissect the precise molecular logic of SUMO-dependent silencing effector recruitment.
Our understanding of the functional role of H3K9me3 as a strong transcriptional silencer also merits further investigation because, at least in some cases, H3K9me3 is permissive to – or even required for – transcription. Given that active heterochromatic genes have H3K9me3 on their bodies but not their TSSs, it is tempting to speculate that H3K9me3 is repressive only at promoter regions, while being compatible with, or even promoting, transcription on gene bodies. Elucidating how the same histone mark can lead to different functional outcomes will improve our understanding of the role of histone modifications in epigenetic regulation.
Finally, it is clear that the regulation of heterochromatin homeostasis is essential to maintain appropriate gene expression patterns while keeping TEs in check. H3K9me3-associated heterochromatin can self-maintain and spread, presumably thanks to coupling of H3K9me3 reader and writer complexes. Heterochromatin spreading is thought to be limited by the dose of silencing factors (Elgin and Reuter, 2013), but how this is controlled to ensure proper silencing is not known. The recently proposed auto-regulation of heterochromatin through a negative-feedback mechanism is thus an exciting new direction for future studies of heterochromatin maintenance and H3K9me3 function.
Acknowledgements
We thank the three reviewers for the insightful comments and suggestions that helped us to improve this article. The role of H3K9me3 in various aspects of gene regulation is a broad topic and we apologize to colleagues whose relevant work was not discussed due to space limitations.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
The authors' research is supported by grants from the National Institutes of Health (R01 GM097363 to A.A.A. and R01 GM110217 to K.F.T.) and Minobrnauka of the Russian Federation (14.W03.31.0007), and by David and Lucile Packard Foundation Awards to A.A.A. Deposited in PMC for release after 12 months.
References
- Aagaard L., Laible G., Selenko P., Schmid M., Dorn R., Schotta G., Kuhfittig S., Wolf A., Lebersorger A., Singh P. B. et al. (1999). Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 18, 1923-1938. 10.1093/emboj/18.7.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkouche A., Mugat B., Barckmann B., Varela-Chavez C., Li B., Raffel R., Pélisson A. and Chambeyron S. (2017). Piwi is required during Drosophila embryogenesis to license dual-strand piRNA clusters for transposon repression in adult ovaries. Mol. Cell 66, 411-419.e4. 10.1016/j.molcel.2017.03.017 [DOI] [PubMed] [Google Scholar]
- Allan R. S., Zueva E., Cammas F., Schreiber H. A., Masson V., Belz G. T., Roche D., Maison C., Quivy J.-P., Almouzni G. et al. (2012). An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature 487, 249-253. 10.1038/nature11173 [DOI] [PubMed] [Google Scholar]
- Allis C. D. and Jenuwein T. (2016). The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487-500. 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- Allshire R. C. and Madhani H. D. (2018). Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 19, 229-244. 10.1038/nrm.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloia L., Di Stefano B. and Di Croce L. (2013). Polycomb complexes in stem cells and embryonic development. Development 140, 2525-2534. 10.1242/dev.091553 [DOI] [PubMed] [Google Scholar]
- Andersen P. R., Tirian L., Vunjak M. and Brennecke J. (2017). A heterochromatin-dependent transcription machinery drives piRNA expression. Nature 549, 54-59. 10.1038/nature23482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A. A., Sachidanandam R., Bourc'his D., Schaefer C., Pezic D., Toth K. F., Bestor T. and Hannon G. J. (2008). A piRNA pathway primed by individual transposons is linked to De Novo DNA methylation in mice. Mol. Cell 31, 785-799. 10.1016/j.molcel.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A., Sapetschnig A., Weick E.-M., Mitchell J., Bagijn M. P., Cording A. C., Doebley A.-L., Goldstein L. D., Lehrbach N. J., Le Pen J. et al. (2012). piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150, 88-99. 10.1016/j.cell.2012.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aucott R., Bullwinkel J., Yu Y., Shi W., Billur M., Brown J. P., Menzel U., Kioussis D., Wang G., Reisert I. et al. (2008). HP1-β is required for development of the cerebral neocortex and neuromuscular junctions. J. Cell Biol. 183, 597-606. 10.1083/jcb.200804041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyanathan K., Lechner M. S., Bell P., Maul G. G., Schultz D. C., Yamada Y., Tanaka K., Torigoe K. and Rauscher F. J. III (2003). Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 17, 1855-1869. 10.1101/gad.1102803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A. J. and Kouzarides T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381-395. 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., Allshire R. C. and Kouzarides T. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120-124. 10.1038/35065138 [DOI] [PubMed] [Google Scholar]
- Becker J. S., Nicetto D. and Zaret K. S. (2016). H3K9me3-Dependent heterochromatin: barrier to cell fate changes. Trends Genet. 32, 29-41. 10.1016/j.tig.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J. S., McCarthy R. L., Sidoli S., Donahue G., Kaeding K. E., He Z., Lin S., Garcia B. A. and Zaret K. S. (2017). Genomic and proteomic resolution of heterochromatin and its restriction of alternate fate genes. Mol. Cell 68, 1023-1037.e15. 10.1016/j.molcel.2017.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuchle D., Struhl G. and Muller J. (2001). Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128, 993-1004. [DOI] [PubMed] [Google Scholar]
- Bilodeau S., Kagey M. H., Frampton G. M., Rahl P. B. and Young R. A. (2009). SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 23, 2484-2489. 10.1101/gad.1837309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkent M., Bennett B. D., Lackford B., Bar-Nur O., Brumbaugh J., Wang L., Du Y., Fargo D. C., Apostolou E., Cheloufi S. et al. (2016). A serial shRNA screen for roadblocks to reprogramming identifies the protein modifier SUMO2. Stem Cell Reports 6, 704-716. 10.1016/j.stemcr.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Aravin A. A., Stark A., Dus M., Kellis M., Sachidanandam R. and Hannon G. J. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089-1103. 10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- Brower-Toland B., Riddle N. C., Jiang H., Huisinga K. L. and Elgin S. C. R. (2009). Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster. Genetics 181, 1303-1319. 10.1534/genetics.108.100271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulut-Karslioglu A., Perrera V., Scaranaro M., de la Rosa-Velazquez I. A., van de Nobelen S., Shukeir N., Popow J., Gerle B., Opravil S., Pagani M. et al. (2012). A transcription factor–based mechanism for mouse heterochromatin formation. Nat. Struct. Mol. Biol. 19, 1023-1030. 10.1038/nsmb.2382 [DOI] [PubMed] [Google Scholar]
- Canzio D., Chang E. Y., Shankar S., Kuchenbecker K. M., Simon M. D., Madhani H. D., Narlikar G. J. and Al-Sady B. (2011). Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell 41, 67-81. 10.1016/j.molcel.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H. and Bergman Y. (2009). Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10, 295 10.1038/nrg2540 [DOI] [PubMed] [Google Scholar]
- Chalker D. L., Meyer E. and Mochizuki K. (2013). Epigenetics of ciliates. Cold Spring Harb. Perspect. Biol. 5, a017764 10.1101/cshperspect.a017764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S., Elling U., Hopfgartner B., Jung Y. L., Murn J., Ninova M., Hubmann M., Badeaux A. I., Ang C. E., Tenen D. et al. (2015). The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528, 218 10.1038/nature15749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Liu H., Liu J., Qi J., Wei B., Yang J., Liang H., Chen Y., Chen J., Wu Y. et al. (2013). H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 45, 34-42. 10.1038/ng.2491 [DOI] [PubMed] [Google Scholar]
- Chen Y.-C. A., Stuwe E., Luo Y., Ninova M., Le Thomas A., Rozhavskaya E., Li S., Vempati S., Laver J. D., Patel D. J. et al. (2016). Cutoff suppresses RNA polymerase II termination to ensure expression of piRNA precursors. Mol. Cell 63, 97-109. 10.1016/j.molcel.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong E. B., Elde N. C. and Feschotte C. (2017). Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 18, 71-86. 10.1038/nrg.2016.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos P. A. C., Christensen J., Agger K. and Helin K. (2008). Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 22, 1115-1140. 10.1101/gad.1652908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E., Tedeschi T. and Hazelrigg T. (2014). Epigenetic regulation of oogenesis and germ stem cell maintenance by the Drosophila histone methyltransferase Eggless/dSetDB1. Dev. Biol. 388, 181-191. 10.1016/j.ydbio.2014.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E., Moon W., Wang S., Smith K. and Hazelrigg T. (2007). Histone methylation is required for oogenesis in Drosophila. Development 134, 157-165. 10.1242/dev.02698 [DOI] [PubMed] [Google Scholar]
- Collins R. E., Tachibana M., Tamaru H., Smith K. M., Jia D., Zhang X., Selker E. U., Shinkai Y. and Cheng X. (2005). In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J. Biol. Chem. 280, 5563-5570. 10.1074/jbc.M410483200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P. L., Kyle K. E., Egawa T., Shinkai Y. and Oltz E. M. (2015). The histone methyltransferase SETDB1 represses endogenous and exogenous retroviruses in B lymphocytes. Proc. Natl. Acad. Sci. USA 112, 8367-8372. 10.1073/pnas.1422187112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossec J.-C., Theurillat I., Chica C., Búa Aguín S., Gaume X., Andrieux A., Iturbide A., Jouvion G., Li H., Bossis G. et al. (2018). SUMO safeguards somatic and pluripotent cell identities by enforcing distinct chromatin states. Cell Stem Cell 23, 742-757.e8. 10.1016/j.stem.2018.10.001 [DOI] [PubMed] [Google Scholar]
- Coux R.-X., Teixeira F. K. and Lehmann R. (2018). L(3)mbt and the LINT complex safeguard cellular identity in the Drosophila ovary. Development 145, dev160721 10.1242/dev.160721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeñas-Potts C. and Matunis M. J. (2013). SUMO: a multifaceted modifier of chromatin structure and function. Dev. Cell 24, 1-12. 10.1016/j.devcel.2012.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B., Schotta G., Hülsmann B. B., Brehm A., Becker P. B., Reuter G. and Imhof A. (2001). Physical and functional association of SU(VAR)3-9 and HDAC1 in Drosophila. EMBO Rep. 2, 915-919. 10.1093/embo-reports/kve210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge J. E., Kang Y.-K., Beppu H., Lei H. and Li E. (2004). Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. Biol. 24, 2478-2486. 10.1128/MCB.24.6.2478-2486.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecco G., Cassano M., Kauzlaric A., Duc J., Coluccio A., Offner S., Imbeault M., Rowe H. M., Turelli P. and Trono D. (2016). Transposable elements and their KRAB-ZFP controllers regulate gene expression in adult tissues. Dev. Cell 36, 611-623. 10.1016/j.devcel.2016.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., James T. C., Foster-Hartnett D. M., Hartnett T., Ngan V. and Elgin S. C. (1990). Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 87, 9923-9927. 10.1073/pnas.87.24.9923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., Morris G. D., Reuter G. and Hartnett T. (1992). The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics 131, 345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C. R. and Reuter G. (2013). Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 5, a017780 10.1101/cshperspect.a017780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M., Endo T. A., Shinga J., Hayashi K., Farcas A., Ma K.-W., Ito S., Sharif J., Endoh T., Onaga N. et al. (2017). PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. Elife 6, e21064 10.7554/eLife.21064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymery A., Liu Z., Ozonov E. A., Stadler M. B. and Peters A. H. F. M. (2016). The methyltransferase Setdb1 is essential for meiosis and mitosis in mouse oocytes and early embryos. Development 143, 2767-2779. 10.1242/dev.132746 [DOI] [PubMed] [Google Scholar]
- Fasching L., Kapopoulou A., Sachdeva R., Petri R., Jönsson M. E., Männe C., Turelli P., Jern P., Cammas F., Trono D. et al. (2015). TRIM28 represses transcription of endogenous retroviruses in neural progenitor cells. Cell Rep. 10, 20-28. 10.1016/j.celrep.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner G. J., Kimura Y., Daub C. O., Wani S., Plessy C., Irvine K. M., Schroder K., Cloonan N., Steptoe A. L., Lassmann T. et al. (2009). The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 41, 563-571. 10.1038/ng.368 [DOI] [PubMed] [Google Scholar]
- Friedman J. R., Fredericks W. J., Jensen D. E., Speicher D. W., Huang X. P., Neilson E. G. and Rauscher F. J. (1996). KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 10, 2067-2078. 10.1101/gad.10.16.2067 [DOI] [PubMed] [Google Scholar]
- Frietze S., O'Geen H., Blahnik K. R., Jin V. X. and Farnham P. J. (2010). ZNF274 recruits the histone methyltransferase SETDB1 to the 39 ends of ZNF genes. PLoS ONE 5 10.1371/journal.pone.0015082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K., Okuda A., Yusa K. and Shinkai Y. (2018). A CRISPR knockout screen identifies SETDB1-target retroelement silencing factors in embryonic stem cells. Genome Res. 28, 846-858. 10.1101/gr.227280.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauchier M., Kan S., Barral A., Sauzet S., Agirre E., Bonnell E., Saksouk N., Barth T. K., Ide S., Urbach S. et al. (2019). SETDB1-dependent heterochromatin stimulates alternative lengthening of telomeres. Sci. Adv. 5, eaav3673 10.1126/sciadv.aav3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T. and Elgin S. C. R. (2013). Maternal depletion of Piwi, a component of the RNAi system, impacts heterochromatin formation in Drosophila. PLoS Genet. 9, e1003780 10.1371/journal.pgen.1003780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall I. M. (2002). Establishment and maintenance of a heterochromatin domain. Science 297, 2232-2237. 10.1126/science.1076466 [DOI] [PubMed] [Google Scholar]
- Heitz E. (1928). Das heterochromatin der moose. Jahrb Wiss Bot. 762-818. [Google Scholar]
- Hiragami-Hamada K., Soeroes S., Nikolov M., Wilkins B., Kreuz S., Chen C., De La Rosa-Velázquez I. A., Zenn H. M., Kost N., Pohl W. et al. (2016). Dynamic and flexible H3K9me3 bridging via HP1β dimerization establishes a plastic state of condensed chromatin. Nat. Commun. 7, 11310 10.1038/ncomms11310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D. and Moazed D. (2015). RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16, 71-84. 10.1038/nrg3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeault M., Helleboid P.-Y. and Trono D. (2017). KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543, 550-554. 10.1038/nature21683 [DOI] [PubMed] [Google Scholar]
- Ishiuchi T., Enriquez-Gasca R., Mizutani E., Bošković A., Ziegler-Birling C., Rodriguez-Terrones D., Wakayama T., Vaquerizas J. M. and Torres-Padilla M.-E. (2015). Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 22, 662 10.1038/nsmb.3066 [DOI] [PubMed] [Google Scholar]
- Ivanov A. V., Peng H., Yurchenko V., Yap K. L., Negorev D. G., Schultz D. C., Psulkowski E., Fredericks W. J., White D. E., Maul G. G. et al. (2007). PHD Domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell 28, 823-837. 10.1016/j.molcel.2007.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S. A., Taverna S. D., Zhang Y., Briggs S. D., Li J., Eissenberg J. C., Allis Cd. and Khorasanizadeh S. (2001). Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 20, 5232-5241. 10.1093/emboj/20.18.5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A., Colmenares S. U. and Karpen G. H. (2018). Heterochromatin: guardian of the genome. Annu. Rev. Cell Dev. Biol. 34, 265-288. 10.1146/annurev-cellbio-100617-062653 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Loh Y.-H. E., Rajarajan P., Hirayama T., Liao W., Kassim B. S., Javidfar B., Hartley B. J., Kleofas L., Park R. B. et al. (2017). The methyltransferase SETDB1 regulates a large neuron-specific topological chromatin domain. Nat. Genet. 49, 1239-1250. 10.1038/ng.3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A., Kronenberg Z., Lynch V. J., Zhuo X., Ramsay L., Bourque G., Yandell M. and Feschotte C. (2013). Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 9, e1003470 10.1371/journal.pgen.1003470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M. M. M., Goyal P., Maksakova I. A. A., Bilenky M., Leung D., Tang J. X. X., Shinkai Y., Mager D. L. L., Jones S., Hirst M. et al. (2011). DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8, 676-687. 10.1016/j.stem.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Takemoto K. and Shinkai Y. (2018). A somatic role for the histone methyltransferase Setdb1 in endogenous retrovirus silencing. Nat. Commun. 9, 1683 10.1038/s41467-018-04132-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R. and Alberts B. M. (1995). Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J. Cell Sci. 108, 1419-1431. [DOI] [PubMed] [Google Scholar]
- Kim J., Zhao H., Dan J., Kim S., Hardikar S., Hollowell D., Lin K., Lu Y., Takata Y., Shen J. et al. (2016). Maternal Setdb1 is required for meiotic progression and preimplantation development in mouse. PLoS Genet. 12, e1005970 10.1371/journal.pgen.1005970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov M. S., Sokolova O. A., Yakushev E. Y., Stolyarenko A. D., Mikhaleva E. A., Lavrov S. A. and Gvozdev V. A. (2011). Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc. Natl. Acad. Sci. USA 108, 18760-18765. 10.1073/pnas.1106676108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G., Chia N.-Y., Jeyakani J., Hwang C., Lu X., Chan Y.-S., Ng H.-H. and Bourque G. (2010). Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 42, 631-634. 10.1038/ng.600 [DOI] [PubMed] [Google Scholar]
- Lachner M., O'Carroll D., Rea S., Mechtler K. and Jenuwein T. (2001). Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116-120. 10.1038/35065132 [DOI] [PubMed] [Google Scholar]
- Lehnertz B., Ueda Y., Derijck A. A. H. A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T. and Peters A. H. F. M. (2003). Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13, 1192-1200. 10.1016/S0960-9822(03)00432-9 [DOI] [PubMed] [Google Scholar]
- LeThomas A., Rogers A. K., Webster A., Marinov G. K., Liao S. E., Perkins E. M., Hur J. K., Aravin A. A. and Tóth K. F. (2013). Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 27, 390-399. 10.1101/gad.209841.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Danzer J. R., Alvarez P., Belmont A. S. and Wallrath L. L. (2003). Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development 130, 1817-1824. 10.1242/dev.00405 [DOI] [PubMed] [Google Scholar]
- Liu S., Brind'Amour J., Karimi M. M., Shirane K., Bogutz A., Lefebvre L., Sasaki H., Shinkai Y. and Lorincz M. C. (2014). Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 28, 2041-2055. 10.1101/gad.244848.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Magri L., Zhang F., Marsh N. O., Albrecht S., Huynh J. L., Kaur J., Kuhlmann T., Zhang W., Slesinger P. A. et al. (2015). Chromatin landscape defined by repressive histone methylation during oligodendrocyte differentiation. J. Neurosci. 35, 352-365. 10.1523/JNEUROSCI.2606-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomberk G., Wallrath L. and Urrutia R. (2006a). The heterochromatin protein 1 family. Genome Biol. 7, 228 10.1186/gb-2006-7-7-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomberk G., Bensi D., Fernandez-Zapico M. E. and Urrutia R. (2006b). Evidence for the existence of an HP1-mediated subcode within the histone code. Nat. Cell Biol. 8, 407-415. 10.1038/ncb1383 [DOI] [PubMed] [Google Scholar]
- Lupo A., Cesaro E., Montano G., Zurlo D., Izzo P. and Costanzo P. (2013). KRAB-Zinc finger proteins: a repressor family displaying multiple biological functions. Curr. Genomics 14, 268-278. 10.2174/13892029113149990002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan T. S., Gifford W. D., Driscoll S., Lettieri K., Rowe H. M., Bonanomi D., Firth A., Singer O., Trono D. and Pfaff S. L. (2012). Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57-63. 10.1038/nature11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida S., Takizawa Y., Ishimaru M., Sugita Y., Sekine S., Nakayama J., Wolf M. and Kurumizaka H. (2018). Structural basis of heterochromatin formation by human HP1. Mol. Cell 69, 385-397.e8. 10.1016/j.molcel.2017.12.011 [DOI] [PubMed] [Google Scholar]
- Maeda I., Okamura D., Tokitake Y., Ikeda M., Kawaguchi H., Mise N., Abe K., Noce T., Okuda A. and Matsui Y. (2013). Max is a repressor of germ cell-related gene expression in mouse embryonic stem cells. Nat. Commun. 4, 1754 10.1038/ncomms2780 [DOI] [PubMed] [Google Scholar]
- Maison C., Bailly D., Roche D., de Oca R. M., Probst A. V., Vassias I., Dingli F., Lombard B., Loew D., Quivy J.-P. et al. (2011). SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat. Genet. 43, 220-227. 10.1038/ng.765 [DOI] [PubMed] [Google Scholar]
- Maison C., Quivy J.-P. and Almouzni G. (2016). Suv39h1 links the SUMO pathway to constitutive heterochromatin. Mol. Cell Oncol. 3, e1225546 10.1080/23723556.2016.1225546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J. H. A., O'Sullivan R. J., Braunschweig U., Opravil S., Radolf M., Steinlein P. and Jenuwein T. (2005). The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 24, 800-812. 10.1038/sj.emboj.7600545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S., Liu Y., Lu F., Iwabuchi K. A., Shen L., Inoue A. and Zhang Y. (2014). Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 159, 884-895. 10.1016/j.cell.2014.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Leung D., Miyashita H., Maksakova I. A., Miyachi H., Kimura H., Tachibana M., Lorincz M. C. and Shinkai Y. (2010). Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927-931. 10.1038/nature08858 [DOI] [PubMed] [Google Scholar]
- Matsumura Y., Nakaki R., Inagaki T., Yoshida A., Kano Y., Kimura H., Tanaka T., Tsutsumi S., Nakao M., Doi T. et al. (2015). H3K4/H3K9me3 bivalent chromatin domains targeted by lineage-specific DNA methylation pauses adipocyte differentiation. Mol. Cell 60, 584-596. 10.1016/j.molcel.2015.10.025 [DOI] [PubMed] [Google Scholar]
- Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.-K., Koche R. P. et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553-560. 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K., Tando Y., Sekinaka T., Otsuka K., Hayashi Y., Kobayashi H., Kamio A., Ito-Matsuoka Y., Takehara A., Kono T. et al. (2018). SETDB1 is essential for mouse primordial germ cell fate determination by ensuring BMP signaling. Development 145, dev164160 10.1242/dev.164160 [DOI] [PubMed] [Google Scholar]
- Mohn F., Sienski G., Handler D. and Brennecke J. (2014). The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 157, 1364-1379. 10.1016/j.cell.2014.04.031 [DOI] [PubMed] [Google Scholar]
- Muerdter F., Guzzardo P. M., Gillis J., Luo Y., Yu Y., Chen C., Fekete R. and Hannon G. J. (2013). A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in drosophila. Mol. Cell 50, 736-748. 10.1016/j.molcel.2013.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J.-i., Rice J. C., Strahl B. D., Allis C. D. and Grewal S. I. (2001). Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110-113. 10.1126/science.1060118 [DOI] [PubMed] [Google Scholar]
- Nicetto D., Donahue G., Jain T., Peng T., Sidoli S., Sheng L., Montavon T., Becker J. S., Grindheim J. M., Blahnik K. et al. (2019). H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science 363, 294-297. 10.1126/science.aau0583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S. J., Schneider R., Bauer U.-M., Bannister A. J., Morrison A., O'Carroll D., Firestein R., Cleary M., Jenuwein T., Herrera R. E. et al. (2001). Rb targets histone H3 methylation and HP1 to promoters. Nature 412, 561-565. 10.1038/35087620 [DOI] [PubMed] [Google Scholar]
- Ninova M., Chen Y.-C. A., Godneeva B., Rogers A. K., Luo Y., Aravin A. A. and Fejes Tóth K. (2019a). The SUMO ligase Su(var)2-10 links piRNA-guided target recognition to chromatin silencing. bioRxiv 533091 10.1101/533091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninova M., Godneeva B., Chen Y.-C. A., Luo Y., Prakash S. J., Jankovics F., Erdélyi M., Fejes Tóth K. and Aravin A. A. (2019b). The SUMO ligase Su(var)2-10 controls eu- and heterochromatic gene expression via establishment of H3K9 trimethylation and negative feedback regulation. Mol. Cell (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottke A., Colaiácovo M. P. and Shi Y. (2009). Developmental roles of the histone lysine demethylases. Development 136, 879-889. 10.1242/dev.020966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Geen H., Squazzo S. L., Iyengar S., Blahnik K., Rinn J. L., Chang H. Y., Green R. and Farnham P. J. (2007). Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 3, 0916-0926. 10.1371/journal.pgen.0030089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder T. T., Kara N., Cherry A., Sinha A. U., Zhu N., Bernt K. M., Cahan P., Mancarci B. O., Unternaehrer J., Gupta P. B. et al. (2012). Chromatin-modifying enzymes as modulators of reprogramming. Nature 483, 598 10.1038/nature10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M., Leibovitch B. A., Gandhi S. G., Chikka M. R., Rao M., Bhadra U., Birchler J. A. and Elgin S. C. R. (2004). Heterochromatic silencing and HP1 localization in drosophila are dependent on the RNAi machinery. Science 303, 669-672. 10.1126/science.1092653 [DOI] [PubMed] [Google Scholar]
- Peng H., Begg G. E., Schultz D. C., Friedman J. R., Jensen D. E., Speicher D. W. and Rauscher F. J. (2000). Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J. Mol. Biol. 295, 1139-1162. 10.1006/jmbi.1999.3402 [DOI] [PubMed] [Google Scholar]
- Peters A. H. F. M., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A. et al. (2001). Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323-337. 10.1016/S0092-8674(01)00542-6 [DOI] [PubMed] [Google Scholar]
- Peters A. H. F. M., Kubicek S., Mechtler K., O'Sullivan R. J., Derijck A. A. H. A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y. et al. (2003). Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577-1589. 10.1016/S1097-2765(03)00477-5 [DOI] [PubMed] [Google Scholar]
- Pezic D., Manakov S. A., Sachidanandam R., Castro-diaz N., Ecco G., Coluccio A. and Aravin A. A. (2014). piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev. 28, 1410-1428. 10.1101/gad.240895.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero J. S., Hartnett T. and Eissenberg J. C. (2004). Functional analysis of the chromo domain of HP1. EMBO J. 14, 3977-3986. 10.1002/j.1460-2075.1995.tb00069.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K., Fang J., Mlynarczyk-Evans S. K., Cao R., Worringer K. A., Wang H., de la Cruz C. C., Otte A. P., Panning B. and Zhang Y. (2003). Role of histone H3 lysine 27 methylation in X inactivation. Science 300, 131-135. 10.1126/science.1084274 [DOI] [PubMed] [Google Scholar]
- Psakhye I. and Jentsch S. (2012). Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151, 807-820. 10.1016/j.cell.2012.10.021 [DOI] [PubMed] [Google Scholar]
- Ragunathan K., Jih G. and Moazed D. (2015). Epigenetic inheritance uncoupled from sequence-specific recruitment. Science 348, 1258699-1258699 10.1126/science.1258699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P., Malone C. D., Navarro C., Newbold S. P., Hayes P. S., Sachidanandam R., Hannon G. J. and Lehmann R. (2011). piRNA production requires heterochromatin formation in drosophila. Curr. Biol. 21, 1373-1379. 10.1016/j.cub.2011.06.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S., Eisenhaber F., O'Carroll D., Strahl B. D., Sun Z.-W., Schmid M., Opravil S., Mechtler K., Ponting C. P., Allis C. D. et al. (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593-599. 10.1038/35020506 [DOI] [PubMed] [Google Scholar]
- Rebollo R., Karimi M. M., Bilenky M., Gagnier L., Miceli-Royer K., Zhang Y., Goyal P., Keane T. M., Jones S., Hirst M. et al. (2011). Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet. 7, e1002301 10.1371/journal.pgen.1002301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W., Dean W. and Walter J. (2001). Epigenetic reprogramming in mammalian development. Science 293, 1089-1093. 10.1126/science.1063443 [DOI] [PubMed] [Google Scholar]
- Rice J. C., Briggs S. D., Ueberheide B., Barber C. M., Shabanowitz J., Hunt D. F., Shinkai Y. and Allis C. D. (2003). Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12, 1591-1598. 10.1016/S1097-2765(03)00479-9 [DOI] [PubMed] [Google Scholar]
- Richards E. J. and Elgin S. C. R. (2002). Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108, 489-500. 10.1016/S0092-8674(02)00644-X [DOI] [PubMed] [Google Scholar]
- Riddle N. C., Minoda A., Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B., Tolstorukov M. Y., Gorchakov A. A., Jaffe J. D., Kennedy C., Linder-Basso D. et al. (2011). Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 21, 147-163. 10.1101/gr.110098.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L. and Paro R. (2004). Epigenetic regulation of cellular memory by the polycomb and trithorax group proteins. Annu. Rev. Genet. 38, 413-443. 10.1146/annurev.genet.38.072902.091907 [DOI] [PubMed] [Google Scholar]
- Robertson K. D., Ait-Si-Ali S., Yokochi T., Wade P. A., Jones P. L. and Wolffe A. P. (2000). DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25, 338-342. 10.1038/77124 [DOI] [PubMed] [Google Scholar]
- Rowe H. M., Jakobsson J., Mesnard D., Rougemont J., Reynard S., Aktas T., Maillard P. V., Layard-Liesching H., Verp S., Marquis J. et al. (2010). KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463, 237 10.1038/nature08674 [DOI] [PubMed] [Google Scholar]
- Rowe H. M., Kapopoulou A., Corsinotti A., Fasching L., Macfarlan T. S., Tarabay Y., Viville S., Jakobsson J., Pfaff S. L. and Trono D. (2013). TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 23, 452-461. 10.1101/gr.147678.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov N. V., Aravin A. A., Zelentsova E. S., Schostak N. G., Sachidanandam R., Mccombie W. R., Hannon G. J. and Ev M. B. E. (2010). Small RNA-based silencing strategies for transposons in the process of invading Drosophila species. RNA 16, 1634-1645. 10.1261/rna.2217810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov N. V., Hammell M. and Hannon G. J. (2013). Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 27, 400-412. 10.1101/gad.209767.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G., Ebert A., Krauss V., Fischer A., Hoffmann J., Rea S., Jenuwein T., Dorn R. and Reuter G. (2002). Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 21, 1121-1131. 10.1093/emboj/21.5.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B., Chourrout D., Vervoort M., Leblanc B. and Cavalli G. (2007). Genome regulation by polycomb and trithorax proteins. Cell 128, 735-745. 10.1016/j.cell.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Bourbon H.-M., Di Croce L. and Cavalli G. (2017). Genome regulation by polycomb and trithorax: 70 Years and counting. Cell 171, 34-57. 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Schultz D. C., Friedman J. R. and Rauscher F. J. III (2001). Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev. 15, 428 10.1101/gad.869501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D. C., Ayyanathan K., Negorev D., Maul G. G. and Rauscher Iii F. J. (2002). SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16, 919-932. 10.1101/gad.973302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seller C. A., Cho C.-Y. and O'farrell P. H. (2019). Rapid embryonic cell cycles defer the establishment of heterochromatin by Eggless/SetDB1 in Drosophila. 1 running title: cell cycle slowing times heterochromatin formation 2. 1-15. 10.1101/450155<AQ: Please provide journal title and volume number in reference Seller et al. (2019).> [DOI] [Google Scholar]
- Seum C., Reo E., Peng H., Rauscher F. J., Spierer P. and Bontron S. (2007). Drosophila SETDB1 is required for chromosome 4 silencing. PLoS Genet. 3, 709-719. 10.1371/journal.pgen.0030076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiio Y. and Eisenman R. N. (2003). Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA 100, 13225-13230. 10.1073/pnas.1735528100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. A., Eun S. C., Hyun S. K., Ho J. C. Y., Watts F. Z., Sang D. P. and Jang Y. K. (2005). SUMO modification is involved in the maintenance of heterochromatin stability in fission yeast. Mol. Cell 19, 817-828. 10.1016/j.molcel.2005.08.021 [DOI] [PubMed] [Google Scholar]
- Sienski G., Dönertas D. and Brennecke J. (2012). Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151, 964-980. 10.1016/j.cell.2012.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G., Batki J., Senti K.-A., Dönertas D., Tirian L., Meixner K. and Brennecke J. (2015). Silencio/CG9754 connects the Piwi–piRNA complex to the cellular heterochromatin machinery. Genes Dev. 29, 1-14. 10.1101/gad.271908.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R. K. and Martienssen R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272-285. 10.1038/nrg2072 [DOI] [PubMed] [Google Scholar]
- Smith Z. D., Chan M. M., Mikkelsen T. S., Gu H., Gnirke A., Regev A. and Meissner A. (2012). A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484, 339-344. 10.1038/nature10960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolko A. E., Shapiro-Kulnane L. and Salz H. K. (2018). The H3K9 methyltransferase SETDB1 maintains female identity in Drosophila germ cells. Nat. Commun. 9, 4155 10.1038/s41467-018-06697-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., Donahue G. and Zaret K. S. (2012). Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151, 994-1004. 10.1016/j.cell.2012.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R., Gonzales-Cope M., Chronis C., Bonora G., McKee R., Huang C., Patel S., Lopez D., Mishra N., Pellegrini M. et al. (2013). Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat. Cell Biol. 15, 872 10.1038/ncb2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabell M., Bjørkmo M., Aalen R. B. and Lambertsson A. (2006). The Drosophila SET domain encoding gene dEset is essential for proper development. Hereditas 143, 177-188. 10.1111/j.2006.0018-0661.01970.x [DOI] [PubMed] [Google Scholar]
- Stielow B., Sapetschnig A., Wink C., Krüger I. and Suske G. (2008). SUMO-modified Sp3 represses transcription by provoking local heterochromatic gene silencing. EMBO Rep. 9, 899-906. 10.1038/embor.2008.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stielow B., Finkernagel F., Stiewe T., Nist A. and Suske G. (2018). MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6. PLoS Genet. 14, e1007193 10.1371/journal.pgen.1007193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Sugimoto K., Fukushima T. and Shinkai Y. (2001). SET Domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276, 25309-25317. 10.1074/jbc.M101914200 [DOI] [PubMed] [Google Scholar]
- Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H. et al. (2002). G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16, 1779-1791. 10.1101/gad.989402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S.-L., Nishi M., Ohtsuka T., Matsui T., Takemoto K., Kamio-Miura A., Aburatani H., Shinkai Y. and Kageyama R. (2012). Essential roles of the histone methyltransferase ESET in the epigenetic control of neural progenitor cells during development. Development 139, 3806-3816. 10.1242/dev.082198 [DOI] [PubMed] [Google Scholar]
- Tatsumi D., Hayashi Y., Endo M., Kobayashi H., Yoshioka T., Kiso K., Kanno S., Nakai Y., Maeda I., Mochizuki K. et al. (2018). DNMTs and SETDB1 function as co-repressors in MAX-mediated repression of germ cell–related genes in mouse embryonic stem cells. PLoS ONE 13, e0205969 10.1371/journal.pone.0205969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd C. D., Deniz Ö., Taylor D. and Branco M. R. (2019). Functional evaluation of transposable elements as enhancers in mouse embryonic and trophoblast stem cells. Elife 8, e44344 10.7554/eLife.44344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch B., Hofmann A., Krauss V., Dorn R., Korge G. and Reuter G. (1994). The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 13, 3822-3831. 10.1002/j.1460-2075.1994.tb06693.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng T.-Y., Lee C.-H., Chan L.-W. and Shen C.-K. J. (2007). Epigenetic regulation of the Drosophila chromosome 4 by the histone H3K9 methyltransferase dSETDB1. Proc. Natl. Acad. Sci. USA 104, 12691-12696. 10.1073/pnas.0705534104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura Y., Ichimura T., Uwada J., Tachibana T., Sugahara S., Nakao M. and Saitoh H. (2006). Involvement of SUMO modification in MBD1- and MCAF1-mediated heterochromatin formation. J. Biol. Chem. 281, 23180-23190. 10.1074/jbc.M602280200 [DOI] [PubMed] [Google Scholar]
- Vassen L., Fiolka K. and Möröy T. (2006). Gfi1b alters histone methylation at target gene promoters and sites of γ-satellite containing heterochromatin. EMBO J. 25, 2409-2419. 10.1038/sj.emboj.7601124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D. and Malik H. S. (2009). Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu. Rev. Genet. 43, 467-492. 10.1146/annurev-genet-102108-134802 [DOI] [PubMed] [Google Scholar]
- Vogel M. J., Guelen L., de Wit E., Peric-Hupkes D., Lodén M., Talhout W., Feenstra M., Abbas B., Classen A.-K. and van Steensel B. (2006). Human heterochromatin proteins form large domains containing KRAB-ZNF genes. Genome Res. 16, 1493-1504. 10.1101/gr.5391806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T. A., Kidner C., Hall I. M., Teng G., Grewal S. I. S. and Martienssen R. A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833-1837. 10.1126/science.1074973 [DOI] [PubMed] [Google Scholar]
- Wakimoto B. T., Hearn M. G., Berloco M., Wakimoto B. T., Eissenberg J. C., Henikoff S., Rosen C. and Dorsett D. (1990). The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics 125, 141-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. H. and Elgin S. C. R. (2011). Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc. Natl. Acad. Sci. USA 108, 21164-21169. 10.1073/pnas.1107892109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Reddy B. D. and Jia S. (2015). Rapid epigenetic adaptation to uncontrolled heterochromatin spreading. Elife 4, e06179 10.7554/eLife.06179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu X., Gao Y., Yang L., Li C., Liu W., Chen C., Kou X., Zhao Y., Chen J. et al. (2018). Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol. 20, 620-631. 10.1038/s41556-018-0093-4 [DOI] [PubMed] [Google Scholar]
- Wolf D. and Goff S. P. (2007). TRIM28 Mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131, 46-57. 10.1016/j.cell.2007.07.026 [DOI] [PubMed] [Google Scholar]
- Wolf G., Greenberg D. and Macfarlan T. S. (2015a). Spotting the enemy within: targeted silencing of foreign DNA in mammalian genomes by the Krüppel-associated box zinc finger protein family. Mob. DNA 6, 17 10.1186/s13100-015-0050-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G., Yang P., Füchtbauer A. C., Füchtbauer E.-M., Silva A. M., Park C., Wu W., Nielsen A. L., Pedersen F. S. and Macfarlan T. S. (2015b). The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 29, 538-554. 10.1101/gad.252767.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Huang B., Chen H., Yin Q., Liu Y., Xiang Y., Zhang B., Liu B., Wang Q., Xia W. et al. (2016). The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652-657. 10.1038/nature18606 [DOI] [PubMed] [Google Scholar]
- Yamashita K., Sato A., Asashima M., Wang P.-C. and Nishinakamura R. (2007). Mouse homolog of SALL1, a causative gene for Townes?Brocks syndrome, binds to A/T-rich sequences in pericentric heterochromatin via its C-terminal zinc finger domains. Genes Cells 12, 171-182. 10.1111/j.1365-2443.2007.01042.x [DOI] [PubMed] [Google Scholar]
- Yang B. X., El Farran C. A., Guo H. C., Yu T., Fang H. T., Wang H. F., Schlesinger S., Seah Y. F. S., Goh G. Y. L., Neo S. P. et al. (2015). Systematic identification of factors for provirus silencing in embryonic stem cells. Cell 163, 230-245. 10.1016/j.cell.2015.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara J. C. and Wakimoto B. T. (2006). Oxymoron no more: the expanding world of heterochromatic genes. Trends Genet. 22, 330-338. 10.1016/j.tig.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Yeap L.-S., Hayashi K. and Surani M. A. (2009). ERG-associated protein with SET domain (ESET)-Oct4 interaction regulates pluripotency and represses the trophectoderm lineage. Epigenetics Chromatin 2, 12 10.1186/1756-8935-2-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Gu J., Jin Y., Luo Y., Preall J. B., Ma J., Czech B. and Hannon G. J. (2015). Panoramix enforces piRNA-dependent cotranscriptional silencing. Science 350, 339-342. 10.1126/science.aab0700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P., Han J., Guo G., Orlov Y. L., Huss M., Loh Y.-H., Yaw L.-P., Robson P., Lim B. and Ng H.-H. (2009). Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 23, 2507-2520. 10.1101/gad.1831909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Mosch K., Fischle W. and Grewal S. I. S. (2008). Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 15, 381-388. 10.1038/nsmb.1406 [DOI] [PubMed] [Google Scholar]
- Zilberman D., Cao X. and Jacobsen S. E. (2003). ARGONAUTE4 Control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299, 716-719. 10.1126/science.1079695 [DOI] [PubMed] [Google Scholar]