ABSTRACT
As embryos mature, cells undergo remarkable transitions that are accompanied by shifts in transcription factor regulatory networks. Mechanisms driving developmental transitions are incompletely understood. The embryonic intestine transitions from a rapidly proliferating tube with pseudostratified epithelium prior to murine embryonic day (E) 14.5 to an exquisitely folded columnar epithelium in fetal stages. We sought to identify factors driving mouse fetal intestinal maturation by mining chromatin accessibility data for transcription factor motifs. ATAC-seq accessible regions shift during tissue maturation, with CDX2 transcription factor motifs abundant at chromatin-accessible regions of the embryo. Hepatocyte nuclear factor 4 (HNF4) transcription factor motifs are the most abundant in the fetal stages (>E16.5). Genetic inactivation of Hnf4a and its paralog Hnf4g revealed that HNF4 factors are redundantly required for fetal maturation. CDX2 binds to and activates Hnf4 gene loci to elevate HNF4 expression at fetal stages. HNF4 and CDX2 transcription factors then occupy shared genomic regulatory sites to promote chromatin accessibility and gene expression in the maturing intestine. Thus, HNF4 paralogs are key components of an intestinal transcription factor network shift during the embryonic to fetal transition.
KEY WORDS: HNF4 transcription factors, Developing intestine, Maturation, Chromatin
Summary: A shift in transcription factor regulatory networks accompanies transition of the mouse embryonic intestine to its mature form, with HNF4 paralogs required to drive expression of differentiation genes.
INTRODUCTION
The developing embryo is a collection of partially fated cells that expand exponentially during the stages of organogenesis, at approximately embryonic day (E) 9.5 to E13.5 in mice (Cao et al., 2019). As development proceeds into fetal stages, specified cells undergo transitions to acquire the characteristics of mature tissues. The mechanisms of these developmental transitions are not completely understood but are of great importance in understanding the basic mechanisms of development and developmental disorders, and in facilitating the efforts of regenerative medicine.
The embryonic gut tube arises from endoderm after gastrulation and is specified along the anterior–posterior axis into distinct derivative endodermal organs. The primitive gut divides into foregut, midgut and hindgut; the small intestine develops primarily from the midgut. Murine gut tube formation is completed by E9.5 and its inner lining consists of a highly proliferative pseudostratified epithelium as the gut tube elongates. From E14.5 to E18.5, the tissue undergoes a remarkable transition to a columnar epithelium and the processes of villus morphogenesis, elongation and maturation occur (Chin et al., 2017; Walton et al., 2016a; Wells and Spence, 2014). The transcriptional regulatory mechanisms triggering this embryonic (defined here as the time prior to villus morphogenesis) to fetal (defined here as stages post villus emergence) transition in the developing gut remain unclear.
A shift in transcription factor regulatory networks accompanies the majority of known cellular transitions (Niwa, 2018; Wilkinson et al., 2017). Transcription factors function at distal genomic regulatory regions known as enhancers, which are the primary drivers of tissue-specific gene expression (Consortium, 2012; Nord et al., 2013; Shen et al., 2012; Visel et al., 2009). In our efforts to understand the mechanisms driving developmental transitions in the gut, we recently mapped chromatin profiles of the esophagus, forestomach, hindstomach and small intestine over developmental time. We noted a clear transition in chromatin accessibility within the developing intestine that corresponds to the stage in which the morphogenetic events reshaping the intestine occur (∼E15.5). The transcription factor CDX2 operates on both sides of this developmental transition. In the early embryonic intestine (prior to E13.5), CDX2 is required for intestinal specification and loss of CDX2 leads to ectopic features of stomach and esophageal tissues in the intestine (Banerjee et al., 2018; Gao et al., 2009; Grainger et al., 2010; Kumar et al., 2019). Acute inactivation of CDX2 at later developmental timepoints (post E13.5) compromises mature tissue functions, but intestinal identity is maintained (Banerjee et al., 2018; Kumar et al., 2019; Verzi et al., 2011). Although CDX2 is a clear driver of intestinal specification and plays an additional role in driving fetal maturation, it remains unclear how this developmental transition occurs.
In this study, we aimed to find transcription factors that could function specifically in intestinal maturation. We found that genomic regions that become increasingly accessible at fetal stages are most enriched for DNA-binding motifs known to bind to the HNF4 transcription factor family. In adult intestine, HNF4A and HNF4G have been shown to drive expression of genes important for digestive physiology (Boyd et al., 2009; Cattin et al., 2009; Lindeboom et al., 2018). HNF4A binding activity is modulated by the microbiome (Davison et al., 2017) and suppressed in a mouse model of ulcerative colitis (Chahar et al., 2014). We recently demonstrated that HNF4A and HNF4G redundantly drive enterocyte identity in adult tissues (Chen et al., 2019). Although HNF4 transcription factors have been studied in a number of developmental contexts, their redundant functions have not been assayed in the context of the developing gut.
Hnf4a deletion is embryonically lethal, with defects in the visceral endoderm (VE) (Chen et al., 1994; Duncan et al., 1997). Complementation of Hnf4a−/− embryos with Hnf4a+/+ VE by tetraploid aggregation (Duncan et al., 1997; Li et al., 2000) or conditional deletion of Hnf4a (Babeu et al., 2009; Garrison et al., 2006; Parviz et al., 2003) allowed for the investigation of HNF4 function in later developmental stages. In the liver, Hnf4a was dispensable for liver specification but was essential for hepatocyte differentiation (Hayhurst et al., 2001; Li et al., 2000; Parviz et al., 2003). However, no consequence of HNF4A loss in the developing small intestinal epithelium has been observed. This could be a result of inefficient depletion in the small intestine caused by the mosaic action of the Foxa3-Cre driver, as suggested by Garrison et al. (2006). Inactivation of conditional Hnf4a alleles using the Villin-Cre driver resulted in no embryonic phenotype (Babeu et al., 2009), possibly a result of the relatively late onset of Villin-Cre expression (Madison et al., 2002) or unappreciated genetic redundancy with Hnf4g. Here, we demonstrate that intestinal expression of Hnf4 paralogs is elevated during embryonic to fetal transition and that Hnf4 genes depend on CDX2 for expression in both mice and humans. Analysis of chromatin accessibility and data from chromatin immunoprecipitation combined with DNA sequencing (ChIP-seq) in the developing and adult gut supports a model in which the transcription factor CDX2 activates Hnf4 gene expression, and HNF4 and CDX2 together drive tissue maturation by activating genes important for intestinal function in post-natal life. We have generated a mouse model lacking HNF4 factors in the embryonic intestine to show that although HNF4 factors are dispensable for intestinal specification and villus morphogenesis, they are crucial for maturation of the fetal intestine.
RESULTS
Chromatin landscapes indicate that HNF4 factors are more likely to function in intestinal maturation than specification
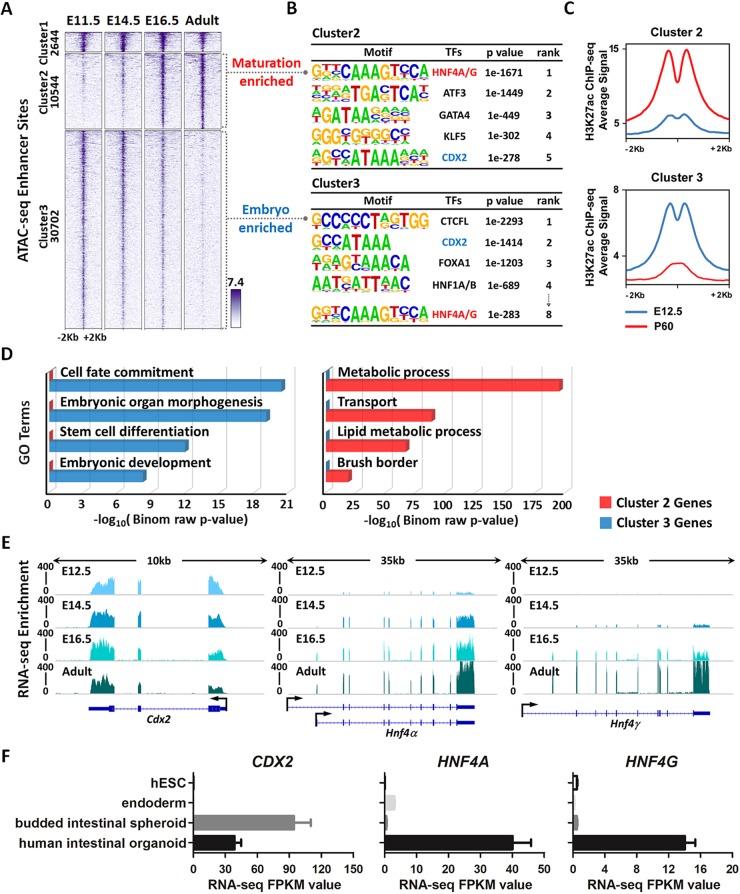
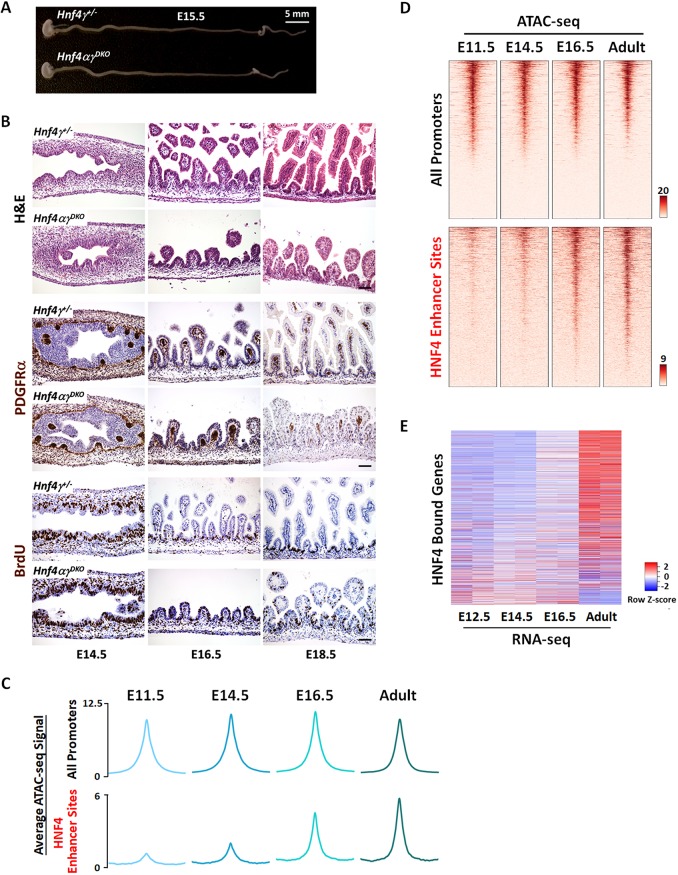
Deciphering the mechanisms of tissue maturation is crucial in understanding developmental disorders and facilitating efforts in regenerative medicine. Developmental transitions are accompanied by changes in chromatin accessibility and transcription factor regulatory networks. To help understand the regulatory mechanisms driving intestinal maturation, we re-analyzed ATAC-seq (assay for transposase-accessible chromatin using sequencing) data (Banerjee et al., 2018) to identify chromatin-accessible regions in isolated intestinal epithelial cells (Fig. 1A). Three categories of accessible regions were identified (MACS P value ≤10−5): 2644 regions (cluster 1) were accessible at E11.5 and remained accessible at all stages examined; 10,544 regions (cluster 2, ‘maturation-enriched regions’) of intestinal chromatin were progressively accessible; and 30,702 regions (cluster 3, ‘embryo-enriched regions’) lost accessibility from E11.5 to adult (Fig. 1A; Table S1). Notably, a gain in accessibility at maturation-enriched regions coincided with the loss of accessibility at embryo-enriched regions. This E14.5-16.5 transition stage is defined by villus morphogenesis and maturation. To identify the regulatory complexes probably operating during this developmental transition, we applied DNA-binding motif analysis. Regions selectively accessible in the maturing gut exhibited HNF4A/G as the top-scoring motif (Fig. 1B; Table S1), suggesting that HNF4 factors function in maturation of the developing gut. Conversely, CDX2 was among the most prevalent transcription factor motifs in the 30,702 regions that are more accessible in embryonic epithelium (Fig. 1B; Table S1). H3K27ac (Kazakevych et al., 2017), which marks active enhancer regions, was enriched at stage-specific accessible chromatin regions defined by ATAC-seq (Fig. 1C), corroborating the putative regulatory functions of these regions. Gene ontology analysis suggested that genes near the accessible regions of the early embryonic intestine function in intestinal specification and morphogenesis, whereas genes near accessible chromatin in the maturing intestine exhibit function consistent with mature tissue (Fig. 1D; Table S1). DNA-binding motif enrichment analysis at each timepoint revealed that although CDX2 binding motifs were enriched as early as E11.5, HNF4 binding motifs were first detected at E14.5 and became more prevalent at accessible chromatin regions of fetal and adult stages (Fig. S1A). Mirroring their motif enrichment profiles, we found that Cdx2 transcripts were highly and equally expressed across early embryonic to adult stages, whereas Hnf4 transcripts were not robustly expressed until fetal and adult stages (Fig. 1E). Protein expression levels of HNF4 factors (Fig. S1B,C) appeared to increase over developmental time, consistent with the increase in transcript levels. Similarly, human CDX2 expression was observed upon intestinal specification of human embryonic stem cell (ESC)-derived endoderm by FGF and WNT, whereas elevated expression of HNF4A and HNF4G was not observed until cultures were further matured into human intestinal organoids (Fig. 1F; Fig. S1D). Taken together, the chromatin landscapes, DNA motif enrichment and expression profiles of intestinal epithelial cells across developmental time suggest that HNF4 factors are more likely to function in tissue maturation than in intestinal specification.
Fig. 1.
Chromatin landscapes indicate that HNF4 factors function in maturation of the developing gut. (A) ATAC-seq (GSE115541, n=2 biological replicates per timepoint; isolated embryonic epithelium was collected from the entire small intestine) data defined regions of accessible chromatin across indicated stages of mouse intestinal epithelium development (MACS P value ≤10−5). K-means clustering analysis reveals a shift in accessible enhancer chromatin of intestinal epithelial cells across developmental time. (B) HOMER de novo DNA-motif enrichment analysis of ATAC-seq regions (MACS P value ≤10−5) shows that CDX2 binding sequences are more prevalent in accessible regions of the early embryonic state (embryo-enriched, cluster 3), whereas HNF4 binding sequences are more prevalent in accessible regions of the fetal and adult states (maturation-enriched, cluster 2). (C) H3K27ac ChIP-seq (GSE89684, n=2 biological replicates per timepoint) profiles demonstrate that the active chromatin marker is enriched in a stage-specific manner, corresponding to embryo-specific or P60 adult-specific accessible regions. (D) GREAT GO term analysis shows distinct gene ontologies of target genes linked to stage-specific ATAC-seq sites within 20 kb. (E) RNA-seq of purified epithelium (GSE115541, n=2 biological replicates per timepoint; isolated embryonic epithelium was collected from the entire small intestine) shows that Cdx2 is highly and equally expressed across developmental time, whereas Hnf4a and Hnf4g are not robustly expressed until E14.5 and E16.5, respectively. (F) RNA-seq (E-MTAB-4168) shows that CDX2 expression is induced when human endoderm is specified to intestine by treatment with FGF4 (500 ng/ml) and CHIR99021 (2 µM; WNT agonist). By contrast, HNF4A/G expression is not induced until human intestinal organoids are formed by subsequent differentiation steps. See schematic in Fig. S1D for details on human intestinal organoid differentiation conditions. Data are presented as mean±s.e.m. At least three biological replicates are included per stage, and samples from the same stage are grouped and presented.
HNF4 factors are dispensable for intestinal specification and villus morphogenesis, and CDX2 functions upstream of HNF4 factors
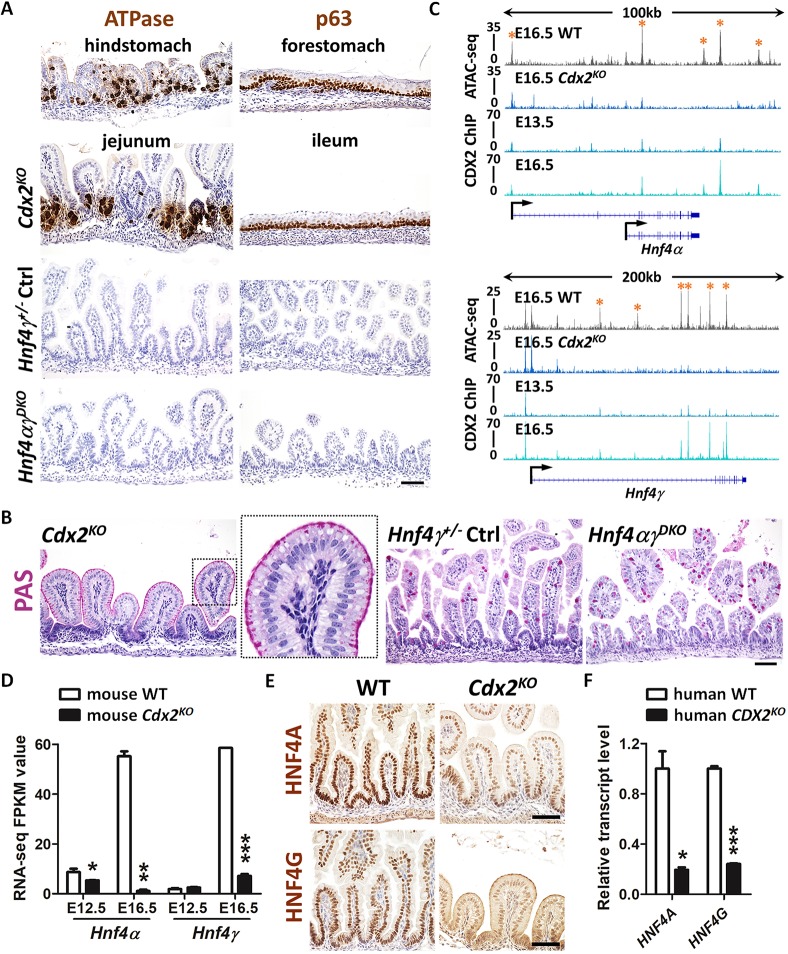
To test the function of HNF4 in the developing gut, we inactivated conditional alleles of Hnf4a (Hayhurst et al., 2001) in the gut endoderm using the Shh-Cre driver (Harfe et al., 2004), which is activated in the intestinal epithelium starting at ∼E9.5. We also examined germline null embryos lacking Hnf4g mediated by CRISPR knockout (Chen et al., 2019), and embryos with both HNF4 paralogs simultaneously deleted (hereafter referred to Hnf4αKO, Hnf4γKO and Hnf4αγDKO; Fig. S1E,F). As reported by this group and others (Banerjee et al., 2018; Gao et al., 2009; Grainger et al., 2010; Kumar et al., 2019), Cdx2KO in the early embryonic gut epithelium leads to an intestine exhibiting hindstomach characteristics (ATPase- and foveolar PAS-positive cells) in the jejunum and stratified squamous esophageal characteristics (p63-positive cells) in the ileum. However, similar characteristics were not observed in the Hnf4αγDKO embryos (Fig. 2A,B), suggesting that HNF4 factors are dispensable for intestinal specification. These findings are consistent with the analysis of accessible chromatin (see above, Fig. 1), in which CDX2 but not HNF4 motifs are enriched at regulatory regions most active in the E11.5 embryonic intestine (Fig. 1B; Fig. S1A).
Fig. 2.
HNF4 is activated by CDX2 in the developing gut, but HNF4 is not required for intestinal specification. (A) Immunostaining of ATPase and p63 (representative of four biological replicates) in E18.5 control, Shh-Cre;Cdx2f/f (Cdx2KO) and Shh-Cre;Hnf4af/f;Hnf4gCrispr/Crispr (Hnf4αγDKO) embryos. The hindstomach marker ATPase is ectopically expressed in the Cdx2KO jejunum but not in the control and Hnf4αγDKO jejunum. The forestomach marker p63 is ectopically expressed in the Cdx2KO ileum (squamous mucosa) but not in the control and Hnf4αγDKO ileum. (B) PAS staining indicates that normal intestinal goblet cells are replaced with cells resembling gastric foveolar cells (PAS-positive cells at apical cell surface) in the Cdx2KO jejunum of E18.5 embryos, but not in the control or Hnf4αγDKO jejunum (representative of four biological replicates). (C) CDX2 binds to Hnf4a and Hnf4g loci at E13.5 and E16.5 (CDX2 ChIP, GSE115314, n=2 biological replicates; whole small intestine epithelium), and accessible chromatin is compromised at gene loci of Hnf4a and Hnf4g upon CDX2 loss (ATAC-seq, GSE115314, n=2 biological replicates; whole small intestine epithelium). Asterisks denote putative regulatory regions. (D) The transcript levels of Hnf4a and Hnf4g are significantly downregulated in the intestinal epithelial cells of the Shh-Cre;Cdx2KO. Data are presented as mean±s.e.m (RNA-seq, GSE115541, n=2 or 3 biological replicates; whole small intesitne epithelium). Statistical tests are embedded in DESeq2 at ***P<0.001, **P<0.01 and *P<0.05. (E) Immunostaining of HNF4A and HNF4G shows reduced protein levels of HNF4 paralogs in E18.5 Cdx2KO (representative of four biological replicates). (F) qRT-PCR shows reduced transcript levels of HNF4A and HNF4G in human intestinal organoids derived from CDX2CrisprKO human ESCs compared with organoids derived from control cells. Data are presented as mean±s.e.m. (n=3 biological replicates, Student's t-test, two-sided at P<0.001*** and P<0.05*). Scale bars: 50 μm in A,B,E.
Because CDX2 is required for intestinal specification but HNF4 factors appear dispensable, we hypothesized that CDX2 might function upstream of HNF4 factors. Examination of CDX2 ChIP-seq data revealed that CDX2 bound to loci of Hnf4a and Hnf4g at E13.5 and the ChIP-seq signal strengthened at E16.5 (ChIP-seq panel in Fig. 2C). Loss of CDX2 resulted in loss of chromatin accessibility at the gene loci of Hnf4 factors (ATAC-seq panel in Fig. 2C) and reduced Hnf4 transcript (Fig. 2D) and protein levels (Fig. 2E), suggesting that CDX2 is a direct activator of Hnf4a, which is consistent with previous findings by Gao et al. (2009). Hnf4g expression was similarly dependent on CDX2 (Fig. 2C-E); however, because of the later onset of Hnf4g expression, these differences first become apparent at E16.5 (Fig. 2D). In developing human intestine, HNF4 factor expression is similarly dependent on CDX2, as reflected by measurement of HNF4 transcript levels in human intestinal organoids derived from control or CDX2CrisprKO human ESCs (Fig. 2F). These results indicate that expression of HNF4 factors is dependent on CDX2 during fetal maturation of the intestine.
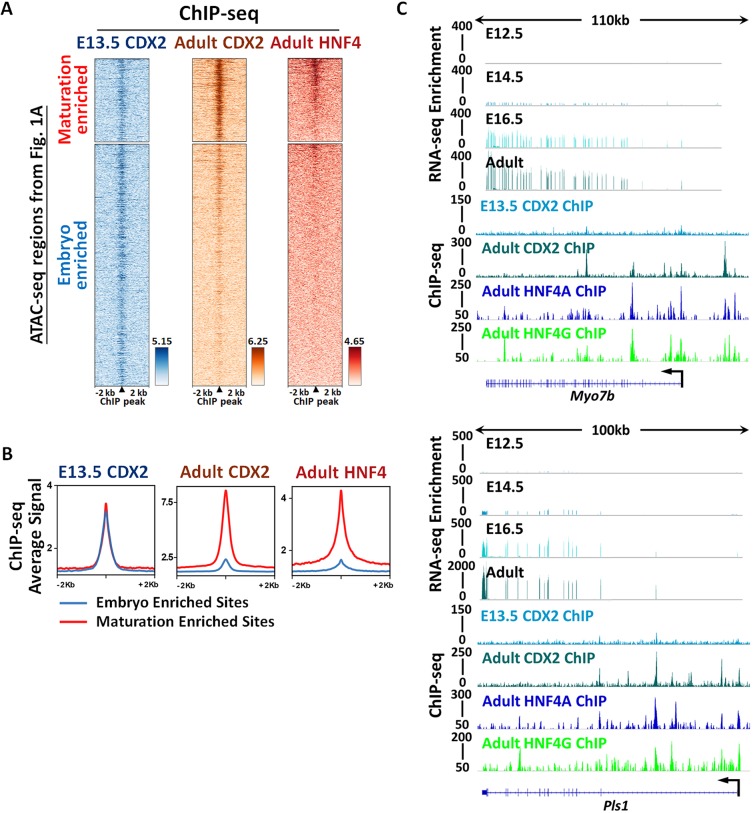
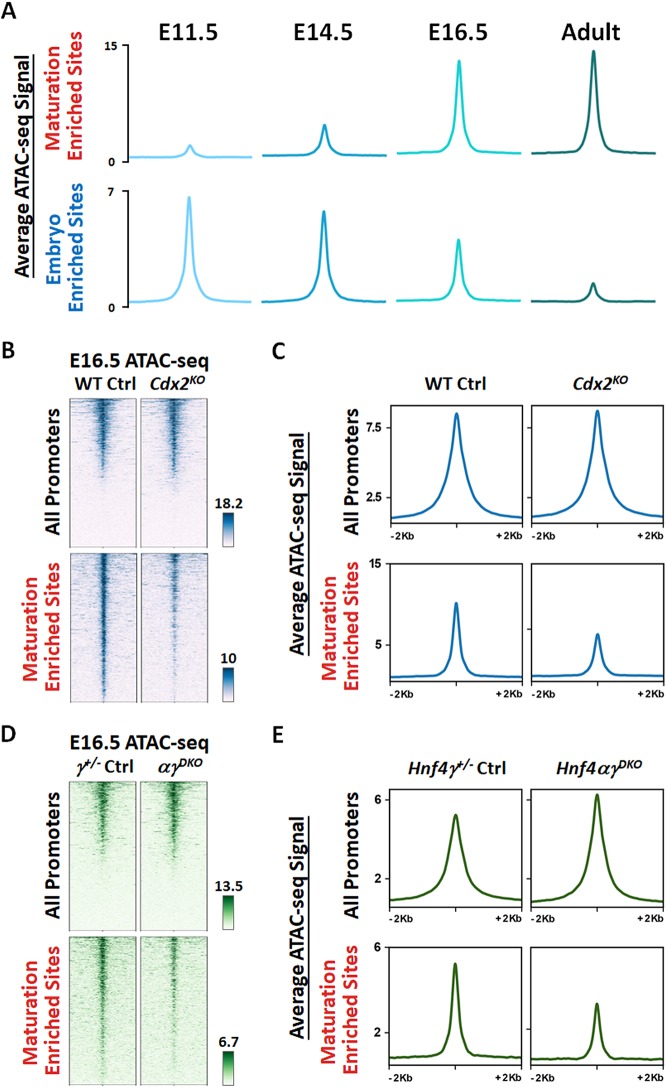
CDX2 bound to both maturation-enriched and embryo-enriched regions of intestinal accessible chromatin (ATAC-seq regions in Fig. 1A) at E13.5. When the intestine is mature, the CDX2 and HNF4 ChIP-seq signal was most robust at maturation-enriched regions, with comparatively less binding at embryonic accessible regions (Fig. 3A,B), suggesting that HNF4 and CDX2 work to activate maturation-specific gene expression. Consistent with this idea, genes such as Myo7b and Pls1 (Fig. 3C), which are bound by both HNF4 and CDX2, exhibited increased transcript expression during tissue maturation. To test whether HNF4 and CDX2 function is required for chromatin accessibility at regions that become increasingly accessible in the fetal stages, we compared the ATAC-seq signal in control and mutant embryos. Chromatin accessibility was indeed compromised at maturation-enriched regions from the intestinal epithelium of both E16.5 Cdx2KO and Hnf4αγDKO embryos (Fig. 4A-E). Chromatin accessibility loss in these mutants is selective to maturation-enriched chromatin, as chromatin accessibility at promoters is unaltered, and serves as an internal control. Taken together, these data suggest an active role of CDX2 and HNF4 factors in binding and maintaining accessibility at maturation-accessible regions.
Fig. 3.
Chromatin regions that become accessible in the fetal tissue are bound by CDX2 and HNF4. Genes near these regions are activated at fetal stages. (A,B) ChIP-seq profiles show that CDX2 (GSE115314, n=2 biological replicates; whole small intestine epithelium) binds to both maturation-enriched (cluster 2 in Fig. 1A) and embryo-enriched (cluster 3 in Fig. 1A) regions of intestinal accessible enhancer chromatin in E13.5 embryos. When tissues become mature, CDX2 (GSE34568 and GSE115314, n=2 biological replicates) and HNF4 (GSE112946, n=2 biological replicates per HNF4A and HNF4G ChIP; adult duodenum epithelium) bind more robustly to the maturation-enriched regions rather than to the embryo-enriched regions. The peaks of CDX2 ChIP in the heatmaps are aligned to the binding events of HNF4 ChIP. (C) Examples of genes located at maturation-enriched regions are visualized using IGV. Brush border genes, such as Myo7b and Pls1, are expressed when maturation occurs at E16.5 and are robustly expressed when intestines become mature in the adult (RNA-seq panels, GSE115541, n=2 biological replicates). These genes are directly bound by CDX2 (GSE34568 and GSE115314, n=2 biological replicates) and HNF4 (GSE112946, n=2 biological replicates per HNF4A and HNF4G ChIP).
Fig. 4.
Maturation-enriched regions of accessible enhancer chromatin show loss of accessibility in the intestinal epithelium of Cdx2KO and Hnf4αγDKO mutants. (A) The intestinal chromatin (GSE115541, n=2 biological replicates) becomes more accessible at maturation-enriched regions and less accessible at embryo-enriched regions (ATAC-seq regions defined in Fig. 1A) from E11.5 embryo to adult. (B-E) The intestinal chromatin accessibility at the maturation-enriched sites is compromised upon depletion (Shh-Cre) of Cdx2 (B,C; GSE115314) or Hnf4 genes (D,E) in E16.5 embryos, whereas promoter regions are relatively unaffected and serve as an internal control (n=2 wild-type controls, 1 Hnf4g+/− control, 2 Cdx2KO mutants and 2 Hnf4αγDKO mutants; whole small intestine epithelium).
HNF4 factors thus appear dispensable for tissue specification, but their expression pattern and control of maturation-enriched regulatory regions suggest potential roles during later developmental stages. From E14.5 to E15.5, the pseudostratified intestinal epithelium resolves to a columnar epithelium and mesenchymal cells crosstalk with epithelial cells to initiate villus formation (Chin et al., 2017; Walton et al., 2016a; Wells and Spence, 2014). For example, platelet-derived growth factor receptor alpha (PDGFRα)-expressing mesenchymal clusters serve as signaling centers for villus morphogeneis (Walton et al., 2012). We investigated whether villus morphogenesis is disrupted upon HNF4 loss. At E15.5, Hnf4αγDKO intestines were similar to their littermate controls (Fig. 5A) and there were no obvious defects in epithelial morphology, mesenchymal condensations (PDGFRα-positive cells) or proliferation [bromodeoxyuridine (BrdU)-positive cells] upon HNF4 loss at E14.5 (Fig. 5B). These results suggest that proliferation and tissue morphogenesis initiate properly in the absence of HNF4 paralogs. Consistent with the observation of normal villus morphogenesis in HNF4 mutants, adult HNF4-bound regions (as defined by ChIP-seq; Chen et al., 2019) were not very accessible during embryonic stages, but became markedly more accessible following villus morphogenesis (Fig. 5C,D). Chromatin accessibility at promoter regions served as an internal control for each timepoint (Fig. 5C,D). Genes linked to HNF4 ChIP-seq regions also exhibited increased expression after villus morphogenesis, as the tissue matures (RNA-seq, Fig. 5E). Together, these findings suggest that HNF4 probably functions after villus morphogenesis to drive gene expression during maturation of the developing fetal gut.
Fig. 5.
Villus morphogenesis initiates despite the loss of HNF4; chromatin accessibility and gene regulation at HNF4-binding sites suggest a role for HNF4 after villus morphogenesis. (A) Whole mount images of representative intestines dissected from Hnf4αγDKO and littermate Hnf4g+/− control embryos at E15.5 (n=4 biological replicates per genotype). (B) H&E staining and immunostaining of PDGFRα and BrdU across developmental time show no striking differences, indicating that HNF4 factors are dispensable for the onset of villus morphogenesis (representative of four biological replicates). The pregnant female mice were injected with 1 mg BrdU at 1 h before euthanasia. Scale bars: 50 μm. (C,D) Chromatin becomes accessible during fetal and adult stages at enhancer regions bound by HNF4 in the adult epithelium (MACS P value ≤10−3, GSE112946, n=2 biological replicates per ChIP), indicating a potential role for HNF4 in activating genes near these regions during intestinal maturation. (E) Increased transcript levels of HNF4-bound genes in intestinal epithelial cells are observed across developmental time from E12.5 to adult villi. Genes with transcriptional start sites within 20 kb of HNF4 enhancer bound sites (GSE112946, n=2 biological replicates per ChIP) were used for plotting a heatmap showing the relative transcript expression levels over the indicated developmental stages (GSE115541, n=2 biological replicates per timepoint).
HNF4 factors are redundantly required for fetal maturation of the intestine
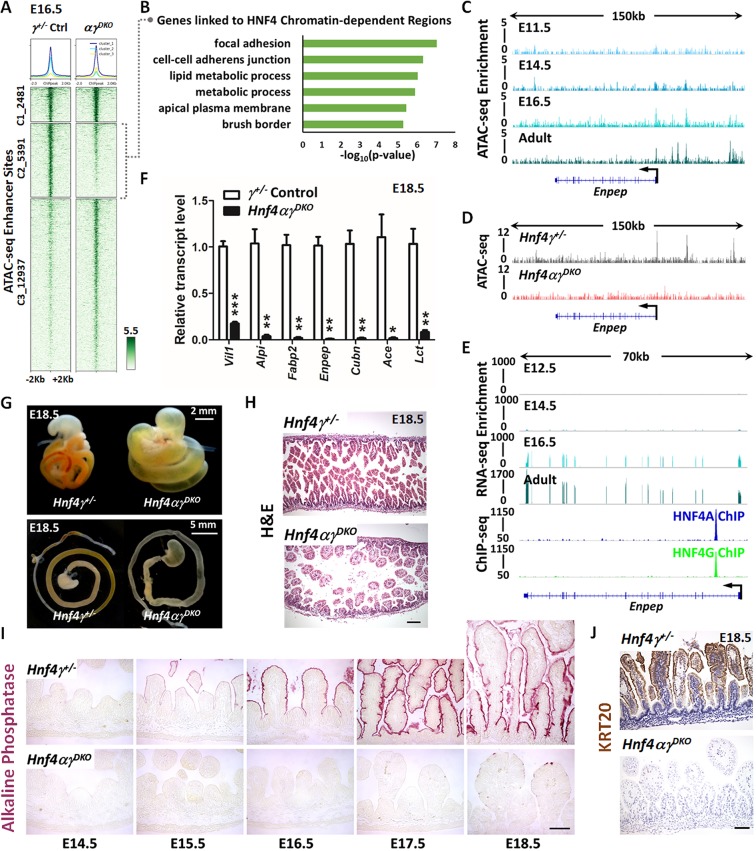
From E15.5 to E18.5, villi elongate into the lumen and cells that leave the intervillus regions exit the cell cycle and begin to express markers of differentiation. To appreciate the function of HNF4 factors during this time, we compared ATAC-seq data from E16.5 intestinal epithelial cells isolated from Hnf4αγDKO with data from control embryos. 5391 accessible chromatin regions lost accessibility upon HNF4 loss (cluster 2 in Fig. 6A; Table S2). Genes near these HNF4-dependent regions were associated with mature intestinal functions, such as adhesion, brush border formation and lipid metabolism (Fig. 6B; Table S2). For example, chromatin accessibility increased at regions of the brush border gene Enpep as the tissue matured (Fig. 6C), and accessibility was almost completely lost in the Hnf4αγDKO epithelium (Fig. 6D). HNF4 paralogs directly bound to many maturation-specific genes (ChIP-seq in Fig. 6E and Fig. S2), and transcript levels of these maturation-specific genes were dramatically reduced in Hnf4αγDKO (Fig. 6F). Conversely, genes near chromatin regions gaining accessibility upon HNF4 loss (cluster 3 in Fig. 6A) were associated with cellular stress (Table S2). Consistent with the loss of expression of maturation genes, Hnf4αγDKO intestines exhibited a translucent and distended lumen at E18.5, suggesting an underdeveloped intestine compared to their littermate controls (Fig. 6G). The gross morphological defects in intestines were more severe with loss of three or four Hnf4 alleles compared with loss of one or two Hnf4 alleles (Fig. S3A). After birth, Hnf4αγDKO pups were not able to survive, whereas Hnf4αKOγ+/− pups survived but exhibited growth retardation (Fig. S3B,C), which could be the result of incomplete intestinal maturation. At E18.5, villi were noticeably shorter in mutants lacking three or four Hnf4 alleles but not in the Hnf4 single mutants or controls (Fig. 6H; Fig. S3D), suggesting that HNF4 paralogs are redundantly required for villus elongation and extension into the developing gut lumen. There was no obvious increase in apoptosis in Hnf4αγDKO, as evaluated by cleaved caspase 3 staining (Fig. S4A). The differentiation marker, alkaline phosphatase, which is localized to the apical surface of villus enterocytes, was normally expressed in the controls beginning at E15.5 in the maturing tissue and increased over developmental time (top panels in Fig. 6I). However, alkaline phosphatase was not detectable in Hnf4 double mutants (bottom panels in Fig. 6I). As expected for genetic redundancy between Hnf4 paralogs, Hnf4 single mutants showed normal expression of alkaline phosphatase (Fig. S4B). Proliferative cells, which are normally restricted to the intervillus regions of the fetal gut, were expanded into the villi of Hnf4αγDKO (Fig. S4C), which could be attributed to the loss of villus differentiation in mutants lacking HNF4 factors (Fig. 6I,J). Taken together, our results indicate that HNF4 paralogs are dispensable for specification and villus morphogenesis in the developing gut, but are redundantly required for fetal maturation through direct binding to fetal maturation genes.
Fig. 6.
HNF4 paralogs are redundantly required for fetal maturation of the intestine. (A) K-means clustering of ATAC-seq data collected from E16.5 intestinal epithelial cells isolated from Hnf4αγDKO and control embryos identify 5391 regions (cluster 2) that are dependent on HNF4 factors for chromatin accessibility (n=2 biological Hnf4αγDKO replicates and 1 Hnf4g+/− control in this study; MACS P value ≤10−5). (B) Functional annotation of the genes linked to HNF4-dependent accessible chromatin by DAVID. Genes with transcriptional start sites within 20 kb of ATAC-seq sites of cluster 2 (loss of chromatin accessibility upon HNF4 loss) from A were used for analysis. (C-E) Examples of chromatin accessibility at HNF4-dependent ATAC-seq sites are visualized using IGV. (C) ATAC-seq (GSE115541) shows a time-dependent increase of chromatin accessibility at the locus of the brush border gene Enpep from E11.5 embryo to adult. (D) ATAC-seq shows compromised chromatin accessibility at the Enpep locus in E16.5 Hnf4αγDKO. (E) RNA-seq (GSE115541) shows corresponding increase in transcript levels of Enpep over developmental time. HNF4A and HNF4G directly bind to Enpep (GSE112946, ChIP-seq panel), suggesting direct regulation. (F) qPCR shows that the transcript levels of genes known to be expressed in the mature intestine are dramatically reduced in isolated E18.5 intestinal epithelial cells from Hnf4αγDKO compared with those from littermate Hnf4g+/− controls. Data are presented as mean±s.e.m. (n=4 controls and 5 mutants, Student's t-test, two-sided at ***P<0.001, **P<0.01 and *P<0.05). (G) Whole mount images of E18.5 intestine indicate that loss of HNF4 paralogs leads to an underdeveloped intestine with distended and translucent lumen (representative of six biological replicates). (H) Strikingly stunted villi are observed in E18.5 Hnf4αγDKO embryos compared with littermate Hnf4g+/− controls, as revealed by H&E staining (representative of four biological replicates, E18.5 duodenum; see expanded panel in Fig. S3D). (I) Hnf4αγDKO exhibits diminished alkaline phosphatase staining (differentiation marker) across developmental time (representative of four biological replicates; E14.5-E18.5 duodenum). (J) Keratin 20 immunostaining also indicates compromised intestinal differentiation in Hnf4αγDKO embryos (representative of four biological replicates; E18.5 duodenum). Scale bars: 50 μm in H,I,J. See Figs S2 and S3 for additional metrics of intestinal maturation.
DISCUSSION
The regulatory mechanisms governing the transition of embryonic tissue to mature tissue is a significant frontier for both developmental biology and regenerative medicine. Chromatin accessibility dynamics across intestinal development can provide new insights into the fundamental molecular basis of intestinal specification and maturation. HNF4 motifs are most prevalent in the accessible chromatin during the maturation of the developing gut, and we provide evidence that HNF4 transcription factors are indeed important for maturation of the fetal intestine. Here, we build a model in which CDX2 functions in gut specification, then binds to and activates HNF4 factors in the maturing gut. Together, these factors are required to mature the fetal tissue, ultimately achieving a stabilized and mature intestine.
Interestingly, lower-level CDX2 binding in the embryo was observed at regions that became accessible in fetal stages (Fig. 3A). Lower-level CDX2 ChIP-seq signal at these poorly accessible regions could reflect a ‘low-level sampling’ behavior that has recently been described for the FOXA1 transcription factor (Donaghey et al., 2018). When ectopically expressed in fibroblasts, FOXA1 exhibits low ChIP-seq signal at binding sites typically exclusive to other cell lineages, such as liver or endoderm, a behavior that could relate to the relatively strong interaction of FOXA1 with chromatin and its slow nuclear mobility compared with other transcription factors (Sekiya et al., 2009). Low-level FOXA1 binding is strengthened at these sites by co-expression of partner factor GATA4. HNF4 factors could similarly be stabilizing CDX2 at maturation-specific regions. A potential partner for CDX2 at embryo-specific regions remains elusive. It will be interesting to test whether CDX2 exhibits similar nuclear mobility as FOXA1. We also note that although expression levels of HNF4A were markedly lower in embryonic intestine (Fig. 1E, Fig. S1B), HNF4A motifs were present at embryo-enriched accessible regions, although at a lower statistical threshold. These motifs could foreshadow HNF4 binding at later developmental timepoints, reflecting a function for HNF4A during endoderm development (Fig. 1F), or indicate another unappreciated role for HNF4 prior to villus morphogenesis. However, using the Shh-Cre driver in this study, no overt differences were observed between intestines of control and Hnf4αγDKO embryos prior to villus morphogenesis.
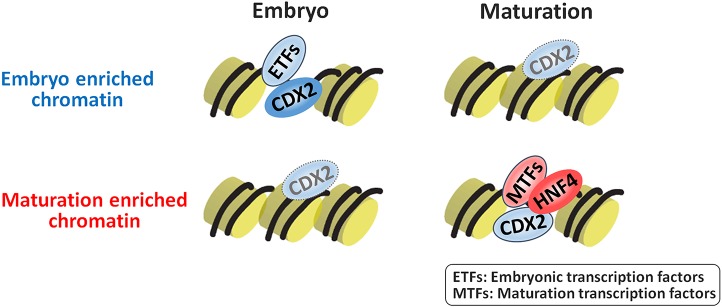
It is also interesting to note that CDX2 appears to function in both the embryonic and fetal transcription factor regulatory networks (Fig. 7). Transitions between regulatory networks could more seamlessly occur when certain factors are present across the transition, such as the presence of Sox2 and Esrrb in both embryonic stem cells and trophoblast stem cells (Adachi et al., 2013). Rather than shutting down an entire set of transcription factors and establishing an entirely new set of factors, the common presence of CDX2 might function as a placeholder to transition from the embryonic to fetal networks. Inhibitory factors such as Blimp1 (Prdm1) have also been shown to play a role in tissue maturation (Harper et al., 2011; Mould et al., 2015).
Fig. 7.
Potential model of how intestinal transcription factor networks shift as embryos mature to adults. Left: CDX2 functions in gut specification, and robustly binds to embryo-enriched accessible chromatin regions with other embryonic transcription factors. In the embryo, lower-level CDX2 binding might also occur at regions that will become accessible in the fetus. Right: As intestine matures to fetal stages, maturation-enriched regions become more accessible. A new transcription factor network, highlighted by HNF4, works together to stabilize enhancer chromatin and drive intestinal maturation.
Transcription factors and developmental signaling pathways function in complex and collaborative networks to promote proper tissue development and function. BMP signals control intestinal villus patterning (Walton et al., 2016b) and intestinal looping (Nerurkar et al., 2017), but the functions and mechanisms of BMP signals in villus maturation are not fully explored. In a recent study, we identified a reinforcing feed-forward loop of BMP/SMAD signaling and HNF4, which promotes and stabilizes enterocyte cell identity in the adult intestine (Chen et al., 2019). Future studies could also investigate the presence of this reinforcing loop of BMP/SMAD signaling and HNF4 in the developing gut, and whether it promotes the maturation of the developing intestine or stabilization of mature transcription factor networks. These findings would build on the field's knowledge of intestinal development and help influence the efforts of regenerative medicine to provide healthy intestinal tissue to patient populations.
MATERIALS AND METHODS
Mice
The Shh-Cre transgene (Harfe et al., 2004), Cdx2f/f (Verzi et al., 2010), Hnf4af/f (Hayhurst et al., 2001) and Hnf4gCrispr/Crispr (Chen et al., 2019) alleles were bred to achieve the indicated genotypes. The Shh-Cre− embryos served as littermate controls unless otherwise indicated. The day on which a vaginal plug was found was considered to be E0.5. Embryonic tail biopsies were used for rapid genotyping using KAPA Mouse Genotyping Kits (Kapa Biosystems, KK7352). All mouse protocols and experiments were approved by the Rutgers Institutional Animal Care and Use Committee. All samples were collected between 12:00 and 14:00 to avoid circadian variability.
Histology and immunostaining
Intestinal tissues were fixed with 4% paraformaldehyde at 4°C overnight, washed with PBS and dehydrated through ascending alcohols prior to xylene processing and paraffin embedding. Paraffin sections (5 μm thick) were used for histological staining. Staining with Hematoxylin (VWR, 95057-858) and Eosin (Sigma-Aldrich, HT110180) was performed using standard procedures. Alkaline phosphatase activity was detected using the AP Staining Kit II (Stemgent). For periodic acid-Schiff (PAS) staining, slides were incubated in 0.5% periodic acid and stained with Schiff's reagent (Alfa Aesar, J612171). In the case of tissue prepared for BrdU immunohistochemistry, pregnant female mice were injected with 1 mg BrdU and embryos harvested 1 h after injection. Immunohistochemistry was performed using primary antibodies against HNF4A (Santa Cruz Biotechnology, sc-6556 X, 1:2000), HNF4G (Santa Cruz Biotechnology, sc-6558 X, 1:2000), ATPase (MBL International D032-3, 1:200), P63 (Santa Cruz Biotechnology, sc-8343, 1:500), BrdU (Bio-Rad MCA2060, 1:500), Ki67 (Abcam ab16667, 1:300), PDGFRα (Santa Cruz Biotechnology, sc-338, 1:1000), cleaved caspase 3 (Cell Signaling Technology, 9661, 1:200) and keratin 20 (Cell Signaling Technology, 13063, 1:2500). After incubating with secondary antibody and the Vectastain ABC HRP Kit (Vector Labs), slides were developed using 0.05% diaminobenzidine (DAB, Amresco 0430) and 0.015% hydrogen peroxide in 0.1 M Tris. After mounting, the slides were viewed on a Nikon Eclipse E800 microscope. Images were photographed with a Retiga 1300 CCD (QImaging) camera using QCapture imaging software. When adjustments of sharpness, contrast or brightness were made, they were applied uniformly for comparative images.
Human embryonic stem cell culture and differentiation
All human ESC work was reviewed and approved by the University of Michigan human pluripotent stem cell research oversight committee (HPSCRO). The human ESC cell line H9 (WA09, NIH stem registry #0062) was obtained from the WiCell Research Institute. CDX2CrisprKO human ESCs were generated as described previously (Kumar et al., 2019). Human ESCs were maintained and differentiated into endoderm, hind/midgut and human intestinal organoids as previously described (Kumar et al., 2019; Tsai et al., 2017).
Intestinal epithelial cell isolation
Embryos were collected at E18.5. The freshly harvested embryonic small intestine (caudal stomach to rostral caecum) was opened longitudinally with forceps, cut into 1 cm pieces and then rotated in 3 mM EDTA in PBS at 4°C for 40 min. To release epithelial cells from underlying muscular tissue, the tissue was vigorously shaken after EDTA incubation. The supernatant was collected as the whole epithelium fraction. Cells were pelleted by centrifugation at 170 g at 4°C and then washed with cold PBS and processed for RNA extraction using Trizol (Invitrogen, 15596018) according to the manufacturer's protocols.
RNA extraction and qRT-PCR
The RNA was reverse transcribed using SuperScript III First-Strand Synthesis SuperMix (Invitrogen, 18080-400) with Oligo(dT)20 primers to prepare cDNA. qRT-PCR analysis was performed using gene-specific primers and SYBR Green PCR Master Mix (Applied Biosystems, 4309155). The sequences of the primers used are available upon request. The 2−ΔΔCt method was applied to calculate the fold change of relative transcript levels; Hprt1 was used for normalization.
Intestinal epithelial cell isolation and ATAC-seq
Embryos were collected at E16.5, and the freshly harvested embryonic small intestine (caudal stomach to rostral caecum) was opened longitudinally with forceps and cut into 1 cm pieces. Intestinal tissues were treated with prewarmed 0.25% trypsin for 8 min at 37°C on a vortex station (speed set between 6 and 7), neutralized with 10% FBS, and passed through a 70 μm cell strainer. Cells were stained with anti-CD326 (EpCAM) magnetic microbeads antibody (Miltenyi Biotec, 130-105-958) for 40 min on ice. To obtain single magnetic antibody conjugated EpCAM-positive epithelial cells, stained cells were passed through a 40 μm cell strainer and then collected over a MS column (Miltenyi Biotec, 130-042-201) in a magnetic field. Approximately 20,000 cells were used for ATAC-seq, as described previously (Buenrostro et al., 2013, 2015) with slight modifications. Briefly, cells were resuspended in ice-cold lysis buffer (10 mM Tris, pH 7.4, 10 mM NaCl, 3 mM MgCl2 and 0.1% NP-40) and then centrifuged at 500 g for 10 min at 4°C. The isolated nuclei were incubated with Nextera Tn5 Transposase (Illumina FC-121-1030) at 37°C for 30 min. The transposed chromatin was purified with QIAquick PCR Purification Kit (Qiagen); PCR was amplified with high-fidelity 2X PCR Master Mix (New England Biolabs M0541). One-third of the maximum fluorescent intensity was used to determine the additional cycles. The PCR amplified libraries were purified, fragment size selected using Pippin Prep and sequencing carried out on Illumina NextSeq 550.
Bioinformatics analysis
For ATAC-seq and ChIP-seq analysis, raw sequencing reads (fastq) were quality checked using fastQC (v0.11.3) and were further aligned to mouse (mm9) genomes using bowtie2 (v2.2.6) (Langmead and Salzberg, 2012) to obtain bam files. Deeptools bamCoverage (Ramírez et al., 2016) (v2.4.2, duplicate reads ignored, RPKM normalized and extended reads) was used to generate bigwig files from bam files, and BigWigMerge (v2) was used to merge the bigwig files of different replicates. MACS (v1.4.1) (Zhang et al., 2008) was used for peak calling and to generate bed files from bam files. BEDTools (v2.17.0) (Quinlan, 2014) was used to merge, intersect or subtract the intervals of bed files. Promoters were defined within 2 kb of the transcription start sites of RefSeq genes; enhancers were defined by excluding promoters. Haystack (v0.4.0) (Pinello et al., 2018) quantile normalized bigwigs were used to create k-means clustering heatmaps of ATAC-seq using computeMatrix and plotHeatmap from Deeptools (v2.4.2) (Ramírez et al., 2016). Genomic regions of desired k-means clusters were extracted from bed files generated by plotHeatmap for further analysis. Homer findMotifsGenome.pl (v4.8.3, homer de novo Results) (Heinz et al., 2010) was used to identify transcription factor motifs enriched at peaks. Genes associated with peaks were identified using the peak2gene/BETA-minus function (v1.0.2) in Cistrome tools (Liu et al., 2011). Enriched gene ontologies were identified from genomic regions (bed file) using GREAT analysis (v3.0.0) (McLean et al., 2010) or DAVID (v6.8) (Huang da et al., 2009). SitePro (v1.0.2) (Shin et al., 2009) was used to visualize the average signals of ChIP-seq in the desired genomic regions. The Integrative Genomics Viewer (IGV 2.4.13) (Robinson et al., 2011) was used to visualize bigwig tracks.
For RNA-seq analysis, raw sequencing reads (fastq) were quality checked using fastQC (v0.11.3) and were further aligned to mouse (mm9) genomes using Tophat2 (v2.1.0) (Kim et al., 2013) to generate bam files. Kallisto (v0.44.0) (Bray et al., 2016) was utilized to quantify the transcript abundances of the RNA-seq samples through pseudoalignment, using single-end reads and an Ensembl mm9 transcriptome build index. Then, the tximport (v1.8.0) (Soneson et al., 2015) package was run in R (v3.5.2) to create gene-level count matrices for use with DESeq2 (v1.20) (Love et al., 2014) by importing quantification data obtained from Kallisto. DESeq2 was then used to generate RPKM values per kilobase of gene length per million mapped fragments at each time-course point, with comparison of Cdx2KO replicates and wild-type replicates. DESeq2 was also used to generate P values for gene matches. Genes with FPKM>1, a commonly used minimal expression threshold, were used for further analysis. Heatmapper (Babicki et al., 2016) was used to display relative transcript levels of genes of interest by using normalized RPKM values. IGV (Robinson et al., 2011) was used to visualize bam tracks.
Statistical analysis
The data are presented as mean±s.e.m. and statistical comparisons were performed using the two-sided Student's t-test at ***P<0.001, **P<0.01 or *P<0.05. Bioinformatics-related statistical analysis was done with the embedded statistics in each package, including HOMER (Heinz et al., 2010), GREAT (McLean et al., 2010), DAVID (Huang da et al., 2009) and DESeq2 (Love et al., 2014). P<0.05 (95% confidence interval) was considered statistically significant.
Supplementary Material
Acknowledgements
We thank Madhurima Saxena and Ramesh Shivdasani for helpful suggestions and curating published data. We thank Kenneth Zaret for helpful discussions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: L.C., M.P.V.; Methodology: L.C.; Validation: L.C., N.H.T.; Formal analysis: L.C., R.A., Y.-H.T.; Investigation: L.C., N.H.T., S.L., R.P.V., A.P., Y.-H.T.; Resources: J.R.S.; Data curation: L.C.; Writing - original draft: L.C., M.P.V.; Writing - review & editing: L.C., R.P.V., R.A., A.P., J.R.S., M.P.V.; Visualization: L.C.; Supervision: M.P.V.; Project administration: M.P.V.; Funding acquisition: J.R.S., M.P.V.
Funding
This research was funded by a grant from the National Cancer Institute (R01CA190558 to M.P.V.). J.R.S and M.P.V are also supported by the Intestinal Stem Cell Consortium from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and National Institute of Allergy and Infectious Diseases (NIAID) (U01 DK103141). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Support was also received from the Sequencing Facility of the Rutgers Cancer Institute of New Jersey funded by the National Cancer Institute (P30CA072720) and the University of Michigan Center for Gastrointestinal Research (UMCGR) (NIDDK 5P30DK034933). L.C. was supported by New Jersey Commission on Cancer Research grant (DFHS18PPC051). N.H.T., S.L., R.P.V. and A.P. were supported by MacMillan Summer Undergraduate Research Fellowships. Deposited in PMC for release after 12 months.
Data availability
All ATAC-seq data of this study have been deposited in GEO under accession number GSE128674. The following datasets from GEO were reanalyzed. The accession numbers for the transcriptome and chromatin accessibility of time-course wild type and Cdx2KO from our previous studies are GSE115314 (Kumar et al., 2019) and GSE115541 (Banerjee et al., 2018). The accession numbers for the CDX2 ChIP-seq and HNF4 ChIP-seq from our previous studies are GSE34568 (Verzi et al., 2013), GSE115314 (Kumar et al., 2019) and GSE112946 (Chen et al., 2019). GSE89684 (Kazakevych et al., 2017) was used to mark active chromatin with the time-course H3K27ac ChIP-seq. The accession number for the RNA-seq data of differentiation of human ESCs into human intestinal organoids is E-MTAB-4168 (Tsai et al., 2017) in the ArrayExpress database. Data curated from public sources are also listed in Table S3.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.179432.supplemental
References
- Adachi K., Nikaido I., Ohta H., Ohtsuka S., Ura H., Kadota M., Wakayama T., Ueda H. R. and Niwa H. (2013). Context-dependent wiring of Sox2 regulatory networks for self-renewal of embryonic and trophoblast stem cells. Mol. Cell 52, 380-392. 10.1016/j.molcel.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Babeu J.-P., Darsigny M., Lussier C. R. and Boudreau F. (2009). Hepatocyte nuclear factor 4alpha contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 297, G124-G134. 10.1152/ajpgi.90690.2008 [DOI] [PubMed] [Google Scholar]
- Babicki S., Arndt D., Marcu A., Liang Y., Grant J. R., Maciejewski A. and Wishart D. S. (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44, W147-W153. 10.1093/nar/gkw419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K. K., Saxena M., Kumar N., Chen L., Cavazza A., Toke N. H., O'Neill N. K., Madha S., Jadhav U., Verzi M. P. et al. (2018). Enhancer, transcriptional, and cell fate plasticity precedes intestinal determination during endoderm development. Genes Dev. 32, 1430-1442. 10.1101/gad.318832.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd M., Bressendorff S., Møller J., Olsen J. and Troelsen J. T. (2009). Mapping of HNF4alpha target genes in intestinal epithelial cells. BMC Gastroenterol. 9, 68 10.1186/1471-230X-9-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N. L., Pimentel H., Melsted P. and Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525-527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y. and Greenleaf W. J. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213-1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J. D., Wu B., Chang H. Y. and Greenleaf W. J. (2015). ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21-29. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D. M., Hill A. J., Zhang F., Mundlos S., Christiansen L., Steemers F. J. et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496-502. 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattin A.-L., Le Beyec J., Barreau F., Saint-Just S., Houllier A., Gonzalez F. J., Robine S., Pincon-Raymond M., Cardot P., Lacasa M. et al. (2009). Hepatocyte nuclear factor 4alpha, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol. Cell. Biol. 29, 6294-6308. 10.1128/MCB.00939-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahar S., Gandhi V., Yu S., Desai K., Cowper-Sal-lari R., Kim Y., Perekatt A. O., Kumar N., Thackray J. K., Musolf A. et al. (2014). Chromatin profiling reveals regulatory network shifts and a protective role for hepatocyte nuclear factor 4alpha during colitis. Mol. Cell. Biol. 34, 3291-3304. 10.1128/MCB.00349-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Toke N. H., Luo S., Vasoya R. P., Fullem R. L., Parthasarathy A., Perekatt A. O. and Verzi M. P. (2019). A reinforcing HNF4-SMAD4 feed-forward module stabilizes enterocyte identity. Nat. Genet. 51, 777-785. 10.1038/s41588-019-0384-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. S., Manova K., Weinstein D. C., Duncan S. A., Plump A. S., Prezioso V. R., Bachvarova R. F. and Darnell J. E. Jr. (1994). Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 8, 2466-2477. 10.1101/gad.8.20.2466 [DOI] [PubMed] [Google Scholar]
- Chin A. M., Hill D. R., Aurora M. and Spence J. R. (2017). Morphogenesis and maturation of the embryonic and postnatal intestine. Semin. Cell Dev. Biol. 66, 81-93. 10.1016/j.semcdb.2017.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium E. P. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57-74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison J. M., Lickwar C. R., Song L., Breton G., Crawford G. E. and Rawls J. F. (2017). Microbiota regulate intestinal epithelial gene expression by suppressing the transcription factor Hepatocyte nuclear factor 4 alpha. Genome Res. 27, 1195-1206. 10.1101/gr.220111.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaghey J., Thakurela S., Charlton J., Chen J. S., Smith Z. D., Gu H., Pop R., Clement K., Stamenova E. K., Karnik R. et al. (2018). Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat. Genet. 50, 250-258. 10.1038/s41588-017-0034-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S. A., Nagy A. and Chan W. (1997). Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of Hnf-4(−/−) embryos. Development 124, 279-287. [DOI] [PubMed] [Google Scholar]
- Gao N., White P. and Kaestner K. H. (2009). Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev. Cell 16, 588-599. 10.1016/j.devcel.2009.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison W. D., Battle M. A., Yang C., Kaestner K. H., Sladek F. M. and Duncan S. A. (2006). Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology 130, 1207-1220. 10.1053/j.gastro.2006.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger S., Savory J. G. A. and Lohnes D. (2010). Cdx2 regulates patterning of the intestinal epithelium. Dev. Biol. 339, 155-165. 10.1016/j.ydbio.2009.12.025 [DOI] [PubMed] [Google Scholar]
- Harfe B. D., Scherz P. J., Nissim S., Tian H., McMahon A. P. and Tabin C. J. (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517-528. 10.1016/j.cell.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Harper J., Mould A., Andrews R. M., Bikoff E. K. and Robertson E. J. (2011). The transcriptional repressor Blimp1/Prdm1 regulates postnatal reprogramming of intestinal enterocytes. Proc. Natl. Acad. Sci. USA 108, 10585-10590. 10.1073/pnas.1105852108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst G. P., Lee Y.-H., Lambert G., Ward J. M. and Gonzalez F. J. (2001). Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 21, 1393-1403. 10.1128/MCB.21.4.1393-1403.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H. and Glass C. K. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576-589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T. and Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Kazakevych J., Sayols S., Messner B., Krienke C. and Soshnikova N. (2017). Dynamic changes in chromatin states during specification and differentiation of adult intestinal stem cells. Nucleic Acids Res. 45, 5770-5784. 10.1093/nar/gkx167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R. and Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Tsai Y. H., Chen L., Zhou A., Banerjee K. K., Saxena M., Huang S., Toke N. H., Xing J., Shivdasani R. A. et al. (2019). The lineage-specific transcription factor CDX2 navigates dynamic chromatin to control distinct stages of intestine development. Development 146, dev172189 10.1242/dev.172189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. and Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357-359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Ning G. and Duncan S. A. (2000). Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 14, 464-474. [PMC free article] [PubMed] [Google Scholar]
- Lindeboom R. G. H., van Voorthuijsen L., Oost K. C., Rodríguez-Colman M. J., Luna-Velez M. V., Furlan C., Baraille F., Jansen P. W. T. C., Ribeiro A., Burgering B. M. et al. (2018). Integrative multi-omics analysis of intestinal organoid differentiation. Mol. Syst. Biol. 14, e8227 10.15252/msb.20188227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Ortiz J. A., Taing L., Meyer C. A., Lee B., Zhang Y., Shin H., Wong S. S., Ma J., Lei Y. et al. (2011). Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 12, R83 10.1186/gb-2011-12-8-r83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison B. B., Dunbar L., Qiao X. T., Braunstein K., Braunstein E. and Gumucio D. L. (2002). Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 277, 33275-33283. 10.1074/jbc.M204935200 [DOI] [PubMed] [Google Scholar]
- McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., Wenger A. M. and Bejerano G. (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495-501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould A. W., Morgan M. A. J., Nelson A. C., Bikoff E. K. and Robertson E. J. (2015). Blimp1/Prdm1 functions in opposition to Irf1 to maintain neonatal tolerance during postnatal intestinal maturation. PLoS Genet. 11, e1005375 10.1371/journal.pgen.1005375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerurkar N. L., Mahadevan L. and Tabin C. J. (2017). BMP signaling controls buckling forces to modulate looping morphogenesis of the gut. Proc. Natl. Acad. Sci. USA 114, 2277-2282. 10.1073/pnas.1700307114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H. (2018). The principles that govern transcription factor network functions in stem cells. Development 145, dev157420 10.1242/dev.157420 [DOI] [PubMed] [Google Scholar]
- Nord A. S., Blow M. J., Attanasio C., Akiyama J. A., Holt A., Hosseini R., Phouanenavong S., Plajzer-Frick I., Shoukry M., Afzal V. et al. (2013). Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell 155, 1521-1531. 10.1016/j.cell.2013.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parviz F., Matullo C., Garrison W. D., Savatski L., Adamson J. W., Ning G., Kaestner K. H., Rossi J. M., Zaret K. S. and Duncan S. A. (2003). Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat. Genet. 34, 292-296. 10.1038/ng1175 [DOI] [PubMed] [Google Scholar]
- Pinello L., Farouni R. and Yuan G.-C. (2018). Haystack: systematic analysis of the variation of epigenetic states and cell-type specific regulatory elements. Bioinformatics 34, 1930-1933. 10.1093/bioinformatics/bty031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R. (2014). BEDTools: the swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 11.12.1-11.12.34. 10.1002/0471250953.bi1112s47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Ryan D. P., Gruning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F. and Manke T. (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160-W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G. and Mesirov J. P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24-26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T., Muthurajan U. M., Luger K., Tulin A. V. and Zaret K. S. (2009). Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 23, 804-809. 10.1101/gad.1775509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Yue F., McCleary D. F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V. V. et al. (2012). A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116-120. 10.1038/nature11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Liu T., Manrai A. K. and Liu X. S. (2009). CEAS: cis-regulatory element annotation system. Bioinformatics 25, 2605-2606. 10.1093/bioinformatics/btp479 [DOI] [PubMed] [Google Scholar]
- Soneson C., Love M. I. and Robinson M. D. (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 4, 1521 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.-H., Nattiv R., Dedhia P. H., Nagy M. S., Chin A. M., Thomson M., Klein O. D. and Spence J. R. (2017). In vitro patterning of pluripotent stem cell-derived intestine recapitulates in vivo human development. Development 144, 1045-1055. 10.1242/dev.138453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi M. P., Shin H., He H. H., Sulahian R., Meyer C. A., Montgomery R. K., Fleet J. C., Brown M., Liu X. S. and Shivdasani R. A. (2010). Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell 19, 713-726. 10.1016/j.devcel.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi M. P., Shin H., Ho L.-L., Liu X. S. and Shivdasani R. A. (2011). Essential and redundant functions of caudal family proteins in activating adult intestinal genes. Mol. Cell. Biol. 31, 2026-2039. 10.1128/MCB.01250-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi M. P., Shin H., San Roman A. K., Liu X. S. and Shivdasani R. A. (2013). Intestinal master transcription factor CDX2 controls chromatin access for partner transcription factor binding. Mol. Cell. Biol. 33, 281-292. 10.1128/MCB.01185-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A., Blow M. J., Li Z., Zhang T., Akiyama J. A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F. et al. (2009). ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854-858. 10.1038/nature07730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton K. D., Freddo A. M., Wang S. and Gumucio D. L. (2016a). Generation of intestinal surface: an absorbing tale. Development 143, 2261-2272. 10.1242/dev.135400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton K. D., Whidden M., Kolterud A., Shoffner S. K., Czerwinski M. J., Kushwaha J., Parmar N., Chandhrasekhar D., Freddo A. M., Schnell S. et al. (2016b). Villification in the mouse: Bmp signals control intestinal villus patterning. Development 143, 427-436. 10.1242/dev.130112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton K. D., Kolterud A., Czerwinski M. J., Bell M. J., Prakash A., Kushwaha J., Grosse A. S., Schnell S. and Gumucio D. L. (2012). Hedgehog-responsive mesenchymal clusters direct patterning and emergence of intestinal villi. Proc. Natl. Acad. Sci. USA 109, 15817-15822. 10.1073/pnas.1205669109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. M. and Spence J. R. (2014). How to make an intestine. Development 141, 752-760. 10.1242/dev.097386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson A. C., Nakauchi H. and Göttgens B. (2017). Mammalian transcription factor networks: recent advances in interrogating biological complexity. Cell Syst. 5, 319-331. 10.1016/j.cels.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W. et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.