ABSTRACT
Pluripotent stem cells give rise to all cells of the adult organism, making them an invaluable tool in regenerative medicine. In response to differentiation cues, they can activate markedly distinct lineage-specific gene networks while turning off or rewiring pluripotency networks. Recent innovations in chromatin and nuclear structure analyses combined with classical genetics have led to novel insights into the transcriptional and epigenetic mechanisms underlying these networks. Here, we review these findings in relation to their impact on the maintenance of and exit from pluripotency and highlight the many factors that drive these processes, including histone modifying enzymes, DNA methylation and demethylation, nucleosome remodeling complexes and transcription factor-mediated enhancer switching.
KEY WORDS: Epigenetics, Pluripotency, Transcriptional regulation, Embryonic stem cells, Epiblast-like cells, Mouse
Summary: In this Review, we discuss emerging concepts of the various levels of epigenetic control of transcriptional regulation in the maintenance and exit from mouse pluripotency.
Introduction
Cell-fate transitions are ultimately driven by changes in gene expression. Gene expression is regulated by the binding of transcription factors (TFs) to specific and conserved DNA motifs within enhancers and promoters, which then, in collaboration with chromatin-modifying enzymes, leads to target gene activation or repression (Long et al., 2016). Although promoters are directly adjacent to the genes they regulate, enhancers can be thousands to millions of bases away.
Enhancers and promoters show a range of activities. Inactive enhancers and promoters show minimal expression of their target genes and tend to have a high degree of nucleosome occupancy and the presence of repressive histone marks such as histone H3 lysine 27 trimethylation (H3K27me3) and H3K9 di- and trimethylation (H3K9me2/3). A subset of TFs can reverse this inactive state by recruiting nucleosome remodeling complexes and histone modifying enzymes, resulting in an accumulation of activating marks and ultimately increased transcription. These activating marks include histone H3K4 trimethylation (H3K4me3) and monomethylation (H3K4me1), which are found at active promoters and enhancers, respectively. Active enhancers and promoters also correlate with actively transcribing RNA polymerase II (Pol II) and histone H3K27 acetylation (H3K27ac). In addition, Pol II transcribes a class of non-coding RNAs termed enhancer RNAs (eRNAs) at active enhancers, which are proposed to participate in enhancer-promoter communication (Li et al., 2016). Intermediate enhancer states such the ‘primed’ and ‘poised’ states are also observed, and are associated with subsets of marks that are thought to allow for rapid activation or repression in response to cell signaling cues. Although the different marks correlate well with the activity of enhancers and the expression of their cognate genes, the sequence of events required to interconvert between activity states is only partially understood. Furthermore, it remains to be clarified mechanistically how enhancers and promoters work in concert to modulate gene expression and to what degree physical contacts between these elements matter.
Pluripotent stem cells of the mammalian epiblast, which give rise to all cells in the embryo, have proven to be valuable models for understanding transcriptional regulation. This transient in vivo population of cells can be cultured indefinitely in vitro under defined conditions. These cells, termed embryonic stem cells (ESCs), enable the study of mechanisms involved in the earliest fate decisions in embryonic development. During early differentiation both in vivo and in vitro, ESCs undergo molecular and morphogenetic changes, while retaining their ability to contribute to all cells of the adult animal (Smith, 2017). These changes occur over a span of several days and have led to the definition of distinct pluripotent states. However, there is much confusion in the literature regarding the terminology used to describe these states. In a recent review, Austin Smith aimed to clarify this issue by specifically defining distinct pluripotent states (Smith, 2017). In agreement with this Review, we use the following terminology. ESCs cultured in the presence of leukemia inhibitory factor (LIF) and 2i (glycogen synthase kinase-3 beta and mitogen-activated protein kinase kinase inhibitors) (Ying et al., 2008) will be considered as naïve pluripotent cells. They are similar to early pre-implantation epiblast cells (Hayashi et al., 2011; Nakamura et al., 2016). Epiblast-like cells (EpiLCs) differentiated in either serum alone or FGF/activin will be defined as exhibiting the formative pluripotent state; these cells are similar to late epiblast cells (Krishnakumar et al., 2016; Hayashi et al., 2011). We also discuss results from studies of mouse ESCs cultured in serum plus LIF (S/L; also described as metastable ESCs) and mouse epiblast stem cells (EpiSCs) cultured in FGF/activin, both of which are more heterogeneous cell populations. S/L-cultured ESCs converge on a bulk transcriptome comprising a mixture of naïve and formative markers (Guo et al., 2016), whereas EpiSCs converge on a bulk transcriptome similar to late gastrula-stage ectoderm and are distinct from EpiLCs (Kojima et al., 2014; Tsakiridis et al., 2014).
A remarkable feature of transitions between naïve and other pluripotency states is the large degree of epigenetic reprogramming that occurs in the absence of major changes in the expression of many of core pluripotency genes (Buecker et al., 2014; Ficz et al., 2013; Galonska et al., 2015; Habibi et al., 2013; Krishnakumar et al., 2016; Marks et al., 2012). This epigenetic reorganization is thought to rewire the ESC genome in preparation for rapid lineage diversification with the onset of gastrulation (Fig. 1) (Smith, 2017). Global levels of DNA methylation markedly increase when ESCs exit from naïve pluripotency, accompanied by increased expression of DNA methyltransferases (DNMTs) 1, 3A and 3B (Von Meyenn et al., 2016a; Ficz et al., 2013; Habibi et al., 2013; Hackett et al., 2013; Leitch et al., 2013). Furthermore, bivalent H3K27me3 and H3K4me3 marks are established at CpG-rich island (CGI)-positive promoters (Marks et al., 2012). In contrast, the expression of most pluripotency genes, including octamer-binding transcription factor 4 (OCT4, also known as POU5F1) and SRY-Box 2 (SOX2), remains unchanged, although the enhancers driving expression of these genes do change (Buecker et al., 2014; Chen et al., 2018; Galonska et al., 2015). Understanding how these various epigenetic and chromatin-based changes regulate the transition between pluripotent states, and the various molecular factors that mediate them, will contribute to our understanding of cell-fate decisions in development and disease, and could help us to direct cell-fate decisions in the context of regenerative medicine.
Fig. 1.
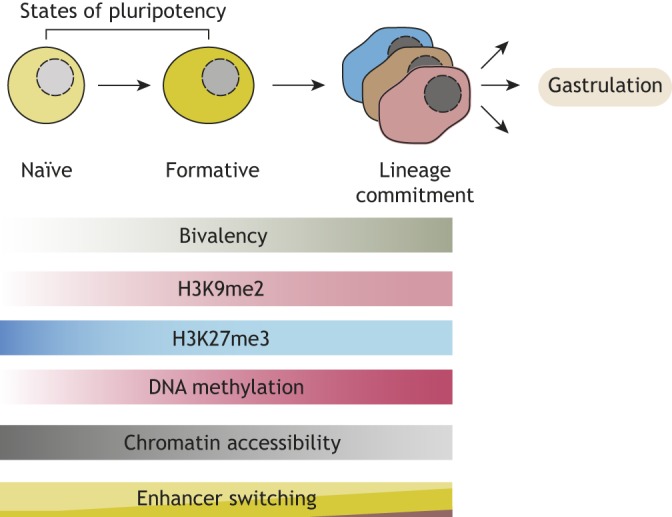
States of pluripotency. Embryonic stem cells transition through distinct states of pluripotency (naïve, formative) before they undergo lineage commitment (and then gastrulation). This transition is accompanied by various changes in chromatin, including gain of bivalent domains and global DNA methylation, global gain of H3K9 dimethylation (H3K9me2) and loss of chromatin accessibility. In addition, many genes are regulated by changing sets of enhancers.
In this Review, we focus on studies that have used state-of-the-art technologies combined with classical genetics to tease out the mechanistic relationships underlying the regulation of transcription in pluripotent states, as well as during the exit from pluripotency. Using the terminologies defined above, we present emerging concepts of transcriptional regulation in pluripotency at the levels of histone modifications, DNA methylation, nucleosome accessibility, chromatin looping and enhancer switching. We also highlight how crosstalk between these chromatin modifications and TFs can influence pluripotency.
Histone modifying enzymes
Histone modifying enzymes are a diverse group of proteins catalyzing the covalent post-translational modification of histones, most prominently at their N-terminal ‘tail’ regions. Most of these enzymes are ubiquitously expressed and have no direct DNA-binding domains, and thus need to be recruited in a cell type-specific and dynamic manner. Although the list of histone modifications and their associated enzymes continues to grow, we focus here on a subset of these enzymes that catalyze the addition of marks with well-known functions in transcriptional activation and repression.
MLL proteins
In mammals, mixed lineage leukemia (MLL)/Set1A/B proteins are responsible for H3K4 methylation, which is a marker of poised, primed and active enhancers and promoters. The H3K4 mono- and dimethyltransferases MLL3 (KMT2C) and MLL4 (KMT2D) are enriched at enhancers (Hu et al., 2013). In ESCs, loss of both MLL3/4 or MLL4 alone results in comparably reduced H3K4me1 levels, suggesting that MLL4 is the major driver of H3K4me1 (Fig. 2). Indeed, MLL4 knockout (KO) mice show early embryonic lethality at around embryonic day (E)9.5, whereas MLL3 KO mice survive until birth (Lee et al., 2013). Surprisingly, even in the absence of MLL3/4, expression of core pluripotency genes including Sox2, Nanog and Oct4 remains unchanged when ESCs are cultured in naïve or S/L conditions. In addition, colony formation assays suggest no major defects in self-renewal. However, levels of H3K27ac at MLL3/4-bound sites are decreased, as is eRNA transcription, Pol II density and expression of nearby genes (Dorighi et al., 2017; Wang et al., 2016).
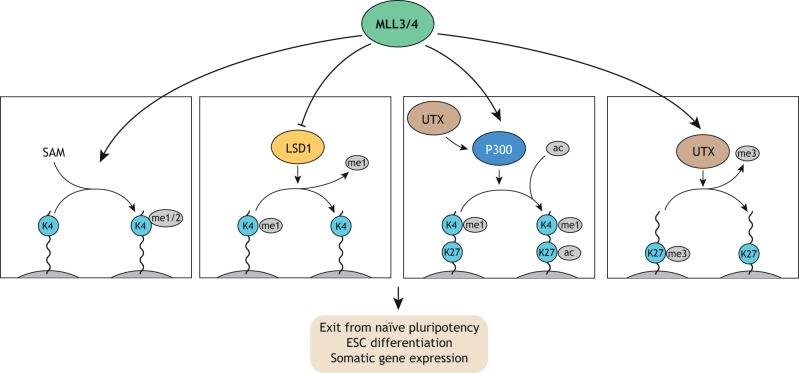
Fig. 2.
MLL3/4 complex functions in ESCs. The MLL3/4 histone methyltransferases function as part of the COMPASS complex to promote H3K4 mono- and dimethylation (me1/2), using S-Adenosyl methionine (SAM) as a cofactor. In addition, MLL4 counteracts LSD1 to preserve H3K4me1. MLL3/4 also co-operate with P300 and UTX to promote H3K27 demethylation and acetylation (ac). Biologically, MLL3/4 proteins are required for exiting naïve pluripotency and proper differentiation into somatic lineages.
Upon removal of 2i, MLL4-deficient ESCs fail to adopt a transcriptome characteristic of S/L-cultured ESCs but maintain a naïve-like ESC state (Fig. 2) (Cao et al., 2018). Molecularly, this is confirmed by decreased levels of H3K4me1 and H3K27ac levels at S/L-induced genes. Furthermore, MLL3/4-deficient ESCs show an impaired capacity to give rise to somatic lineages during embryoid body differentiation, as evidenced by persistence of many pluripotency markers and a failure to induce expression of differentiation markers (Wang et al., 2016). Interestingly, the failure in enhancer activation and exit of naïve pluripotency in MLL4-deficient ESCs can be reversed by depletion of the H3K4 demethylase LSD1 (KDM1A), which co-occupies MLL4-bound sites (Cao et al., 2018). This finding suggests that MLL4 counteracts the repressive role of LSD1 at bound enhancers and that its departure from specific enhancers during differentiation is associated with their decommissioning via LSD1. In line with this interpretation, impairing MLL4 recruitment to chromatin by deletion of its N-terminal PHD finger cluster mimics the MLL4 KO phenotype (Cao et al., 2018).
The effect of MLL3/4 on H3K27ac levels and gene expression appears to be independent of their methyltransferase activity, as point mutations or complete deletion of their catalytic SET domain results in few changes (Cao et al., 2018; Dorighi et al., 2017). This surprising finding questions the importance of H3K4me1 in enhancer activity. However, given H3K4me1 is reduced but not abolished in MLL3/4 mutant cell lines, compensation by other MLL proteins might explain this finding. H3K4me1 does appear to play an important role in chromatin looping. Loss of MLL3/4 protein or of just its methyltransferase activity results in a decrease in cohesin occupancy and long-distance enhancer-promoter interactions, particularly at large clusters of enhancers termed super-enhancers and enhancers that gain MLL3/4-dependent H3K4me1 during early ESC differentiation (Yan et al., 2018). Another study identified an interaction between H3K4me1-containing nucleosomes and chromatin remodeling complexes, which suggests H3K4me1 might have a role in recruiting these factors (Local et al., 2018). These data suggest that H3K4me1 might confer robustness to enhancer activation by creating a local environment that favors cohesin recruitment and H3K27 acetylation.
MLL3/4 are not the only methyltransferases found at enhancers (Dorighi et al., 2017). A recent study demonstrated that MLL1 is required for EpiSC maintenance and is also predominantly bound at enhancers (Zhang et al., 2016). Genetic loss or pharmacological inhibition of MLL1 results in reprogramming to a naïve-like ESC state along with a reduction of H3K4me1 at MLL1-bound sites in EpiSCs. Thus, MLL1-dependent H3K4 methylation appears to be required for maintenance of the EpiSC state. Another recent study found that MLL2 and MLL4 deletion in ESCs results in loss of H3K4me1 at partially overlapping sets of distal regulatory elements, suggesting redundant and non-redundant functions of these MLL proteins (Morgan et al., 2017). A careful dissection of the relative contributions of MLL proteins to H3K4me1 and target gene transcription at different classes of enhancers and promoters in well-defined pluripotency conditions is necessary to clarify the role of MLL proteins and H3K4me1 in ESC transcriptional regulation. Of particular interest would be to determine the extent to which MLL function is required for maintenance of enhancer activity as opposed to de novo activation during cell state transitions.
MLL proteins also act in concert with other activating chromatin-modifying enzymes to shape enhancer activity in ESCs. For example, the H3K27 demethylase UTX (KDM6A) cooperates with MLL4 to recruit P300 and enhance H3K27 acetylation (Fig. 2) (Wang et al., 2017), which in turn further promotes H3K4 methylation. The cooperativity of MLL4, UTX and P300 ultimately leads to greatly enhanced transcriptional activation. The catalytic activity of UTX is dispensable for this mechanism. Furthermore, during ESC differentiation, UTX suppresses enhancer of zeste 2 (EZH2)-dependent H3K27 methyltransferase activity following DNA replication, providing a transient state of open chromatin allowing binding of fate-specifying TFs to the nascent DNA strand (Petruk et al., 2017). It will be interesting to see whether this function of UTX occurs in cooperation with P300 and MLL3/4, and to assess the impact of this mechanism on target gene transcription.
Histone deacetylases
As ESCs differentiate, global levels of H3K27ac decrease, correlating with a globally more closed chromatin state and decreased gene expression (Efroni et al., 2008; Lee et al., 2004; Meshorer et al., 2006). This deacetylation of histones, including that of H3K27ac, is catalyzed by class I histone deacetylases (HDAC1-3), which assemble into large multi-subunit complexes including other chromatin-modifying enzymes. Sin3-Hdac, NuRD and CoREST are examples of such complexes that are essential during early embryonic development (Kelly and Cowley, 2013).
To investigate the redundancy between ESC-expressed class I HDACs, a recent study generated inducible HDAC1/2 double KOs in S/L-cultured ESCs and demonstrated that HDAC1/2 loss results in defects in chromosomal segregation, pronounced cell death and decreased core pluripotency factor expression, which is accompanied by globally increased levels of histone acetylation (Jamaladdin et al., 2014). Although these data highlight HDAC1/2 as crucial regulators of pluripotency, many of the observed defects could be indirect. It remains unclear whether the observed phenotype is due to increased histone acetylation levels or linked to catalytic-independent functions similar to transcriptional repression by class IIa HDACs, which involves inhibition of TF activity through direct interaction and recruitment of co-repressors (Dressel et al., 2001; Zhang et al., 2001, 2002). This could be addressed by generating catalytically dead HDAC mutants. In addition, the role and impact of the opposing activities of the histone acetyltransferases P300/CBP should be investigated at HDAC1/2-dependent loci.
Using a proteomics approach, a recent study identified SIN3-HDAC Complex Associated Factor (FAM60A, also known as SINHCAF) as a new ESC-specific subunit of the Sin3a-HDAC complex (Streubel et al., 2017). FAM60A colocalizes genome-wide with other complex members at promoters and is required for SIN3A localization. SIN3A and FAM60A depletion results in a common set of deregulated target genes, including genes associated with transforming growth factor (TGF) beta signaling and G1 cell cycle phase length. As with the HDAC1/2 double KO cells, it would be important to address the role of H3K27 deacetylation in the regulation of SIN3A-HDAC target genes and how it could regulate the short and unique G1 phase of pluripotent cells.
Polycomb repressive complexes 1 and 2
Polycomb repressive complexes 1 (PRC1) and 2 (PRC2) are epigenetic repressors (Di Croce and Helin, 2013) that are essential for early embryonic development, in which they function to maintain proper gene expression patterns (Boyer et al., 2006; O'Carroll et al., 2001). Both complexes comprise a catalytic subunit as well as multiple regulatory subunits. The core of canonical PRC1 is formed by chromobox-domain (CBX) proteins, PCGF family members 2 or 4 (PCGF2/4) and the ubiquitin E3 ligase subunits RING1A/B, which catalyze the ubiquitylation of H2AK119 (H2AK119ub) (Fig. 3A). Non-canonical PRC1 complexes in turn are devoid of CBX proteins, but instead contain RING1 and YY1 Binding Protein (RYBP) or YY1 Associated Factor 2 (YAF2) (Di Croce and Helin, 2013; Rose et al., 2016). They also contain PCGF1, PCGF3, PCGF5 or PCGF6. By contrast, PRC2 consists of the core subunits EZH1/2, suppressor of zeste 12 (SUZ12) and embryonic ectoderm development (EED), with EZH1/2 catalyzing H3K27 methylation (Fig. 3B). Non-core subunits segregate PRC2 into two major subtypes, PRC2.1 [containing PCL homolog (PCL1-3) alongside EPOP or LCOR] and PRC2.2 (containing JARID2 and AEBP2). Both PRC1 and PRC2 have the capacity to bind H3K27me3 and H2AK119ub, respectively, reinforcing each other's binding to chromatin (Blackledge et al., 2014; Cooper et al., 2014; Min et al., 2003; Wang et al., 2004).
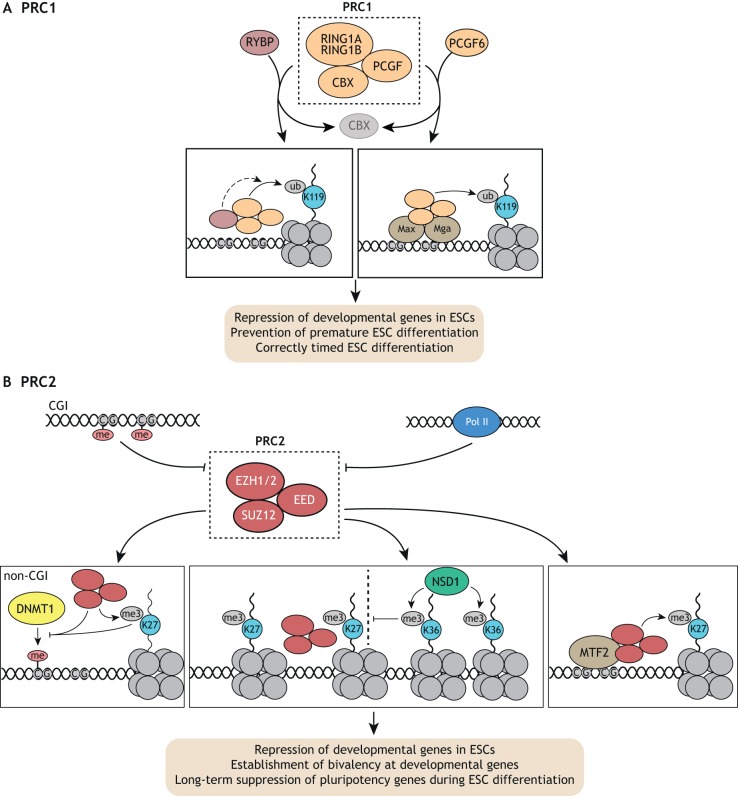
Fig. 3.
PRC1 and PRC2 functions in ESCs. (A) The PRC1 complex, consisting of core members RING1A/B, CBX and PCGF proteins, is responsible for ubiquitylation (ub) of H2AK119. Additional regulatory proteins such as RYBP can tune its ubiquitylation activity and PCGF6 can promote its recruitment by MAX/MGA. PRC1 is required for proper repression of developmental genes and premature differentiation in ESCs, and for correctly timed ESC differentiation. (B) The PRC2 complex, consisting of core members EZH1/2, EED and SUZ12, is responsible for trimethylation (me3) of H3K27. DNA methylation at CGIs and active Pol II oppose PRC2 recruitment, whereas MTF2 recruits PRC2 to unmethylated CG-rich regions. PRC2 activity in turn opposes DNA methylation by DNMT1 at non-CGI regions. Furthermore, NSD1-dependent H3K36 trimethylated domains can restrict PRC2-dependent H2K27me3 domains. In S/L-cultured ESCs, PRC2 is responsible for the establishment of bivalent domains at developmental genes before lineage commitment and long-term suppression of pluripotency genes upon differentiation.
PRC2 has been extensively studied in ESCs and is responsible for genome-wide H3K27me2 and H3K27me3 (Chamberlain et al., 2008; Ferrari et al., 2014; Margueron et al., 2008; Montgomery et al., 2005; Pasini et al., 2004, 2007; Schoeftner et al., 2006; Shen et al., 2008). Accordingly, reintroduction of the PRC2 subunit EZH2 in EZH1/2 double KO ESCs is sufficient to accurately re-establish wild-type H3K27me2/3 patterns, even after those marks have all been lost (Højfeldt et al., 2018). This suggests that PRC2 binding and de novo methylation occur independent of previous H3K27 methylation. In line with this, it has been shown that H3K27 methylation is a stepwise process involving transient mono- and dimethylated states (Højfeldt et al., 2018). Recent evidence suggests EZH1 preferentially deposits H3K27 monomethyl marks, whereas EZH2 shows a preference for depositing K27 di- and trimethyl marks (Lee et al., 2018). Interestingly, histone linker length enhances methyltransferase activity of EZH2. Future studies should address the extent to which differential substrate preference of EZH1/2 and histone linker length relates to local K27 methylation patterns.
PRC2 preferably binds CGIs, however DNA methylation and active transcription oppose PRC2 binding at these sites (Di Croce and Helin, 2013) (Fig. 3B). Remarkably, inhibition of Pol II binding or elongation in naïve ESCs alone rapidly induces global ectopic PRC2 recruitment to nucleosome-depleted CGIs associated with PRC2 binding in differentiated tissues (Riising et al., 2014). This effect is reversible following Pol II inhibitor washout, suggesting PRC2 can be displaced by Pol II. Similar to Pol II recruitment, dCas9-mediated recruitment of the SWI/SNF nucleosome remodeling complex in S/L-cultured ESCs is sufficient to oppose local PRC1/2 function (Braun et al., 2017). These results suggest that factors promoting a local environment permissive for transcription are sufficient to oppose PRC2, and that the absence of such factors is a prerequisite for local PRC2 activity.
A recent study profiled an array of epigenetic modifications upon transitioning ESCs from naïve to S/L-cultured conditions and reported a global enrichment of PRC2-dependent H3K27me3 in naïve ESCs (van Mierlo et al., 2019). This enrichment is mainly attributed to extended eu- and heterochromatic regions outside of CGIs, which were previously shown to gain H3K27me3 in S/L-cultured compared with naïve ESCs (Marks et al., 2012). Interestingly, deletion of the PRC2 component EED in naïve ESCs results in a gain of DNA methylation over these non-CGI regions, suggesting a requirement of PRC2 in naïve ESCs for opposing widespread DNA methylation. Overall, these studies highlight extensive competitive crosstalk between PRC2 activity, transcription and DNA methylation, which is essential for the epigenetic landscape of naïve and S/L-cultured ESCs. Future studies are needed to better characterize the epistasis of this crosstalk, the nature of the factors involved and how these activities link to pluripotency gene expression and activation of fate-specifying gene programs.
The nature of the factors that recruit PRC2 in ESCs is beginning to come to light. Recent studies have identified a crucial role for the polycomb-like protein MTF2 in the recruitment of PRC2 to unmethylated CGIs (Li et al., 2017; Perino et al., 2018) (Fig. 3B). Crystal structures of MTF2 suggest that it specifically binds unmethylated CpG-containing DNA via its winged-helix domain (Li et al., 2017). In support of this finding, a proteomic screen using an unmethylated CpG-containing PRC2 motif as bait enriched for MTF2 at stoichiometric levels compared with core PRC2 components (Perino et al., 2018). Interestingly, in addition to a CpG-rich context, MTF2 binding preferably occurs at sites with a specific DNA helical shape involving an increased minor groove width and a decreased propeller twist. PRC2 activity is further restricted by the H3K36 methyltransferase NSD1, as indicated by the finding that NSD1 depletion results in genome-wide accumulation of H3K27me3 domains (Streubel et al., 2018) (Fig. 3B). Thus, sequence-specific DNA binding proteins and epigenetic context are crucial factors for defining PRC2-binding domains.
The core PRC1 subunits RING1A and RING1B also promote the recruitment of PRC2. Loss of RING1B catalytic activity results in a depletion of H2AK119ub and negatively affects PRC2 recruitment and activity in S/L-cultured ESCs, albeit to a lesser extent than does loss of RING1B altogether (Illingworth et al., 2015). Similarly, mutation of RING1A/B ubiquitylation sites in the histone variant H2A.Z.1 decreases PRC1/2 recruitment to bivalent promoters in ESCs (Surface et al., 2016). Therefore, RING1B has both catalytic-dependent and catalytic-independent roles in PRC2 recruitment and gene silencing, the relative importance of which varies in a locus-specific manner (Rose et al., 2016). Notably, whereas EZH2, SUZ12 and EED are dispensable for ESC self-renewal in naïve and S/L conditions, RING1A/B are required to prevent premature differentiation in S/L-cultured ESCs (Chamberlain et al., 2008; Endoh et al., 2008; Pasini et al., 2004; Shen et al., 2008; van Mierlo et al., 2019). This suggests that RING1A/B have essential functions in ESCs that extend beyond cooperative binding with PRC2. Such functions could depend on their roles in protein complexes that are distinct from PRC1, such as the BCL6 co-repressor complex (Gearhart et al., 2006).
PRC2 targets in ESCs are enriched for developmental genes (Boyer et al., 2006), suggesting that PRC2 is involved in the timely expression of these genes upon ESC differentiation. However, transcriptomic analysis of SUZ12 KO cells shows only minor differences during early differentiation compared with wild-type cells (Riising et al., 2014). In contrast, PRC2 is required at later stages of differentiation for maintaining correct expression levels of pluripotency and differentiation genes. Future studies should address which factors initiate transcriptional repression during ESC differentiation and why PRC2 is required for maintenance of repression.
Recent studies have also highlighted lineage-specific roles for both PRC1 and PRC2 activity. For example, one study revealed dynamic changes in PRC1 and PRC2 interactomes during differentiation of ESCs to neural precursor cells (NPCs), (Kloet et al., 2016). Core PRC2 remains intact in NPCs, with a switch of substoichiometric non-core interactors such as EPOP and LCOR. In contrast, PRC1 core subunit composition undergoes a switch in CBX and PCGF isoforms. Furthermore, H3K27me1/2/3 show dynamic redistribution during early ESC differentiation both at enhancers and at promoters (Juan et al., 2016). Interestingly, shifting the balance of H3K27me2 to H3K27me3 using a hyperactive EZH2 mutant results in ESC differentiation biased toward a neuronal program and renders cells refractory to 2i conditions.
Non-canonical PRC1 complexes often show distinct but also overlapping functions compared with canonical PRC1. For example, the PCGF6-containing non-canonical PRC1 complex (PRC1.6) is required for ESC self-renewal through suppression of germ cell-related genes (Endoh et al., 2017; Stielow et al., 2018; Zhao et al., 2017b) (Fig. 3A). Strikingly, the interaction of PCGF6 with RING1B is required for H2AK119ub and H2K27me3 marks at bound sites. The recruitment of PCGF6 is not dependent on H3K27me3 but on the TFs MGA/MAX, highlighting the importance of TFs in the recruitment of these repressive complexes (Endoh et al., 2017). PCGF1, a member of another non-canonical PRC1 complex, is dispensable for ESC self-renewal, but required for differentiation. PCGF1 is necessary for PRC1 recruitment and surprisingly results in gene activation rather than repression (Yan et al., 2017). Similarly, PCGF3 and 5 positively influence gene expression via P300 recruitment during mesoderm specification (Zhao et al., 2017a).
Other evidence for the importance of non-canonical PRC1 configurations in ESCs was shown in a recent study addressing the role of RYBP and YAF2 (Rose et al., 2016). Loss of both RYBP and YAF2 results in a reduction of H2AK119ub at PRC1 target sites, particularly ones with low PRC1 occupancy, in the absence of any change in global H2AK119ub levels or RING1B recruitment (Fig. 3A). Thus, RYBP/YAF2 shape PRC1 activity rather than influence its recruitment, and this activity is required for normal H2AK119ub levels at sites with low PRC1 occupancy. In addition, the reduction of H2AK119ub in RYBP/YAF2 double KO cells correlates well with changes in H3K27me3 levels, without major changes in SUZ12 occupancy, supporting a model in which PRC1 activity dictates local PRC2 activity. At a subset of PRC1/2 sites experiencing the strongest changes in H2AK119ub and H3K27me3 upon RYBP/YAF2 deletion, PRC1/2 occupancy is reduced, and cognate genes are de-repressed. This suggests there is a PRC1/2 activity-based threshold below which PRC1/2 occupancy cannot be sustained, allowing gene-activating cues to take control. Overall, these results highlight that non-canonical PRC1 complexes are important regulators of ESC pluripotency and differentiation, and show that PRC complex subunit composition dictates whether it acts as an activator or repressor within the same cell type.
JmjC domain histone demethylases
Jumonji C (JmjC) domain-containing proteins are a family of iron- and alpha-ketoglutarate-dependent histone demethylases that act on methylated H3K4, H3K9, H3K27, H3K36, H3R2 and H4R3, which are generally associated with transcriptionally repressed chromatin. They phylogenetically separate into distinct clusters with different substrate specificities (Cloos et al., 2008). These clusters include the JMJD2 cluster proteins, which consecutively demethylate H3K9me3/me2 as well as H3K36me3/2, and the JMJD1 cluster proteins, which demethylate H3K9me2/me1 (Fig. 4).
Fig. 4.
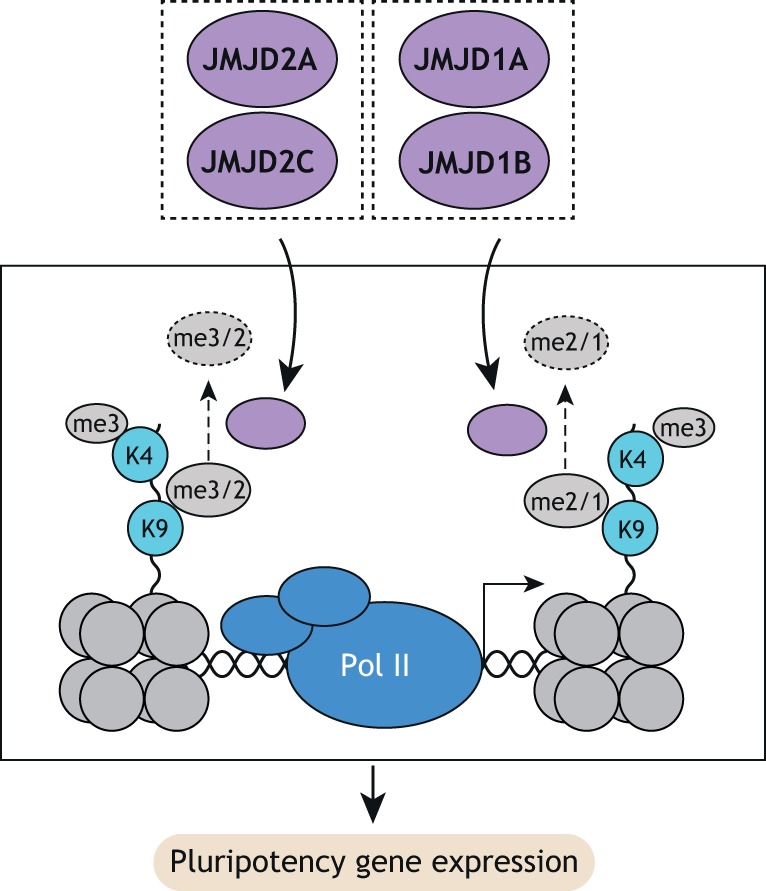
JmjC-containing histone demethylase functions in ESCs. JMJD2A/C predominantly demethylate H3K9me3/2, whereas JMJD1A/B demethylate H3K9me2/1 at H3K4me3-positive domains, which are enriched at promoters. Both JMJD clusters are required for pluripotency gene expression in ESCs.
The activity of the JMJD2 proteins JMJD2A (KDM4A) and JMJD2C (KDM4C), but not that of JMJD2B (KDM4B), is crucially required for naïve pluripotency (Pedersen et al., 2016). Specifically, JMJD2A/C act redundantly to suppress H3K9 methylation, and to a lesser extent H3K36 methylation, at H3K4me3-marked promoters in naïve ESCs. These data suggest that JMJD2 proteins are actively recruited to promoters to oppose default repression with H3K9 and H3K36 methylation. Surprisingly, JMJD2C alone may play a role in differentiation towards somatic lineages by regulating distal regulatory elements (Tomaz et al., 2017). However, the physiological relevance of this function is unclear given that JMJD2C KO is compatible with mouse development (Pedersen et al., 2014). Further studies are needed to clarify this apparent contradiction.
The JMJD1 proteins JMJD1A (KDM3A) and JMJD1B (KDM3B) play a crucial role in blocking H3K9 methylation at gene-dense chromosomal regions (Kuroki et al., 2018). Loss of JMJD1A and B results in perturbed gene expression, including decreased OCT4 levels and rapid cell death. This phenotype can be rescued by additional deletion of the H3K9 methyltransferase G9A (EHMT2), suggesting that JMJD1A/B counteract excessive G9A activity to ensure both pluripotency gene expression and ESC viability.
Collectively, these studies highlight that the active recruitment of H3K9 demethylation machinery to regulatory elements acts as an essential mechanism for maintaining pluripotency and enabling differentiation of ESCs (Fig. 4).
DNA methylation and demethylation
DNA methylation is a well-characterized heritable epigenetic modification of the DNA molecule itself that shows dynamic regulation in development and disease, and that typically has gene repressive functions. During early embryonic development, DNA is initially demethylated in several waves and, in mice, reaches its lowest methylation level in the blastocyst-stage embryo, after which point it sharply increases. DNA methylation is also one of the most distinguishing epigenetic differences between naïve and later pluripotency stages. Below, we discuss key enzymes involved in the deposition and removal of this modification and highlight recent insights into their impact on the transition between pluripotent states.
DNMT-driven DNA methylation
DNA methylation at the fifth position of cytosine (5mC) is a well-characterized epigenetic mark with important functions in mammalian development. 5mC is enriched at CpG dinucleotides but largely absent at regions of high CpG density (CpG islands) (Kim and Costello, 2017). The enzymes catalyzing DNA methylation are DNMT family members including DNMT1, DNMT3A and DNMT3B (Fig. 5A). DNMT3A and DNMT3B are the predominant de novo methylation enzymes (Okano et al., 1999), whereas DNMT1 is responsible for the maintenance of DNA methylation, acting via its capacity to recognize, with the help of UHRF1, hemimethylated DNA at replication forks and methylate the opposing strand (Bostick et al., 2007; Hermann et al., 2004; Sharif et al., 2007).
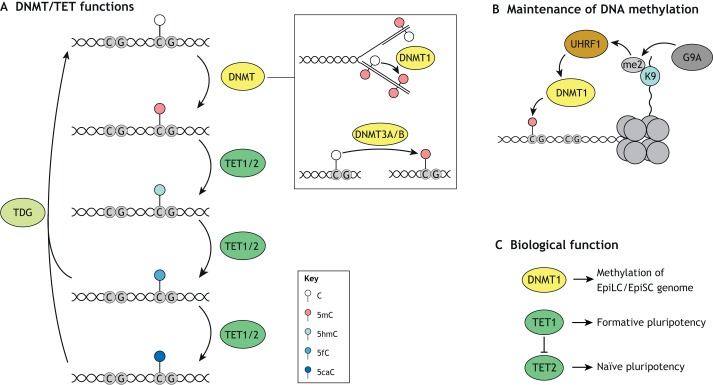
Fig. 5.
DNMT and TET functions in ESCs. (A) DNMT proteins catalyze the methylation of cytosine (5mC) at CG-rich regions. DNMT1 converts hemimethylated DNA to fully methylated DNA at replication forks during DNA replication, whereas DNMT3A/B catalyze de novo methylation. TET proteins, in turn, are responsible for the stepwise oxidation of 5mC via the intermediates 5-hydroxymethyl-cytosine (5hmC), 5-formyl-cytosine (5fC) and 5-carboxyl-cytosine (5caC). 5fC and 5caC are depurinated by TDG, which is followed by restoration of unmethylated cytosine by the base excision pathway. (B) Maintenance of DNA methylation via DNMT1 is a major driver of global DNA methylation in S/L-cultured ESCs. DNMT1 was shown to be recruited by UHRF1, which in turn localizes to G9A-dependent H3K9me2-positive domains. (C) DNMT1 is required for global methylation of the EpiLC/EpiSC genome. TET1 and TET2 have antagonistic roles in promoting formative and naïve pluripotency, respectively.
Naïve ESCs are globally hypomethylated relative to most other cell types including S/L-cultured ESCs, EpiLCs and EpiSCs (Von Meyenn et al., 2016a; Ficz et al., 2013; Habibi et al., 2013; Hackett et al., 2013; Leitch et al., 2013). The conversion of S/L-cultured ESCs into naïve ESCs is associated with diminishment of 5mC, which can be accelerated by deleting UHRF1 or DNMT1 (Von Meyenn et al., 2016b). Interestingly, the loss of DNMT3A/B or the demethylation pathway components TET1/2/3 or TDG (discussed below) has little effect on the kinetics of this loss of methylation. The stability of UHRF1 and its recruitment of DNMT1 to DNA replication forks is also reduced in naïve compared with S/L ESCs. This UHRF1-dependent DNMT1 recruitment partially depends on the binding of UHRF1 to methylated H3K9 residues (Liu et al., 2013) (Fig. 5B). Accordingly, loss of G9A leads to globally reduced 5mC levels (Dong et al., 2008; Myant et al., 2011). Although H3K9me3 levels are not changed in the S/L to 2i transition, H3K9me2 levels are significantly reduced, possibly underlying impaired recruitment of UHRF1 and subsequently DNMT1 (Von Meyenn et al., 2016b). In addition, expression of PRAMEL7, a member of the preferentially expressed antigen in melanoma (PRAME) family, in S/L ESCs was shown to induce a naïve-like ESC expression signature, which was dependent on its induction of proteosomal degradation of UHRF1, resulting in DNA hypomethylation (Graf et al., 2017).
Together, these data show that a reduction in 5mC maintenance rather than active depletion likely underlies the global DNA hypomethylation status associated with naïve ESCs (Fig. 5C). Furthermore, although these results highlight UHRF1 and DNMT1 as central players in regulating the difference in DNA methylation between naïve and S/L-cultured ESCs, an increase in their levels is unlikely to fully explain the gain of DNA methylation that is observed during differentiation of naïve ESCs to EpiLCs or EpiSCs. Future studies should address the epistasis of de novo versus maintenance methylation during ESC differentiation. In addition, it will be important to address how these global methylation changes affect the activity of individual regulatory elements and which other regulatory factors cooperate with DNMTs to modulate DNA methylation and subsequently gene expression. In this context, a recent study demonstrated that promoter, genic and intergenic CGIs retain low levels of DNA methylation in the transition from naïve to formative pluripotency, whereas intergenic and repeat elements followed the global trend of gained methylation (Von Meyenn et al., 2016a). DNA methylation is also thought to contribute to the increased transcriptional heterogeneity characteristic of S/L-cultured ESCs, specifically by increasing the fraction of cells with reduced levels of naïve ESC-specific TFs REX1 (ZFP42) and estrogen related receptor beta (ESRRB) (Singer et al., 2014).
TET-mediated DNA demethylation
DNA methylation is enzymatically reversed in a stepwise oxidation process. The enzymes ten-eleven translocation methylcytosine dioxygenase 1 and 2 (TET1 and TET2) catalyze the iterative oxidation of 5mC to 5-carbonylcytosine (5caC) via 5-hydroxymethylcytosine (5hmC) and 5-formylcytosine (5fC) intermediates (Kohli and Zhang, 2013) (Fig. 5A). Subsequently, thymine DNA glycosylase (TDG) excises 5fC and 5caC, generating an abasic site that is restored to a cytosine through the base excision repair pathway. Alternatively, 5hmC can be removed by proliferative dilution.
TET1 and TET2 are highly expressed in S/L-cultured and naïve ESCs (Dawlaty et al., 2011; Koh et al., 2011). Although complete loss of TET activity is compatible with self-renewal in S/L-cultured cells, TET activity is essential for proper regulation of gene expression during ESC differentiation (Dawlaty et al., 2014). Recent evidence suggests a more complex scenario in ESCs whereby TET2 promotes expression of naïve ESC genes and TET1 promotes EpiLC-enriched genes (Fig. 5C) (Fidalgo et al., 2016). This switch in TET requirement was shown to depend on the zinc-finger TF ZFP281, which can recruit TET1 and repress TET2 indirectly through a microRNA. Another study demonstrated that TET1/2 could serve as transcriptional activators alongside core pluripotency factors in naïve ESCs (Finley et al., 2018). Future studies are needed to address potential opposing roles of TET1/2 in the transition between different pluripotency states, what factors recruit TET2 and how this links to DNA methylation at bound regulatory elements.
Nucleosome remodeling
Nucleosome remodeling is the energy-dependent process by which histones, and hence nucleosomes, are repositioned along the chromatin fiber. Frequently, nucleosome remodeling results in increased chromatin accessibility, a feature that facilitates the binding of sequence-specific factors (Kadoch and Crabtree, 2015). However, a recent study challenges the view that chromatin accessibility indicates the local depletion of nucleosomes (Voong et al., 2016). Using a chemical mapping technique, this study revealed that active regulatory elements in ESCs that were previously designated as nucleosome-depleted, including those bound by OCT4 and CCCTC-binding factor (CTCF), retain nucleosomes that are loosely bound or fragile. Although the link between nucleosome occupancy and chromatin accessibility requires further investigation, it is clear that nucleosome remodeling remains an important regulatory mechanism in pluripotent and differentiating ESCs, and involves a number of remodeling factors.
The SWI/SNF family
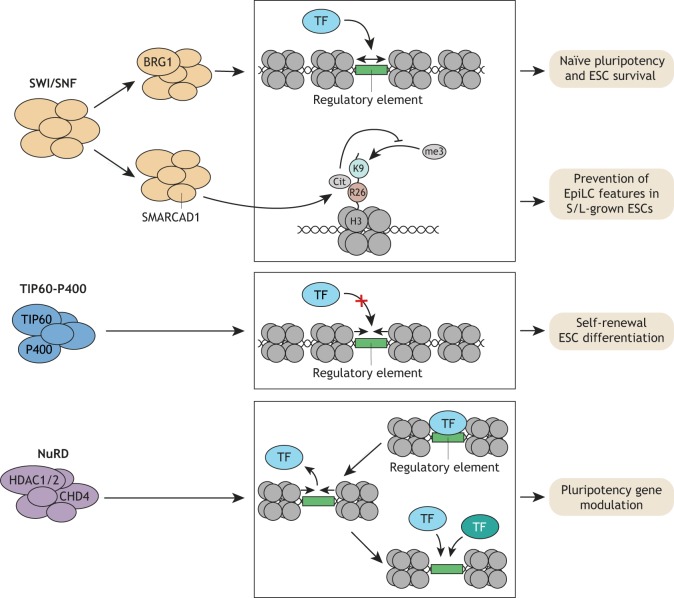
The ATPase-dependent SWI/SNF (or BAF) nucleosome remodeling complex is a large multi-subunit complex that is thought to facilitate the binding of sequence-specific factors by evicting nucleosomes and thereby increasing chromatin accessibility (Kadoch and Crabtree, 2015). The catalytic subunit of the ESC-expressed SWI/SNF complex BRG1 (SMARCA4), is essential for maintaining pluripotency (Ho et al., 2009) and is strongly enriched at pluripotency TF binding sites, at which it was recently shown to be required together with topoisomerase II for the recruitment of pluripotency TFs (Miller et al., 2017) (Fig. 6). An unexpected role of another embryonic tissue-enriched SWI/SNF family member SMARCAD1 was discovered in a screen for factors binding deiminated histone 3 arginine 26 (H3R26Cit), a mark important for early embryonic development (Christophorou et al., 2014). SMARCAD1 is enriched in S/L-cultured ESCs compared with EpiSCs and its depletion results in the appearance of some EpiSC features (Xiao et al., 2017). SMARCAD1 chromatin binding and H3R26Cit counteract H3K9me3 deposition; however, it is unclear how this mechanism links to gene expression changes observed upon SMARCAD1 depletion and to what degree nucleosome remodeling is involved. These results highlight SWI/SNF nucleosome remodeling complexes as integral parts of pluripotency TF recruitment and maintenance of pluripotency states (Fig. 6). Future studies are needed to identify the subunit composition of these complexes, the factors regulating their activity and localization, and their exact mechanism of action.
Fig. 6.
Nucleosome remodeling complexes in ESCs. Three major families of nucleosome remodeling complexes function in ESCs. The SWI/SNF complex member BRG1 promotes the recruitment of pluripotency TFs and is therefore required for naïve pluripotency and ESC survival. SMARCAD1 links SWI/SNF complexes to deimination of H3R26 and repression of H3K9me3. SMARCAD1 prevents acquisition of EpiLC features. The TIP60-P400 complex counteracts TF recruitment by decreasing chromatin accessibility, and thereby is required for ESC self-renewal and differentiation. Finally, the NuRD complex can transiently decrease chromatin accessibility at transcriptionally active promoters, which allows for reestablishment of altered TF binding patterns. NuRD modulates pluripotency gene expression under naïve conditions and during exit from naïve pluripotency.
The TIP60-P400 complex
The TIP60-P400 complex, which is part of the INO80 family, exhibits lysine acetyltransferase (KAT) and ATPase-dependent nucleosome remodeling activities. In ESCs, it is largely dispensable for transcriptional activation and appears rather to repress developmental genes (Chen et al., 2013; Fazzio et al., 2008). A recent paper described KAT/ATPase-dependent and -independent functions of TIP60-P400 in ESCs (Acharya et al., 2017). Specifically, although TIP60-P400 proteins are essential for self-renewal, their catalytic activities appear to be surprisingly dispensable. Instead, TIP60-P400 occupancy at promoter proximal sites reduces chromatin accessibility resulting in gene repression (Fig. 6). TIP60 KAT activity is, however, required for proper ESC differentiation, acting to regulate the timing of mesodermal and endodermal marker expression (Fig. 6). Future studies are needed to decipher the molecular mechanism of how catalytic and non-catalytic functions of TIP60/P400 impact gene expression and chromatin accessibility in such an opposing manner during ESC self-renewal and differentiation.
The NuRD complex
The nucleosome remodeling and deacetylation (NuRD) complex acts in many cellular contexts as a repressor. It is composed of enzymatic and non-enzymatic subunits. The two enzymatic activities are lysine deacetylation activity mediated via HDAC1/2, and SWI/SNF-type ATPase-based nucleosome remodeling activity mediated via chromodomain helicase DNA-binding protein 4 (CHD4). The non-enzymatic subunits include the general histone chaperones RBBP4/7, as well as proteins that are specific to NuRD, namely GATAD2A/B, MTA proteins (MTA1,2 and/or 3), CDK2AP1 and MBD2/3. A recent study in which the NuRD subunit MBD3 was reintroduced into MBD3-depleted ESCs (via tamoxifen-dependent rapid nuclear translocation) suggests that NuRD has both activating and repressing gene modulatory functions in ESCs (Bornelöv et al., 2018). Interestingly, this restoration of NuRD activity results in enriched binding at active promoters and enhancers, where it temporarily decreases chromatin accessibility, displaces Pol II and reduces nascent transcription. As a result, pluripotency TFs are temporarily displaced at a subset of sites and they then rebind to different degrees (Fig. 6). This change could potentially account for the active or repressive gene modulation seen in response to NuRD restoration. During exit from naïve pluripotency, NuRD-dependent changes in nucleosome and Pol II occupancy are associated with differential binding of the pluripotency TFs KLF4, ZFP42 and OTX2 to select enhancers and expression of their cognate genes (Bornelöv et al., 2018). Collectively, these data imply a role for NuRD-dependent nucleosome remodeling in resetting TF binding at active regulatory elements, allowing a transcriptional state permissive for activation or repression to evolve (Fig. 6). Although more studies are needed to demonstrate whether this proposed function of NuRD occurs in other cellular contexts, it suggests that nucleosome remodeling may be continuously involved in modulating the activity of regulatory elements in ESCs.
Chromatin looping
Chromatin exhibits a dynamic structure that enables interactions between distal elements via the formation of loops. Some of these loops occur stereotypically across development and across species, as in the case of topologically associated domains (TADs), whereas others exhibit cell-type specificity such as super enhancers in contact with their target gene promoters. A major factor involved in these loops is the cohesin complex, a ring-like multi-subunit complex that is essential for sister chromatid cohesion through its capability to topologically enclose two chromatin fibers (Merkenschlager and Odom, 2013). A subset of cohesin sites is co-occupied by the Mediator complex, which is a global regulator of Pol II-dependent transcription (Allen and Taatjes, 2015). Although both cohesin and the Mediator complex are essential for chromatin architecture and pluripotency gene expression in ESCs (Kagey et al., 2010), a number of studies have highlighted additional factors that play a role.
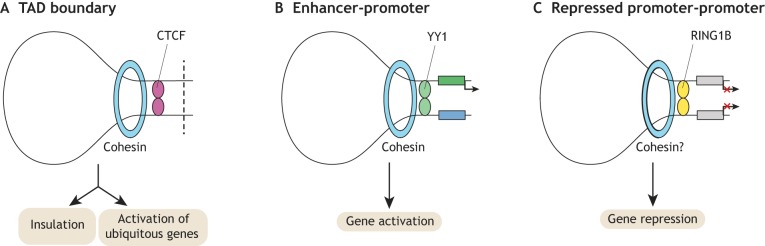
CTCF
The dimerizing sequence-specific factor CTCF is significantly enriched at TAD boundaries and occupies a large subset of cohesin-bound sites (Fig. 7A). Acute depletion of CTCF in ESCs using auxin-induced degron systems triggers rapid loss of TAD insulation (Nora et al., 2017), without a loss of higher order chromosome folding into open and closed compartments (A/B compartments). Importantly, loss of TAD insulation results in prominent changes in gene expression, with upregulated genes more likely to be within the same TAD and the majority of downregulated genes having a CTCF binding site nearby. Interestingly, even though CTCF binding can be inhibited by DNA methylation (Phillips and Corces, 2009), a recent study demonstrated that the absence of DNA methylation in ESCs does not alter global CTCF binding, TAD structure or higher order chromatin folding (Nothjunge et al., 2017). Thus, other factors likely collaborate with CTCF to allow for the precise establishment of TAD boundaries and higher order chromatin folding in ESCs. Future studies should aim to identify these factors.
Fig. 7.
Chromatin looping factors in ESCs. Three factors were recently shown to be responsible, together with cohesin, for the formation of distinct chromatin loops in ESCs. (A,B) CTCF and YY1 both function together with cohesin in the formation of largely distinct loops, TAD boundaries (A) and enhancer-promoter loops (B), respectively. TAD boundaries are required for insulation of TADs and activation of ubiquitously expressed genes. Enhancer-promoter loops regulate gene transcription more directly. (C) RING1B plays a role in controlling contacts between repressed promoters, thereby contributing to repression of cognate genes.
YY1
The specific enrichment of CTCF at TAD boundaries raised the question of whether there is an analogous dimerizing factor mediating enhancer-promoter loops. Two recent studies identified the TF YY1 as such a factor, demonstrating that YY1 binding at promoters and enhancers is necessary for looping and proper cognate gene expression in naïve ESCs and during ESC to NPC differentiation (Beagan et al., 2017; Weintraub et al., 2017). The fact that ubiquitously expressed YY1 occupies only a subset of all its potential motifs at enhancers and promoters in ESCs suggests that other factors such as lineage-dependent TFs may promote local accessibility of YY1 to specific regulatory elements to shape cell type-specific gene expression. These results highlight YY1 as a novel defining feature of enhancer-promoter loops in addition to cohesin and Mediator (Fig. 7B).
PRC1/2
A recent unbiased screen (using promoter-capture HiC) for regulatory elements contacting promoters through long-range interactions in ESCs revealed an enrichment for PRC1-bound promoters (Schoenfelder et al., 2015a). Given the positive reinforcement of PRC1 and PRC2 binding, these results are in agreement with previous results implicating the PRC2 core subunit EED in the formation of distinct chromatin interaction domains in ESCs (Denholtz et al., 2013). Subsequent analyses in S/L-cultured ESCs showed that promoters of all four HOX gene clusters and early developmental TF genes are enriched among these PRC1-bound interaction sites (Schoenfelder et al., 2015b). Removal of RING1A and RING1B results in dissolution of these interactions (Fig. 7C). Almost all PRC1-bound interaction sites also show PRC2 occupancy. However, in comparison with RING1B loss, most of the tested HOX gene cluster interactions remained detectable, albeit reduced, upon EED depletion. Similarly, a previous study showed a reduction but no loss of long-range interactions of HOX gene clusters upon EED loss (Joshi et al., 2015). RING1B-bound HOX gene clusters are enriched for contacts with poised enhancers, many of which are also occupied by RING1B. Surprisingly though, and in contrast to RING1B-bound promoter-promoter interactions, these promoter-enhancer interactions remain intact in the absence of RING1A/B (Schoenfelder et al., 2015b). At the level of transcription, loss of RING1B-bound promoter-promoter contacts results in upregulation of cognate genes without a significant gain of contacts with active enhancers. Similarly, the RING1B-bound genes in contact with poised enhancers get upregulated upon RING1A/B deletion, alongside a gain in active enhancer marks (Schoenfelder et al., 2015b). A candidate factor that may restrict and shape PRC-dependent long-range interactions could be MLL2; a recent study showed that loss of MLL2 results in increased PRC1/2 occupancy at bivalent promoters and redistribution of their long-range interactions, including those at HOX genes (Mas et al., 2018). Together, these studies highlight PRC1, PRC2 and MLL2 as central players in the formation and maintenance of defined long-range promoter-promoter interactions, which are enriched at promoters of most HOX gene clusters and other developmental genes. Future studies should address whether the formation of such PRC-dependent promoter-promoter contacts are a general feature of PRC-mediated gene repression in ESCs and subsequent stages of development.
A recent study also discovered that some of the poised enhancers in ESCs are near genes that gain activity in anterior neural progenitors (AntNPCs), correlating with accumulation of H3K27ac at those enhancers (Cruz-Molina et al., 2017). Using circularized chromosome conformation capture (4C) analysis, this study showed that those enhancers are looped to their target gene promoters in both ESCs and AntNPCs. The deletions of selected poised enhancers, or of EED, results in failure to induce target genes in AntNPCs. In addition, 4C analysis of EED-null ESCs suggests a requirement for PRC2 for the looping of these poised enhancers to target promoters. CG-rich motifs are overrepresented in poised versus active enhancers, and deletion of selected poised enhancers harboring such elements results in locally diminished H3K27me3 levels. These results implicate PRC2 in the formation of poised enhancer-promoter contacts, which become active enhancer-promoter loops later in development. However, given the broad epigenetic changes upon EED deletion, it is not clear whether looping of the selected poised enhancer elements in ESCs is required for the activation of their cognate genes in AntNPCs. Similar to PRC2, PRC1 is enriched at poised enhancers (Cruz-Molina et al., 2017), however its requirement for AntNPC target gene induction and loop formation was not tested. Overall, these results extend the physiologically relevant spectrum of PRC2-dependent long-range interactions to poised enhancer-promoter contacts. These studies highlight that inactive regulatory elements adapt complex looped 3D structures in ESCs that might be permissive of cell type-specific gene expression upon differentiation cues.
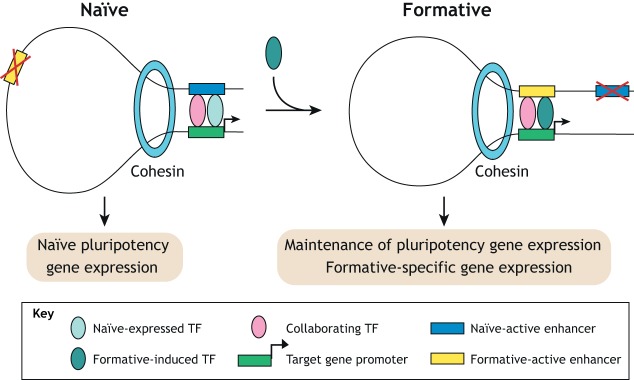
Enhancer switching
Marked changes in enhancer activity occur in the transition from naïve to formative pluripotency, with comparably fewer pronounced changes in cognate gene expression (Fig. 8). TFs are essential regulators of enhancer activity and are likely involved in this rewiring of enhancer networks. An overview of these changes was provided by a study of core pluripotency factors (Galonska et al., 2015). This study revealed that exiting naïve pluripotency is associated with widespread differential OCT4, SOX2 and NANOG binding, which correlates with differential enhancer activity at selected sites. Mechanistically, reorganization of OCT4 enhancer binding depends on collaboration with the EpiLC-specific TF OTX2 (Buecker et al., 2014). Even in the absence of differentiation cues, ectopic OTX2 expression is sufficient to redirect OCT4 to EpiLC-specific sites, highlighting the context-dependent nature of TF enhancer selection. Similarly, another study demonstrated that GRHL2, a TF that is upregulated in EpiLCs, activates new enhancers upon exit of naïve pluripotency to maintain, not alter, expression of a subset of expressed ESC genes (Chen et al., 2018). These same genes are regulated by distinct TFs at alternative enhancers in the naïve ESC state. Given that GRHL2 is involved in downstream differentiation to epithelial lineages, its maintenance of expression of a subset of ground-state pluripotency genes could contribute to a network that is permissive to cues driving epithelial fate as EpiLCs differentiate and diversify into the germ layers.
Fig. 8.
Enhancer switching between ESC pluripotency states. The transition from naïve to formative pluripotency is associated with numerous changes in enhancer activity, TF and cohesin occupancy. Naïve-expressed TFs collaborate to drive naïve-specific gene expression. Upon exit from naïve pluripotency they can be redirected through collaboration with formative-specific TFs to activate other enhancers and induce formative-specific genes or contribute to maintenance of expression of certain pluripotency genes.
Together, these findings highlight that, in general, lineage-specifying and signal-dependent TFs are likely involved in the rewiring of pluripotency enhancer-promoter networks, creating a transcriptional environment permissive to lineage diversification.
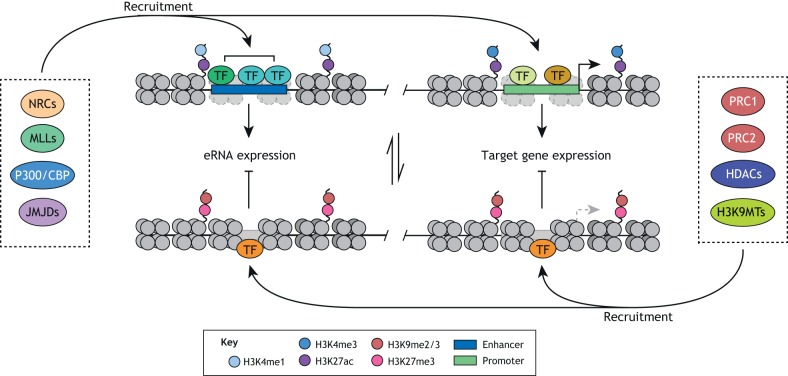
Crosstalk between transcription factors and chromatin-modifying enzymes
TFs are sequence-specific DNA-binding proteins that can recruit chromatin-modifying enzymes and thus create enhancer and promoter states that are either permissive or repressive for transcription (Fig. 9). For example, NANOG-dependent recruitment of TET1 enhances establishment of pluripotency during reprogramming (Costa et al., 2013). A subset of TFs named ‘pioneer factors’ are thought to bind nucleosome-bound DNA and facilitate the binding of other collaborating TFs and chromatin remodeling enzymes (Long et al., 2016). An exemplar pioneer factor is the core pluripotency factor OCT4. Indeed, the binding of OCT4 to DNA is required for the binding of BRG1, the catalytical subunit of the SWI/SNF chromatin-remodeling complex, resulting in open chromatin at co-bound sites (King and Klose, 2017). These open chromatin conditions are required for the binding of other pluripotency TFs including SOX2 and NANOG. Similarly, the binding of FOXD3, a TF essential for early embryonic development (Hanna et al., 2002), increases chromatin accessibility at its newly bound sites in EpiLCs, potentially through its interaction with BRG1 (Krishnakumar et al., 2016). FOXD3 also interacts with HDAC1/2 and LSD1, suggesting it can recruit these factors to simultaneously suppress its target genes (Krishnakumar et al., 2016; Respuela et al., 2016).
Fig. 9.
Regulation of enhancer and promoter activity through TF-mediated recruitment of chromatin-modifying enzymes. Collaborative binding of multiple TFs creates an environment favoring the recruitment of chromatin-modifying enzymes. Enzymes catalyzing modifications that correlate with enhancer and gene activity include nucleosome remodeling complexes (NRCs), MLLs, P300/CBP and JMJDs. Enzymes catalyzing modifications that correlate with enhancer and gene repression include PRC1/2, HDACs and H3K9 methyltransferases (H3K9MTs). Active enhancers and promoters typically correlate with local chromatin accessibility and H3K27ac signal. In addition, active enhancers and promoters carry H3K4me1 and H3K4me3, respectively. Inactive enhancers and promoters typically lose chromatin accessibility and gain H3K9 and H3K27 methylation.
Alternatively, TFs can promote silencing of specific loci. Activity-dependent neuroprotective protein (ADNP), a protein associated with neurological disorders, was recently shown to be essential for naïve pluripotency (Ostapcuk et al., 2018). This function is based on its sequence-specific binding and recruitment of heterochromatin protein 1 (HP1, also known as CBX5) and CHD4 to euchromatic H3K9me3-depleted regions, leading to diminished chromatin accessibility and cognate gene expression. These data highlight the necessity of TFs to actively suppress chromatin accessibility, thereby preventing premature expression of lineage-specifying genes. Another unexpected mechanism of TF regulation by epigenetic enzymes was demonstrated by a recent study showing the requirement of lysyl oxidase-like 2 (LOXL2)-dependent oxidation of lysine residues in the transcription initiation factor TFIID subunit TAF10 during ESC differentiation (Iturbide et al., 2015). In this context, oxidation results in degradation of TAF10, release of core TFIID complex members from core pluripotency promoters and, consequently, decreased transcription.
Conclusions and future directions
Transcriptional regulation of maintenance and exit of pluripotency exemplifies the requirement of the interplay between sequence-specific TFs and chromatin-modifying enzymes to regulate gene activation and repression. This interplay is often transient and multivalent, requiring experimental approaches with appropriate temporal and molecular resolution in order to dissect and characterize outstanding key questions (Box 1). A prerequisite to reliably apply such approaches is to account for transcriptional heterogeneity derived from cell-to-cell variation and variation between culture conditions. The extent of this heterogeneity can be addressed by single cell analyses. For example, single cell RNA sequencing shows that S/L-cultured ESCs have a higher degree of transcriptional heterogeneity than 2i-cultured ESCs (Guo et al., 2016). Therefore, it is hard to interpret whether differences in population-based assays using S/L culture conditions co-occur in the same cells or occur in distinct subsets of cells. Although transcriptional heterogeneity might have important functions during early embryonic development, such as during the naïve-to-formative pluripotency transition, 2i-cultured ESCs correlate better with the transcriptome of early epiblast cells, making 2i a more biologically meaningful culture method (Hayashi et al., 2011; Nakamura et al., 2016). However, long-term culture in 2i can lead to a loss of imprints (Choi et al., 2017; Yagi et al., 2017). Similarly, EpiLCs are more similar to late epiblast cells than EpiSCs are (Hayashi et al., 2011). In general, the field would profit from more standardized culture conditions (Mulas et al., 2019) that would facilitate data comparisons across multiple studies. The ultimate goal of these culture models should be to develop truly physiologically relevant models of epigenetic control of transcriptional regulation that better inform our understanding of pluripotency and early cell fate decisions in the embryo.
Box 1. Key questions.
Is MLL-dependent H3K4me1 and P300/CBP-dependent H3K27ac required for maintenance of active ESC enhancers and/or de novo enhancer activation during differentiation?
What is the impact of differential DNA methylation and PRC activity on the activity of enhancer and gene sets, and what is the link to maintenance and exit of pluripotency?
What is the epistatic relationship of nucleosome remodeling, histone and DNA modifications, and TF binding during de novo enhancer activation and decommissioning of active enhancers?
How do changes in chromatin architecture link to transcriptional activity in pluripotency and differentiation, and which factors are involved?
Besides more standardized culture conditions, several recent technological advances have proven suitable to address the mechanisms underlying the dynamic interplay of chromatin structure and transcriptional regulation. These include implementations of single cell-based analyses of chromatin accessibility, structure and epigenetic marks (reviewed by Ludwig and Bintu, 2019), which could be applied to early stages of ESC differentiation. In addition, tools such as the auxin-inducible degron system allow for increased temporal resolution and have been fundamental in highlighting the essential structural role of CTCF in TAD boundary maintenance (Nora et al., 2017). Such tools could be applied to analyze other complexes that, when deleted, are associated with deleterious phenotypes, such as the SWI/SNF complex. In addition, multi-subunit complexes with changing subunit composition and no apparent sequence specificity are frequently localized to defined loci, raising the question of what provides the specificity. Locus-specific mass spectrometry approaches such as ChIP-SICAP or CAPTURE have proven to be valuable in specifically identifying novel factors and complex compositions associated with known chromatin-associated factors (Liu et al., 2017; Rafiee et al., 2016). In addition, imaging technologies such as live-cell super-resolution imaging and electron tomography are valuable to resolve the structural dynamics associated with different ESC states (Cho et al., 2018; Ou et al., 2017).
Overall, the combination of optimized culture conditions with the rapidly evolving technologies for chromatin structure analysis makes the ESC culture system ideal to address the impact of transcriptional control, epigenetic modifications and nuclear architecture on developmental cell fate decisions.
Acknowledgements
We thank Ryan Boileau, Bryan Marsh and Brian DeVeale for critical reading of the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
D.G. is supported by a research fellowship from the Deutsche Forschungsgemeinschaft and R.B. is supported by grants from the National Institutes of Health (R01GM125089-01 and R01GM122439-01). Deposited in PMC for release after 12 months.
References
- Acharya D., Hainer S. J., Yoon Y., Wang F., Bach I., Rivera-Pérez J. A. and Fazzio T. G. (2017). KAT-independent gene regulation by Tip60 promotes ESC self-renewal but not pluripotency. CELREP 19, 671-679. 10.1016/j.celrep.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen B. L. and Taatjes D. J. (2015). The mediator complex: a central integrator of transcription. Nature Publishing Group 16, 155-166. 10.1038/nrm3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagan J. A., Duong M. T., Titus K. R., Zhou L., Cao Z., Ma J., Lachanski C. V., Gillis D. R. and Phillips-Cremins J. E. (2017). YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res. 27, 1139-1152. 10.1101/gr.215160.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge N. P., Farcas A. M., Kondo T., King H. W., McGouran J. F., Hanssen L. L. P., Ito S., Cooper S., Kondo K., Koseki Y. et al. (2014). Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157, 1445-1459. 10.1016/j.cell.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornelöv S., Reynolds N., Xenophontos M., Gharbi S., Johnstone E., Floyd R., Ralser M., Signolet J., Loos R., Dietmann S. et al. (2018). The nucleosome remodeling and deacetylation complex modulates chromatin structure at sites of active transcription to fine-tune gene expression. Mol. Cell 71, 56-72.e4. 10.1016/j.molcel.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M., Kim J. K., Estève P.-O., Clark A., Pradhan S. and Jacobsen S. E. (2007). UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760-1764. 10.1126/science.1147939 [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K. et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature Publishing Group 441, 349-353. 10.1038/nature04733 [DOI] [PubMed] [Google Scholar]
- Braun S. M. G., Kirkland J. G., Chory E. J., Husmann D., Calarco J. P. and Crabtree G. R. (2017). Rapid and reversible epigenome editing by endogenous chromatin regulators. Nat Comms 8, 560 10.1038/s41467-017-00644-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buecker C., Srinivasan R., Wu Z., Calo E., Acampora D., Faial T., Simeone A., Tan M., Swigut T. and Wysocka J. (2014). Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 14, 838-853. 10.1016/j.stem.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K., Collings C. K., Morgan M. A., Marshall S. A., Rendleman E. J., Ozark P. A., Smith E. R. and Shilatifard A. (2018). An Mll4/COMPASS-Lsd1 epigenetic axis governs enhancer function and pluripotency transition in embryonic stem cells. Sci. Adv. 4, eaap8747 10.1126/sciadv.aap8747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain S. J., Yee D. and Magnuson T. (2008). Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 26, 1496-1505. 10.1634/stemcells.2008-0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. B., Hung J.-H., Hickman T. L., Coles A. H., Carey J. F., Weng Z., Chu F. and Fazzio T. G. (2013). Hdac6 regulates Tip60-p400 function in stem cells. Elife 2, e01557 10.7554/eLife.01557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A. F., Liu A. J., Krishnakumar R., Freimer J. W., DeVeale B. and Blelloch R. (2018). GRHL2-dependent enhancer switching maintains a pluripotent stem cell transcriptional subnetwork after exit from naive pluripotency. Cell Stem Cell 23, 226-238.e4. 10.1016/j.stem.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W.-K., Spille J.-H., Hecht M., Lee C., Li C., Grube V. and Cisse I. I. (2018). Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412-415. 10.1126/science.aar4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Huebner A. J., Clement K., Walsh R. M., Savol A., Lin K., Gu H., Di Stefano B., Brumbaugh J., Kim S.-Y. et al. (2017). Prolonged Mek1/2 suppression impairs the developmental potential of embryonic stem cells. Nature 548, 219-223. 10.1038/nature23274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou M. A., Castelo-Branco G., Halley-Stott R. P., Oliveira C. S., Loos R., Radzisheuskaya A., Mowen K. A., Bertone P., Silva J. C. R., Zernicka-Goetz M. et al. (2014). Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature Publishing Group 507, 104-108. 10.1038/nature12942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos P. A. C., Christensen J., Agger K. and Helin K. (2008). Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 22, 1115-1140. 10.1101/gad.1652908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S., Dienstbier M., Hassan R., Schermelleh L., Sharif J., Blackledge N. P., De Marco V., Elderkin S., Koseki H., Klose R. et al. (2014). Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. CELREP 7, 1456-1470. 10.1016/j.celrep.2014.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y., Ding J., Theunissen T. W., Faiola F., Hore T. A., Shliaha P. V., Fidalgo M., Saunders A., Lawrence M., Dietmann S. et al. (2013). NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature Publishing Group 495, 370-374. 10.1038/nature11925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Molina S., Respuela P., Tebartz C., Kolovos P., Nikolić M., Fueyo R., van Ijcken W. F. J., Grosveld F., Frommolt P., Bazzi H. et al. (2017). PRC2 facilitates the regulatory topology required for poised enhancer function during pluripotent stem cell differentiation. Cell Stem Cell 20, 689-705.e9. 10.1016/j.stem.2017.02.004 [DOI] [PubMed] [Google Scholar]
- Dawlaty M. M., Ganz K., Powell B. E., Hu Y.-C., Markoulaki S., Cheng A. W., Gao Q., Kim J., Choi S.-W., Page D. C. et al. (2011). Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166-175. 10.1016/j.stem.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty M. M., Breiling A., Le T., Barrasa M. I., Raddatz G., Gao Q., Powell B. E., Cheng A. W., Faull K. F., Lyko F. et al. (2014). Loss of tet enzymes compromises proper differentiation of embryonic stem cells. Dev. Cell 29, 102-111. 10.1016/j.devcel.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denholtz M., Bonora G., Chronis C., Splinter E., de Laat W., Ernst J., Pellegrini M. and Plath K. (2013). Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell 13, 602-616. 10.1016/j.stem.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L. and Helin K. (2013). Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20, 1147-1155. 10.1038/nsmb.2669 [DOI] [PubMed] [Google Scholar]
- Dong K. B., Maksakova I. A., Mohn F., Leung D., Appanah R., Lee S., Yang H. W., Lam L. L., Mager D. L., Schübeler D. et al. (2008). DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 27, 2691-2701. 10.1038/emboj.2008.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorighi K. M., Swigut T., Henriques T., Bhanu N. V., Scruggs B. S., Nady N., Still C. D. II, Garcia B. A., Adelman K. and Wysocka J. (2017). Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol. Cell 66, 568-576.e4. 10.1016/j.molcel.2017.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressel U., Bailey P. J., Wang S.-C., Downes M., Evans R. M. and Muscat G. E. (2001). A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J. Biol. Chem. 276, 17007-17013. 10.1074/jbc.M101508200 [DOI] [PubMed] [Google Scholar]
- Efroni S., Duttagupta R., Cheng J., Dehghani H., Hoeppner D. J., Dash C., Bazett-Jones D. P., Le Grice S., McKay R. D. G., Buetow K. H. et al. (2008). Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2, 437-447. 10.1016/j.stem.2008.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M., Endo T. A., Endoh T., Fujimura Y.-I., Ohara O., Toyoda T., Otte A. P., Okano M., Brockdorff N., Vidal M. et al. (2008). Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 135, 1513-1524. 10.1242/dev.014340 [DOI] [PubMed] [Google Scholar]
- Endoh M., Endo T. A., Shinga J., Hayashi K., Farcas A., Ma K.-W., Ito S., Sharif J., Endoh T., Onaga N. et al. (2017). PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. Elife 6, 1513 10.7554/eLife.27970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio T. G., Huff J. T. and Panning B. (2008). An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134, 162-174. 10.1016/j.cell.2008.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari K. J., Scelfo A., Jammula S., Cuomo A., Barozzi I., Stützer A., Fischle W., Bonaldi T. and Pasini D. (2014). Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol. Cell 53, 49-62. 10.1016/j.molcel.2013.10.030 [DOI] [PubMed] [Google Scholar]
- Ficz G., Hore T. A., Santos F., Lee H. J., Dean W., Arand J., Krueger F., Oxley D., Paul Y.-L., Walter J. et al. (2013). FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Stem Cell 13, 351-359. 10.1016/j.stem.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo M., Huang X., Guallar D., Sánchez-Priego C., Valdes V. J., Saunders A., Ding J., Wu W.-S., Clavel C. and Wang J. (2016). Zfp281 coordinates opposing functions of Tet1 and Tet2 in pluripotent states. Cell Stem Cell 19, 355-369. 10.1016/j.stem.2016.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley L. W. S., Vardhana S. A., Carey B. W., Alonso-Curbelo D., Koche R., Chen Y., Wen D., King B., Radler M. R., Rafii S. et al. (2018). Pluripotency transcription factors and Tet1/2 maintain Brd4-independent stem cell identity. Nat. Cell Biol. 20, 565-574. 10.1038/s41556-018-0086-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galonska C., Ziller M. J., Karnik R. and Meissner A. (2015). Ground state conditions induce rapid reorganization of core pluripotency factor binding before global epigenetic reprogramming. Cell Stem Cell 17, 462-470. 10.1016/j.stem.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearhart M. D., Corcoran C. M., Wamstad J. A. and Bardwell V. J. (2006). Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell. Biol. 26, 6880-6889. 10.1128/MCB.00630-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf U., Casanova E. A., Wyck S., Dalcher D., Gatti M., Vollenweider E., Okoniewski M. J., Weber F. A., Patel S. S., Schmid M. W. et al. (2017). Pramel7 mediates ground-state pluripotency through proteasomal-epigenetic combined pathways. Nat. Cell Biol. 19, 763-773. 10.1038/ncb3554 [DOI] [PubMed] [Google Scholar]
- Guo G., Pinello L., Han X., Lai S., Shen L., Lin T.-W., Zou K., Yuan G.-C. and Orkin S. H. (2016). Serum-based culture conditions provoke gene expression variability in mouse embryonic stem cells as revealed by single-cell analysis. CELREP 14, 956-965. 10.1016/j.celrep.2015.12.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi E., Brinkman A. B., Arand J., Kroeze L. I., Kerstens H. H. D., Matarese F., Lepikhov K., Gut M., Brun-Heath I., Hubner N. C. et al. (2013). Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 13, 360-369. 10.1016/j.stem.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Hackett J. A., Dietmann S., Murakami K., Down T. A., Leitch H. G. and Surani M. A. (2013). Synergistic mechanisms of DNA demethylation during transition to ground-state pluripotency. Stem Cell Reports 1, 518-531. 10.1016/j.stemcr.2013.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna L. A., Foreman R. K., Tarasenko I. A., Kessler D. S. and Labosky P. A. (2002). Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes Dev.. 16, 2650-2661. 10.1101/gad.1020502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Ohta H., Kurimoto K., Aramaki S. and Saitou M. (2011). Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146, 519-532. 10.1016/j.cell.2011.06.052 [DOI] [PubMed] [Google Scholar]
- Hermann A., Goyal R. and Jeltsch A. (2004). The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 279, 48350-48359. 10.1074/jbc.M403427200 [DOI] [PubMed] [Google Scholar]
- Ho L., Jothi R., Ronan J. L., Cui K., Zhao K. and Crabtree G. R. (2009). An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. USA 106, 5187-5191. 10.1073/pnas.0812888106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Højfeldt J. W., Laugesen A., Willumsen B. M., Damhofer H., Hedehus L., Tvardovskiy A., Mohammad F., Jensen O. N. and Helin K. (2018). Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat. Struct. Mol. Biol. 25, 225-232. 10.1038/s41594-018-0036-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D., Gao X., Morgan M. A., Herz H. M., Smith E. R. and Shilatifard A. (2013). The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell. Biol. 33, 4745-4754. 10.1128/MCB.01181-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth R. S., Moffat M., Mann A. R., Read D., Hunter C. J., Pradeepa M. M., Adams I. R. and Bickmore W. A. (2015). The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev. 29, 1897-1902. 10.1101/gad.268151.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbide A., Pascual-Reguant L., Fargas L., Cebrià J. P., Alsina B., García de Herreros A. and Peiró S. (2015). LOXL2 oxidizes methylated TAF10 and controls TFIID-dependent genes during neural progenitor differentiation. Mol. Cell 58, 755-766. 10.1016/j.molcel.2015.04.012 [DOI] [PubMed] [Google Scholar]
- Jamaladdin S., Kelly R. D. W., O'Regan L., Dovey O. M., Hodson G. E., Millard C. J., Portolano N., Fry A. M., Schwabe J. W. R. and Cowley S. M. (2014). Histone deacetylase (HDAC) 1 and 2 are essential for accurate cell division and the pluripotency of embryonic stem cells. Proc. Natl. Acad. Sci. USA 111, 9840-9845. 10.1073/pnas.1321330111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi O., Wang S.-Y., Kuznetsova T., Atlasi Y., Peng T., Fabre P. J., Habibi E., Shaik J., Saeed S., Handoko L. et al. (2015). Dynamic reorganization of extremely long-range promoter-promoter interactions between two states of pluripotency. Cell Stem Cell 17, 748-757. 10.1016/j.stem.2015.11.010 [DOI] [PubMed] [Google Scholar]
- Juan A. H., Wang S., Ko K. D., Zare H., Tsai P.-F., Feng X., Vivanco K. O., Ascoli A. M., Gutierrez-Cruz G., Krebs J. et al. (2016). Roles of H3K27me2 and H3K27me3 examined during fate specification of embryonic stem cells. CELREP 17, 1369-1382. 10.1016/j.celrep.2016.09.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C. and Crabtree G. R. (2015). Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci. Adv. 1, e1500447 10.1126/sciadv.1500447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey M. H., Newman J. J., Bilodeau S., Zhan Y., Orlando D. A., van Berkum N. L., Ebmeier C. C., Goossens J., Rahl P. B., Levine S. S. et al. (2010). Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430-435. 10.1038/nature09380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R. D. W. and Cowley S. M. (2013). The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochm. Soc. Trans. 41, 741-749. 10.1042/BST20130010 [DOI] [PubMed] [Google Scholar]
- Kim M. and Costello J. (2017). DNA methylation: an epigenetic mark of cellular memory. Exp. Mol. Med. 49, e322-e322. 10.1038/emm.2017.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King H. W. and Klose R. J. (2017). The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. Elife 6, 380 10.7554/eLife.22631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloet S. L., Makowski M. M., Baymaz H. I., van Voorthuijsen L., Karemaker I. D., Santanach A., Jansen P. W. T. C., Di Croce L. and Vermeulen M. (2016). The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat. Struct. Mol. Biol. 23, 682-690. 10.1038/nsmb.3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K. P., Yabuuchi A., Rao S., Huang Y., Cunniff K., Nardone J., Laiho A., Tahiliani M., Sommer C. A., Mostoslavsky G. et al. (2011). Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8, 200-213. 10.1016/j.stem.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli R. M. and Zhang Y. (2013). TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472-479. 10.1038/nature12750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima Y., Kaufman-Francis K., Studdert J. B., Steiner K. A., Power M. D., Loebel D. A. F., Jones V., Hor A., de Alencastro G., Logan G. J. et al. (2014). The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell 14, 107-120. 10.1016/j.stem.2013.09.014 [DOI] [PubMed] [Google Scholar]
- Krishnakumar R., Chen A. F., Pantovich M. G., Danial M., Parchem R. J., Labosky P. A. and Blelloch R. (2016). FOXD3 regulates pluripotent stem cell potential by simultaneously initiating and repressing enhancer activity. Cell Stem Cell 18, 104-117. 10.1016/j.stem.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki S., Nakai Y., Maeda R., Okashita N., Akiyoshi M., Yamaguchi Y., Kitano S., Miyachi H., Nakato R., Ichiyanagi K. et al. (2018). Combined loss of JMJD1A and JMJD1B reveals critical roles for H3K9 demethylation in the maintenance of embryonic stem cells and early embryogenesis. Stem Cell Reports 10, 1340-1354. 10.1016/j.stemcr.2018.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-E., Wang C., Xu S., Cho Y.-W., Wang L., Feng X., Baldridge A., Sartorelli V., Zhuang L., Peng W. et al. (2013). H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife 2, 2817-2825. 10.7554/eLife.01503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-H., Holder M., Grau D., Saldaña-Meyer R., Yu J.-R., Ganai R. A., Zhang J., Wang M., LeRoy G., Dobenecker M.-W. et al. (2018). Distinct stimulatory mechanisms regulate the catalytic activity of polycomb repressive complex 2. Mol. Cell 70, 435-448.e5. 10.1016/j.molcel.2018.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Hart S. R. L. and Skalnik D. G. (2004). Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis 38, 32-38. 10.1002/gene.10250 [DOI] [PubMed] [Google Scholar]
- Leitch H. G., McEwen K. R., Turp A., Encheva V., Carroll T., Grabole N., Mansfield W., Nashun B., Knezovich J. G., Smith A. et al. (2013). Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 20, 311-316. 10.1038/nsmb.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Notani D. and Rosenfeld M. G. (2016). Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet. 17, 207-223. 10.1038/nrg.2016.4 [DOI] [PubMed] [Google Scholar]
- Li H., Liefke R., Jiang J., Kurland J. V., Tian W., Deng P., Zhang W., He Q., Patel D. J., Bulyk M. L. et al. (2017). Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 549, 287-291. 10.1038/nature23881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Gao Q., Li P., Zhao Q., Zhang J., Li J., Koseki H. and Wong J. (2013). UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat Comms 4, 1563 10.1038/ncomms2562 [DOI] [PubMed] [Google Scholar]
- Liu X., Zhang Y., Chen Y., Li M., Zhou F., Li K., Cao H., Ni M., Liu Y., Gu Z. et al. (2017). In situ capture of chromatin interactions by biotinylated dCas9. Cell 170, 1-36. 10.1016/j.cell.2017.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Local A., Huang H., Albuquerque C. P., Singh N., Lee A. Y., Wang W., Wang C., Hsia J. E., Shiau A. K., Ge K. et al. (2018). Identification of H3K4me1-associated proteins at mammalian enhancers. Nat. Genet. 50, 73-82. 10.1038/s41588-017-0015-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H. K., Prescott S. L. and Wysocka J. (2016). Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167, 1170-1187. 10.1016/j.cell.2016.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig C. and Bintu L. (2019). Mapping chromatin modifications at the single cell level. Development 146, dev170217 10.1242/dev.170217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., Li G., Sarma K., Blais A., Zavadil J., Woodcock C. L., Dynlacht B. D. and Reinberg D. (2008). Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 32, 503-518. 10.1016/j.molcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A. F., Smith A. et al. (2012). The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149, 590-604. 10.1016/j.cell.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas G., Blanco E., Ballaré C., Sansó M., Spill Y. G., Hu D., Aoi Y., Le Dily F., Shilatifard A., Marti-Renom M. A. et al. (2018). Promoter bivalency favors an open chromatin architecture in embryonic stem cells. Nat. Genet. 50, 1452-1462. 10.1038/s41588-018-0218-5 [DOI] [PubMed] [Google Scholar]
- Merkenschlager M. and Odom D. T. (2013). CTCF and cohesin: linking gene regulatory elements with their targets. Cell 152, 1285-1297. 10.1016/j.cell.2013.02.029 [DOI] [PubMed] [Google Scholar]
- Meshorer E., Yellajoshula D., George E., Scambler P. J., Brown D. T. and Misteli T. (2006). Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell 10, 105-116. 10.1016/j.devcel.2005.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E. L., Hargreaves D. C., Kadoch C., Chang C.-Y., Calarco J. P., Hodges C., Buenrostro J. D., Cui K., Greenleaf W. J., Zhao K. et al. (2017). TOP2 synergizes with BAF chromatin remodeling for both resolution and formation of facultative heterochromatin. Nat. Struct. Mol. Biol. 24, 344-352. 10.1038/nsmb.3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J., Zhang Y. and Xu R.-M. (2003). Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 17, 1823-1828. 10.1101/gad.269603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery N. D., Yee D., Chen A., Kalantry S., Chamberlain S. J., Otte A. P. and Magnuson T. (2005). The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr. Biol. 15, 942-947. 10.1016/j.cub.2005.04.051 [DOI] [PubMed] [Google Scholar]
- Morgan M. A. J., Rickels R. A., Collings C. K., He X., Cao K., Herz H.-M., Cozzolino K. A., Abshiru N. A., Marshall S. A., Rendleman E. J. et al. (2017). A cryptic Tudor domain links BRWD2/PHIP to COMPASS-mediated histone H3K4 methylation. Genes Dev. 31, 2003-2014. 10.1101/gad.305201.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulas C., Kalkan T., von Meyenn F., Leitch H. G., Nichols J. and Smith A. (2019). Defined conditions for propagation and manipulation of mouse embryonic stem cells. Development 146, dev173146 10.1242/dev.173146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myant K., Termanis A., Sundaram A. Y. M., Boe T., Li C., Merusi C., Burrage J., de Las Heras J. I. and Stancheva I. (2011). LSH and G9a/GLP complex are required for developmentally programmed DNA methylation. Genome Res. 21, 83-94. 10.1101/gr.108498.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Okamoto I., Sasaki K., Yabuta Y., Iwatani C., Tsuchiya H., Seita Y., Nakamura S., Yamamoto T. and Saitou M. (2016). A developmental coordinate of pluripotency among mice, monkeys and humans. Nature Publishing Group 537, 57-62. 10.1038/nature19096 [DOI] [PubMed] [Google Scholar]
- Nora E. P., Goloborodko A., Valton A.-L., Gibcus J. H., Uebersohn A., Abdennur N., Dekker J., Mirny L. A. and Bruneau B. G. (2017). Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930-933.e22. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothjunge S., Nührenberg T. G., Grüning B. A., Doppler S. A., Preissl S., Schwaderer M., Rommel C., Krane M., Hein L. and Gilsbach R. (2017). DNA methylation signatures follow preformed chromatin compartments in cardiac myocytes. Nat. Comms. 8, 1667 10.1038/s41467-017-01724-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D., Erhardt S., Pagani M., Barton S. C., Surani M. A. and Jenuwein T. (2001). The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21, 4330-4336. 10.1128/MCB.21.13.4330-4336.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M., Bell D. W., Haber D. A. and Li E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247-257. 10.1016/S0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- Ostapcuk V., Mohn F., Carl S. H., Basters A., Hess D., Iesmantavicius V., Lampersberger L., Flemr M., Pandey A., Thomä N. H. et al. (2018). Activity-dependent neuroprotective protein recruits HP1 and CHD4 to control lineage-specifying genes. Nature 557, 739-743. 10.1038/s41586-018-0153-8 [DOI] [PubMed] [Google Scholar]
- Ou H. D., Phan S., Deerinck T. J., Thor A., Ellisman M. H. and O'Shea C. C. (2017). ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357, eaag0025 10.1126/science.357.6346.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Bracken A. P., Jensen M. R., Lazzerini Denchi E. and Helin K. (2004). Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 23, 4061-4071. 10.1038/sj.emboj.7600402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Bracken A. P., Hansen J. B., Capillo M. and Helin K. (2007). The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 27, 3769-3779. 10.1128/MCB.01432-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen M. T., Agger K., Laugesen A., Johansen J. V., Cloos P. A. C., Christensen J. and Helin K. (2014). The demethylase JMJD2C localizes to H3K4me3-positive transcription start sites and is dispensable for embryonic development. Mol. Cell. Biol. 34, 1031-1045. 10.1128/MCB.00864-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen M. T., Kooistra S. M., Radzisheuskaya A., Laugesen A., Johansen J. V., Hayward D. G., Nilsson J., Agger K. and Helin K. (2016). Continual removal of H3K9 promoter methylation by Jmjd2 demethylases is vital for ESC self-renewal and early development. EMBO J. 35, 1550-1564. 10.15252/embj.201593317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perino M., van Mierlo G., Karemaker I. D., van Genesen S., Vermeulen M., Marks H., van Heeringen S. J. and Veenstra G. J. C. (2018). MTF2 recruits polycomb repressive complex 2 by helical-shape-selective DNA binding. Nat. Genet. 50, 1002-1010. 10.1038/s41588-018-0134-8 [DOI] [PubMed] [Google Scholar]
- Petruk S., Cai J., Sussman R., Sun G., Kovermann S. K., Mariani S. A., Calabretta B., McMahon S. B., Brock H. W., Iacovitti L. et al. (2017). Delayed accumulation of H3K27me3 on nascent DNA is essential for recruitment of transcription factors at early stages of stem cell differentiation. Mol. Cell 66, 247-257.e5. 10.1016/j.molcel.2017.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. E. and Corces V. G. (2009). CTCF: master weaver of the genome. Cell 137, 1194-1211. 10.1016/j.cell.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiee M.-R., Girardot C., Sigismondo G. and Krijgsveld J. (2016). Expanding the circuitry of pluripotency by selective isolation of chromatin-associated proteins. Mol. Cell 64, 624-635. 10.1016/j.molcel.2016.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Respuela P., Nikolić M., Tan M., Frommolt P., Zhao Y., Wysocka J. and Rada-Iglesias A. (2016). Foxd3 promotes exit from naive pluripotency through enhancer decommissioning and inhibits germline specification. Cell Stem Cell 18, 118-133. 10.1016/j.stem.2015.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riising E. M., Comet I., Leblanc B., Wu X., Johansen J. V. and Helin K. (2014). Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol. Cell 55, 347-360. 10.1016/j.molcel.2014.06.005 [DOI] [PubMed] [Google Scholar]
- Rose N. R., King H. W., Blackledge N. P., Fursova N. A., Ember K. J., Fischer R., Kessler B. M. and Klose R. J. (2016). RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes. Elife 5, 524 10.7554/eLife.18591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S., Sengupta A. K., Kubicek S., Mechtler K., Spahn L., Koseki H., Jenuwein T. and Wutz A. (2006). Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 25, 3110-3122. 10.1038/sj.emboj.7601187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S., Furlan-Magaril M., Mifsud B., Tavares-Cadete F., Sugar R., Javierre B.-M., Nagano T., Katsman Y., Sakthidevi M., Wingett S. W. et al. (2015a). The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 25, 582-597. 10.1101/gr.185272.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S., Sugar R., Dimond A., Javierre B.-M., Armstrong H., Mifsud B., Dimitrova E., Matheson L., Tavares-Cadete F., Furlan-Magaril M. et al. (2015b). Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat. Genet. 47, 1179-1186. 10.1038/ng.3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J., Muto M., Takebayashi S.-I., Suetake I., Iwamatsu A., Endo T. A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K. et al. (2007). The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature Publishing Group 450, 908-912. 10.1038/nature06397 [DOI] [PubMed] [Google Scholar]
- Shen X., Liu Y., Hsu Y.-J., Fujiwara Y., Kim J., Mao X., Yuan G.-C. and Orkin S. H. (2008). EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491-502. 10.1016/j.molcel.2008.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer Z. S., Yong J., Tischler J., Hackett J. A., Altinok A., Surani M. A., Cai L. and Elowitz M. B. (2014). Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol. Cell 55, 319-331. 10.1016/j.molcel.2014.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. (2017). Formative pluripotency: the executive phase in a developmental continuum. Development 144, 365-373. 10.1242/dev.142679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stielow B., Finkernagel F., Stiewe T., Nist A. and Suske G. (2018). MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6. PLoS Genet. 14, e1007193 10.1371/journal.pgen.1007193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel G., Fitzpatrick D. J., Oliviero G., Scelfo A., Moran B., Das S., Munawar N., Watson A., Wynne K., Negri G. L. et al. (2017). Fam60a defines a variant Sin3a-Hdac complex in embryonic stem cells required for self-renewal. EMBO J. 36, 2216-2232. 10.15252/embj.201696307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel G., Watson A., Jammula S. G., Scelfo A., Fitzpatrick D. J., Oliviero G., McCole R., Conway E., Glancy E., Negri G. L. et al. (2018). The H3K36me2 methyltransferase Nsd1 demarcates PRC2-mediated H3K27me2 and H3K27me3 domains in embryonic stem cells. Mol. Cell 70, 371-379.e5. 10.1016/j.molcel.2018.02.027 [DOI] [PubMed] [Google Scholar]
- Surface L. E., Fields P. A., Subramanian V., Behmer R., Udeshi N., Peach S. E., Carr S. A., Jaffe J. D. and Boyer L. A. (2016). H2A.Z.1 monoubiquitylation antagonizes BRD2 to maintain poised chromatin in ESCs. CELREP 14, 1142-1155. 10.1016/j.celrep.2015.12.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaz R. A., Harman J. L., Karimlou D., Weavers L., Fritsch L., Bou-Kheir T., Bell E., del Valle Torres I., Niakan K. K., Fisher C. et al. (2017). Jmjd2c facilitates the assembly of essential enhancer-protein complexes at the onset of embryonic stem cell differentiation. Development 144, 567-579. 10.1242/dev.142489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakiridis A., Huang Y., Blin G., Skylaki S., Wymeersch F., Osorno R., Economou C., Karagianni E., Zhao S., Lowell S. et al. (2014). Distinct Wnt-driven primitive streak-like populations reflect in vivo lineage precursors. Development 141, 1209-1221. 10.1242/dev.101014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mierlo G., Dirks R. A. M., De Clerck L., Brinkman A. B., Huth M., Kloet S. L., Saksouk N., Kroeze L. I., Willems S., Farlik M. et al. (2019). Integrative proteomic profiling reveals PRC2-dependent epigenetic crosstalk maintains ground-state pluripotency. Cell Stem Cell 24, 123-137.e8. 10.1016/j.stem.2018.10.017 [DOI] [PubMed] [Google Scholar]
- Von Meyenn F., Berrens R. V., Andrews S., Santos F., Collier A. J., Krueger F., Osorno R., Dean W., Rugg-Gunn P. J. and Reik W. (2016a). Comparative principles of DNA methylation reprogramming during human and mouse in vitro primordial germ cell specification. Dev. Cell 39, 104-115. 10.1016/j.devcel.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Meyenn F., Iurlaro M., Habibi E., Liu N. Q., Salehzadeh-Yazdi A., Santos F., Petrini E., Milagre I., Yu M., Xie Z. et al. (2016b). Impairment of DNA methylation maintenance is the main cause of global demethylation in naive embryonic stem cells. Mol. Cell 62, 983 10.1016/j.molcel.2016.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voong L. N., Xi L., Sebeson A. C., Xiong B., Wang J.-P. and Wang X. (2016). Insights into nucleosome organization in mouse embryonic stem cells through chemical mapping. Cell 167, 1555-1570.e15. 10.1016/j.cell.2016.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Brown J. L., Cao R., Zhang Y., Kassis J. A. and Jones R. S. (2004). Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 14, 637-646. 10.1016/j.molcel.2004.05.009 [DOI] [PubMed] [Google Scholar]
- Wang C., Lee J.-E., Lai B., Macfarlan T. S., Xu S., Zhuang L., Liu C., Peng W. and Ge K. (2016). Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition. Proc. Natl. Acad. Sci. USA 113, 11871-11876. 10.1073/pnas.1606857113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-P., Tang Z., Chen C.-W., Shimada M., Koche R. P., Wang L.-H., Nakadai T., Chramiec A., Krivtsov A. V., Armstrong S. A. et al. (2017). A UTX-MLL4-p300 transcriptional regulatory network coordinately shapes active enhancer landscapes for eliciting transcription. Mol. Cell 67, 308-321.e6. 10.1016/j.molcel.2017.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub A. S., Li C. H., Zamudio A. V., Sigova A. A., Hannett N. M., Day D. S., Abraham B. J., Cohen M. A., Nabet B., Buckley D. L. et al. (2017). YY1 is a structural regulator of enhancer- promoter loops. Cell 171, 1573-1579.e28. 10.1016/j.cell.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Lu J., Sridhar B., Cao X., Yu P., Zhao T., Chen C.-C., McDee D., Sloofman L., Wang Y. et al. (2017). SMARCAD1 contributes to the regulation of naive pluripotency by interacting with histone citrullination. CELREP 18, 3117-3128. 10.1016/j.celrep.2017.02.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi M., Kishigami S., Tanaka A., Semi K., Mizutani E., Wakayama S., Wakayama T., Yamamoto T. and Yamada Y. (2017). Derivation of ground-state female ES cells maintaining gamete-derived DNA methylation. Nature Publishing Group 548, 224-227. 10.1038/nature23286 [DOI] [PubMed] [Google Scholar]
- Yan J., Chen S.-A. A., Local A., Liu T., Qiu Y., Dorighi K. M., Preissl S., Rivera C. M., Wang C., Ye Z. et al. (2018). Histone H3 lysine 4 monomethylation modulates long-range chromatin interactions at enhancers. Cell Res. 28, 204-220. 10.1038/cr.2018.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Zhao W., Huang Y., Tong H., Xia Y., Jiang Q. and Qin J. (2017). Loss of polycomb group protein Pcgf1 severely compromises proper differentiation of embryonic stem cells. Sci. Rep. 7, 46276 10.1038/srep46276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.-L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P. and Smith A. (2008). The ground state of embryonic stem cell self-renewal. Nature Publishing Group 453, 519-523. 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. L., McKinsey T. A., Lu J. R. and Olson E. N. (2001). Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J. Biol. Chem. 276, 35-39. 10.1074/jbc.M007364200 [DOI] [PubMed] [Google Scholar]
- Zhang C. L., McKinsey T. A. and Olson E. N. (2002). Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol. Cell. Biol. 22, 7302-7312. 10.1128/MCB.22.20.7302-7312.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Gayen S., Xiong J., Zhou B., Shanmugam A. K., Sun Y., Karatas H., Liu L., Rao R. C., Wang S. et al. (2016). MLL1 inhibition reprograms epiblast stem cells to naive pluripotency. Cell Stem Cell 18, 481-494. 10.1016/j.stem.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Huang Y., Zhang J., Liu M., Ji H., Wang C., Cao N., Li C., Xia Y., Jiang Q. et al. (2017a). Polycomb group RING finger proteins 3/5 activate transcription via an interaction with the pluripotency factor Tex10 in embryonic stem cells. J. Biol. Chem. 292, 21527-21537. 10.1074/jbc.M117.804054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Tong H., Huang Y., Yan Y., Teng H., Xia Y., Jiang Q. and Qin J. (2017b). Essential role for polycomb group protein Pcgf6 in embryonic stem cell maintenance and a noncanonical polycomb repressive complex 1 (PRC1) integrity. J. Biol. Chem. 292, 2773-2784. 10.1074/jbc.M116.763961 [DOI] [PMC free article] [PubMed] [Google Scholar]