ABSTRACT
The BAF (SWI/SNF) chromatin remodeling complex plays a crucial role in modulating spatiotemporal gene expression during mammalian development. Although its remodeling activity was characterized in vitro decades ago, the complex actions of BAF in vivo have only recently begun to be unraveled. In living cells, BAF only binds to and remodels a subset of genomic locations. This selectivity of BAF genomic targeting is crucial for cell-type specification and for mediating precise responses to environmental signals. Here, we provide an overview of the distinct molecular mechanisms modulating BAF chromatin binding, including its combinatory assemblies, DNA/histone modification-binding modules and post-translational modifications, as well as its interactions with proteins, RNA and lipids. This Review aims to serve as a primer for future studies to decode the actions of BAF in developmental processes.
KEY WORDS: BAF, SWI/SNF, Chromatin, DNA, RNA, Histone, Transcription factor
Summary: A comprehensive Review covering the molecular mechanisms modulating the selectivity of genomic targeting for the BAF chromatin remodeling complex.
Introduction
A complex higher organism, such as a human being, comprises a myriad of morphologically and functionally distinctive cell types. All of these cells contain the same genomic DNA, which underlies a broad spectrum of developmental and physiological processes. However, in a given cell at a given time, only a subset of these genomic sequences is accessible to transcriptional machineries, allowing highly specialized biological functions to be performed across different cell types. The exact molecular mechanisms governing this gene expression selectivity, which is vital for cell-type specification, remain incompletely understood.
The majority of genomic DNA wraps around nucleosomes, which not only package the genome but also serve as barriers to prevent promiscuous transcription. The BAF (SWI/SNF) chromatin remodeling complex, which uses energy from ATP hydrolysis to actively reposition nucleosomes, is a crucial player for modulating developmental processes. Indeed, altered BAF function is implicated in many human diseases, including neurodevelopmental disorders and different cancer types (Helsmoortel et al., 2014; Kadoch et al., 2013; Mathur et al., 2017; Nakayama et al., 2017). The roles of BAF in development and disease have been discussed in several recent reviews (Alfert et al., 2019; Hodges et al., 2016; Hota and Bruneau, 2016). Here, we focus on the molecular mechanisms that contribute to the specificity of BAF genomic targeting. We discuss different BAF combinatorial assemblies, various BAF modules that can bind to histone modifications and DNA, transcription factors that can recruit BAF to specific genomic sites, as well as the unique characteristics of distinct BAF assemblies in genomic targeting. We also summarize the impact of post-translational modifications, non-coding RNAs and lipids on controlling BAF targeting. These fundamental molecular mechanisms serve as the basis for understanding the actions of BAF in regulating development and disease.
The composition and assembly of BAF
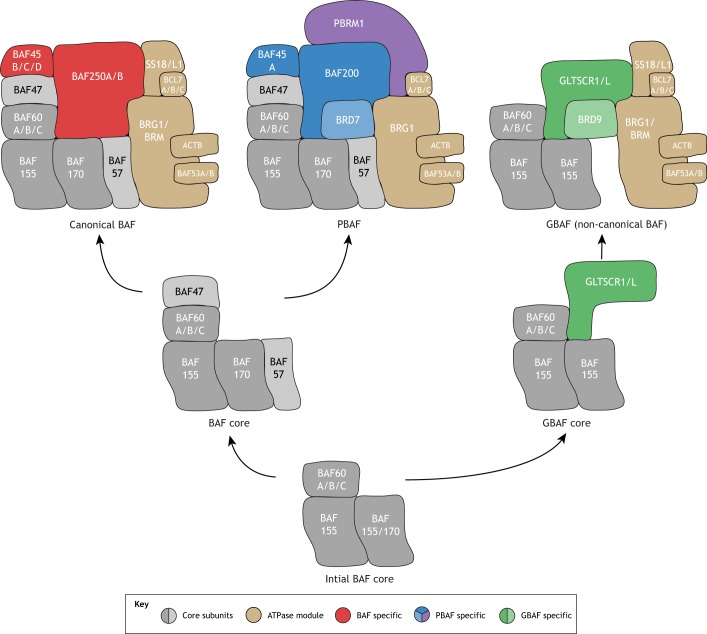
About 30 genes in the human genome have been identified thus far to encode BAF subunits. These subunits are differentially expressed in various cell types and assemble into biochemically stable complexes ranging from 0.87 MDa to 1.41 MDa in size (Mashtalir et al., 2018). Each BAF complex includes one ATPase catalytic subunit, BRG1 (SMARCA4) or BRM (SMARCA2), and several additional regulatory subunits. Based on their distinct biochemical properties, three major BAF assemblies have been described: canonical BAF, PBAF and GBAF (sometimes termed non-canonical BAF). The reported compositions of these BAF assemblies vary slightly in the literature, as they can be influenced by different biochemical purification strategies as well as by the specific cell type being studied. Representative compositions are summarized in Fig. 1.
Fig. 1.
BAF complex composition and assembly. Schematic representation of the subunit composition and assembly of canonical BAF, PBAF and GBAF (also called non-canonical BAF). The core subunits are indicated in gray, the subunits of the ATPase module are indicated in beige, and the unique subunits pertaining to each complex are highlighted with different colors (red for BAF, blue/purple for PBAF and green for GBAF). Exact subunit composition appears to influenced by the specific cell type as well as by the complex purification method. As such, the images shown here are simplified representations that attempt to accommodate recent findings.
In mammals, the highly conserved canonical BAF complex features BAF250A (ARID1A) or BAF250B (ARID1B). Loss of Baf250a or Baf250b (Arid1a and Arid1b) in mouse models leads to early embryonic lethality (Celen et al., 2017; Lei et al., 2012), indicating that both genes are essential for early development despite their homology. In addition to BAF250A/B, BAF45B/C/D (DPF1/2/3) is specific to canonical BAF (Middeljans et al., 2012). The other subunits found in canonical BAF are shared with those found in PBAF and/or GBAF; the functional features of these subunits are discussed in more detail in later sections.
The PBAF complex was initially identified as ‘SWI/SNF complex B’, which was observed to be distinct from canonical BAF (‘SWI/SNF complex A’) based on the different salt concentration required to elute the complex from a phosphocellulose column (Ward et al., 1991; Xue et al., 2000). Notably, PBAF represents the BAF assembly with the largest molecular weight. Four subunits are exclusive to PBAF: BAF200 (ARID2), PBRM1 (BAF180), BAF45A (PHF10) and BRD7 (Kadoch et al., 2013; Kaeser et al., 2008; Middeljans et al., 2012; Wang et al., 1996).
The GBAF complex, which is named after its unique subunit GLTSCR1/1L, was recently isolated as a distinct BAF complex with a smaller molecular weight. GBAF lacks four canonical BAF subunits (BAF47, BAF57, BAF170 and BAF250) but features the unique incorporation of GLTSCR1/1L and BRD9 (Alpsoy and Dykhuizen, 2018).
Recent elegant biochemical experiments have provided key insights into how these different BAF complexes are assembled inside living mammalian cells (Mashtalir et al., 2018). The earliest assembled subunits are the BAF155/170 (SMARCC1/2) dimers, consistent with studies showing that loss of both BAF155 and BAF170 leads to complex destabilization (Chen and Archer, 2005; Narayanan et al., 2015; Sohn et al., 2007). BAF60 then binds to this initial dimer where it can diverge towards GBAF by integrating GLTSCR1/1L or towards the two larger BAF assemblies by incorporating BAF47 and BAF57. BAF200 or BAF250 is subsequently added to further distinguish between canonical BAF and PBAF. Notably, the ATPase BRG1/BRM forms a module together with β-actin (ACTB), BAF53A/B (ACTL6A/B), BCL7A/B/C and SS18/L1 to finalize the assembly process for canonical BAF, and PBRM1 is further added as the last subunit for PBAF. As purified ATPase alone can remodel nucleosomes in vitro (Phelan et al., 1999), its late addition as a module is likely to be a mechanism for preventing promiscuous chromatin remodeling.
BAF subunit gene expression varies across different cell types, thereby contributing to distinct and cell-specific representations of canonical BAF, PBAF and GBAF. Further complexity is provided by subunits that are encoded by multiple genes, including BAF45A/B/C/D, BAF60A/B/C and BCL7A/B/C (Fig. 2). Likewise, the BAF53 subunit can be encoded by BAF53A (ACTL6A) or BAF53B (ACTL6B), with the latter being highly restricted to differentiated neurons (Lessard et al., 2007). The tissue-specific expression of distinct BAF subunits is instrumental for cell type-specific gene expression. For example, BAF60C is highly expressed in muscle cells, but not in keratinocytes (Bao et al., 2013; Forcales et al., 2012; Lickert et al., 2004). This is in agreement with the findings that BAF60C is specifically expressed in the heart and somites of early developing embryos, and is required to recruit BAF to heart-specific enhancers (Lickert et al., 2004). The unique combinatory assemblies of BAF – merging various nucleosomal recognition modules of BAF subunits and producing unique interacting surfaces for other molecules – can strongly influence the binding selectivity as well as the functionality of BAF complexes across the genome (Fig. 3).
Fig. 2.
Relative expression of BAF genes in distinct cell types. Cartoon illustration showing the relative expression of BAF subunits in different cell types. Different font sizes indicate the relative gene expression, based on RNA-seq data generated by the Roadmap Epigenomics Project (Kundaje et al., 2015); larger font size indicates a higher expression level. The expression of each BAF gene is normalized based on the total expression of all BAF subunits across these cell types. The relative expression is calculated using the ratio of normalized expression versus the average expression of each BAF gene.
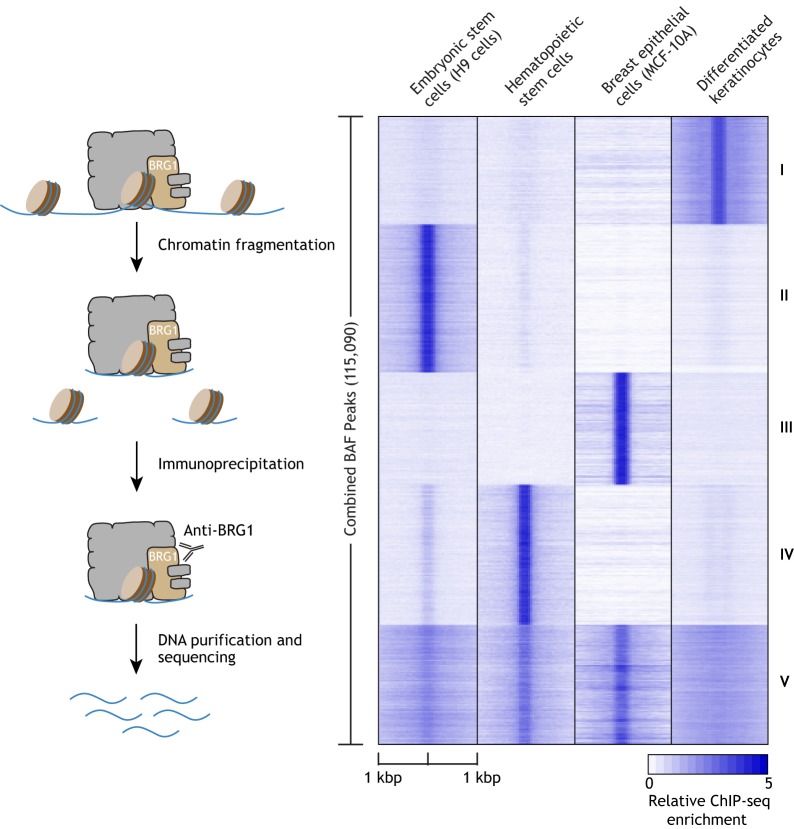
Fig. 3.
Comparison of BAF genomic binding in different cell types. Summit-centered heatmap of BRG1 (SMARCA4) ChIP-seq data illustrating the binding of BRG1 to chromatin in four different human cell types. A simple schematic of a ChIP-seq workflow is also included on the left side of the heatmap. Darker shade of blue indicates enrichment of BRG1 binding. The heatmap is clustered into five groups according to K-means. Groups I-IV demonstrate BRG1-binding sites that are highly specific to the individual cell types, while group V represents BRG1-binding sites that are shared among multiple cell types. The raw data were acquired from previously published studies (Abraham et al., 2013; Bao et al., 2015; Barutcu et al., 2016; Rada-Iglesias et al., 2011) and were normalized based on sequencing depth.
Nucleosome-binding modules of canonical BAF
Recognition of specific histone modifications
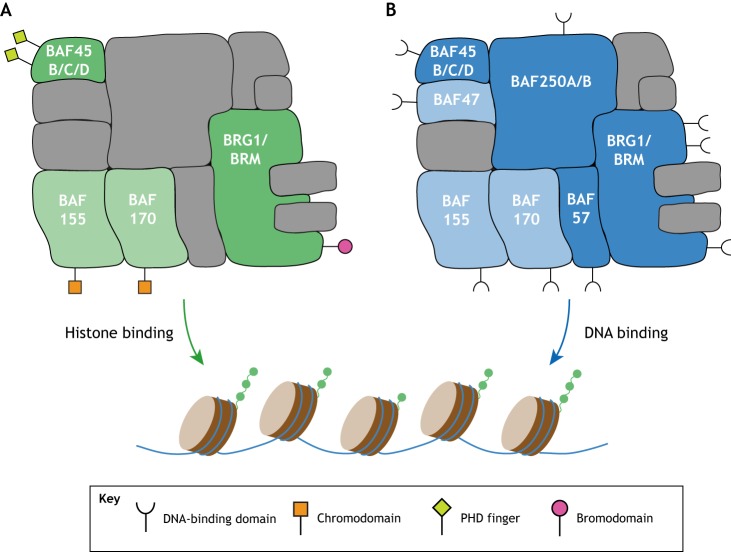
The canonical BAF complex features two plant homeodomain (PHD) fingers in BAF45, a bromodomain in BRG1/BRM, as well as two chromodomains in BAF155 and BAF170. These modules enable BAF to recognize specific post-translational modifications on histone tails, such as acetylation and methylation (Fig. 4).
Fig. 4.
Histone-/DNA-binding domains of the canonical BAF complex. Cartoon illustration highlighting BAF subunits that can bind to histone modifications (green) and BAF subunits that can bind to DNA (blue); subunits with experimentally validated binding abilities are shown in a dark shade, and those with predicted binding abilities shown in a light shade. Symbols for specific DNA/histone binding domains are indicated in the key.
The tandem PHD fingers in BAF45 allow this single subunit to simultaneously sample two post-translational modifications on the same histone tail to modulate BAF binding. In canonical BAF, BAF45 can be encoded by BAF45B (DPF1), BAF45C (DFP3) or BAF45D (DPF2). The PHD fingers in BAF45B and BAF45D are highly similar (∼90% similarity), but they are more divergent in BAF45C (∼40% similarity). Based on in vitro peptide assays, the double PHD finger of BAF45C was initially found to recognize H3K14 acetylation in combination with unmethylated H3K4 (Zeng et al., 2010). It was subsequently identified that H3K4 monomethylation, but not trimethylation, can also be accommodated by the PHD fingers of BAF45C to augment BAF binding at enhancers (Local et al., 2018). In the same study, BAF45D was found to associate with H3K4me1 in a similar way to BAF45C, although in vitro evidence further revealed higher affinity of BAF45D towards crotonylated H3K14 than to acetylated H3K14 (Xiong et al., 2016). Considering the sequence differences between the PHD fingers in BAF45C and BAF45D, it is not impossible to imagine that these two proteins recognize different histone modifications. However, a side-by-side comparison using the same techniques and experimental conditions is still currently lacking.
The bromodomain in BRG1/BRM is highly conserved in eukaryotes. This domain is capable of recognizing histone acetylation in vitro in the context of histone-tail peptides and reconstituted nucleosomes (Agalioti et al., 2002; Morrison et al., 2017; Shen et al., 2007). However, the role of this bromodomain in a cellular context in vivo, and in developmental processes, still remains controversial. A mutation in the bromodomain of BRM, which abolishes its interaction with histone acetylation, weakens BRM-chromatin association based on fluorescence recovery after photobleaching (FRAP) assays in cultured cells pre-treated with an HDAC inhibitor (Fedorov et al., 2015; Sutherell et al., 2016); however, a bromodomain mutation in BRG1 does not appear to alter its global chromatin association based on differential salt extraction (Morrison et al., 2017). Deletion of the bromodomain in the Drosophila homolog also does not appear to noticeably affect viability or fertility (Elfring et al., 1998). Yet a recently developed bromodomain inhibitor for BRG1 (PFI-3), which also has cross-reactivity with a bromodomain in the PBAF-specific subunit PBRM1, was shown to induce embryonic and trophoblast stem cell differentiation (Fedorov et al., 2015). Interestingly, several phosphorylation sites near this bromodomain have been identified to regulate BAF binding to specific sites, suggesting that this bromodomain may play a role in sensing specific cellular signals rather than being essential for general BAF chromatin association. These phosphorylation sites are discussed in more detail in a later section.
Diverse use of DNA-binding domains
A total of seven canonical BAF subunits contain DNA-binding domains, with those in BAF47, BAF57, BAF250 and BRG1/BRM having been experimentally tested. BAF155, BAF170 and BAF45 also feature domains with putative DNA-binding capacities (Fig. 4).
BAF47 features an N-terminal winged-helix DNA-binding domain that is characterized by a helix-turn-helix motif followed by a β-hairpin, with the loop in the hairpin constituting a ‘wing’ (Allen et al., 2015). Nuclear magnetic resonance (NMR) analysis indicates that the BAF47 winged-helix domain can strongly bind to double-stranded DNA in vitro. Consistent with this observation, biallelic inactivation of BAF47 in malignant rhabdoid tumors and epithelioid sarcomas significantly impairs BAF chromatin binding without affecting complex stability; re-expression of BAF47 drives genome-wide gain of BAF binding, especially at enhancers and bivalent promoters (Nakayama et al., 2017). These pieces of evidence suggest that the DNA-binding capacity of BAF47 is likely to be crucial for stabilizing BAF chromatin association at enhancers and promotors.
BAF250A (ARID1A) and BAF250B (ARID1B), which are mutually exclusive in canonical BAF assembly, contain AT-rich interacting domains (ARIDs) that share 90% similarity. Crosslinking mass spectrometry of BAF purified from human cells identified minimal association of this ARID with other BAF subunits, suggesting that it plays a primary role in DNA binding rather than in complex assembly (Mashtalir et al., 2018). Despite its name, this domain has no strong preference towards AT-rich DNA sequences (Dallas et al., 2000; Nie et al., 2000; Wilsker et al., 2004). BAF250A itself has been found to be essential for sustaining global BAF chromatin association in colorectal carcinoma cell lines, where depletion of BAF250A strongly impairs BAF occupancy at enhancers (Mathur et al., 2017). Strikingly, a single point mutation in the BAF250A ARID domain (V1068G) is sufficient to disrupt BAF binding in vitro and also in vivo in mice, where it results in heart and extra-embryonic vasculature defects followed by embryonic lethality by E13.5 (Chandler et al., 2013). In the context of BAF250A loss, the binding of BAF250B-containing BAF to chromatin remains intact at distinct sites to preserve residual BAF binding (Mathur et al., 2017), consistent with observations that BAF250B is a synthetic lethal vulnerability in BAF250A-deficient cancers (Helming et al., 2014). Taken together, these studies suggest that the ARID domain in BAF250A/B is essential for mediating BAF targeting to regulatory regions.
The HMG domain in BAF57 (SMARCE1) was actually the first identified DNA-binding domain in BAF. This domain has high affinity in vitro for cruciform DNA, a four-way-junction DNA structure that mimicks the topology of DNA as it enters and exits the nucleosome (Wang et al., 1998). Expression of a BAF57 mutant with a deleted HMG domain impairs T cell sub-lineage specification during thymic development, dysregulating both CD4 gene activation as well as CD8 suppression without affecting BAF binding (Chi et al., 2002). Thus, it is likely that this HMG DNA-binding domain plays a role in modulating gene expression instead of controlling site-specific BAF binding.
The catalytic subunit BRG1/BRM includes at least three regions with DNA-binding capacity, including a helicase ATPase core, a surface basic patch and an AT-hook DNA-binding motif (Bourachot et al., 1999; Han et al., 2014; Morrison et al., 2017). The helicase ATPase core alone displays high affinity for chromatinized, but not naked, DNA in vitro (Han et al., 2014). The AT-hook motif in BRG1/BRM, which is sandwiched between the ATPase core and the bromodomain, has HMG-like DNA-binding properties with higher affinity for linear DNA than cruciform DNA in vitro (Bourachot et al., 1999; Singh et al., 2006). In addition, a surface basic patch identified within the bromodomain of BRG1/BRM displays affinity for both linear and nucleosomal DNA in vitro, although this region does not appear to be essential for global BAF chromatin targeting when tested using differential salt extraction (Morrison et al., 2017). Thus, the BAF catalytic subunit alone features multivalent DNA-binding capacity through different regions that can cooperate to enhance BAF binding and function.
In addition to these experimentally characterized DNA-binding domains, several BAF subunits feature putative DNA-binding domains based on functional prediction. For example, BAF155 and BAF170 include leucine zipper, SANT and SWIRM domains, while BAF45B/C/D feature a Krüppel-like zinc-finger motif. The essentiality of these potential DNA-binding activities in the genome-wide association of BAF complexes still remains to be characterized in cellular and developmental contexts.
Distinct nucleosome-binding activities of PBAF and GBAF
Unique chromatin association mechanisms of PBAF
PBAF is highly conserved in eukaryotes from yeast, fly and worm to human, and shows distinct biological functions and chromatin association patterns from canonical BAF. Its yeast counterpart, RSC, is essential for mitotic growth (Cairns et al., 1996). In C. elegans, PBAF is required for gonad development (Shibata et al., 2011). In Drosophila, PBAF (termed PBAP) and the canonical complex localize to both distinct and overlapping bands on polytene chromosomes (Mohrmann et al., 2004), with PBAP being essential for eggshell formation and metamorphosis (Carrera et al., 2008). Similarly in mammals, PBAF and canonical BAF localize to distinct and overlapping regions (Nakayama et al., 2017; Raab et al., 2015; Wei et al., 2018). Furthermore, salt fractionation experiments suggest that PBAF binds more tightly to chromatin than does canonical BAF (Porter and Dykhuizen, 2017). PBAF-specific subunits are implicated in multiple developmental processes in mammals, including cardiovascular development, glucose metabolism and osteoblast differentiation (Kim et al., 2016; Wang et al., 2004; Xu et al., 2012). Thus, these findings indicate that PBAF plays distinct roles from canonical BAF in development. But what confers the specific genomic binding patterns of PBAF? As we highlight below, recent studies indicate that the protein/DNA-binding properties of subunits that are unique to PBAF may play a role.
PBRM1, a key defining subunit of PBAF, boasts a total of six tandem bromodomains. Functional dissection of these bromodomains indicates that at least five of them contribute to PBAF chromatin binding, with the second bromodomain being the most crucial (Porter and Dykhuizen, 2017). The second bromodomain can recognize acetylated or propionylated H3K14 peptides in vitro (Kebede et al., 2017; Porter and Dykhuizen, 2017). When multiple lysines are acetylated within the same H3 peptide (H3K14, K18, K23 and K27), the affinity of this peptide for PBRM1 is strongly increased (Porter and Dykhuizen, 2017). These pieces of evidence suggest that the bromodomains function cooperatively to recruit PBAF to highly acetylated chromatin regions. In addition to the bromodomains, PBRM1 includes a highly conserved HMG domain (Xue et al., 2000), which is predicted to bind to nucleosomal DNA.
BAF200 is another subunit unique to PBAF that shares ∼16% similarity with BAF250A and BAF250B. BAF200 is essential for PBAF assembly and acts by stabilizing the incorporation of PBRM1 (Yan et al., 2005). BAF200 contains an ARID domain, an RFX DNA-binding domain and two zinc fingers. The ARID domain, which is similar to the one in BAF250A/B, does not appear to recognize specific DNA sequences in vitro (Patsialou et al., 2005). It is unclear how these domains contribute to the specific genomic targeting of PBAF.
The bromodomain protein BRD7, which is also unique to PBAF, can bind to the H3K14ac peptide in vitro (Cong et al., 2006) and can also recognize acetylation on non-histone proteins. A recent study revealed a very interesting phenomenon with regard to the BRD-mediated association of PBAF versus BAF with chromatin. This study showed that, in β cells, both BRD7 and BRD9 recognize and bind to acetylated (K91Ac) vitamin D receptor (VDR). However, association with BRD9 corresponds to an inactive state. The ligand vitamin D then shifts the binding of VDR to BRD7, which recruits PBAF to promote chromatin accessibility and enhancer activation in response to inflammatory stress (Wei et al., 2018).
BAF45A is also recognized as a PBAF-specific subunit (Middeljans et al., 2012). A winged-helix domain is unique to BAF45A, but is absent in other BAF45B/C/D proteins (Allen et al., 2015). BAF45A also lacks the C2H2-type Krüppel-like zinc-finger motif that is present in the other BAF45 proteins. These features may underlie the essentiality of BAF45A in maintaining hematopoietic stem cells and neural stem cells, whereas BAF45B/C are associated with tissue differentiation (Krasteva et al., 2017; Lessard et al., 2007).
Unique characteristics of the GBAF complex
The GBAF complex defines a smaller, non-canonical BAF complex in which GLTSCR1 or its paralogue GLTSCR1L is the defining subunit. These two proteins share only 33% similarity; however, both of them contain a GLTSCR domain, which serves as a GBAF-specific binding region required for association with other GBAF subunits (Michel et al., 2018).
When compared with canonical BAF, GBAF localizes to both distinct and shared genomic regions. GBAF is more enriched at sites marked with H3K4me3, and it also uniquely localizes to topologically associating domain (TAD) boundaries (Gatchalian et al., 2018; Michel et al., 2018). In BAF-perturbed synovial sarcoma and malignant rhabdoid tumors, GBAF is necessary to maintain gene expression at retained promoter-proximal and CTCF sites. Through the study of these BAF47-deficient tumors, GBAF-specific subunits were identified as synthetic lethal cancer targets that can significantly attenuate cell proliferation (Michel et al., 2018).
The bromodomain protein BRD9 was recently recognized as a component of the GBAF complex (Alpsoy and Dykhuizen, 2018; Gatchalian et al., 2018; Michel et al., 2018; Wang et al., 2019). Among several inhibitors developed for targeting BRD9, dBRD9 was found to selectively target and chemically degrade BRD9, but not BRD4 or BRD7 (Remillard et al., 2017). BRD9 inhibition or knockdown impairs GBAF genomic targeting, indicating that BRD9 is a key mediator in facilitating GBAF chromatin association (Gatchalian et al., 2018; Michel et al., 2018). Although BRD9 can bind to acetylated or butyrylated histone peptides in vitro (Flynn et al., 2015), its direct targets in vivo still remain incompletely understood. One target, in addition to VDR (as mentioned above), could be BRD4. In mouse embryonic stem cells (ESCs), BRD9 chromatin binding is impaired in the presence of the BRD4 inhibitor JQ1 or the BRD9 inhibitor I-BRD9 (Gatchalian et al., 2018; Theodoulou et al., 2016), suggesting a model whereby BRD9 associates with BRD4 to tether GBAF to chromatin. As GLTSCR1 can also interact with BRD4 (Rahman et al., 2011), it remains to be confirmed whether BRD9 or GLTSCR1 bind directly to BRD4.
The association of BAF with transcription factors
General and cell-type specific transcription factors
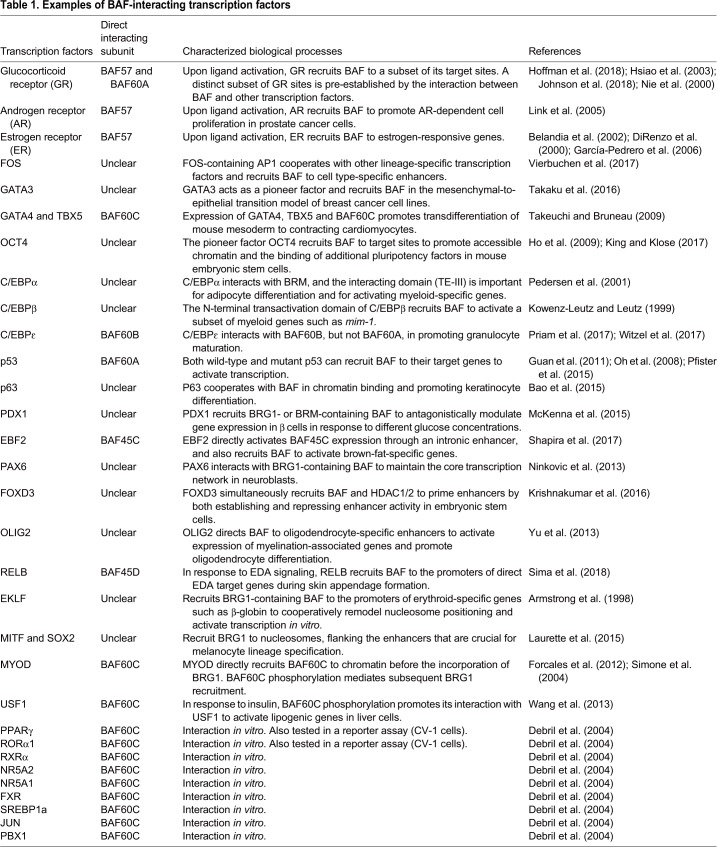
The transactivator GAL4 was identified as one of the first BAF-interacting transcription factors. GAL4, which comprises a sequence-specific DNA-binding domain and an acidic transactivation domain, can recruit BAF to elicit site-specific nucleosome eviction in both yeast and mammalian cells (Côté et al., 1994; Kwon et al., 1994). A number of additional transcription factors containing a transactivation domain have subsequently been identified as BAF interactors that harbor the ability to recruit BAF to distinct genomic sites and drive specific developmental processes. Below (and in Table 1), we provide some examples of such factors and discuss how they modulate BAF recruitment and activity.
Table 1.
Examples of BAF-interacting transcription factors
A subset of the ubiquitously expressed activating protein 1 (AP1) transcription factors can interact with BAF. AP1 factors function as dimers of JUN, FOS, ATF or MAF family proteins. Based on in vitro studies, BAF60A (SMARCD1) can bind to FOS/JUN, but not to FOS-related antigen 2 (FRA2)/JUND heterodimers (Ito et al., 2001). This selectivity has been corroborated in a recent study in which BAF was found to interact with FOS heterodimers, but not with JUND homodimers (Vierbuchen et al., 2017). Notably, FOS but not JUN possesses a C-terminal transactivating domain. These FOS-containing AP1 dimers cooperate with other lineage-specific transcription factors and recruit BAF to cell-type-specific enhancers.
BAF has also been reported to interact with the CCAAT enhancer-binding protein (C/EBP) family of transcription factors. C/EBPα is expressed across many tissue types and functions as a lineage-instructive transcription factor. It features a total of three transactivation elements. Domain mapping analysis identified that transactivation element 3 (TE-III) of C/EBPα is both necessary and sufficient to interact with BAF, and is also essential for promoting adipogenesis and for activating myeloid-specific genes (Pedersen et al., 2001). In the same C/EBP family, C/EBPβ, which features a complex N terminus transactivation domain, was found to be essential for mediating BAF-dependent myeloid gene activation (Kowenz-Leutz and Leutz, 1999). Furthermore, the interaction between C/EBPɛ and BAF was reported to regulate neutrophil granulocyte differentiation, through direct binding to BAF60B (Priam et al., 2017; Witzel et al., 2017).
The tumor suppressor p53, which features a transactivation domain in its N terminus, can interact and recruit BAF even when p53 is mutated in cancer (Guan et al., 2011; Lee et al., 2002; Pfister et al., 2015). The BAF subunit directly mediating this interaction was identified as BAF60A (Oh et al., 2008). Using a human ovarian epithelial cell line, p53 was found to directly recruit BAF to the promoters of p53 target genes, including CDKN1A and SMAD3 (Guan et al., 2011). In breast cancer cells, mutant p53 (R282W) recruits BAF to activate VEGFR2 (Pfister et al., 2015), a key receptor for promoting angiogenesis. In differentiated keratinocytes, BAF binding is highly enriched at binding sites for p63 (Bao et al., 2015), a p53 family member that acts as a master transcriptional regulator of epidermal keratinocytes. Knockdown of p63 reduces BAF binding at these sites and BRG1/BRM knockdown also impairs p63 binding, suggesting cooperative binding between BAF and p63. RNA-sequencing (RNA-seq) analysis has further revealed that epidermal differentiation is suppressed following knockdown of either BRG1/BRM or p63 (Bao et al., 2015), suggesting that the interaction between BAF and a lineage-specific transcription factor is essential for activating terminal tissue differentiation.
Multiple lineage-specific transcription factors with transactivation domains have been reported to interact with BAF in other specialized cell types. In islet β cells, for example, PDX1 recruits BAF to target genes where BRG1- and BRM-BAF complexes dynamically interact with PDX1 and antagonistically modulate gene expression dependent on high versus low glucose concentrations (McKenna et al., 2015). In ESCs, OCT4 – a transcription factor that contains two transactivation domains in its N and C termini – recruits BAF to its target sites to promote chromatin accessibility in regulating the pluripotency network (King and Klose, 2017). This is in agreement with previous observations that BAF colocalizes extensively with pluripotency transcription factors, including OCT4, at a genome-wide level (Ho et al., 2009). In brown adipocytes, EBF2 directly interacts with BAF45C and recruits BAF to activate brown-fat-specific genes (Shapira et al., 2017). GATA3, which is a key regulator of T lymphocyte development and mammary epithelial cell differentiation, was found to recruit BRG1 to GATA3-bound chromatin motifs in multiple breast cancer cell lines, promoting the expression of genes associated with mesenchymal-to-epithelial transition (MET). Notably, GATA3 lacking an N-terminal transactivating domain can still bind to chromatin but is no longer able to recruit BAF and generate accessible chromatin (Takaku et al., 2016).
TBX5 and GATA4, which are two transactivators that function cooperatively in cardiogenesis (Ang et al., 2016), have also been identified as BAF-interacting transcription factors. The ectopic expression of TBX5 and GATA4 together with BAF60C in cultured mouse embryos is sufficient to transdifferentiate mesoderm into beating cardiac myocytes (Takeuchi and Bruneau, 2009). In this context, BAF60C is required for the binding of GATA4 to cardiac loci in order to initiate the cardiac program, where TBX5 is then required for full differentiation into contracting cardiomyocytes. PAX6 also physically interacts with BAF and enhances the expression of neurogenic transcription factors, forming a cross-regulatory network and driving a neurogenic fate (Ninkovic et al., 2013). In melanocytes, MITF and SOX2 recruit BAF to MITF-associated enhancers (Laurette et al., 2015). In addition, erythroid Krüppel-like factor (EKLF) can also recruit BRG1, but not BRM, to the promoters of erythroid-specific genes such as β-globin to cooperatively remodel nucleosome positioning and activate transcription in vitro (Armstrong et al., 1998). These studies underscore the crucial roles of BAF recruitment by specific transcription factors in cell fate determination and differentiation.
Notably, not all transcription factors with transactivation domains can directly recruit BAF. The Rel/NF-κB family transcription factor RELB, a potent transactivator, requires a specific linker protein TRK-fusion gene (TFG) to recruit BAF to the promoters of ectodysplasin A (EDA) targets in response to signaling during skin appendage formation. TFG directly interacts with BAF45D (Sima et al., 2018). Thus, the ‘transactivation domain’ itself is not necessarily sufficient to mediate direct interactions between BAF and transcription factors. It has also been shown that transcription factors without a transactivation domain can interact with BAF. One example is the helix-loop-helix transcription factor Olig2, which can recruit BAF to myelination-associated genes to control oligodendrocyte differentiation (Yu et al., 2013). Another example is FOXD3, which serves as a priming factor during the process of ESC differentiation into epiblast cells, and recruits BRG1 and HDAC1/2 to FOXD3-binding motifs at enhancers to simultaneously promote nucleosome depletion and suppress maximal activation (Krishnakumar et al., 2016). These findings indicate that a transactivation domain is not always essential to mediate an interaction between BAF and transcription factors.
Association with nuclear hormone receptors
Nuclear hormone receptors are a group of specialized transcription factors that, when activated by their respective ligands, bind to specific DNA sequences via two zinc fingers in their DNA-binding domain. Both BAF57 and BAF60 have been reported to directly interact with nuclear hormone receptors.
In yeast, the glucocorticoid receptor (GR) was the first example of a nuclear hormone receptor that co-immunoprecipitated with a BAF subunit, namely swi3 – the yeast homolog of BAF155/170 (Yoshinaga et al., 1992). This phenomenon was subsequently confirmed in mammalian cells leveraging a chromosome-integrated mouse mammary tumor virus (MMTV) promoter, with multiple classes of nuclear hormone receptors being identified as BAF-interacting partners (Fryer and Archer, 1998). BAF57 and BAF60A were then both found to directly mediate the interactions between BAF and GR (Hsiao et al., 2003; Nie et al., 2000). Using an ultrafast ultraviolet laser crosslinking assay in vitro, the binding kinetics of GR and BAF to chromatin were found to be highly transient and periodic. This process is initiated by GR binding and is followed by BAF recruitment and transient nucleosome remodeling. Chromatin then reverts to its initial state and the cycle repeats (Nagaich et al., 2004). This study suggests that the continuous cooperation between BAF and transcription factors, as well as the repetitive ATP-driven remodeling action of BAF, are required to actively maintain genome accessibility. Using imaging approaches, BAF recruitment was also observed to be regulated in a hormone-dependent manner (Johnson et al., 2008). Recent genome-wide analyses have further uncovered the complexity of BAF-GR interactions at their ‘native’ binding sites. These highlight that, while GR can indeed recruit BAF to remodel nucleosomes at a subset of binding sites, a different subset of GR binding sites is pre-established by the interaction between BAF and other pioneer factors (Hoffman et al., 2018; Johnson et al., 2018), revealing the complexity of BAF recruitment in the presence of multiple transcription factors in a cellular context.
BAF also interacts with androgen receptor (AR) or estrogen receptor (ER) in specialized cell types. The BAF57 subunit, which is unique to mammals, directly mediates the interaction between AR/ER and BAF. In prostate cancer cells, BAF is recruited to AR targets through BAF57 upon ligand activation, promoting AR-dependent cell proliferation (Link et al., 2005). In breast cancer cells, BAF is also recruited by ER through its hormone-binding domain, as well as through a DNA-binding region to estrogen-responsive promoters, in a similar ligand-dependent manner (Belandia et al., 2002; DiRenzo et al., 2000; García-Pedrero et al., 2006).
In addition, BAF60C has been identified to interact with several other nuclear receptors in vitro, including peroxisome proliferator-activated receptor γ (PPARγ), bile acid receptor farnesoid X receptor (FXR), retinoic acid-related orphan receptor α1 (RORα1), the liver receptor homolog 1 (LRH-1 or NR5A2) and the steroidogenic factor 1 (SF1 or NR5A1) (Debril et al., 2004). Whether these interactions also occur, and control BAF targeting, in cells in vivo still remains unclear.
Other mechanisms modulating BAF targeting
Global or site-specific modulation by post-translational modifications
Post-translational modifications play crucial roles in modulating how proteins interact with RNA, DNA and other proteins. To date, phosphorylation, ubiquitylation and methylation have all been identified as post-translational modifications that occur on the BAF complex and modulate its activity. Phosphorylation marks crucial switches that influence global or site-specific BAF targeting, arginine methylation modulates a subset of BAF targeting sites and ubiquitylation degrades excess BAF subunits to encourage the targeting of intact BAF complexes.
High levels of phosphorylation have been identified on both BRG1 and BRM during mitosis in HeLa cells. This mitotic phosphorylation was found to not only weaken BAF chromatin association (Muchardt et al., 1996), but also to inactivate BAF catalytic activity (Sif et al., 1998). In in vitro assays, ERK1 can efficiently phosphorylate and inactivate BAF, while PP2A can de-phosphorylate and re-activate BAF (Sif et al., 1998); however, the true identities of the kinase(s) and phosphatase(s) controlling BAF phosphorylation in vivo during the cell cycle still remain unclear.
The ATM kinase, which plays a key role in the DNA damage response, also phosphorylates BRG1 (at serine 721). This stimulates BRG1 nucleosome binding by enhancing affinity of the BRG1 bromodomain for H3K14ac. Moreover, phosphorylated BRG1 forms a repair foci that colocalizes with γ-H2Ax. This transient phosphorylation of BRG1 is essential for DNA damage repair, as mutation of serine 721 to an alanine results in defective double-stranded break repair (Kwon et al., 2015).
In skeletal muscle cells, multiple phosphorylation events on BAF have been identified. For example, BRG1 in proliferative myoblasts is phosphorylated by protein kinase C βI (PKCβI) on residues flanking the bromodomain, which inhibits BAF association with the myogenin promoter. During differentiation, the phosphatase calcineurin opposes PKCβI function and de-phosphorylates BAF. This phenomenon promotes the binding of BAF to the myogenin promoter and activates myogenic gene expression (Nasipak et al., 2015). BRG1 also interacts with casein kinase 2 (CK2) in myoblasts. Phosphomimetic mutations on the predicted CK2 phosphorylation sites of BRG1 impair both cell proliferation and viability by inhibiting BAF binding to the Pax7 promoter (Padilla-Benavides et al., 2017).
Phosphorylation also occurs on the regulatory subunits of BAF. BAF60C phosphorylation, for example, plays critical roles in terminal differentiation. During skeletal myogenesis, BAF60C independently forms a preassembled complex with the muscle-specific transcription factor MyoD. Subsequently, p38α/β phosphorylates BAF60C at threonine 229, which then promotes BAF recruitment to the regulatory regions of myogenic genes (Forcales et al., 2012; Simone et al., 2004). Similarly, BAF60C is phosphorylated at serine 247 by atypical protein kinase C (aPKC) in liver cells in response to insulin. This phosphorylation event causes BAF60C to translocate to the nucleus and interact with upstream stimulatory factor 1 (USF1), which subsequently recruits BAF to lipogenic genes to activate gene expression (Wang et al., 2013).
BAF can also be regulated by methylation. BAF155 was identified as a substrate of the arginine methyltransferase CARM1 in breast cancer cell lines. In particular, methylation of BAF155 at arginine 1064 was found to be a biomarker for cancer recurrence and metastasis. This arginine methylation is crucial for directing BAF to specific targets, including MYC pathway genes (Wang et al., 2014). BAF155 mRNA is also regulated by methylation. In the developing mouse cortex, the m6A methyltransferase RBM15 interacts with BAF155 mRNA to regulate BAF155 expression levels, which in turn control the expression of adherens junction genes (Xie et al., 2019).
In addition to phosphorylation and methylation, ubiquitylation has been identified on several BAF subunits, and several E3 ubiquitin ligases have been shown to interact with BAF complex subunits. TRIP12 interacts with BAF57 (Keppler and Archer, 2010), Unkempt interacts with BAF60B (Lorès et al., 2010), and CHFR interacts with BRG1, BAF47 and BAF60A (Jung et al., 2012). In these studies, ubiquitylation was shown to lead to degradation of BAF subunits that are not incorporated into the complex, suggesting a potential mechanism to inhibit binding of individual BAF subunits to chromatin when they are not integrated in the BAF complex.
Recruitment or antagonism by non-coding RNAs
In addition to protein-protein interactions, BAF chromatin targeting can be regulated by interactions with non-coding RNAs (Fig. 5). The long non-coding RNA, lncTCF7, which is highly expressed in hepatocellular carcinoma, can directly tether BAF to the promoter of its neighboring gene TCF7 to activate its expression (Wang et al., 2015). This subsequently results in activation of the Wnt signaling pathway for priming liver cancer stem cell self-renewal. The small nuclear RNA 7SK also physically interacts with BAF and recruits it to suppress pervasive transcription in ESCs and HeLa cells (Flynn et al., 2016; Hainer et al., 2015). The cardioprotective lncRNA Mhrt (myosin heavy-chain-associated RNA transcripts), which is abundantly expressed from Myh7 loci specifically in the heart, also interacts with BRG1; when BRG1 is aberrantly induced by pathological stress, Mhrt binds to the helicase domain of BRG1 to block it from binding to the promoters of Myh6, Myh7 and Opn to inhibit their gene expression and protect the heart from cardiomyopathy (Han et al., 2014).
Fig. 5.
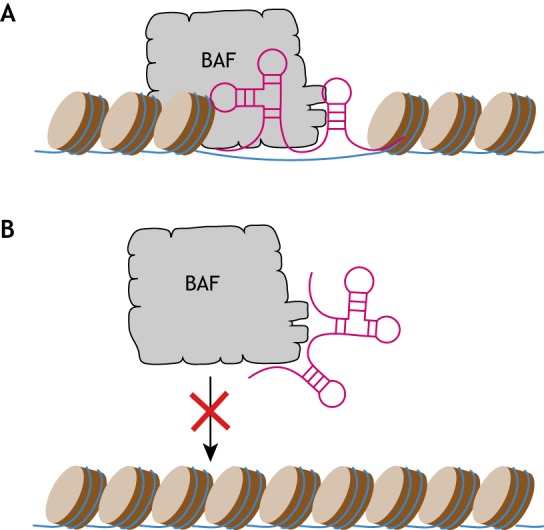
Roles of non-coding RNAs in modulating BAF targeting. Schematic illustration showing two different mechanisms by which non-coding RNAs (ncRNAs; purple) can modulate BAF targeting. (A) BAF can be tethered to chromatin through ncRNAs, as seen in the cases of lncTCF7 and the small nuclear RNA 7SK. (B) ncRNAs can also bind directly to BAF and inhibit the binding of BAF to chromatin, as seen in the case of Myhrt.
It should be noted, however, that the interaction between BAF and RNA has recently been re-evaluated. Previous studies demonstrated that the lncRNA SChLAP1 (second chromosome locus associated with prostate 1), which is highly expressed in a subset of metastatic prostate cancers, interacts with BAF47 (Prensner et al., 2013). However, detailed re-examination using RNA immunoprecipitation followed by high-throughput sequencing (RIP-seq) revealed that BAF displays uniform enrichment throughout many primary transcripts (Raab et al., 2019), which raises the issue of whether BAF-RNA interactions function broadly to modulate BAF activity across many genes or whether BAF only associates non-specifically with RNAs. These results pave the way for further experimentation to determine the exact functional role of BAF-RNA interactions in regulating gene expression.
Control of BAF targeting by lipids
Finally, the association of BAF with chromatin has also been shown to be regulated by lipids, namely phosphatidylinositol 4,5-bisphosphate (PIP2). In lymphocytes, global BAF-chromatin association is modulated by PIP2. Resting lymphocytes are characterized by small and compact nuclei, and the majority of BAF complexes in these cells are not tightly associated inside the nucleus. However, within 10 min of antigenic activation, BAF becomes tightly associated within the nucleus, and the nucleus volume increases 5- to 10-fold within hours (Rando et al., 2002; Zhao et al., 1998). This BAF-nucleus association is induced by PIP2, a major mediator of antigen receptor signaling in lymphocytes. BAF can directly bind PIP2, and PIP2 further stimulates BAF binding to the pointed ends and branch points of actin filaments, as well as chromatin, by relieving intramolecular capping of β-actin (Rando et al., 2002; Zhao et al., 1998). BAF-PIP2 binding is mediated by two BAF regulatory subunits: β-actin and BAF53A. Structural analysis indicates that BAF53A and β-actin form a heterodimer, using different interfaces from the regions involved in actin polymerization (Nishimoto et al., 2012). A similar observation was made with the two actin-related subunits in the yeast BAF (SWI/SNF) complex (Schubert et al., 2013). In the presence of PIP2, BAF has the capacity to bind actin filaments in addition to accommodating a BAF53-β-actin dimer (Rando et al., 2002; Zhao et al., 1998). Notably, a recent study reported global loss of BAF genomic binding in β-actin-knockout mouse embryonic fibroblasts (Xie et al., 2018), highlighting a crucial role for β-actin in BAF-chromatin association. It is unclear whether lipids are directly involved in BAF-chromatin association in other cell types in addition to lymphocytes and whether lipids other than PIP2 could be involved. It is also unclear whether the incorporation of BAF53B, which shares 87% amino acid identity with BAF53A (Wu et al., 2007), may affect BAF-lipid interaction.
Conclusions and future perspectives
The BAF complex functions as a highly versatile and tunable regulator for controlling specific gene expression as a result of its capacity to interact broadly with DNA, RNA, proteins and even lipids. Among these different interactions, one emerging theme is cooperativity. Within individual subunits, such as BRG1 and PBRM1, multiple bromodomains or DNA-binding modules establish multivalent interactions to enhance chromatin association. Cooperativity also occurs among different BAF subunits. For example, disruption of BAF47, BAF57 or BAF250 can all lead to impaired BAF binding at enhancers, suggesting that these subunits may cooperate to establish stable BAF binding. Furthermore, this enhancer binding is dependent on the interaction between BAF and transcription factors, which guide BAF to bind to specific DNA sequences. Thus, the selectivity of BAF chromatin binding is most likely a reflection of high-level cooperativity and positive reinforcement across multiple targeting mechanisms.
The flexibility of BAF binding, controlled by signaling, is another major feature modulating global or specific BAF chromatin association. As discussed above, BAF can be switched ‘on’ and ‘off’ on chromatin during the cell cycle in response to its phosphorylation, while PIP2 rapidly induces global BAF chromatin association during the process of T cell activation. DNA damage also signals BAF recruitment to a repair foci, while numerous post-translational modifications and non-coding RNAs can modulate BAF binding to specific sites. These interactions present another layer of regulation to control BAF-chromatin association, beyond the intrinsic binding capacities of DNA/histone-binding domains and recruitment by transcription factors.
It is noteworthy that the binding of BAF to chromatin is not always accompanied by chromatin remodeling on the site. Indeed, although BRD9 can recruit BRG1 to VDR-binding sites prior to ligand association, chromatin remodeling is not initiated until this BRD9-containing BAF is replaced by BRD7-containing PBAF upon ligand binding (Wei et al., 2018). Moreover, in differentiated keratinocytes, BRG1/BRM loss only impairs chromatin accessibility at a subset of BAF-binding sites (Bao et al., 2015), although this could be explained by substantial colocalization among other chromatin remodelers at many genomic sites (Morris et al., 2014). Therefore, BAF may require additional signals to fully activate its catalytic action, and may also play redundant roles with other remodelers in governing chromatin accessibility at a subset of genomic regions.
As well as binding to chromatin, BAF can influence chromatin modifications. In differentiated keratinocytes, BRG1/BRM knockdown leads to the downregulation of H3K27Ac on sites that are dependent on BAF to sustain genome accessibility (Bao et al., 2015). Similarly, deletion of BRG1, BAF250A or BAF47 in mouse embryonic fibroblasts globally decreases H3K27Ac (Alver et al., 2017), while, during cortical development, BAF155 and BAF170 knockout causes global upregulation of H3K27me3 as well as reduction of H3K9Ac (Narayanan et al., 2015). The physical interactions between BAF and histone modifiers could explain this epigenomic dysregulation. Indeed, BAF reportedly interacts with the histone acetyltransferase p300 to sustain H3K27Ac at enhancers (Alver et al., 2017). Complex purification of BAF in the developing pallium during late neurogenesis also identified that the demethylases KDM6A/B and KDM1A associate with BAF to suppress both H3K27me2/3 as well as H3K4me2 in promoting neuronal differentiation and inhibiting cell proliferation, respectively (Nguyen et al., 2018). These studies indicate that BAF functions beyond simply recognizing and binding to histone modifications, and actively cooperates together with other histone modifiers to control the epigenomic landscape.
Leveraging BAF-ATPase-deficient cell lines, the BAF catalytic subunit was recently found to play a major role in defining BAF/PBAF identity as well as in promoting chromatin association. Furthermore, the catalytic activity of BAF is required for the complex to bind to a considerable subset of genomic sites (Pan et al., 2019). This study is in agreement with the highly periodic nature of BAF binding observed in vitro (Nagaich et al., 2004), and it indicates that BAF may require completion of each catalytic cycle to regain binding to at least a subset of its genomic targets.
Overall, these studies highlight the sophisticated action of BAF in binding and remodeling chromatin. It is highly plausible that many molecular mechanisms are cell and tissue specific, especially when considering the variations in both the subunit composition of BAF as well as its potential interacting partners. Thus, future research delineating the action as well as the interacting networks of BAF, in different cell and tissue types, should provide more insights for understanding how BAF dysregulation impacts the pathogenesis of diseases such as neurological disorders and various cancers.
Acknowledgements
We apologize to colleagues whose original publications and reviews related to the topic are not directly referenced due to limited space.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
The authors’ research is supported by a National Institutes of Health K99/R00 Award (R00AR065480) and by the Searle Leadership Fund (X.B.); by a National Institutes of Health Carcinogenesis T32 training grant (T32CA009560) to P.J.H.; and by a National Institutes of Health CMBD T32 training grant (T32GM008061) to S.M.L. Deposited in PMC for release after 12 months.
References
- Abraham B. J., Cui K., Tang Q. and Zhao K. (2013). Dynamic regulation of epigenomic landscapes during hematopoiesis. BMC Genomics 14, 193 10.1186/1471-2164-14-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agalioti T., Chen G. and Thanos D. (2002). Deciphering the transcriptional histone acetylation code for a human gene. Cell 111, 381-392. 10.1016/S0092-8674(02)01077-2 [DOI] [PubMed] [Google Scholar]
- Alfert A., Moreno N. and Kerl K. (2019). The BAF complex in development and disease. Epigenet. Chromatin 12, 19 10.1186/s13072-019-0264-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M. D., Freund S. M. V., Zinzalla G. and Bycroft M. (2015). The SWI/SNF subunit INI1 contains an N-terminal winged helix DNA binding domain that is a target for mutations in Schwannomatosis. Structure 23, 1344-1349. 10.1016/j.str.2015.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpsoy A. and Dykhuizen E. C. (2018). Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J. Biol. Chem. 293, 3892-3903. 10.1074/jbc.RA117.001065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alver B. H., Kim K. H., Lu P., Wang X., Manchester H. E., Wang W., Haswell J. R., Park P. J. and Roberts C. W. M. (2017). The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat. Commun. 8, 14648 10.1038/ncomms14648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang Y.-S., Rivas R. N., Ribeiro A. J. S., Srivas R., Rivera J., Stone N. R., Pratt K., Mohamed T. M. A., Fu J.-D., Spencer C. I. et al. (2016). Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell 167, 1734-1749.e22. 10.1016/j.cell.2016.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J. A., Bieker J. J. and Emerson B. M. (1998). A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95, 93-104. 10.1016/S0092-8674(00)81785-7 [DOI] [PubMed] [Google Scholar]
- Bao X., Tang J., Lopez-Pajares V., Tao S., Qu K., Crabtree G. R. and Khavari P. A. (2013). ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell 12, 193-203. 10.1016/j.stem.2012.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X., Rubin A. J., Qu K., Zhang J., Giresi P. G., Chang H. Y. and Khavari P. A. (2015). A novel ATAC-seq approach reveals lineage-specific reinforcement of the open chromatin landscape via cooperation between BAF and p63. Genome Biol. 16, 284 10.1186/s13059-015-0840-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barutcu A. R., Lajoie B. R., Fritz A. J., McCord R. P., Nickerson J. A., van Wijnen A. J., Lian J. B., Stein J. L., Dekker J., Stein G. S. et al. (2016). SMARCA4 regulates gene expression and higher-order chromatin structure in proliferating mammary epithelial cells. Genome Res. 26, 1188-1201. 10.1101/gr.201624.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belandia B., Dauvois S., L'Horset F., Lopez G., Hoare S., Kushner P. J. and Parker M. G. (2002). Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 21, 4094-4103. 10.1093/emboj/cdf412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourachot B., Yaniv M. and Muchardt C. (1999). The activity of mammalian brm/SNF2alpha is dependent on a high-mobility-group protein I/Y-like DNA binding domain. Mol. Cell. Biol. 19, 3931-3939. 10.1128/MCB.19.6.3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B. R., Lorch Y., Li Y., Zhang M., Lacomis L., Erdjument-Bromage H., Tempst P., Du J., Laurent B. and Kornberg R. D. (1996). RSC, an essential, abundant chromatin-remodeling complex. Cell 87, 1249-1260. 10.1016/S0092-8674(00)81820-6 [DOI] [PubMed] [Google Scholar]
- Carrera I., Zavadil J. and Treisman J. E. (2008). Two subunits specific to the PBAP chromatin remodeling complex have distinct and redundant functions during Drosophila development. Mol. Cell. Biol. 28, 5238-5250. 10.1128/MCB.00747-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celen C., Chuang J.-C., Luo X., Nijem N., Walker A. K., Chen F., Zhang S., Chung A. S., Nguyen L. H., Nassour I. et al. (2017). Arid1b haploinsufficient mice reveal neuropsychiatric phenotypes and reversible causes of growth impairment. eLife 6, e25730 10.7554/eLife.25730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler R. L., Brennan J., Schisler J. C., Serber D., Patterson C. and Magnuson T. (2013). ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol. Cell. Biol. 33, 265-280. 10.1128/MCB.01008-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. and Archer T. K. (2005). Regulating SWI/SNF subunit levels via protein-protein interactions and proteasomal degradation: BAF155 and BAF170 limit expression of BAF57. Mol. Cell. Biol. 25, 9016-9027. 10.1128/MCB.25.20.9016-9027.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi T. H., Wan M., Zhao K., Taniuchi I., Chen L., Littman D. R. and Crabtree G. R. (2002). Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418, 195-199. 10.1038/nature00876 [DOI] [PubMed] [Google Scholar]
- Cong P., Jie Z., Ying L. H., Ming Z., Li W. L., Hong Z. Q., Xin Y. Y., Wei X., Rong S. S., Ling L. X. et al. (2006). The transcriptional regulation role of BRD7 by binding to acetylated histone through bromodomain. J. Cell. Biochem. 97, 882-892. 10.1002/jcb.20645 [DOI] [PubMed] [Google Scholar]
- Côté J., Quinn J., Workman J. L. and Peterson C. L. (1994). Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265, 53-60. 10.1126/science.8016655 [DOI] [PubMed] [Google Scholar]
- Dallas P. B., Pacchione S., Wilsker D., Bowrin V., Kobayashi R. and Moran E. (2000). The human SWI-SNF complex protein p270 is an ARID family member with non-sequence-specific DNA binding activity. Mol. Cell. Biol. 20, 3137-3146. 10.1128/MCB.20.9.3137-3146.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debril M.-B., Gelman L., Fayard E., Annicotte J.-S., Rocchi S. and Auwerx J. (2004). Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J. Biol. Chem. 279, 16677-16686. 10.1074/jbc.M312288200 [DOI] [PubMed] [Google Scholar]
- DiRenzo J., Shang Y., Phelan M., Sif S., Myers M., Kingston R. and Brown M. (2000). BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol. Cell. Biol. 20, 7541-7549. 10.1128/MCB.20.20.7541-7549.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfring L. K., Daniel C., Papoulas O., Deuring R., Sarte M., Moseley S., Beek S. J., Waldrip W. R., Daubresse G., DePace A. et al. (1998). Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics 148, 251-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov O., Castex J., Tallant C., Owen D. R., Martin S., Aldeghi M., Monteiro O., Filippakopoulos P., Picaud S., Trzupek J. D. et al. (2015). Selective targeting of the BRG/PB1 bromodomains impairs embryonic and trophoblast stem cell maintenance. Sci. Adv. 1, e1500723-e1500723. 10.1126/sciadv.1500723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn E. M., Huang O. W., Poy F., Oppikofer M., Bellon S. F., Tang Y. and Cochran A. G. (2015). A subset of human bromodomains recognizes butyryllysine and crotonyllysine histone peptide modifications. Structure 23, 1801-1814. 10.1016/j.str.2015.08.004 [DOI] [PubMed] [Google Scholar]
- Flynn R. A., Do B. T., Rubin A. J., Calo E., Lee B., Kuchelmeister H., Rale M., Chu C., Kool E. T., Wysocka J. et al. (2016). 7SK-BAF axis controls pervasive transcription at enhancers. Nat. Struct. Mol. Biol. 23, 231-238. 10.1038/nsmb.3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcales S. V., Albini S., Giordani L., Malecova B., Cignolo L., Chernov A., Coutinho P., Saccone V., Consalvi S., Williams R. et al. (2012). Signal-dependent incorporation of MyoD-BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. EMBO J. 31, 301-316. 10.1038/emboj.2011.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer C. J. and Archer T. K. (1998). Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 393, 88-91. 10.1038/30032 [DOI] [PubMed] [Google Scholar]
- García-Pedrero J. M., Kiskinis E., Parker M. G. and Belandia B. (2006). The SWI/SNF chromatin remodeling subunit BAF57 is a critical regulator of estrogen receptor function in breast cancer cells. J. Biol. Chem. 281, 22656-22664. 10.1074/jbc.M602561200 [DOI] [PubMed] [Google Scholar]
- Gatchalian J., Malik S., Ho J., Lee D.-S., Kelso T. W. R., Shokhirev M. N., Dixon J. R. and Hargreaves D. C. (2018). A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat. Commun. 9, 5139 10.1038/s41467-018-07528-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B., Wang T.-L. and Shih I.-M. (2011). ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 71, 6718-6727. 10.1158/0008-5472.CAN-11-1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer S. J., Gu W., Carone B. R., Landry B. D., Rando O. J., Mello C. C. and Fazzio T. G. (2015). Suppression of pervasive noncoding transcription in embryonic stem cells by esBAF. Genes Dev. 29, 362-378. 10.1101/gad.253534.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P., Li W., Lin C.-H., Yang J., Shang C., Nurnberg S. T., Jin K. K., Xu W., Lin C.-Y., Lin C.-J. et al. (2014). A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514, 102-106. 10.1038/nature13596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helming K. C., Wang X., Wilson B. G., Vazquez F., Haswell J. R., Manchester H. E., Kim Y., Kryukov G. V., Ghandi M., Aguirre A. J. et al. (2014). ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat. Med. 20, 251-254. 10.1038/nm.3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsmoortel C., Vulto-van Silfhout A. T., Coe B. P., Vandeweyer G., Rooms L., van den Ende J., Schuurs-Hoeijmakers J. H. M., Marcelis C. L., Willemsen M. H., Vissers L. E. L. M. et al. (2014). A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat. Genet. 46, 380-384. 10.1038/ng.2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Jothi R., Ronan J. L., Cui K., Zhao K. and Crabtree G. R. (2009). An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. USA 106, 5187-5191. 10.1073/pnas.0812888106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges C., Kirkland J. G. and Crabtree G. R. (2016). The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb. Perspect. Med. 6, a026930 10.1101/cshperspect.a026930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman J. A., Trotter K. W., Ward J. M. and Archer T. K. (2018). BRG1 governs glucocorticoid receptor interactions with chromatin and pioneer factors across the genome. eLife 7, e35073 10.7554/eLife.35073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hota S. K. and Bruneau B. G. (2016). ATP-dependent chromatin remodeling during mammalian development. Development 143, 2882-2897. 10.1242/dev.128892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao P.-W., Fryer C. J., Trotter K. W., Wang W. and Archer T. K. (2003). BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol. Cell. Biol. 23, 6210-6220. 10.1128/MCB.23.17.6210-6220.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Yamauchi M., Nishina M., Yamamichi N., Mizutani T., Ui M., Murakami M. and Iba H. (2001). Identification of SWI·SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers. J. Biol. Chem. 276, 2852-2857. 10.1074/jbc.M009633200 [DOI] [PubMed] [Google Scholar]
- Johnson T. A., Elbi C., Parekh B. S., Hager G. L. and John S. (2008). Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor–regulated promoter. Mol. Biol. Cell 19, 3308-3322. 10.1091/mbc.e08-02-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. A., Chereji R. V., Stavreva D. A., Morris S. A., Hager G. L. and Clark D. J. (2018). Conventional and pioneer modes of glucocorticoid receptor interaction with enhancer chromatin in vivo. Nucleic Acids Res.. 46, 203-214. 10.1093/nar/gkx1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung I., Sohn D. H., Choi J., Kim J. M., Jeon S., Seol J. H. and Seong R. H. (2012). SRG3/mBAF155 stabilizes the SWI/SNF-like BAF complex by blocking CHFR mediated ubiquitination and degradation of its major components. Biochem. Biophys. Res. Commun. 418, 512-517. 10.1016/j.bbrc.2012.01.057 [DOI] [PubMed] [Google Scholar]
- Kadoch C., Hargreaves D. C., Hodges C., Elias L., Ho L., Ranish J. and Crabtree G. R. (2013). Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592-601. 10.1038/ng.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser M. D., Aslanian A., Dong M.-Q., Yates J. R., Emerson B. M. and Emerson B. M. (2008). BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J. Biol. Chem. 283, 32254-32263. 10.1074/jbc.M806061200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebede A. F., Nieborak A., Shahidian L. Z., Le Gras S., Richter F., Gómez D. A., Baltissen M. P., Meszaros G., Magliarelli H. D. F., Taudt A. et al. (2017). Histone propionylation is a mark of active chromatin. Nat. Struct. Mol. Biol. 24, 1048-1056. 10.1038/nsmb.3490 [DOI] [PubMed] [Google Scholar]
- Keppler B. R. and Archer T. K. (2010). Ubiquitin-dependent and ubiquitin-independent control of subunit stoichiometry in the SWI/SNF complex. J. Biol. Chem. 285, 35665-35674. 10.1074/jbc.M110.173997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Andrés Salazar Hernández M., Herrema H., Delibasi T. and Park S. W. (2016). The role of BRD7 in embryo development and glucose metabolism. J. Cell. Mol. Med. 20, 1561-1570. 10.1111/jcmm.12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King H. W. and Klose R. J. (2017). The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. eLife 6, e22631 10.7554/eLife.22631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowenz-Leutz E. and Leutz A. (1999). A C/EBPβ isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4, 735-743. 10.1016/S1097-2765(00)80384-6 [DOI] [PubMed] [Google Scholar]
- Krasteva V., Crabtree G. R. and Lessard J. A. (2017). The BAF45a/PHF10 subunit of SWI/SNF-like chromatin remodeling complexes is essential for hematopoietic stem cell maintenance. Exp. Hematol. 48, 58-71.e15. 10.1016/j.exphem.2016.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R., Chen A. F., Pantovich M. G., Danial M., Parchem R. J., Labosky P. A. and Blelloch R. (2016). FOXD3 regulates pluripotent stem cell potential by simultaneously initiating and repressing enhancer activity. Cell Stem Cell 18, 104-117. 10.1016/j.stem.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M. J. et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317-330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H., Imbalzano A. N., Khavari P. A., Kingston R. E. and Green M. R. (1994). Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370, 477-481. 10.1038/370477a0 [DOI] [PubMed] [Google Scholar]
- Kwon S.-J., Park J.-H., Park E.-J., Lee S.-A., Lee H.-S., Kang S. W. and Kwon J. (2015). ATM-mediated phosphorylation of the chromatin remodeling enzyme BRG1 modulates DNA double-strand break repair. Oncogene 34, 303-313. 10.1038/onc.2013.556 [DOI] [PubMed] [Google Scholar]
- Laurette P., Strub T., Koludrovic D., Keime C., Le Gras S., Seberg H., Van Otterloo E., Imrichova H., Siddaway R., Aerts S. et al. (2015). Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. eLife 2015, e06857 10.7554/eLife.06857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Kim J. W., Seo T., Hwang S. G., Choi E.-J. and Choe J. (2002). SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J. Biol. Chem. 277, 22330-22337. 10.1074/jbc.M111987200 [DOI] [PubMed] [Google Scholar]
- Lei I., Gao X., Sham M. H. and Wang Z. (2012). SWI/SNF protein component BAF250a regulates cardiac progenitor cell differentiation by modulating chromatin accessibility during second heart field development. J. Biol. Chem. 287, 24255-24262. 10.1074/jbc.M112.365080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J., Wu J. I., Ranish J. A., Wan M., Winslow M. M., Staahl B. T., Wu H., Aebersold R., Graef I. A. and Crabtree G. R. (2007). An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55, 201-215. 10.1016/j.neuron.2007.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H., Takeuchi J. K., von Both I., Walls J. R., McAuliffe F., Lee Adamson S., Mark Henkelman R., Wrana J. L., Rossant J. and Bruneau B. G. (2004). Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 432, 107-112. 10.1038/nature03071 [DOI] [PubMed] [Google Scholar]
- Link K. A., Burd C. J., Williams E., Marshall T., Rosson G., Henry E., Weissman B. and Knudsen K. E. (2005). BAF57 governs androgen receptor action and androgen-dependent proliferation through SWI/SNF. Mol. Cell. Biol. 25, 2200-2215. 10.1128/MCB.25.6.2200-2215.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Local A., Huang H., Albuquerque C. P., Singh N., Lee A. Y., Wang W., Wang C., Hsia J. E., Shiau A. K., Ge K. et al. (2018). Identification of H3K4me1-associated proteins at mammalian enhancers. Nat. Genet. 50, 73-82. 10.1038/s41588-017-0015-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorès P., Visvikis O., Luna R., Lemichez E. and Gacon G. (2010). The SWI/SNF protein BAF60b is ubiquitinated through a signalling process involving Rac GTPase and the RING finger protein Unkempt. FEBS J.. 277, 1453-1464. 10.1111/j.1742-4658.2010.07575.x [DOI] [PubMed] [Google Scholar]
- Mashtalir N., D'Avino A. R., Michel B. C., Luo J., Pan J., Otto J. E., Zullow H. J., McKenzie Z. M., Kubiak R. L., St, Pierre R., et al. (2018). Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell 175, 1272-1288.e20. 10.1016/j.cell.2018.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur R., Alver B. H., San Roman A. K., Wilson B. G., Wang X., Agoston A. T., Park P. J., Shivdasani R. A. and Roberts C. W. M. (2017). ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat. Genet. 49, 296-302. 10.1038/ng.3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna B., Guo M., Reynolds A., Hara M. and Stein R. (2015). Dynamic recruitment of functionally distinct Swi/Snf chromatin remodeling complexes modulates Pdx1 activity in Islet β cells. Cell Rep. 10, 2032-2042. 10.1016/j.celrep.2015.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B. C., D'Avino A. R., Cassel S. H., Mashtalir N., McKenzie Z. M., McBride M. J., Valencia A. M., Zhou Q., Bocker M., Soares L. M. M. et al. (2018). A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat. Cell Biol. 20, 1410-1420. 10.1038/s41556-018-0221-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middeljans E., Wan X., Jansen P. W., Sharma V., Stunnenberg H. G. and Logie C. (2012). SS18 together with animal-specific factors defines human BAF-type SWI/SNF complexes. PLoS ONE 7, e33834 10.1371/journal.pone.0033834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrmann L., Langenberg K., Krijgsveld J., Kal A. J., Heck A. J. R. and Verrijzer C. P. (2004). Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol. Cell. Biol. 24, 3077-3088. 10.1128/MCB.24.8.3077-3088.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S. A., Baek S., Sung M.-H., John S., Wiench M., Johnson T. A., Schiltz R. L. and Hager G. L. (2014). Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat. Struct. Mol. Biol. 21, 73-81. 10.1038/nsmb.2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison E. A., Sanchez J. C., Ronan J. L., Farrell D. P., Varzavand K., Johnson J. K., Gu B. X., Crabtree G. R. and Musselman C. A. (2017). DNA binding drives the association of BRG1/hBRM bromodomains with nucleosomes. Nat. Commun. 8, 16080 10.1038/ncomms16080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C., Reyes J. C., Bourachot B., Leguoy E. and Yaniv M. (1996). The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J.. 15, 3394-3402. 10.1002/j.1460-2075.1996.tb00705.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaich A. K., Walker D. A., Wolford R. and Hager G. L. (2004). Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol. Cell 14, 163-174. 10.1016/S1097-2765(04)00178-9 [DOI] [PubMed] [Google Scholar]
- Nakayama R. T., Pulice J. L., Valencia A. M., McBride M. J., McKenzie Z. M., Gillespie M. A., Ku W. L., Teng M., Cui K., Williams R. T. et al. (2017). SMARCB1 is required for widespread BAF complex–mediated activation of enhancers and bivalent promoters. Nat. Genet. 49, 1613-1623. 10.1038/ng.3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan R., Pirouz M., Kerimoglu C., Pham L., Wagener R. J., Kiszka K. A., Rosenbusch J., Seong R. H., Kessel M., Fischer A. et al. (2015). Loss of BAF (mSWI/SNF) complexes causes global transcriptional and chromatin state changes in forebrain development. Cell Rep. 13, 1842-1854. 10.1016/j.celrep.2015.10.046 [DOI] [PubMed] [Google Scholar]
- Nasipak B. T., Padilla-Benavides T., Green K. M., Leszyk J. D., Mao W., Konda S., Sif S., Shaffer S. A., Ohkawa Y. and Imbalzano A. N. (2015). Opposing calcium-dependent signalling pathways control skeletal muscle differentiation by regulating a chromatin remodelling enzyme. Nat. Commun. 6, 7441 10.1038/ncomms8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H., Kerimoglu C., Pirouz M., Pham L., Kiszka K. A., Sokpor G., Sakib M. S., Rosenbusch J., Teichmann U., Seong R. H. et al. (2018). Epigenetic regulation by BAF complexes limits neural stem cell proliferation by suppressing Wnt signaling in late embryonic development. Stem Cell Rep. 10, 1734-1750. 10.1016/j.stemcr.2018.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z., Xue Y., Yang D., Zhou S., Deroo B. J., Archer T. K. and Wang W. (2000). A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 20, 8879-8888. 10.1128/MCB.20.23.8879-8888.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninkovic J., Steiner-Mezzadri A., Jawerka M., Akinci U., Masserdotti G., Petricca S., Fischer J., von Holst A., Beckers J., Lie C. D. et al. (2013). The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell 13, 403-418. 10.1016/j.stem.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto N., Watanabe M., Watanabe S., Sugimoto N., Yugawa T., Ikura T., Koiwai O., Kiyono T. and Fujita M. (2012). Heterocomplex formation by Arp4 and β-actin is involved in the integrity of the Brg1 chromatin remodeling complex. J. Cell Sci. 125, 3870-3882. 10.1242/jcs.104349 [DOI] [PubMed] [Google Scholar]
- Oh J., Sohn D. H., Ko M., Chung H., Jeon S. H. and Seong R. H. (2008). BAF60a interacts with p53 to recruit the SWI/SNF complex. J. Biol. Chem. 283, 11924-11934. 10.1074/jbc.m705401200 [DOI] [PubMed] [Google Scholar]
- Padilla-Benavides T., Nasipak B. T., Paskavitz A. L., Haokip D. T., Schnabl J. M., Nickerson J. A. and Imbalzano A. N. (2017). Casein kinase 2-mediated phosphorylation of Brahma-related gene 1 controls myoblast proliferation and contributes to SWI/SNF complex composition. J. Biol. Chem. 292, 18592-18607. 10.1074/jbc.M117.799676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., McKenzie Z. M., D'Avino A. R., Mashtalir N., Lareau C. A., St. Pierre R., Wang L., Shilatifard A. and Kadoch C. (2019). The ATPase module of mammalian SWI/SNF family complexes mediates subcomplex identity and catalytic activity–independent genomic targeting. Nat. Genet. 51, 618-626. 10.1038/s41588-019-0363-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsialou A., Wilsker D. and Moran E. (2005). DNA-binding properties of ARID family proteins. Nucleic Acids Res.. 33, 66-80. 10.1093/nar/gki145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen T. Å., Kowenz-Leutz E., Leutz A. and Nerlov C. (2001). Cooperation between C/EBPα TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev.. 15, 3208-3216. 10.1101/gad.209901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister N. T., Fomin V., Regunath K., Zhou J. Y., Zhou W., Silwal-Pandit L., Freed-Pastor W. A., Laptenko O., Neo S. P., Bargonetti J. et al. (2015). Mutant p53 cooperates with the SWI/SNF chromatin remodeling complex to regulate VEGFR2 in breast cancer cells. Genes Dev. 29, 1298-1315. 10.1101/gad.263202.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan M. L., Sif S., Narlikar G. J. and Kingston R. E. (1999). Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3, 247-253. 10.1016/S1097-2765(00)80315-9 [DOI] [PubMed] [Google Scholar]
- Porter E. G. and Dykhuizen E. C. (2017). Individual bromodomains of polybromo-1 contribute to chromatin association and tumor suppression in clear cell renal carcinoma. J. Biol. Chem. 292, 2601-2610. 10.1074/jbc.M116.746875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner J. R., Iyer M. K., Sahu A., Asangani I. A., Cao Q., Patel L., Vergara I. A., Davicioni E., Erho N., Ghadessi M. et al. (2013). The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 45, 1392-1398. 10.1038/ng.2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priam P., Krasteva V., Rousseau P., D'Angelo G., Gaboury L., Sauvageau G. and Lessard J. A. (2017). SMARCD2 subunit of SWI/SNF chromatin-remodeling complexes mediates granulopoiesis through a CEBPɛ dependent mechanism. Nat. Genet. 49, 753-764. 10.1038/ng.3812 [DOI] [PubMed] [Google Scholar]
- Raab J. R., Resnick S. and Magnuson T. (2015). Genome-wide transcriptional regulation mediated by biochemically distinct SWI/SNF complexes. PLoS Genet.. 11, e1005748 10.1371/journal.pgen.1005748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab J. R., Smith K. N., Spear C. C., Manner C. J., Calabrese J. M. and Magnuson T. (2019). SWI/SNF remains localized to chromatin in the presence of SCHLAP1. Nat. Genet. 51, 26-29. 10.1038/s41588-018-0272-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A. and Wysocka J. (2011). A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279-283. 10.1038/nature09692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S., Sowa M. E., Ottinger M., Smith J. A., Shi Y., Harper J. W. and Howley P. M. (2011). The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol. Cell. Biol. 31, 2641-2652. 10.1128/MCB.01341-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando O. J., Zhao K., Janmey P. and Crabtree G. R. (2002). Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc. Natl. Acad. Sci. USA 99, 2824-2829. 10.1073/pnas.032662899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remillard D., Buckley D. L., Paulk J., Brien G. L., Sonnett M., Seo H.-S., Dastjerdi S., Wühr M., Dhe-Paganon S., Armstrong S. A. et al. (2017). Degradation of the BAF complex factor BRD9 by Heterobifunctional ligands. Angew. Chemie Int. Ed. 56, 5738-5743. 10.1002/anie.201611281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert H. L., Wittmeyer J., Kasten M. M., Hinata K., Rawling D. C., Héroux A., Cairns B. R. and Hill C. P. (2013). Structure of an actin-related subcomplex of the SWI/SNF chromatin remodeler. Proc. Natl. Acad. Sci. USA 110, 3345-3350. 10.1073/pnas.1215379110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira S. N., Lim H.-W., Rajakumari S., Sakers A. P., Ishibashi J., Harms M. J., Won K.-J. and Seale P. (2017). EBF2 transcriptionally regulates brown adipogenesis via the histone reader DPF3 and the BAF chromatin remodeling complex. Genes Dev.. 31, 660-673. 10.1101/gad.294405.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Xu C., Huang W., Zhang J., Carlson J. E., Tu X., Wu J. and Shi Y. (2007). Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry 46, 2100-2110. 10.1021/bi0611208 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Uchida M., Takeshita H., Nishiwaki K. and Sawa H. (2012). Multiple functions of PBRM-1/Polybromo- and LET-526/Osa-containing chromatin remodeling complexes in C. elegans development. Dev. Biol. 361, 349-357. 10.1016/j.ydbio.2011.10.035 [DOI] [PubMed] [Google Scholar]
- Sif S., Stukenberg P. T., Kirschner M. W. and Kingston R. E. (1998). Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev.. 12, 2842-2851. 10.1101/gad.12.18.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sima J., Yan Z., Chen Y., Lehrmann E., Zhang Y., Nagaraja R., Wang W., Wang Z. and Schlessinger D. (2018). Eda-activated RelB recruits an SWI/SNF (BAF) chromatin-remodeling complex and initiates gene transcription in skin appendage formation. Proc. Natl. Acad. Sci. USA 115, 8173-8178. 10.1073/pnas.1800930115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone C., Forcales S. V., Hill D. A., Imbalzano A. N., Latella L. and Puri P. L. (2004). p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat. Genet. 36, 738-743. 10.1038/ng1378 [DOI] [PubMed] [Google Scholar]
- Singh M., D'Silva L. and Holak T. A. (2006). DNA-binding properties of the recombinant high-mobility-group-like AT-hook-containing region from human BRG1 protein. Biol. Chem. 387, 1469-1478. 10.1515/BC.2006.184 [DOI] [PubMed] [Google Scholar]
- Sohn D. H., Lee K. Y., Lee C., Oh J., Chung H., Jeon S. H. and Seong R. H. (2007). SRG3 interacts directly with the major components of the SWI/SNF chromatin remodeling complex and protects them from proteasomal degradation. J. Biol. Chem. 282, 10614-10624. 10.1074/jbc.M610563200 [DOI] [PubMed] [Google Scholar]
- Sutherell C. L., Tallant C., Monteiro O. P., Yapp C., Fuchs J. E., Fedorov O., Siejka P., Müller S., Knapp S., Brenton J. D. et al. (2016). Identification and development of 2,3-dihydropyrrolo[1,2-a]quinazolin-5(1H)-one inhibitors targeting bromodomains within the switch/sucrose nonfermenting complex. J. Med. Chem. 59, 5095-5101. 10.1021/acs.jmedchem.5b01997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaku M., Grimm S. A., Shimbo T., Perera L., Menafra R., Stunnenberg H. G., Archer T. K., Machida S., Kurumizaka H. and Wade P. A. (2016). GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol. 17, 36 10.1186/s13059-016-0897-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi J. K. and Bruneau B. G. (2009). Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459, 708-711. 10.1038/nature08039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoulou N. H., Bamborough P., Bannister A. J., Becher I., Bit R. A., Che K. H., Chung C.-W., Dittmann A., Drewes G., Drewry D. H. et al. (2016). Discovery of I-BRD9, a selective cell active chemical probe for bromodomain containing protein 9 inhibition. J. Med. Chem. 59, 1425-1439. 10.1021/acs.jmedchem.5b00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., Ling E., Cowley C. J., Couch C. H., Wang X., Harmin D. A., Roberts C. W. M. and Greenberg M. E. (2017). AP-1 transcription factors and the BAF complex mediate signal-dependent enhancer selection. Mol. Cell 68, 1067-1082.e12. 10.1016/j.molcel.2017.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Côté J., Xue Y., Zhou S., Khavari P. A., Biggar S. R., Muchardt C., Kalpana G. V., Goff S. P., Yaniv M. et al. (1996). Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J.. 15, 5370-5382. 10.1002/j.1460-2075.1996.tb00921.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Chi T., Xue Y., Zhou S., Kuo A. and Crabtree G. R. (1998). Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl. Acad. Sci. USA 95, 492-498. 10.1073/pnas.95.2.492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhai W., Richardson J. A., Olson E. N., Meneses J. J., Firpo M. T., Kang C., Skarnes W. C. and Tjian R. (2004). Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 18, 3106-3116. 10.1101/gad.1238104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wong R. H. F., Tang T., Hudak C. S., Yang D., Duncan R. E. and Sul H. S. (2013). Phosphorylation and recruitment of BAF60c in chromatin remodeling for lipogenesis in response to insulin. Mol. Cell 49, 283-297. 10.1016/j.molcel.2012.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhao Z., Meyer M. B., Saha S., Yu M., Guo A., Wisinski K. B., Huang W., Cai W., Pike J. W. et al. (2014). CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell 25, 21-36. 10.1016/j.ccr.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., He L., Du Y., Zhu P., Huang G., Luo J., Yan X., Ye B., Li C., Xia P. et al. (2015). The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 16, 413-425. 10.1016/j.stem.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Wang X., Wang S., Troisi E. C., Howard T. P., Haswell J. R., Wolf B. K., Hawk W. H., Ramos P., Oberlick E. M., Tzvetkov E. P. et al. (2019). BRD9 defines a SWI/SNF sub-complex and constitutes a specific vulnerability in malignant rhabdoid tumors. Nat. Commun. 10, 1881 10.1038/s41467-019-09891-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. M., Wallace L. J., Cowan D., Shadbolt P. and Levison P. R. (1991). Phosphocellulose as a tool for rapid purification of DNA-modifying enzymes. Anal. Chim. Acta 249, 195-200. 10.1016/0003-2670(91)87022-Y [DOI] [Google Scholar]
- Wei Z., Yoshihara E., He N., Hah N., Fan W., Pinto A. F. M., Huddy T., Wang Y., Ross B., Estepa G. et al. (2018). Vitamin D switches BAF complexes to protect β cells. Cell 173, 1135-1149.e15. 10.1016/j.cell.2018.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsker D., Patsialou A., Zumbrun S. D., Kim S., Chen Y., Dallas P. B. and Moran E. (2004). The DNA-binding properties of the ARID-containing subunits of yeast and mammalian SWI/SNF complexes. Nucleic Acids Res. 32, 1345 10.1093/nar/gkh277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzel M., Petersheim D., Fan Y., Bahrami E., Racek T., Rohlfs M., Puchałka J., Mertes C., Gagneur J., Ziegenhain C. et al. (2017). Chromatin-remodeling factor SMARCD2 regulates transcriptional networks controlling differentiation of neutrophil granulocytes. Nat. Genet. 49, 742-752. 10.1038/ng.3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. I., Lessard J., Olave I. A., Qiu Z., Ghosh A., Graef I. A. and Crabtree G. R. (2007). Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 56, 94-108. 10.1016/j.neuron.2007.08.021 [DOI] [PubMed] [Google Scholar]
- Xie X., Almuzzaini B., Drou N., Kremb S., Yousif A., Farrants A.-K. Ö., Gunsalus K. and Percipalle P. (2018). β-Actin-dependent global chromatin organization and gene expression programs control cellular identity. FASEB J.. 32, 1296-1314. 10.1096/fj.201700753R [DOI] [PubMed] [Google Scholar]
- Xie Y., Castro-Hernández R., Sokpor G., Pham L., Narayanan R., Rosenbusch J., Staiger J. F. and Tuoc T. (2019). RBM15 modulates the function of chromatin remodeling factor BAF155 through RNA methylation in developing cortex. Mol. Neurobiol., 1-16. 10.1007/s12035-019-1595-1 [DOI] [PubMed] [Google Scholar]
- Xiong X., Panchenko T., Yang S., Zhao S., Yan P., Zhang W., Xie W., Li Y., Zhao Y., Allis C. D. et al. (2016). Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat. Chem. Biol. 12, 1111-1118. 10.1038/nchembio.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Flowers S. and Moran E. (2012). Essential role of ARID2 protein-containing SWI/SNF complex in tissue-specific gene expression. J. Biol. Chem. 287, 5033-5041. 10.1074/jbc.M111.279968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Canman J. C., Lee C. S., Nie Z., Yang D., Moreno G. T., Young M. K., Salmon E. D. and Wang W. (2000). The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. USA 97, 13015-13020. 10.1073/pnas.240208597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Cui K., Murray D. M., Ling C., Xue Y., Gerstein A., Parsons R., Zhao K. and Wang W. (2005). PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev.. 19, 1662-1667. 10.1101/gad.1323805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga S. K., Peterson C. L., Herskowitz I. and Yamamoto K. R. (1992). Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258, 1598-1604. 10.1126/science.1360703 [DOI] [PubMed] [Google Scholar]
- Yu Y., Chen Y., Kim B., Wang H., Zhao C., He X., Liu L., Liu W., Wu L. M. N., Mao M. et al. (2013). Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 152, 248-261. 10.1016/j.cell.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Zhang Q., Li S. D., Plotnikov A. N., Walsh M. J. and Zhou M.-M. (2010). Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature 466, 258-262. 10.1038/nature09139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Wang W., Rando O. J., Xue Y., Swiderek K., Kuo A. and Crabtree G. R. (1998). Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95, 625-636. 10.1016/S0092-8674(00)81633-5 [DOI] [PubMed] [Google Scholar]