ABSTRACT
Tissue-specific establishment of repressive chromatin through creation of compact chromatin domains during development is necessary to ensure proper gene expression and cell fate. Caenorhabditis elegans synMuv B proteins are important for the soma/germline fate decision and mutants demonstrate ectopic germline gene expression in somatic tissue, especially at high temperature. We show that C. elegans synMuv B proteins regulate developmental chromatin compaction and that the timing of chromatin compaction is temperature sensitive in both wild type and synMuv B mutants. Chromatin compaction in mutants is delayed into developmental time periods when zygotic gene expression is upregulated and demonstrates an anterior-to-posterior pattern. Loss of this patterned compaction coincides with the developmental time period of ectopic germline gene expression, which leads to a developmental arrest in synMuv B mutants. Finally, accelerated cell division rates at elevated temperature may contribute to a lack of coordination between expression of tissue specific transcription programs and chromatin compaction at high temperature. Thus, chromatin organization during development is regulated both spatially and temporally by synMuv B proteins to establish repressive chromatin in a tissue-specific manner to ensure proper gene expression.
KEY WORDS: H3K9me2, Chromatin, Gene repression, SynMuv B, Temperature stress
Summary: C. elegans synMuv B proteins regulate chromatin compaction preceding upregulation of zygotic gene expression to ensure proper gene repression and cell fate decisions, especially at high temperature.
INTRODUCTION
Establishment of proper chromatin organization during development is required for fate specification. As cells go from a totipotent one-cell zygote to a multicellular differentiated organism, chromatin transitions from a highly dynamic and non-ordered state to a more static state, with clear euchromatic and heterochromatic domains within the nucleus (Mutlu et al., 2018; Politz et al., 2013; Yuzyuk et al., 2009). These changes in chromatin are concurrent with the developmental transition from no/very low zygotic gene expression to zygotic, fate-specific gene expression (Levin et al., 2012; Robertson and Lin, 2015; Spencer et al., 2011). Closed or compacted chromatin domains are often associated with genes that demonstrate low/no expression. Chromatin compaction is thought to have an influence on the level of gene expression, in part by regulating the ability of transcription factors and polymerase to have access to genes (Elgin and Reuter, 2013; Simon and Kingston, 2013).
Repressive chromatin domains are formed during early embryogenesis (Gonzalez-Sandoval, et al., 2015; Mutlu et al., 2018; Politz et al., 2013; Yuzyuk et al., 2009). There are two types of canonical repressive chromatin: constitutive heterochromatin, which covers gene-poor and repeat-rich regions of the genome, and facultative heterochromatin, which is found in gene-rich regions of the genome and acts as a spatio-temporal regulator of gene repression (Ahringer and Gasser, 2018; Latorre et al., 2015). Caenorhabditis elegans have holocentric chromosomes with gene-rich, active centers and gene-poor, repeat-rich arms (Ahringer and Gasser, 2018; Albertson and Thomson, 1982; Gerstein et al., 2010). Constitutive heterochromatin is found along chromosome arms in C. elegans, and facultative heterochromatin can be found in the more euchromatic centers of chromosomes (Liu et al., 2011; Evans et al., 2016). Although these classical designations of repressed chromatin have been characterized in worms, it has become increasingly clear that chromatin dynamics are complex and cannot always be classified into these two designations (Ahringer and Gasser, 2018; Liu et al., 2011). Repressive chromatin is established in active regions of the genome by tissue-specific mechanisms in other organisms, but the regulators and establishment of facultative heterochromatin during crucial developmental windows for proper fate specification have yet to be elucidated (Gaertner et al., 2012; Smolko et al., 2018).
synMuv B proteins are a group of conserved transcriptional repressors important in the tissue-specific repression of a large set of genes. Loss of a subset of synMuv B proteins, including members of the DREAM complex, LIN-15B and MET-2, results in ectopic expression of germline genes in the somatic intestine (Andersen and Horvitz, 2007; Petrella et al., 2011; Wang et al., 2005; Wu et al., 2012). Gene misexpression in the intestine leads to high temperature L1 larval Arrest (HTA) in DREAM complex and lin-15B mutants (Petrella et al., 2011). The DREAM complex and LIN-15B bind to promoter regions of target genes throughout the genome where they are thought to repress their expression (Goetsch et al., 2017; Rechtsteiner et al., 2019). The highly conserved DREAM complex has eight known subunits, including LIN-35 and LIN-54 (Fay and Yochem, 2007; Harrison et al., 2006). LIN-35, the single worm homolog of the mammalian pocket protein retinoblastoma, mediates the interaction of the subcomplex portions of the DREAM complex, and in it its absence the DREAM complex has highly reduced binding and repression capabilities (Goetsch et al., 2017). LIN-54 acts as one of the DNA-binding subunits of the DREAM complex and in its absence the DREAM complex is lost from 80-90% of target loci (Tabuchi et al., 2011). LIN-15B has not been shown to be part of the DREAM complex, but lin-15B mutants exhibit the same phenotypes as seen in DREAM complex mutants, including ectopic germline gene expression and HTA (Petrella et al., 2011). MET-2 catalyzes mono- and di-methylation of histone H3 lysine 9 (H3K9me1 and H3K9me2), a histone modification associated with repressive chromatin (Andersen and Horvitz, 2007; Towbin et al., 2012). met-2 mutants lose ∼80% of H3K9me2 (Towbin et al., 2012). Although loss of the DREAM complex, LIN-15B, or H3K9me2 catalyzed by MET-2 all lead to ectopic expression of germline genes, the role and order of each of these proteins during development remains unknown.
The phenotypes demonstrated by synMuv B mutants are temperature sensitive. Larval arrest happens only at high temperature and ectopic germline gene expression is more extensive at high temperatures (Petrella et al., 2011). Chromatin has been shown to be affected by changes in temperature in other organisms. For example, early studies in Drosophila revealed that high temperature causes incomplete polytene heterochromatin formation (Hartmann-Goldstein, 1967). Additionally, work in plants has shown that flowering time is linked to changes in chromatin that occur in response to increased temperature (Zografos and Sung, 2012). Therefore, the temperature sensitivity of synMuv B mutants may be associated with temperature sensitivity of chromatin. We hypothesized that synMuv B proteins regulate gene expression at the chromatin level throughout development and are particularly necessary for proper chromatin regulation and buffering of gene expression during temperature stress.
Here, we investigate the loss of synMuv B proteins on chromatin compaction during development and during times of temperature stress. The temperature sensitivity of chromatin, and its regulating proteins, has never been examined in a developmental context in C. elegans before this study. We report that the timing of chromatin compaction during embryogenesis based on cell number is delayed at high temperatures, even in wild-type embryos. We demonstrate that synMuv B mutants have an increased amount of open chromatin, both in general chromatin assays and at specific germline gene loci, especially at high temperature. We find that the delay in chromatin compaction in synMuv B mutants occurs in an anterior-to-posterior pattern. This patterned developmental delay occurs during the temperature-sensitive periods for the high-temperature arrest phenotype and ectopic germline gene expression. Our data support a model in which synMuv B proteins are necessary to regulate the proper spatial and temporal formation of repressive chromatin throughout development to promote fate-specific gene expression programs.
RESULTS
synMuv B proteins regulate developmental chromatin compaction
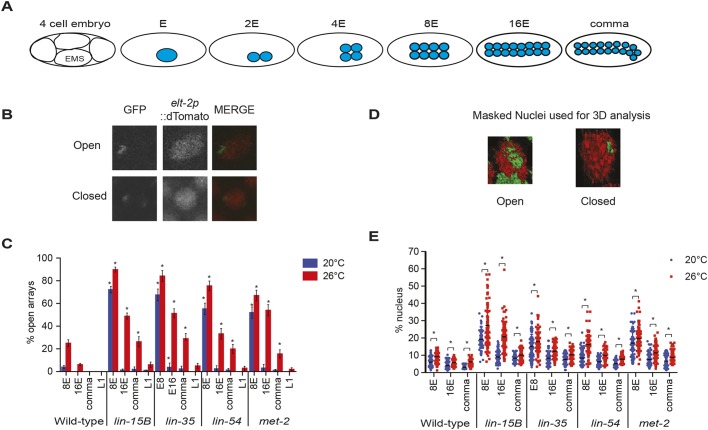
Loss of the histone H3 K9 (H3K9me1/2) mono- and di-methyltransferase met-2 results in loss of ∼80-90% of H3K9me2 and delayed general chromatin compaction during development (Fakhouri et al., 2010; Mutlu et al., 2018; Yuzyuk et al., 2009). As H3K9me2 localization to synMuv B target genes is lost in the soma of lin-15B mutants, and to a lesser extent in lin-35 mutants (Rechtsteiner et al., 2019), we investigated whether chromatin compaction is compromised more broadly in synMuv B mutants. To assess general chromatin compaction during development, we utilized the nuclear spot assay (Fakhouri et al., 2010; Yuzyuk et al., 2009). The nuclear spot assay uses an extra-chromosomal array containing numerous lacO sites that are bound by a ubiquitously expressed GFP-LacI protein. This assay has been previously utilized in C. elegans to visualize chromatin compaction throughout development as differentiation is achieved (Gonzalez-Sandoval et al., 2015; Yuzyuk et al., 2009). We visualized chromatin compaction in intestinal cells in wild type, lin-15B, lin-35, lin-54 and met-2 mutants at three embryonic stages and in L1 larvae at 20°C and 26°C. These specific embryonic stages were chosen based on the state of zygotic gene expression and ease of staging. First, 8E embryos have eight intestinal cells and represent an early embryonic stage (∼50- to 100-cell embryo), in which the zygotic gene expression program has yet to be fully activated. Second, 16E embryos have 16 intestinal cells and represent a mid-embryonic stage (∼150- to 200-cell embryo), in which zygotic gene expression has been upregulated. Finally, comma-stage embryos have 20 intestinal cells and represent a mid-to-late embryonic stage (>200 cells), in which zygotic gene expression is fully underway (Fig. 1A). We scored intestinal cells as having either open arrays, which have a de-condensed floret or crescent shape within the nucleus, or closed arrays, which have a tight ellipse/puncta shape within the nucleus, to determine differences between strains and temperatures (Fig. 1B) (Yuzyuk et al., 2009). In wild-type embryos, we observed that at 20°C by the 8E stage chromatin was already in a primarily compact state in intestinal cells (Fig. 1C). However, in wild type at 26°C at the 8E and 16E stages, we saw that there were significantly more intestinal nuclei that displayed open arrays, which by the comma stage were closed (Fig. 1C). This suggests that temperature causes a delay in developmental chromatin compaction even in a wild-type state.
Fig. 1.
Loss of synMuv B proteins causes a delay in chromatin compaction in the intestinal lineage during development. (A) Schematic representing the number of intestinal cells and the approximate number of total cells in the stages scored throughout this study. EMS, the precursor cell to the E lineage. (B) Representative images of intestinal nuclei with open and closed extra-chromosomal arrays. In merged image, green is LacI::GFP and red is elt-2p::dTomato. (C) Intestinal nuclei were scored as open or closed based on array morphology (Yuzyuk et al., 2009) in worms raised at 20°C and 26°C at the 8E, 16E, comma and L1 stages. Asterisks represent significant difference between the wild-type population at that stage and temperature (P<0.01, two-way ANOVA). Error bars indicate s.e.m. (D) Representative masked images of intestinal cell nuclei with open or closed extra-chromosomal arrays using the isosurface function in Metamorph; green is LacI::GFP and red is elt-2p::dTomato. (E) The array volume to nuclear volume ratio was measured using the isosurface function in Metamorph at 8E, 16E and comma at 20°C and 26°C. Each dot represents a single intestinal cell and the line represents the median. Asterisk represents a significant difference between 20°C and 26°C (P<0.01, two-way ANOVA).
In all four synMuv B mutants at 20°C at the 8E stage, we saw that there were significantly more intestinal nuclei with open arrays compared with wild-type embryos at 20°C (Fig. 1C). However, for all synMuv B mutants at 20°C by the 16E stage, there were very few intestinal cells with open arrays, and only in lin-35 mutants was the number of cells with open arrays significantly higher than wild type at 20°C (Fig. 1C). At 26°C, mutants show a more drastic delay in chromatin compaction displaying significantly more open arrays through all stages examined, except for lin-54 and met-2 mutants at the L1 stage (Fig. 1C). Furthermore, we saw that within an individual mutant background at 26°C compared with 20°C, there were significantly more intestinal nuclei with open arrays at each stage of development, except for met-2 mutants at the L1 stage (Fig. 1C). Overall, synMuv B mutant intestinal cells showed a developmental delay in compaction at 26°C compared with 20°C, and, at high temperature, compaction was delayed into developmental stages at which zygotic gene expression has started.
To confirm our scoring method, we performed 3D volumetric analysis of the intestinal nucleus by measuring the volume of an array compared with the volume of the nucleus. Open arrays should take up more volumetric space, and therefore have a larger array volume to nuclear volume ratio. 3D volumetric data showed similar trends to those described above; intestinal cells had a larger array volume to nuclear volume ratio in mutants versus wild type, and at 26°C versus 20°C (Fig. 1D,E). In contrast to the open/closed scoring in wild type at 26°C compared with 20°C, even at stages where we scored most or all of the arrays as closed, we saw a small but significant increase in the array volume to nuclear volume ratio (Fig. 1E). This indicated that arrays take up a variable amount of nuclear volume even when they appear compacted. In mutants, there was a striking increase in not only the percentage of nuclear volume taken up by the array compared with wild type, but also an increase in the variance of the array volume within the same genotype, especially at 26°C. In general, as development proceeded, mutant cells showed a decreased array volume to nuclear volume ratio, decreased variance of array volume within a genotype, and a significant difference between temperatures (Fig. 1E). Overall, the 3D volumetric analysis supports our open/closed data as we see a temperature-sensitive chromatin compaction phenotype throughout development, predominantly in synMuv B mutants.
Wild-type embryos show an increased rate of cell division throughout development at 26°C
Developmental speed increases at elevated temperatures based on the first cell division and time to hatching (Begasse et al., 2015; Wood et al., 1980). However, the developmental timing of the intestinal lineage has not been described at 26°C. The delay in chromatin compaction in wild-type embryos at 26°C compared with 20°C at 8E and 16E may be due to chromatin compaction requiring a minimum absolute time to occur whereas cell division rates can be accelerated. To test this hypothesis, we measured the time to each stage of intestinal development based on E cell number at 20°C and 26°C. To perform these timing experiments, two-cell embryos expressing a fluorescent intestinal cell marker were dissected from gravid adults, plated at 20°C or 26°C, and observed throughout development. The time in minutes was recorded for each embryo as they reached 8E, 16E, comma, pretzel and L1 stages. We found that the time to 8E in wild-type embryos was on average 3.9 h at 20°C and 3.09 h at 26°C (Fig. 2). The decreased developmental time at high temperature in wild-type embryos was observed at all stages examined, as embryos plated at 26°C reached each division approximately 1 h before embryos plated at 20°C (Fig. 2; Tables S1 and S2). Thus, the absolute time at which an embryo reached 8E or 16E was significantly earlier at 26°C versus 20°C.
Fig. 2.
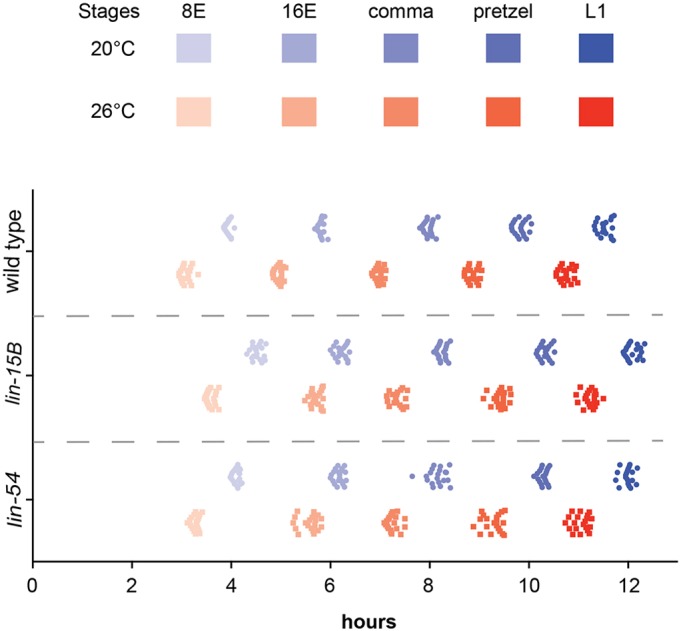
synMuv B mutants have delayed developmental timing compared with wild-type embryos. Wild-type, lin-15B and lin-54 L4 P0 hermaphrodites were placed at 20°C or 26°C overnight. The next day, F1 two-cell embryos were dissected, plated, and returned to the same temperature. The time to each intestinal stage (8E, 16, comma, pretzel and L1) was observed and recorded for each temperature. Each point represents an individual embryo (n=20 embryos/condition).
synMuv B mutants have delayed development compared with wild-type embryos
We found that synMuv B mutants have open chromatin in later stages of development, especially at high temperature. To determine whether synMuv B mutants have delayed chromatin compaction due to changes in the overall rate of embryonic development, we observed the time to each intestinal division at 20°C and 26°C for lin-15B and lin-54 mutants using the same methods as described above for wild type. If synMuv B mutants had a much faster rate of cell cycle compared with wild type at a given temperature, then that could explain a delay in chromatin compaction into later stages of intestinal development. However, synMuv B mutants were slower to each intestinal division compared with wild-type embryos (Fig. 2). synMuv B mutant embryos reached a specific developmental intestinal stage on average 30 and 21 min (lin-15B and lin-54, respectively) after wild-type embryos reached the same stage at the same temperature (Fig. 2; Tables S1 and S3). The developmental delay in reaching each embryonic cell stage based on intestinal cell number in synMuv B mutants stayed relatively constant throughout embryonic development (Fig. 2, Table S3). However, like wild-type embryos, synMuv B mutant embryos also showed accelerated development of about 1 h less time to each stage at 26°C compared with 20°C (Fig. 2, Table S2). Thus, an acceleration of cell division rate in mutants compared with wild-type embryos cannot explain the general delay in chromatin compaction in synMuv B mutants. However, similar to in wild type, an acceleration in development at 26°C compared with 20°C may contribute to the exacerbated delay in chromatin compaction observed at elevated temperature.
There is not a delay in MET-2 nuclear localization in lin-15B mutants
The timing of general chromatin compaction during development is concordant with the increased nuclear localization of MET-2 (Mutlu et al., 2018). Therefore, if the timing of MET-2 nuclear localization is delayed in lin-15B or DREAM complex mutants this could lead to a delay in chromatin compaction. We looked at the localization of MET-2:FLAG in lin-15B mutants to determine whether mutants have a delay in MET-2 nuclear localization beyond the 8E stage. lin-15B mutants demonstrated normal MET-2 nuclear accumulation compared with wild-type embryos with clear accumulation of MET-2 by the ∼50-cell stage, which coincides with the beginning of the 8E stage (Fig. S1).
Individual embryos contain nuclei with both open chromatin and closed chromatin
While scoring array compaction, we observed that, at a population-wide level, the number of intestinal cells with open arrays decreases gradually through development in mutants. For example, in pooled lin-15B mutant intestinal cells at 26°C at the 16E stage about 50% of the cells contained open arrays (Fig. 1C). We wanted to determine whether, at a particular stage of development, the partial compaction seen in the population of intestinal cells was due primarily to variance in compaction of individual intestinal cells within an embryo (Fig. 3A, model 1) or variance in compaction of intestinal cells between different embryos (Fig. 3B, model 2). If model 1 is supported, we would expect to see a population of embryos with a mosaic of open and closed cells within each embryo. If model 2 is supported, we would expect to see a binary distribution of embryos within the population containing either all open or all closed intestinal cells. We found that in a population of embryos, each embryo contained a mix of intestinal cells with both open and closed arrays that was close to the population mean (Fig. 3C). For example, in lin-15B mutants at 26°C at 8E, when about 90% of intestinal cells were scored as open (Fig. 1C), embryos had a range of 6/8 to 8/8 cells with open arrays. At 16E, when about 50% of intestinal cells were scored as open (Fig. 1C), lin-15B mutant embryos had between 6/16 and 10/16 cells with open arrays. We did not find a genotype or temperature with a bimodal distribution in which the embryos within a population fell into a mix of 100% closed and 100% open arrays. These data support model 1. As development proceeded and chromatin compaction was achieved, the number of open cells within an embryo decreased. The pattern of compaction within an embryo was similar between mutant genotypes at the same stage and temperature (Fig. 3C).
Fig. 3.
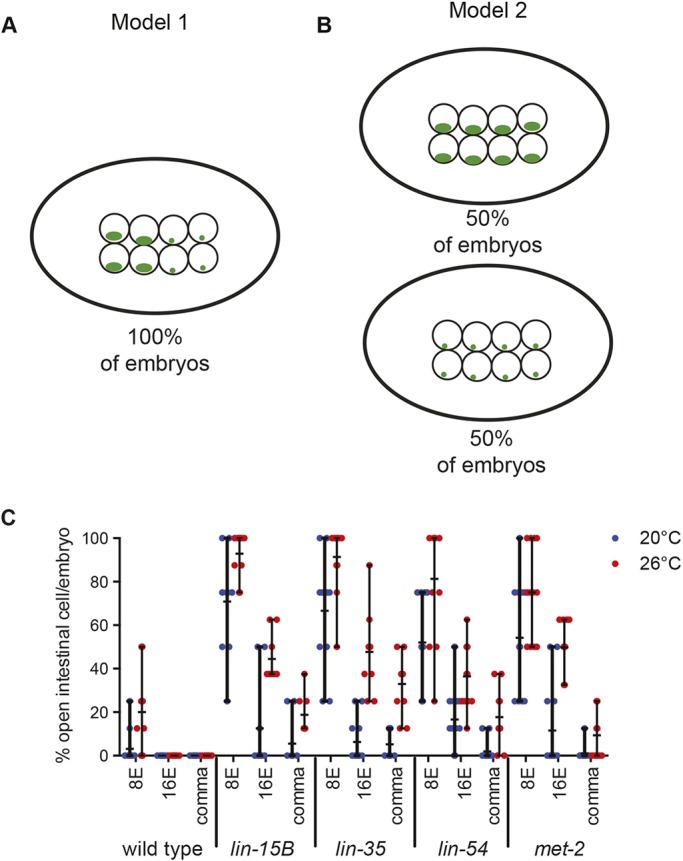
The number of intestinal cells with open chromatin varies within a single nucleus. (A,B) Models that explain a variance in compaction of intestinal cells within an embryo (A) or a variance in compaction of intestinal cells between different embryos (B). Green represents LacO::GFP bound to extra-chromomal arrays. (C) The percentage of intestinal cells with an array in an open configuration per embryo was determined for 8E-, 16E- and comma-stage embryos at 20°C and 26°C. Each dot represents an embryo and the horizontal black line represents the mean percentage of open cells per embryo. Vertical black line indicates the range.
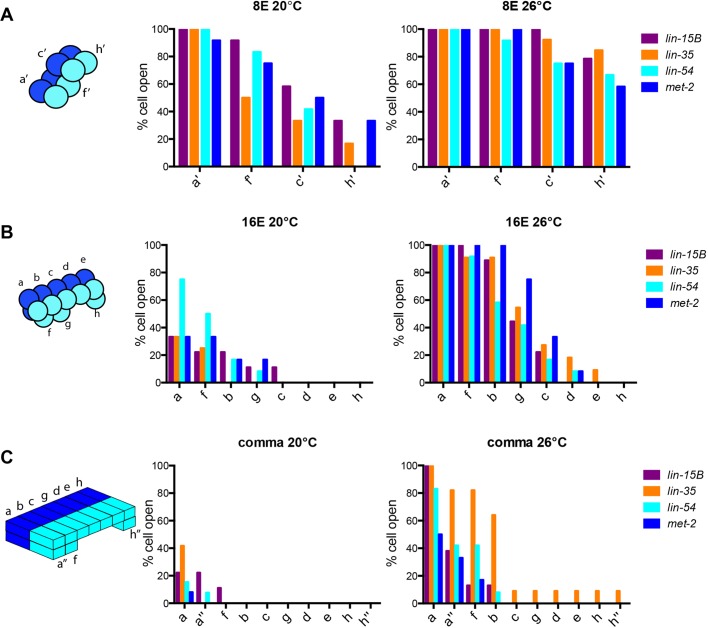
synMuv B proteins regulate chromatin compaction spatially in an anterior-to-posterior direction
While scoring the number of open cells within an embryo, we observed an anterior-to-posterior pattern of chromatin compaction. The intestinal cell lineage descends from a single cell, E. The first cell division is an anterior-posterior division in which E becomes 2E. These two intestinal cells divide in a left-right division (2E to 4E) to establish the bilateral symmetry of the intestine. The resulting cells are designated as sister cells with a set consisting of a left and right pair. The third division, when 4E gives rise to 8E, is an anterior-posterior division although the cells are slightly displaced in the dorsal-ventral plane (Fig. 4A, sister cells labeled as a′, c′, f′, h′). The fourth division is an anterior-posterior division that gives rise to 16E with some cells more dorsal than others (Fig. 4B, sister cells labeled as a-h). These 16 cells subsequently polarize and change shape before there is a final round of cell division in which only the most anterior pair of sister cells (a) divide in a dorsal-ventral direction, and the most posterior pair of sister cells (h) divide in an anterior-posterior direction leading to the final arrangement of cells within the nucleus (Fig. 4C, sister cells labeled a, a″, b, c, d, e, f, g, h, h″). We found that there was a clear pattern in the anterior-posterior location of cells with open arrays within an embryo. At 8E, when mutants have varying numbers of cells open at 20°C and 26°C, we found that the anterior pair of sister cells maintained open arrays more often in all four genotypes, whereas the most posterior cells most often had closed arrays (Fig. 4). For example, at 20°C, when lin-15B mutants have a range of 2/8 to 8/8 cells open, the a′ sister cells were open 100% of the time. The following pair of sister cells, f′, had the next highest percentage of open cells (90%). The more posterior sister cells, c′, had a decreasing number of open cells (60%), and the two most posterior sister cells, h′, had the lowest percentage of open cells (30%) (Fig. 4A). This pattern of open arrays being more often in the anterior and less often in the posterior was observed in all four mutants, at 8E, 16E and comma stages, at both 20°C and 26°C (Fig. 4). Overall, if an embryo had any intestinal cells with an open array, they were always the cells found at the anterior section of the intestine (Fig. 4). This suggests that, as development proceeds and chromatin compaction is achieved, compaction occurs first in most posterior cells and then proceeds in a posterior-to-anterior direction. This pattern seems to be independent of proliferative state, as there is a final cell division at both anterior and posterior positions to give rise to 20E (Fig. 4C, a and a″, h and h″).
Fig. 4.
In synMuv B mutants anterior intestinal cells are more delayed in compaction than posterior intestinal cells. (A-C) The position of open cells within a single embryo was monitored throughout development at 8E (A), 16E (B) and 20E (C) at both 20°C and 26°C. The anterior-to-posterior location of a cell was noted and that cell was recorded as having either an open or closed array. The percentage of cells with open arrays for each pair of sister cells was plotted to in an anterior-to-posterior directionality. The schematics to the left of the graphs represent the spatial organization of the intestinal cells at the 8E, 16E and comma stages.
synMuv B proteins regulate compaction of chromosomal germline gene loci
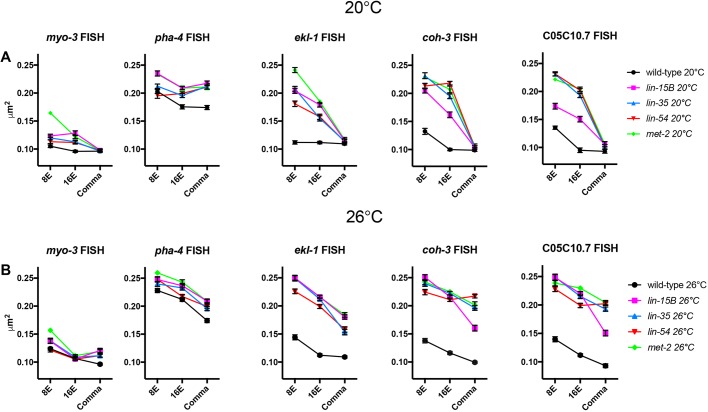
To determine whether chromatin compaction is also altered at chromosomal loci whose expression is regulated by synMuv B proteins, we performed fluorescent in situ hybridization (FISH). Pairs of red and green probes, ∼100 kb apart, were created flanking endogenous loci to determine the level of compaction at those loci (Yuzyuk et al., 2009). All loci assessed were located in central regions of autosomes, which are generally euchromatic, to control for autosomal location. As controls, we used the pha-4 gene, which should be open and expressed in the intestine, and the myo-3 gene, which should be closed and not expressed in the intestine (Ardizzi and Epstein, 1987; Horner et al., 1998). We investigated three synMuv B-regulated genes, ekl-1, coh-3 and CO5C10.7, which are germline genes that are upregulated in synMuv B mutants (Petrella et al., 2011). Intestinal cells were imaged in 3D stacks at the 8E, 16E and comma stages, and we calculated the level of chromatin compaction of the loci based on the 3D distance between the centroid of each probe (Yuzyuk et al., 2009). We found that, as predicted, the non-expressed myo-3 control locus was more compact than the expressed pha-4 control locus in all genotypes at both temperatures in all embryonic stages examined (Fig. 5, Fig. S2). The myo-3 locus demonstrated compaction reminiscent of compaction of extra-chromosomal arrays, which are also not expressed (Fig. 5, Fig. S2). In wild type at 20°C, myo-3 is completely compacted by the 8E stage (Fig. 5, Fig. S2). However, in wild type at 26°C, there was a significant difference in the level of compaction of the myo-3 locus between the 8E and 16E stages, representing a delay in compaction timing compared with 20°C (Fig. 5, Fig. S2). Thus, although there may be slight delays in chromatin compaction of an unexpressed locus at elevated temperatures or in mutants, cellular mechanisms can overcome these delays by late embryogenesis.
Fig. 5.
synMuv B-regulated chromosomal loci are open chromatin in synMuv B mutants. (A,B) 8E-, 16E- and comma-stage wild-type, lin-15B, lin-35, lin-54 and met-2 embryos at 20°C (A) and 26°C (B) were labeled with 568-5-dUTP (red) and Alexa Fluor 488 (green) probes 50 kb upstream and 50 kb downstream of myo-3, pha-4, ekl-1, coh-3 and C05C10.7. Plots represent the distribution of 3D distances between centroids (μm2). Error bars represent s.e.m. See also Fig. S1.
We next analyzed the three synMuv B target genes (ekl-1, coh-3, C05C10.7), which have higher expression in mutants than in wild type and higher expression at 26°C within mutants (Petrella et al., 2011). In wild type, all three of these loci showed similar patterns to the myo-3 locus with loci reaching maximal compaction between the 8E or 16E stage at both temperatures (Fig. 5, Fig. S2). Thus, in wild type at both 20°C and 26°C the synMuv B target loci showed a pattern of compaction like that of a non-expressed locus. However, in synMuv B mutants the three target genes showed two different patterns of developmental compaction depending on the temperature being assayed. In synMuv B mutants at 20°C at the 8E stage, the ekl-1, coh-3 and C05C10.7 loci were all less compacted than the equivalent locus in wild type, but showed wild-type levels of compaction by the comma stage (Fig. 5A). Thus, at 20°C the three synMuv B-regulated loci showed a delay in compaction but by late embryogenesis their level of compaction matched that seen in wild type. On the other hand, in synMuv B mutants at 26°C the three synMuv B target loci were more open than the same locus in wild type at all developmental stages examined (Fig. 5B). In lin-35, lin-54 and met-2 mutants, both the coh-3 and C05C10.7 loci were as open as the pha-4 locus in the same genotype (Fig. 5B, Fig. S2). Thus, synMuv B target loci demonstrate a level of chromatin compaction similar to that of highly expressed genes late into development, but only at 26°C. The presence of open chromatin around synMuv B targets during later developmental time periods at high temperature may leave these genes vulnerable to the ectopic misexpression found in synMuv B mutants.
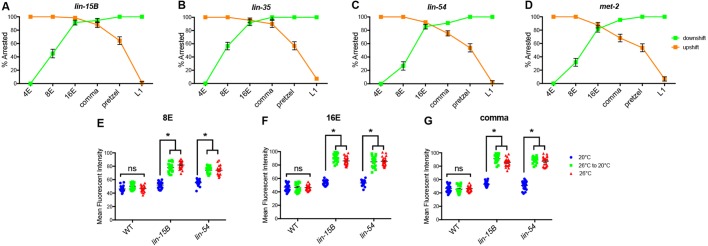
The critical developmental time period for larval high-temperature arrest in synMuv B mutants is from 16E to the comma stage
synMuv B mutants arrest at the L1 larval stage, which is at least in part due to ectopic expression of germline genes (Petrella et al., 2011). To determine whether the developmental time period important for the HTA phenotype is consistent with the embryonic stages in which we see prolonged open chromatin, we performed temperature-shift experiments on lin-15B, lin-35, lin-54 and met-2 mutants. Previous studies determined that the temperature-sensitive period for HTA is in a broad developmental range during embryogenesis and early larval development (Petrella et al., 2011), but did not look at specific embryonic stages. To perform HTA experiments, four-cell embryos expressing a fluorescent intestinal cell marker were dissected from gravid adults and downshifted or upshifted at 4E, 8E, 16E, comma, pretzel and L1 developmental stages. The pretzel stage was included as an easily distinguishable later developmental stage to tease apart differences seen in worms shifted at comma and L1 stages. Wild-type embryos were also dissected and shifted at all stages and never arrested (data not shown). All mutant embryos upshifted to 26°C from 20°C at 4E or 8E arrested at the L1 stage (Fig. 6A-D). When upshifted at 16E all mutant genotypes except lin-15B showed slightly fewer arrested animals than at earlier stages (Fig. 6A-D). A smaller number of embryos upshifted at comma and pretzel stages arrested (Fig. 6A-D). Finally, embryos upshifted at the larval L1 stage did not arrest (Fig. 6A-D). Together, embryos shifted early in development arrested whereas embryos shifted in mid-to-late embryogenesis sometimes arrested, suggesting the critical time period for HTA to be after 16E. We also performed downshift experiments to hone in on the developmental window crucial for HTA. Embryos downshifted from 26°C to 20°C at 4E did not arrest and about half of embryos downshifted at 8E arrested (Fig. 6A-D). Strikingly, most embryos downshifted at 16E and comma arrested (Fig. 6A-D). Downshifting at the pretzel or L1 stage resulted in 100% of larvae arresting at the L1 stage. (Fig. 6A-D). These data combined suggest that 16E to comma is the most crucial temperature-sensitive time period for HTA. However, upshifting after or downshifting before this time period can still cause some animals to undergo HTA, suggesting that there may be stochastic chromatin compaction leading to varying amounts of ectopic gene expression, and a leaky arrest phenotype.
Fig. 6.
16E to comma stages of embryogenesis is the temperature-sensitive period crucial for larval HTA. (A-D) lin-15B (A), lin-35 (B), lin-54 (C) and met-2 (D) L4 P0 hermaphrodites were placed at 20°C for upshift or 26°C for downshift experiments. Two-cell embryos were dissected from P0 worms, and upshifted (orange) or downshifted (green) at 4E, 8E, 16E, comma, pretzel or L1 stage and scored for L1 larval arrest (n=60-78/point). Error bars represent standard error of the proportion. (E-G) Mean fluorescent intensity of PGL-1 expression was determined for L1 wild-type, lin-15B and lin-54 embryos that were downshifted at 8E (E), 16E (F) or comma (G) stage (n=20 L1/stage and temperature). Each symbol represents an individual worm, black lines represent the mean fluorescent intensity of the population. (*P<0.01, two-way ANOVA.) See also Fig. S2.
Downshifting from 26°C to 20°C at the pretzel or L1 stages resulted in 100% larval arrest (Fig. 6A-D), suggesting that gene misexpression programs leading to HTA are activated and their effects are irreversible. Although it was previously reported that HTA was irreversible upon downshifting, whether germline genes continued to be ectopically expressed after downshift was not assessed (Petrella et al., 2011). We set out to determine if germline gene ectopic expression was at similar levels after downshift as in worms that continually experienced 26°C. We analyzed the level of the germline-specific P-granule protein PGL-1 by using strains with the endogenous pgl-1 locus tagged with GFP (Andralojc et al., 2017). pgl-1 is a synMuv B-regulated germline gene that has been used to show ectopic germline gene expression in synMuv B mutants (Petrella et al., 2011; Wang et al., 2005; Wu et al., 2012). lin-15B and lin-54 mutant embryos were downshifted at 8E, 16E and comma stages and imaged at the L1 stage to determine the level of ectopic PGL-1 expression. We found that L1s downshifted as embryos resembled L1s kept at 26°C at all stages examined with no significant difference in average maximum fluorescent intensity (Fig. 6E-G, Fig. S3). There was a significant difference when comparing shifted embryos and those kept at 26°C with mutant embryos raised at 20°C (Fig. 6E-G, Fig. S3). Because GFP protein is a long-lived protein it cannot be determined whether the level of PGL-1::GFP seen in L1s represents actively ongoing PGL-1 expression or previously expressed protein. However, as the PGL-1::GFP expression level was indistinguishable between L1s grown continually at 26°C and those down-shifted from 26°C to 20°C, those embryos down-shifted during embryonic development did reach higher levels of expression than those embryos kept continually at 20°C. This suggests that the ectopic gene expression program that leads to HTA was activated in early embryogenesis under temperature stress and that the effects of this misexpression are irreversible.
DISCUSSION
Chromatin compaction during development is necessary to achieve proper gene expression and differentiation. Previous studies have identified that H3K9 methylation is essential for chromatin compaction and organization (Ahringer and Gasser, 2018; Mutlu et al., 2018; Towbin et al., 2012). Although methylation of H3K9 is necessary, the pathways and proteins needed to establish areas of compaction in a tissue-specific way remain unclear. Here, we show that synMuv B proteins are necessary for timely chromatin compaction during development. Loss of any of these proteins – the DREAM complex DNA-binding protein LIN-54; the pocket protein that helps maintain DREAM complex binding, LIN-35; LIN-15B; or the H3K9me1/2 methyltransferase MET-2 – causes impaired chromatin compaction and ectopic germline gene expression in early development. This suggests that binding of LIN-15B, the DREAM complex, and methylation of H3K9 are each necessary for proper repression at germline gene loci in somatic tissue. Loss of any one of these proteins causes ectopic expression of germline genes and improper organismal development during temperature stress.
Methylation of H3K9 is necessary to achieve chromatin compaction at the right time and place during development. Loss of MET-2 causes loss of ∼80% of H3K9me1/2 across the genome. Previous studies have shown that met-2 mutants ectopically express germline genes and have reduced embryonic chromatin compaction (Fakhouri et al., 2010; Towbin et al., 2012). We have shown that met-2 mutants also exhibit the HTA phenotype (Rechtsteiner et al., 2019). Interestingly, the DREAM complex binds only ∼1400 and LIN-15B binds to ∼2100 locations throughout the genome, primarily on autosomes (Goetsch et al., 2017; Latorre et al., 2015; Rechtsteiner et al., 2019). Loss of LIN-15B or DREAM binding, at relatively few places throughout the genome, causes the same phenotypes as losing ∼80% of H3K9me1/2. One explanation for this similarity is that LIN-15B and the DREAM complex may be tissue-specific regulators of H3K9 methylation at synMuv B-regulated loci and this modification may be important for repression of germline genes in somatic tissue (Rechtsteiner et al., 2019). Another explanation is that loss of individual mechanisms that contribute to closed chromatin (broad H3K9me2 or DREAM complex/LIN-15B regulation at specific loci) each lead to an increased temperature sensitivity of compact chromatin formation globally. Therefore, the similarity in phenotype is a secondary consequence to general changes in chromatin compaction at many loci across the genome. Further analysis of the interaction of the DREAM complex/LIN-15B with MET-2 and H3K9me2 are needed to distinguish these possibilities.
Chromatin is found in a generally unorganized and open state early in development (Politz et al., 2013; Yuzyuk et al., 2009). As development proceeds and differentiation occurs, chromatin becomes compacted and organized in a tissue-specific manner (Mutlu et al., 2018; Politz et al., 2013; Yuzyuk et al., 2009). Organization of chromatin within the nucleus generally precedes upregulation of zygotic gene expression and is important for correct cell fate decisions (Gaertner et al., 2012; Mutlu et al., 2018; Politz et al., 2013; Yuzyuk et al., 2009). Because poised Pol II accumulates at promoters in a developmental stage-specific, but not tissue-specific manner (Gaertner et al., 2012; Bowman et al., 2013; Hsu et al., 2015), upregulation of gene expression during the maternal-to-zygotic switch occurs globally throughout the embryo (Gaertner et al., 2012; Bowman et al., 2013; Hsu et al., 2015). Therefore, establishment of tissue-specific repression, including establishment of repressive chromatin, must occur before global gene expression is upregulated to achieve cell fate-specific expression patterns (Gaertner et al., 2012; Politz et al., 2013; Yuzyuk et al., 2009). Loss of synMuv B proteins leads to global changes in chromatin compaction that continue into the developmental time of the maternal-to-zygotic switch, especially at high temperature, leaving their target loci vulnerable to expression when zygotic gene expression is upregulated. synMuv B mutants demonstrate three different temperature-sensitive phenotypes: ectopic germline gene expression, delayed chromatin compaction, and HTA. Because changes in chromatin compaction and gene expression are so intimately linked, it is difficult to distinguish the more primary of these phenotypes. It may be that synMuv B protein binding blocks the access of transcription factors or Pol II binding/activation to synMuv B target loci. Thus, in the absence of these synMuv B proteins these genes are ectopically expressed, which in turn leads to a loss of chromatin compaction. On the other hand, synMuv B proteins may help establish repressive chromatin environments before the maternal-to-zygotic switch, such that when synMuv B proteins are mutated the cell experiences an inability to compact chromatin, especially at synMuv B target loci. In this case, it is the loss of compaction that allows these genes to be expressed either through changes in activator accessibility or conformational changes to the chromatin that allow active transcription. Because chromatin compaction can eventually reach wild-type levels in synMuv B mutants at 20°C, despite the near complete loss of DREAM complex binding and H3K9me2 at germline gene promoters (Goetsch et al., 2017; Rechtsteiner et al., 2019), temperature appears to be key to both increased gene misexpression and loss of chromatin compaction.
The synMuv B phenotypes of delayed chromatin compaction, ectopic germline gene expression, and HTA are all temperature sensitive (Petrella et al., 2011). However, this temperature sensitivity is not likely to be due to misfolding of mutant proteins or complex disassociation as a result of a missing member. First, even mutants that produce no protein, such as lin-35(n745) and lin-15B(n744), demonstrate temperature sensitivity. Second, the binding of the DREAM complex is significantly impaired in both lin-35(n745) and lin-54(n2231) mutants even at 20°C. Finally, double mutants between different components of the pathway, for example lin-15B; met-2 or lin-15B; lin-35, do not demonstrate enhanced phenotypes at lower temperatures (M.E.C. and L.N.P., unpublished data). Therefore, it appears that there is something about temperature effects on the broader system that is integral to these phenotypes. Early studies in Drosophila revealed that high temperature causes incomplete heterochromatin formation (Hartmann-Goldstein, 1967). Our data in wild-type embryos indicates that formation of closed chromatin may require a minimum absolute time to occur, which is demonstrated by the delay in chromatin compaction to later developmental stages when cell division rates are accelerated at high temperature. In synMuv B mutants, the loss of a single protein/complex that helps to reorganize chromatin into compact domains results in a short delay in chromatin compaction compared with wild type at 20°C, but compensatory mechanisms by redundant players appear to allow for generally normal development. That synMuv B mutants take longer to reach each developmental stage compared with wild type may aid in coordination of formation of closed chromatin and upregulation of zygotic gene expression in the mutants at 20°C. However, synMuv B mutant embryos, similar to wild-type embryos, take ∼60 min less time to reach each stage of development at 26°C versus 20°C. Therefore, the increase in cell cycle speed combined with a slowed rate of formation of compact chromatin formation in synMuv B mutants at 26°C may result in disruption of the coordination in the timing of compact chromatin formation, and either Pol II activation or transcription factor binding during development. This lack of coordination may be the primary cause of failure of synMuv B target loci to compact chromatin, ectopic gene expression, and ultimately HTA at elevated temperature.
The experiments in this study were all performed with a high-temperature condition at the end of the temperature range at which laboratory experiments are routinely carried out for C. elegans. This raises the question of whether these temperature stresses reflect levels that these organisms would experience under more natural conditions. Work in the last 10-15 years has greatly expanded our understanding of the ecology of naturally occurring C. elegans populations (Schulenburg and Félix, 2017). Unlike what was previously thought, it is now known that the reproductive lifespan and embryogenesis of C. elegans occur at the soil surface, often within rotting plant vegetation (Félix and Duveau, 2012; Frezal and Felix, 2015; Kiontke et al., 2011). In our temperature-shift experiments, a window of 6-8 h within embryogenesis at 26°C elicited a severe chromatin delay in developing embryos resulted in HTA in mutants. Given the short time period needed, it seems likely that there are circumstances in which C. elegans embryos in natural habitats would experience the temperatures tested here. Thus, having mechanisms that ensure that chromatin compaction occurs during the correct developmental window at these temperatures would be necessary for species survival.
We found that delayed chromatin compaction in synMuv B mutants demonstrated an anterior-to-posterior pattern within intestinal cells. The anterior cells of the intestine were the last to adopt closed chromatin, a pattern independent of replication capacity, as there is both a final anterior and a final posterior division to form the C. elegans intestine. The C. elegans intestine is patterned anteriorly to posteriorly by Wnt signaling proteins (Fukushige et al., 1996; Lin et al., 1998; Schroeder and McGhee, 1998). Wnt signaling-associated transcription factors are loaded into the most anterior cells of the intestine at each division (Schroeder and McGhee, 1998). It is possible that anteriorly loaded transcriptional factors are able to bind ectopically and express vulnerable germline genes in the anterior intestine, leading to high-temperature arrest.
One of the hallmark phenotypes of synMuv B mutants is the ectopic expression of germline genes in the soma. It is important to understand how germline genes are repressed in somatic tissue as many human somatic cancers that are highly proliferative and metastatic show upregulation of germline genes (Gure et al., 2005; Maine et al., 2016; Xu et al., 2014). Like C. elegans, mammals also repress germline gene expression during embryogenesis in somatic cells and recent work has indicated that this repression is dependent on H3K9me2 (Ebata et al., 2017; Lian et al., 2018). Given that the DREAM complex is completely conserved between worms and mammals (Litovchick, et al., 2007), its role in establishment of developmental repressive chromatin at germline genes may also be conserved. Further studies of synMuv B regulation will provide evidence of how organisms establish and maintain chromatin structure and gene expression states to ensure vitality.
MATERIALS AND METHODS
Strains and nematode culture
C. elegans were cultured on NGM plates seeded with the Escherichia coli strain AMA1004 at 20°C unless otherwise noted. N2 (Bristol) was used as wild type. Strain LNP0050 containing the transgenes bnEx80(68xlacO+myo-3::mCherry+worm genomic DNA); gwls39(baf- 1p::GFP::lacI::let-8583\′UTR;vit-5::GFP); caIs79(elt-2p::dTomato;pRF4)″ was created for this study and crossed into mutant worms. Mutant alleles used were lin-15B(n744), lin-35(n745), lin-54(n2231), lin-37(n758), and met-2(n4256). Strain EL364 has a CRISPR-tagged 3XFlag::met-2 (Mutlu et al., 2018). The complete strain list can be found in Table S4.
Nuclear spot assay
Wild-type and mutant embryos and L1s with gwls39[baf-1p::GFP::lacI;;let-858 3\′UTR; vit-5::GFP]; bnEx85 (68xlacO+myo-3::mcherry+genomic C05C10.7+worm genomic DNA) and caIs79[elt-2p::dTomato, pRF4 (rol-6+)] arrays were collected and fixed with methanol and acetone and mounted using Gelutol (Petrella et al., 2011). Each genotype was scored at 8E, 16E, comma and L1 stages. High-temperature samples were collected by upshifting L4 P0 worms to 26°C before collecting F1 embryos or L1s. L1 larvae were obtained by hatching embryos in the absence of food in M9 buffer. Images were acquired in z-stacks using a Nikon A1R laser-scanning confocal unit controlled by NIS-Elements fitted on a Nikon inverted Eclipse Ti-E microscope with a Nikon DS-Qi1Mc camera and Plan Apo 100×/1.4 numerical aperture oil objective. For chromatin compaction analysis, each intestinal cell's array was scored by eye as either open or closed as described by Yuzyuk et al. (2009). Briefly, arrays that had a hazy, distended, floret or crescent configuration were scored as ‘open’ as this morphology suggests that these arrays were partially de-compacted. As development proceeded, arrays became compacted and gained a clear, tight, ellipsoid morphology. These ellipsoid arrays were scored as ‘closed’ as this suggested globally compacted and repressed arrays. Nuclear volumetric array compaction analysis was performed using the isosurface function in Metamporph V7.8.8.0. ND2 files were opened in Metamorph, a rectangular box was drawn around a single embryo, and the stack was opened in 4D viewer. Inclusive threshold and the isosurface function were used to create a mask of the two channels selected: the red intestinal cells and the green arrays. Each individual intestinal cell and array was selected, and the volume was calculated using the measure function. Specifically, the volume of both the nucleus and GFP+ LacO arrays of each individual intestinal cell was measured and used to determine the percentage of the nucleus that the array represented. Statistical analysis was performed by two-way ANOVA using Prism GraphPad.
Embryo E cell location lacO array scoring
Embryos scored for chromatin compaction analysis were also scored for percentage of open cells within an embryo. The intestinal cells at each developmental stage were lettered anterior-to-posterior as in WormBook (McGhee, 2007) so that chromatin compactions in each specific intestinal cell could be compared between embryos. The exact intestinal cells with open arrays in each embryo were recorded and the percentages of open intestinal cells were calculated. Statistical analysis was performed by two-way ANOVA using Prism GraphPad.
Fluorescence in situ hybridization
Fluorescent probes were made by PCR labeling as described by Lanctot et al. (2013) using fluorescent 568-5-dUPT (Thermo Fisher, C11399) and ULYSIS Nucleic Acid Labeling Kits using Alexa Fluor 488 (Molecular Probes/Life Technologies, U21650) (primer sequences available in Table S5). Worm genomic DNA was used as the template to create five ∼500 bp probes ∼50 kbp upstream and downstream of pha-4, myo-3, ekl-1, coh-3 and C05C10.7 loci. High-temperature samples were collected by upshifting L4 P0 worms to 26°C before collecting F1 embryos. Embryos were dissected in 1× M9 on slides and then frozen in liquid nitrogen. After freeze-cracking, slides were fixed in methanol (on ice, 2 min) followed by 4% formaldehyde (4°C, 10 min) and then washed twice in 1× PBS (room temperature, 2 min). Embryos were permeabilized in 0.5% Triton X-100 in PBS (room temperature, 5 min), washed twice in 1× PBS (room temperature, 2 min), rinsed in 0.01 N HCl, and incubated in 0.1 N HCl (room temperature, 2 min). Slides were then washed once with 1× PBS (room temperature, 3 min), once with 2× SSC (room temperature, 3 min) and then treated with 50 μg/ml of RNase A (VWR, 1247C393) in 2× SSC for 45 min at 37°C by overlaying the sample with 500 μl RNAse solution and incubating in a humidified chamber. Slides were then washed once with 2× SSC (room temperature, 2 min), and then incubated in 2× SSC/50% formamide for 2 h at room temperature. The probe was diluted to a concentration of 2-5 ng/µl in 100% deionized formamide and an equal volume of hybridization buffer was added. 25 µl of the probe solution was added to the sample, and overlaid with a glass chamber sealed with cement glue. The slides were pre-hybridized overnight at 37°C in a humidified chamber. FISH probes and samples were denatured at 76°C by placing the slides on a heat block, and then incubated for hybridization for 3 days at 37°C. Hybridization was followed by three washes in 2× SSC (37°C, 5 min) and two in 0.2× SSC (55°C, 5 min). Images were captured using a Nikon A1R laser-scanning confocal unit controlled by NIS-Elements fitted on a Nikon inverted Eclipse Ti-E microscope with a Nikon DS-Qi1Mc camera and Plan Apo 100×/1.4 numerical aperture oil objective. Analysis was performed using Metamorph isosurface function to determine the 3D distance between the two centroids as described by Yuzyuk et al. (2009). ND2 files were opened in Metamorph, a rectangular box was drawn around a single embryo, and the stack was opened in 4D viewer. Inclusive threshold and the isosurface function were used to create a mask of both probes. Each masked probe was selected and the x, y and z coordinates were calculated using the measure function to determine the centroid of the probe in 3D space. Statistical analysis was performed by two-way ANOVA using Prism GraphPad.
Embryo shift assays
For downshifting experiments: P0 L4 wild-type worms and mutant strains expressing caIs79[elt-2p::dTomato, pRF4 (rol-6+)] were upshifted to 26°C. The next day, two-cell embryos were dissected out of adults in 1× M9, plated on NGM, and the plate was returned to 26°C. Developmental stages were determined based on the number of intestinal cells made visible by the caIs79 transgene. Embryos were downshifted to 20°C when the majority of embryos on the plate were at 4E (∼2 h), 8E (∼3 h), 16E (∼5 h), comma (∼7 h), pretzel ∼(9 h) and L1 (∼14 h) stages. The HTA phenotype was scored 2 days after shifting as described by Petrella et al. (2011). Each downshift was done four to six times with 10-20 embryos plated per replicate (n=55-77). For upshifting experiments, a similar protocol was used: two-cell embryos were dissected out of adults and maintained continually at 20°C. After dissection, embryos were plated and kept at 20°C until being upshifted to 26°C at 4E, 8E, 16E, comma, pretzel and L1 stages. Plates were scored for HTA 2 days after shifting. Each upshift was done four to six times with 10-20 embryos plated per replicate (n=58-101).
PGL expression analysis
P0 L4 wild-type worms and mutant strains expressing caIs79[elt-2p::dTomato, pRF4 (rol-6+)]; pgl-1(sam33[pgl-1::gfp::3xFlag]) were upshifted to 26°C. The next day, adults were dissected in 1× M9 and two-cell embryos placed in 1× M9 on polylysine-coated slides in humid chambers and returned to 26°C. Embryos were downshifted on slides to 20°C at 8E, 16E and comma stages. Arrested L1 larva were fixed in methanol and acetone and imaged in z-stacks using a Nikon A1R laser-scanning confocal unit controlled by NIS-Elements fitted on a Nikon inverted Eclipse Ti-E microscope with a Nikon DS-Qi1Mc camera and Plan Apo 60×/1.2 numerical aperture oil objective. Mean fluorescent intensity of PGL-1:GFP was determined using ImageJ. Statistical analysis was performed by two-way ANOVA using Prism GraphPad.
Embryonic timing assays
P0 L4 wild-type worms and mutant strains expressing caIs79[elt-2p::dTomato, pRF4 (rol-6+)] were placed at 20°C or 26°C. The next day, two-cell embryos were dissected out of adults in 1× M9, plated on NGM, a timer was started, and the plate was returned to 20°C or 26°C. Plates were removed from the incubator and monitored for ∼15 min at predetermined times based on approximate timing of stages known for each stage scored (8E, 16E, comma, pretzel and L1). During a given window of time, the time at which each embryo within the population reached the specific development stage was recorded. The plate was then returned to the incubator and the process repeated for each subsequent stage. Developmental stages were determined based on the number of intestinal cells in which elt-2p::dTomato expression was detected. n=20 embryos/condition.
MET-2::FLAG localization
Immunostaining of embryos was adapted from Strome and Wood (1983). Embryos were collected by picking gravid 3xFlag::met-2 adults (Mutlu et al., 2018) into 20 µl bubbles of 1× M9 on hydrophobic polylysine-covered slides. Adults were dissected in half at the vulva site to release embryos from inside the worm. Samples were then overlaid with a coverslip, excess liquid was wicked away, and the slide was immersed in liquid nitrogen for at least 5 min. Slides were removed from liquid nitrogen, the coverslip was removed, and the samples were fixed in methanol at 4°C for 10 min and acetone at 4°C for 10 min. Slides were air-dried and blocked in 1.5% BSA, 1.5% OVA in PBS for 30 min at room temperature. Slides were incubated with anti-FLAG primary antibody (Sigma-Aldrich, F1804-200 μg) for ∼18 h at 4°C at a 1:100 dilution. Slides were washed twice in PBS for 10 min, blocked for 15 min at room temperature, and incubated with Alexa Fluor 488-conjugated secondary antibody (Thermo Fisher Scientific, A-10680) at a 1:500 dilution and DAPI at a 1:500 dilution for 2 h at room temperature. Slides were washed four times for 10 min each in PBS at room temperature and were mounted in Gelutol mounting media. Images were acquired with Nikon A1R laser-scanning confocal controlled by NIS-Elements fitted on a Nikon inverted Eclipse Ti-E microscope with a Nikon DS-Qi1Mc camera and Plan Apo 100×/1.4 numerical aperture oil objective.
Supplementary Material
Acknowledgements
We thank Jim McGhee for JM163, Eleanor Maine for EL634, and Dustin Updike for DUP75 strains. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Many thanks to Anita Manogaran for comments and discussion of the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.E.C., L.N.P.; Methodology: M.E.C.; Investigation: M.E.C.; Writing - original draft: M.E.C.; Writing - review & editing: M.E.C., L.N.P.; Supervision: L.N.P.; Funding acquisition: L.N.P.
Funding
This work was supported by National Institutes of Health grants (R00GM98436 and R15GM122005 to L.N.P.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.174383.supplemental
References
- Ahringer J. and Gasser S. M. (2018). Repressive chromatin in Caenorhabditis elegans: establishment, composition, and function. Genetics 208, 491-511. 10.1534/genetics.117.300386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson D. G. and Thomson J. N. (1982). The kinetochores of Caenorhabditis elegans . Chromosoma 86, 409-428. 10.1007/BF00292267 [DOI] [PubMed] [Google Scholar]
- Andersen E. C. and Horvitz H. R. (2007). Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development 134, 2991-2999. 10.1242/dev.009373 [DOI] [PubMed] [Google Scholar]
- Andralojc K. M., Campbell A. C., Kelly A. L., Terrey M., Tanner P. C., Gans I. M., Senter-Zapata M. J., Khokhar E. S. and Updike D. L. (2017). ELLI-1, a novel germline protein, modulates RNAi activity and P-granule accumulation in Caenorhabditis elegans. PLoS Genet. 12, e1006611 10.1371/journal.pgen.1006611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardizzi J. P. and Epstein H. F. (1987). Immunochemical localization of myosin heavy chain isoforms and paramyosin in developmentally and structurally diverse muscle cell types of the nematode Caenorhabditis elegans . J. Cell Biol. 105, 2763-2770. 10.1083/jcb.105.6.2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begasse M. L., Leaver M., Vazquez F., Grill S. W. and Hyman A. A. (2015). Temperature dependence of cell division timing accounts for a shift in the thermal limits of C. elegans and C. briggsae. Cell Rep. 10, 647-653. 10.1016/j.celrep.2015.01.006 [DOI] [PubMed] [Google Scholar]
- Bowman E. A., Bowman C. R., Ahn J. H. and Kelly W. G. (2013). Phosphorylation of RNA polymerase II is independent of P-TEFb in the C. elegans germline. Development 140, 3703-3713. 10.1242/dev.095778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebata K. T., Mesh K., Liu S., Bilenky M., Fekete A., Acker M. G., Hirst M., Garcia B. A. and Ramalho-Santos M. (2017). Vitamin C induces specific demethylation of H3K9me2 in mouse embryonic stem cells via Kdm3a/b. Epigenet. Chromat. 10, 36 10.1186/s13072-017-0143-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C. R. and Reuter G. (2013). Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 5, a017780 10.1101/cshperspect.a017780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K. J., Huang N., Stempor P., Chesney M. A., Down T. A. and Ahringer J. (2016). Stable Caenorhabditis elegans chromatin domains separate broadly expressed and developmentally regulated genes. Proc. Natl. Acad. Sci. USA 113, E7020-E7029. 10.1073/pnas.1608162113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhouri T. H. I., Stevenson J., Chisholm A. D. and Mango S. E. (2010). Dynamic chromatin organization during foregut development mediated by the organ selector gene PHA-4/FoxA. PLoS Genet. 6, e1001060 10.1371/journal.pgen.1001060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay D. S. and Yochem J. (2007). The synMuv genes of Caenorhabditis elegans in vulval development and beyond. Dev. Biol. 306, 1-9. 10.1016/j.ydbio.2007.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M.-A. and Duveau F. (2012). Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 10, 59 10.1186/1741-7007-10-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezal L. and Felix M.-A. (2015). C. elegans outside the Petri dish. eLife 4, e05849 10.7554/eLife.05849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T., Schroeder D. F., Allen F. L., Goszczynski B. and McGhee J. D. (1996). Modulation of gene expression in the embryonic digestive tract of C. elegans. Dev. Biol. 178, 276-288. 10.1006/dbio.1996.0218 [DOI] [PubMed] [Google Scholar]
- Gaertner B., Johnston J., Chen K., Wallaschek N., Paulson A., Garruss A. S., Gaudenz K., Kumar B. D., Krumlauf R. and Zeitlinger J. (2012). Poised RNA Polymerase II changes over developmental time and prepares genes for future expression. Cell Rep. 2, 1670-1683. 10.1016/j.celrep.2012.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M. B., Lu Z. J., Van Nostrand E. L., Cheng C., Arshinoff B. I., Liu T., Yip K. Y., Robilotto R., Rechsteiner A., Ikegami K. et al. (2010). Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330, 1775-1787. 10.1126/science.1196914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetsch P. D., Garrigues J. M. and Strome S. (2017). Loss of the Caenorhabditis elegans pocket protein LIN-35 reveals MuvB's innate function as the repressor of DREAM target genes. PLoS Genet. 13, e1007088 10.1371/journal.pgen.1007088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sandoval A., Towbin B. D., Kalck V., Cabianca D. S., Gaidatzis D., Hauer M. H., Geng L., Wang L., Yang T., Wang X. et al. (2015). Perinuclear anchoring of H3K9-methylated chromatin stabilizes induced cell fate in C. elegans embryos. Cell 163, 1333-1347. 10.1016/j.cell.2015.10.066 [DOI] [PubMed] [Google Scholar]
- Gure A. O., Chua R., Williamson B., Gonen M., Ferrera C. A., Gnjatic S., Ritter G., Simpson A. J. G., Chen Y.-T., Old L. J. et al. (2005). Cancer-testis genes are coordinately expressed and are markers of poor outcome in non-small cell lung cancer. Clin. Cancer Res. 11, 8055-8062. 10.1158/1078-0432.CCR-05-1203 [DOI] [PubMed] [Google Scholar]
- Harrison M. M., Ceol C. J., Lu X. and Horvitz H. R. (2006). Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc. Natl. Acad. Sci. USA 103, 16782-16787. 10.1073/pnas.0608461103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann-Goldstein I. J. (1967). On the relationship between heterochromatization and variegation in Drosophila, with special reference to temperature sensitive periods. Genet. Res. 10, 143-159. 10.1017/S0016672300010880 [DOI] [PubMed] [Google Scholar]
- Horner M. A., Quintin S., Domeier M. E., Kimble J., Labouesse M. and Mango S. E. (1998). pha-4, an HNF-3I homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev. 12, 1947-1952. 10.1101/gad.12.13.1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H. T., Chen H. M., Yang Z., Wang J., Lee N. K., Burger A., Zaret K., Liu T., Levine E. and Mango S. E. (2015). Recruitment of RNA polymerase II by the pioneer transcription factor PHA-4. Science 348, 1372-1376. 10.1126/science.aab1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K. C., Félix M.-A., Ailion M., Rockman M. V., Braendle C., Pénigault J.-B. and Fitch D. H. A. (2011). A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol. Biol. 11, 339 10.1186/1471-2148-11-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C. and Meister P. (2013). Microscopic analysis of chromatin localization and dynamics in C. elegans. Methods Mol. Biol. 1042, 153-172. 10.1007/978-1-62703-526-2_11 [DOI] [PubMed] [Google Scholar]
- Latorre I., Chesney M. A., Garrigues J. M., Stempor P., Appert A., Francesconi M., Strome S. and Ahringer J. (2015). The DREAM complex promotes gene body H2A.Z for target repression. Genes Dev. 29, 495-500. 10.1101/gad.255810.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M., Hashimshony T., Wagner F. and Yanai I. (2012). Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev. Cell 22, 1101-1108. 10.1016/j.devcel.2012.04.004 [DOI] [PubMed] [Google Scholar]
- Lian Y., Meng L., Ding P. and Sang M. (2018). Epigenetic regulation of MAGE family in human cancer progression-DNA methylation, histone modification and non-coding RNAs. Clin. Epigenet. 10, 115 10.1186/s13148-018-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Hill R. J. and Priess J. R. (1998). POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell 92, 229-239. 10.1016/S0092-8674(00)80917-4 [DOI] [PubMed] [Google Scholar]
- Litovchick L., Sadasivam S., Florens L., Zhu X., Swanson S. K., Velmurugan S., Chen R., Washburn M. P., Liu X. S. and DeCaprio J. A. (2007). Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell 26, 539-551. 10.1016/j.molcel.2007.04.015 [DOI] [PubMed] [Google Scholar]
- Liu T., Rechsteiner A., Egelhofer T. A., Vielle A., Latorre I., Cheung M.-S., Ercan S., Ikegami K., Jensen M., Kolasinska-Zwierz P. et al. (2011). Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 21, 227-236. 10.1101/gr.115519.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine E. A., Westcott J. W., Prechtl A. M., Dang T. T., Whitehurst A. W. and Pearson G. W. (2016). The cancer-testis antigens SPANX-A/C/D and CTAG2 promote breast cancer invasion. Oncotarget 7, 14708-14726. 10.18632/oncotarget.7408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D. (2007). The C. elegans intestine. In WormBook (ed. The C. elegans Research Community), pp. 1-36. 10.1895/wormbook.1.133.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu B., Chen H.-M., Moresco J. J., Orelo B. D., Yang B., Gaspar J., Kepller-Ross S., Yates J. R. III, Hall D. H., Maine E. M. et al. (2018). Regulated nuclear accumulation of a histone methyltransferase times the onset of heterochromatin formation in C. elegans embryos. Sci. Adv. 4, eaat6224 10.1101/326231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella L. N., Wang W., Spike C. A., Rechtsteiner A., Reinke V. and Strome S. (2011). synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development 138, 1069-1079. 10.1242/dev.059501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz J. C. R., Scalzo D. and Groudine M. (2013). Something silent this way forms: the functional organization of the repressive nuclear compartment. Annu. Rev. Cell Dev. Biol. 29, 241-270. 10.1146/annurev-cellbio-101512-122317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechtsteiner A., Costello M. E., Egelhofer T. A., Garrigues J. M., Strome S. and Petrella L. N. (2019). Repression of germline genes in Caenorhabditis elegans somatic tissues by H3K9 Dimethylation of their promoters. Genetics 12, 125-140. 10.1534/genetics.118.301878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson S. and Lin R. (2015). The maternal-to-zygotic transition in C. elegans. Curr. Top. Dev. Biol. 113, 1-42. 10.1016/bs.ctdb.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Schroeder D. F. and McGhee J. D. (1998). Anterior-posterior patterning within the Caenorhabditis elegans endoderm. Development 125, 4877-4887. [DOI] [PubMed] [Google Scholar]
- Schulenburg H. and Félix M.-A. (2017). The natural biotic environment of Caenorhabditis elegans. Genetics 206, 55-86. 10.1534/genetics.116.195511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J. A. and Kingston R. E. (2013). Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49, 808-824. 10.1016/j.molcel.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolko A. E., Shapiro-Kulnane L. and Salz H. K. (2018). The H3K9 methyltransferase SETDB1 maintains female identity in Drosophila germ cells. Nat. Commun. 9, 4155 10.1038/s41467-018-06697-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer W. C., Zeller G., Watson J. D., Henz S. R., Watkins K. L., McWhirter R. D., Petersen S., Sreedharan V. T., Widmer C., Jo J. et al. (2011). A spatial and temporal map of C. elegans gene expression. Genome Res. 21, 325-341. 10.1101/gr.114595.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S. and Wood W. B. (1983). Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35, 15-25. 10.1016/0092-8674(83)90203-9 [DOI] [PubMed] [Google Scholar]
- Tabuchi T. M., Deplancke B., Osato N., Zhu L. J., Barrasa M. I., Harrisons M. M., Horvitz H. R., Walhout A. J. M. and Hagstrom K. A. (2011). Chromosome-biased binding and gene regulation by the Caenorhabditis elegans DRM complex. PLoS Genet. 7, e1002074 10.1371/journal.pgen.1002074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin B. D., González-Aguilera C., Sack R., Gaidatzis D., Kalck V., Meister P., Askjaer P. and Gasser S. M. (2012). Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150, 934-947. 10.1016/j.cell.2012.06.051 [DOI] [PubMed] [Google Scholar]
- Wang D., Kennedy S., Conte D. Jr, Kim J. K., Gabel H. W., Kamath R. S., Mello C. C. and Ruvkun G. (2005). Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature 436, 593-597. 10.1038/nature04010 [DOI] [PubMed] [Google Scholar]
- Wood W. B., Hecht R., Carr S., Vanderslice R., Wolf N. and Hirsh D. (1980). Parental effects and phenotypic characterization of mutations that affect early development in Caenorhabditis elegans. Dev. Biol. 74, 446-469. 10.1016/0012-1606(80)90445-5 [DOI] [PubMed] [Google Scholar]
- Wu X., Shi Z., Cui M., Han M. and Ruvkin G. (2012). Repression of germline RNAi pathways in somatic cells by retinoblastoma pathway chromatin complexes. PLoS Genet. 8, e1002542 10.1371/journal.pgen.1002542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Tang X., Lu M., Tang Q., Zhang H., Zhu H., Xu N., Zhang D., Xiong L., Mao Y. et al. (2014). Overexpression of MAGE-A9 predicts unfavorable outcome in breast cancer. Exp. Mol. Pathol. 97, 579-584. 10.1016/j.yexmp.2014.11.001 [DOI] [PubMed] [Google Scholar]
- Yuzyuk T., Fakhouri T. H. I., Kiefer J. and Mango S. E. (2009). The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Dev. Cell 16, 699-710. 10.1016/j.devcel.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zografos B. R. and Sung S. (2012). Vernalization-mediated chromatin changes. J. Exp. Bot. 63, 4343-4348. 10.1093/jxb/ers157 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.