ABSTRACT
A deficiency in BRG1, the catalytic subunit of the SWI/SNF chromatin remodeling complex, results in a meiotic arrest during spermatogenesis. Here, we explore the causative mechanisms. BRG1 is preferentially enriched at active promoters of genes essential for spermatogonial pluripotency and meiosis. In contrast, BRG1 is also associated with the repression of somatic genes. Chromatin accessibility at these target promoters is dependent upon BRG1. These results favor a model in which BRG1 coordinates spermatogenic transcription to ensure meiotic progression. In spermatocytes, BRG1 interacts with SCML2, a testis-specific PRC1 factor that is associated with the repression of somatic genes. We present evidence to suggest that BRG1 and SCML2 concordantly regulate genes during meiosis. Furthermore, BRG1 is required for the proper localization of SCML2 and its associated deubiquitylase, USP7, to the sex chromosomes during pachynema. SCML2-associated mono-ubiquitylation of histone H2A lysine 119 (H2AK119ub1) and acetylation of histone lysine 27 (H3K27ac) are elevated in Brg1cKO testes. Coincidentally, the PRC1 ubiquitin ligase RNF2 is activated while a histone H2A/H2B deubiquitylase USP3 is repressed. Thus, BRG1 impacts the male epigenome by influencing the localization and expression of epigenetic modifiers. This mechanism highlights a novel paradigm of cooperativity between SWI/SNF and PRC1.
KEY WORDS: SWI/SNF chromatin remodeling, BRG1, Transcriptional regulation, SCML2
Summary: Epigenetic studies of BRG1 reveal for the first time that SWI/SNF and PRC1 cooperate during gametogenesis to regulate transcription.
INTRODUCTION
Spermatogenesis is a developmental cascade in which genetic information is passed on from mitotic precursors to meiotically derived haploid gametes. This process is particularly sensitive to the activity of several epigenetic regulators known to influence meiotic recombination. Examples include the role of the meiosis-specific H3K4 methyltransferase PR domain zinc-finger protein9 (PRDM9) in double stranded break (DSB) formation, the roles of polycomb repressive complex2 (PRC2), and the H3K9 methyl transferases EHMT2 [euchromatic histone lysine N-methyltransferase 2 (G9a)] and suppressor of variegation 3-9 1 or 2 (SUV39H1/H2) in homolog pairing (synapsis) (Brick et al., 2012; Diagouraga et al., 2018; Hayashi et al., 2005; Mu et al., 2014; Tachibana et al., 2007; Takada et al., 2011). Other crucial activities include the regulation of spermatogenic transcription by the polycomb repressive complexes PRC1 and PRC2 (Hasegawa et al., 2015; Mu et al., 2014). Furthermore, the ATP-dependent family of nucleosome remodelers, such as INO80, SWI/SNF and CHD5, which are known to modulate chromatin accessibility, are also essential for spermatogenesis (Kim et al., 2012; Li et al., 2014; Serber et al., 2015). In summary, chromatin modifiers play a vital role in germline development.
Our lab has previously reported the role of BRG1 (SMARCA4 – SWI/SNF catalytic subunit) in male meiosis (Kim et al., 2012). Briefly, the germline depletion of BRG1 results in pachytene arrest. Mutant spermatocytes display unrepaired DNA DSBs, evidenced by persistent γH2Ax, chromosomal asynapsis and reduced MLH1 foci, a marker of crossovers (Kim et al., 2012; Wang et al., 2012). Coincidentally, an enhanced level of repressive chromatin is observed in mutant spermatocytes. As SWI/SNF modulates chromatin accessibility by either sliding or evicting nucleosomes (reviewed by Clapier et al., 2017), it is plausible that changes in chromatin structure seen in the Brg1cKO testes might result in meiotic defects by potentially influencing transcription or DNA repair. Both processes are influenced by SWI/SNF.
SWI/SNF is associated with both gene activation and repression. In mouse embryonic fibroblasts, BRG1 and the core SWI/SNF subunit SNF5 coordinate nucleosome occupancy at promoters to achieve transcriptional regulation (Tolstorukov et al., 2013). The transcriptional outcome is often dictated by the subunit composition of the complex, which in turn can influence its genome-wide association and interactions with other gene regulators (Euskirchen et al., 2011; Raab et al., 2015; Raab et al., 2017). This is a theme that features frequently in the regulation of tissue and cell type-specific transcriptional programs that impact crucial processes such as embryonic stem (ES) cell pluripotency and differentiation, and neuronal and cardiac cell fate specification (reviewed by Ho and Crabtree, 2010). Thus, SWI/SNF directed gene regulation plays a crucial role during development.
Apart from gene regulation, SWI/SNF has also been implicated in several DNA repair mechanisms. In yeast and cell culture models, SWI/SNF is recruited to sites of DNA damage to promote accessibility and stimulate γH2Ax, a key component of the DNA damage response (DDR) signaling pathway (Kwon et al., 2015; Lee et al., 2010; Ogiwara et al., 2011). Other activities include the recruitment of homologous recombination (HR) and non-homologous end-joining (NHEJ) repair factors (Ogiwara et al., 2011; Qi et al., 2014; Watanabe et al., 2014), and the transcriptional silencing of regions adjacent to DNA DSBs (Kakarougkas et al., 2013). Although BRG1 is dispensable for γH2Ax formation during meiosis, it has been reported to influence the distribution of DDR factors like RAD51 (DNA recombinase) and RPA (replication protein A1) (Kim et al., 2012).
In this study, we present evidence to show that BRG1 coordinates spermatogenic transcription. BRG1 activates genes essential for maintaining spermatogonial pluripotency and meiotic progression. In contrast it represses somatic genes. Our data suggest that somatic gene repression is achieved through an interaction with SCML2 (sex comb on midleg-like 2), a known testis-specific PRC1 (polycomb repressive complex 1) member (Luo et al., 2015). BRG1 is required for the normal localization of SCML2, suggesting a role in recruitment. Furthermore, histone modifications associated with SCML2, such as the repressive mono-ubiquitylation of histone H2A lysine 119 (H2AK119ub1) and activating acetylation of histone lysine 27 (H3K27ac), are perturbed in Brg1cKO testes. Coincidentally, BRG1 also activates the expression of the H2A ubiquitin ligase, RNF2 (ring finger protein 2) and represses the histone H2A/H2B deubiquitylase USP3 (ubiquitin specific peptidase 3). Thus, SWI/SNF can epigenetically regulate germline transcription by SCML2-dependent and -independent mechanisms.
RESULTS
BRG1 associates with transcriptionally active and poised chromatin
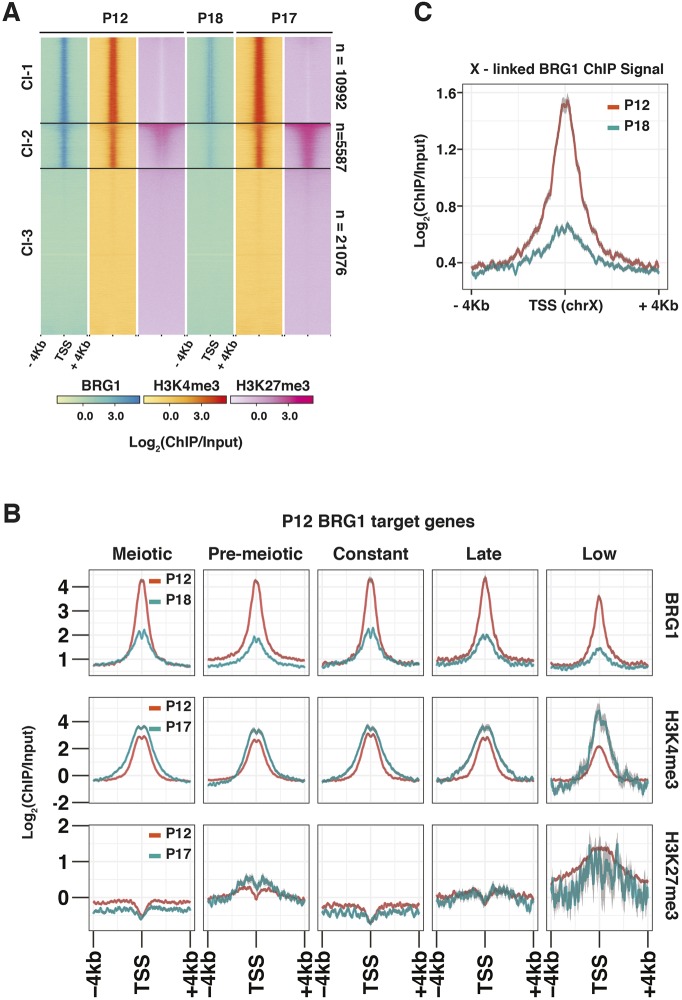
To understand the functions of SWI/SNF activity during meiosis, we determined the genome-wide association of BRG1 by ChIP-seq. We reasoned that the examination of BRG1 occupancy concurrent to crucial meiotic processes such as DSB repair and homologous chromosome synapsis would yield insight into the molecular mechanisms underlying the meiotic defects observed in the Brg1fl/Δ; Mvh-creTg/0 (Brg1cKO; see Materials and Methods) males. For this purpose we isolated chromatin from spermatogenic cells obtained from P12 (mostly pre-pachytene germ cells) and P18 testes (predominantly pachytene spermatocytes) (Bellve et al., 1977; Goetz et al., 1984). At these stages, BRG1 appears to be enriched promoter proximally (Fig. 1A, panels 1 and 4). This is consistent with the fact that more than 50% of BRG1 peaks associate with promoters at P12 and P18, whereas only a minority maps to distal sites (Fig. S1A). By performing K-means clustering, we categorized transcription start sites (TSS) into three different classes (class 1, Cl-1; class 2, Cl-2; class, Cl-3) based upon their association with BRG1. These TSSs feature high (Cl-1), medium (Cl-2) and insignificant (Cl-3) BRG1 enrichment.
Fig. 1.
BRG1 is enriched at transcriptionally active and poised regions. (A,B) The relative enrichment of BRG1, H3K4me3 and H3K27me3 from P12, P17 and P18 testes at (A) RefSeq gene, TSS±4 kb shown using heatmap with K-means clustering, and (B) TSS±4 kb associated with BRG1 target (P12 peaks) genes categorized by their temporal expression profile. (C) P12 and P18 BRG1 enrichment at TSS±4 kb on chromosome X. TSS, transcription start site.
To gain insight into the activity of genes associated with BRG1 in spermatogenic cells, we surveyed the chromatin environment surrounding their TSSs. We monitored the enrichment of activating trimethylation of histone H3 lysine 4 (H3K4me3) and repressive trimethylation of histone H3 lysine 27 (H3K27me3) associated with Cl-1 to Cl-3 TSSs at P12 and P17 (Mu et al., 2014) (Fig. 1A). Robust H3K4me3 enrichment appeared to be associated only with the K-means clusters from P12 to P17 (Fig. 1A, panels 2 and 5). In contrast, H3K27me3 levels at P12 and P17 appeared depleted at Cl-1 TSSs featuring high BRG1 occupancy (Fig. 1A, panels 3 and 6). Such antagonism is a well-established feature of SWI/SNF and PRC2 genomic associations (Wilson et al., 2010). Cl-1 and Cl-3 TSSs only displayed basal levels of H3K27me3, which was particularly discernable at P17. Unlike Cl-1 and Cl-3 TSSs, Cl-2 TSSs displayed significantly higher levels of H3K27me3, which appeared progressively enhanced from P12 to P17 (Fig. 1A, panels 3 and 6). The co-occurrence of H3K27me3 and H3K4me3 at Cl-2 TSSs resemble features of bivalent promoters, which are usually associated with transcriptionally quiescent genes poised to be reactivated at later stages of development (Bernstein et al., 2006; Hammoud et al., 2014; Lesch et al., 2013). Therefore, genes associated with Cl-2 TSSs likely represent repressed BRG1 target genes. In contrast, potential activated gene targets appear to be associated with Cl-1 TSSs.
To understand BRG1-directed gene regulation in the context of spermatogenesis, we categorized genes associated with each K-means cluster (Fig. 1A) by their temporal expression profiles, previously determined from whole-testes RNA-seq data (Margolin et al., 2014). These include genes maximally expressed in testes, at P6 (pre-meiotic, Pmei), from P8-P20 (meiotic, Mei), at P38 (late, adult testis), from P6-P38 (constantly; Const) and those expressed at low levels in testes from P6-P38 (low, <2 reads per kilobase per million of reads; RPKM). The majority of Cl-1-associated genes were meiotic with relatively fewer pre-meiotic genes (Fig. S1B). In contrast, Cl-2 and Cl-3 mostly comprised pre-meiotic and low genes. This is particularly interesting given that Cl-1 TSSs are exclusively marked by H3K4me3, while Cl-2 promoters display bivalent chromatin modifications. We observed similar trends when monitoring H3K4me3 and H3K27me3 dynamics at TSSs of BRG1 target genes (P12 peaks) categorized by their temporal expression profile during meiosis (P12 to P17) (Fig. 1B). Over this duration, all gene categories experienced a fourfold decrease in BRG1 enrichment in the presence of abundant H3K4me3 (Fig. 1B, top and middle row). Only pre-meiotic and low gene targets displayed elevated levels of H3K27me3 from P12 to P17 (Fig. 1B, bottom row), distinguishing them from meiotic, constant and late gene targets, which appeared exclusively marked by H3K4me3 (Fig. 1B, middle and bottom row). Candidate pre-meiotic targets such as Zbtb16 and Id4, which are markers of undifferentiated spermatogonial cells along with Pdgfra, a somatic signaling receptor, that are normally repressed during meiosis, displayed bivalent promoters (Basciani et al., 2002; Green et al., 2018; Hammoud et al., 2014). In contrast, meiotic target, Sycp1, which is essential for synaptonemal complex assembly, displayed a H3K4me3-enriched promoter (Fig. S1C) (De Vries et al., 2005). Thus, BRG1 might coordinate the expression of genes over the measured course of spermatogenesis.
Given its association with active (H3K4me3) and poised (H3K4me3/H3K37me3) chromatin, we were curious to examine BRG1 localization to the sex chromosomes, which are transcriptionally silenced during pachynema (reviewed by Turner, 2007). We examined BRG1 occupancy at the TSSs of X-linked genes at P12 (pre-pachytene stages) and P18 (pachytene stages). BRG1 enrichment at X-linked TSSs appeared reduced at P18 relative to P12, but not absent (Fig. 1C). Therefore, BRG1 associates with meiotically inactivated sex chromosome.
Apart from TSSs, H3K4me3 is also enriched at DSB/recombination hotspots, known to be associated with a meiosis-specific histone methyl transferase, PRDM9 (Brick et al., 2012; Diagouraga et al., 2018; Hayashi et al., 2005). We therefore examined BRG1 association at these PRDM9 sites previously mapped by ChIP-seq in P12 testes (Baker et al., 2015). The lack of enrichment at PRDM9 peaks makes it unlikely that BRG1 directly affects DSB formation (Fig. S1D). Thus, BRG1 might play a major role in gene regulation during meiosis.
BRG1 coordinates spermatogenic gene expression
The promoter-centric association of BRG1 prompted us to examine its influence on the transcription of target genes by RNA-seq. We compared transcript abundance between spermatogenic cells isolated from P12 Brg1fl/+ (Brg1WT) and Brg1fl/Δ; Mvh-creTg/0 (Brg1cKO) testes, where the germ cell populations are mostly pre-pachytene and therefore unlikely to be influenced by pachytene arrest. In fact, the loss of BRG1 did not appear to affect the development of pre-pachytene spermatocytes, as staged by γH2Ax (meiotic marker) at P10 and P13 (Fig. S2A). Incidence of pachytene arrest only manifested at P14 (Fig. S2A). In agreement with these results, the abundance of pre-pachytene protein-coding transcripts (Ball et al., 2016) appeared similar between P12 Brg1WT and Brg1cKO testes (Fig. S2B). Only early and late pachytene-specific transcripts were slightly less abundant upon the loss of BRG1 at P12, which is predictive of pachytene arrest at later stages (Fig. S2B). Overall, we do not expect the analysis of gene expression to be impacted significantly by secondary effects such as developmental delays.
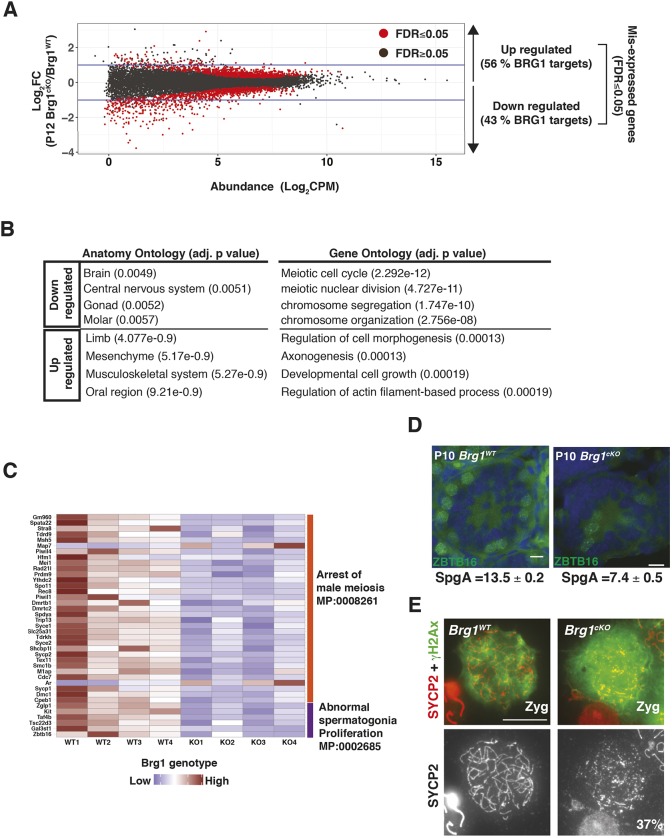
To identify genes significantly mis-expressed (FDR≤0.05) upon the loss of BRG1, we performed an edgeR analysis on the RNA-seq data. An equivalent number of genes was either transcriptionally downregulated (n=1100) or upregulated (n=983) in P12 Brg1cKO relative to Brg1WT testes (Fig. 2A). More genes were downregulated (n=310) by a magnitude of twofold or higher, relative to those upregulated (n=75), upon the loss of BRG1 (Fig. 2A). Nearly half of these differentially regulated genes were associated with BRG1 peaks (P12 peaks). The downregulated genes appear normally expressed in gonadal tissue and the nervous system, and were enriched for gene ontology (GO) terms relevant to meiotic processes (Fig. 2B). In contrast, the upregulated genes were normally expressed in limb and muscle and were enriched for GO terms relevant to somatic developmental processes (Fig. 2B). BRG1 therefore coordinates germline transcription by activating meiotic genes while concomitantly repressing somatic genes. As BRG1 is essential for meiotic progression (Kim et al., 2012), we monitored the expression of genes associated with abnormal spermatogonia proliferation and meiotic arrest phenotypes, curated from the mouse genome database (Blake et al., 2003). A heatmap displaying transcript abundance (z-score) of candidate genes, measured across P12 Brg1WT and Brg1cKO replicates revealed reduced transcript levels associated with most candidates with the exception of androgen receptor (Ar) and microtubule-associated protein 7 (Map7) (Fig. 2C). Next, we adopted a reverse genetic approach to test whether the mis-expression of candidate genes was associated with specific phenotypes in the Brg1cKO. To achieve this, we chose a pre-meiotic candidate, Zbtb16, and meiotic candidate, Sycp2.
Fig. 2.
BRG1 influences transcription during spermatogenesis. (A) Log2 fold-change (Brg1cKO/ Brg1WT, y-axis) in transcript abundance (CPM, counts per million, x-axis) of genes at P12. Dots represent genes and horizontal blue lines indicate twofold change. (B) Table listing anatomy and gene ontology terms associated with BRG1-regulated genes. Benjamini-Hochberg, adjusted P-values are reported in parenthesis. (C) Heatmap showing transcript abundance (z-scores) of genes (rows) associated with mouse phenotype ontologies in P12 BrgWT (WT1-WT4) and BrgcKO (KO1-KO4) (columns). (D) P10 BrgWT and Brg1cKO testes cryosections (25× objective) immunolabeled for ZBTB16 (green) and counterstained with DAPI (blue). The average numbers of SpgA/tubule and standard error of measurement (SEM) are indicated. Scale bar: 20 μm. (E) BrgWT and Brg1cKO zygotene spermatocytes (100× objective) immunolabeled for SYCP2 (red) and γH2Ax (green). Scale bar: 10 μm.
Zbtb16 is essential for the maintenance of a pool of undifferentiated type A spermatogonia (SpgA) (Buaas et al., 2004). Although BRG1 is dispensable for the establishment of spermatogonia, its role in SpgA maintenance remains unknown (Kim et al., 2012). We quantified the number of SpgA in P10 Brg1WT and Brg1cKO testes cryosections by immunostaining for ZBTB16. The Brg1cKO testes contained 46% fewer SpgA, when compared with Brg1WT testes (Fig. 2D). This reduction was not associated with a proliferative defect, indicated by unchanged proliferating cell nuclear antigen (PCNA) levels (Fig. S3A). To validate the transcriptional basis of this defect, we performed qRT-PCR to determine the expression of Zbtb16 in purified THY1+ spermatogonia (enriched for SpgA) isolated from P8 Brg1WT, Brg1Het (Brg1fl/+;Mvh-creTg/0) and Brg1cKO testes. We also monitored the expression of other established stem cell factors, such as: inhibitor of DNA binding 4 (Id4); POU domain, class 5, transcription factor 1 (Pou5f1, also known as Oct4); POU domain, class 3, transcription factor 1 (Pou3f1, also known as Oct6); and forkhead box O1 (Foxo1) (Dann et al., 2008; Goertz et al., 2011; Oatley et al., 2011; Wu et al., 2010). Transcripts associated with Zbtb16, Id4 and Pou5f1 were downregulated upon the loss of BRG1 in the purified THY1+ fractions (Fig. S3B). The fact that Thy1 mRNA levels remain abundant argues against a loss of spermatogonial cells early in development (Fig. S3B). Thus, BRG1 regulates the maintenance of undifferentiated spermatogonial cells by activating the expression of crucial stem cell factors.
The meiotic gene candidate Sycp2 constitutes the structural component of the lateral element of meiotic chromosomal axes and is essential for synapsis (Yang et al., 2006). Coincidentally, Brg1cKO spermatocytes display an increase in asynapsis (Kim et al., 2012). In the Brg1cKO, SCYP2 levels were distinctly lower relative to the controls (Fig. S3C) and abnormally assembled into short lateral filaments and aggregates in 37% of the mutant zygotene spermatocytes (total scored=212) (Fig. 2E). Whereas Sycp2−/− spermatocytes fail to form SYCP3 elements, we did not observe a similar defect in the Brg1cKO spermatocytes (Fig. S3D) (Yang et al., 2006). We reasoned that the reduced levels of SYCP2 might be sufficient to facilitate apparently normal SYCP3 assembly. A paucity in SYCP2 may therefore limit meiotic progression by potentially compromising the formation of a functional synaptonemal complex. In addition to the mis-regulation of essential germ cell factors, we also identified a significant increase in the expression of several somatic genes in the Brg1cKO (Table S1). Pdgfra, a signaling receptor, generally associated with somatic cells (Basciani et al., 2002; Schmahl et al., 2008) and pre-meiotic spermatogonia (Hammoud et al., 2014), was upregulated by more than twofold over a period spanning meiosis I in the Brg1cKO (Fig. S3E). Interestingly, Ar, the expression of which is limited to Sertoli, Leydig and myoid cells (Zhou et al., 2002), was abnormally expressed in germ cell nuclei devoid of BRG1 (Fig. S3F).
BRG1 is required to maintain chromatin accessibility at promoters
Chromatin remodelers reposition nucleosomes, thereby regulating accessibility to transcription factors. Thus, BRG1 might influence transcription by modulating the structure of the germline epigenome. To investigate this possibility, we performed an assay for transposase-accessible chromatin (ATAC)-seq to map open chromatin in pre-pachytene and pachytene spermatogenic cells isolated from P12 and P18 testes, respectively. Similar to the RNA-seq data, we surveyed differences in chromatin accessibility between P12 Brg1WT and Brg1cKO testes (Fig. S4A, left panel). As P18 Brg1cKO testes are characterized by severe pachytene arrest, we compared chromatin accessibility between P18 Brg1WT and Brg1Het (Brg1fl/Δ) testes (Fig. S4A, right panel). In normal spermatogenic cells, strong ATAC read coverage was detected promoter proximally at both P12 and P18 (Fig. S4A). This promoter accessibility was significantly diminished in P12 Brg1cKO testes and also under conditions of haploinsufficiency in P18 Brg1Het testes (Fig. S4A).
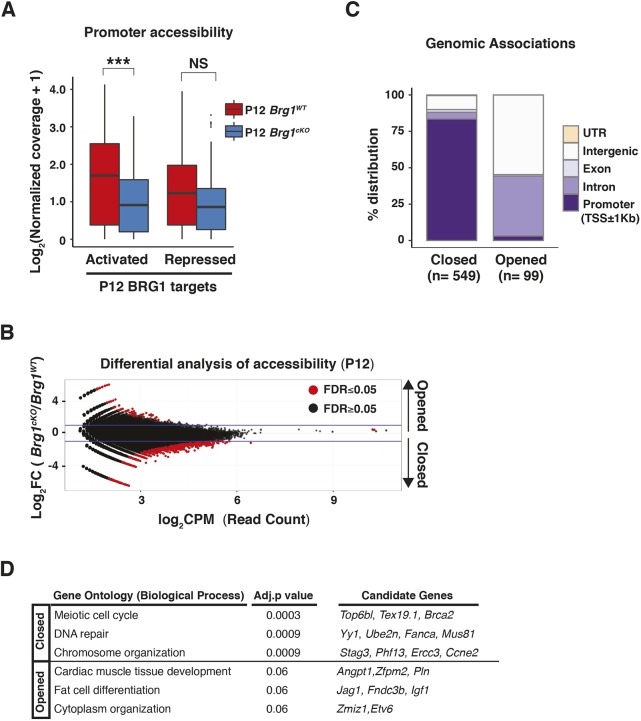
As promoter-proximal chromatin responds to the loss of BRG1, we first monitored the changes in chromatin accessibility at promoters of target genes differentially regulated by BRG1 (Fig. 3A). Promoters of BRG1 target genes that were normally activated (downregulated upon BRG1 loss) displayed a significant decrease in promoter accessibility in the Brg1cKO relative to Brg1WT testes. In contrast, promoters of repressed gene targets (upregulated upon BRG1 loss) failed to display a significant difference in chromatin accessibility upon the loss of BRG1 (Fig. 3A).
Fig. 3.
BRG1 directly regulates chromatin accessibility at promoters. (A) Log2 normalized ATAC-seq read coverage (y-axis, pseudocount added) at promoters (TSS±0.5 kb) of BRG1 targets (x-axis) in P12 Brg1wt (red) and Brg1cKO (blue) spermatogenic cells. ***P<0.001; NS, not significant, as calculated by Wilcoxon rank sum test. (B) Log2 fold-change (Brg1cKO/Brg1WT, y-axis) in read counts (CPM, counts per million, x-axis) at P12. Dots represent 300 bp binned regions. Horizontal blue lines indicate a twofold change. (C,D) Genomic associations (C) and gene ontology (D) (with Benjamini-Hochberg, adjusted P-values) of closed and opened regions.
To identify genome-wide changes in chromatin accessibility at P12, we performed edgeR on the ATAC read counts obtained from wild-type and mutant samples. The vast majority of regions that displayed significant differences in chromatin accessibility appeared less accessible (closed; n=549), leaving only a few regions that acquired greater accessibility (opened; n=99) upon the loss of BRG1 at P12 (Fig. 3B). Consistent with the general decrease in promoter accessibility in the Brg1cKO (Fig. S4A), the closed regions were overwhelmingly associated with promoters (Fig. 3C and Fig. S4B, panel 1). In contrast, the opened regions were prominently featured within introns and intergenic regions (Fig. 3C and Fig. S4B panels 2 and 3). This overall decrease in chromatin accessibility genome-wide is consistent with the previously observed increase in repressive epigenetic modifications in Brg1cKO spermatocytes (Kim et al., 2012). The genes associated with closed promoters in Brg1cKO testes were mostly meiotic in function (Fig. 3D, Table S2). Thus, BRG1 activates meiotic genes likely by maintaining chromatin accessibility at cognate promoters. In contrast, a few genes associated with the distal sites that appeared more accessible in the Brg1cKO testes represent somatic factors (Fig. 3D, Table S2).
BRG1 physically interacts with SCML2, a non-canonical PRC1 factor
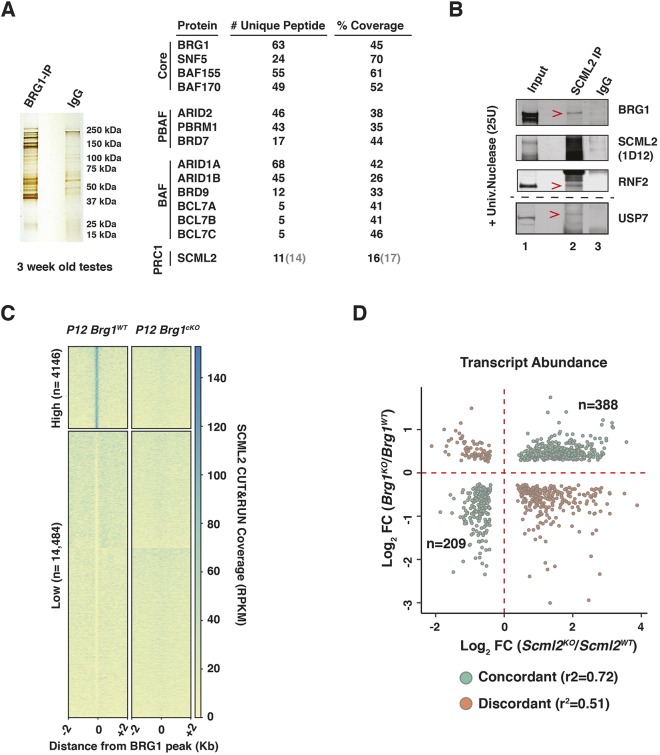
To investigate further mechanisms governing SWI/SNF-mediated epigenetic regulation, we monitored BRG1 interactions in testes nuclear extracts from 3-week-old mice by performing immuno-pulldowns (IP) (Fig. 4A). Proteins isolated from a BRG1 IP and control nonspecific (ns) IgG pulldown were identified by mass spectrometry (MS). Known SWI/SNF subunits were specifically identified in the BRG1 IP, thus demonstrating the efficacy of our method (Fig. 4A, Fig. S5A). Furthermore, the presence of both SWI/SNF sub-complexes, brahma associated factor (BAF) and polybromo-BAF (PBAF), were detected in the germ line (Fig. 4A, Fig. S5A) (reviewed by Masliah-Planchon et al., 2015). More interestingly, we identified peptides associated with SCML2, a known testes-specific PRC1 factor (Hasegawa et al., 2015; Luo et al., 2015) (Fig. 4A). As candidate peptides were also detected in the non-specific IgG IP, we validated these interactions by performing a reverse IP with an antibody against SCML2 (Fig. 4B). The IP was conducted on nuclear lysates treated with universal nuclease (benzonase) to eliminate non-specific, DNA-mediated interactions. BRG1 was specifically detected in the SCML2 IP, compared with nsIgG (Fig. 4B; lanes 2 and 3). Additionally, SNF5 (SMARCB1), a core SWI/SNF subunit, and RNF2, a known SCML2-interacting partner, were detected by co-IP (Fig. S5C, Fig. 4B) (Hasegawa et al., 2015). The smearing of the SCML2 band in the nuclear extracts (Fig. 4B) prompted us to confirm the specificity of the SCML2 antibody. We did this by performing western blots on nuclear extracts obtained from spermatogenic cells and ovaries. Consistent with its known male-specific expression pattern, we fail to detect a SCML2 signal in nuclear extracts obtained from ovaries (Fig. S5B, lane 2). Hence, the smearing might be a product of protein instability or indicative of isoforms.
Fig. 4.
BRG1 physically interacts with SCML2 to regulate gene expression. (A) Silver stained gel (left) and table (right) summarizing BRG1 IP-MS results. Numbers within parentheses are derived from IgG IP. (B) SCML2 co-IP analysis. Red arrowheads label interacting proteins. Lane numbers are labeled. (C) Heatmap with K-means clustering showing normalized CUT&RUN read coverage associated with SCML2 at P12 BRG1 peaks±2 kb. RPKM, reads per kilobase per million mapped reads. (D) Log2 fold-change (KO/WT) in transcript abundance upon the loss of BRG1 (y-axis) or SCML2 (x-axis). Dots represent concordantly (green) or discordantly (orange) misregulated genes (common genes, FDR<0.05). r2 was calculated using Pearson's correlation test.
In pachytene spermatocytes, SCML2 is also known to interact with USP7, a deubiquitylase and non-canonical member of the mammalian PRC1.4 complex (Lecona et al., 2015; Luo et al., 2015). In agreement with these previous studies, we also detected an association between SCML2 and USP7 (Fig. 4B). However, USP7 did not appear to interact directly with BRG1 (Fig. S5D, lanes 2-4). Thus, BRG1 only associates with SCML2 during meiosis.
BRG1 and SCML2 concordantly regulate genes during meiosis
In pachytene spermatocytes, SCML2 is known to repress somatic and spermatogonial genes, while concurrently activating certain meiotic and late spermatogenic genes (Hasegawa et al., 2015). This pattern is similar to the epigenetic role of BRG1 in the germ line. Interestingly, we observe robust enrichment of BRG1 at TSSs of genes differentially regulated (FDR<0.05) by SCML2 during pachynema (Fig. S6A). Furthermore, the chromatin accessibility at these TSSs is reduced upon the loss of BRG1 (Fig. S6B). Hence, BRG1 might interact with SCML2 to mutually regulate transcription.
To test this possibility, we probed SCML2 localization in P12 Brg1WT and Brg1cKO spermatogenic cells using a modified version of the CUT&RUN method (Hainer et al., 2019; Skene et al., 2018). We monitored SCML2 enrichment at BRG1 peaks (Table S3) by K-means clustering to find regions with robust (high, n=4146) and near-absent SCML2 occupancy (low, n=14,484) in Brg1WT spermatogenic cells. Interestingly, the ‘high’ sites were devoid of SCML2 in Brg1cKO spermatogenic cells (Fig. 4C). Thus, ‘high’ sites represent loci at which BRG1 recruits SCML2. They are associated with genes that are differentially regulated by BRG1 (FDR<0.05) (activated, n=226; repressed n=263).
We then compared the P12 BRG1 and pachytene SCML2 RNA-seq data sets to look for genes that may be commonly regulated (FDR<0.05). About 65% (n=597) of genes commonly mis-expressed in the absence of either BRG1 or SCML2 exhibited concordant expression changes that were highly correlated (r2=0.72). The remaining 35% (n=330) showed discordant changes (r2=0.51) (Fig. 4D). Of the concordantly regulated genes, 64% (n=388) were repressed, while the remainder (n=209) were activated. The concordantly repressed genes accounted for 40% of the genes upregulated in the P12 Brg1cKO testes. In contrast, only 19% of genes downregulated in the P12 Brg1cKO testes overlapped with concordantly activated genes. The commonly repressed genes were mostly somatic in function (Fig. S6C) and included the somatic signaling receptor PDGFRA, which is associated with BRG1 (Figs S1C and S2E). Thus, during normal prophase I, BRG1 appears to achieve gene repression by recruiting SCML2. The co-activated genes were associated with GO terms relevant to the mitotic spindle checkpoint (Fig. S6B). Evidence for such checkpoint mechanisms have been reported late in meiosis (Eaker et al., 2001; Lee et al., 2011).
BRG1 influences SCML2 and USP7 localization to the sex body
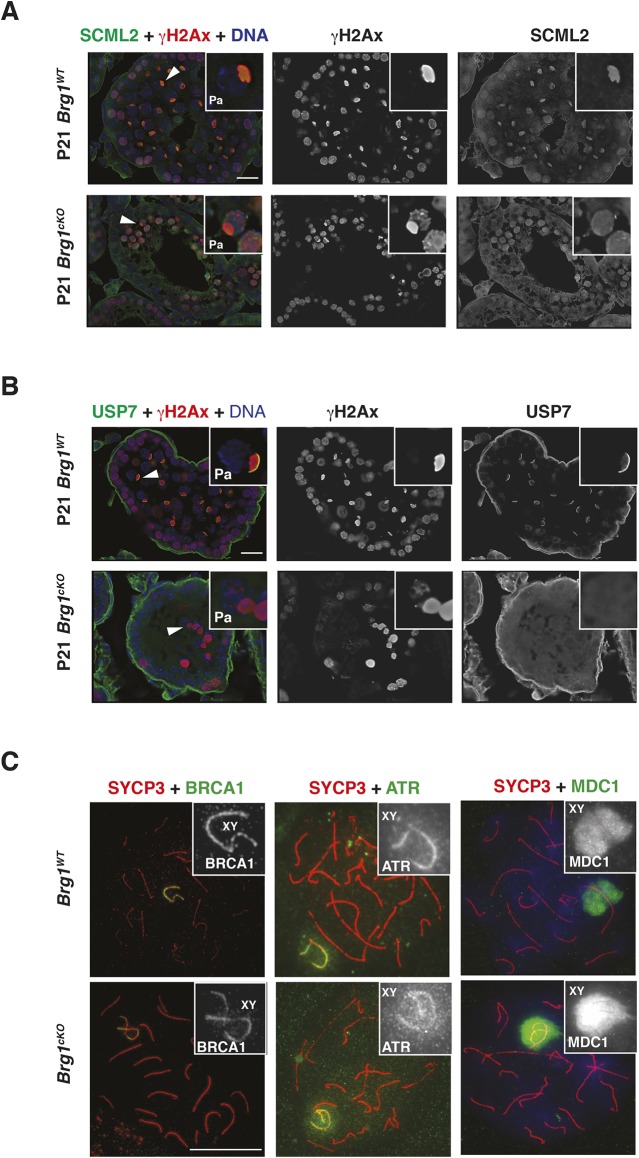
During pachynema, both SCML2 and its interacting partner, USP7, paint the sex body: a γH2Ax-enriched sub-nuclear compartment containing the sex-linked chromosomes (Hasegawa et al., 2015; Luo et al., 2015). Therefore, we determined whether SCML2 localization to the sex body was dependent on BRG1.
We first compared SCML2 localization in Brg1WT and Brg1cKO testes cryosections from 2- and 3-week-old males by immunofluorescence (Fig. 5A, Fig. S7A). Mutant pachytene spermatocytes were identified by staining for γH2Ax, given that its association with the sex body remains unperturbed in the Brg1cKO (Kim et al., 2012). The loss of BRG1 appeared to impact the localization of SCML2 in Brg1cKO pachytene spermatocytes (Fig. 5A). Here, SCML2 appeared abnormally distributed genome-wide without normally accumulating on the sex body (Fig. 5A, panel 3 insets). We confirmed these defects by co-staining for ATR (ataxia telangiectasia and Rad3 related), a DDR factor enriched on the sex body (Royo et al., 2013), and also stained for BRG1, to demonstrate protein loss in mutant pachytene spermatocytes (Fig. S7A). Given that SCML2 physically associates with γH2Ax (Hasegawa et al., 2015; Luo et al., 2015), it may be possible that SCML2 localizes to autosomal sites harboring persistent γH2Ax in Brg1cKO spermatocytes (Fig. 5A, panel insets). Surprisingly, subtle defects in SCML2 localization were also seen in Brg1Het pachytene spermatocytes, where it appeared more homogenously distributed genome-wide (Fig. S7B, panel insets).
Fig. 5.
BRG1 influences the localization of SCML2 to the sex body. (A,B) P21 Brg1WT and Brg1cKO testes cryosections (63× objective) co-stained for γH2Ax (red) and (A) SCML2 (green) or (B) USP7 (green). DNA (blue) is stained with DAPI. Arrowheads label the sex body. Insets show representative pachytene (Pa) spermatocytes. Scale bars: 20 μm. (C) Brg1WT and Brg1cKO pachytene spermatocytes spreads (100× objective) co-stained for SYCP3 (red) and BRCA1 (left, green), ATR (middle, green) and MDC1 (right, green). Insets highlight sex chromosomes. Scale bar: 20 μm.
We next examined SCML2 localization in Brg1WT and Brg1cKO meiotic spreads co-stained with a synaptonemal complex marker, SYCP3, to visualize chromosomes. Mutant meiotic spreads were obtained from Brg1cKO testes generated using two independent germline-specific CREs: Mvh-cre and Stra8-cre. Similar to the cryosections, SCML2 association with the sex chromosomes was perturbed in Brg1cKO pachytene spreads (Fig. S8). However, the phenotype appeared less severe in Brg1cKO pachytene spermatocytes generated with Stra8-cre, relative to Mvh-cre (Fig. S8). Such differences might be a consequence of the distinct temporal activity of each CRE (see Materials and Methods) (Gallardo et al., 2007; Sadate-Ngatchou et al., 2008).
In addition to SCML2, we also monitored the localization of USP7 in pachytene spermatocytes obtained from Brg1WT and Brg1cKO testes. Even though BRG1 does not directly interact with USP7 (Fig. S5C), we posited that the mis-localization of SCML2 in Brg1cKO pachytene spermatocytes might affect USP7 enrichment on the sex body. In fact, this seems to be the case in Brg1cKO pachytene spermatocytes immunofluorescently stained for USP7 and γH2Ax (Fig. 5B).
From previous studies it is clear that the mislocalization of SCML2 does not affect processes that are crucial to the initiation of meiotic sex chromosome inactivation (MSCI) (Hasegawa et al., 2015). As the loss of BRG1 has been previously reported to influence MSCI (Wang et al., 2012), we examined its impact on the localization of known MSCI factors, such as BRCA1, ATR and MDC1, to the sex body (Ichijima et al., 2011; Turner et al., 2004). By immunofluorescence, BRCA1, ATR and MDC1 appeared stably associated with the sex body in Brg1cKO pachytene spermatocytes (Fig. 5C). Thus, BRG1-like SCML2 does not affect the initiation of MSCI. Interestingly, it has been previously reported that the loss of MDC1 abrogates SCML2 recruitment to the sex chromosomes (Hasegawa et al., 2015). Therefore, the stable association of MDC1 with the sex body in Brg1cKO spermatocytes (Fig. 5C) suggests that SWI/SNF may function downstream MDC1 in the recruitment of SCML2 to the sex body.
BRG1 influences the abundance of SCML2-associated histone modifications
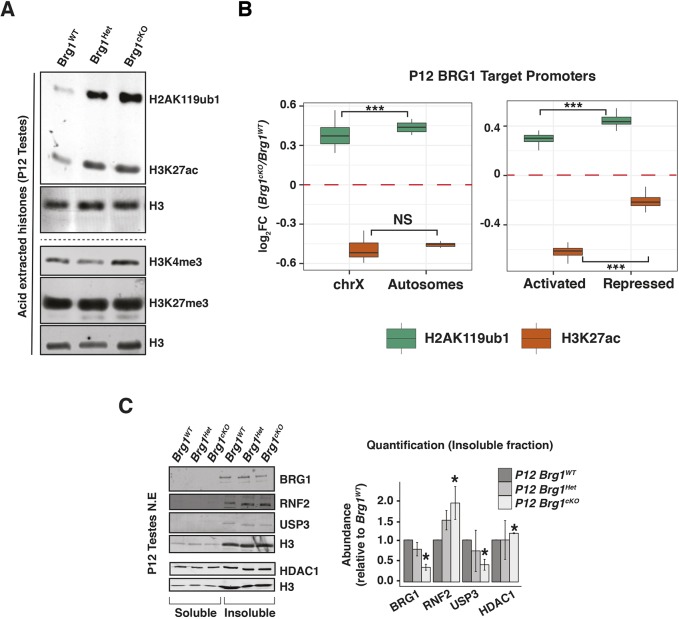
As SCML2 is known to regulate both H2AK119ub1 and H3K27ac during meiosis (Adams et al., 2018; Hasegawa et al., 2015), we investigated whether they are also affected by BRG1. We monitored the abundance of H2AK119ub1 and H3K27ac in acid extracts obtained from P12 Brg1WT, Brg1Het and Brg1cKO spermatogenic nuclei (Fig. 6A, top panel). We also examined the abundance of H3K4me3 and H3K27me3, which are associated with BRG1 target promoters (Fig. 6A, bottom panel). Although neither H3K4me3 nor H3K27me3 levels were perturbed, both H2AK119ub1 and H3K27ac were dramatically elevated in P12 Brg1Het and Brg1cKO, relative to the Brg1WT, spermatogenic cells (Fig. 6A, top panel). Interestingly, these epigenetic perturbations were undetectable at P8 and P10, which coincide with the initiation of meiosis and pairing (leptonema to zygonema) (Fig. S9A). Thus, BRG1 suppresses H2AK119ub1 and H3K27ac prior to the onset of pachynema. The epigenetic response to the partial loss of BRG1 (in P12 Brg1Het) prompted us to verify whether it was a consequence of the Mvh-cre transgene. This scenario seems unlikely, given that H2AK119ub1 was not globally elevated in males expressing the CRE relative to littermate controls without the CRE (Fig. S9B). Another cause for concern was that the H2AK119ub1 antibody (clone E6C5) used in this study was previously reported to recognize non-histone epitopes (Hasegawa et al., 2015). Hence, we validated the specificity of clone E6C5 by performing western blots on acid-extracted histones obtained from RNF2 (PRC1 E3-ubiquitin ligase) knockout (Rnf2KO) embryonic stem (ES) cells engineered using CRISPR-CAS9. The near-absence of H2AK119ub1 signal in the Rnf2KO relative to Rnf2WT ES cell histone extracts confirms the specificity of clone E6C5 (Fig. S9C). Furthermore, H2AK119ub1 is clearly detected in chromatin fractions obtained from P12 Brg1WT, Brg1Het and Brg1cKO testes (Fig. S9D).
Fig. 6.
BRG1 regulates H2AK119ub1 and H3K27ac during spermatogenesis. (A) Western blot analysis of H2AK119ub1, H3K27ac, H3K4me3 and H3K27me3 from P12 Brg1WT, Brg1Het and Brg1cKO testis acid extracts. Loading control is histone H3. (B) Log2 fold-change (cKO/WT, y-axis) in H2AK119ub1 (green box) and H3K27ac (orange box) at BRG1 occupied promoters (TSS±0.5 kb, x-axis) at P12, categorized by chromosomal location (left) and transcriptional status (right). ***P<0.001; NS, not significant, as calculated by Wilcoxon rank sum test. (C) Western blot analysis (left) and quantification (right) of BRG1, RNF2, USP3 and HDAC1 in P12 BrgWT, BrgHet and Brg1cKO spermatogenic cells. Loading control is H3. Normalized protein abundances are determined from at least two independent trials. Data are mean±s.e.m. *P<0.05 (Student's t-test).
BRG1 suppresses H2AK119ub1 and enhances H3K27ac at target promoters
Based on the genome-wide increase in H2AK119ub1 and H3K27ac in Brg1cKO testes, we surveyed changes in these modifications at promoters occupied by BRG1 (P12 peaks) by ChIP-seq in P12 Brg1WT and Brg1cKO spermatogenic cells. First, we categorized the BRG1 targets by their chromosomal location, the rationale being that both H2AK119ub1 and H3K27ac are known to be differentially regulated between the autosomes and sex chromosomes at pachynema (Adams et al., 2018). The BRG1 target promoters displayed contrasting changes in H2AK119ub1 and H3K27ac enrichment in the Brg1cKO spermatogenic cells. Irrespective of their location on either the X chromosome or autosomes, target promoters displayed enhanced H2AK119ub1 combined with reduced H3K27ac in the Brg1cKO relative to Brg1WT spermatogenic cells (Fig. 6B, left panel). This mirrors changes in H2AK119ub1 and H3K27ac that occur on the sex chromosomes in Scml2KO pachytene spermatocytes (Adams et al., 2018; Hasegawa et al., 2015). Thus, SCML2 might influence histone modifications at BRG1 loci.
Next, we analyzed the changes in H2AK119ub1 and H3K27ac at promoters of BRG1 target genes that are differentially expressed. We hypothesized that these histone modifications might dictate the activity of associated genes. Both activated and repressed target promoters displayed elevated H2AK119ub1 and reduced H3K27ac in the Brg1cKO relative to Brg1WT spermatogenic cells. Such epigenetic changes are typically associated with gene silencing and accounts for the perturbation in normal gene activation. At the same time, the failure to repress gene targets appears to occur independently of H2AK119ub1 or H3K27ac. One possibility is that bivalently modified TSSs dictate the repression of BRG1 target genes (Fig. 1A, Cl-2).
Interestingly, SCML2 is known to facilitate H3K27me3 at bivalent domains (Maezawa et al., 2018). We re-analyzed the ChIP-seq data to monitor H3K4me3 and H3K27me3 enrichment from Scml2WT and Scml2KO pachytene spermatocytes at BRG1 target TSSs. Whereas H3K4me3 was unperturbed, H3K27me3 levels at TSSs of repressed targets decreased twofold upon the loss of SCML2 during pachynema (Fig. S10). Thus, a loss of repressive bivalent modifications might underlie the mis-expression of BRG1 repressed genes.
The loss of H3K27ac at promoters normally occupied by BRG1 in Brg1cKO spermatogenic cells appeared inconsistent with its genome-wide increase (Fig. 6A; top panel, Fig. 6B). To determine whether other genomic regions acquire greater H3K27ac enrichment upon the loss of BRG1, we performed a differential peak calling analysis using the macs2 bdgdiff algorithm (Zhang et al., 2008). This analysis revealed that several intronic and intergenic regions that lost, gained or maintained (common) H3K27ac peaks in Brg1cKO relative to the Brg1WT spermatogenic cells (Fig. S11A; left panel, Fig. S11B). Compared with the common peaks, regions that lost or gained H3K27ac were normally depleted of BRG1 (Fig. S11A, right panel). Thus, although total H3K27ac levels are elevated, its local distribution appears heterogeneous upon the loss of BRG1.
BRG1 can also influence the epigenome in a SCML2-independent manner
Although our data suggest that BRG1 regulates H2AK119ub1 and H3K27ac, through its interaction with SCML2, we cannot rule out the possibility that BRG1 might also influence these histone modifications by directly regulating the expression of cognate epigenetic modifiers. In fact, our ChIP-seq data identified BRG1 peaks at promoters of epigenetic modifiers known to influence H2AK119ub1, H2BK120ub1 and H3K27ac (Fig. S10A). These include the H2A ubiquitin ligase RNF2 (Wang et al., 2004), USP3, a H2A/H2B deubiquitylase (Nicassio et al., 2007), and the histone deacetylases HDAC1 and HDAC2 (Gallinari et al., 2007) (Fig. S12A). The transcript abundances of all these targets were significantly altered in response to the loss of BRG1 (Fig. S12B).
In the case of the histone ubiquitin modifiers Rnf2 and Usp3, the former displayed an increase whereas the latter displayed a decrease in transcript abundance in Brg1cKO relative to the Brg1WT spermatogenic cells at P12 (Fig. S12B, row 1). This is consistent with significant changes in protein levels in Brg1cKO (RNF2, 88% increase; USP3, 59% decrease) relative to the Brg1WT chromatin fractions prepared from testes (Fig. 6C, lanes 4-6). Thus, BRG1 may influence H2AK119ub1 by maintaining a balanced expression of RNF2 and USP3. Furthermore, these expression patterns represent an SCML2-independent mechanism of epigenetic regulation. As USP3 has also been associated with mono-ubiquitylated H2B (Nicassio et al., 2007), we monitored the levels of mono-ubiquitylation of H2B lysine 120 (H2BK120ub1), a modification associated with gene activation and chromatin relaxation (Fierz et al., 2011; Minsky et al., 2008; Pavri et al., 2006). At P12, H2BK120ub1 appeared elevated genome-wide in Brg1cKO, relative to Brg1WT spermatogenic cells (Fig. S12C).
Similar to the histone ubiquitin modifiers, H3K27ac-associated modifiers Hdac1 and Hdac2 displayed reduced transcript abundance in the Brg1Het and Brg1cKO (Fig. S12B, row 2). At least in the case of Hdac1, we were unable to identify a corresponding depletion in protein levels by western blot (Fig. 6C, lanes 1-6). As HDAC1 has been shown to compensate for HDAC2 in various developmental scenarios (Ma et al., 2012; Montgomery et al., 2009; Yamaguchi et al., 2010), it is unlikely that the perturbation in H3K27ac levels in the Brg1cKO spermatogenic cells occurs via mis-expression of HDAC1/2.
DISCUSSION
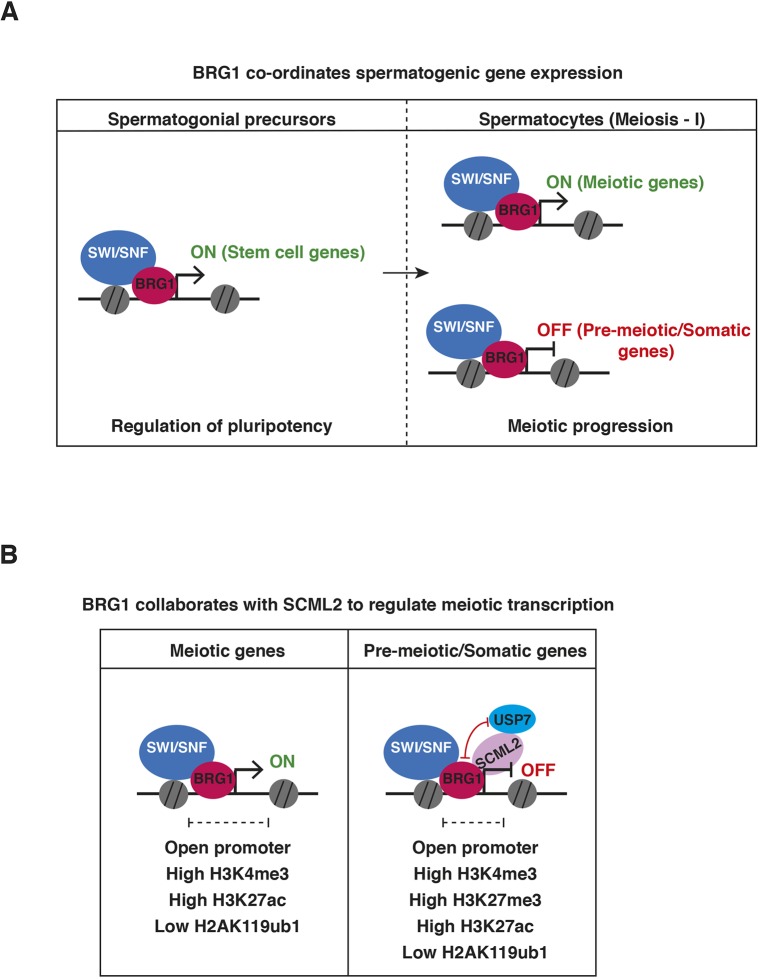
In this study, we have integrated genomic and proteomic approaches to show that SWI/SNF-directed regulation of transcription influences meiotic progression in males. In spermatogenic cells, BRG1 was overwhelmingly promoter associated, which is distinct from what has been observed in other mammalian cell types and embryonic tissue (Alexander et al., 2015; Alver et al., 2017; Attanasio et al., 2014). This localization re-enforces the notion that cell- or tissue-specific associations influence SWI/SNF function during development (reviewed by Ho and Crabtree, 2010). We propose a model in which the SWI/SNF ATPase activates essential spermatogenic genes while maintaining the repression of somatic genes (Fig. 7). We showed that BRG1 facilitates promoter accessibility of differentially regulated genes, which is consistent with the generally accepted mechanism of SWI/SNF (reviewed by Clapier et al., 2017). The activated genes play crucial roles in the maintenance of undifferentiated spermatogonial cell populations and facilitate meiotic progression. Target stem cell factors include Zbtb16 and Id4. The latter gene specifically identifies spermatogonial stem cells (SSCs) (Green et al., 2018; Oatley et al., 2011). The influence of BRG1 on SSC maintenance demonstrates a conserved role for SWI/SNF across various stem cell lineages (Ho et al., 2009; Lessard et al., 2007). The regulation of Sycp2, which is associated with synaptonemal complex assembly and homolog synapsis (Yang et al., 2006), potentially explains the incomplete synapsis and subsequent meiotic arrest seen in Brg1cKO spermatocytes (Kim et al., 2012). Thus, the male sterility associated with Brg1cKO adults is a consequence of a shortage of germline progenitors and essential meiotic factors.
Fig. 7.
Model describing the role of SWI/SNF in spermatogenic gene regulation. (A) During spermatogenesis, BRG1 activates genes essential for the maintenance of undifferentiated spermatogonia and ensures meiotic progression by activating meiotic genes and repressing pre-meiotic and somatic genes in spermatocytes. (B) During meiosis, activated gene promoters display H3K4me3, while repressed gene promoters are bivalently modified (H3K4me3 and H3K27me3). BRG1 maintains promoter accessibility, suppresses H2AK119ub1 and enhances H3K27ac at target genes. We propose that BRG1 recruits SCML2 and its associated deubiquitylase, USP7, to epigenetically silence somatic genes.
BRG1-mediated repression of the somatic transcriptome during meiosis is a feature shared with two other epigenetic regulators: PRC2 and SCML2. The latter protein is a testes-specific PRC1 factor (Hasegawa et al., 2015; Mu et al., 2014). Here, we propose that BRG1 achieves the repression of its target genes by recruiting SCML2 activity. This is supported by the following observations: (1) BRG1 physically interacts with SCML2; (2) BRG1 is enriched at the promoters of genes regulated by SCML2; (3) BRG1 concordantly represses most genes commonly regulated by SCML2; and (4) BRG1 influences SCML2 occupancy at BRG1 peaks prior to pachynema and SCML2 association with sex chromosomes during pachynema.
In contrast to the repressed genes, BRG1 also concordantly activates a smaller proportion of SCML2-regulated genes. These include key mitotic spindle checkpoint regulators. Whether SWI/SNF influences similar pathways during meiosis remains to be tested.
In addition to these observations, BRG1 also influences the known SCML2 histone modifications H2AK119ub1 and H3K27ac (Adams et al., 2018; Hasegawa et al., 2015). However, unlike SCML2, BRG1 does not appear to differentially regulate autosomal and sex-linked chromatin (Hasegawa et al., 2015). It is possible that such differential regulation does not manifest at pre-pachytene stages. Alternatively, this regulation may be indicative of distinct epigenetic outcomes associated with SCML2-dependent and -independent mechanisms. The latter is demonstrated by the effect of BRG1 on the expression of potent H2AK119ub1 modifiers, such as RNF2 and USP3.
As counter-intuitive as it may seem, repressed BRG1 targets genes are associated with low H2AK119ub1 and higher H3K27ac levels. This pattern indicates that repression occurs by a distinct mechanism. One possibility that we explored was that bivalent promoters (H3K4me3/H3K27me3) established the repression of BRG1 target genes. Such modifications are a hallmark of somatic gene silencing and have been shown to be influenced by SCML2 during spermatogenesis (Hammoud et al., 2014; Lesch et al., 2013; Lesch et al., 2016; Maezawa et al., 2018). In fact, H3K27me3 levels at BRG1-repressed gene promoters were depleted upon the loss of SCML2 at pachynema. Thus, SCML2 might potentially govern the repression of BRG1 target genes by regulating bivalent modifications.
In addition to H2AK119ub1, BRG1 also suppresses H2BK120ub1, which is associated with transcriptional activation and chromatin relaxation (Fierz et al., 2011; Minsky et al., 2008; Pavri et al., 2006). Hence, an increase in H2BK120ub1 might potentially de-repress genes in Brg1cKO spermatogenic cells. This change may be due to the misregulation of USP3 or mislocalization of USP7, two deubiquitylases whose activities have been associated with H2BK120ub1 (Nicassio et al., 2007; Van Der Knaap et al., 2005). The evidence for USP7 is based on its function in Drosophila (Van Der Knaap et al., 2005). In contrast, mammalian USP7 appears to influence H2BK120ub1 in a non-catalytic fashion (Lecona et al., 2015; Maertens et al., 2010; Huether et al., 2014).
In conclusion, we have revealed the transcriptional basis for the meiotic defects previously described in Brg1cKO males, and present a new paradigm for studying cooperation between SWI/SNF and PRC1 factors in the regulation of the epigenome. Although recent studies have illustrated the propensity of BRG1 to evict canonical PRC1 members from chromatin in normal and oncogenic cell culture models (Kadoch et al., 2017; Stanton et al., 2017), the relationship between SWI/SNF and variant PRC1 factors remain unexplored.
MATERIALS AND METHODS
Generation of Brg1 conditional deletion and genotyping
Brg1 floxed (Sumi-Ichinose et al., 1997), Mvh-Cre (activated at ∼E15) (Gallardo et al., 2007) and Stra8-Cre (activated only in males at P3) (Sadate-Ngatchou et al., 2008) were maintained on an outbred genetic background using CD-1 mice. Brg1fl/fl females were crossed to Brg1fl+;Mvh-CreTg/0 males to obtain Brg1flΔ;Mvh-CreTg/0 (Brg1cKO), Brg1fl+;Mvh-CreTg/0 (Brg1Het) and Brg1fl+ (Brg1WT) littermate controls. Similar crosses were made to generate the Stra8 Cre-induced conditional knockouts and littermate controls. Genotyping primers used in this study include: Brg1fl/+ alleles, (forward) 5′-CCTAGCCAAGGTAGCGTGTCCTCAT-3′ and (reverse) 5′-CCAGGACCACATACAAGGCCTTGTCT-3′; the excised allele (Δ), reverse primer used above in combination with (forward) 5′-CTAACCGTGTATGTAGCCAGTTCTGCCT-3′; Mvh-Cre, (forward) 5′-CACGTGCAGCCGTTTAAGCCGCGT-3′ and (reverse) 5′-TTCCCATTCTAAACAACACCCTGAA-3′; and Stra8-Cre, (forward) 5′-GTGCAAGCTGAACAACAGGA-3′ and (reverse) 5′-AGGGACACAGCATTGGAGTC-3′. All animal work was carried out in accordance with approved IACUC protocols at the University of North Carolina at Chapel Hill.
Disruption of Rnf2 by CRISPR-Cas9
The sequences of sgRNAs for Rnf2 are 5′-CACCGTGTTTACATCGGTTTTGCG-3′ and 5′-AAACCGCAAAACCGATGTAAACAC-3′. sgRNAs were cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (42230, Addgene) using the Golden Gate assembly cloning strategy (Bauer et al., 2015). Briefly, 5×104 E14 ES cells were cultured on 60 mm dishes for 1 day and then transfected with plasmids expressing Cas9 and sgRNAs, along with a plasmid expressing PGK-PuroR (Addgene, 31937) using FuGENE HD reagent (Promega) according to the manufacturer's instructions. The cells were treated with 2 μg/ml puromycin for 2 days and recovered in normal culture medium until ES cell colonies grew up. Rnf2-targeted colonies were verified by DNA sequencing.
Immunofluorescence staining
Spermatocyte spreads were prepared as described previously (Peters et al., 1997) or by using a protocol adapted from Wojtasz et al. (2009) to generate ‘3D-preserved’ spermatocytes. The latter protocol entails a detergent-spreading technique in which single cell preparations (obtained as described by Biswas, 2018) were treated with 0.25% NP-40 for not more than 2 min. These spreads were used to view SYCP2 staining. All spermatocyte spreads were generated from 2- to 3-week-old mice. Prepared slides were either dried down and stored at −80°C or stored in phosphate-buffered saline (PBS) at 4°C in the case of ‘3D-preserved’ spermatocytes.
Testis cryosections were prepared as described previously (Kim et al., 2012) with a few modifications. Briefly, juvenile/adult testes were fixed in 10% neutral buffered formalin at 4°C. After 20 min of incubation in NBF, the tissue was halved and then fixed for up to 1 h. Fixed tissue was washed three times in PBS at room temperature and then saturated through a sucrose series: 10% (30 min), 20% (30 min) and 30% (1 h). The tissues were then incubated overnight in 30% sucrose/optimum cutting temperature (OCT) formulation at 4°C and subsequently embedded in OCT. Frozen sections were cut at 9 μm. Antigen retrieval was performed for all antibodies used in this study. Briefly, slides were incubated in boiling citrate buffer (10 mM citric acid, pH 6.0) for 10 min. Over this period, citrate buffer was replaced every 2 min with fresh boiling buffer and then allowed to cool down gradually for up to 20 min.
Tissue sections and spreads were washed in PBS followed by permeabilization in 0.1% Triton-X 100 and then blocked in antibody dilution buffer (10% bovine serum albumin, 10% goat/donkey serum, 0.05% Triton-X 100) diluted 1:10 in PBS for 20 min before incubation with primary antibody overnight at 4°C. The following day, samples were again washed, permeabilized and blocked in ADB/PBS after which they were incubated for 1 h with Alexa Fluor-conjugated secondary antibodies. Immunostained slides were finally washed twice in PBS/0.32% photoflo (Kodak), once in H2O/0.32% photoflo and then counterstained with DAPI before mounting in Prolong Gold anti-fade medium (P-36931; Life Technologies). A list of all the primary antibodies used in this study is provided (Table S4). We used Alexa Fluor-conjugated secondary antibodies (Invitrogen; A-11029, A-11036, A-11055, A10042 and A31571) at a dilution of 1:500. All imaging in this study was carried out on a Zeiss AxioImager-M2.
Isolation of spermatogonial stem cells and RNA extraction
Testes cell suspensions were generated from 8-day-old Brg1WT (n=5), Brg1Het (n=1) and Brg1cKO (n=2) mice as described previously (Kubota and Brinster, 2008). Conditional deletions were generated using the Mvh-Cre transgene. Spermatogonial stem cells (SSCs) were enriched using THY1+ microbeads (Miltenyi Biotec; 131-049-101) followed by their isolation on magnetic activated cell sorting (MACS) columns (Miltenyi Biotec; 131-090-312). RNA from SSCs were isolated and purified using the PicoPure RNA isolation kit (Life Technologies; KIT0204).
RT-PCR and qPCR
cDNA was synthesized using random primer mix (NEB) and ProtoScript II reverse transcriptase (NEB). Real-time qPCR was performed using Sso Fast EvaGreen supermix (Bio-Rad) on a CFX96 thermocycler (Bio-Rad). A list of qRT-PCR primers used in this study is provided (Table S5).
Isolation of spermatogenic cells
Spermatocyte enriched populations were isolated by methods described previously (Chang et al., 2011; Mu et al., 2014) using percoll and cell strainers. Cell populations were then used for downstream applications such as RNA-seq, ATAC-seq, ChIP-seq and nuclear lysate preparations for immunoprecipitations.
RNA-seq
RNA was extracted from spermatogenic cells obtained from four biological replicates of P12 Brg1WT and Brg1cKO mice each. Conditional deletions were generated using the Mvh-Cre transgene. Cells were treated with the TRIzol reagent (Invitrogen) and total RNA was isolated and cleaned up using the Direct-zol RNA kit (Zymo). Sequencing libraries were prepared using a Kapa mRNA library kit as per the manufacturer's instruction and then sequenced on an Illumina Hiseq 4000 (50 bp reads, single end). Scml2 RNA-seq data have previously been published and are available in GEO under accession number GSE55060 (Hasegawa et al., 2015).
RNA-seq data analysis
Gene expression was quantified using kallisto (Bray et al., 2016). Transcript levels (counts) were summarized per gene using tximport (Soneson et al., 2016) and then imported to perform a differential analysis of gene expression using edgeR (Robinson et al., 2010). The mouse (mm9) gene/transcript annotations were retrieved using the ensembldb R package (https://github.com/jotsetung/ensembldb). Low abundance genes (counts per million <1 across 4 replicates) were filtered out in edgeR and significant differences in counts were called at a false discovery rate (FDR)≤0.05. The lists of differentially expressed genes are provided (Table S1). Anatomy ontology terms were curated from EMAPA (The Edinburgh Mouse Atlas Project) (Hayamizu et al., 2013) and the analysis was done on MouseMine (www.mousemine.org) (Motenko et al., 2015). Gene ontology analysis were performed using clusterProfiler (Yu et al., 2012).
ChIP-seq
BRG1 ChIP was performed exactly as described previously (Raab et al., 2015). We performed the ChIP in duplicates on 4×107 wild-type (CD-1) spermatogenic cells obtained from P12 and P18 mice each. H2AK119ub1 and H3K27ac ChIPs were also performed in duplicate on spermatogenic cells obtained from P12 Brg1WT and Brg1cKO mice using a method described previously for low chromatin inputs (Brind'Amour et al., 2015), with minor modifications (see supplemental materials for details). The BRG1 ChIP samples were sequenced on a Hiseq 2500 using v4 chemistry (50 bp reads, single end), whereas the H3K27ac and H2AK119ub1 ChIP samples were sequenced on an Illumina Hiseq 4000 (50 bp reads, single end). The antibodies used for ChIP are listed in Table S4. H3K4me3, H3K27me3 ChIP-seq data have previously been published and are available under GEO accession numbers GSE61902 (P12 and P18 testes ChIP-seq) (Mu et al., 2014) and GSE89502 (Scml2WT and Scml2KO pachytene spermatocyte ChIP-seq) (Maezawa et al., 2018). ChIP data analysis methods can be found in the supplementary Materials and methods.
ATAC-seq
ATAC-seq was performed on spermatogenic cells isolated from two biological replicates of P12 Brg1WT and Brg1cKO mice each. Only a single sample from 18-day-old Brg1WT and Brg1Het mice was processed for ATAC-seq. Conditional deletions were generated using the Mvh-Cre transgene. ATAC-seq libraries were made as previously reported (Buenrostro et al., 2013) with the exception of a double-sided SPRI bead size selection step of each library, using 0.5× and 1× ratio of SPRI beads to obtain a library size range of ∼150 bp to ∼2 kb. All libraries were combined and sequenced on a single lane of Illumina Hiseq 2500 using v4 chemistry (50 bp reads, single). For data analysis methods, see supplementary Materials and methods.
Differential analysis of chromatin accessibility
For the differential analysis of chromatin accessibility, we adopted a method described for analyzing significant differences in counts between DNA hypersensitive sites identified by DNase seq (Shibata et al., 2012). We compared differences in chromatin accessibility between 12-day-old (P12) Brg1WT and Brg1cKO spermatogenic cells. Here, the pairwise comparison was performed using edgeR, after obtaining the read counts from each replicate across defined 300 bp windows generated from a union set of the top 100,000 peaks (ranked by F-Seq). We called windows with significantly different counts from the pairwise comparison at a FDR≤0.05. The significantly altered regions were annotated using HOMER, peakannotate.pl (Heinz et al., 2010). Regions with significant differences in open chromatin are provided in Table S2.
CUT&RUN (cleavage under targets and release using nuclease)
SCML2 localization in P12 Brg1WT and Brg1cKO spermatogenic cells (700,000 cells/sample) were determined by performing a modified version of CUT&RUN as described previously (Hainer et al., 2019). The samples were sequenced using an Illumina Hiseq 4000 (50 bp reads, single end). Details of the antibodies used are in Table S4. Data analysis details can be found in the supplementary Materials and methods.
Preparation of nuclear lysates
Nuclear extracts were prepared from spermatocyte enriched preparations as previously described (Chandler et al., 2013; Li et al., 1991) with minor modifications (see supplementary Materials and methods). These nuclear lysates were used for co-immunoprecipitations (co-IP) and for the mass spectrometric analysis of BRG1 immunopulldowns (see supplementary Materials and methods).
Identification of BRG1-interacting proteins by mass spectrometry
Proteins isolated from BRG1 IP and IP with a non-specific rabbit IgG were run ∼2 cm below the bottom of the well of a precast SDS polyacrylamide gel (short gel). The short gel was stained with GelCode blue protein stain (ThermoFisher) and the lanes containing each sample were cut out and subjected to in-gel digestion using trypsin, following which the resulting peptides were analyzed by mass spectrometry (Liquid Chromatography-MS/MS). Candidate peptides were identified using Mascot (version 2.5; Matrix Science). Additional details are provided in the supplementary Materials and methods.
Preparation of sub cellular protein fractions
Cytosolic, nucleoplasmic (soluble) and chromatin (insoluble) fractions were prepared as described previously (Méndez and Stillman, 2000) from spermatogenic cells obtained from P12 and P21 Brg1WT, Brg1Het and Brg1cKO mice. Conditional deletions were generated using the Mvh-Cre transgene.
Preparation of acid extracted histones
Histones were extracted from spermatogenic cells obtained from P12 Brg1WT, Brg1Het and Brg1cKO mice using acid extraction protocol described previously (Shechter et al., 2007). Conditional deletions were generated using the Mvh-Cre transgene.
Western blotting
Protein samples were separated by polyacrylamide gel electrophoresis and then transferred to PVDF (polyvinylidene difluoride) membranes (Bio-Rad) using wet/semi-dry transfer apparatus (Bio-Rad). Western blots were generated using the Li-COR Bioscience Odyssey fluorescent western blotting reagents. All the antibodies used in this study and their corresponding dilutions are listed in Table S4.
Supplementary Material
Acknowledgements
We thank Magnuson lab members for helpful comments on manuscript preparation. Next-generation sequencing was performed at the University of North Carolina High-Throughput sequencing facility and Duke Center for Genomic and Computational Biology. Mass spectrometry analysis was performed at the University of Massachusetts Medical School Mass Spectrometry Core Facility.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: D.U.M., Y.S., T.M.; Methodology: D.U.M.; Validation: D.U.M.; Formal analysis: D.U.M., T.M.; Investigation: D.U.M., Y.S., W.M.; Data curation: D.U.M.; Writing - original draft: D.U.M., T.M.; Writing - review & editing: D.U.M., Y.S., W.M., T.M.; Supervision: T.M.; Project administration: T.M.; Funding acquisition: T.M.
Funding
This work was supported by National Institutes of Health grants R01GM101974 and U42OD010924. Deposited in PMC for release after 12 months.
Data availability
Raw and processed RNA-seq, ChIP-seq, ATAC-seq and CUT&RUN data have been deposited in GEO under accession number GSE119179.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.174094.supplemental
References
- Adams S. R., Maezawa S., Alavattam K. G., Abe H., Sakashita A., Shroder M., Broering T. J., Sroga Rios J., Thomas M. A., Lin X. et al. (2018). RNF8 and SCML2 cooperate to regulate ubiquitination and H3K27 acetylation for escape gene activation on the sex chromosomes. PLoS Genet. 14 e1007233 10.1371/journal.pgen.1007233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J. M., Hota S. K., He D., Thomas S., Ho L., Pennacchio L. A. and Bruneau B. G. (2015). Brg1 modulates enhancer activation in mesoderm lineage commitment. Development 142, 1418-1430. 10.1242/dev.109496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alver B. H., Kim K. H., Lu P., Wang X., Manchester H. E., Wang W., Haswell J. R., Park P. J. and Roberts C. W. M. (2017). The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat. Commun. 8, 14648 10.1038/ncomms14648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attanasio C., Nord A. S., Zhu Y., Blow M. J., Biddie S. C., Mendenhall E. M., Dixon J., Wright C., Hosseini R., Akiyama J. A. et al. (2014). Tissue-specific SMARCA4 binding at active and repressed regulatory elements during embryogenesis. Genome Res. 24, 920-929. 10.1101/gr.168930.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. L., Kajita S., Walker M., Saxl R. L., Raghupathy N., Choi K., Petkov P. M. and Paigen K. (2015). PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. 11, PLoS Genet. 11, e1004916 10.1371/journal.pgen.1004916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball R. L., Fujiwara Y., Sun F., Hu J., Hibbs M. A., Handel M. A. and Carter G. W. (2016). Regulatory complexity revealed by integrated cytological and RNA-seq analyses of meiotic substages in mouse spermatocytes. BMC Genomics 17, 1-17. 10.1186/s12864-015-2294-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basciani S., Mariani S., Arizzi M., Ulisse S., Rucci N., Jannini E. A., Rocca C. D., Manicone A., Carani C., Spera G. et al. (2002). Expression of platelet-derived growth factor-A (PDGF-A), PDGF-B, and PDGF receptor-α and -β during human testicular development and disease. J. Clin. Endocrinol. Metab. 87, 2310-2319. 10.1210/jcem.87.5.8476 [DOI] [PubMed] [Google Scholar]
- Bauer D. E., Canver M. C. and Orkin S. H. (2015). Generation of genomic deletions in mammalian cell lines via CRISPR/Cas9. J. Vis. Exp. 95, e52118.2 10.3791/52118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellve A. R., Cavicchia J. C., Millette C. F., O'Brien D. A, Bhatnagar Y. M. and Dym M. (1977). Spermatogenic cells of the prepubertal mouse: isolation and morphological characterization. J. Cell Biol. 74, 68-85. 10.1083/jcb.74.1.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315-326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Biswas U., Stevense M. and Jessberger R. (2018). SMC1α substitutes for many meiotic functions of SMC1β but cannot protect telomeres from damage. Curr. Biol. 28, 249-261. 10.1016/j.cub.2017.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake J. A., Richardson J. E., Bult C. J., Kadin J. A., Eppig J. T., Baldarelli R. M., Beal J. S., Bradt D. W., Burkart D. L., Butler N. E. et al. (2003). MGD: the mouse genome database. Nucleic Acids Res. 31, 193-195. 10.1093/nar/gkg047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N. L., Pimentel H., Melsted P. and Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525-527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Brick K., Smagulova F., Khil P., Camerini-Otero R. D. and Petukhova G. V. (2012). Genetic recombination is directed away from functional genomic elements in mice. Nature 485, 642-645. 10.1038/nature11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brind'Amour J., Liu S., Hudson M., Chen C., Karimi M. M. and Lorincz M. C. (2015). An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat. Commun. 6, 6033 10.1038/ncomms7033 [DOI] [PubMed] [Google Scholar]
- Buaas F. W., Kirsh A. L., Sharma M., McLean D. J., Morris J. L., Griswold M. D., De Rooij D. G. and Braun R. E. (2004). Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 36, 647-652. 10.1038/ng1366 [DOI] [PubMed] [Google Scholar]
- Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y. and Greenleaf W. J. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213-1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler R. L., Brennan J., Schisler J. C., Serber D., Patterson C. and Magnuson T. (2013). ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol. Cell. Biol. 33, 265-280. 10.1128/MCB.01008-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.-F., Lee-Chang J. S., Panneerdoss S., MacLean J. A. and Rao M. K. (2011). Isolation of Sertoli, Leydig, and spermatogenic cells from the mouse testis. BioTechniques 51, 341-344. 10.2144/000113764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C. R., Iwasa J., Cairns B. R. and Peterson C. L. (2017). Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407-422. 10.1038/nrm.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann C. T., Alvarado A. L., Molyneux L. A., Denard B. S., Garbers D. L. and Porteus M. H. (2008). Spermatogonial stem cell self-renewal requires OCT4, a factor downregulated during retinoic acid-induced differentiation. Stem Cells 26, 2928-2937. 10.1634/stemcells.2008-0134 [DOI] [PubMed] [Google Scholar]
- De Vries F. A. T., De Boer E., Van Den Bosch M., Baarends W. M., Ooms M., Yuan L., Liu J. G., Van Zeeland A. A., Heyting C. and Pastink A. (2005). Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19, 1376-1389. 10.1101/gad.329705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diagouraga B., Clément J. A. J., Duret L., Kadlec J., de Massy B. and Baudat F. (2018). PRDM9 methyltransferase activity is essential for meiotic DNA double-strand break formation at its binding sites. Mol. Cell 69, 853-865.e6. 10.1016/j.molcel.2018.01.033 [DOI] [PubMed] [Google Scholar]
- Eaker S., Pyle A., Cobb J. and Handel M. A. (2001). Evidence for meiotic spindle checkpoint from analysis of spermatocytes from Robertsonian-chromosome heterozygous mice. J. Cell Sci. 114, 2953-2965. [DOI] [PubMed] [Google Scholar]
- Euskirchen G. M., Auerbach R. K., Davidov E., Gianoulis T. A., Zhong G., Rozowsky J., Bhardwaj N., Gerstein M. B. and Snyder M. (2011). Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 7 10.1371/journal.pgen.1002008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierz B., Chatterjee C., McGinty R. K., Bar-Dagan M., Raleigh D. P. and Muir T. W. (2011). Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 7, 113-119. 10.1038/nchembio.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo T., Shirley L., John G. B. and Castrillon D. H. (2007). Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45, 413-417. 10.1002/dvg.20310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinari P., Di Marco S., Jones P., Pallaoro M. and Steinkühler C. (2007). HDACs, histone deacetylation and gene transcription: From molecular biology to cancer therapeutics. Cell Res. 17, 195-211. 10.1038/sj.cr.7310149 [DOI] [PubMed] [Google Scholar]
- Goertz M. J., Wu Z., Gallardo T. D., Hamra F. K. and Castrillon D. H. (2011). Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. J. Clin. Invest. 121, 3456-3466. 10.1172/JCI57984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz P., Chandley A. C. and Speed R. M. (1984). Morphological and temporal sequence of meiotic prophase development at puberty in the male mouse. J. Cell Sci. 65, 249-263. [DOI] [PubMed] [Google Scholar]
- Green C. D., Ma Q., Manske G. L., Shami A. N., Zheng X., Marini S., Moritz L., Sultan C., Gurczynski S. J., Moore B. B. et al. (2018). A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-seq. Dev. Cell 46, 651-667.e10. 10.1016/j.devcel.2018.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer S. J., Boskovic A., McCannell K. N., Rando O. J. and Fazzio T. G. (2019). Profiling of pluripotency factors in single cells and early embryos. Cell (in press). 10.1016/j.cell.2019.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud S. S., Low D. H. P., Yi C., Carrell D. T., Guccione E. and Cairns B. R. (2014). Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 15, 239-253. 10.1016/j.stem.2014.04.006 [DOI] [PubMed] [Google Scholar]
- Hasegawa K., Sin H.-S., Maezawa S., Broering T. J., Kartashov A. V., Alavattam K. G., Ichijima Y., Zhang F., Bacon W. C., Greis K. D. et al. (2015). SCML2 establishes the male germline epigenome through regulation of histone H2A ubiquitination article SCML2 establishes the male germline epigenome through regulation of histone H2A ubiquitination. Dev. Cell 32, 574-588. 10.1016/j.devcel.2015.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayamizu T. F., Wicks M. N., Davidson D. R., Burger A., Ringwald M. and Baldock R. A. (2013). EMAP/EMAPA ontology of mouse developmental anatomy: 2013 update. J. Biomed. Semantics 4, 15 10.1186/2041-1480-4-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Yoshida K. and Matsui Y. (2005). A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438, 374-378. 10.1038/nature04112 [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H. and Glass C. K. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576-589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L. and Crabtree G. R. (2010). Chromatin remodelling during development. Nature 463, 474-484. 10.1038/nature08911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Ronan J. L., Wu J., Staahl B. T., Chen L., Kuo A., Lessard J., Nesvizhskii A. I., Ranish J. and Crabtree G. R. (2009). An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. USA 106, 5181-5186. 10.1073/pnas.0812889106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huether R., Dong L., Chen X., Wu G., Parker M., Wei L., Ma J., Edmonson M. N., Hedlund E. K., Rusch M. C., et al. (2014). The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat. Commun. 5, 1-7. 10.1038/ncomms4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichijima Y., Ichijima M., Lou Z., Nussenzweig A., Daniel Camerini-Otero R., Chen J., Andreassen P. R. and Namekawa S. H. (2011). MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev. 25, 959-971. 10.1101/gad.2030811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C., Williams R. T., Calarco J. P., Miller E. L., Weber C. M., Braun S. M. G., Pulice J. L., Chory E. J. and Crabtree G. R. (2017). Dynamics of BAF–Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat. Genet. 49, 213-222. 10.1038/ng.3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarougkas A., Ismail A., Chambers A., Riballo E., Herbert A., Künzel J., Löbrich M., Jeggo P. and Downs J. (2013). Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol. Cell 55, 723-732. 10.1016/j.molcel.2014.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Fedoriw A. M. and Magnuson T. (2012). An essential role for a mammalian SWI/SNF chromatin-remodeling complex during male meiosis. Development 139, 1133-1140. 10.1242/dev.073478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H. and Brinster R. L. (2008). Culture of rodent spermatogonial stem cells, male germline stem cells of the postnatal animal. Methods Cell Biol. 86, 59-84. 10.1016/S0091-679X(08)00004-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S.-J., Park J.-H., Park E.-J., Lee S.-A., Lee H.-S., Kang S. W. and Kwon J. (2015). ATM-mediated phosphorylation of the chromatin remodeling enzyme BRG1 modulates DNA double-strand break repair. Oncogene 34, 303-313. 10.1038/onc.2013.556 [DOI] [PubMed] [Google Scholar]
- Lecona E., Narendra V. and Reinberg D. (2015). USP7 cooperates with SCML2 to regulate the activity of PRC1. Mol. Cell. Biol. 35, 1157-1168. 10.1128/MCB.01197-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-S., Park J.-H., Kim S.-J., Kwon S.-J. and Kwon J. (2010). A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 29, 1434-1445. 10.1038/emboj.2010.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Ogushi S., Saitou M. and Hirano T. (2011). Condensins I and II are essential for construction of bivalent chromosomes in mouse oocytes. Mol. Biol. Cell 22, 3465-3477. 10.1091/mbc.e11-05-0423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch B. J., Dokshin G. A., Young R. A., McCarrey J. R. and Page D. C. (2013). A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proc. Natl. Acad. Sci. USA 110, 16061-16066. 10.1073/pnas.1315204110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch B. J., Silber S. J., McCarrey J. R. and Page D. C. (2016). Parallel evolution of male germline epigenetic poising and somatic development in animals. Nat. Genet. 48, 1-10. 10.1038/ng.3483 [DOI] [PubMed] [Google Scholar]
- Lessard J., Wu J. I., Ranish J. A., Wan M., Winslow M. M., Staahl B. T., Wu H., Aebersold R., Graef I. A. and Crabtree G. R. (2007). An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55, 201-215. 10.1016/j.neuron.2007.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. C., Ross J., Scheppler J. A. and Franza B. R. Jr. (1991). An in vitro transcription analysis of early responses of the human immunodeficiency virus type 1 long terminal repeat to different transcriptional activators. Mol. Cell. Biol. 11, 1883-1893. 10.1128/MCB.11.4.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wu J., Kim S.-Y., Zhao M., Hearn S. A., Zhang M. Q., Meistrich M. L. and Mills A. A. (2014). Chd5 orchestrates chromatin remodelling during sperm development. Nat. Commun. 5, 3812 10.1038/ncomms4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Zhou J., Leu N. A., Abreu C. M., Wang J., Anguera M. C., de Rooij D. G., Jasin M. and Wang P. J. (2015). Polycomb protein SCML2 associates with USP7 and counteracts histone H2A ubiquitination in the XY chromatin during male meiosis. PLoS Genet. 11, e1004954 10.1371/journal.pgen.1004954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P., Pan H., Montgomery R. L., Olson E. N. and Schultz R. M. (2012). Compensatory functions of histone deacetylase 1 (HDAC1) and HDAC2 regulate transcription and apoptosis during mouse oocyte development. Proc. Natl. Acad. Sci. 109, E481-E489. 10.1073/pnas.1118403109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens G. N., El Messaoudi-Aubert S., Elderkin S., Hiom K. and Peters G. (2010). Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. EMBO J. 29, 2553-2565. 10.1038/emboj.2010.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maezawa S., Hasegawa K., Yukawa M., Kubo N. and Sakashita A., Alavattam K. G., Sin H. S., Kartashov A. V., Sasaki H., Barski A. et al. (2018). Polycomb protein SCML2 facilitates H3K27me3 to establish bivalent domains in the male germline. Proc. Natl. Acad. Sci. USA 115, 4957-4962. 10.1073/pnas.1804512115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin G., Khil P. P., Kim J., Bellani M. A. and Camerini-Otero R. D. (2014). Integrated transcriptome analysis of mouse spermatogenesis. BMC Genomics 15, 39 10.1186/1471-2164-15-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah-Planchon J., Bièche I., Guinebretière J.-M., Bourdeaut F. and Delattre O. (2015). SWI/SNF chromatin remodeling and human malignancies. Annu. Rev. Pathol. Mech. Dis. 10, 145-171. 10.1146/annurev-pathol-012414-040445 [DOI] [PubMed] [Google Scholar]
- Méndez J. and Stillman B. (2000). Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602-8612. 10.1128/MCB.20.22.8602-8612.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minsky N., Shema E., Field Y., Schuster M., Segal E. and Oren M. (2008). Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 10, 483-488. 10.1038/ncb1712 [DOI] [PubMed] [Google Scholar]
- Montgomery R. L., Hsieh J., Barbosa A. C., Richardson J. A. and Olson E. N. (2009). Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc. Natl. Acad. Sci. 106, 7876-7881. 10.1073/pnas.0902750106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motenko H., Neuhauser S. B., O'Keefe M. and Richardson J. E. (2015). MouseMine: a new data warehouse for MGI. Mamm. Genome 26, 325-330. 10.1007/s00335-015-9573-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu W., Starmer J., Fedoriw A. M., Yee D. and Magnuson T. (2014). Repression of the soma-specific transcriptome by Polycomb-repressive complex 2 promotes male germ cell development. Genes Dev. 28, 2056-2069. 10.1101/gad.246124.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicassio F., Corrado N., Vissers J. H. A., Areces L. B., Bergink S., Marteijn J. A., Geverts B., Houtsmuller A. B., Vermeulen W., Di Fiore P. P. et al. (2007). Human USP3 is a chromatin modifier required for s phase progression and genome stability. Curr. Biol. 17, 1972-1977. 10.1016/j.cub.2007.10.034 [DOI] [PubMed] [Google Scholar]
- Oatley M. J., Kaucher A. V., Racicot K. E. and Oatley J. M. (2011). Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol. Reprod. 85, 347-356. 10.1095/biolreprod.111.091330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara H., Ui A., Otsuka A., Satoh H., Yokomi I., Nakajima S., Yasui A., Yokota J. and Kohno T. (2011). Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 30, 2135-2146. 10.1038/onc.2010.592 [DOI] [PubMed] [Google Scholar]
- Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A. and Reinberg D. (2006). Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125, 703-717. 10.1016/j.cell.2006.04.029 [DOI] [PubMed] [Google Scholar]
- Peters A. H. F. M., Plug A. W., Van Vugt M. J. and De Boer P. (1997). A drying-down technique for the spreading of mammalian melocytes from the male and female germline. Chromosom. Res. 5, 66-68. 10.1023/A:1018445520117 [DOI] [PubMed] [Google Scholar]
- Qi W., Wang R., Chen H., Wang X., Xiao T., Boldogh I., Ba X., Han L. and Zeng X. (2014). BRG1 promotes the repair of DNA double-strand breaks by facilitating the replacement of RPA with RAD51. J. Cell Sci. 128, 317-330. 10.1242/jcs.159103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab J. R., Resnick S. and Magnuson T. (2015). Genome-wide transcriptional regulation mediated by biochemically distinct SWI/SNF complexes. PLoS Genet. 11, 1-26, e1005748 10.1371/journal.pgen.1005748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab J. R., Runge J. S., Spear C. C. and Magnuson T. (2017). Co-regulation of transcription by BRG1 and BRM, two mutually exclusive SWI/SNF ATPase subunits. Epigenetics Chromatin 10, 1-15. 10.1186/s13072-016-0108-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. and Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139-140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo H., Prosser H., Ruzankina Y., Mahadevaiah S. K., Cloutier J. M., Baumann M., Fukuda T., Höög C., Tóth A., de Rooij D. G. et al. (2013). ATR acts stage specifically to regulate multiple aspects of mammalian meiotic silencing. Genes Dev. 27, 1484-1494. 10.1101/gad.219477.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadate-Ngatchou P. I., Payne C. J., Dearth A. T. and Braun R. E. (2008). Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 46, 738-742. 10.1002/dvg.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahl J., Rizzolo K. and Soriano P. (2008). The PDGF signaling pathway controls multiple steroid-producing lineages. Genes Dev. 22, 3255-3267. 10.1101/gad.1723908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serber D. W., Runge J. S., Menon D. U. and Magnuson T. (2015). The mouse INO80 chromatin remodeling complex is an essential meiotic factor for spermatogenesis. Biol. Reprod. 94, 8 10.1095/biolreprod.115.135533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D., Dormann H. L., Allis C. D. and Hake S. B. (2007). Extraction, purification and analysis of histones. Nat. Protoc. 2, 1445-1457. 10.1038/nprot.2007.202 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Sheffield N. C., Fedrigo O., Babbitt C. C., Wortham M., Tewari A. K., London D., Song L., Lee B. K., Iyer V. R. et al. (2012). Extensive evolutionary changes in regulatory element activity during human origins are associated with altered gene expression and positive selection. PLoS Genet. 8, e1002789 10.1371/journal.pgen.1002789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene P. J., Henikoff J. G. and Henikoff S. (2018). Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006-1019. 10.1038/nprot.2018.015 [DOI] [PubMed] [Google Scholar]
- Soneson C., Love M. I. and Robinson M. D. (2016). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 4, 1521 10.12688/f1000research.7563.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton B. Z., Hodges C., Calarco J. P., Braun S. M. G., Ku W. L., Kadoch C., Zhao K. and Crabtree G. R. (2017). Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat. Genet. 49, 282-288. 10.1038/ng.3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi-Ichinose C., Ichinose H., Metzger D. and Chambon P. (1997). SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol. Cell. Biol. 17, 5976-5986. 10.1128/MCB.17.10.5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Nozaki M., Takeda N. and Shinkai Y. (2007). Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 26, 3346-3359. 10.1038/sj.emboj.7601767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y., Naruse C., Costa Y., Shirakawa T., Tachibana M., Sharif J., Kezuka-Shiotani F., Kakiuchi D., Masumoto H., Shinkai Y. et al. (2011). HP1 links histone methylation marks to meiotic synapsis in mice. Development 138, 4207-4217. 10.1242/dev.064444 [DOI] [PubMed] [Google Scholar]
- Tolstorukov M. Y., Sansam C. G., Lu P., Koellhoffer E. C., Helming K. C., Alver B. H., Tillman E. J., Evans J. A., Wilson B. G., Park P. J. et al. (2013). Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc. Natl. Acad. Sci. 110, 10165-10170. 10.1073/pnas.1302209110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J. M. A. (2007). Meiotic sex chromosome inactivation. Development 134, 1823-1831. 10.1242/dev.000018 [DOI] [PubMed] [Google Scholar]
- Turner J. M. A., Aprelikova O., Xu X., Wang R., Kim S., Chandramouli G. V. R., Barrett J. C., Burgoyne P. S. and Deng C. X. (2004). BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr. Biol. 14, 2135-2142. 10.1016/j.cub.2004.11.032 [DOI] [PubMed] [Google Scholar]
- Van Der Knaap J. A., Kumar B. R. P., Moshkin Y. M., Langenberg K., Krijgsveld J., Heck A. J. R., Karch F. and Verrijzer C. P. (2005). GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol. Cell 17, 695-707. 10.1016/j.molcel.2005.02.013 [DOI] [PubMed] [Google Scholar]
- Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S. and Zhang Y. (2004). Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873-878. 10.1038/nature02985 [DOI] [PubMed] [Google Scholar]
- Wang J., Gu H., Lin H. and Chi T. (2012). Essential roles of the chromatin remodeling factor Brg1 in spermatogenesis in mice. Biol. Reprod. 86, 186-186. 10.1095/biolreprod.111.097097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R., Ui A., Kanno S. I., Ogiwara H., Nagase T., Kohno T. and Yasui A. (2014). SWI/SNF factors required for cellular resistance to dna damage include arid1a and arid1b and show interdependent protein stability. Cancer Res. 74, 2465-2475. 10.1158/0008-5472.CAN-13-3608 [DOI] [PubMed] [Google Scholar]
- Wilson B. G., Wang X., Shen X., McKenna E. S., Lemieux M. E., Cho Y. J., Koellhoffer E. C., Pomeroy S. L., Orkin S. H. and Roberts C. W. M. (2010). Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18, 316-328. 10.1016/j.ccr.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L., Daniel K., Roig I., Bolcun-Filas E., Xu H., Boonsanay V., Eckmann C. R., Cooke H. J., Jasin M., Keeney S. et al. (2009). Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 5, e1000702 10.1371/journal.pgen.1000702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Oatley J. M., Oatley M. J., Kaucher A. V., Avarbock M. R. and Brinster R. L. (2010). The POU domain transcription factor POU3F1 is an important intrinsic regulator of GDNF-induced survival and self-renewal of mouse spermatogonial stem cells1. Biol. Reprod. 82, 1103-1111. 10.1095/biolreprod.109.083097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Cubizolles F., Zhang Y., Reichert N., Kohler H., Seiser C. and Matthias P. (2010). Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev. 24, 455-469. 10.1101/gad.552310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., De La Fuente R., Leu N. A., Baumann C., McLaughlin K. J. and Wang P. J. (2006). Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J. Cell Biol. 173, 497-507. 10.1083/jcb.200603063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y. and He Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol. 16, 284-287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W. et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Nie R., Prins G. S., Saunders P. T. K., Katzenellenbogen B. S. and Hess R. A. (2002). Localization of androgen and estrogen receptors in adult male mouse reproductive tract. J. Androl. 23, 870-881. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.