Abstract
Gestational diabetes mellitus (GDM) is associated with adverse pregnancy outcomes in pregnant women and it’s prevalence is increasing worldwide. A total 100 cases of GDM and 100 healthy controls were included in a study and blood samples were collected in plain vials from all study participant. Difference among several variables which included BMI, glycemia, insulinaemia, HbA1c, total cholesterol, triglycerides, urinary albumin were compared between GDM cases and controls and were found to be statistically significant (p < 0.0001). Additionally, GDM cases showed 4.0 fold increase in miRNA-19a and 4.7 mean increase in miRNA-19b expression compared to healthy control individuals. A positive correlation was observed between miRNA-19a and miRNA-19b among GDM cases. However the correlation coefficient was 0.13 between miRNA-19a and miRNA-19b. This suggested that with the increase in miRNA-19a, miRNA-19b also increased. The findings of this study concludes that an increase in microRNA-19a and microRNA-19b is observed in GDM cases and could be linked with increased risk factor for worsening of the disease. MicroRNA-19a and 19b have been linked to alcoholism and smoking and could also be the factors in GDM.
Keywords: Cell free, MicroRNA expression, Gestational diabetes mellitus
Introduction
Gestational diabetes mellitus (GDM) is related to altered carbohydrate metabolism or potentially abridged or reduced glucose tolerance before pregnancy and gestational diabetes usually confirmed during pregnancy (Djelmis et al. 2016). GDM is linked with adverse pregnancy outcomes for the pregnant women and it affects roughly 7–14% of obese pregnant females (El-Chaar et al. 2013). GDM prevalence rate is mounting day by day all over the world, approximately 14% of pregnancies exaggerated by GDM (International Diabetes Federation 2017). It has been showed to be associated with prenatal difficulties and increases risk of future metabolic disorder in both mothers and their children. The oral glucose tolerance test (OGTT) was used to confirm the GDM (Metzger et al. 2010). Later on both mothers and their children have more chance to develop metabolic disorders such as obesity, type 2 diabetes mellitus (T2D) and cardiovascular related disorder (Mitanchez et al. 2015). Several factors are implicated in the pathogenesis of GDM (Yan and Yang 2014). Approximately 18–20 nucleotide long MicroRNAs are noncoding nucleotide sequence of RNA linked to control post-transcriptional gene regulation through interfering with translation machinery and lead to mRNA cleavage or inhibition of translational (Sayed and Abdellatif 2011). Upregulation of several microRNAs was observed in 16–19 weeks of pregnant women’s sample (Zhu et al. 2015a, b). But it’s still not clear that GDM could be diagnosed by microRNAs and could be used as GDM biomarkers. Evidences are emerging that unusual expression of miRNAs is associated with complications in women’s pregnancy, such as preeclampsia and GDM (Zhao et al. 2013). Increasing evidence revealed miRNAs in the cause and development of GDM (Poirier et al. 2017) and proposed that miRNA regulation in mother may be used as biomarkers to diagnose the GDM. Several miRNAs expressed in women’s placentas T2DM, confirming that miRNAs expressed at stage of GDM play an essential candidate in metabolic disorder (Collares et al. 2013). A research by Zhu et al. (2015a, b) revealed that sample drawn at 16th–19th gestational weeks showed upregulation of miR-16, microRNA-17-5p, microRNA-19a, microRNA-19b, and microRNA-20a in plasma samples of GDM pregnant women compared to healthy individuals (Zhu et al. 2015a, b). A research by Cao et al. (2017) said that expression differences of miR-19a and miR-19b. More recently, Pheiffer et al. (2018) reported decreased miR-19a and miR-19b expression in South African women with GDM. Therefore, present study aimed to study the association of microRNA 19a and 19b expression among Gestational Diabetes patients.
Materials and methods
Blood sample collection and serum separation
Present study recruited 100 confirmed cases of GDM after screening by performing oral glucose tolerance test (75 g) at the stage of second trimester (24–28 weeks of gestation) and equal number of female healthy controls. 2.5 ml of patient’s peripheral blood samples were drawn in plain vials from GDM cases and 2 ml of venous blood from healthy individuals in red vials. Blood samples drawn in plain vials were centrifuged at 1500 rpm to separate the serum were collected and stored at − 70 °C. This study was ethically approved and conducted at Department of Obstetrics, The Second Hospital of Shandong University, Jinan, Shandong, China. Informed consent was obtained from study participants.
Total RNA extraction
Total cell free RNA extraction from serum samples was done using Trizol method from GDM patients as well as from healthy controls individuals and stored at − 70 °C in RNase-free eppendorf tubes. The quality and purity of RNA were determined by the A260/280 ratio by spectrophotometer method.
Polyadenylation and cDNA synthesis
Total 10 ng from total extracted RNA was used for Polyadenylation and cDNA synthesis using advanced microRNA cDNA Synthesis Kit (A28007, TaqMan, Thermo Scientific) by following manufacturer protocol. Reverse Transcriptase enzyme and other essential reagents were added subsequently for cDNA synthesis to switch in poly (A)—tailed miRNAs into cDNA using universal RT primer supplied with the manufacturer kit.
Quantitative real time PCR for miRNA-19a and miRNA-19b expression
Relative quantification was performed by using Quantitative real-time PCR (qPCR) to compute the cell free based microRNA-19a and microRNA-19b expression level in GDM patients compared to healthy controls. QRT-PCR was performed in Quant Studio 6 using advanced Taqman master mix (4444556), Taqman probes for miRNA-19a (479228_mir) and 19b (477962_mir) were used for quantification and U6 was used as internal control.
Statistical analysis
All the data analysis was done by Graph Pad Prism 5.03 version of software. Relative quantification method 2−(∆∆ct) was applied to compute the fold change in expression of miRNA-19a and miRNA-19b in GDM patients. All the data presented in mean and standard deviation. Mann–Whitney U test and student ‘T’ test were used to compare the groups and p value < 0.05 was considered to be statistically significant.
Results
Demographic characteristic
All the demographic characteristic of GDM cases and healthy controls were depicted in Table 1. In brief, All 100 the GDM cases were females and 100 healthy female subjects were included and more details were depicted in Table 1.
Table 1.
Clinical and demographic characteristic of GDM cases and healthy controls
| Variables | GDM cases | Healthy controls |
|---|---|---|
| Age | ||
| < 35 years | 45 (45%) | 40 (40%) |
| > 35 years | 55 (55%) | 60 (60%) |
| Alcoholism | ||
| Yes | 48 (48%) | 50 (50%) |
| No | 52 (52%) | 50 (50%) |
| Smoking | ||
| Yes | 43 (43%) | 45 (45%) |
| No | 57 (57%) | 55 (55%) |
| Hypertension | ||
| Yes | 40 (40%) | 28 (28%) |
| No | 60 (60%) | 72 (72%) |
Association of biochemical parameters among GDM cases and healthy controls
Several biochemical parameters were included in present study and it was analysed to observe differences in biochemical parameters among GDM cases and controls (Table 2). BMI (body mass index) of GDM cases was observed 28.41 while healthy controls BMI was 23.56 and differences between them was found to be statistically significant (p < 0.0001). Other parameters such as glycemia, insulinaemia, HbA1c, total cholesterol, triglycerides, urinary albumin were analysed and differences among GDM cases and controls were found to be significant (p < 0.0001).
Table 2.
Comparison of biochemical parameters among GDM cases and healthy controls
| Biochemical parameters | GDM cases (Mean ± SD) |
Healthy controls (Mean ± SD) |
p value |
|---|---|---|---|
| Body mass index (BMI) | 28.41 ± 2.18 | 23.56 ± 1.52 | < 0.0001 |
| Glycemia (mg/dl) | 79.48 ± 2.95 | 68.05 ± 9.29 | < 0.0001 |
| Insulinaemia (mUI/ml) | 13.48 ± 2.50 | 7.08 ± 0.97 | < 0.0001 |
| HbA1C | 6.06 ± 0.60 | 4.96 ± 0.76 | < 0.0001 |
| Total cholesterol (mg/dl) | 284.2 ± 34.74 | 205.2 ± 19.05 | < 0.0001 |
| Triglycerides (mg/dl) | 219.2 ± 37.12 | 139.5 ± 24.92 | < 0.0001 |
| Urinary Albumin (mg/l) | 16.28 ± 2.44 | 7.26 ± 0.84 | < 0.0001 |
MicroRNA-19a expression and GDM cases
GDM cases showed increased cell free miRNA-19a expression (4.0 mean fold) compared to healthy control. Patients who were < 35 years of age showed 4.22 fold miRNA-19a expression while > 35 years of age group showed 3.82 fold miRNA-19a expression and differences among them was found to be significant (p = 0.03). It was observed that GDM patients had history of alcoholism habit showed 4.31 fold miRNA-19a expression while those who were non alcoholic had 3.72 fold miRNA-19a expression and differences among them was found to be significant (p = 0.001). However no such association of miRNA-19a expression was observed with smoking and hypertension (Table 3).
Table 3.
Association of miR-19a expression with respect to different variables
| Variables | miR-19a expression (fold change in mean ± SD) |
p value |
|---|---|---|
| Overall expression | 4.0 ± 0.92 | – |
| Age | ||
| < 35 years | 4.22 ± 0.84 | 0.03 |
| > 35 years | 3.82 ± 0.95 | |
| Alcoholism | ||
| Yes | 4.31 ± 0.91 | 0.001 |
| No | 3.72 ± 0.84 | |
| Smoking | ||
| Yes | 4.01 ± 0.98 | 0.91 |
| No | 3.99 ± 0.85 | |
| Hypertension | ||
| Yes | 4.01 ± 0.91 | 0.97 |
| No | 4.00 ± 0.95 | |
MicroRNA-19b expression and GDM cases
GDM cases showed 4.77 mean fold increased miRNA-19b expression compared to healthy control. It has been observed that patients who were < 35 years of age showed 5.12 fold miRNA-19a expression while > 35 years of age group showed 4.48 fold miRNA-19a expression and differences between the groups was found to be significant (p = 0.02). GDM patients had history of smoking habit showed 5.04 fold miRNA-19b expression while those who were not in habit of smoking had 4.41 fold miRNA-19b expression and differences between groups was found to be significant (p = 0.03). However no such effect of alcoholism and hypertension was observed on miRNA-19b expression (Table 4).
Table 4.
Association of miR-19b expression with respect to different variables
| Variables | miR-19b expression (fold change in mean ± SD) |
p value |
|---|---|---|
| Overall expression | 4.77 ± 1.55 | – |
| Age | ||
| < 35 years | 5.12 ± 1.84 | 0.02 |
| > 35 years | 4.48 ± 1.57 | |
| Alcoholism | ||
| Yes | 4.69 ± 1.38 | 0.59 |
| No | 4.84 ± 1.71 | |
| Smoking | ||
| Yes | 5.04 ± 1.47 | 0.03 |
| No | 4.41 ± 1.61 | |
| Hypertension | ||
| Yes | 4.98 ± 1.62 | 0.06 |
| No | 4.46 ± 1.42 | |
Correlation between miRNA 19a and 19b expression in GDM patients
A positive correlation was observed between miRNA-19a and miRNA-19b among GDM cases, However the correlation was not very strong (correlation coefficient = 0.13). correlation coefficient indicated that that with the increase of miRNA-19a, miRNA-19b was also increased (Fig. 1).
Fig. 1.
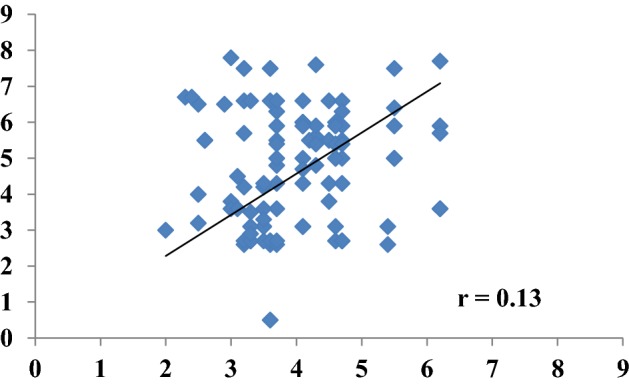
Correlation between miRNA-19a and 19b among GDM patients
Discussion
The pathophysiology of patients with GDM is not yet completely known. Insufficient β-cell adjustment to peripheral resistance to insulin and characterization of 2nd and 3rd gestation trimester is expected to be key cause of GDM, However the molecular signalling behind such crash is still under study (Di Cianni et al. 2003). Negative regulation of gene expression and translation can be controlled by microRNA (Lai 2003) and each miRNA can modulate various genes as well as only gene can be controlled by numerous miRNAs (Lewis et al. 2003). miRNAs virtually involved in each cellular process and involved in cell cycle development, differentiation, and regulation (Wilczynska and Bushell 2015). Several studies confirmed that miRNAs are not exclusively intracellular nucleic acid but also cell free and present in a circulatory system as circulating in biological fluids, including plasma or serum (Fehlmann et al. 2016). Essentially, cell free circulating microRNAs were abnormally expressed in the circulatory system during the pathogenesis or development of diseases (Turchinovich et al. 2012). It has been observed that GDM patients showed increased in biochemical parameters compared to healthy controls. Several parameters such as Body Mass Index, Glycemia, Insulinaemia, HbA1c, Total cholesterol, Triglycerides and Urinary Albumin were high in GDM patients. A study by Guido Sebastiani observed increased biochemical parameters such as Glycemia, Insulinaemia, HbA1c, Total cholesterol, Triglycerides and Urinary Albumin were high in GDM cohort (Sebastiani et al. 2017). Increased miRNA-19a and miRNA-19b was observed among GDM patients in contrast to healthy controls. In the same way a study by Zhu et al. (2015a, b) found that miR-19a, miR-19b was found to be unregulated in 16th–19th gestational week’s plasma samples in pregnant diabetic females with respect to control. Several miRNAs such as miR-17-5p, miR-19a-3p, miR-19b-3p, and miR-20a-5p belong to the miRNA-17-92 cluster. This plays key role in physiological process during pregnancy and increased expression of these miRNAs may lead to an abnormal pregnancy (Zhu et al. 2015a, b). miRNAs 19a-19b expression was found to be associated with age, alcohol and smoking among GDM patients. A positive correlation was observed between miRNA-19a, miRNA-19b that shows both are linked together and with the GDM. A study in streptozotocin induced diabetic mice showed increased miRNA-19b expression reported by Costantino and colleagues (2016). Present study showed microRNA-19a and 19b were positively correlated among GDM patients showed possible involvement in the pathogenesis of disease in female pregnant patients. It has been observed that alcoholism and smoking linked with microRNA-19a and 19b expression in GDM patients. However, increased microRNA-19a and microRNA-19b was found to be associated with GDM.
Conclusion
Present study concluded that increased microRNA-19a and microRNA-19b was observed in GDM patients and could be linked with the pathogenesis and worseness of disease. Increased microRNA-19a was linked with age and alcoholism as well as increased microRNA-19b expression was linked with age and smoking could be the reason for worseness of disease.
Acknowledgements
Authors thanks to all participants who involved in study and given precious time and significant contribution to complete this research work.
Author contributions
FW: sample collection and experimentation. XZ: data Analysis and writing. HZ: research plan, finalizing manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Obtained.
References
- Cao Y-L, Jia Y-J, Xing B-H, Shi D-D, Dong X-J. Plasma microRNA-16-5p, -17-5p and -20a-5p: novel diagnostic biomarkers for gestational diabetes mellitus. J Obstet Gynaecol Res. 2017;43:974–981. doi: 10.1111/jog.13317. [DOI] [PubMed] [Google Scholar]
- Collares CV, Evangelista AF, Xavier DJ, Rassi DM, Arns T, Foss-Freitas MC, et al. Identifying common and specific microRNAs expressed in peripheral blood mononuclear cell of type 1, type 2, and gestational diabetes mellitus patients. BMC Res Notes. 2013;6:491. doi: 10.1186/1756-0500-6-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino S, Paneni F, Luscher TF, Cosentino F. MicroRNA profiling unveils hyperglycaemic memory in the diabetic heart. Eur Heart J. 2016;37:572–576. doi: 10.1093/eurheartj/ehv599. [DOI] [PubMed] [Google Scholar]
- Di Cianni G, Miccoli R, Volpe L, Lencioni C, Del Prato S. Intermediate metabolism in normal pregnancy and in gestational diabetes. Diabetes Metab Res Rev. 2003;19:259–270. doi: 10.1002/dmrr.390. [DOI] [PubMed] [Google Scholar]
- Djelmis J, Pavic M, Mulliqi Kotori V, Pavlic Renar I, Ivanisevic M, Oreskovic S. Prevalence of gestational diabetes mellitus according to IADPSG and NICE criteria. Int J Gynaecol Obstet. 2016;135:250–254. doi: 10.1016/j.ijgo.2016.07.005. [DOI] [PubMed] [Google Scholar]
- El-Chaar D, Finkelstein SA, Tu X, et al. The impact of increasing obesity class on obstetrical outcomes. J Obstet Gynaecol Can. 2013;35:224–233. doi: 10.1016/S1701-2163(15)30994-4. [DOI] [PubMed] [Google Scholar]
- Fehlmann T, Ludwig N, Backes C, Meese E, Keller A. Distribution of microRNA biomarker candidates in solid tissues and body fluids. RNA Biol. 2016;13:1084–1088. doi: 10.1080/15476286.2016.1234658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Diabetes Federation (2017) IDF diabetes atlas—across the globe. http://diabetesatlas.org/across-the-globe.html. Accessed 6 July 2018
- Lai EC. microRNAs: runts of the genome assert themselves. Curr Biol. 2003;13:R925–R936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih I, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Metzger BE, Gabbe SG, Persson B, Buchanan TA, Catalano PA, Damm P, Dyer AR, Leive A, Hod M, et al. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care. 2010;33:676–682. doi: 10.2337/dc10-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitanchez D, Yzydorczyk C, Siddeek B, Boubred F, Benahmed M, Simeoni U. The offspring of the diabetic mother-short- and long-term implications. Best Pract Res Clin Obstet Gynaecol. 2015;29:256–269. doi: 10.1016/j.bpobgyn.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Pheiffer C, Dias S, Rheeder P, Adam S. Decreased expression of circulating miR-20a-5p in south African women with gestational diabetes mellitus. Mol Diagn Ther. 2018;22:345–352. doi: 10.1007/s40291-018-0325-0. [DOI] [PubMed] [Google Scholar]
- Poirier C, Desgagné V, Guérin R, Bouchard L. MicroRNAs in pregnancy and gestational diabetes mellitus: emerging role in maternal metabolic regulation. Curr Diabetes Rep. 2017;17:35. doi: 10.1007/s11892-017-0856-5. [DOI] [PubMed] [Google Scholar]
- Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- Sebastiani G, Guarino E, Grieco GE, Formichi C, Poggi CD, Ceccarelli E, Dotta F. Circulating microrna (mirna) expression profiling in plasma of patients with gestational diabetes mellitus reveals upregulation of mirna mir-330-3p. Front Endocrinol. 2017;8:345. doi: 10.3389/fendo.2017.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A, Weiz L, Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem Sci. 2012;37:460–465. doi: 10.1016/j.tibs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Wilczynska A, Bushell M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015;22:22–33. doi: 10.1038/cdd.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Yang H. Gestational diabetes mellitus, programing and epigenetics. J Matern Fetal Neonatal Med. 2014;27:1266–1269. doi: 10.3109/14767058.2013.853733. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Moley KH, Gronowski AM. Diagnostic potential for miRNAs as biomarkers for pregnancy-specific diseases. Clin Biochem. 2013;46(10–11):953–960. doi: 10.1016/j.clinbiochem.2013.01.026. [DOI] [PubMed] [Google Scholar]
- Zhu Yanan, Tian Fei, Li Hailing, Zhou Youxia, Jiafeng Lu, Ge Qinyu, Zhu Yanan, Tian Fei, Li Hailing, Zhou Youxia, Jiafeng Lu, Ge Qinyu. Profiling maternal plasma microRNA expression in early pregnancy to predict gestational diabetes mellitus. Int J Gynecol Obstet. 2015;130(1):49–53. doi: 10.1016/j.ijgo.2015.01.010. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Tian F, Li H, Zhou Y, Lu J, et al. Profiling maternal plasma microRNA expression in early pregnancy to predict gestational diabetes mellitus. Int J Gynaecol Obstet. 2015;130:49–53. doi: 10.1016/j.ijgo.2015.01.010. [DOI] [PubMed] [Google Scholar]


