Abstract
Background
The NAC (NAM, AFAT, and CUC) transcription factors play critical roles in rice (Oryza sativa) development and stress regulation. Overexpressing a microRNA (miR164b)-resistant OsNAC2 mutant gene, which generates transcripts that cannot be targeted by miR164b, improves rice plant architecture and yield; however, the performance of these mOsNAC2-overexpressing lines, named ZUOErN3 and ZUOErN4, under abiotic stress conditions such as drought have not yet been fully characterized.
Results
In this study, we showed that the germination of ZUOErN3 and ZUOErN4 seeds was delayed in comparison with the wild-type (WT) seeds, although the final germination rates of all lines were over 95%. The quantification of the endogenous ABA levels revealed that the germinating mOsNAC2-overexpressing seeds had elevated ABA levels, which resulted in their slower germination. The mOsNAC2-overexpressing plants were significantly more drought tolerance than the WT plants, with the survival rate increasing from 11.2% in the WT to nearly 70% in ZUOErN3 and ZUOErN4 plants after a drought treatment. Salt (NaCl) tolerance was also increased in the ZUOErN3 and ZUOErN4 plants due to significantly increased ABA levels. A reverse transcription quantitative PCR (RT-qPCR) analysis showed a significant increase in the expression of the ABA biosynthesis genes OsNCED1 and OsNCED3 in the mOsNAC2-overexpressing lines, and the expression levels of the stress-responsive genes OsP5CS1, OsLEA3, and OsRab16 were significantly increased in these plants. Moreover, OsNAC2 directly interacted with the promoters of OsLEA3 and OsNCED3 in yeast one-hybrid assays.
Conclusions
Taken together, our results show that OsNAC2 plays a positive regulatory role in drought and salt tolerance in rice through ABA-mediated pathways.
Keywords: Rice (Oryza sativa), OsNAC2, ABA, Drought tolerance, Salt tolerance, microRNA
Background
Rice (Oryza sativa L.) is a major food crop worldwide, and as the population increases, so does the demand for this crop; however, rice is often exposed to drought, salt, and other stresses, which may severely affect its yield (Valliyodan and Nguyen, 2006). The cultivation of abiotic stress-resistant rice is therefore important for generating sufficient rice yields and ensuring food security.
To maintain growth under environmental stresses, plants often depend on hormones such as abscisic acid (ABA), which plays a critical role in integrating a wide range of stress signals and controlling the downstream stress responses (Zhu, 2016). ABA biosynthesis and the resulting endogenous ABA levels are increased under abiotic stresses, with drought and salt stresses representing the most important environmental signals upregulating the transcription of the ABA biosynthesis genes (Xiong et al., 2003).
The NAC (NAM, AFAT, and CUC) family of transcription factors (TFs) are plant-specific, and 151 of them have been detected in rice (Nuruzzaman et al., 2010). The rice NAC TFs play vital roles in diverse aspects of plant growth, development, and stress responses (Puranik et al., 2012). The involvement of the NAC TFs in rice abiotic stress responses has been extensively explored, and some rice NAC genes have been reported to function in abiotic stress tolerance; for example, the overexpression of OsNAC5 (Takasaki et al., 2010; Song et al., 2011; Jeong et al., 2013), OsNAC6 (Hu et al., 2008), OsNAC9 (Redillas et al., 2012), OsNAC10 (Jeong et al., 2010), or OsNAP (Chen et al., 2014) improves drought and salinity tolerance in transgenic rice.
Several studies have reported that OsNAC2 has important functions in rice (Mao et al., 2007; Fang et al., 2014; Chen et al., 2015; Mao et al., 2017; Jiang et al., 2018). In a promoter activation-tagging mutant, an increase in OsNAC2 expression results in an increased tiller angle in rice (Mao et al., 2007). In addition, OsNAC2 is associated with plant height (Chen et al., 2015), the promotion of leaf senescence (Mao et al., 2017), and increased yields (Jiang et al., 2018), highlighting the important roles this gene plays in rice growth and development. Several NAC transcripts are cleaved by miR164 to modulate developmental processes (Mallory et al., 2004; Guo et al., 2005; Kim et al., 2009); for example, post-transcriptional modifications of the NAC domain TFs by miR164 function in the postembryonic regulation of the shoot meristem (Li and Zhang, 2016).
Some NAC TFs can improve abiotic-stress tolerance (Hu et al., 2006; Saada et al., 2013; Chen et al., 2014; Hong et al., 2016; Huang et al., 2016), with some even increasing yields under abiotic-stress conditions (Jeong et al., 2010; Redillas et al., 2012); however, there have been no reports of a single NAC gene regulated by a microRNA positively promoting plant architecture, yield, and abiotic-stress tolerance. Previously, we found that OsNAC2 is a target of the microRNA miR164b (Jiang et al., 2018). When miR164b-resistant OsNAC2 was overexpressed in rice, the transgenic plants exhibited ideotype plant architecture traits and had dramatically increased yields (Jiang et al., 2018); however, the performance of these transgenic plants in response to stress conditions has not yet been elucidated.
Here, we found that the plants overexpressing the miR164b-resistant form of OsNAC2 had higher levels of drought and salt tolerance than the wild type (WT), and that the ABA content was increased in the transgenic plants. We also found that the expression levels of the ABA biosynthesis genes and the stress-responsive genes were increased in these lines relative to the WT. Our study provides new insights that may enable the development of high-yielding rice varieties with increased drought tolerance using genetic engineering.
Results
The overexpression of OsNAC2 slows down germination
In our previous study, we showed the miR164b-resistant mOsNAC2-overexpressing lines, ZUOErN3 and ZUOErN4, had a significant high-yield potential (Jiang et al., 2018). In this study, we found that the germination of ZUOErN3 and ZUOErN4 seeds was substantially slower than that of the WT seeds (Fig. 1a–c). During the imbibition for three days, we observed that more seeds germinated from WT (80.1 ± 1.1%) than from the overexpressing plants (76.8 ± 0.3% and 76.9 ± 1.7% in ZUOErN3 and ZUOErN4, respectively; Fig. 1a, d). Five (Fig. 1b) and seven (Fig. 1c) days after germination, obvious differences were observed between the seedlings; for example, the WT seedlings were taller than the overexpression plants. Further observation after 48 h of imbibition showed that the germination percentage of the WT seeds was 7.6 ± 0.7%, whereas only 1.5 ± 0.6% and 2.1 ± 0.8% of the ZUOErN3 and ZUOErN4 seeds, respectively, had germinated. After 57 h of imbibition, 36.4 ± 0.5% of the WT seeds had germinated, whereas the germination percentages of the ZUOErN3 and ZUOErN4 seeds were 25.5 ± 0.5% and 21.2 ± 0.8%, respectively. After 90 h of imbibition, no significant differences were observed in the overall germination rates of the WT and transgenic seeds, with all lines reaching over 95% germination (Fig. 1d). The above results showed that the overexpressing seeds germinated more slowly than the WT seeds, but no difference was observed in their overall germination rates.
Fig. 1.
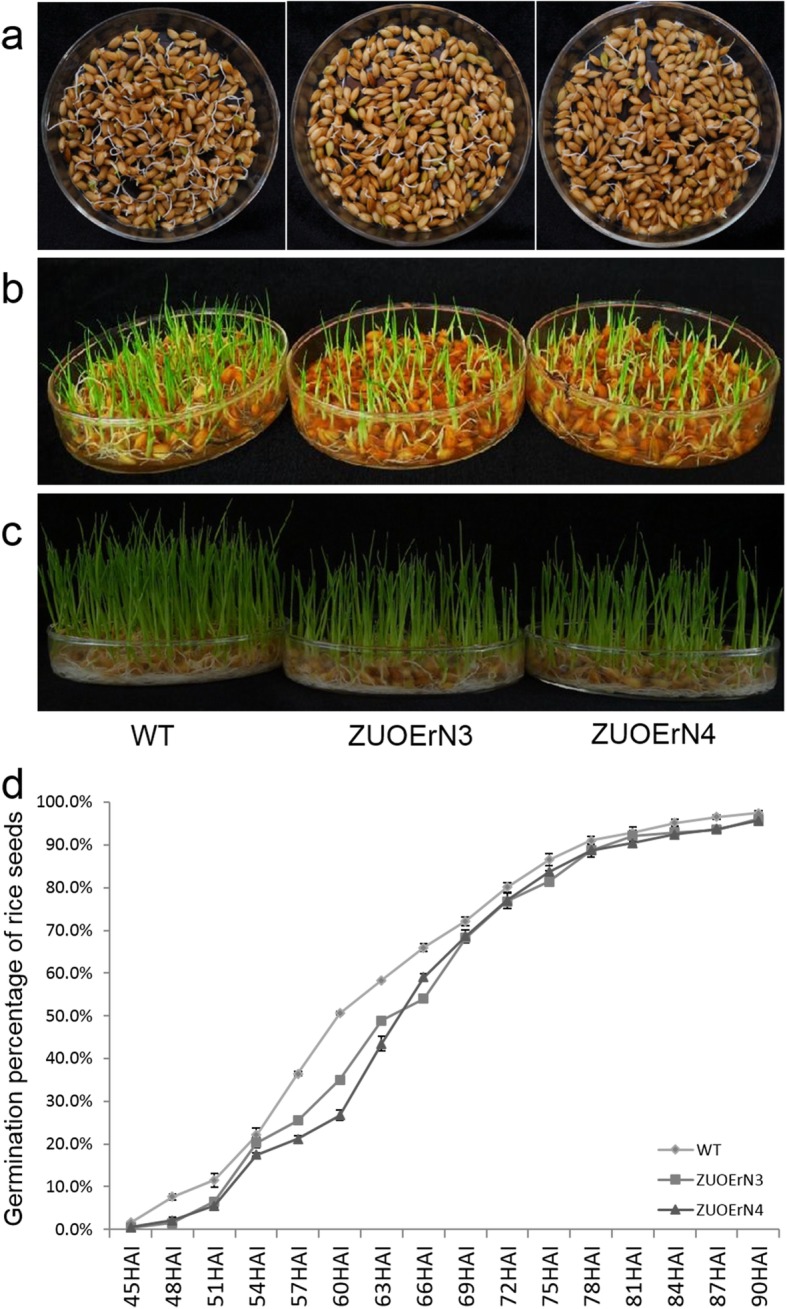
mOsNAC2-overexpressing seeds germinate more slowly than those of WT. The seeds were presoaked with distilled water for 48 h and then germinated at 25/23 °C (day/night) in petri dishes. The germination percentage was calculated at different times after imbibition. The images were taken at 3 d (a), 7 d (b), and 9 d (c) after imbibition in water. (d) Germination percentage after imbibition in water. WT: wild-type seeds; ZUOErN3 and ZUOErN4: OsNAC2 overexpression lines; HAI: hours after imbibition. Error bars represent SD of three biological replicates (n > 200). ** P < 0.01 (t-test)
The delayed germination in mOsNAC2-overexpressing seeds is associated with an increased ABA content
Studies have shown that an increased ABA content in seeds slows germination, whereas a reduction in ABA content accelerates germination (Zhu et al., 2009). To investigate whether changes in the seed germination rates observed here were due to differences in the ABA levels, we applied different amounts of ABA to the WT seeds during germination. When an ABA concentration of 2.5 μmol/L was applied, the germination rate of the WT seeds decreased to a level similar to that of the transgenic seeds. On days 3, 5, and 7 after germination, the seedling growth potential was almost the same as the transgenic seedlings (Additional file 1: Fig. S1a–c). These results showed that the application of exogenous ABA to the WT seeds decreased their germination rate.
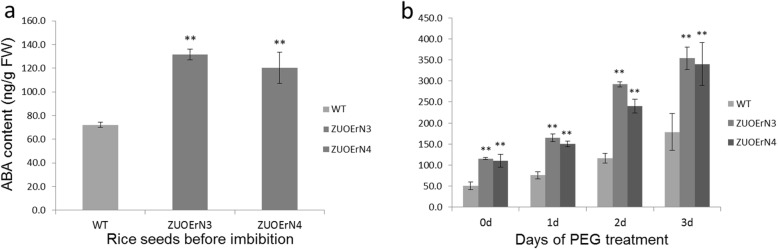
ABA is a carotenoid-derived plant hormone. Fluridone (FLU) can inhibit the biosynthesis of carotenoids, and thus affects ABA biosynthesis (Yoshioka et al., 1998). When 80 μmol/L FLU was applied to the seeds during germination, the phenotypes of the WT and overexpressing plants were not significantly different after two days (Additional file 1: Fig. S2a, b); the ZUOErN3 and ZUOErN4 seedlings were 0.54 ± 0.15 cm and 0.55 ± 0.07 cm in length, respectively, while the WT seedlings were 0.53 ± 0.07 cm long. At three and five days after germination, the seedlings lengths were still not significantly different (Additional file 1: Fig. S2b; S3a, b); for example, the ZUOErN3 and ZUOErN4 seedlings were 2.11 ± 0.18 cm and 2.08 ± 0.21 cm in length, respectively, while the WT seedlings were 2.14 ± 0.31 cm long at three days after germination (Additional file 1: Fig. S2b). Thus, the inhibition of endogenous ABA biosynthesis results in a similar germination speed and seedling growth in the WT and overexpressing seeds. We then used the radioimmunoassay (RIA) method to investigate the endogenous ABA content in the seeds (Quarrie et al., 1988; Zhu et al., 2009). The ABA level in the WT seeds was 72.1 ± 2.2 ng/g, whereas it was significantly higher (131.6 ± 4.4 ng/g in ZUOErN3 and 120.4 ± 13.3 ng/g in ZUOErN4) in the transgenic seeds (Fig. 2a). This further supports the conclusion that increased ABA contents in the mOsNAC2-overexpressing plants delays germination.
Fig. 2.
ABA levels are elevated in transgenic seeds and drought-treated seedlings. ABA levels in rice seeds (a) and seedlings with PEG treatment (b). Seedlings were cultured in hydroponic solution with 10% PEG 6000 to simulate drought. Error bars show the SD of three biological replicates. ** P < 0.01 (t-test)
The mOsNAC2-overexpressing plants have higher drought tolerance
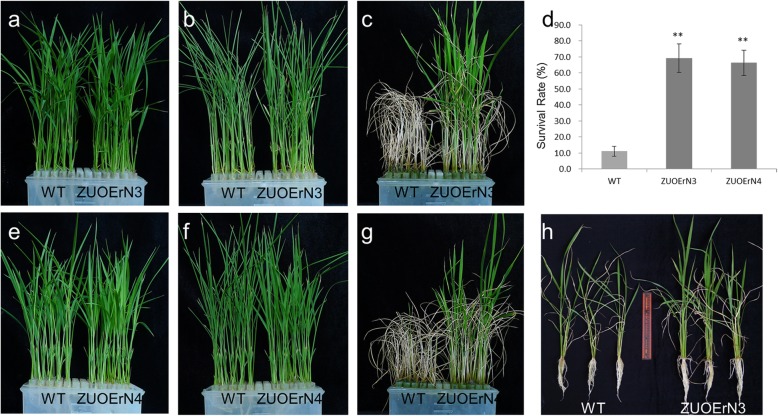
Plant ABA contents are related to the tolerance of stresses such as drought (Zhu et al., 2009). We detected higher ABA contents in the mOsNAC2-overexpressing seeds (Fig. 2a); therefore, we explored whether the mOsNAC2-overexpressing plants have increased drought tolerance by subjecting two-week-old seedlings to polyethylene glycol (PEG)-induced drought conditions. The phenotypes of the WT, ZUOErN3, and ZUOErN4 seedlings were similar before PEG treatment (Fig. 3a, e), while after a three-day treatment, the leaves of the WT plants were curled and wilted, showing obvious symptoms of dehydration (Fig. 3b, f), whereas the leaves of ZUOErN3 and ZUOErN4 showed limited symptoms of drought stress (Fig. 3b, f). After an eight-day drought treatment and an eight-day recovery, most of the WT plants were shriveled and dead, while the overexpressing plants had green leaves (Fig. 3c, g). At the same time, the survival rates of the ZUOErN3 and ZUOErN4 plants were 69.2 ± 8.8% and 66.3 ± 7.9%, respectively, while that of the WT plants was only 11.2 ± 3.1% (Fig. 3d). These results showed that the overexpression of OsNAC2 significantly increases the drought tolerance and survival rate at the seedling stage.
Fig. 3.
Drought tolerance of WT and OsNAC2 overexpression plants. Seeds were germinated in water and transferred to Kimura B nutrient solution. Two-week-old seedlings were treated in Kimura B nutrient solution containing 10% PEG 6000. Plants before treatment (a, e), 10% PEG-treated for 3 d (b, f), and after 8 d treatment and then recovery for 8 d (c, g), are shown. d: The survival rate of WT and transgenic lines treated with 10% PEG 6000 in c and g. Error bars show the SD of three biological replicates (n = 60). ** P < 0.01 (t-test). h: The phenotype of WT and OsNAC2 overexpression line ZUOErN3 plants treated with 15% PEG 6000 for 5 d and allowed to recover for 7 d. Three independent experiments were performed. ZUONrE3, ZUOErN4: OsNAC2 overexpression plants
We further studied drought tolerance at the tillering stage. The plants were treated with 15% PEG for five days and allowed to recover for seven days, after which the transgenic plants appeared healthier than the WT. The transgenic plants grew new leaves, whereas the WT only recovered their green color (Fig. 3h). These results indicated that drought tolerance in the transgenic plants at the tillering stage is also higher than the WT.
mOsNAC2-overexpressing plants have increased salt tolerance
To study the response of the mOsNAC2-overexpressing plants to salinity stress, the seedlings were treated with 50 mM sodium chloride (salt). After eight days of treatment, most of the WT leaves were withered and dead, whereas only a few of the transgenic plants leaves had died, and new leaves had grown (Fig. 4a, b, d, e). After an eight-day recovery period, most of the WT plants were dead, whereas new leaves had grown in all the transgenic plants (Fig. 4c, f).
Fig. 4.
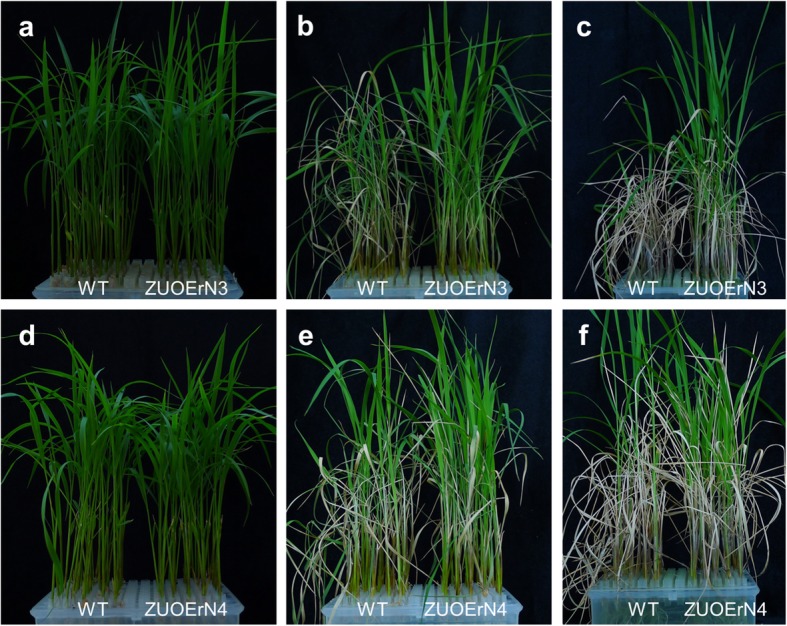
Salt tolerance of WT and OsNAC2 overexpression plants. Seeds were germinated in water and transferred to Kimura B nutrient solution. Two-week-old seedlings were treated in Kimura B nutrient solution with 50 mM sodium chloride. WT and OsNAC2 overexpression plants (ZUONrE3 and ZUOErN4) before treatment (a, d), 50 mM sodium chloride treated for 8 d (b, e), and then recovery for 8 d (c, f) are shown
We explored the expression pattern of OsNAC2 in the WT during salt stress responses using RT-qPCR. No obvious differences in the OsNAC2 expression level were detected after a 1-h salt treatment, but the expression levels were greatly increased after 3 to 48 h of treatment (Additional file 1: Fig. S4). These results showed that overexpression of OsNAC2 increased salinity tolerance in rice.
ABA biosynthesis gene expression is increased in the mOsNAC2-overexpressing plants
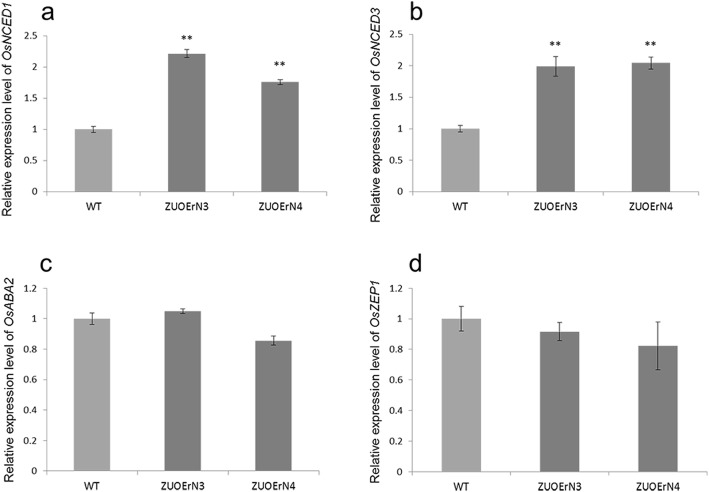
The nine-cis-epoxycarotenoid dehydrogenases (NCEDs) are the key enzymes in ABA biosynthesis (Zhu et al., 2009). OsNCED1 is mainly expressed in the rice leaves under normal conditions (Ye et al., 2011). Our RT-qPCR analysis showed that OsNCED1 was significantly more highly expressed in the transgenic plants than in the WT under normal growth conditions (Fig. 5a). OsNCED3 is a major gene promoting ABA biosynthesis during drought stress in rice (Tan et al., 2003; Ye et al., 2011). Our RT-qPCR analysis revealed that the OsNCED3 expression levels in the ZUOErN3 and ZUOErN4 lines were 0.99- and 1.04-fold higher than in the WT, respectively (Fig. 5b). We also analyzed the expression of genes encoding another two important enzymes in ABA biosynthesis, xanthoxin dehydrogenase (OsABA2) (Endo et al., 2014) and zeaxanthin epoxidase (OsZEP1) (Oliver et al., 2007); However, no significant differences were detected in the expression levels of either of these genes between the transgenic and WT plants (Fig. 5c, d). These results suggest that the increased expression of some ABA biosynthesis genes, such as OsNCED1 and OsNCED3, may result in the higher ABA content observed in the transgenic plants.
Fig. 5.
Expression levels of ABA biosynthesis-related genes. The relative expression levels of OsNCED1 (a), OsNCED3 (b), OsABA2 (c), and OsZEP1 (d) were assessed using RT-qPCR with leaves at the tillering stage. The data were normalized using the rice UBI gene and are shown relative to WT. Error bars represent SD of three biological replicates. ** P < 0.01 (t-test)
PEG increases the ABA content in the mOsNAC2-overexpressing seedlings
To determine whether ABA was involved in the enhanced drought tolerance observed in ZUOErN3 and ZUOErN4, the ABA concentrations were measured in PEG-treated seedlings. Before this drought treatment, the ABA contents in the transgenic (ZUOErN3 and ZUOErN4) and WT plants were 115.6 ± 2.5, 110.6 ± 15.1, and 51.0 ± 9.3 ng/g fresh weight (FW), respectively (Fig. 2b). After the PEG treatment, the ABA contents increased in both the transgenic and WT plants, although the ABA contents in the transgenic plants remained significantly higher than in the WT plants after 1-, 2-, and 3-day treatments (Fig. 2b). These results suggest that the higher ABA levels in transgenic plants may contribute to their increased drought tolerance.
Stress-response genes are more highly expressed in the mOsNAC2-overexpressing plants
To further investigate the possible mechanisms by which OsNAC2 regulates the expression of stress-related genes, we employed RT-qPCR to evaluate the expression of several important abiotic stress-responsive genes in the WT and mOsNAC2-overexpressing lines. The expression of the drought-induced gene OsLEA3 (encoding a group 3 late-embryogenesis abundant protein) was more than two-fold higher in the transgenic plants, while the expression of the ABA-responsive gene OsRAB16 (encoding a small GTP-binding protein) was more than four-fold greater in these plants (Fig. 6). Proline is one of the most well-known osmoprotectants, and its accumulation has been widely observed in organisms under abiotic stress (Sripinyowanich et al., 2013). The expression of the proline biosynthesis gene OsP5CS1 was about 0.7-fold higher in the transgenic plants compared with the WT, while the expression of OsProt did not significantly change (Fig. 6). These results suggest that OsLEA3, OsRAB16, and OsP5CS1 are involved in the drought and salt stress responses.
Fig. 6.
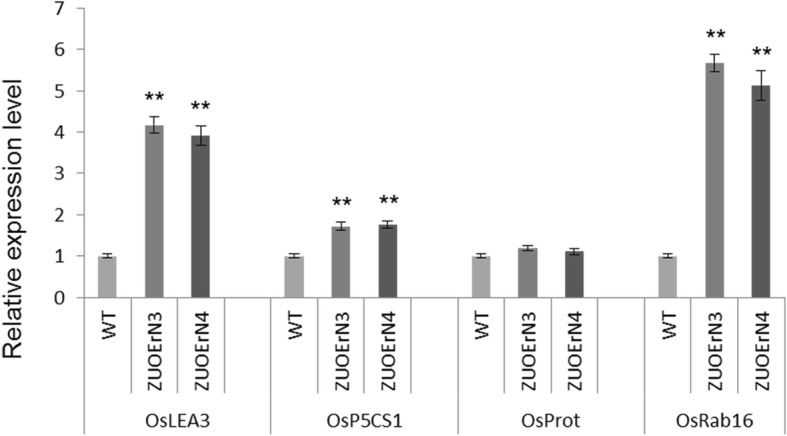
Expression level of abiotic stress-related genes. The relative expression levels of OsLEA3, encoding a group 3 late-embryogenesis abundant protein; OsP5CS, involved in biosynthesis of proline, encoding Δ1-pyrroline-5-carboxylate synthetase; OsProt, encoding a proline transporter; and OsRab16, encoding a small GTP-binding protein, were assayed using RT-qPCR with seedlings. The data were normalized using the rice UBI gene and are shown relative to WT. Error bars represent SD of three biological replicates. ** P < 0.01 (t-test)
OsNAC2 directly regulates expression of OsNCED3 and OsLEA3
In this study, the expression levels of OsNCED3 and OsLEA3 were significantly increased in the mOsNAC2-overexpressing seedlings than in WT. We speculate that these two genes may be targets of OsNAC2. To further investigate the candidate targets of OsNAC2, we performed yeast one-hybrid assays to test the interaction of OsNAC2 with the promoters of OsLEA3 and OsNCED3. The full-length OsNAC2 cDNA was fused in-frame to the GAL4 activation domain in the pGADT7 vector. Promoter sequence regions of different lengths were ligated into the pAbAi vector. The yeast one-hybrid assays showed that OsNAC2 directly interacts with the promoters of OsLEA3 and OsNCED3 (Fig. 7).
Fig. 7.
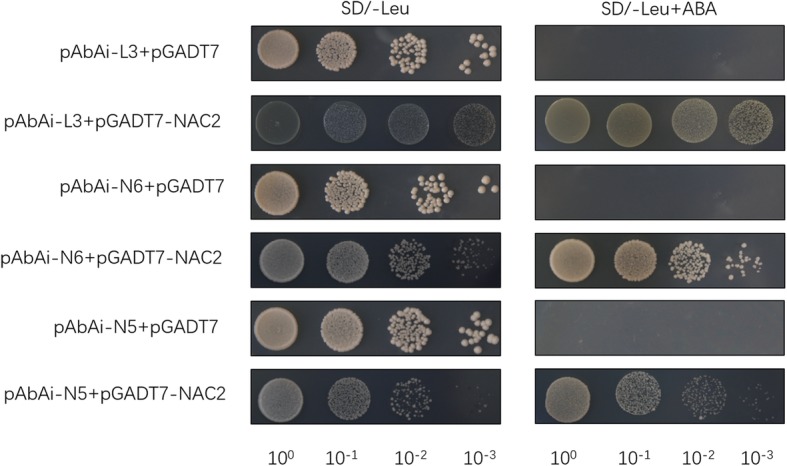
OsLEA3 and OsNCED3 are direct target genes of OsNAC2. The full-length OsNAC2 cDNA was fused to the GLA4 activation domain in the prey vector. The promoter sequence regions of different lengths were fused to an AbAi reporter gene in the bait vector. For yeast one-hybrid assay, Yeast cells were cotransformed with a bait vector and a prey vector and grown in liquid medium and diluted in a 10× dilution series (from 100 to 10− 3). Each dilution was spotted on both SD/−leu and SD/−leu with 300 ng/ml AbA to suppress the background and to test the strength of the interaction. L3, partial OsLEA3 promoter sequence; N6 and N5, partial OsNCED3 promoter sequences
Discussion
NAC TFs play important functions in rice stress tolerance. In this study, we identified OsNAC2 as a positive regulatory factor of abiotic-stress tolerance, as the mOsNAC2-overexpressing rice plants exhibited an increased tolerance to drought and salt stress. Previous studies have shown that the expression levels of SNAC1 (STRESS-RESPONSIVE NAC1) and OsNAC6 are induced by drought and salinity, and that transgenic rice plants overexpressing these genes also display increased drought and salt tolerance (Hu et al., 2006; Lee et al., 2017). Taken together, the results of these studies and our present work point to similar roles for the NAC TFs in abiotic stress responses. In addition, our previous study revealed that mOsNAC2-overexpressing plants have an ideotype plant architecture, with thick stems, well-developed root systems, and high yields (Jiang et al., 2018). A highly developed root system can absorb more moisture and nutrients from the soil, thereby conferring better drought tolerance and facilitating higher yields (Henry et al., 2012). Several other NAC TFs can increase rice yields under abiotic stress conditions (Jeong et al., 2010, 2013). OsNAC10 increases rice yields under stress conditions. OsNAC5-overexpressing rice also enhanced drought tolerance and increased grain yields under drought conditions. OsNAC5 interacts with OsLEA3, an abiotic stress-induced gene in rice that plays vital role in drought and salt tolerance (Jeong et al., 2013), similar to the direct interaction of OsNAC2 with the promoter of OsLEA3 observed in our current study. We also found the miR164b-resistant mOsNAC2-overexpressing plants to have enhanced tolerance of drought and salt stress, partly because of their direct regulation of OsLEA3.
ABA plays an important role in the regulation of stress responses in plants. Many genes in the NAC gene family can increase stress tolerance, which is intimately associated with ABA (Chen et al., 2014; Huang et al., 2016; Hong et al., 2016; Shen et al., 2017). In this study, the mOsNAC2-overexpressing plants exhibited an increased tolerance of drought and salt, and further measurements showed that their ABA contents were significantly higher than in the WT. The expression levels of OsNCED1 and OsNCED3, encoding rate-limiting enzymes in ABA biosynthesis, were also significantly higher in the transgenic plants than in the WT. These findings suggest that OsNAC2 regulates the expression of ABA biosynthesis genes to increase the ABA content, thereby increasing abiotic-stress tolerance. It was previously reported that OsNAC2 directly promotes the expression of OsNCED3 to increase the ABA content (Mao et al., 2017). We also illustrated that OsNAC2 directly interacts with the promoter of OsNCED3 in the yeast one-hybrid system, supporting the results of the previous report.
The expression levels of several abiotic stress-responsive genes were significantly increased in the mOsNAC2-overexpressing plants compared with the WT. These genes included OsLEA3, encoding a group 3 late-embryogenesis abundant protein (Xiao et al., 2007); OsRAB16, encoding a small GTP-binding protein (Yong et al., 2009); and OsP5CS1, encoding Δ1-pyrroline-5-carboxylate synthetase, an enzyme involved in the biosynthesis of proline (Sripinyowanich et al., 2013). This increased expression suggests that the enhanced stress tolerance in the mOsNAC2-overexpressing plants may also be related to the proline levels and these candidate proteins, which mediate abiotic stress tolerance. Indeed, it has been reported that proline accumulation is a possible indicator of osmotic stress tolerance, as it participates in regulation of the osmotic pressure of the cytoplasm in plants (Do et al., 2003). In addition, Rab family proteins play crucial roles in the plant tolerance of environmental stresses. OsRAB16 is an ABA-responsive gene (Mohamed and Aisha, 2019) and exhibited greater than four-fold higher expression in the mOsNAC2-overexpressing plants than in the WT, consistent with the increased ABA levels observed in the transgenic plants. Finally, OsNAC5 directly interacts with the promoter of OsLEA3 to regulate salt tolerance (Jeong et al., 2013); here, we showed that OsNAC2 directly interacts with the promoter of OsLEA3 to regulate drought and salt tolerance.
NAC TF proteins have important functions in the growth and development of rice, with each NAC TF playing many roles; for example, the NAC TF OsNAP regulates leaf senescence (Liang et al., 2014) and increases abiotic stress tolerance during the vegetative and reproductive stages (Chen et al., 2014). OsNAC5 and OsNAC10, when specifically expressed in the roots, can modify root structure and increase drought tolerance, thereby increasing yields under drought conditions (Jeong et al., 2010, 2013). Our study, along with others, indicates that OsNAC2 has several important functions in the growth and development of rice, especially in enhancing yields (Jiang et al., 2018). Here, we have shown that OsNAC2 plays a positive regulatory role in drought- and salt-tolerance in rice through ABA-mediated pathways. These findings may enable the production of new stress-tolerant and high-yielding rice varieties.
Conclusions
Our results demonstrated that OsNAC2 positively regulates rice tolerance of drought and salt stress via an ABA-mediated pathway. This study increases our understanding of the molecular mechanisms by which rice responds to drought and salt stress and broadens our knowledge of NAC TF function in rice. This information will be helpful for breeding drought- and salt-tolerant rice varieties using genetic engineering.
Methods
Rice germination and growth conditions
Seeds of rice (Oryza sativa L.) WT and mOsNAC2-overexpressing lines were harvested during the same season. The seeds were imbibed for 2 d in water with or without 80 μmol/L FLU or 2.5 μmol/L ABA at 25 °C/23 °C (day/night) in a growth chamber. After four days, the germinated seeds were transferred into Kimura B complete nutrient solution (Yoshida et al., 1976) under photosynthetically active radiation 600–1500 μmol m− 2 s− 1. The two-week-old seedlings were used for further experiments.
PEG and salt treatments
For salt stress conditions, 2-week-old seedlings were treated with Kimura B complete nutrient solution supplemented with 100 mM sodium chloride and maintained in a greenhouse with a 13/11-h day/night cycle (25/23 °C). After stress treatments, the seedlings were harvested separately at 0-, 1-, 3-, 6-, 12-, 24-, and 48-h intervals. The tissues were frozen in liquid nitrogen and stored at − 80 °C until RNA extraction.
To analyze drought tolerance of the transgenic plants, WT and transgenic homozygous seeds (ZUOErN3 and ZUOErN4) were germinated in water and transferred to Kimura B nutrient solution 4 days after germination. The 2-week-old seedlings were then transferred into new Kimura B nutrient solution with 10% PEG 6000. The plants were photographed before treatment, at 3 d, and at 8 d, and then following an 8-d recovery period, during which they were treated with Kimura B nutrient solution only. The survival rate was measured after seedling recovery. PEG-treated seedlings, simulating drought treatment, were collected after treatment in the 10% PEG solution for 0 h, 1 d, 2 d, and 3 d, and stored at − 80 °C for ABA determination.
To analyze salt stress, the seedlings were treated with Kimura B nutrient solution with 50 mM sodium chloride. The plants were photographed before treatment, after treatment for 8 d, and after recovery for 8 d.
Germination treatment
Rice seeds were imbibed in water for 2 d. The seeds were then directly sown in petri dishes on filter paper to allow determination of percentage germination. To observe the effects of ABA and fluridone (FLU) on germination, 2.5 μmol/L ABA and 80 μmol/L FLU were added to the water before imbibition. Seeds were placed in a growth chamber with a 13/11-h day/night cycle (25/23 °C) to facilitate germination. The germination rate was recorded or photographed, depending on the experiments. Each experiment was repeated three times.
Measurement of the endogenous ABA level
For estimation of the endogenous ABA level in seeds and seedlings, 0.5 g of rice seeds or 1.0 g of seedlings was homogenized in 3 ml of distilled water and then shaken at 4 °C overnight. The homogenates were centrifuged at 12,000 g for 10 min at 4 °C and the supernatants used for an RIA assay. The subsequent steps were conducted as previously described (Quarrie et al., 1988; Zhu et al., 2009).
RNA isolation and RT-qPCR
RNA was isolated using an RNA extraction kit with TRIzol reagent (Invitrogen, USA) and quantified with a DU730 spectrophotometer (Beckman Coulter, Germany). cDNA was synthesized with a 5 × iScript RT Supermix kit (TaKaRa, Dalian, China) using about 2 μg of total RNA. RT-qPCR was performed with SYBR premix Ex Taq II in a total volume of 20 μL on the Bio-Rad CFX 96 following the manufacturer’s protocol. Data were normalized to the internal rice UBIQUITIN (UBI) gene, and relative quantification was used for data analysis.
The primers for rice UBI and OsNAC2 amplification are described in Jiang et al. (2018). The primers for OsNCED1 and OsNCED3 are described in Zhu et al. (2009). The primers for OsLEA3, OsP5C91, OsProt, and OsRab16 are described in Yin et al. (2017). The primers used for OsABA2 and OsZEP1 are as follows: ABA2F, 5′-TGTGGATCTGCTACCTAAGG-3′; ABA2R, 5′-GTAAAGCCACCATCCACCATG-3′; ZEPF, 5′-CGGATGCCATTGAGTTTGGTTC-3′; and ZEPR, 5′-CAGATTCATATGGGAGCGTGC-3′.
Yeast one-hybrid
For yeast one-hybrid assays, the The full-length OsNAC2 cDNA was fused in-frame to the GAL4 activation domain in the pGADT7 vector. The different promoter sequences regions of OsNCED3 and OsLEA3 were ligated into the pAbAi vector. The assays were performed according to the manufacturer’s instructions (CLONTECH Laboratories, USA; cat. # 630491) or the reported publication (Xie et al., 2018). The L3, partial OsLEA3 promoter sequence; N6 and N5, partial OsNCED3 promoter sequences were followed the previous publication (Jeong et al., 2013; Mao et al., 2017).
Supplementary information
Additional file 1: Figure S1. Effects of ABA on rice seed germination. WT seeds were imbibed in water containing 2.5 μmol/L ABA at 25/23 °C (day/night). Seeds from mOsNAC2-overexpressing lines (ZUOErN3 and ZUOErN4) were imbibed in water only. Images were taken after germination for 3 d (a), 5 d (b), and 8 d (c). Figure S2. Effects of fluridone (FLU) on rice seed germination. WT seeds were imbibed in water at 25/23 °C (day/night). Seeds from OsNAC2 overexpression lines (ZUOErN3 and ZUOErN4) were imbibed in water containing 80 μmol/L FLU at 25/23 °C (day/night). The images were taken on day 2 after germination (a). The seedling length was measured after germination for 2 d, 3d, and 5 d (b). Figure S3. Effects of FLU on rice seedling growth. WT seedlings were cultured in water at 25/23 °C (day/night). OsNAC2 overexpression lines (ZUOErN3 and ZUOErN4) were cultured in water containing 80 μmol/L FLU. The images were taken on day 3 (a) and day 5 (b) after germination. Figure S4. Expression level of OsNAC2 under salt stress. The expression of OsNAC2 in rice seedlings grown in 50 mM sodium chloride was assayed by RT-qPCR. RT-qPCR data were normalized using the rice UBI gene and are shown relative to 0 h. Error bars represent SD of three biological replicates.
Acknowledgments
We thank Dr. Xiangjun Zhou (Purdue University) for providing helpful comments on our manuscript.
Abbreviations
- ABA
Abscisic acid
- ABA2
xanthoxin dehydrogenase
- FLU
fluridone
- LEA3
3 late-embryogenesis abundant protein
- miRNA
microRNA
- NAC
NAM, AFAT, and CUC transcription factor
- NCEDs
Nine-cis-epoxycarotenoid dehydrogenases
- P5CS1
Δ1-pyrroline-5-carboxylate synthetase
- PEG
Polyethylene glycol
- RAB16
a small GTP-binding protein
- RIA
Radioimmunoassay
- RT-qPCR
Reverse transcription quantitative PCR
- WT
wild-type
- ZEP1
Zeaxanthin epoxidase
Authors’ contributions
DJ and CZ conceived the study and designed the experiments. DJ, LZ, WC, NY, and JX contributed to the performance of the experiments and the data analysis. DJ and CZ drafted the manuscript. NY contributed good advice on the design of the experiment. All authors read and approved the final manuscript.
Funding
This work was supported by National Key Research and Development Program of China (Grant No. 2016YFD0100400) and the National Natural Science Foundation of China (Grant No. 31100872).
Availability of data and materials
All data supporting the conclusions of this article are provided within the article (and its additional files).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dagang Jiang, Email: dagangj@scau.edu.cn.
Lingyan Zhou, Email: lingyanzh@163.com.
Weiting Chen, Email: chenweit@126.com.
Nenghui Ye, Email: nye@hunau.edu.cn.
Jixing Xia, Email: xiajx@gxu.edu.cn.
Chuxiong Zhuang, Email: zhuangcx@scau.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12284-019-0334-6.
References
- Chen X, Lu SC, Wang YF, Zhang X, Lv B, Luo LQ, Xi DD, Chen JB, Ma H, Ming F. OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. Plant J. 2015;82:302–314. doi: 10.1111/tpj.12819. [DOI] [PubMed] [Google Scholar]
- Chen X, Wang YF, Lv B, Li J, Luo LQ, Lu SC, Zhang X, Ma H, Ming F. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 2014;55:604–619. doi: 10.1093/pcp/pct204. [DOI] [PubMed] [Google Scholar]
- Do TH, Michel J, Geert A, Christian H, Tran TT, Le VS, Nancy HR. Proline accumulation and Δ1-pyrroline-5-carboxylate synthetase gene properties in three rice cultivars differing in salinity and drought tolerance. Plant Sci. 2003;165(5):1059–1068. doi: 10.1016/S0168-9452(03)00301-7. [DOI] [Google Scholar]
- Endo A, Nelson KM, Thoms K, Abrams SR, Nambara E, Sato Y. Functional characterization of xanthoxin dehydrogenase in rice. J Plant Physiol. 2014;171(14):1231–1240. doi: 10.1016/j.jplph.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Fang YJ, Xie KB, Xiong LZ. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J Exp Bot. 2014;65(8):2119–2135. doi: 10.1093/jxb/eru072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF, Chua NH. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to Downregulate Auxin signals for Arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry A, Cal AJ, Batoto TC, Torres RO, Serraj R. Root attributes affecting water uptake of rice (Oryza sativa) under drought. J Exp Bot. 2012;63(3):4751–4763. doi: 10.1093/jxb/ers150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Zhang H, Huang L, Li D, Song F. Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in Rice. Front Plant Sci. 2016;7:4. doi: 10.3389/fpls.2016.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HH, Dai MQ, Yao JL, Xiao BZ, Li XH, Zhang QF, Xiong LZ. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci U S A. 2006;103:12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HH, You J, Fang YJ, Zhu XY, Qi ZY, Xiong LZ. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol. 2008;67:169–181. doi: 10.1007/s11103-008-9309-5. [DOI] [PubMed] [Google Scholar]
- Huang L, Hong YB, Zhang HJ, Li DY, Song MF. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 2016;16:203. doi: 10.1186/s12870-016-0897-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Choi YD, Kim MY, Reuzeau C, Kim JK. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010;153:185–197. doi: 10.1104/pp.110.154773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JS, Kim YS, Redillas MC, Jang J, Jung H, Bang SW, Choi YD, Ha SH, Reuzeau C, Kim JK. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotech J. 2013;11:101–114. doi: 10.1111/pbi.12011. [DOI] [PubMed] [Google Scholar]
- Jiang DG, Chen WT, Dong JF, Li J, Yang F, Wu ZC, Zhou H, Wang WS, Zhuang CX. Overexpression of OsmiR164b-resistant OsNAC2 improves plant architecture and grain yield in rice. J Exp Bot. 2018;69(7):1533–1543. doi: 10.1093/jxb/ery017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science. 2009;323:1053–1057. doi: 10.1126/science.1166386. [DOI] [PubMed] [Google Scholar]
- Lee DK, Chung PJ, Jeong JS, Jang G, Bang SW, Jung H, Kim YS, Ha SH, Choi YD, Kim JK. The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant Biotech J. 2017;15:754–764. doi: 10.1111/pbi.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhang BH. MicroRNAs in control of plant development. J Cell Physiol. 2016;231:303–313. doi: 10.1002/jcp.25125. [DOI] [PubMed] [Google Scholar]
- Liang CZ, Wang YQ, Zhu YN, Tanga JY, Hua B, Liu LC, Ou SJ, Wu HK, Sun XH, Chu JF, Chu CC. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci U S A. 2014;111(27):10013–10018. doi: 10.1073/pnas.1321568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Dugas DV, Bartel DP, Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol. 2004;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Mao CJ, Lu SC, Lv B, Zhang B, Shen JB, He JM, Luo LQ, Xi DD, Chen X, Ming F. A Rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017;174:1747–1763. doi: 10.1104/pp.17.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao CZ, Ding WN, Wu YN, Yu J, He XW, Shou HX, Wu P. Overexpression of a NAC-domain protein promotes shoot branching in rice. New Phytol. 2007;176:288–298. doi: 10.1111/j.1469-8137.2007.02177.x. [DOI] [PubMed] [Google Scholar]
- Mohamed AE, Aisha AA (2019) Overexpression of Rice Rab7 gene improves drought and heat tolerance and increases grain yield in Rice (Oryza sativa L.). genes 10(1):56 [DOI] [PMC free article] [PubMed]
- Nuruzzaman M, Manimekalai R, Sharonia AM, Satoh K, Kondoh H, Ooka H, Kikuchi S. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Oliver SN, Dennis ES, Dolferus R. ABA regulates Apoplastic sugar transport and is a potential signal for cold-induced pollen sterility in Rice. Plant Cell Physiol. 2007;48:1319–1330. doi: 10.1093/pcp/pcm100. [DOI] [PubMed] [Google Scholar]
- Puranik S, Sahu PP, Srivastava PS, Prasad M. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17:369–381. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Quarrie SA, Whitford PN, Appleford NEJ, Wang TL, Cook SK, Henson IE, Loveys BR. A monoclonal antibody to (S)-abscisic acid: its characterization and use in a radioimmunoassay for measuring abscisic acids in crude extracts of cereal and lupin leaves. Planta. 1988;173:330–339. doi: 10.1007/BF00401020. [DOI] [PubMed] [Google Scholar]
- Redillas MC, Jeong JS, Kim YS, Jung H, Bang SW, Choi YD, Ha SW, Reuzeau C, Kim J. The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotech J. 2012;10:792–805. doi: 10.1111/j.1467-7652.2012.00697.x. [DOI] [PubMed] [Google Scholar]
- Saada AS, Li X, Li HP, Huang T, Gao CS, Guo MW, Cheng W, Zhao GY, Liao YC. A rice stress-responsive NAC gene enhances tolerance of transgenic wheat to drought and salt stresses. Plant Sci. 2013;203–204:33–40. doi: 10.1016/j.plantsci.2012.12.016. [DOI] [PubMed] [Google Scholar]
- Shen JB, Lv B, Luo LQ, He JM, Mao CJ, Xi DD, Ming F. The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci Rep. 2017;7:40641. doi: 10.1038/srep40641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SY, Chen Y, Chen J, Dai XY, Zhang WH. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta. 2011;234:331–345. doi: 10.1007/s00425-011-1403-2. [DOI] [PubMed] [Google Scholar]
- Sripinyowanich S, Klomsakul P, Boonburapong B, Bangyeekhun T, Asami T, Gu HY, Buaboocha T, Chadchawan S. Exogenous ABA induces salt tolerance in indica rice (Oryza sativa L.): the role of OsP5CS1 and OsP5CR gene expression during salt stress. Environ Exp Bot. 2013;86:94–105. doi: 10.1016/j.envexpbot.2010.01.009. [DOI] [Google Scholar]
- Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K, Nakashima K. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol Gen Genomics. 2010;284:173–183. doi: 10.1007/s00438-010-0557-0. [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu LJ, Li QB, Cline K, Mccarty DR. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003;35:44–56. doi: 10.1046/j.1365-313X.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- Valliyodan B, Nguyen HT. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr Opin Plant Biol. 2006;9(2):189–195. doi: 10.1016/j.pbi.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Xiao BZ, Huang YM, Tang N, Xiong LZ. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet. 2007;115(1):35–46. doi: 10.1007/s00122-007-0538-9. [DOI] [PubMed] [Google Scholar]
- Xie Y, Zhang Y, Han J, Luo J, Li G, Huang J, Wu H, Tian Q, Zhu Q, Chen Y, Kawano Y, Liu YG, Chen L. The Intronic cis element SE1 recruits trans-acting repressor complexes to repress the expression of ELONGATED UPPERMOST INTERNODE1 in Rice. Mol Plant. 2018;11:720–735. doi: 10.1016/j.molp.2018.03.001. [DOI] [PubMed] [Google Scholar]
- Xiong LM, Zhu JK. Regulation of abscisic acid biosynthesis. Plant Physiol. 2003;133:29–36. doi: 10.1104/pp.103.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye NH, Zhu GH, Liu YG, Li YX, Zhang JH. ABA controls H2O2 accumulation through the induction of OsCATB in Rice leaves under water stress. Plant Cell Physiol. 2011;52:689–698. doi: 10.1093/pcp/pcr028. [DOI] [PubMed] [Google Scholar]
- Yin X, Cui Y, Wang M, Xia X. Overexpression of a novel MYB-related transcription factor, OsMYBR1, confers improved drought tolerance and decreased ABA sensitivity in rice. Biochem Biophys Res Commun. 2017;490:1355–1361. doi: 10.1016/j.bbrc.2017.07.029. [DOI] [PubMed] [Google Scholar]
- Yong CY, Chao YY, Yang MY, Cheng SY, Cho SC, Kao CH. NaCl-induced expression of glutathione reductase in roots of rice (Oryza sativa L.) seedlings is mediated through hydrogen peroxide but not abscisic acid. Plant Soil. 2009;320:103–115. doi: 10.1007/s11104-008-9874-z. [DOI] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice. International Rice Research Institute, Manila, Philippines. p 63–64
- Yoshioka T, Endo T, Satoh S. Restoration of Seed Germination at Supraoptimal Temperatures by Fluridone, an Inhibitor of Abscisic Acid Biosynthesis. Plant Cell Physiol. 1998;39(3):307–312. doi: 10.1093/oxfordjournals.pcp.a029371. [DOI] [Google Scholar]
- Zhu G, Ye N, Zhang J. Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol. 2009;50:644–651. doi: 10.1093/pcp/pcp022. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Effects of ABA on rice seed germination. WT seeds were imbibed in water containing 2.5 μmol/L ABA at 25/23 °C (day/night). Seeds from mOsNAC2-overexpressing lines (ZUOErN3 and ZUOErN4) were imbibed in water only. Images were taken after germination for 3 d (a), 5 d (b), and 8 d (c). Figure S2. Effects of fluridone (FLU) on rice seed germination. WT seeds were imbibed in water at 25/23 °C (day/night). Seeds from OsNAC2 overexpression lines (ZUOErN3 and ZUOErN4) were imbibed in water containing 80 μmol/L FLU at 25/23 °C (day/night). The images were taken on day 2 after germination (a). The seedling length was measured after germination for 2 d, 3d, and 5 d (b). Figure S3. Effects of FLU on rice seedling growth. WT seedlings were cultured in water at 25/23 °C (day/night). OsNAC2 overexpression lines (ZUOErN3 and ZUOErN4) were cultured in water containing 80 μmol/L FLU. The images were taken on day 3 (a) and day 5 (b) after germination. Figure S4. Expression level of OsNAC2 under salt stress. The expression of OsNAC2 in rice seedlings grown in 50 mM sodium chloride was assayed by RT-qPCR. RT-qPCR data were normalized using the rice UBI gene and are shown relative to 0 h. Error bars represent SD of three biological replicates.
Data Availability Statement
All data supporting the conclusions of this article are provided within the article (and its additional files).