Abstract
The treatment of bacterial infections is hindered by the presence of biofilms and metabolically inactive persisters. Here, we report the synthesis of an enantiomeric block co-beta-peptide, poly(amido-D-glucose)-block-poly(beta-L-lysine), with high yield and purity by one-shot one-pot anionic-ring opening (co)polymerization. The co-beta-peptide is bactericidal against methicillin-resistant Staphylococcus aureus (MRSA), including replicating, biofilm and persister bacterial cells, and also disperses biofilm biomass. It is active towards community-acquired and hospital-associated MRSA strains which are resistant to multiple drugs including vancomycin and daptomycin. Its antibacterial activity is superior to that of vancomycin in MRSA mouse and human ex vivo skin infection models, with no acute in vivo toxicity in repeated dosing in mice at above therapeutic levels. The copolymer displays bacteria-activated surfactant-like properties, resulting from contact with the bacterial envelope. Our results indicate that this class of non-toxic molecule, effective against different bacterial sub-populations, has promising potential for the treatment of S. aureus infections.
Subject terms: Antimicrobials, Bacteria, Pathogens, Polymer chemistry
The authors report the synthesis of an enantiomeric block co-beta-peptide that kills methicillin-resistant Staphylococcus aureus, including biofilm and persister bacterial cells, and disperses biofilms. The copolymer displays antibacterial activity in human ex vivo and mouse in vivo infection models without toxicity.
Introduction
Antimicrobial resistance in bacteria is a serious and growing clinical problem, eroding the therapeutic armamentarium and leaving limited treatment options for certain infections. Compounding the difficulty of treating antibiotic-resistant strains is the presence of persisters, subpopulations that are antibiotic-tolerant due to metabolic inactivity1,2, and the capacity of bacteria to develop biofilms3, both of which lead to chronic and recurrent infections4–7. The World Health Organization (WHO) recently published a priority list of bacteria for which new antibiotics are urgently needed8. Methicillin-resistant Staphylococcus aureus (MRSA), a WHO high-priority pathogen, is a leading cause of mortality due to antibiotic-resistant infections9,10. Initially restricted to hospitals and healthcare settings, MRSA is causing an increasing number of infections in the community11,12. MRSA is associated with poor clinical outcomes13: it causes frequent skin and soft tissue infections14 and can disseminate, resulting in life-threatening bloodstream infections, endocarditis, bone and joint infections, as well as pneumonia15,16. S. aureus is prone to form biofilms and also exists in the form of metabolically inactive antibiotic-tolerant persister phenotype3. Last-resort antibiotics such as vancomycin are largely ineffective against S. aureus persisters and biofilms17. New therapeutics are needed to combat the spread of difficult to treat drug-resistant S. aureus infections. Alternative antibacterial agents should have bactericidal activity against replicating cells, persisters, and established biofilms. Cationic alpha-peptides and membrane-active agents have been investigated as alternative antimicrobials to combat biofilms and persisters18,19, but unselective toxicity is a complicating factor20.
Amongst the various synthetic polymer families being explored as peptidomimetics21–25, beta-peptides are promising because they can exhibit biological activity comparable to natural peptides, but have better proteolytic stability26, and are usually amphiphilic and non-mutagenic27. Beta-peptides have been considered for use in diverse therapeutic applications such as antimicrobial agents28–30, vaccine drugs31, protein–protein interaction inhibitors32,33, and drug delivery34,35. Alpha-peptide antimicrobials are known to form facially amphiphilic (FA) structures that enhance the bactericidal properties but tend to be hemolytic and toxic36. Compared to alpha-peptides, beta-peptides have an extra methylene group in the backbone. The hydrophobicity of beta-peptides may be tuned by the structure of the side chains. Further, beta-peptides may be designed to form foldamers exhibiting diverse secondary structures, such as helices and beta-sheets37–39 and complex tertiary and quaternary structures40.
Munoz-Guerra and colleagues reported the first research on nylon-3 and analogs, which included the synthesis and helical propensity of these beta-peptides41–43. In the development of antimicrobial beta-peptides, previous efforts focus mainly on random co-beta-peptides and optimization of their cationic versus hydrophobic beta-lactam residues to reduce hemolysis whilst maintaining a good bactericidal effect44–47. There is no reported work on glycosylated block co-beta-peptides. Block co-poly(beta-peptides) are interesting as they may show unique combinations of properties displayed by the individual blocks, which are as yet under-exploited for the development of next-generation antibacterials. Also, a strategy for the facile synthesis of block co-beta-peptides has not been previously reported.
In this study, we report a simple one-shot one-pot anionic ring opening (co)polymerization (AROP) strategy to synthesize a series of enantiomeric block co-beta-peptides, which cannot be made by sequential copolymerization. Two beta-lactam monomers with contrasting reactivities—a protected d-glucose (DGup) beta-lactam and a protected cationic beta-l-lysine (BLKp) beta-lactam—can be block copolymerized in one shot. The resulting optimized block co-beta-peptide, PDGu(7)-block-PBLK(13), is non-cytotoxic and non-hemolytic in vitro. Further, the block co-beta-peptide has interesting biological properties. Unlike classical antibiotics, PDGu(7)-b-PBLK(13) retains potency against MRSA persister cells and biofilms. It is active against both the community-acquired (CA-) and hospital-associated (HA-) MRSA strains. The block copolymer also effectively removes biofilm biomass but the homocationic beta-peptide (PBLK(20)) cannot. The block copolymer is bactericidal against MRSA in various murine models of systemic acute and established infections, and also in an ex vivo human skin infection model, while having no in vivo acute toxicity in murine repeated dosing studies. This study opens up possibilities of treatment for recalcitrant MRSA infections.
Results
Synthesis of the (co)polymers via one-shot one-pot AROP
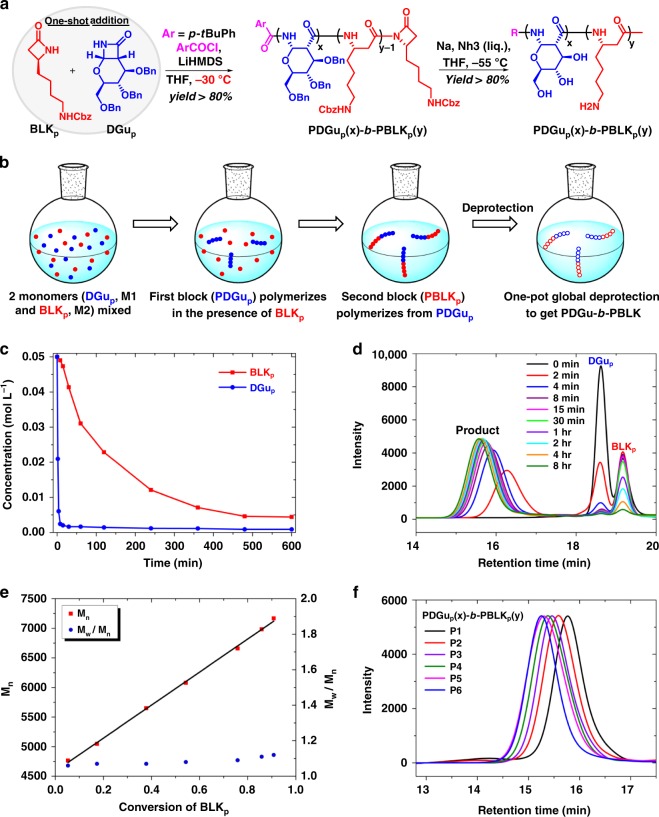
The monomers N-Cbz-β-lactam-l-lysine (BLKp) and O-Bn-β-lactam-d-glucose (DGup) were synthesized and verified by nuclear magnetic resonance (NMR) spectroscopy (Supplementary Methods, Supplementary Figs. 1–3). The copolymer synthetic strategy relies on the observation that the homopolymerization of N-Cbz-β-lactam-l-lysine monomer (BLKp) is much slower than the homopolymerization of O-Bn-β-lactam-d-glucose monomer (DGup) (Supplementary Table 1). When DGup and BLKp monomers (10:10, mole/mole) were mixed together in tetrahydrofuran, DGup was totally consumed in 8 min while the BLKp monomer required 8 h for complete reaction (Fig. 1a–c, Supplementary Figs. 4, 5). The molecular weight of the product increased linearly over an 8-h period during which DGup disappeared rapidly in the first few minutes, while BLKp was consumed gradually over the next few hours (Fig. 1d). A plot of molecular weight (Mn) versus BLKp conversion (Fig. 1e) shows a linear relationship and the Đ values of the products remain small (1.06–1.12). These results are consistent with the growth of a single copolymer chain through rapid consumption of DGup followed by slower but contiguous incorporation of BLKp, and thus provide evidence for a ‘block-like’ structure of the resulting copolymer. A series of poly(Bn-amido-d-glucose)-block-poly(Cbz-beta-l-lysine) (PDGup(x)-b-PBLKp(y)) block copolymers, with varying ratios of x to y but constant target total degree of polymerization of 20, i.e. (x + y) = 20, was synthesized (Fig. 1a, Supplementary Fig. 6). The molecular weights are close to the design values based on gel permeation chromatography (GPC) relative to polystyrene standards, confirming that the AROP process is well-controlled (Fig. 1f, Supplementary Table 2).
Fig. 1.
Facile one-shot one-pot synthesis of PDGu(x)-b-PBLK(y) block copolymer. a Synthetic scheme of PDGu(x)-b-PBLK(y). b One-shot addition of both monomers (DGup and BLKp) leads to block copolymerization when the monomers have contrasting reactivities. c–e Kinetic studies and f GPC measurements verify the well-controlled single chain block architecture of PDGup(x)-b-PBLKp(y). c Remaining monomer concentration vs time. d GPC curves of partially polymerized products at selected quenching times. e Molecular weight (Mn) and molecular weight distribution (Đ) as a function of conversion of BLKp. f GPC of protected-(co)polymers
After one-step deprotection, the final products PDGu(x)-b-PBLK(y) were obtained with overall yields greater than 65% (Supplementary Fig. 7). NMR spectroscopy measurements of PDGu(x)-b-PBLK(y) show two sets of signals belonging to PDGu and PBLK, respectively, corroborating their block rather than random structures (Supplementary Figs. 8–22). NMR spectra also show that the ratios of DGu to BLK in PDGu(x)-b-PBLK(y) after purification deviate slightly from the stoichiometric ratios of added monomers. For example, the actual composition of DGu and BLK in PDGu(10)-b-PBLK(10) is PDGu(7)-b-PBLK(13); the PBLK block is 66 mol% versus the design value of 50 mol%. This trend is repeatable and can be seen in other compositions (Table 1). The molecular weights (Mn) of the homocationic PBLK(20), homosugar PDGu(20), and copolymer PDGu(7)-b-PBLK(13) were, respectively, 3012 Da, 3159 Da, and 3391 Da, as measured by the Matrix Assisted Laser Desorption/Ionization-Time of Flight (MALDI-TOF) mass spectroscopy (Supplementary Fig. 23).
Table 1.
Design and actual ratios of DGu to BLK before and after deprotection
| Sample | Design ratio of DGup to BLKp | Actual ratioa of DGup to BLKp | Actual ratiob of DGu to BLK after deprotection |
|---|---|---|---|
| P1 | PDGu p (6.7)- b -PBLK p (13.3) | PDGu p (6)- b -PBLK p (14) | PDGu(5)- b -PBLK(15) |
| P2 | PDGu p (8)- b -PBLK p (12) | PDGu p (8)- b -PBLK p (12) | PDGu(6)- b -PBLK(14) |
| P3 | PDGu p (10)- b -PBLK p (10) | PDGu p (10)- b -PBLK p (10) | PDGu(7)- b -PBLK(13) |
| P4 | PDGu p (12)- b -PBLK p (8) | PDGu p (12)- b -PBLK p (8) | PDGu(9)- b -PBLK(11) |
| P5 | PDGu p (13.3)- b -PBLK p (6.7) | PDGu p (14)- b -PBLK p (6) | PDGu(10)- b -PBLK(10) |
| P6 | PDGu p (20) | PDGu p (20) | PDGu(20) |
aRatios were calculated based on 1H NMR integrations of (protected) PDGup(x)-b-PBLKp(y)
bRatios were calculated based on 1H NMR integrations of (deprotected) PDGu(x)-b-PBLK(y)
When we attempted the synthesis of PDGup(10)-b-PBLKp(10) by the sequential addition of DGup (M1) followed by BLKp (M2) at −30 °C (Supplementary Fig. 24a), the amount of isolated undesired homopolymer PDGup after the reaction could be more than 50% of the yield (based on DGup). This sequential copolymerization of higher reactivity DGup followed by lower reactivity BLKp could be finished rapidly in <1 h, but achieved only low purity block copolymer with substantial PDGup homopolymer. We expected that by reversing the order of addition of monomers, i.e. first BLKp and then DGup (Supplementary Fig. 24b), in which the first block (PBLKp) has a lower reactivity and also a higher transfer rate to DGup, the block copolymerization would successfully occur. However, the first step requires up to 8 h to reach ~90% conversion (based on BLKp). In addition, the final mixture was very viscous and contained a large proportion of pre-mature terminated PBLKp. Regardless of the sequence of monomer addition, sequential copolymerization cannot successfully synthesize cationic glycosylated block copoly(beta-peptide) with good yield and purity. Unexpectedly, with one-shot AROP with simultaneous feed of the two beta-lactams, we could achieve successful synthesis of the block copolymers PDGup-b-PBLKp.
PDGu(7)-b-PBLK(13) is antibacterial and non-cytotoxic
The PDGu(x)-b-PBLK(y) series was tested against a panel of Gram-positive bacteria. The homopolymer PBLK(20) was active against most tested bacteria, but was unselective and cytotoxic to eukaryotic cells and also hemolytic (Table 2, Supplementary Figs. 25 and 26). The block copolymerization process decreased cytotoxicity while maintaining potency against S. aureus. The copolymer PDGu(7)-b-PBLK(13) shows the most balanced profile, combining potency against S. aureus with good selectivity index (>25) and no hemolysis (HC10 > 20,000 µg mL−1) (Table 2, Supplementary Figs. 25 and 26). The copolymer shows good activity against USA300 (Table 2), the predominant CA-MRSA11. Further profiling demonstrated that the copolymer is also potent against other MRSA strains from major lineages of global epidemiology48, including (HA-)MRSA strains resistant to multiple conventional antibiotics (including vancomycin, daptomycin) (Table 3). Kill-kinetics experiments revealed that PDGu(7)-b-PBLK(13) killed replicating MRSA faster than vancomycin (Supplementary Fig. 27). The selection of escape mutants to PDGu(7)-b-PBLK(13) at 10× its minimum inhibitory concentration (MIC) was unsuccessful, showing that the propensity for emergence of resistance is extremely low (frequency below 3 × 10−10, which is much lower than reported values for antibiotics49,50). We then tried to select mutants by the continued pressure of sub-inhibitory concentrations of the block copolymer for up to 14 days (as described previously51). This approach also did not select for copolymer-resistant MRSA colonies. As a control, escape mutants resistant to the antibiotic ciprofloxacin were easily selected (Supplementary Fig. 28).
Table 2.
Antimicrobial and hemolytic activity and biocompatibility of (co)polymers
| Sample | MIC90 (μg mL−1) | HC10 (μg mL−1) RBCa | IC50 (μg mL−1) 3T3b | ||||
|---|---|---|---|---|---|---|---|
| SA 25923 | SA 29213 | MRSA BAA40 | MRSA USA300 | Bacillus subtilis 6633 | |||
| PBLK(20) | 8 | 8 | 8 | 8 | 4 | 5000 | 18 |
| PDGu(5)- b -PBLK(15) | 8 | 8 | 8 | 8 | 4 | 3300 | 100 |
| PDGu(6)- b -PBLK(14) | 16 | 8 | 8 | 8 | 4 | 4800 | 150 |
| PDGu(7)- b -PBLK(13) | 16 | 8 | 8 | 8 | 4 | >20,000 | 430 |
| PDGu(9)- b -PBLK(11) | 32 | 16 | 16 | 16 | 8 | >20,000 | 395 |
| PDGu(10)- b -PBLK(10) | 64 | 32 | 32 | 32/64 | 16 | >20,000 | 630 |
| PDGu(20) | >512 | >512 | >512 | >512 | >512 | >20,000 | >1024 |
aRBC: red blood cells
b3T3: mouse fibroblast cells
Table 3.
Antimicrobial activity against multi-drug-resistant clinically isolated MRSA
| Serial no. | Designation | MIC (μg mL−1) | Multi-drug resistance | Major lineage/clonal complex48 | |||
|---|---|---|---|---|---|---|---|
| PDGu(7)-b-PBLK(13) | Resistant antibiotic | ||||||
| VAN-resistant S. aureus | 1 | HIP11714 | 16 | VAN | 512 | CIP, CLI, ERY, GEN, LVX, MXF, OXA,RIF, TEC | 5 |
| 2 | HIP11983 | 16 | 16 | CIP, CLI, ERY, GEN, LVX, MXF, OXA, TET | 5 | ||
| 3 | HIP13170 | 16 | 128 | CIP, CLI, ERY, GEN, LVX, MXF, OXA, TEC, TET | 5 | ||
| 4 | HIP13419 | 16 | 64 | CIP, CLI, ERY, GEN, LVX, MXF, OXA, TEC, TET | 5 | ||
| 5 | HIP14300 | 16 | 32 | CIP, CLI, ERY, LVX, MXF, OXA,TEC | 5 | ||
| 6 | HIP15178 | 16 | 512 | CIP, CLI, ERY, LVX, MXF, OXA, TEC | 5 | ||
| 7 | AIS2006032 | 16 | >512 | CIP, CLI, ERY, LVX, MXF, OXA, TEC | 5 | ||
| DAP non-susceptible VAN intermediate S. aureus | 8 | HIP09433 | 16 | DAP | 4 | CIP, ERY, GEN, LVX, MXF, OXA, PEN, TMP | 45 |
| 9 | SAMER-S6 | 16 | 16 | TMP, PEN, TEC | 5 | ||
| 10 | 6820 | 16 | 8 | OXA, RIF, TEI | 5 | ||
| 11 | TTSH-478700 | 8 | 16 | CIP, LVX | 22 | ||
| 12 | TTSH-671549 | 16 | 8 | CIP, ERY, LVX | 22 | ||
| 13 | TTSH-478701 | 8 | 4 | CIP, ERY, LVX, RIF | 22 | ||
| 14 | ATCC 700789 | 16 | 4 | CIP, ERY, LVX, RIF, TOB | 5 | ||
| MDR MRSA | 15 | ATCC BAA38 | 16 | TET | 128 | PEN, STR | 8 |
| 16 | ATCC BAA39 | 16 | 128 | CIP, ERY, GEN, IPM, LVX, PEN, TMP, TOB | 8 | ||
| 17 | ATCC BAA44 | 16 | 32 | CIP, ERY, GEN, LVX, PEN, TOB | 8 | ||
CIP ciprofloxacin, CLI clindamycin, DAP daptomycin, ERY erythromycin, GEN gentamicin, IPM imipenem, LVX levofloxacin, MXF moxifloxacin, OXA oxacillin, PEN penicillin, RIF rifampicin, STR streptomycin, TEC teicoplanin, TET tetracycline, TMP trimethoprim, TOB tobramycin, VAN vancomycin
PDGu(7)-b-PBLK(13) targets the bacterial envelope
The structure of the cationic block co-beta-peptide suggests a possible mechanism of action involving membrane interaction. Confocal microscopy of fluorescently labeled bacterial cells showed that the rhodamine-labeled PDGu(7)-b-PBLK(13) accumulated preferentially in the bacteria envelope (i.e. cell wall and cell membrane) (Fig. 2a, Supplementary Fig. 29). Membrane damage was confirmed using propidium iodide (PI) as a marker of plasma membrane integrity (Fig. 2b). Results showed that both the block copolymer and cationic homopolymer are membrane active, but the copolymer induces less PI staining, suggesting that it is less membrane-lytic (Fig. 2b). DiSC35 dye assay, which probes plasma membrane potential changes, corroborated the finding that PDGu(7)-b-PBLK(13) mildly depolarized the bacterial plasma membrane, unlike the homocationic PBLK(20) that had a more pronounced effect (Fig. 2c). Together, the PI staining and DiSC35 assay results indicate that the copolymer disturbs the bacterial membrane without causing severe leakage.
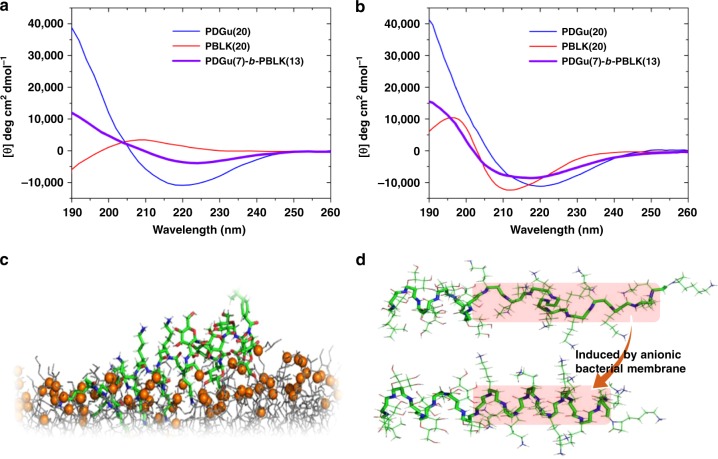
Fig. 2.
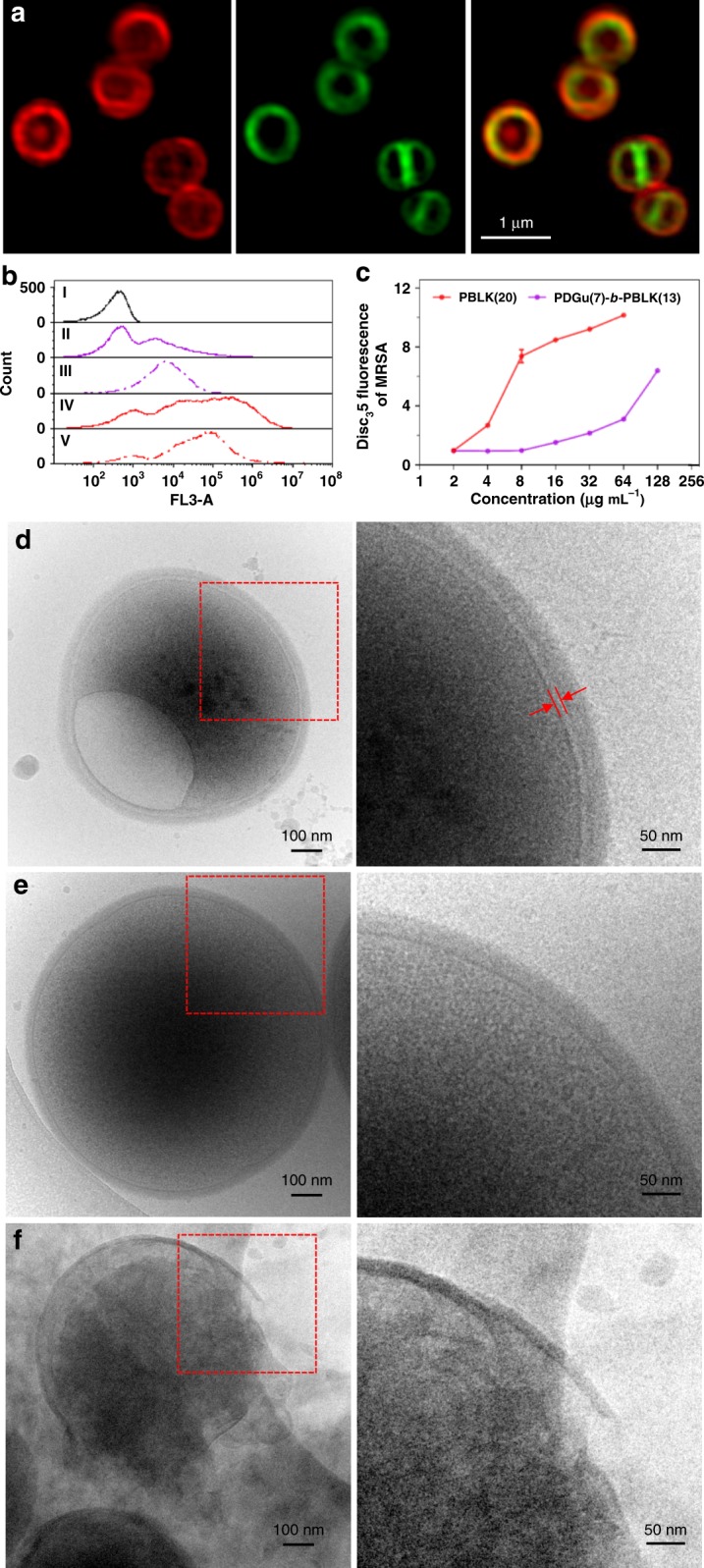
PDGu(7)-b-PBLK(13) targets bacterial cell envelope. It accumulates in MRSA USA300 cell envelope, mildly (at MIC) permeabilizing the membrane but significantly weakening cell wall/membrane attachment. a Confocal microscopy images of copolymer-treated MRSA USA300. From left to right: rhodamine-labeled copolymer channel, FM1-43-labeled bacteria membrane channel, superimposed images from both channels, respectively. b Flow cytometry study of propidium iodide-stained MRSA USA300. From top to bottom: live bacteria control, bacteria treated with 1× MIC PDGu(7)-b-PBLK(13), 4× MIC PDGu(7)-b-PBLK(13), 1× MIC PBLK(20), and 4× MIC PBLK(20). c DiSC35 membrane depolarization assay. Data are presented as mean ± standard deviation. d–f Cryo-TEM image of polymer treated MRSA USA300. d PDGu(7)-b-PBLK(13) treated bacteria with enlarged periplasmic space and vacuole structure formation; e untreated control; f PBLK(20)-treated bacteria with cell lysis
The effect of PDGu(7)-b-PBLK(13) on the morphology of S. aureus was also visualized by cryo-transmission electron microscopy (cryo-TEM), which revealed a much larger periplasmic space gap (of about 7–8 nm, Fig. 2d; indicated by red arrows), together with bleb and vacuole formation (Fig. 2d, Supplementary Fig. 30). In contrast, periplasmic gap widening, blebs, and vacuoles were not observed in untreated bacteria (Fig. 2e, Supplementary Fig. 31). Treatment with PBLK(20) led to significant bacterial envelope deformation, cell leakage, and lysis (Fig. 2f, Supplementary Fig. 32). The copolymer with its hydrophilic sugar block aggregates at the membrane interface, leading to the observed larger periplasmic gap.
Circular dichroism (CD) spectropolarimetry showed that in free solution, the block co-beta-peptide likely adopts a helix–coil conformation attributed respectively to the sugar52 and cationic53 blocks (Fig. 3a). However, in the presence of model vesicles containing anionic bacterial lipids, CD spectrum shows that the cationic block of the copolymer, like the cationic homopolymer (PBLK(20)), undergoes a transition to likely a left-handed helix structure54 (Fig. 3b, Supplementary Fig. 33a–f). Computer simulation shows that the binding of PDGu(7)-b-PBLK(13) to bacterial membrane is provided mainly by the PBLK block while the PDGu block protrudes into the water–membrane interface because of its weaker binding to the membrane and its strong hydrophilicity (Fig. 3c, Supplementary Fig. 34). In free solution, electrostatic repulsion between the lysine side chains of the cationic block causes the cationic block to exist as a random coil conformation (Supplementary Fig. 34a–d). At the anionic bacterial lipid surface, the positive charges in the PBLK block of the copolymer are neutralized by anionic bacterial lipids so that the lysine side chain charge–charge repulsion causing the distortion of the helical conformation of the copolymer PBLK block is substantially reduced and the copolymer transitions from a helix–coil structure to a helix–helix structure (Fig. 3d, Supplementary Fig. 34e–g). The resulting helix–helix structure of the sugar-cationic block copolymer binds to the anionic bacterial membrane (Supplementary Fig. 34h–k). It is known that beta peptides containing cyclic beta-amino acids adopt different helical structures to those containing non-cyclic amino acids39. From the CD data, we see that the (cyclic) PDGu spectrum exhibited a minimum at 220 nm while the (non-cyclic) PBLK spectrum exhibited a minimum at 213 nm (Fig. 3b). Our computer simulation corroborated that the PDGu and PBLK blocks adopt different helical conformations, with 3.5 residues/turn and 3 residues/turn, respectively (Supplementary Fig. 34j, Supplementary Note). (In the presence of zwitterionic lipids (model vesicles representative of mammalian membrane), the copolymer retained its helix–coil conformation (Supplementary Fig. 33c)).
Fig. 3.
Bacterial-induced secondary structure transition of PDGu(7)-b-PBLK(13). a, b Molar ellipticity [θ] CD spectra of PDGu(20) (blue), PBLK(20) (red), and PDGu(7)-b-PBLK(13) (purple) in DI (a) and in the presence of anionic POPG liposomes (b). c A snapshot of computer simulation of PDGu(7)-b-PBLK(13) binding to anionic bacterial membrane. The membrane model is colored as gray lines with the head groups of the lipid molecules shown as orange spheres. PDGu(7)-b-PBLK(13) is shown as a stick model, its carbon, oxygen, nitrogen, and hydrogen atoms are colored as green, red, blue, and white, respectively. d Computer simulation of transition from helix–coil in solution to helix–helix induced by anionic membrane
The accumulation of the block co-beta-peptide at the outer leaflet of the cytoplasmic membrane causes the increased periplasmic space visible in the cryo-TEM (Fig. 2d), which led to detachment of the cell wall from the cytoplasmic membrane and a weakened membrane–cell wall interface. The copolymer also aggregates inside the cell wall leading to defects in the cell wall function (Fig. 2a, d). The blebs observed with copolymer treatment using cryo-TEM (Supplementary Fig. 30) may be formed by membrane-bound cytoplasm herniating through cell wall defects as intracellular water expands during the freezing process of the cryo-TEM preparation55,56. The vacuoles observed (Supplementary Fig. 30) may be ice pockets formed during the cryo-TEM process as water migrates to the polymer-rich periplasmic space since the cytoplasmic membrane is detached from the cell wall. Taken together, the copolymer disturbs the cell envelope which includes the membrane, the membrane–cell wall interface, and also the cell wall.
PDGu(7)-b-PBLK(13) eradicates persister bacteria and biofilms
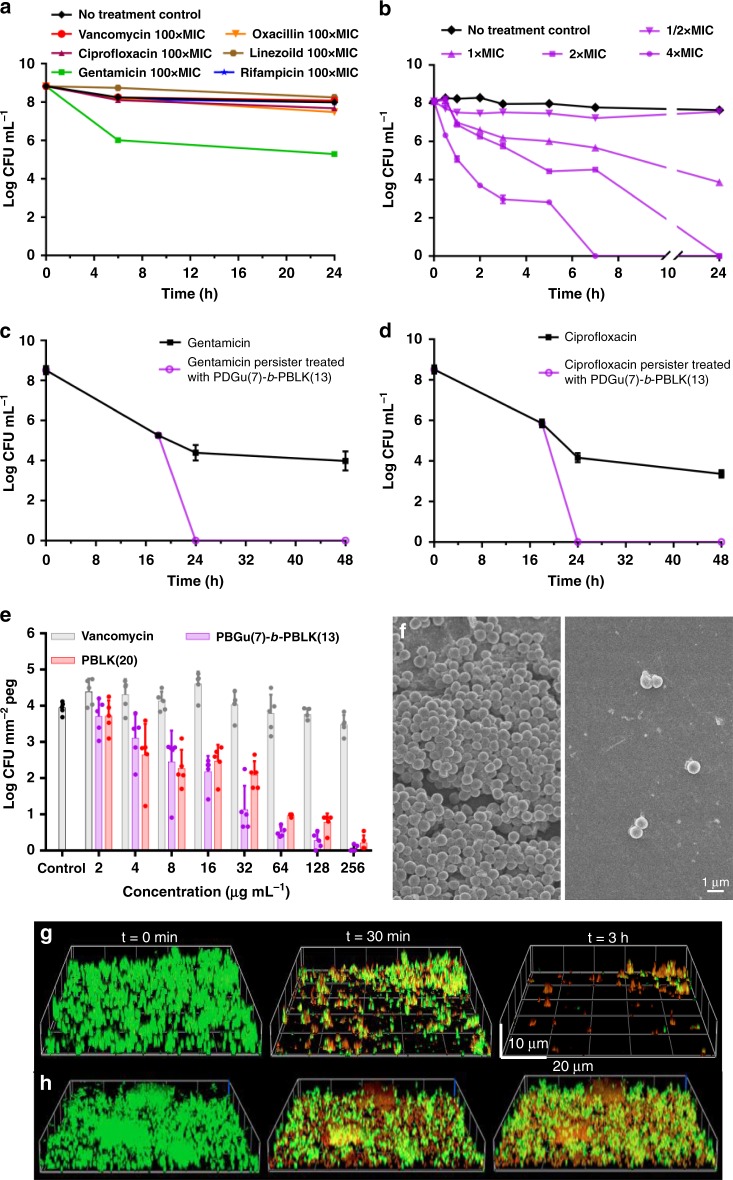
Classical antibiotics are usually significantly less potent against persisters/non-replicating bacteria1,2,6,7. Since PDGu(7)-b-PBLK(13) kills S. aureus by surface contact-induced membrane/envelope damage, we hypothesized that the block copolymer may retain potency against persisters and S. aureus biofilms18. Nutrient-starved persisters were generated by passaging S. aureus in PBS medium, a condition under which the bacteria can survive for extended periods of time without replicating. Consistent with published literature1, non-replicating S. aureus was phenotypically resistant to antibiotics from various categories (including vancomycin, oxacillin, rifampicin, etc.) up to a dose 100× MIC (Fig. 4a). Conversely, PDGu(7)-b-PBLK(13) was highly potent against non-replicating starved persister S. aureus at a concentration as low as twofold its MIC (Fig. 4b). Furthermore, PDGu(7)-b-PBLK(13) effectively eradicated antibiotic-induced persisters that escaped killing by 10× MIC gentamicin and ciprofloxacin treatment (Fig. 4c, d). PDGu(7)-b-PBLK(13) was also effective at dispersing preformed MRSA biofilms, achieving a reduction of more than 99.9% of the biofilm bacteria, greatly outperforming vancomycin, which had an insignificant effect on biofilm bacteria (Fig. 4e).
Fig. 4.
PDGu(7)-b-PBLK(13) is bactericidal toward MRSA USA300 persisters and biofilms in vitro. a Kill-kinetics of various antibiotics at 100× MIC; and b PDGu(7)-b-PBLK(13) against non-replicating MRSA USA300. c, d Kill-kinetics of PDGu(7)-b-PBLK(13) at 4× MIC against persisters generated by 10× MIC gentamicin (c) and ciprofloxacin (d) treatment. e Activity of PDGu(7)-b-PBLK(13) and PBLK(20) on established MRSA biofilms using the MBEC™ Assay. Data are presented as mean ± standard deviation. f FESEM image of MBEC™ microtiter plate pegs: (left) control peg without treatment and (right) peg treated with PDGu(7)-b-PBLK(13). g, h Confocal microscopy images of PDGu(7)-b-PBLK(13) (g) and PBLK(20) (h) treated MRSA biofilm at t = 0 min, 30 min, and 3 h. Biofilms were stained with Live/Dead BacLight™ kit
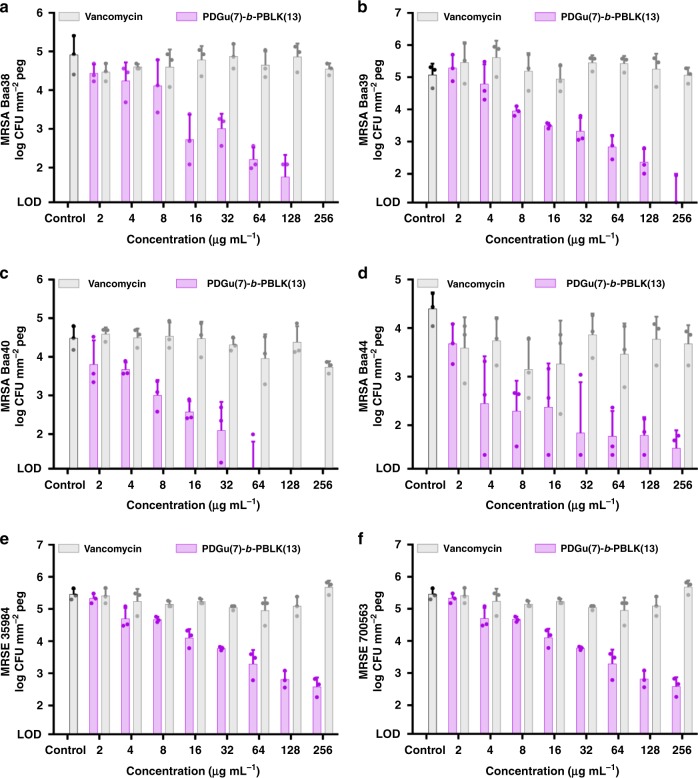
In addition to killing bacteria in biofilm, the block copolymer effectively dispersed the biofilm itself (as shown by confocal microscopy and FESEM) and the dispersed bacteria were shown to be dead (Fig. 4f, g, Supplementary Fig. 35a, b). The homocationic PBLK(20) kills biofilm bacteria but does not disperse them (Fig. 4e, h, Supplementary Fig. 35c). Copolymer aggregation at the cell wall accounts for its ability to detach bacteria from biofilm biomass since the sugar block would form a non-fouling coating around the bacteria. (CA-)MRSA USA300 maintained in a broth medium supplemented with glucose typically forms biofilm involving cell-wall anchored protein (fibronectin-binding proteins)57, whilst many (HA-)MRSA58,59 and Staphylococcus epidermidis60,61 strains form biofilms involving the polysaccharide intercellular adhesin encoded by the ica locus62. To determine if the block copolymer is active against other types of biofilms, biofilms formed by various HA-MRSA and methicillin-resistant S. epidermidis (MRSE) strains under conditions promoting the ica locus expression63 were treated with the copolymer. Our copolymer PDGu(7)-b-PBLK(13) was more active than vancomycin in eradicating the biofilms of HA-MRSA and MRSE strains (Fig. 5). Hence, our copolymer is effective not only against MRSA biofilms involving fibronectin-binding protein, but also against other major types of biofilm formed by HA-MRSA and MRSE.
Fig. 5.
PDGu(7)-b-PBLK(13) eradicates biofilms of HA-MRSA and MRSE strains. It shows dose-dependent eradication of biofilm bacteria under conditions that promote polysaccharide intercellular adhesion; y-axis: biofilm bacteria (CFU mm−2 peg) formed by different HA-MRSA strains (a ATCC BAA38, b ATCC BAA39, c ATCC BAA40, d ATCC BAA44) and MRSE strains (e ATCC 35984, f ATCC 700563). (Vancomycin is used as antibiotic control.) Data are presented as mean ± standard deviation
Copolymer is efficacious in murine and ex vivo human skin models
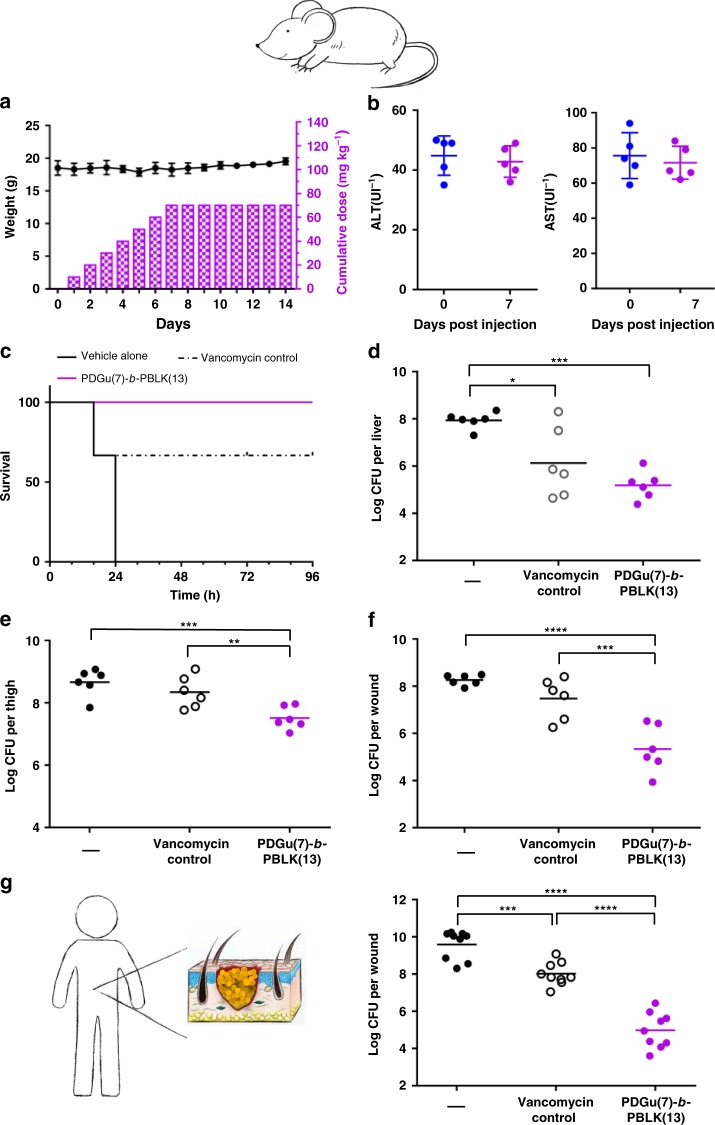
Before in vivo efficacy testing, acute toxicity of the block copolymer was evaluated in mice. Intravenous injection of PDGu(7)-b-PBLK(13) at a cumulative dose of 70 mg kg−1 (10 mg kg−1 per day × 7 days) was well tolerated in all mice, with no death observed up to 7 days post-injection (Fig. 6a). PDGu(7)-b-PBLK(13) induced no liver and kidney toxicity, confirming its low in vivo acute toxicity (Fig. 6b, Supplementary Fig. 36, Supplementary Table 3).
Fig. 6.
PDGu(7)-b-PBLK(13) is efficacious in vivo against MRSA USA300 with no toxicity. a, b In vivo repetitive toxicity of daily 10 mg kg−1 i.v. injection of PDGu(7)-b-PBLK(13) for 7 consecutive days. a Mice weight (left y-axis) and cumulative dosage (right y-axis) over 14 days. b ALT and AST biomarker changes at t = 0 and 7 days. Data are presented as mean ± standard deviation. c Survival% and d bacteria log reduction in liver in a systemic infection model. Vehicle alone (–), PDGu(7)-b-PBLK(13), or vancomycin control at 5 mg kg−1 were applied at a single dose, 2-h post infection. e In vivo antimicrobial activity of PDGu(7)-b-PBLK(13) against MRSA USA300 in a deep-seated neutropenic thigh infection model. First treatment was applied 24-h post infection at 20 mg kg−1, with a second dose at 20 mg kg−1 applied 3 h later. f In vivo antimicrobial activity of PDGu(7)-b-PBLK(13) against MRSA USA300 in an established murine excision wound model. Vehicle alone (–), PDGu(7)-b-PBLK(13), or vancomycin control at the same dosing (i.e. 2.5 mg kg−1) were applied six times over 2 days, starting 72-h post infection. g Ex vivo antimicrobial activity of PDGu(7)-b-PBLK(13) against MRSA USA300 in an established wounded human skin model. Vehicle alone (–), PDGu(7)-b-PBLK(13), or vancomycin control at 100 µg were applied three times with 3-h interval between treatments, starting 48 h post infection; **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001 by one-way ANOVA followed by Dunnett test
The in vivo efficacy of PDGu(7)-b-PBLK(13) was then evaluated in a mouse model of acute systemic infection. Mice were infected with MRSA USA300 at a lethal dose (100% death within 24 h in untreated controls). At 2 h post infection, a single 5 mg kg−1 dose of intraperitoneally (i.p.) injected copolymer resulted in 100% rescue of the mice (6/6 mice) and significantly reduced bacterial loads in major organs (Fig. 6c, d, Supplementary Fig. 37). In contrast, vancomycin treatment at the same dosage achieved only 67% survival (4/6 mice). We further evaluated the efficacy of the copolymer against persisters/biofilm with a deep-seated thigh infection model in neutropenic mice known to be particularly resistant to antibiotic treatment18,64. In this model, the copolymer achieved a 93.7% (1.2 log10) reduction in bacteria load, whereas vancomycin was ineffective (Fig. 6e). We also evaluated the efficacy of the co-beta-peptide against biofilm bacteria in a murine excision wound model. A biofilm was established in the wound with a 72-h infection period, by which time the bacteria have developed stable biofilms65,66. After the 72-h infection, copolymer treatment was given and achieved 99.87% (2.9 log10) reduction in bacterial load, which was significantly better than vancomycin (83.8%, i.e. 0.8 log10, reduction) (Fig. 6f), showing that the block copolymer has high activity even against an established S. aureus infection known to be recalcitrant to antibiotic treatment.
In addition to the murine models, we also demonstrated the efficacy of the copolymer with an ex vivo human skin model with severely established (48 h) infection (Fig. 6g). The copolymer treatment achieved 99.998% (4.6 log10) reduction of bacterial burden in contrast to the 97.3% (1.6 log10) reduction of vancomycin treatment. Copolymer treated ex vivo human wound sites were also clear of pus/debris corroborating its anti-fouling/biofilm dispersal properties (Supplementary Fig. 38).
Discussion
Eradication of persisters and biofilms remains one of the biggest challenges in antibacterial drug discovery. Antibiotic-tolerant bacteria are associated with longer treatment time and relapse of infection. Because most antibiotics target macromolecular machinery only essential for active replication, they are significantly less potent against non-replicating persisters or established biofilms. PDGu(7)-b-PBLK(13) kills non-replicating, antibiotic-tolerant persisters, and biofilm-associated MRSA, both in vitro and in vivo. We show that it can eradicate the clinically relevant CA-MRSA (USA 300). We also showed that our copolymer is just as effective against HA-MRSA strains with resistance to multiple conventional antibiotics (Table 3, Fig. 5). Multi-drug-resistant (MDR) HA-MRSA bacteria cause the majority of nosocomial bacteremia/septicemia and device-related infections involving biofilm formation. The ability of our co-beta-peptide to kill all the sub-populations (planktonic, persister and biofilm states) of MRSA bacteria is attributable to its mechanism(s) of kill—membrane disruption and interface weakening effects which are not related to metabolism. The reduced tendency of the block copolymer to bind mammalian membranes is linked to their less negatively charged surface36,67. This co-beta-peptide shows eradication of persister and biofilm MRSA and has ultra-low toxicity, both of which were shown using in vivo murine models. Further, it also shows eradication of an established infection with an ex vivo human skin model.
Upon surface-contact with bacterial membrane, the cationic block undergoes transition from a random coil in free solution to a helix. The block copolymer possesses a unique bacteria-triggered surfactant effect that contributes to biofilm dispersal—the cationic block adsorbs onto the negatively charged bacterial envelope while the hydrophilic sugar block has a strong tendency to promote dissolution, resulting in a “surfactant-like” solvation of bacteria from biofilm. The block copolymer forms an anti-fouling PDGu layer around the bacteria. Conversely, the homocationic PBLK(20) led to pore formation (Fig. 2b, c, f), like other AMPs36, but without promoting biofilm detachment (Fig. 4h); this is probably linked to the inability of the homocationic polymer to form an anti-fouling layer around the bacteria. The amine group of the cationic block dominates the interactive topology with erythrocytes but its hydrophilicity minimizes hemolysis. Further, the neutral sugar block also increases the hydrophilicity of the block copolymer. Other antimicrobial peptides and biosurfactants (such as surfactin, rhamnolipid, or phenol-soluble modulins) are intrinsically amphiphilic with exposed hydrophobic domains in free solution and are typically hemolytic since their freely exposed hydrophobic moieties would interact with erythrocytes68.
Biofilm eradication using conventional antibiotics is typically challenging69,70. The limited efficacy of vancomycin against biofilms is not an exception; many other antibiotics that are commonly used for MRSA infection have significantly reduced efficacy against biofilm bacteria71. Besides antibiotics, our copolymer outperforms many cationic antimicrobial peptides and conventional antiseptic therapeutics in established wound infections that have been previously reported19,72, not to mention its superior safety profile that makes it suitable for translation into clinics. The copolymer eradicates biofilm MRSA and also disperses the biomass. Since the block copolymer forms an anti-fouling PDGu coating around the bacterial cell envelope, the adhesion of the bacteria to extracellular polymeric substances (EPSs) and to substrates is reduced, explaining the strong dispersing effect of PDGu(7)-b-PBLK(13) on biofilms (Fig. 4e). The coated bacteria can effectively detach due to reduced surface hydrophobicity and interaction with biofilm matrix, leading to biofilm dispersal73–75. Moreover, the copolymer biofilm eradication effect is observed in the major types of biofilms formed by different MRSA (and MRSE) strains under various conditions, which include the types involving fibronectin binding protein as well as polysaccharide intercellular adhesin. This is clinically significant since MRSA is a common pathogen that forms biofilms during infection, as well as on medical devices15,76.
The block co-beta-peptide (PDGu(7)-b-PBLK(13)) demonstrates excellent bactericidal efficacy against all MRSA sub-populations, i.e. replicating, biofilm-associated, and antibiotic-induced persister bacteria. It is active against CA-MRSA (USA300) and numerous other MDR HA-MRSA. The cationic block co-beta-peptide undergoes a bacterial-membrane-triggered conformation change from a random coil to likely a helix. Its antibacterial activity in established MRSA murine infection models is superior to that of vancomycin, and it exhibits no acute in vivo toxicity in repeated dosing studies at levels above those required for therapeutic efficacy. Further, the copolymer effectively eradicates established MRSA infections in an ex vivo human skin model. It also kills biofilm bacteria while effectively dispersing the biofilm mass of CA-MRSA; it also shows efficacy against the major types of biofilms formed by HA-MRSA. It acts as a bacteria-triggered surfactant leading to biofilm dispersal. As resistance toward all classes of antibiotics rapidly evolves and spreads77, the outstanding efficacy of PDGu(7)-b-PBLK(13) against S. aureus persisters and biofilms, as well as its excellent safety window, makes this block co-beta-peptide a valuable candidate to treat MRSA infections.
Methods
General procedure for the polymerization of β-lactams
In a nitrogen-purged glovebox, a mixture of two β-lactams (BLKp and DGup) dissolved in tetrahydrofuran with a defined molar ratio was placed into a Schlenk tube equipped with a magnetic stirrer (Fig. 1a). Then, 4-t-butylbenzoyl chloride (tBuBzCl, 5 mol% with respect to the total amount of β-lactam) was added. The Schlenk tube was sealed, removed from the glove box, and cooled to −30 °C under argon atmosphere. To the stirring reaction solution was then slowly added a premade stock solution of lithium bis(trimethylsilyl)amide (LiHMDS, 12.5 mol% with respect to the total amount of β-lactam). The resulting mixture was stirred at −30 °C for about 8 h until the reaction was finished (monitored by TLC) and was then quenched with methanol. After completion, a white solid was precipitated by adding hexane (40 mL). The mixture was centrifuged and the supernatant solution was decanted. After two more repetitions of the precipitation/centrifugation procedure, the white pellet was dried overnight under a nitrogen stream to yield the protected product PDGup(x)-b-PBLKp(y) as a white powder.
General procedure for the debenzylation of PDGup(10)-b-PBLKp(10)
Polymer PDGup(10)-b-PBLKp(10) (145 mg) and 54 mg (0.48 mmol, ~1.2 equiv. to monomers) of potassium tert-butoxide (KOt-Bu) were dissolved in 5.0 mL of tetrahydrofuran. The polymer solution was added dropwise to a rapidly stirred solution of sodium (160 mg, 7.0 mmol) in liquid ammonia (15 mL) at −78 °C under nitrogen. The reaction mixture was warmed to −55 °C and maintained at this temperature for about 2 h, after which a saturated aqueous solution of ammonium chloride (NH4Cl, 10 mL) was added to quench the reaction. The solution was warmed to room temperature in a water bath to evaporate the ammonia. The resulting clear solution was filtered, washed with DI water, and dialyzed with 1000 MWCO tubing for 36 h with ten water changes. After lyophilization, PDGu(7)-b-PBLK(13) was obtained as an amorphous white solid. Copolymers (PDGup(x)-b-PBLKp(y)) with other design block lengths (x, y) were synthesized by a similar procedure.
Reaction kinetics studies
High-performance liquid chromatography (HPLC) was employed to determine β-lactam consumption. For these measurements, a series of reactions was performed with identical conditions (temperature: −30 °C, initial concentration: [DGup] = 0.05 M, [BLKp] = 0.05 M, activator [ArCOCl] = 0.005 M, inititator [LiHMDS] = 0.0125 M) but quenched at different times. After purification by flash column chromatography, concentrated reaction mixtures were mixed with a certain amount of paraben (internal standard) and diluted with acetonitrile to the same volume. Aliquots of these solutions were transferred to vials and injected into a Shimadzu LC-20AD HPLC workstation equipped with an IB column. Monomer concentration was calculated from the peak area ratio relative to a known amount of internal standard. GPC curves were determined versus polystyrene standards using dimethylformamide (1 mg mL−1 LiBr) as the eluent at a flow rate of 1.0 mL min−1 through two Styragel columns (HR5 and HR5E, 7.8 × 300 mm) in series at 40 °C with a refractive index detector.
Bacterial strains
All bacteria strains of Table 2, Strains #14–17 of Table 3, and all bacteria strains of Fig. 5 were purchased from ATCC. Vancomycin-resistant S. aureus (Strains #1–7 of Table 3) were kindly provided by Prof. Barry N. Kreiswirth and Dr. José R. Mediavilla from the Center for Discovery and Innovation, Hackensack Meridian Health (USA). Daptomycin non-susceptible vancomycin-intermediate MRSA (Strains #8 and 9 of Table 3) were kindly provided by BEIresources.org. Strain #10 of Table 3 was kindly provided by Dr. Adriana Rosato from the Houston Methodist Research Institute (USA). Strains #11–13 of Table 3 were kindly provided by Tan Tock Seng Hospital (TTSH, Singapore).
Multilocus sequence typing (MLST) characterization for the three VISA strains from local hospital (Strains #11–13 of Table 3) were conducted. Overnight culture from single colony was washed with 10 mM Tris buffer, resuspended in 800 µL of lysis buffer containing 5 mg mL−1 lysozyme, 10 mM EDTA, and 10 mM Tris. After 1 h incubation at 37 °C with shaking, the suspension was heated to 95 °C for 10 min and subsequently transferred to ice. 1 mL of ice-cold phenol/chloroform/isoamyl alcohol (25:24:1) was added and mixed thoroughly by inverting the tubes five times, followed by incubation for 5 min on ice. After centrifugation at 20,000×g for 20 min, the aqueous layer was transferred to a fresh tube and DNA was precipitated by adding 1 mL of ice-cold ethanol, followed by incubation for 15 min on ice. The DNA pellet was collected by centrifugation and washed once with ice-cold 70% ethanol, and resuspended in 50 µL of water. The extracted DNA were amplified by PCR using Novagen KOD Hot Start DNA Polymerase, and the amplified products were sequenced by Sanger sequencing. The obtained sequence was submitted to MLST database (http://www.mlst.net/) to obtain the sequence type (ST).
MIC determination
Bacteria in logarithmic phase of growth were diluted to 1 × 106 colony-forming units (CFU) per milliliter in Mueller Hinton Broth (MHB, Difco®). Polymers were dissolved at 10.24 mg mL−1 in deionized water and diluted to desired concentration in MHB using twofold serial dilution in a 96-well plate (NuncTM). A total of 50 µL of bacteria in MHB suspension were added to 50 µL of polymer to achieve a final volume of 100 µL per well. The plate was incubated aerobically at 37 °C for 18 h, and the optical density of each well was measured at a wavelength of 600 nm (TECAN, infinite F200). MIC90 is defined as the lowest concentration that exhibited more than 90% inhibition of the bacteria growth. All tests were performed three times independently with two samples in each test. For tests involving daptomycin, 50 µg mL−1 CaCl2 is supplemented to the medium.
MTT cytotoxicity test
Mouse fibroblasts (3T3 cells) were purchased from ATCC. Cells were seeded at 2 × 104 cells per well in a volume of 200 μL of Dulbecco’s Modified Eagle’s medium (DMEM, GibcoTM) in a 96-well tissue culture plate, and incubated at 37 °C in a humidified incubator with 5% CO2 for 24 h. Polymer stock solution was prepared in PBS (phosphate-buffered saline, GibcoTM) at a concentration of 10 mg mL−1 and diluted to desired concentrations in DMEM complete medium. Polymer in DMEM solution was added into the cell-seeded 96-well plate and incubated at 37 °C for 24 h. Subsequently cells were rinsed with PBS and 1 mg mL−1 MTT in DMEM was added into each well. The plate was incubated for 4 h, after which the MTT solution was aspirated and 100 μL of dimethyl sulfoxide was added into each well. The plate was shaken at 150 rpm for 10 min and the absorbance of each well was measured at 570 nm using a microplate reader spectrophotometer (BIO-RAD, Benchmark Plus). Cell viability was calculated using the following formula and IC50 was interpolated using mean values of triplicate measurements.
| 1 |
Hemolysis assay
The human blood hemolysis experiment was reviewed and approved by the Institutional Review Board of Nanyang Technological University (IRB-2015-03-040). Human blood samples were obtained from a healthy donor (age 23, male) and informed consent was given in accordance with NTU-IRB ethical principles. Fresh human blood was washed with PBS twice and red blood cells were resuspended to 5% v/v in PBS. Polymers were twofold serial diluted in PBS and 50 µL of polymer solution samples were mixed with red blood cell suspension in a 96-well plate. The plate was incubated for 1 h at 37 °C under mild shaking. The microplate was centrifuged at 1000 rpm for 10 min; 80-µL aliquots of the supernatant were then transferred to a new 96-well microplate and diluted with another 80 µL of PBS. Hemolytic activity was calculated from absorbance measured at 540 nm using a microplate reader spectrophotometer (Benchmark Plus, BIO-RAD):
| 2 |
where Op is the absorbance of polymer, Ob is the absorbance of negative control, and Ot is the absorbance of positive control. HC10 values (concentration that causes 10% hemolysis) were interpolated using mean values of triplicate measurements.
Kill kinetics of non-replicating/antibiotic-generated persisters
A culture of MRSA USA300 was washed two times with PBS and resuspended in PBS at a final concentration of 108 CFU mL−1. The bacteria suspension was incubated in PBS for 1 h to adapt the cells to starvation. Polymer and antibiotic were added to 1 mL of bacteria in PBS suspension in Eppendorf tubes to achieve a desired final polymer/antibiotic concentration. The Eppendorf tubes were incubated aerobically under shaking at 37 °C. At desired time points, 20 µL of each sample was serial diluted in PBS, and plated on nutrient agar plates for CFU determination. For killing of persister bacteria that escaped standard antibiotic treatment, 108 CFU log-phase bacteria in 1 mL of MHB were challenged with antibiotics (ciprofloxacin or gentamicin) at 10× MIC for 18 h. Half of the bacteria were washed to remove antibiotics and challenged with copolymer at 4× MIC in MHB. The other half continued under challenge with antibiotics as a control. Aliquots of samples at each time point were washed with PBS twice to remove antibiotics/polymers and serial diluted in PBS to determine CFU. Error bars were produced from two independent tests, with duplicate samples for each test.
Spontaneous mutation frequency
At day 1, initial inocula of 3.5 × 109 CFU exponential-phase MRSA USA300 in 10 mL of MHB were placed in 50-mL falcon tubes and challenged with polymer at 10× MIC under shaking at 37 °C. Polymer was changed every 48 h during the incubation. The OD600nm values were recorded daily over 6 days. At days 3 and 6, 100 μL of the sample was serially diluted in PBS and plated on nutrient agar plates for CFU determination.
Resistance evolution by serial passage
Exponential-phase MRSA USA300 (106 CFU) were grown in 1 mL of MHB containing copolymer or antibiotic control ciprofloxacin at a gradient of concentrations: 0.25× MIC, 0.5× MIC, 1× MIC, 2× MIC, and 4× MIC. At 24-h intervals, the cultures were checked for growth and the MIC value for each day was recorded. Cultures from the second highest concentrations that allowed growth (OD600 ≥ 1) were diluted 1:1000 into fresh MHB containing 0.25× MIC, 0.5× MIC, 1× MIC, 2× MIC, and 4× MIC of copolymer/ciprofloxacin. The serial passaging was repeated daily for 14 days. Three independent biological replicates were conducted for each experiment.
Microscopic studies
Log-phase MRSA USA300 bacteria were washed and diluted to 108 CFU mL−1 in PBS and incubated with polymer at 37 °C for 4 h. The bacteria suspension was centrifuged and resuspended in PBS for cryo-TEM imaging. For confocal imaging, 108 CFU mL−1 log-phase bacteria were incubated with rhodamine-labeled polymer for 1 h and subsequently stained with membrane dye FM1-43FX before confocal microscopy imaging.
Bacterial membrane integrity assays
Log-phase bacteria (MRSA USA300) were washed and diluted to 108 CFU mL−1 in PBS and incubated with polymer at 37 °C for 0.5 or 1.5 h, and stained with PI (L13152 Invitrogen). Samples were washed twice and resuspended in PBS to 107 CFU mL−1 and analyzed using flow cytometry (BD Accuri C6 plus). Data are plotted as normalized histogram of fluorescence intensity from FL3 channel. For DiSC35 membrane depolarization assay, log-phase bacteria were washed and resuspended to 107 CFU mL−1 in 5 mM HEPES buffer (pH 7.8) containing 20 mM glucose and 0.1 M KCl. DiSC35 solution was added to bacteria suspension to achieve a final concentration of 100 nM and allowed to quench for 30 min. Polymer solution was added to achieve the desired concentration in a black 96-well plate (Costar). Fluorescence readings were recorded 5 min after polymer addition with a Tecan reader at an excitation wavelength of 622 nm and an emission wavelength of 670 nm.
Biofilm assays
A total of 150 µL of MRSA USA300 bacteria in tryptic soy broth (TSB) containing 1% glucose (initial inoculum of 106 CFU per well) was added into each well of an MBEC plate (Innovotech, Canada). After 24 h of incubation at 37 °C under mild shaking, the pegs were washed twice using 200 µL of PBS and transferred to a 96-well plate containing a twofold dilution series of polymer in PBS (200 µL per well). MBEC pegs were exposed to the polymer for 3.5 h, and subsequently washed before sonication-releasing the biofilm bacteria into the recovery plate for CFU counting. For FESEM imaging of pegs, untreated control and copolymer-treated (64 µg mL−1 for 3.5 h) pegs were removed aseptically, fixed with 4% paraformaldehyde at 4 °C overnight, and dehydrated using a graded ethanol series. For confocal imaging, 24-h preformed biofilm was established in a collagen-coated glass-bottom Petri dish (MatTek). Biofilm was stained with BacLight™ live/dead kit. Polymer in PBS solution (32 µg mL−1) was dropwise added to avoid physical disturbances to biofilm and confocal images were taken immediately after polymer addition and at defined times thereafter. For biofilms formed with polysaccharide intercellular adhesin, various strains of HA-MRSA and MRSE biofilms were established under high-salt conditions using TSB + 4% NaCl.
Secondary structure study
Polymers were dissolved at 0.05 mg mL−1 in different media, i.e. DI water, 10 mM phosphate buffer (pH 2.6–8.7), 20 mM carbonate buffer (pH 10.8) and in the presence of 1 mg mL−1 POPG or POPC liposomes. (For the POPG liposomes, the polymer:lipid (P:L) molar ratios of PBLK(20), PDGu(20), and PDGu(7)-b-PBLK(13) are 1:79, 1:83, and 1:89, respectively.) CD spectra were measured from 190 to 260 nm with 0.5-nm step size, with each measurement performed twice. The final data are presented as the mean values after background extraction.
Mouse model of MRSA USA300 infection and in vivo toxicity
The animal experiments were reviewed and approved by the Animal Ethics and Welfare Committee (AEWC) of Ningbo University. A single 5-mm-diameter excision wound was created on female C57BL6 mice and inoculated with 2.5 µL of MRSA USA300 suspended in PBS (5 × 105 CFU mL−1). For treatment initiated at 72 h post-infection, treatments were applied in total six times over 2 days, i.e. three times per day with 4 h between each treatment. Samples were harvested 4 h after the last treatment to determine the CFU. For systemic infection, 108 CFU mL−1 MRSA USA300 in PBS (with 5% mucin) were i.p. injected into female balb/c mice and the infections were allowed to develop for 2 h. At 2 h post infection, treatment was injected i.p. at 5 mg kg−1 in 200 μL of PBS. Twenty-four hours post infection, the mice were sacrificed and i.p. fluid, liver, kidney, and spleen were harvested and homogenized to determine the CFU. For survival test, mice were monitored up to 96 h post infection/treatment. For deep-seated thigh infection, neutropenic ICR mice were infected by injecting 50 μL of stationary-phase MRSA USA300 in PBS (105 CFU per thigh) into thigh muscles and infection was developed for 24 h. Mice were treated with 20 mg kg−1 copolymer or antibiotic subcutaneously; 3 h later, a second treatment was given by the same route. Thigh tissues were harvested 24 h post first treatment and homogenized to determine the CFU. For in vivo toxicity determination, 10 mg kg−1 of the block copolymer in 200 μL of PBS was injected into female balb/c via tail vein daily for 7 days. Clinical biomarkers were recorded before, 24 h after, 3 days after, and 7 days after the first injection. For histological analysis, mice were sacrificed at 48 h post final injection and tissues were harvested for H&E staining and examination.
Ex vivo wounded human skin infection model
Human skin samples were purchased from Biopredic International. All samples were obtained from healthy donors undergoing cosmetic surgery and informed consent was given in accordance with French law and ethical principles. Five-millimeter-diameter wounds were created and inoculated with 10 µL of MRSA USA300 (2 × 109 CFU mL−1). Infections were developed for 48 h and wound sites were gently rinsed with PBS to remove planktonic bacteria; PBS vehicle alone, 100 µg of vancomycin, or copolymer were applied three times with a 3-h interval between each treatment. Three hours post last treatment, the samples were harvested and homogenized for CFU determination.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Dr. Adriana Rosato for generously sharing strain 6820. We thank Prof. Angelika Gründling and Dr. Moon Tay for their scientific discussions on the manuscript. We thank the funding support from a Singapore Ministry of Education Tier 3 grant (MOE2013-T3-1-004) and a Singapore Ministry of Health Industry Alignment Fund (NMRC/MOHIAFCAT2/003/2014).
Author contributions
K.Z., Y.D., K.P., and M.B.C. conceived the project and wrote the manuscript. Y.D., Z.S., and C.R. synthesized the polymers. K.Z., S.R., and L.R. conducted the in vitro biological tests. K.Z., S.J., and D.K. conducted the in vivo tests. K.Z. and M.E.T. conducted the ex vivo human skin test. Y.L. conducted the computer simulation study. K.P. and M.B.C. supervised and guided the overall research. M.B.C. and G.C.B. guided chemical synthesis. J.R. and Y.Z. supervised the in vivo toxicity tests. Y.M. supervised the computer simulation study. K.C.T. supervised the physical property study. K.M., P.P.D., and O.N. isolated and provided strains from local hospital TTSH and provided useful suggestions. J.R.M. and B.N.K. isolated relevant vancomycin-resistant MRSA strains and conducted the susceptibility tests. H.D., X.L., Y.R.C., and P.T.H participated in the supervision of the project. All authors discussed the results and commented on the manuscript.
Data availability
The data that support the findings of this study are available from the corresponding authors on request. The source data underlying Figs. 4e, 5a–f, 6a, b and 6d–g, Supplementary Fig. 37a–d and Supplementary Table 3 are provided in the Source Data file.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Timothy Foster and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kaixi Zhang, Yu Du, Zhangyong Si, Yang Liu.
Contributor Information
Kevin Pethe, Email: kevin.pethe@ntu.edu.sg.
Mary B. Chan-Park, Email: mbechan@ntu.edu.sg
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-019-12702-8.
References
- 1.Foster TJ. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017;41:430–449. doi: 10.1093/femsre/fux007. [DOI] [PubMed] [Google Scholar]
- 2.Conlon BP, et al. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 2016;1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 3.Foster TJ, Geoghegan JA, Ganesh VK, Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014;12:49. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harms A, Maisonneuve E, Gerdes K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science. 2016;354:aaf4268. doi: 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 5.Conlon, B. P., Rowe, S. E. & Lewis, K. in Biofilm-Based Healthcare-Associated Infections 1–9 (Springer, 2015).
- 6.Fisher RA, Gollan B, Helaine S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017;15:453. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 7.Maisonneuve E, Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 8.Organization, W. H. Global Priority List of Antibiotic-resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva: World Health Organization; 2017. [Google Scholar]
- 9.Control, C. f. D. & Prevention. Antibiotic Resistance Threats in the United States, 2013. (Centres for Disease Control and Prevention, US Department of Health and Human Services, 2013).
- 10.Organization, W. H. Antimicrobial Resistance: Global Report on Surveillance (World Health Organization, 2014).
- 11.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dantes R, et al. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern. Med. 2013;173:1970–1978. doi: 10.1001/jamainternmed.2013.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Hal SJ, et al. Predictors of mortality in Staphylococcus aureus bacteremia. Clin. Microbiol. Rev. 2012;25:362–386. doi: 10.1128/CMR.05022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daum RS. Skin and soft-tissue infections caused by methicillin-resistant Staphylococcus aureus. New Engl. J. Med. 2007;357:380–390. doi: 10.1056/NEJMcp070747. [DOI] [PubMed] [Google Scholar]
- 15.Boucher H, Miller LG, Razonable RR. Serious infections caused by methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 2010;51:S183–S197. doi: 10.1086/653519. [DOI] [PubMed] [Google Scholar]
- 16.Klein EY, et al. Trends in methicillin-resistant Staphylococcus aureus hospitalizations in the United States, 2010-2014. Clin. Infect. Dis. 2017;65:1921–1923. doi: 10.1093/cid/cix640. [DOI] [PubMed] [Google Scholar]
- 17.Rose WE, Poppens PT. Impact of biofilm on the in vitro activity of vancomycin alone and in combination with tigecycline and rifampicin against Staphylococcus aureus. J. Antimicrob. Chemother. 2008;63:485–488. doi: 10.1093/jac/dkn513. [DOI] [PubMed] [Google Scholar]
- 18.Kim W, et al. A new class of synthetic retinoid antibiotics effective against bacterial persisters. Nature. 2018;556:103–107. doi: 10.1038/nature26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Breij A, et al. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 2018;10:eaan4044. doi: 10.1126/scitranslmed.aan4044. [DOI] [PubMed] [Google Scholar]
- 20.Dostert, M., Belanger, C. R. & Hancock, R. E. Design and assessment of anti-biofilm peptides: steps toward clinical application. J. Innate Immun.11, 1–12 (2018). [DOI] [PMC free article] [PubMed]
- 21.Avan I, Hall CD, Katritzky AR. Peptidomimetics via modifications of amino acids and peptide bonds. Chem. Soc. Rev. 2014;43:3575–3594. doi: 10.1039/c3cs60384a. [DOI] [PubMed] [Google Scholar]
- 22.Chongsiriwatana NP, et al. Peptoids that mimic the structure, function, and mechanism of helical antimicrobial peptides. Proc. Natl. Acad. Sci. USA. 2008;105:2794–2799. doi: 10.1073/pnas.0708254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porter EA, Wang X, Lee H-S, Weisblum B, Gellman SH. Antibiotics: non-haemolytic β-amino-acid oligomers. Nature. 2000;404:565. doi: 10.1038/35007145. [DOI] [PubMed] [Google Scholar]
- 24.Wu H, et al. Design and synthesis of unprecedented cyclic γ-AApeptides for antimicrobial development. Chem. Sci. 2012;3:2570–2575. doi: 10.1039/c2sc20428b. [DOI] [Google Scholar]
- 25.Radzishevsky IS, et al. Improved antimicrobial peptides based on acyl-lysine oligomers. Nat. Biotechnol. 2007;25:657. doi: 10.1038/nbt1309. [DOI] [PubMed] [Google Scholar]
- 26.Seebach D, et al. Biological and pharmacokinetic studies with β-peptides. CHIMIA International. J. Chem. 1998;52:734–739. [Google Scholar]
- 27.Hintermann T, Seebach D. The biological stability of β-peptides: no interactions between α-and β-peptidic structures? CHIMIA International. J. Chem. 1997;51:244–247. [Google Scholar]
- 28.Porter EA, Weisblum B, Gellman SH. Mimicry of host-defense peptides by unnatural oligomers: antimicrobial β-peptides. J. Am. Chem. Soc. 2002;124:7324–7330. doi: 10.1021/ja0260871. [DOI] [PubMed] [Google Scholar]
- 29.Liu D, DeGrado WF. De novo design, synthesis, and characterization of antimicrobial β-peptides. J. Am. Chem. Soc. 2001;123:7553–7559. doi: 10.1021/ja0107475. [DOI] [PubMed] [Google Scholar]
- 30.Epand RF, Raguse L, Gellman SH, Epand RM. Antimicrobial 14-helical β-peptides: potent bilayer disrupting agents. Biochemistry. 2004;43:9527–9535. doi: 10.1021/bi049414l. [DOI] [PubMed] [Google Scholar]
- 31.Webb AI, et al. T cell determinants incorporating β-amino acid residues are protease resistant and remain immunogenic in vivo. J. Immunol. 2005;175:3810–3818. doi: 10.4049/jimmunol.175.6.3810. [DOI] [PubMed] [Google Scholar]
- 32.Seebach D, Gardiner J. β-Peptidic peptidomimetics. Acc. Chem. Res. 2008;41:1366–1375. doi: 10.1021/ar700263g. [DOI] [PubMed] [Google Scholar]
- 33.Checco JW, et al. Targeting diverse protein–protein interaction interfaces with α/β-peptides derived from the Z-domain scaffold. Proc. Natl. Acad. Sci. USA. 2015;112:4552–4557. doi: 10.1073/pnas.1420380112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nadithe V, et al. Screening nylon-3 polymers, a new class of cationic amphiphiles, for siRNA delivery. Mol. Pharm. 2014;12:362–374. doi: 10.1021/mp5004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sparr C, et al. Improved efficacy of fosmidomycin against Plasmodium and Mycobacterium species by combination with the cell-penetrating peptide octaarginine. Antimicrob. Agents Chemother. 2013;57:4689–4698. doi: 10.1128/AAC.00427-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fjell CD, Hiss JA, Hancock REW, Schneider G. Designing antimicrobial peptides: form follows function. Nat. Rev. Drug Discov. 2011;11:37. doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 37.Cheng RP, Gellman SH, DeGrado WF. β-Peptides: from structure to function. Chem. Rev. 2001;101:3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 38.Seebach D, Abele S, Gademann K, Jaun B. Pleated sheets and turns of β‐peptides with proteinogenic side chains. Angew. Chem. Int. Ed. 1999;38:1595–1597. doi: 10.1002/(SICI)1521-3773(19990601)38:11<1595::AID-ANIE1595>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 39.Appella DH, et al. Residue-based control of helix shape in beta-peptide oligomers. Nature. 1997;387:381. doi: 10.1038/387381a0. [DOI] [PubMed] [Google Scholar]
- 40.Gopalan RD, Del Borgo MP, Mechler AI, Perlmutter P, Aguilar M-I. Geometrically precise building blocks: the self-assembly of β-peptides. Chem. Biol. 2015;22:1417–1423. doi: 10.1016/j.chembiol.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Fernandez-Santin J, Aymamí J, Rodríguez-Galán A, Munoz-Guerra S, Subirana J. A pseudo α-helix from poly (α-isobutyl-L-aspartate), a nylon-3 derivative. Nature. 1984;311:53. doi: 10.1038/311053a0. [DOI] [Google Scholar]
- 42.García-Alvarez M, León S, Alemán C, Campos JL, Muñoz-Guerra S. Helical nylons 3. Synthesis and crystal structure of poly (β-L-aspartate) s with branched alkyl side chains. Macromolecules. 1998;31:124–134. doi: 10.1021/ma970665u. [DOI] [Google Scholar]
- 43.Fernandez-Santin J, et al. Helical conformations in a polyamide of the nylon-3 family. Macromolecules. 1987;20:62–68. doi: 10.1021/ma00167a013. [DOI] [Google Scholar]
- 44.Mowery BP, et al. Mimicry of antimicrobial host-defense peptides by random copolymers. J. Am. Chem. Soc. 2007;129:15474–15476. doi: 10.1021/ja077288d. [DOI] [PubMed] [Google Scholar]
- 45.Mowery BP, Lindner AH, Weisblum B, Stahl SS, Gellman SH. Structure− activity relationships among random nylon-3 copolymers that mimic antibacterial host-defense peptides. J. Am. Chem. Soc. 2009;131:9735–9745. doi: 10.1021/ja901613g. [DOI] [PubMed] [Google Scholar]
- 46.Liu R, et al. Structure–activity relationships among antifungal nylon-3 polymers: identification of materials active against drug-resistant strains of Candida albicans. J. Am. Chem. Soc. 2014;136:4333–4342. doi: 10.1021/ja500036r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu R, et al. Tuning the biological activity profile of antibacterial polymers via subunit substitution pattern. J. Am. Chem. Soc. 2014;136:4410–4418. doi: 10.1021/ja500367u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monecke S, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE. 2011;6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kosowska-Shick K, et al. Single-and multistep resistance selection studies on the activity of retapamulin compared to other agents against Staphylococcus aureus and Streptococcus pyogenes. Antimicrob. Agents Chemother. 2006;50:765–769. doi: 10.1128/AAC.50.2.765-769.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’neill AJ, Cove JH, Chopra I. Mutation frequencies for resistance to fusidic acid and rifampicin in Staphylococcus aureus. J. Antimicrob. Chemother. 2001;47:647–650. doi: 10.1093/jac/47.5.647. [DOI] [PubMed] [Google Scholar]
- 51.Ling LL, et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chin SL, et al. Combined molecular dynamics simulations and experimental studies of the structure and dynamics of poly-amido-saccharides. J. Am. Chem. Soc. 2016;138:6532–6540. doi: 10.1021/jacs.6b01837. [DOI] [PubMed] [Google Scholar]
- 53.Abele S, Guichard G, Seebach D. (S)-β3-homolysine-and (S)-β3-homoserine-containing β-peptides: CD spectra in aqueous solution. Helv. Chim. Acta. 1998;81:2141–2156. doi: 10.1002/(SICI)1522-2675(19981216)81:12<2141::AID-HLCA2141>3.0.CO;2-5. [DOI] [Google Scholar]
- 54.Kritzer JA, et al. Relationship between side chain structure and 14-helix stability of β3-peptides in water. J. Am. Chem. Soc. 2005;127:167–178. doi: 10.1021/ja0459375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown L, Wolf JM, Prados-Rosales R, Casadevall A. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015;13:620. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pogliano, J., Pogliano, N. & Silverman, J. Daptomycin mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J. Bacteriol.194, 4494–4504 (2012). [DOI] [PMC free article] [PubMed]
- 57.O’Halloran DP, McCourt J, Geoghegan JA, McCarthy H, O’Gara JP. Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol. Lett. 2014;353:157–164. doi: 10.1111/1574-6968.12424. [DOI] [PubMed] [Google Scholar]
- 58.Vergara-Irigaray M, et al. Relevant role of fibronectin-binding proteins in staphylococcus aureus biofilm-associated foreign-body infections. Infect. Immun. 2009;77:3978–3991. doi: 10.1128/IAI.00616-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Formosa-Dague C, et al. Sticky matrix: adhesion mechanism of the staphylococcal polysaccharide intercellular adhesin. ACS Nano. 2016;10:3443–3452. doi: 10.1021/acsnano.5b07515. [DOI] [PubMed] [Google Scholar]
- 60.Rohde H, et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28:1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 61.Vuong C, et al. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 2004;6:269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 62.Cramton SE, Gerke C, Schnell NF, Nichols WW, Götz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zapotoczna M, O’Neill E, O’Gara JP. Untangling the diverse and redundant mechanisms of Staphylococcus aureus biofilm formation. PLoS Pathog. 2016;12:e1005671. doi: 10.1371/journal.ppat.1005671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conlon B, et al. Killing persister cells and eradicating a biofilm infection by activating the ClpP protease. Nature. 2013;503:365. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Percival SL, et al. A review of the scientific evidence for biofilms in wounds. Wound Repair Regen. 2012;20:647–657. doi: 10.1111/j.1524-475X.2012.00836.x. [DOI] [PubMed] [Google Scholar]
- 66.Roche ED, Renick PJ, Tetens SP, Carson DL. A model for evaluating topical antimicrobial efficacy against methicillin-resistant Staphylococcus aureus biofilms in superficial murine wounds. Antimicrob. Agents Chemother. 2012;56:4508–4510. doi: 10.1128/AAC.00467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsuzaki K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2009;1788:1687–1692. doi: 10.1016/j.bbamem.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 68.Manaargadoo-Catin M, Ali-Cherif A, Pougnas J-L, Perrin C. Hemolysis by surfactants—a review. Adv. Colloid Interface Sci. 2016;228:1–16. doi: 10.1016/j.cis.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 69.Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017;15:740. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Busscher HJ, et al. Biomaterial-associated infection: locating the finish line in the race for the surface. Sci. Transl. Med. 2012;4:153rv110. doi: 10.1126/scitranslmed.3004528. [DOI] [PubMed] [Google Scholar]
- 71.Mottola C, et al. Susceptibility patterns of Staphylococcus aureus biofilms in diabetic foot infections. BMC Microbiol. 2016;16:119. doi: 10.1186/s12866-016-0737-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mohamed MF, Seleem MN. Efficacy of short novel antimicrobial and anti-inflammatory peptides in a mouse model of methicillin-resistant Staphylococcus aureus (MRSA) skin infection. Drug Des. Devel. Ther. 2014;8:1979. doi: 10.2147/DDDT.S72129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 2012;10:39. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- 74.Segev-Zarko L-a, Saar-Dover R, Brumfeld V, Mangoni ML, Shai Y. Mechanisms of biofilm inhibition and degradation by antimicrobial peptides. Biochem. J. 2015;468:259–270. doi: 10.1042/BJ20141251. [DOI] [PubMed] [Google Scholar]
- 75.Li J, et al. Block copolymer nanoparticles remove biofilms of drug-resistant gram-positive bacteria by nanoscale bacterial debridement. Nano Lett. 2018;18:4180–4187. doi: 10.1021/acs.nanolett.8b01000. [DOI] [PubMed] [Google Scholar]
- 76.Arciola, C. R., Campoccia, D. & Montanaro, L. Implant infections: adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 16, 397–409 (2018). [DOI] [PubMed]
- 77.Baym M, Stone LK, Kishony R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science. 2016;351:aad3292. doi: 10.1126/science.aad3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors on request. The source data underlying Figs. 4e, 5a–f, 6a, b and 6d–g, Supplementary Fig. 37a–d and Supplementary Table 3 are provided in the Source Data file.