Abstract
The Calvin-Benson-Bassham (CBB) cycle and the 3-hydroxypropionate/4-hydroxybutyrate (HP/HB) cycle are two inorganic carbon assimilation pathways widely used by prokaryotic autotrophs in lakes. We investigated the effect of mixing periods and stable water stratification patterns on the trajectories of both CO2 fixation strategies in a dimictic lake (Piburger See), because information on the spatiotemporal dynamics of prokaryotes using these pathways in freshwater ecosystems is far from complete. Based on a quantitative approach (droplet digital PCR) of genes coding for key enzymes in different CO2 assimilation pathways, nine depths covering the entire water column were investigated on a monthly basis for one year. Our data show that the abundance of photoautotrophs and obligate chemolithoautotrophs preferentially using form IA RubisCO was determined by seasonal variations. Highest numbers were observed in summer, while a strong decline of prokrayotes using RubisCO form IA was measured between December and May, the period where the lake was mostly covered by ice. The spatiotemporal distribution patterns of genes coding for RubisCO form IC genes, an enzyme usually used by facultative autotrophs for CO2 assimilation, were less pronounced. Bacteria harboring RubisCO form II were dominating the oxygen limited hypolimnion, while nitrifying Thaumarchaeota using the HP/HB cycle were of minor importance in the lake. Our data reveal that the seasonal heterogeneity, which is determined by the dimictic thermal regime of the lake, results in pronounced spatiotemporal changes of different CO2 assimilation pathways with depth-dependent environmental parameters as key factors for their distribution.
Subject terms: Limnology, Microbiology
Introduction
Primary production based on photosynthesis performed by plants, algae, cyanobacteria and anoxygenic bacteria is a major biological process on earth and of fundamental importance in the global carbon cycling1. The presence of light, however, is not a prerequisite for the assimilation of inorganic carbon, as also chemosynthesis can provide energy for this process. The chemoautotrophic fixation of inorganic carbon is accomplished by diverse prokaryotes and biochemical variations among different pathways suggest that this capability evolved convergently2–4. To date, six mechanisms are known by which autotrophic organisms fix carbon3, whereby the Calvin-Benson-Bassham (CBB) cycle in bacteria and the recently discovered 3-hydroxypropionate/4-hydroxybutyrate (HP/HB) cycle in Archaea are widely distributed in the pelagic environment of marine4,5 and freshwater habitats6,7.
A CO2 fixation mechanism used by a variety of organisms is the CBB pathway8,9. The enzyme responsible for the actual fixation of CO2 in the Calvin cycle, ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO), is not only used by photoautotrophs, different forms of RubisCO also occur in ecologically and evolutionary diverse chemoautotrophic bacteria8. Phylogenetic analyses based on large subunit genes and deduced amino acid sequences divide form I RubisCO into two groups (“green” and “red”), which may be further subdivided into types IA, IB, IC, ID8,9. Bacterial genome sequences indicate that there are even two different types of form IA structures, with form IAc being associated with carboxysomes8. Cynaobacteria and obligate chemolithoautotrophs often possess form IA RubisCO, whereas form IC enzymes are often associated with facultative autotrophs8,10. The form II RubisCO enzyme is markedly different from form I. Important biochemical features of form II enzymes are their poor affinity to CO2 and the low specificity factor τ, a measure of the ability of the RubisCO to discriminate between CO2 and O211. Therefore, RubisCO form II enzymes are adapted to environments characterized by low oxygen concentrations. A variant of the HP/HB cycle operates in nitrifying Thaumarchaeota12. This pathway was described as the most energy efficient aerobic carbon fixation cycle. This characteristic of the pathway and physiological adaptations to very low ammonia concentration correspond to the oligotrophic lifestyle of Thaumarchaeota13.
Stratified lakes represent ideal systems to investigate proximate physical and chemical drivers of autotrophic microbial communities in a relatively stable framework of habitat heterogeneity6. Chemolithoautotrophy is often depending on oxygen-labile compounds as electron donors and therefore this metabolism is often detected along chemical gradients and in the oxic/anoxic interfaces, which can be particularly found in meromictic and/or eutrophic lakes. Several studies have already shown that bacteria using different forms of RubisCO in the CBB pathway are widely distributed in different lake types and climate zones6,14–18. Thaumarchaeota using the HP/HP cycle are the dominant group of chemoautotrophs in oligotrophic and deep lakes. However, most studies investigating nitrifying Thaumarchaeota in lakes were targeting the ammonia oxidation potential, based on the analysis of genes coding for ammonia monooxygenase19–23. Investigations focusing on autotrophy related genes are limited to a few studies6,7,24,25.
Although autotrophic prokaryotes play a key role in biogeochemical cycling of nitrogen, sulphur and other elements in lakes, information on seasonal succession of autotrophs using different CO2 fixation strategies is rare. Accordingly, the goal of this investigation was to examine the spatiotemporal trajectories of autotrophic prokaryotes using different CO2 assimilation pathways in Piburger See, focusing on chemoautotrophic organisms. A previous investigation in seasonally stratified lake, based on a single sampling event during summer stratification, revealed a high diversity of sequences coding for three forms of RubisCO in the CBB cycle6. For the current study, we were especially interested to follow the spatiotemporal variations of genes coding for different forms of RubisCO and to examine if the occurrence of these forms is related to seasonal changes of environmental variables. Another specific objective was to evaluate if the lack of autotrophic Thaumarchaeota during summer6 also occurs under ice during winter, when environmental conditions for chemoautotrophic growth are more favourable. For the current investigation, water samples were obtained from nine depths sampled on a monthly basis over one year. Gene abundances were analysed via a set of autotrophic gene markers coding for key enzymes in the different CO2 assimilation pathways based on droplet digital PCR (ddPCR). Functional genes were correlated with limnological observations and environmental parameters in order to identify which physical and chemical drivers and processes are most influential for their quantitative occurrence.
Results and Discussion
Seasonal environmental changes
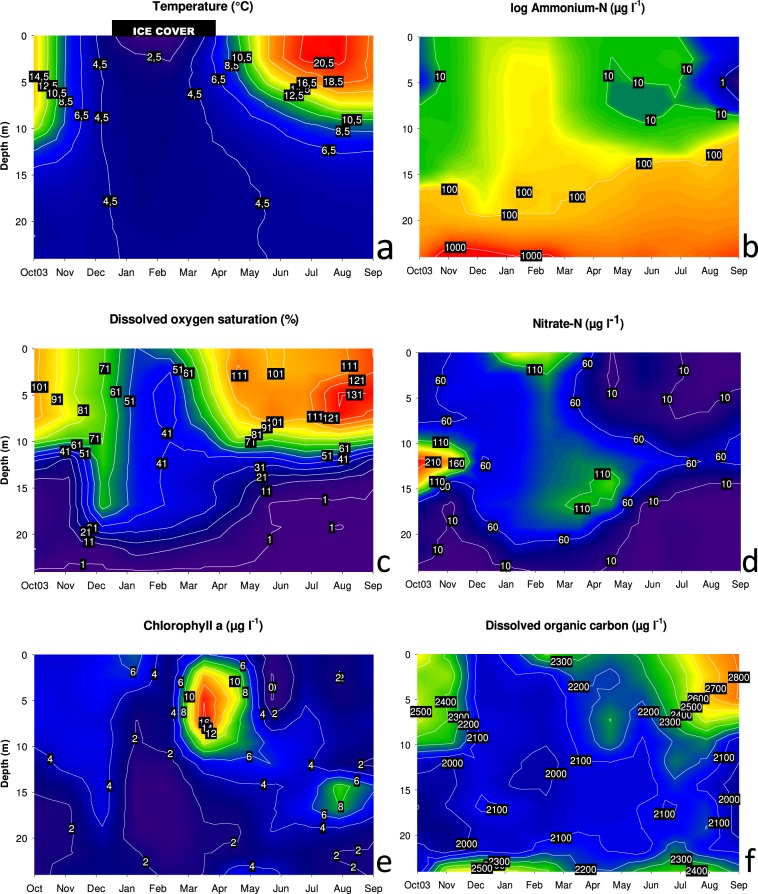
Stratification of Piburger See followed a typical seasonal pattern for a dimictic lake: autumnal mixis occurred at the end of November 2016 and spring thermal overturn shortly after the disappearance of the ice cover at the end of March 2017 (Fig. 1a). An inverse stratification was observed during winter under the ice cover, while from April to early November a stable summer stratification developed. The surface water temperature reached a maximum of 21.1 °C in August. Dissolved oxygen (DO) concentrations suggest that both thermal overturns are very short and not complete, producing a non-mixing water body (monimolimnion) below 21 m depth (Fig. 1c). In the hypolimnion, oxygen was strongly depleted during summer stratification with <10% DO saturation between June and November in depths up to 15 m. Anoxic conditions were observed at the deepest sampling point (24 m) from October to May. From June to September, the anoxic zone reached up to 18 m depth. In the surface water, DO supersaturation was observed during summer stratification with a maximum of 142% in August at 6 m depth. Nitrate concentrations showed the highest values (230 µg NO3-N l−1) in October in the upper hypolimnion (Fig. 1d). In February, under ice, nitrate concentration reached up to 166 µg NO3-N l−1 close to the lake surface. This peak was presumably caused by in-lake nitrification as described for northern oligotrophic and mesotrophic lakes with a winter ice cover26. Nitrate depletion, presumably caused by photoautotrophs, occurred in the epilimnion of the lake. In the hypolimnion, nitrate generally decreased with depth, associated with a pronounced accumulation of ammonium in the reducing environment at the deepest sampling depths (Fig. 1b). Dissolved organic carbon (DOC) measurements demonstrated low spatiotemporal variation, with concentrations constantly higher than 2 mg 1−1. Maximum values were reached in September in the surface water sample (2.82 mg DOC l−1, Fig. 1f). Chlorophyll a (Chl a) levels peaked at the sampling date in April in the epilimnion (Fig. 1e), most probably caused by an algal spring bloom that occurs regularly in Piburger See at this time of the year27–29. The increase of Chl a in the hypolimnion in late summer was also observed in other years and interpreted as a result of sedimentation and light adaptation processes of the photoautotrophic plankton during summer stratification27.
Figure 1.
Spatiotemporal dynamics of temperature (a), ammonium (b), dissolved oxygen saturation (c), nitrate (d), chlorophyll a (e), and dissolved organic carbon (f) measured monthly over a period of one year. Please note the logarithmic scale in the ammonium plot. The black shaded rectangle on the top of the temperature plot represents the approximate duration of the ice cover.
Vertical and temporal distribution of different forms of RubisCO in the Calvin cycle
Autotrophic gene abundance coding for three forms of RubisCO (IA, IC, II) was determined to follow the spatiotemporal dynamics of bacteria using the Calvin cycle for the assimilation of inorganic carbon (Table 1).
Table 1.
Specification of primers applied for the detection of different functional genes related to autotrophic prokaryotes based on ddPCR.
| Pathway, Targeted protein, Phylogenetic coverage | Primer Name | Sequence (5′-3′) | Pconca | Tab | Reference |
|---|---|---|---|---|---|
| CBB-cycle | |||||
| RubisCO form IA | cbbL_IA_f | CGGCACSTGGACCACSGTSTGGAC | 200 | 59 | Alfreider et al.43 |
| Proteobacteria (α, β, γ), Actinobacteria, Cyanobacteria | cbbL_IA_r | GTARTCGTGCATGATGATSGG | |||
| RubisCO form IA in chemoautotrophs | cbbL_IA_CHEM | GARGGNTCNGTNGTYAACGT | 400 | 58 | Alfreider & Bogensperger30 |
| Proteobacteria (α, β, γ), Actinobacteria | cbbL_IA_r | GTARTCGTGCATGATGATSGG | |||
| RubisCO form IC | cbbLR1F | GAACATCAAYTCKCAGCCCTT | 200 | 57 | Alfreider et al.34 |
| Proteobacteria (α, β, γ), Actinobacteria | cbbLR1R | TGGTGCATCTGVCCGGCRTG | |||
| RubisCO form IC dominant in lakes | cbbL_IC_lake_f | AGCCGTMARYAAAGCCAGC | 200 | 60 | This study |
| Burkholderiales | cbbL_IC_lake_r | CGTARAARCCSCGGATCATC | |||
| RubisCO form II | cbbM_f | GGCACCATCATCAAGCCCAAG | 200 | 53 | Alfreider et al.43 |
| Proteobacteria (α, β, γ), Dinophyceae | cbbM_r | TCTTGCCGTAGCCCATGGTGC | |||
| 3-HP/4-HB cycle | |||||
| 4-hydroxybutyryl- CoA dehydratase | qPCR_hcd_f | GACTGATCCWAAAGGDGAYAGAAG | 250 | 56 | Alfreider et al.6 |
| Thaumarchaea 1.1a | qPCR_hcd_r | CCYTTARCATCTGCWGGAATTGC | |||
| Ammonia oxidation pathway | |||||
| Ammonia monooxygenase | GenAOAF | ATAGAGCCTCAAGTAGGAAAGTTCTA | 100 | 55 | Meinhardt et al.40 |
| Thaumarchaea | GenAOAR | CCAAGCGGCCATCCAGCTGTATGTCC | |||
aPrimer concentration (µmol); bTa: Annealing temperature (°C).
RubisCO form IA
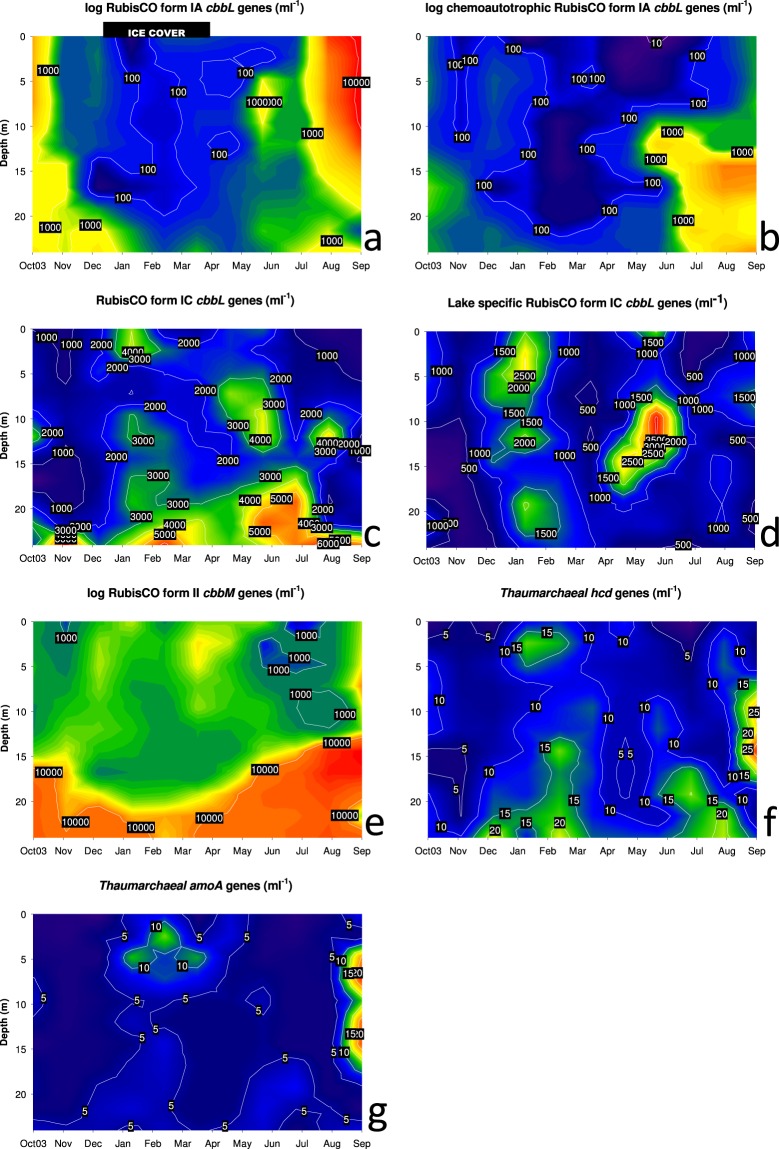
RubisCO form IA (cbbL-IA) gene numbers showed a pronounced maximum in the epilimnion during late summer stratification (Fig. 2a), which was most likely affiliated with cyanobacterial lineages harboring form IA of RubisCO. Sequence analysis of cbbL-IA genes performed in former studies6,30 in Piburger See and other lakes suggest that these RubisCO genes are affiliated with Synechococcus spp. and Paulinella chromatophora chloroplasts. The dominance of coccal cyanobacteria at this time of the year was also observed in an additional investigation, which was focusing on the seasonal succession of the phytoplankton community in Piburger See31. In this study, taxonomic assignments were based on morphological identification and Snowella litoralis, Aphanothece clathrata, as well as Microcystis incerta were the most abundant species. Gene abundances based on ddPCR analysis with primer cbbL_IA_CHEM, which was designed to encompass a functionally wide range of bacterial chemolithoautotrophs and also provides selectivity against oxygenic phototrophs using RubisCO form IA for CO2 fixation (Table 1)30, also peaked during late summer stratification. However, in contrast to the results obtained by the broad range primer pair for cbbL form IA genes, chemoautotrophic RubisCO form IA showed highest values in the hypolimnion of the lake (Fig. 2b). Spatiotemporal distribution patterns of cbbL-IA gene abundances based on both primer pairs showed a similar trend in winter: numbers declined during winter stagnation within almost the entire water column and stayed at a very low level until spring. In former studies, phylogenetic analysis in a variety of lakes including Piburger See showed that chemoautotrophic representatives were mostly Betaproteobacteria6,30, including a considerable part of cbbL-form IA sequences affiliated with the Nitrosomonas oligotropha lineage (cluster 6a). A second major cluster was related to the hydrogen-utilizing Comamonadaceae bacterium H1. Sequence analysis performed with cbbL_IA_CHEM primer additionally revealed a phylotype closely related to Thiobacillus denitrificans, which was detected in 9 m depth during summer stagnation30.
Figure 2.
Spatiotemporal dynamics of cbbL (targeting different RubisCO IA and IC forms; a–d), cbbM (targeting RubisCO form II; e) and thaumarchaeal (hcd, amoA; f, g) gene copy numbers. Samples for analysis were taken monthly along a vertical profile at 3-m intervals. Please note the logarithmic scale in the cbbL-form IA and cbbM plots.
RubisCO form IC
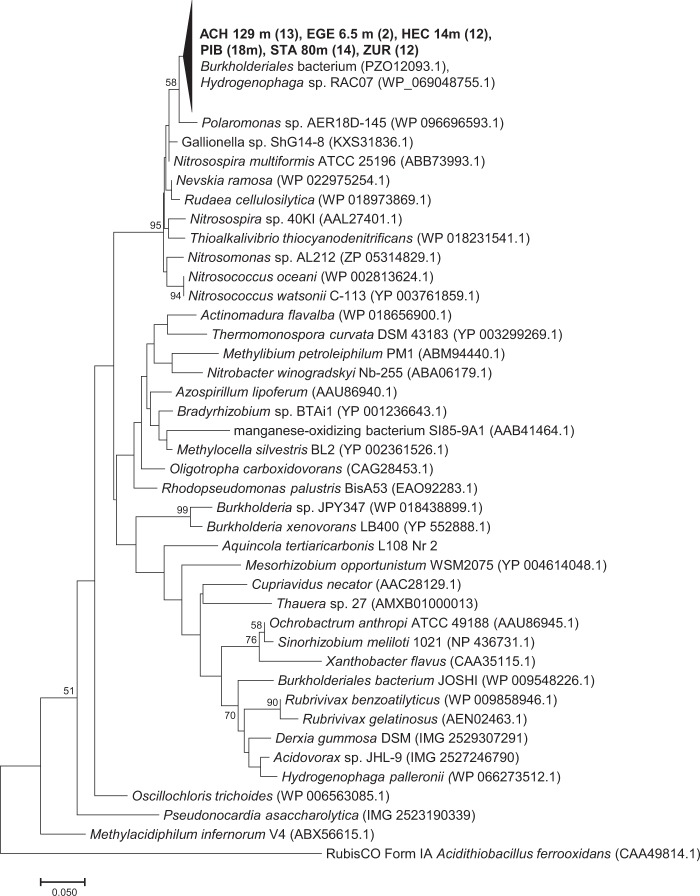
In surface water, bacteria harbouring RubisCO form IC showed maximum values in February under ice and in June and August below the metalimnion. The highest cbbL numbers, however, were detected in the lower hypolimnion throughout the year (Fig. 2c). In agreement with form IA sequences, Alfreider et al.6 showed that based on phylogenetic analysis of form IC sequences Betaproteobacteria were also the taxonomic dominate group of autotrophs. One major clade with highly similar sequences related to different Burkholderiales representatives was detected in a wide range of different lakes and depths including Piburger See6. In order to quantify this important group of autotrophs, for the present work primer pair “cbbL_IC-lake” specifically targeting this cluster of form IC RubisCOs was successfully designed and tested (Fig. 3). Determination of gene abundances related to RubisCO form IC “lake” were in accordance with measurements retrieved with broad range primers of Form IC in February under ice and in May/June at depths between 9 and 15 m (Fig. 2d), indicating that at these depths and dates of the year representatives of the lake cluster were the dominating group of bacteria using RubisCO form IC for CO2 fixation. Information on the ecological properties of the lake cluster bacteria can be derived from the closest cultivated relatives within this cluster (Fig. 3): Hydrogenophaga sp. RAC07, which was isolated from the phycosphere of Chrysochromulina tobin, and a Burkholderiales microbiome obtained from non-axenic cyanobacterial cultures and an associated Burkholderiales microbiome32,33 suggest that members of the RubisCO_form_IC-lake cluster are associated with photoautotrophic planktonic organisms. Furthermore, Hydrogenophaga sp. RAC07 was originally cultivated on organic carbon substrates supporting heterotrophic growth33, proposing that this bacterium is a facultative autotroph exhibiting a metabolic flexibility in relation to the carbon source. In the hypolimnion, cbbL_IC_lake numbers did not correspond with maxima observed by genes detected with broad range primer for Form IC RubisCO. Based on the wide diversity of organism harbouring RubisCO form IC (Fig. 3), the function and taxonomic identity of bacteria not associated with the form IC lake cluster in the hypolimnion remain unclear.
Figure 3.
Phylogenetic tree reflecting the coverage and specificity of qPCR primers cbbl_IC-lake targeting RubisCO form IC in Burkholderiales based on amino acid sequence analysis (shown in bold) and representative relatives obtained from GeneBank. DNA was extracted from water samples of different lakes obtained in a former study (Achensee - ACH, Egelsee - EGE, Hechtsee - HEC, Piburger See - PIB, Starnberger See - STA; Zürichsee - ZUR; Alfreider et al.6. Numbers in brackets indicate the number of clones analyzed.
In contrast to RubisCO form IA gene levels, which showed distinct seasonal and depth-dependent distribution patterns, abundance range and seasonality of organism harbouring form IC genes were less pronounced (Fig. 2a–d). These differences can be explained by the functional diversity of autotrophs using these forms of RubisCO. Most obligate chemolithoautotrophs possessing form IA RubisCO are highly specialized in their physiology and therefore adapted to narrow temporal and spatial ecological niches in the lake. Form IC enzymes, on the other hand, are often associated with facultative autotrophs with an enhanced metabolic flexibility in the utilization of different carbon sources8, resulting in a broader spatial and temporal distribution. This could also explain that the occurrence of bacteria using the RubisCO form IC for CO2 fixation in Piburger See were not clearly associated with individual environmental parameters as revealed by RDA-analysis (Fig. 4).
Figure 4.
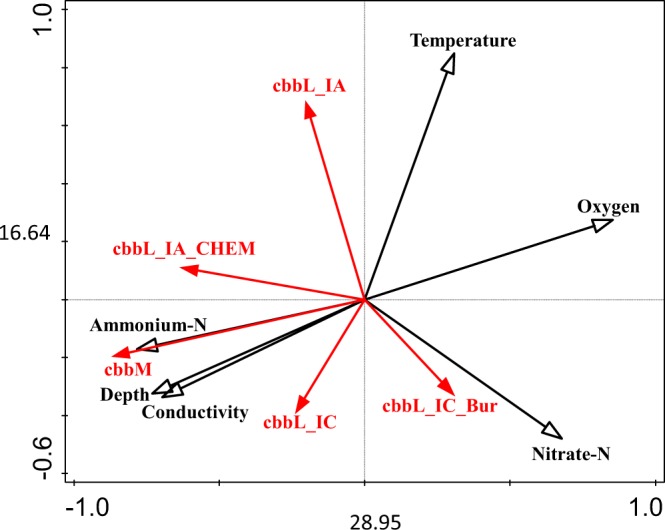
Redundancy analysis biplot of the influence of selected environmental parameters (black arrows) on abundances of different RubisCO genes (cbbL_IA = RubisCO form IA; cbbL_IA_CHEM = RubisCO form IA in chemoautotrophs, cbbL_IC = RubisCO form IC; cbbL_IC_lake = RubisCO form IC in lakes; cbbM = RubisCO form II) obtained from water samples of Piburger See (shown in red, n = 108). Copy numbers of thaumarchaeal genes (hcd, amoA) were not analysed due to the low variability of the data close to the detection limit.
RubisCO form II
Throughout the year, highest abundances of RubisCO form II were observed under anoxic and microaerobic conditions in the hypolimnion of the lake (Fig. 2e). Correspondingly, the distribution of cbbm numbers was correlated with depth-dependent environmental parameters with a negative correlation observed with DO (Fig. 4). A pronounced maximum was observed in August and September below 15 m depth. The strong influence of DO on the distribution of cbbM can be explained by a characteristic biochemical feature of the RubisCO form II protein: a poor affinity to CO2 and a low CO2/O2 substrate specificity9. On average, cbbM was the most abundant autotrophic gene marker observed in Piburger See (avg 6.78 × 103 genes ml−1, n = 106), about three times more numerous than cbbL-IC (avg 2.39 × 103 genes, n = 106) RubisCO forms and almost six times higher than cbbL-IA (avg 1.20 × 103 genes, n = 103). The high levels of form II RubisCO, which are mostly found in the hypolimnion but with a significant amount also in the oxygen rich upper water column of the lake, emphasizes the fact that RubisCO form II is often present in bacteria that also have form I RubisCO8. Based on the dissimilar kinetic properties of different Rubisco forms, chemoautotrophs operating two or even three forms of RubisCO would enable these organisms to efficiently assimilate CO2 under varying aerobic conditions34. The presence of RubisCO form II in the oxygen-saturated pelagic zone can also be explained by the presence of particles and aggregates that provide favorable conditions established by oxygen-depleted microenvironments35. Based on a previous study including Piburger See, the affiliation of cbbM sequences to a wide range of different functional groups allows only a limited understanding on the specific metabolisms and associated ecophysiology of bacteria harbouring this form of RubisCO6.
Lack of Thaumarchaeota using the HP/HB cycle
The seasonal distribution of genes coding for 4-hydroxybutyryl-CoA dehydratase (hcd), the key enzyme in the HP/HB cycle used by nitrifying Thaumarchaeota, was characterized by the absence or very low levels of hcd genes throughout the year (Fig. 2f). Maximum hcd abundances were observed in September at depths between 9 and 15 m (24–33 genes ml−1, Fig. 2f). In agreement with hcd as autotrophic gene marker, also amoA genes coding for thaumarchaeal ammonia monooxygenase were rare or close to the detection limit (Fig. 2g). Highest numbers of amoA genes were also detected between 6 and 15 m in September (13–27 genes ml−1). The absence of Thaumarchaeota in Piburger See was also reported based on sequence analysis of functional genes in the HP/HB cycle6 and other smaller lakes based on qPCR analysis7, where samples were mostly taken during summer stagnation. The reason for the lack of Thaumarchaeota in upper water layers can be explained by surface-related environmental factors, including light inhibition, competition with phototrophs for inorganic carbon and nutrients and potential grazing pressure by mixotrophic organisms6,7,23,25,36,37. In contrast to deep lakes, where Thaumarchaeota are abundant nitrifiers in the oxygenated aphotic zone throughout the year20,21,38,39, the hypolimnion of Piburger See is characterized by low dissolved oxygen concentrations that overlap with inhibiting factors in the euphotic zone during summer. This situation could explain the absence of Thaumarchaeota over the entire water column during summer stagnation7. On the other hand, it has been shown that seasonality has a strong impact on Thaumarchaeota in the epilimnion of lakes, with generally higher abundances of Thaumarchaeota in the winter season than in the summer37. The absence of Thaumarchaeota in the entire water column of Piburger See during the ice-cover period does not confirm the original hypothesis of our study that during this period the pelagic zone of the lake represents a suitable habitat for Thaumarchaeota. The environmental reasons for their absence are unknown, however, methodological issues based on primer coverage and specificity are unlikely. Both primers, GenAOA and qPCR_hcd, were already evaluated in former studies7,40 (Table 1) and produced also highly similar results when applied for the quantification of amoA and hcd genes in Thaumarchaeota in eight different lakes7. Clearly, more research is required to elucidate which additional factors are responsible that Thaumarchaeota are also highly constrained under ice and why niches with potentially favourable conditions for nitrifying Thaumarchaeota are not realised during the winter months.
Conclusions
Bacteria using the CBB cycle with different RubisCO forms showed distinct seasonal and depth-dependent distribution patterns. Cyanobacteria and obligate chemolithoautotrophs using form IA RubisCO are generally highly specialized in their physiology and these organisms developed strong adaptation to narrow temporal ecological niches related to seasonal changes in the lake. In contrast, variations in abundance and seasonality of RubisCO form IC genes, which are often found in facultative autotrophs, were less pronounced. This can be explained by a higher flexibility to changes in environmental conditions by bacteria using form IC RubisCO, based on their ability to utilize different carbon sources. Throughout the entire year, highest numbers of RubisCO Form II genes were measured under anoxic and microaerobic conditions in the hypolimnion of the lake, supporting the biochemical features of form II with a low specificity factor for CO2. Functional genes related to nitrifying Thaumarchaeota were characterized by levels close to the detection limit, which is in accordance with other studies, suggesting that Archaea using the HP/HB cycle are of minor importance in smaller lakes. The reason for the absence of Thaumarchaeota during winter months under ice, when environmental conditions are more favorable, remains unresolved. Overall, this study provides insight on how seasonal limnological changes in a dimictic lake are related to the distribution of prokaryotes based on an autotrophic perspective.
Materials and Methods
Study site and sampling
Piburger See is an oligo-mesotrophic soft water lake situated in a crystalline area in the Tyrolean Alps, Austria (47°11′42″N, 10°53′20″O, 913 m a.s.l.). The lake is dimictic and usually ice-covered from mid-December until the end of March. The catchment is composed of coniferous forest (60%), grazing land and bare rocks (35%) as well as agriculture (5%). The lake has an area of 0.17 km2 and the water retention time is about two years. Detailed information on the lake can be found elsewhere31,41. Water samples were taken monthly over a period of one year (from Oct. 2016 to Sep. 2017) along a vertical profile at 3-m intervals (nine samples from 0 to 24 m depths) at the deepest part of the lake (24.6 m). Samples were collected with a modified Schindler-Patalas sampler (UWITEC, Mondsee, Austria). Depth profiles of temperature, pH, conductivity, and oxygen were determined with a multi-parameter probe (YSI model 6600; Yellow Springs Instruments, USA). In addition to probe measurements, oxygen concentrations were quantified at sampling depths using the Winkler titration method. Secchi transparency was also determined. Lake water nutrients were measured following standard techniques: major cation and anion compositions of water samples were quantified by ion chromatography (Dionex DX-120, Dionex Inc., Sunnyvale, USA). Dissolved organic carbon (DOC) and total organic carbon (TOC) were measured using a TOC/TN-Analyzer (TOC-5000, Shimadzu, Kyoto, Japan). Chlorophyll a was measured spectrophotometrically according to Jeffrey and Humphrey42.
DNA-extraction and quantitative PCR
For DNA analyses, 480–1065 ml of lake water were filtered on polyethersulfone filters (pore size 0.22 µm, Millipore, Bedford, USA) and stored at −20 °C until use. DNA extraction was performed with a PowerWater©DNA Isolation Kit (Qiagen Inc., Hilden, Germany) according to the protocol of the manufacturer. The DNA amount was measured fluorometrically (QuantusTm, QuantiFluor®dsDNA chemistry, Promega Corporation, Fitchburg, USA). Quantitative PCR was accomplished using a ddPCR system (QX200™, Bio-Rad Laboratories, Hercules, USA). Droplet generation was performed with a robotic droplet generator (AutoDG™ Instrument, Bio-Rad). Reactions for ddPCR are based on a QX200 ddPCR EvaGreen Supermix (Bio-Rad), set up to a final volume of 22 µl in 96-well plates following the instructions of the manufacturer. Primers used for ddPCR include genes coding for the enzyme 4-hydroxybutyryl-CoA dehydratase in the thaumarchaeal HP/HB cycle and different forms of RubisCO in the CBB-cycle were designed in former studies6,7,30,34,43. Primers targeting RubisCO form IC genes related to Burkholderiales in lakes were designed for this study (Table 1). Optimal annealing temperatures and primer concentrations for ddPCR were determined based on temperature gradient experiments and testing different primer concentrations. Droplet generation was performed following the protocol provided by the manufacturer (Bio-Rad). For PCR, plates were sealed by heat and PCR amplification was performed using a standard thermal cycler (T100, Bio-Rad). The PCR reaction included the following cycling conditions: initial step of 5 min at 95 °C, 37 cycles including 30 s at 95 °C, 30 s of primer annealing at primer specific temperatures (see Table 1) and 2 min at 72 °C. The final step includes incubation temperatures of 4 °C for 5 min and 90 °C for 5 min for signal stabilization of the PCR. A ramp rate at 2.5 °C/sec was used. Droplet signal measurements were performed using a droplet reader (QX200, Bio-Rad) following the protocol of the manufacturer. Data were analyzed with the software QuantaSoft™ 1.7.4. (Bio-Rad). Samples containing over 100,000 copies were diluted 1:10 or 1:100 and analyzed again. The reliability of the automated threshold settings and fluorescence amplitudes of positive and negative droplets were examined visually.
Evaluation and sequencing of newly designed RubisCO primers q_cbbL_IC-lake
The specificity and coverage of the newly designed qPCR primer pairs q_cbbL_IC-lake (Table 1) were tested using DNA extracts obtained from different lakes for sequence analysis (Fig. 3). Standard PCR-reactions were separated on 1.5% agarose gels6. DNA of proper size was cut out of the gel and purified (MinElute® Gel Extraction Kit, Qiagen Inc.). PCR products selected for cloning were ligated into pGEM-T-Easy Vector plasmid (Promega Corporation) and transformed into JM109 competent cells6. Plasmids were screened for the presence of inserts with the proper length by PCR (GoTaq® G2 Hot Start Master Mix, Promega) using vector-specific primers M13 and inspection of DNA bands on an agarose gel6. Selected PCR-products were Sanger sequenced by a sequencing service enterprise (Eurofins MWG Operon, Ebersberg, Germany). Closest relatives to nucleotide sequences and deduced amino acid sequences were obtained using NCBI’s sequence similarity search tools BLASTN and BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Deduced amino acids were aligned using MUSCLE algorithm as implemented in MEGA X version 10.0.04 software package44, followed by a visual inspection of the alignment. Neighbor-Joining trees applying gamma distribution as the distance method were also computed with the MEGA X program. Bootstrap analysis (1000 replicates) was used to obtain confidence estimates for tree topology. The phylogenetic tree was condensed by compressing subtrees with highly similar sequences.
Statistical analysis
Redundancy analysis (RDA) was used to test and visualize the influence of different environmental factors on the spatiotemporal distribution of functional genes for CO2 fixation in the lake. A forward selection procedure, based on the Monte Carlo permutation test, was used to select meaningful explanatory variables from all environmental parameters measured. The choice of data includes only environmental parameters with a p-value < 0.02.
Sequence data deposition
RubisCO form IC sequences data obtained with primers q_cbbL_IC_lake have been submitted to GenBank databases under accession numbers MK907609 - MK907675.
Acknowledgements
We thank Gry Larson, Elias Dechent and Salvador Morales-Gomez for technical assistance in the field and chemical analysis of lake water samples. Magdalena Jäger, Theresa Bogensperger and Lukas Baumgartner are acknowledged for their support in the lab. This study was financed by the Austrian Science Fund (FWF, Project P 25703) to Alfreider A.
Author contributions
A.A. designed the research and wrote the manuscript. A.A. and B.T. analyzed the data and discussed the results. B.T. commented on the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Raven JA. Contributions of anoxygenic and oxygenic phototrophy and chemolithotrophy to carbon and oxygen fluxes in aquatic environments. Aquat. Microb. Ecol. 2009;56:177–192. doi: 10.3354/ame01315. [DOI] [Google Scholar]
- 2.Pereto JG, Velasco AM, Becerra A, Lazcano A. Comparative biochemistry of CO2 fixation and the evolution of autotrophy. Int. Microbiol. 1999;2:3–10. [PubMed] [Google Scholar]
- 3.Berg IA. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl. Environ. Microbiol. 2011;77:1925–1936. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dang H, Chen CTA. Ecological energetic perspectives on responses of nitrogen-transforming chemolithoautotrophic microbiota to changes in the marine environment. Front Microbiol. 2017;8:91–138. doi: 10.3389/fmicb.2017.01246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hügler M, Sievert SM. Beyond the Calvin Cycle: autotrophic carbon fixation in the ocean. Ann. Rev. Mar. Sci. 2011;3:261–289. doi: 10.1146/annurev-marine-120709-142712. [DOI] [PubMed] [Google Scholar]
- 6.Alfreider A, et al. CO2 assimilation strategies in stratified lakes: diversity and distribution patterns of chemolithoautotrophs. Environ. Microbiol. 2017;19:2754–2768. doi: 10.1111/1462-2920.13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alfreider A, et al. Autotrophic carbon fixation strategies used by nitrifying prokaryotes in freshwater lakes. FEMS Microb. Ecol. 2018;94:fiy163. doi: 10.1093/femsec/fiy163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badger MR, Bek EJ. Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 2008;59:1525–1541. doi: 10.1093/jxb/erm297. [DOI] [PubMed] [Google Scholar]
- 9.Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J. Exp. Bot. 2008;59:1515–1524. doi: 10.1093/jxb/erm361. [DOI] [PubMed] [Google Scholar]
- 10.Nigro LM, King GM. Disparate distributions of chemolithotrophs containing form IA or IC large subunit genes for ribulose-1,5-bisphosphate carboxylase/oxygenase in intertidal marine and littoral lake sediments. FEMS Microbiol. Ecol. 2007;60:113–125. doi: 10.1111/j.1574-6941.2007.00272.x. [DOI] [PubMed] [Google Scholar]
- 11.Tabita FR. Microbial ribulose bisphosphate carboxylase/oxygenase: a different perspective. Photosynth. Res. 1999;60:1–28. doi: 10.1023/A:1006211417981. [DOI] [Google Scholar]
- 12.Könneke M, et al. Ammonia-oxidizing archaea use the most energy efficient aerobic pathway for CO2 fixation. Proc. Natl. Acad. Sci. USA. 2014;111:8239–8244. doi: 10.1073/pnas.1402028111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 14.Giri BJ, Bano N, Hollibaugh J. Distribution of RuBisCO genotypes along a redox gradient in Mono Lake California. Appl. Environ. Microbiol. 2004;70:3443–3448. doi: 10.1128/AEM.70.6.3443-3448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tourova TP, Kovaleva OL, Sorokin DY, Muyzer G. Ribulose-1,5-bisphosphate carboxylase/oxygenase genes as a functional marker for chemolithoautotrophic halophilic sulfur-oxidizing bacteria in hypersaline habitats. Microbiology. 2010;156:2016–2025. doi: 10.1099/mic.0.034603-0. [DOI] [PubMed] [Google Scholar]
- 16.Kong W, Chuchiolo A, Priscu JC, Morgan-Kiss RM. Evidence of form II RubisCO (cbbM) in a perennially ice-covered Antarctic lake. FEMS Microbiol. Ecol. 2012;82:491–500. doi: 10.1111/j.1574-6941.2012.01431.x. [DOI] [PubMed] [Google Scholar]
- 17.Kong W, Ream DC, Priscu JC, Morgan-Kiss RM. Diversity and expression of RubisCO genes in a perennially ice-covered Antarctic lake during the polar night transition. Appl. Environ. Microbiol. 2012;78:4358–4366. doi: 10.1128/AEM.00029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Garcia M, et al. High-throughput single-cell sequencing identifies photoheterotrophs and chemoautotrophs in freshwater bacterioplankton. ISME J. 2012;6:113–123. doi: 10.1038/ismej.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auguet JC, Triadó-Margarit X, Nomokonova N, Camarero L, Casamayor EO. Vertical segregation and phylogenetic characterization of ammonia-oxidizing archaea in a deep oligotrophic lake. ISME J. 2012;6:1786–97. doi: 10.1038/ismej.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vissers, E. W. et al. Temporal and spatial coexistence of archaeal and bacterial amoA genes and gene transcripts in Lake Lucerne. Archaea 289478 (2013). [DOI] [PMC free article] [PubMed]
- 21.Vissers EW, et al. Seasonal and vertical distribution of putative ammonia-oxidizing thaumarchaeotal communities in an oligotrophic lake. FEMS Microbiol. Ecol. 2013;83:515–526. doi: 10.1111/1574-6941.12013. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, et al. AmoA-encoding archaea and thaumarchaeol in the lakes on the northeastern Qinghai-Tibetan Plateau, China. Front. Microbiol. 2013;4:329. doi: 10.3389/fmicb.2013.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hugoni M, et al. Dynamics of ammonia oxidizing archaea and bacteria in contrasted freshwater ecosystems. Res. Microbiol. 2003;16:360–370. doi: 10.1016/j.resmic.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Auguet JC, Borrego CM, Bañeras L, Casamayor EO. Fingerprinting the genetic diversity of the biotin carboxylase gene (accC) in aquatic ecosystems as a potential marker for studies of carbon dioxide assimilation in the dark. Environ. Microbiol. 2008;10:2527–2536. doi: 10.1111/j.1462-2920.2008.01677.x. [DOI] [PubMed] [Google Scholar]
- 25.La Cono V, et al. Partaking of Archaea to biogeochemical cycling in oxygen-deficient zones of meromictic saline Lake Faro (Messina, Italy) Environ. Microbiol. 2013;15:1717–1733. doi: 10.1111/1462-2920.12060. [DOI] [PubMed] [Google Scholar]
- 26.Powers SM, et al. Ice duration drives winter nitrate accumulation in north temperate lakes. Limnol. Oceanogr. Lett. 2017;2:177–186. doi: 10.1002/lol2.10048. [DOI] [Google Scholar]
- 27.Tolotti M, Thies H. Phytoplankton community and limnochemistry of Piburger See (Tyrol, Austria) 28 years after lake restoration. J. Limnol. 2002;61:77–88. doi: 10.4081/jlimnol.2002.77. [DOI] [Google Scholar]
- 28.Prader K. Die Entwicklung des Phytoplanktons im Sommer im Piburger See (Ötztal, Tirol) Ber Nat-med Ver Innsbruck. 1993;80:39–51. [Google Scholar]
- 29.Salcher M, Pernthaler J, Posch T. Spatiotemporal distribution and activity patterns of bacteria from three phylogenetic groups in an oligomesotrophic lake. Limnol. Oceanogr. 2010;55:846–856. doi: 10.4319/lo.2010.55.2.0846. [DOI] [Google Scholar]
- 30.Alfreider A, Bogensperger T. Specific detection of form IA RubisCO genes in chemoautotrophic bacteria. J. Basic Microbiol. 2018;58:712–716. doi: 10.1002/jobm.201800136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thies H, et al. Interactions of temperature and nutrient changes: effects on phytoplankton in the Piburger See (Tyrol, Austria) Freshw. Biol. 2012;57:2057–2075. doi: 10.1111/j.1365-2427.2011.02661.x. [DOI] [Google Scholar]
- 32.Cornet L, et al. Metagenomic assembly of new (sub)polar Cyanobacteria and their associated microbiome from non-axenic cultures. Microb. Genom. 2018;4(9):e000212. doi: 10.1099/mgen.0.000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fixen KR, et al. Genome sequences of eight bacterial species found in coculture with the haptophyte Chrysochromulina tobin. Genome Announc. 2016;4:e01162–16. doi: 10.1128/genomeA.01162-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alfreider. A, et al. Distribution and diversity of autotrophic bacteria in groundwater systems based on the analysis of RubisCO genotypes. System. Appl. Microbiol. 2009;32:140–150. doi: 10.1016/j.syapm.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Dang H, Lovell CR. Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 2016;80:91–138. doi: 10.1128/MMBR.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Small GE, et al. Rates and controls of nitrification in a large oligotrophic lake. Limnol. Oceangr. 2013;58:276–286. doi: 10.4319/lo.2013.58.1.0276. [DOI] [Google Scholar]
- 37.Pollet T, Berdjeb L, Chardon C, Jacquet S. Contrasting temporal patterns in ammonia-oxidizing archaeal community dynamics in two peri-alpine lakes with different trophic status. Aquat. Microb. Ecol. 2018;81:95–108. doi: 10.3354/ame01861. [DOI] [Google Scholar]
- 38.Coci, M., Odermatt, N., Salcher, M., Pernthaler, J. & Corno, G. Ecology and distribution of Thaumarchaea in the deep hypolimnion of Lake Maggiore. Archaea 590434 (2015). [DOI] [PMC free article] [PubMed]
- 39.Callieri C. Micro-players for macro-roles: aquatic microbes in deep lakes. J. Limnol. 2016;75:191–200. doi: 10.4081/jlimnol.2016.1370. [DOI] [Google Scholar]
- 40.Meinhardt KA, et al. Evaluation of revised polymerase chain reaction primers for more inclusive quantification of ammonia‐oxidizing archaea and bacteria. Environ. Microbiol. Rep. 2015;7:354–63. doi: 10.1111/1758-2229.12259. [DOI] [PubMed] [Google Scholar]
- 41.Sommaruga R, Psenner R. Trophic interactions within the microbial food web in Piburgersee (Austria) Arch. Hydrobiol. 1995;132:257–278. [Google Scholar]
- 42.Jeffrey SW, Humphrey GF. New spectrophotometric equations for determining chlorophyll a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem. Physiol. Pflanz. 1975;167:191–194. doi: 10.1016/S0015-3796(17)30778-3. [DOI] [Google Scholar]
- 43.Alfreider A, Vogt C, Hoffmann D, Babel W. Diversity of ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes from groundwater and aquifer microorganisms. Microb. Ecol. 2003;45:317–328. doi: 10.1007/s00248-003-2004-9. [DOI] [PubMed] [Google Scholar]
- 44.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]