Abstract
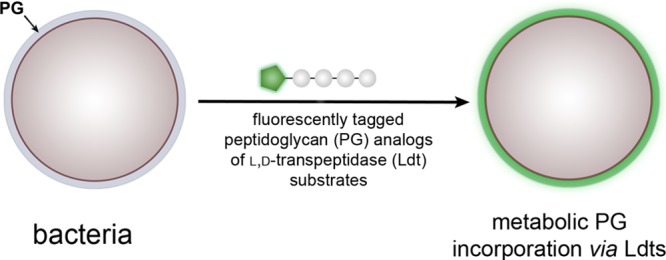
Peptidoglycan (PG) is a cross-linked, meshlike scaffold endowed with the strength to withstand the internal pressure of bacteria. Bacteria are known to heavily remodel their peptidoglycan stem peptides, yet little is known about the physiological impact of these chemical variations on peptidoglycan cross-linking. Furthermore, there are limited tools to study these structural variations, which can also have important implications on cell wall integrity and host immunity. Cross-linking of peptide chains within PG is an essential process, and its disruption thereof underpins the potency of several classes of antibiotics. Two primary cross-linking modes have been identified that are carried out by D,D-transpeptidases and L,D-transpeptidases (Ldts). The nascent PG from each enzymatic class is structurally unique, which results in different cross-linking configurations. Recent advances in PG cellular probes have been powerful in advancing the understanding of D,D-transpeptidation by Penicillin Binding Proteins (PBPs). In contrast, no cellular probes have been previously described to directly interrogate Ldt function in live cells. Herein, we describe a new class of Ldt-specific probes composed of structural analogs of nascent PG, which are metabolically incorporated into the PG scaffold by Ldts. With a panel of tetrapeptide PG stem mimics, we demonstrated that subtle modifications such as amidation of iso-Glu can control PG cross-linking. Ldt probes were applied to quantify and track the localization of Ldt activity in Enterococcus faecium, Mycobacterium smegmatis, and Mycobacterium tuberculosis. These results confirm that our Ldt probes are specific and suggest that the primary sequence of the stem peptide can control Ldt cross-linking levels. We anticipate that unraveling the interplay between Ldts and other cross-linking modalities may reveal the organization of the PG structure in relation to the spatial localization of cross-linking machineries.
Bacterial cell walls are the frontline in controlling how bacteria interact with their environment (or host organisms) and serve to counter high internal turgor pressure. Peptidoglycan (PG), a primary component of bacterial cell walls, is an essential scaffold that provides physical and mechanical stability to bacterial cells (Figure 1A).1−3 Despite the large diversity in bacterial shapes and cell wall configurations, the overall primary PG structure remains relatively constant by having two major structural components. The backbone glycan chain is assembled with disaccharide building blocks that are composed of N-acetyl-glucosamine (GlcNAc) and N-acetyl-muramic acid (MurNAc). A pentapeptide chain (stem peptide) is attached to MurNAc via its N-terminus. Although there are variations within the stem peptide sequence between bacteria, the canonical sequence is L-Ala-D-Glx-(L-Lys/m-DAP)-D-Ala-D-Ala.
Figure 1.
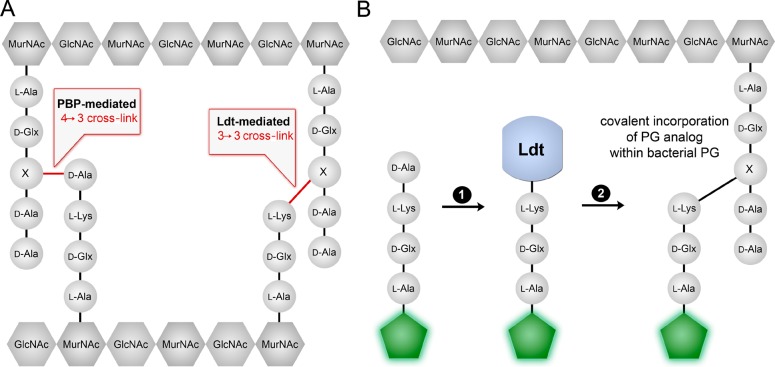
(A) PG cross-linking modes associated with Ltds and PBPs. X represents the third position amino acid (either m-DAP or L-lysine-based amino acids). (B) A synthetic mimic of the stem peptide modified with a fluorescent handle (green hexagon) is covalently incorporated within growing PG scaffold. First, a terminal D-Ala residue is removed by Ltd, leading to a covalent intermediate. Second, this acyl-donor is captured by the third position amino acid within existing PG thus leading to its cross-linking with PG and generating a measurable fluorescent signal.
A growing body of evidence points to the fact that PG undergoes extensive chemical remodeling–both in the glycan and peptide segments–in order to refine its chemical and physical properties.4−6 Modifications include the N-glycolylation of muramic acid in the glycan backbone,7O-acetylation,8,9 or amidation of D-glutamate and m-DAP in the peptide side chain.10 These modifications are critical for the proper integrity and architecture of the PG scaffold. In addition, PG remodeling can have significant influences on drug sensitivity,11−14 interaction with PG sensors on cell surfaces,15,16 and host-microbiota interaction.17−20 The most prominent chemical change to nascent PG scaffold involves covalent cross-linking of neighboring stem peptides by membrane-anchored transpeptidases (TPs). Cell wall cross-linking is essential to bacteria as its inhibition represents a primary mode of action for some of the most potent antibiotics in clinical use. Covalent PG cross-links greatly enhance cell wall strength and define the porosity of this scaffold. Cross-linking levels can vary considerably, ranging from 20 to 90% depending on the organism.21,22
The primary function of PG transpeptidation is to generate an amide bond between the side chain of a stem peptide to the C-terminus of an adjacent stem peptide. Two main classes of enzymes are responsible for PG cross-linking: D,D-transpeptidases and L,D-transpeptidases (Ldts). D,D-transpeptidases (Ldts). D,D-Transpeptidation reactions are carried out by various Penicillin Binding Proteins (PBPs) and are considered to be the predominant mode of PG cross-linking for several classes of bacteria (Figure 1).1 A new class of PG transpeptidases, Ldts, was initially identified in Enterococcus hirae,23 but the enzyme itself was first characterized more recently in Enterococcus faecium.24,25 Since its discovery, Ldts have been shown to be operative in a large number of organisms including Bacillus subtilis,26Mycobacterium tuberculosis,27Clostridium difficile,28 and Escherichia coli.29
There are structurally subtle but functionally important differences between PBP TPs and Ldts. Despite having similar enzymatic functions, the two enzymes have no primary sequence homology. Indeed, Ldts have no similarity to proteins currently in the protein database.14 PBP TPs cross-link PG stem peptides by first removing terminal D-Ala residues on pentapeptide substrates to form covalent intermediates (Figure S1). A neighboring nucleophilic amino group from the third position (L-Lys or m-DAP, depending on the bacterial class) captures the acyl intermediate to generate a 4-3 cross-link. The main variation between PBPs and Ldts is that Ldts generate 3-3 cross-links between PG stem peptides because its substrates are tetrapeptides (Figure 1B). As the enzyme name implies, Ldts substrates are not terminated as D,D-stereocenters but instead as L,D-stereocenters.14,25,30 A majority of bacterial PGs are composed of mostly 4-3 cross-links, with some organisms having a minor component of 3-3 cross-links. A prominent exception is mycobacterial PG, which is composed of mostly 3-3 cross-links.31−34 In the case of drug-sensitive E. faecium, the PG scaffold is composed of mostly 4-3 cross-links, yet both PBPs and Ldts are expressed.35 Exposure of E. faecium to either ampicillin or vancomycin results in a shift to 3-3 cross-links for two different reasons. In vancomycin resistant enterococci (VRE) cells, vancomycin treatment leads to the truncation of the pentapeptide on lipid II.36−38 Tetrapeptide is a substrate for Ldts but not PBPs, resulting in higher levels of 3-3 cross-links. In ampicillin-resistant E. faecium, inactivation of PBPs is compensated by shifting cross-linking substrates from pentapeptide to tetrapeptide.25,39
PG biosynthesis is initiated in the cytoplasm where a series of enzymatic transformations produce lipid II, a lipid-linked disaccharide pentapeptide precursor.2,30 Lipid II is then translocated across the cytoplasmic membrane, and nascent PG is integrated into the existing cell wall by the combination of transglycosylases and TPs. Both steps are critical for proper PG assembly as evidenced by the fact that disruption of these processes can be lethal to bacterial cells. There are two major classes of PG TPs, namely PBPs TPs and Ldts. Both classes play important roles in the assembly of the PG scaffold, although it is unclear whether they are functionally redundant or assume specialized roles. The emergence of single D-amino acid PG probes has been fundamental in advancing live cell PG fluorescence analysis.40−48 As examples of these advances, the presence of PG in Chlamydia trachomatis was established,49 and treadmilling by FtsZ filaments was shown to drive PG synthesis.50−52 Prior studies have demonstrated that structural mimicry of nascent PBP substrates results in PG incorporation in vitro(53) and in live bacterial cells.54,55 Also, during the preparation of our manuscript it was shown, in vitro, that Ldts mediate cross-linking of synthetic Ldt substrates.56 In contrast, there are currently no Ldt-specific probes to tag and visualize Ldt activity in live cells. We assembled a synthetic substrate of Ldt to specifically interrogate Ldt activity and better understand how the primary sequence of the stem peptide can modulate Ldt-mediated cross-linking.
Results
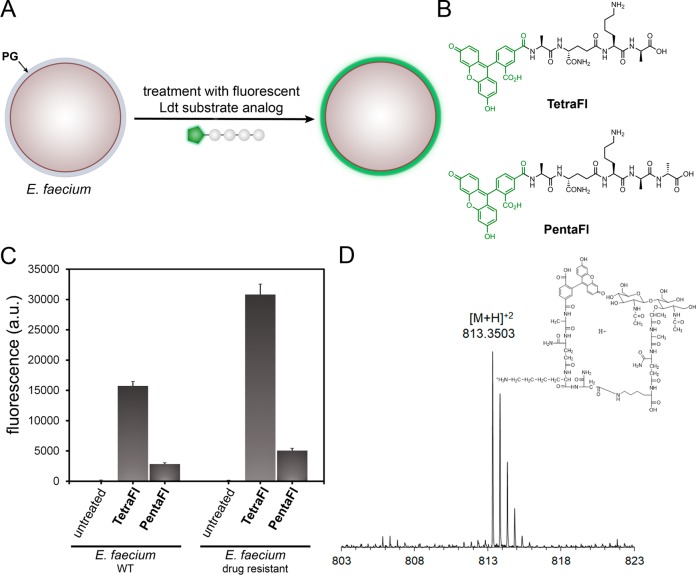
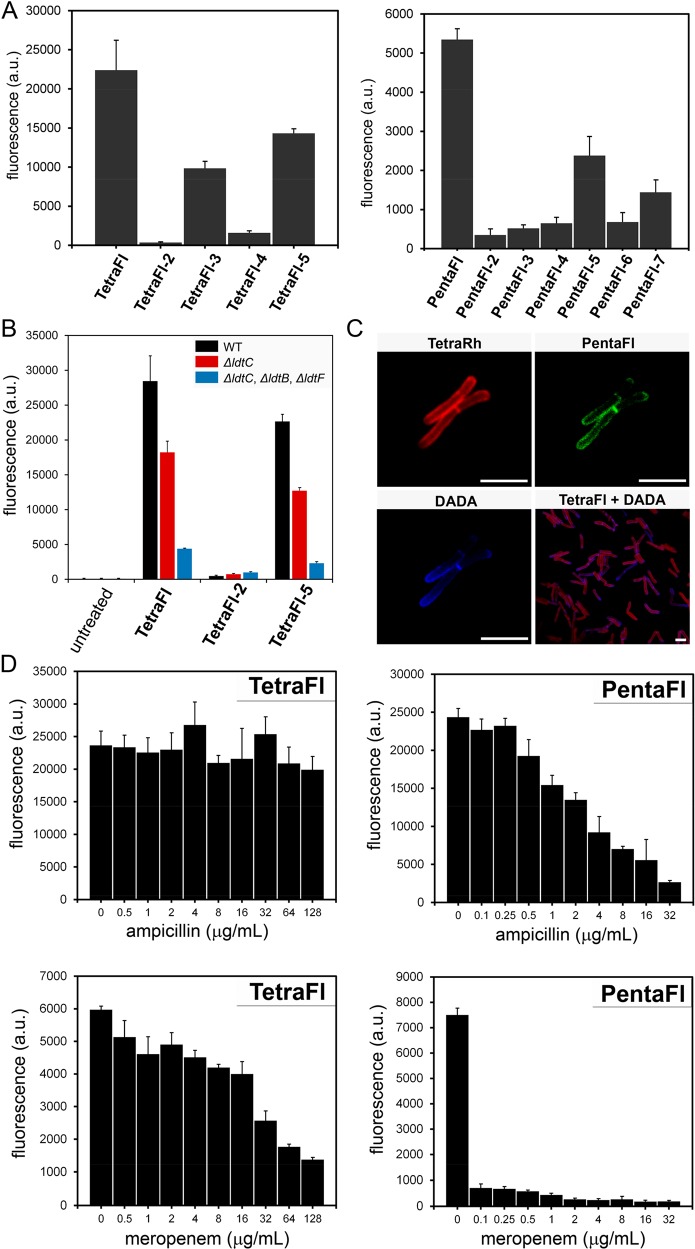
We anticipated that Ldt cross-linking of PG could be quantified by conjugating a fluorescent handle onto the N-terminus of the tetrapeptide PG mimic. Treatment of bacterial cells with the fluorescently tagged stem peptides should lead to their covalent incorporation into the expanding PG scaffold during cell growth (Figure 2A). Cellular fluorescence is subsequently quantified using flow cytometry, and fluorescence levels should correlate with PG cross-linking of synthetic stem peptide mimics. At first, two synthetic stem peptide mimics were synthesized: TetraFl and PentaFl (Figure 2B). Both peptides are structurally similar except for the additional terminal D-Ala in PentaFl, which mimics the endogenous donor substrates of PBP TPs. Drug-sensitive E. faecium cells (WT) at low cell densities (OD600 ∼ 0.05) were treated with either TetraFl or PentaFl, and fluorescence levels were measured after 16 h. In the absence of synthetic stem peptides, background cellular fluorescence levels were low (Figure 2C). Cellular treatment with TetraFl led to an ∼210-fold fluorescence increase over background and an ∼5.5-fold increase over PentaFl. Higher labeling levels for TetraFl relative to PentaFl in E. faecium (WT) likely reflect either a higher overall catalytic efficiency by Ldts or a greater flexibility by Ldts in tolerating synthetic stem peptide mimics. Notably, we did not observe an in situ accumulation of carboxypeptidase products upon overnight incubation of either TetraFl or PentaFl (Figure S2). As expected, treatment of E. faecium with PG probes led to their incorporation into the PG matrix consistent with Ldt processing as revealed by mass spectrometry analysis of PG extracted from cells treated with TetraFl (Figure 2D and Figure S3) and PentaFl (Figure S4). Furthermore, a time-dependent decrease in cellular fluorescence was observed upon lysozyme treatment (Figure S5), and confocal imaging of the isolated sacculi also showed fluorescence consistent with PG incorporation (Figure S6). No apparent effect on cell growth and morphology was observed. These initial results represent the first example of live cell analysis of Ldt activity.
Figure 2.
(A) Schematic diagram delineating incorporation of synthesized fluorescent Ldt substrate and incorporation into bacterial PG. (B) Chemical structure of fluorescein-modified tetrapeptide (TetraFl) and pentapeptide (PentaFl) PG stem mimics. (C) Flow cytometry analysis of E. faecium (WT and drug resistant strain) treated overnight with 100 μM TetraFl or PentaFL. Data are represented as mean + SD (n = 3). (D) Mass spectrum and XIC of TetraFL-PG with 3-3 cross-link with observed [M + H]+2m/z of 818.3503.
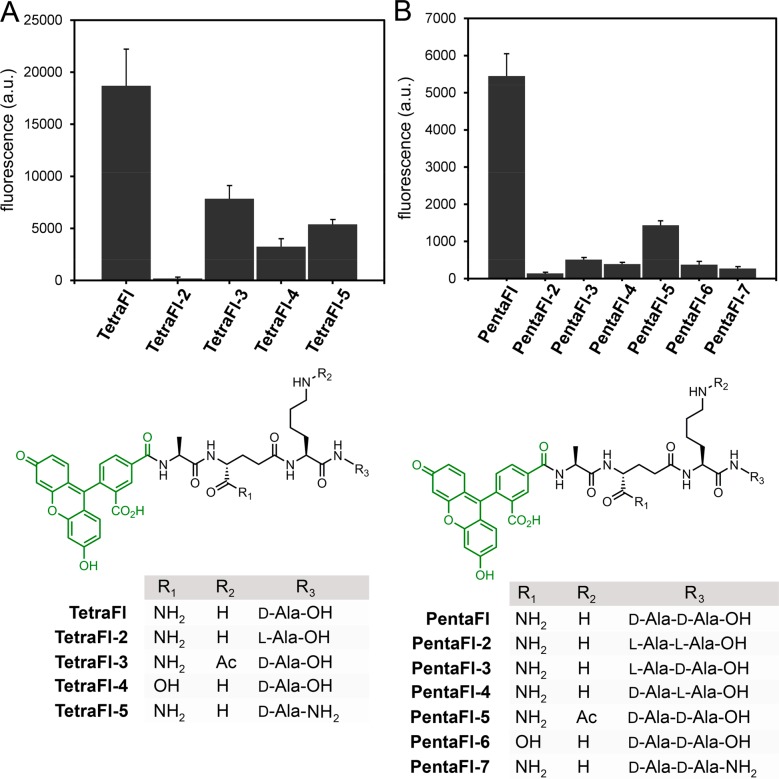
Fluorescence levels were higher for both probes in the drug-resistant strain, which may reflect additional controls in TP cross-linking modalities besides protein expression levels (Figure 2C). Similar trends were found for an additional drug-sensitive and drug-resistant strain of E. faecium further confirming our general strategy of labeling cell surfaces with Ldt analogs (Figure S7). Having established the feasibility of labeling cell surfaces with synthetic stem peptide analogs of Ldt substrates, we set out to extensively map how structural variations can impact cross-linking by surface-bound TPs. Variations of the tetrapeptide sequence were installed within four strategic sites: C-terminus (acid/amide), terminal residue(s) (D-Ala/L-Ala), second position (iso-Gln/iso-Glu), and third position (L-Lys/acetylated L-Lys). Each variation was designed to interrogate specific aspects of substrate recognition by TPs. For the tetrapeptide series, the stereospecificity was evaluated first by cell treatment with TetraFl-2–a variant that has a terminal L-Ala (Figure 3A). Cellular fluorescence levels were reduced to near background levels, thus indicating a strong selection for the correct stereocenter at the terminal Ala position. In TetraFl-3, the third position Lys residue is acetylated to block any potential acyl-transfer reaction to this nucleophilic site. While there was an ∼2.3-fold decrease in fluorescence, labeling levels suggest contribution of the synthetic step peptide as an acyl-acceptor. The introduction of a carboxylic acid at the second position iso-Glu (TetraFl-4), instead of iso-Gln, resulted in a 4.5-fold decrease in surface labeling, a finding that is consistent with recent in vitro analysis that showed reduction in cross-linking.56 Amidation of the C-terminus (TetraFl-5) also led to decreased levels of cell surface labeling, which points to a preference for the endogenous carboxylate at the stem peptide terminus.
Figure 3.
Flow cytometry analysis of E. faecium (drug resistant) treated overnight with 100 μM of tetrapeptide (A) or pentapeptide (B) with variations. Data are represented as mean + SD (n = 3). Chemical series of tetrapeptides and pentapeptides with variations at the C-terminus (acid/amide), terminal residue(s) (D-Ala/L-Ala), second position (iso-Gln/iso-Glu), and third position (L-Lys/acetylated L-Lys).
A similar panel of stem peptide variants was built for the pentapeptide probes (Figure 3B). Overall, the trends were mostly consistent with the tetrapeptide probes including the stereoselectivity at both the fourth and fifth positions. It is interesting that these trends are similar despite the lack of structural similarities between PBP TPs and Ldts. It is worth noting that we were able to recapitulate in E. faecium the in vitro demonstration that the lack of amidation of iso-Glu results in greatly diminished cross-linking by PBPs from Enterococcus faecalis, Streptococcus pneumoniae, and Staphylococcus aureus (S. aureus) (PentaFl-6).31,53,58 Identical patterns of cellular labeling were observed in a second strain of E. faecium across the panels of tetra- and pentapeptide probes (Figure S8), thus reconfirming the necessity for amidation at iso-Glu.
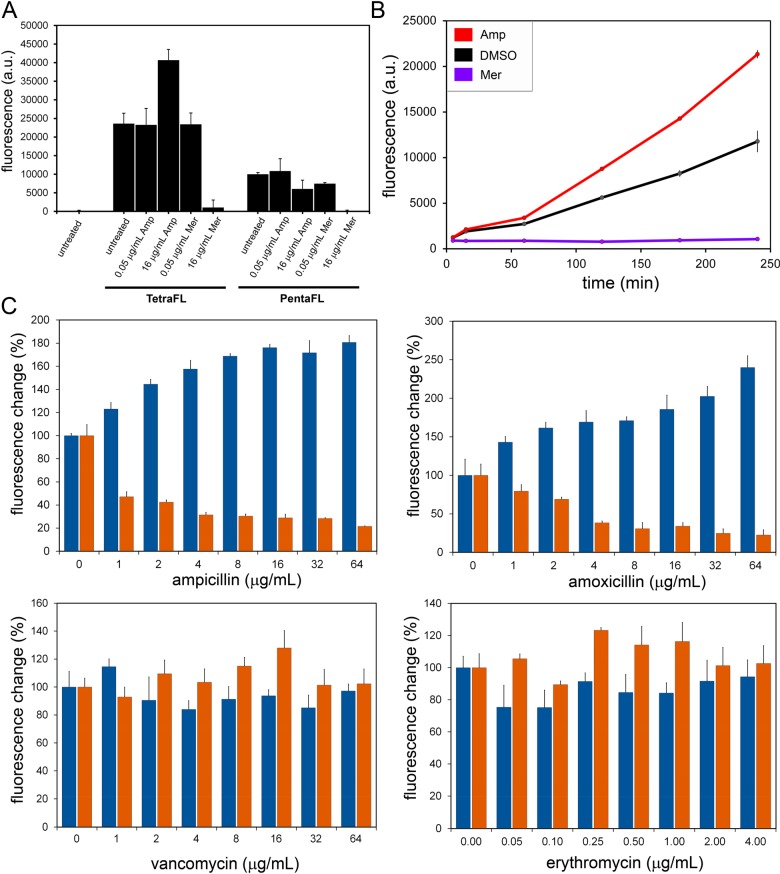
Next, we set out to evaluate how PG cross-linking modes may be affected by various antibiotics in M9 (a multidrug resistant strain of E. faecium)59 (Figure 4A). Initially, we evaluated two β-lactam agents: ampicillin and Meropenem. Whereas ampicillin is not known to inhibit Ldts, Meropenem (along with other carbapenems) has been shown to inhibit both PBPs and Ldts.60,61 At low concentrations (0.05 μg/mL) of Meropenem no change in cellular fluorescence was observed. As expected, treatment at higher concentrations (16 μg/mL) led to reduction in both TetraFl and PentaFl cell labeling. Despite the reduction in cellular fluorescence to basal fluorescence levels, bacterial cells grew similar to untreated cells (MIC ∼ 18 μg/mL). Most interestingly, there was a near 2-fold increase in TetraFl-labeling upon treatment with 16 μg/mL of ampicillin. A similar trend was also observed in a VanA-resistant E. faecium strain (Figure S9). Inclusion of asparagine onto the lysine side chain of TetraFl, which is a closer mimic of E. faecium PG, also demonstrated an ampicillin-induction in surface labeling (Figure S10).
Figure 4.
(A) Flow cytometry analysis of E. faecium (M9) treated overnight with 100 μM TetraFl or PentaFl with or without ampicillin/Meropenem. Data are represented as mean + SD (n = 3). (B) E. faecium (M9) treated with 100 μM TetraFl with 16 μg/mL ampicillin, 8 μg/mL meropenum, or DMSO (vehicle control) at early log phase. Cells were collected at various time points and analyzed by flow cytometry. Data are represented as mean + SD (n = 3). (C) Flow cytometry analysis of E. faecium (M9) treated overnight with 100 μM TetraFl (blue bars) or PentaFl (orange bars) and increasing concentrations of ampicillin, amoxicillin, vancomycin, or erythromycin. Data are represented as mean + SD (n = 3).
To gain further insight into the induction of TetraFl-labeling, a time-course analysis was performed (Figure 4B). E. faecium cells from early log (OD600 ∼ 0.05) were treated with ampicillin, Meropenem, or DMSO and coincubated with TetraFl. Within 60 min, there was a significant difference in fluorescence between DMSO and ampicillin treated cells that became greater over the next 3 h. These results suggest that induction of TetraFl-labeling was observable through the log phase of growth. Finally, we performed a comprehensive concentration-dependency analysis of both TetraFl and PentaFl in the presence of eight antibiotics (Figure 4C and Figure S11). Two agents from the penicillin-class of β-lactams (ampicillin and amoxicillin) yielded similar patterns of response: a concentration-dependent increase in TetraFl labeling and decrease in PentaFl labeling. Critically, reduction in fluorescence levels of bacteria treated with PentaFl suggests that PentaFl is not processed by Ldts. Treatment with two antibiotics that are not β-lactams (vancomycin and erythromycin) led to no significant change in fluorescence across all sublethal concentrations. Moreover, both carbapenems tested (Meropenem and imipenem) led to a reduction of both TetraFl and PentaFl labeling (Figure S11). Likewise, there was a decrease upon treatment with a cephalosporin agent (ceftriaxone), which was previously shown to inhibit Ldt in vitro.(61) Finally, no change in labeling levels was observed upon treatment with a monobactam (aztreonam), and this finding is consistent with prior studies showing insensitivity to this particular agent.62,63
Localization studies were performed next with the two cellular probes that mimic the substrates of the two primary TPs in bacteria. To differentiate the fluorescence signals between the tetra- and pentapeptide probes, the fluorescent moiety in TetraFl was replaced with rhodamine (TetraRh). The goal of this experiment was to establish how PG cross-linking modes are spatially organized within bacterial cells. For these pulse-treatments, all three probes (TetraRh, PentaFl, and DADA) were simultaneously incubated with E. faecium cells. Cells from early log phase (OD600 ∼ 0.1) were labeled for 5 min and subsequently imaged by confocal microscopy (Figure 5A). PentaFl labeling was almost exclusively observed at the septal region of cells. Quite strikingly, Ldt activity showed a clear difference in labeling pattern compared to PBP TP activity. TetraRh labeling was prominent at the septal region but also found throughout the entire cell surface. E. faecium labeling with TetraRh-3, which cannot act as an acyl-acceptor strand, labeled in a similar manner to TetraFl (Figure S12). These results may reflect a difference in localization of Ldt activity relative to PBP TP activity in E. faecium. E. faecium cells were also incubated with a single D-amino acid derivative (Diethyl-Amino-coumarin-D-Alanine, DADAFigure S13), and the isolated PG was digested for LC-MS analysis. Similar levels of incorporation of DADA into the fourth and fifth positions within the PG stem peptide were observed (Figure S14). These results clearly confirm that single D-amino acids can report mixed modes of incorporation and are not universally Ldt-specific. Based on the tetrameric structure of TetraFl, we anticipated it would not get incorporated by D,D-transpeptidases and intracellular pathways and be mediated solely by Ldts.62,63
Figure 5.
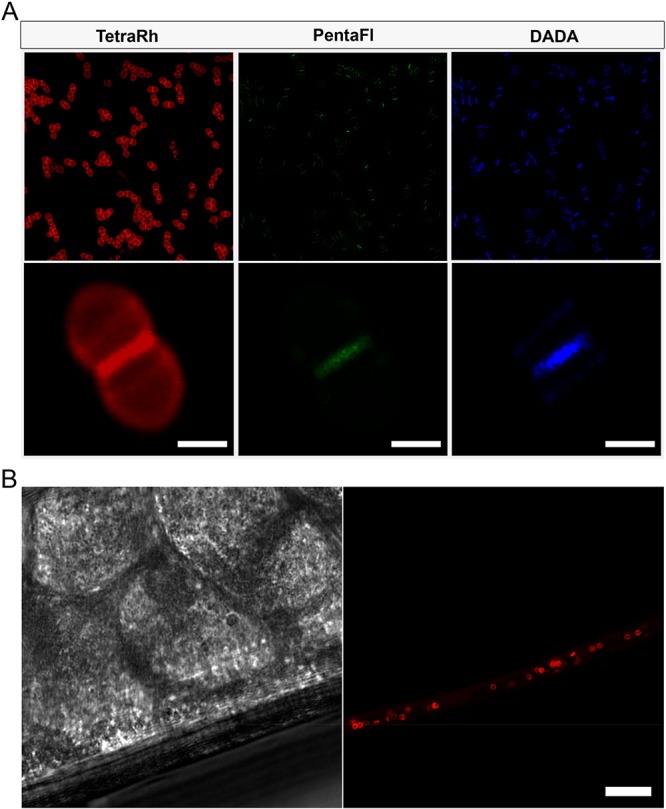
(A) Confocal microscopy image of E. faecium (WT) treated with 5 min pulse of 500 μM TetraRh, 500 μM PentaFl, and 5 mM DADA (scale bar: 1 μm). (B) In vivo labeling of E. faecium in model host. C. elegans were infected with E. faecium for 4 h, washed to remove noncolonized bacteria, and incubated with 50 μM TetraRh for 2 h. The C. elegans were washed, anesthetized, mounted on a bed of agarose, and imaged using confocal microscopy (scale bar: 10 μm).
Toward the goal of assessing Ldt activity in living host animals, we investigated whether TetraRh can label in Caenorhabditis elegans (C. elegans). C. elegans are powerful model animals for studying bacterial pathogenesis.64−66 As an example, C. elegans were recently used to establish how a PG hydrolase from E. faecium can protect C. elegans against Salmonella pathogenesis.17 Moreover, it was previously established that S. aureus cells can be metabolically labeled in live C. elegans by sortase substrates analogs.67,68 For our current work, C. elegans (∼L4 stage) were incubated with E. faecium to establish bacterial colonization. After removing noncolonized bacteria, E. faecium infected C. elegans were incubated with TetraFl for 2 h. Following a washing step, C. elegans were visualized using confocal microscopy (Figure 5B). Remarkably, we were able to specifically label the PG of colonized bacteria in live C. elegans. These results may pave the way to establishing how PG cross-linking is controlled by external factors, including a host response to bacterial infection.
Having established the ability to track Ldt activity in E. faecium, we turned our attention to a different class of pathogens. The PG polymer in mycobacteria69−72 is surrounded by the highly impermeable mycomembrane that endows these organisms with intrinsic resistance to vast types of antibiotics. PG cross-linking in mycobacteria is unique in that proportions of 3-3 cross-linking can reach levels close to 80%. At first, we evaluated the panel of tetra- and pentapeptides for their ability to tag PG of Mycobacterium smegmatis using similar conditions as E. faecium (Figure 6A). Remarkably, high labeling levels were observed for both TetraFl and PentaFl, albeit with lower levels for PentaFl. High labeling levels are unusual considering the well-established permeability barrier imposed by the mycomembranes. Transport via an outer membrane pore may explain the high levels of probe penetration past the mycomembrane layer, a feature that we are currently investigating. The specificity of PG labeling was confirmed by the terminal L-Ala control for both TetraFl and PentaFl. Differences in labeling between TetraFl and PentaFl may reflect the ability of Ldts and PBPs to tolerate the lysine residue on position 3 (as opposed to the natural m-DAP). In addition, iso-Glu amidation was also found to be important for PG incorporation as demonstrated by the reduced fluorescence levels in cells treated with TetraFl-4.
Figure 6.
(A) Flow cytometry analysis of M. smegmatis (WT) treated overnight with 100 μM tetrapeptide or pentapeptide with variations (see Figure 3). Data are represented as mean + SD (n = 3). (B) Flow cytometry analysis of M. smegmatis (WT) and Ldt knockout mutants treated overnight with 100 μM TetraFl, TetraFl-2, or TetraFl-5. Data are represented as mean + SD (n = 3). (C) Confocal microscopy image of M. smegmatis (WT) treated with 30 min pulse of 500 μM TetraRh, 500 μM PentaFl, and 5 mM DADA (scale bar: 2 μm). (D) Flow cytometry analysis of M. smegmatis (WT) treated overnight with 100 μM TetraFl or PentaFl with increasing concentrations of ampicillin or meropenum. Data are represented as mean + SD (n = 3).
We next used Ldt-deletion mutant M. smegmatis strains to establish the contribution of Ldts to cell labeling by tetrapeptide probes. Strains of M. smegmatis were treated with a subset of three tetrapeptides (TetraFl, TetraFl-2, and TetraFl-5) (Figure 6B). A clear reduction in labeling levels was observed in the single Ldt deletion mutant (ΔldtC) across both TetraFl and TetraFl-5 suggestive of this enzyme being involved in incorporation of Ldt probes. Further deletion of Ldts led to a greater than 5-fold reduction.31 The retention of cellular labeling in the triple-deletion strain is most likely a result of the three Ldt genes encoded in the M. smegmatis genome. As expected, treatment with the stereocontrol TetraFl-2 led to basal cell surface labeling levels across all strains. Together, these results implicate Ldts as being the primary mode of PG incorporation by tetrapeptide probes.
The localization of PG cross-linking by the two primary TP modes in M. smegmatis was visualized using confocal microscopy (Figure 6C). Strikingly, clear spatial separation was observed between TetraRh and DADA. DADA-labeling was observed primarily at the pole, and TetraRh-labeling was extensive throughout the cell sidewalls. Pole labeling observed with DADA is similar to single-amino acid probes previously reported for mycobacteria.72 More specifically, a single pole within a dividing cell was labeled more prominently with DADA than the other pole. Co-incubation of M. smegmatis cells with both TetraRh and DADA revealed that primary labeling sites with DADA are mostly devoid of TetraRh labeling.
The sensitivity of the tetra- and pentapeptide probes against a range of antibiotics was also measured in M. smegmatis (Figure 6D). In contrast to our observations with E. faecium, titration of the β-lactam ampicillin led to no observable change in fluorescence levels for TetraFl treated M. smegmatis cells. As expected, ampicillin treatment led to a concentration-dependent decrease in fluorescence in M. smegmatis incubated with PentaFl. Treatment with a carbapenem antibiotic (Meropenem) led to reductions in cellular fluorescence in cells treated with either TetraFl or PentaFl. Together, these results show a lack of response to ampicillin in TetraFl labeling of M. smegmatis and the inhibition of TPs results in reduced labeling levels. The role of iso-Glu amidation in the incorporation of PG probes, and hence PG cross-linking, was also confirmed by treatment of M. smegmatis cells with PentaRh and PentaRh-6 and visualized by fluorescence microscopy (Figure S15). Unmodified iso-Glu in the second position of the stem peptide resulted in background labeling levels. Labeling was shown to be mediated by enzymatic processes as heat-killed M. smegmatis cells did not show any labeling in the presence of PentaRh.
Finally, labeling experiments were extended to M. tuberculosis, the causative agent of tuberculosis. M. tuberculosis cells were incubated with TetraRh and imaged using confocal microscopy at various time points to analyze the progression of surface labeling (Figure 7 and Figure S16). Within 30 min, there was a unique labeling pattern that was contained within segments of cells. At longer incubation times, there was complete labeling throughout the sidewalls of most cells analyzed. Interestingly, treatment with sublethal concentrations of Meropenem resulted in morphological changes that caused bulging of the pole and more accentuated polar labeling (Figure S17). As in the case for both M. smegmatis and E. faecium, two other organisms that express Ltds, labeling of M. tuberculosis cells with TetraRh resulted in higher cellular fluorescence levels than PentaRh (Figure S18). In addition, it was confirmed that amidation of iso-Glu plays a determinant role in PG cross-linking in M. tuberculosis (Figure S19).
Figure 7.
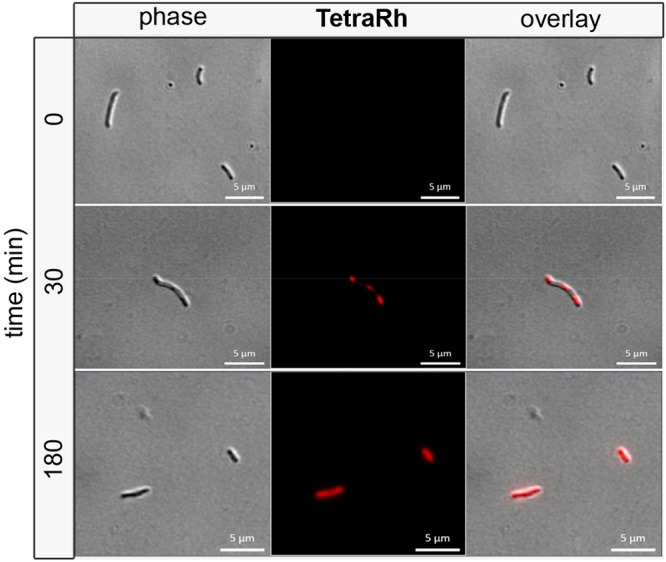
Confocal microscopy image of M. tuberculosis treated with 50 μM TetraRh for 30 min and 3 h (scale bar: 5 μm).
Discussion
Despite the fundamental importance of PG cross-linking to the growth and division of bacterial cells, key questions relating to enzymes that drive this process remain unanswered. These include the following: Are both modes of TP operative at the same time in certain organisms? How are PBP TPs and Ldts organized spatially within a cell? How do structural modifications within the stem peptide control PG cross-linking. Answers to these questions can greatly enhance our current understanding of PG biosynthetic control and dynamics. To address some of these questions, we hypothesized that we can specifically identify Ldt-mediated cell wall cross-linking in live bacterial cells using synthetic nascent PG analogs. We built mimics of the substrate stem peptides for both PBP TPs and Ldts that could serve as surrogates for the endogenous PG substrate in PG cross-linking, thereby becoming covalently imbedded within the PG scaffold in live bacteria.
Structural mimicry of TP substrates served to reveal how the primary structure of PG stem peptides may alter or control PG cross-linking levels in live cells. Our probes may complement existing D-amino acid probes to study cell wall biosynthesis. It is well established that the incubation of bacterial cells with noncanonical D-amino acids can result in PG incorporation at the fourth or fifth position within the stem peptide.6 The site and mode of exogenous D-amino acid incorporation is highly dependent on the bacterial species and can happen via Ldts, D,D-transpeptidases, and also through the intracellular Ddl ligase pathway6 (independent of TPs). The intracellular route was shown to be the preferred mode of incorporation for the alkyne-displaying D-amino acid Listeria monocytogenes (AlkDAla).73 While in some organisms exogenous D-amino acids are exclusively incorporated by Ldts,40,42,52,70,74,75 the mode of incorporation cannot be controlled and is entirely dictated by how the bacteria naturally processes D-amino acids. In other instances, it may be the case that incorporation into the PG is mediated by two or more pathways thus making it impossible to isolate the PG-remodeling mode.6 In contrast, the tetrapeptide based probes should only be processed by Ldts, thus allowing for the specific Ldt-based labeling.
Another distinction in using stem peptide analogs to probe PG processing, compared to single amino acid probes, is that it provides a mode to investigate how the primary structure can control cross-linking levels. In this work, we showed that amidation of iGlu is essential for robust levels of PG cross-linking. These results confirmed that amidation of the stem peptide by MurT/GatD may play a pivotal role in dictating PG cross-linking levels by Ldts. A recent CRISPRi phenotype screen identified that deletion of the enzymes responsible for the amidation of iso-Glu (MurT/GatD) is lethal, which may reflect the lack of PG cross-linking in the absence of iso-Glu amidation.76 These results confirm that structurally subtle changes to the stem peptide structure can potentially impact PG cross-linking levels in live bacterial cells and confirm that MurT/GatD may be a promising antibiotic target. Another example of how we can interrogate PG cross-linking in live cells was demonstrated by the antibiotic challenge in the presence of the PG probes we developed. The exposure of E. faecium cells to ampicillin led to increased labeling of cells. These results suggest that there may be an adaptation response by E. faecium cells when challenged with ampicillin. Bacteria are armed with a number of strategies that allow them to respond to potentially toxic agents, which can be the basis for drug-resistant phenotypes.77 In fact, inducible antibiotic responses have been previously described in enterococci.78−82 We are currently investigating possible response elements that may be responsible for the observed increase in TetraFl labeling.
In conclusion, we have demonstrated for the first time that synthetic tetrapeptide analogs of nascent PG can be incorporated onto PG scaffolds by Ldts in live bacterial cells. The tolerability of N-terminal modification on the synthetic stem peptide allowed for a fluorescent handle to quantify Ldt-based PG incorporation and track the delineation of Ldts across cell surfaces in E. faecium, M. smegmatis, and M. tuberculosis. With these cellular probes in hand, we were able to illustrate how subtle structural modifications to the primary sequence of the stem peptide can control cross-linking efficiency, including recapitulating in vitro results related to iso-Glu amidation. These results are the first live cell confirmation that the enzymes responsible for the amidation of iso-Glu (MurT/GatD) may be potential drug targets. Upon evaluating how cross-linking was altered when challenged with antibiotics, an induction in labeling with the tetra- but not the pentapeptide probe was observed. Additional studies are ongoing to understand if this could represent a drug-resistance mechanism that is related to cellular stress.
Methods
Flow Cytometry Analysis of Bacteria Labeling with TetraFl or PentaFl
Brain heart infusion (BHI) broth containing 100 μM TetraFl or PentaFl was prepared. E. faecium WT (D344s) or drug-resistant E. faecium (M9) from an overnight culture was added to the medium (1:100 dilution) and allowed to grow overnight at 37 °C with shaking at 250 rpm. The bacteria were harvested at 6,000g and washed three times with original culture volume of 1× PBS followed by fixation with 2% formaldehyde in 1× PBS for 30 min at ambient temperature. The cells were washed once more to remove formaldehyde and then analyzed using a BDFacs Canto II flow cytometer using a 488 nm argon laser (L1) and a 530/30 bandpass filter (FL1). A minimum of 10,000 events were counted for each data set. The data was analyzed using the FACSDiva version 6.1.1. For Mycobacterium smegmatis ATCC 14468, the previous procedure was repeated except using LB (0.05% tween) as the growth media.
Flow Cytometry Analysis of E. faecium Labeled with Tetrapeptide or Pentapeptide Variations
Brain heart infusion (BHI) broth containing 100 μM of compounds in Figure 3 was prepared. E. faecium WT (D344s) or drug-resistant E. faecium (M9) from an overnight culture was added to the medium (1:100 dilution) and allowed to grow overnight at 37 °C with shaking at 250 rpm. The bacteria were harvested at 6,000g and washed three times with original culture volume of 1× PBS followed by fixation with 2% formaldehyde in 1× PBS for 30 min at ambient temperature. The cells were washed once more to remove formaldehyde and then analyzed using a BDFacs Canto II flow cytometer using the previously stated parameters. For Mycobacterium smegmatis ATCC 14468, the previous procedure was repeated except using LB (0.05% Tween) as the growth media.
Flow Cytometry Analysis of Antibiotic Treated E. faecium M9 Labeled with TetraFl or PentaFl
Brain heart infusion (BHI) broth containing 100 μM TetraFl or PentaFl was prepared. To the medium was added antibiotics ampicillin, amoxicillin, Meropenem, imipenem, ceftriaxone, aztreonam, vancomycin, or erythromycin at varying submic concentrations. E. faecium (M9) was added to the corresponding medium (1:100 dilution) and allowed to grow overnight at 37 °C with shaking at 250 rpm. The bacteria were harvested at 6,000g and washed three times with original culture volume of 1× PBS followed by fixation with 2% formaldehyde in 1× PBS for 30 min at ambient temperature. The cells were washed once more to remove formaldehyde and then analyzed using a BDFacs Canto II flow cytometer using the previously stated parameters. For Mycobacterium smegmatis ATCC 14468, the previous procedure was repeated except using LB (0.05% Tween) as the growth media.
Time Course Analysis of Antibiotic Treated E. faecium M9 Labeled with TetraFl
Brain heart infusion (BHI) broth containing 100 μM TetraFl was prepared. To the medium was added antibiotics ampicillin (final concentration 16 μg/mL) or Meropenem (final concentration 8 μg/mL) or DMSO (final concentration 1%). E. faecium (M9) was added to the corresponding medium (1:10 dilution) and incubated at 37 °C with shaking at 250 rpm. Samples were collected at various time points, washed three times with 1× PBS, and fixed with 2% formaldehyde in 1× PBS for 30 min at ambient temperature. The cells were washed once more to remove formaldehyde and then analyzed using a BDFacs Canto II flow cytometer using the previously stated parameters.
Acknowledgments
This study was supported by the National Institutes of Health grant GM124893-01 (M.M.P.). M.S.P. was supported by the National Institutes of Health grant AI073772. B.D.K. was supported by the Bill and Melinda Gates Foundation, the Howard Hughes Medical Institute, the South African Medical Research Council with funds received from the National Department of Health, and the South African National Research Foundation. M9 and D344S strains in E. faecium were kind gifts from M. Arthur (UPMC Université Paris).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.9b00427.
Additional experimental details (methods, characterization, and synthesis of PG analogs) and figures (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Vollmer W.; Blanot D.; de Pedro M. A. (2008) Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167. 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- Vollmer W.; Bertsche U. (2008) Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta, Biomembr. 1778, 1714–1734. 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Typas A.; Banzhaf M.; Gross C. A.; Vollmer W. (2012) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136. 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez L.; Espaillat A.; Hermoso J. A.; de Pedro M. A.; Cava F. (2014) Peptidoglycan remodeling by the coordinated action of multispecific enzymes. Microb. Drug Resist. 20, 190–198. 10.1089/mdr.2014.0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H.; Oh D. C.; Cava F.; Takacs C. N.; Clardy J.; de Pedro M. A.; Waldor M. K. (2009) D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325, 1552–1555. 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava F.; de Pedro M. A.; Lam H.; Davis B. M.; Waldor M. K. (2011) Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 30, 3442–3453. 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond J. B.; Mahapatra S.; Crick D. C.; Pavelka M. S. Jr. (2005) Identification of the namH gene, encoding the hydroxylase responsible for the N-glycolylation of the mycobacterial peptidoglycan. J. Biol. Chem. 280, 326–333. 10.1074/jbc.M411006200. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Lazor K. M.; DeMeester K. E.; Liang H.; Heiss T. K.; Grimes C. L. (2017) Postsynthetic Modification of Bacterial Peptidoglycan Using Bioorthogonal N-Acetylcysteamine Analogs and Peptidoglycan O-Acetyltransferase B. J. Am. Chem. Soc. 139, 13596–13599. 10.1021/jacs.7b06820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W. (2008) Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 32, 287–306. 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- Mahapatra S.; Yagi T.; Belisle J. T.; Espinosa B. J.; Hill P. J.; McNeil M. R.; Brennan P. J.; Crick D. C. (2005) Mycobacterial lipid II is composed of a complex mixture of modified muramyl and peptide moieties linked to decaprenyl phosphate. J. Bacteriol. 187, 2747–2757. 10.1128/JB.187.8.2747-2757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis K. M.; Weiser J. N. (2011) Modifications to the peptidoglycan backbone help bacteria to establish infection. Infect. Immun. 79, 562–570. 10.1128/IAI.00651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. D.; Foster E. E.; Wallace A. G.; Kim S. J. (2017) Peptidoglycan O-acetylation increases in response to vancomycin treatment in vancomycin-resistant Enterococcus faecalis. Sci. Rep. 7, 46500. 10.1038/srep46500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin P. (2006) Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 42 (Suppl. 1), S25–34. 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- Mainardi J. L.; Villet R.; Bugg T. D.; Mayer C.; Arthur M. (2008) Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol. Rev. 32, 386–408. 10.1111/j.1574-6976.2007.00097.x. [DOI] [PubMed] [Google Scholar]
- Boudreau M. A.; Fisher J. F.; Mobashery S. (2012) Messenger functions of the bacterial cell wall-derived muropeptides. Biochemistry 51, 2974–2990. 10.1021/bi300174x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah I. M.; Laaberki M. H.; Popham D. L.; Dworkin J. (2008) A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135, 486–496. 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan K. J.; Pedicord V. A.; Wang Y. C.; Kim B.; Lu Y.; Shaham S.; Mucida D.; Hang H. C. (2016) A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science 353, 1434–1437. 10.1126/science.aaf3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin S. E.; Boneca I. G.; Viala J.; Chamaillard M.; Labigne A.; Thomas G.; Philpott D. J.; Sansonetti P. J. (2003) Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278, 8869–8872. 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- Girardin S. E.; Travassos L. H.; Herve M.; Blanot D.; Boneca I. G.; Philpott D. J.; Sansonetti P. J.; Mengin-Lecreulx D. (2003) Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J. Biol. Chem. 278, 41702–41708. 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- Wolf A. J.; Underhill D. M. (2018) Peptidoglycan recognition by the innate immune system. Nat. Rev. Immunol. 18, 243–254. 10.1038/nri.2017.136. [DOI] [PubMed] [Google Scholar]
- Sharif S.; Kim S. J.; Labischinski H.; Chen J.; Schaefer J. (2013) Uniformity of glycyl bridge lengths in the mature cell walls of fem mutants of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 195, 1421–1427. 10.1128/JB.01471-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W.; Holtje J. V. (2004) The architecture of the murein (peptidoglycan) in gram-negative bacteria: vertical scaffold or horizontal layer(s)?. J. Bacteriol. 186, 5978–5987. 10.1128/JB.186.18.5978-5987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyette J.; Perkins H. R.; Polacheck I.; Shockman G. D.; Ghuysen J. M. (1974) Membrane-bound DD-carboxypeptidase and LD-transpeptidase of Streptococcus faecalis ATCC 9790. Eur. J. Biochem. 44, 459–468. 10.1111/j.1432-1033.1974.tb03504.x. [DOI] [PubMed] [Google Scholar]
- Lecoq L.; Dubee V.; Triboulet S.; Bougault C.; Hugonnet J. E.; Arthur M.; Simorre J. P. (2013) Structure of Enterococcus faeciuml,d-transpeptidase acylated by ertapenem provides insight into the inactivation mechanism. ACS Chem. Biol. 8, 1140–1146. 10.1021/cb4001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainardi J. L.; Legrand R.; Arthur M.; Schoot B.; van Heijenoort J.; Gutmann L. (2000) Novel mechanism of beta-lactam resistance due to bypass of DD-transpeptidation in Enterococcus faecium. J. Biol. Chem. 275, 16490–16496. 10.1074/jbc.M909877199. [DOI] [PubMed] [Google Scholar]
- Magnet S.; Arbeloa A.; Mainardi J. L.; Hugonnet J. E.; Fourgeaud M.; Dubost L.; Marie A.; Delfosse V.; Mayer C.; Rice L. B.; Arthur M. (2007) Specificity of L,D-transpeptidases from gram-positive bacteria producing different peptidoglycan chemotypes. J. Biol. Chem. 282, 13151–13159. 10.1074/jbc.M610911200. [DOI] [PubMed] [Google Scholar]
- Lavollay M.; Arthur M.; Fourgeaud M.; Dubost L.; Marie A.; Veziris N.; Blanot D.; Gutmann L.; Mainardi J. L. (2008) The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. J. Bacteriol. 190, 4360–4366. 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J.; Courtin P.; El Meouche I.; Lemee L.; Chapot-Chartier M. P.; Pons J. L. (2011) Clostridium difficile has an original peptidoglycan structure with a high level of N-acetylglucosamine deacetylation and mainly 3–3 cross-links. J. Biol. Chem. 286, 29053–29062. 10.1074/jbc.M111.259150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet S.; Dubost L.; Marie A.; Arthur M.; Gutmann L. (2008) Identification of the L,D-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J. Bacteriol. 190, 4782–4785. 10.1128/JB.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka M. S. Jr.; Mahapatra S.; Crick D. C. (2014) Genetics of Peptidoglycan Biosynthesis. Microbiol. Spectrum 2, MGM2-0034-2013–2013. 10.1128/microbiolspec.MGM2-0034-2013. [DOI] [PubMed] [Google Scholar]
- Squeglia F.; Ruggiero A.; Berisio R. (2018) Chemistry of Peptidoglycan in Mycobacterium tuberculosis Life Cycle: An off-the-wall Balance of Synthesis and Degradation. Chem. - Eur. J. 24, 2533–2546. 10.1002/chem.201702973. [DOI] [PubMed] [Google Scholar]
- Kieser K. J.; Baranowski C.; Chao M. C.; Long J. E.; Sassetti C. M.; Waldor M. K.; Sacchettini J. C.; Ioerger T. R.; Rubin E. J. (2015) Peptidoglycan synthesis in Mycobacterium tuberculosis is organized into networks with varying drug susceptibility. Proc. Natl. Acad. Sci. U. S. A. 112, 13087–13092. 10.1073/pnas.1514135112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.; Lavollay M.; Mainardi J. L.; Arthur M.; Bishai W. R.; Lamichhane G. (2010) The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat. Med. 16, 466–469. 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders A. N.; Wright L. F.; Pavelka M. S. Jr. (2014) Genetic characterization of mycobacterial L,D-transpeptidases. Microbiology 160, 1795–1806. 10.1099/mic.0.078980-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco E.; Hugonnet J. E.; Josseaume N.; Cremniter J.; Dubost L.; Marie A.; Patin D.; Blanot D.; Rice L. B.; Mainardi J. L.; Arthur M. (2010) Activation of the L,D-transpeptidation peptidoglycan cross-linking pathway by a metallo-D,D-carboxypeptidase in Enterococcus faecium. Mol. Microbiol. 75, 874–885. 10.1111/j.1365-2958.2009.07014.x. [DOI] [PubMed] [Google Scholar]
- de Jonge B. L.; Handwerger S.; Gage D. (1996) Altered peptidoglycan composition in vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 40, 863–869. 10.1128/AAC.40.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M.; Depardieu F.; Snaith H. A.; Reynolds P. E.; Courvalin P. (1994) Contribution of VanY D,D-carboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob. Agents Chemother. 38, 1899–1903. 10.1128/AAC.38.9.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann L.; Billot-Klein D.; al-Obeid S.; Klare I.; Francoual S.; Collatz E.; van Heijenoort J. (1992) Inducible carboxypeptidase activity in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 36, 77–80. 10.1128/AAC.36.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco E.; Cortes M.; Josseaume N.; Rice L. B.; Mainardi J. L.; Arthur M. (2014) Serine/threonine protein phosphatase-mediated control of the peptidoglycan cross-linking L,D-transpeptidase pathway in Enterococcus faecium. mBio 5, e01446-14. 10.1128/mBio.01446-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru E.; Hughes H. V.; Brown P. J.; Hall E.; Tekkam S.; Cava F.; de Pedro M. A.; Brun Y. V.; VanNieuwenhze M. S. (2012) In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew. Chem., Int. Ed. 51, 12519–12523. 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru E.; Tekkam S.; Hall E.; Brun Y. V.; Van Nieuwenhze M. S. (2015) Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat. Protoc. 10, 33–52. 10.1038/nprot.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist M. S.; Whiteside S.; Jewett J. C.; Aditham A.; Cava F.; Bertozzi C. R. (2013) (D)-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem. Biol. 8, 500–505. 10.1021/cb3004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist M. S.; Swarts B. M.; Fox D. M.; Lim S. A.; Bertozzi C. R. (2015) Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface. FEMS Microbiol. Rev. 39, 184–202. 10.1093/femsre/fuu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebar M. D.; May J. M.; Meeske A. J.; Leiman S. A.; Lupoli T. J.; Tsukamoto H.; Losick R.; Rudner D. Z.; Walker S.; Kahne D. (2014) Reconstitution of peptidoglycan cross-linking leads to improved fluorescent probes of cell wall synthesis. J. Am. Chem. Soc. 136, 10874–10877. 10.1021/ja505668f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupoli T. J.; Tsukamoto H.; Doud E. H.; Wang T. S.; Walker S.; Kahne D. (2011) Transpeptidase-mediated incorporation of D-amino acids into bacterial peptidoglycan. J. Am. Chem. Soc. 133, 10748–10751. 10.1021/ja2040656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fura J. M.; Pidgeon S. E.; Birabaharan M.; Pires M. M. (2016) Dipeptide-Based Metabolic Labeling of Bacterial Cells for Endogenous Antibody Recruitment. ACS Infect. Dis. 2, 302–309. 10.1021/acsinfecdis.6b00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S.; Libby E. A.; Pidgeon S. E.; Dworkin J.; Pires M. M. (2016) In Vivo Probe of Lipid II-Interacting Proteins. Angew. Chem., Int. Ed. 55, 8401–8404. 10.1002/anie.201603441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidgeon S. E.; Fura J. M.; Leon W.; Birabaharan M.; Vezenov D.; Pires M. M. (2015) Metabolic Profiling of Bacteria by Unnatural C-terminated D-Amino Acids. Angew. Chem., Int. Ed. 54, 6158–6162. 10.1002/anie.201409927. [DOI] [PubMed] [Google Scholar]
- Liechti G. W.; Kuru E.; Hall E.; Kalinda A.; Brun Y. V.; VanNieuwenhze M.; Maurelli A. T. (2014) A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506, 507–510. 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro J. M.; Pereira A. R.; Reichmann N. T.; Saraiva B. M.; Fernandes P. B.; Veiga H.; Tavares A. C.; Santos M.; Ferreira M. T.; Macario V.; VanNieuwenhze M. S.; Filipe S. R.; Pinho M. G. (2018) Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554, 528–532. 10.1038/nature25506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.; Lyu Z.; Miguel A.; McQuillen R.; Huang K. C.; Xiao J. (2017) GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747. 10.1126/science.aak9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson-Filho A. W.; Hsu Y. P.; Squyres G. R.; Kuru E.; Wu F.; Jukes C.; Sun Y.; Dekker C.; Holden S.; VanNieuwenhze M. S.; Brun Y. V.; Garner E. C. (2017) Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743. 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh M. A.; Taguchi A.; Schaefer K.; Van Tyne D.; Lebreton F.; Gilmore M. S.; Kahne D.; Walker S. (2017) Identification of a Functionally Unique Family of Penicillin-Binding Proteins. J. Am. Chem. Soc. 139, 17727–17730. 10.1021/jacs.7b10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam S.; Kim T.; Shoda T.; Sen S.; Deep D.; Luthra R.; Ferreira M. T.; Pinho M. G.; Spiegel D. A. (2015) An Activity-Based Probe for Studying Crosslinking in Live Bacteria. Angew. Chem., Int. Ed. 54, 10492–10496. 10.1002/anie.201503869. [DOI] [PubMed] [Google Scholar]
- Gautam S.; Kim T.; Spiegel D. A. (2015) Chemical probes reveal an extraseptal mode of cross-linking in Staphylococcus aureus. J. Am. Chem. Soc. 137, 7441–7447. 10.1021/jacs.5b02972. [DOI] [PubMed] [Google Scholar]
- Ngadjeua F.; Braud E.; Saidjalolov S.; Iannazzo L.; Schnappinger D.; Ehrt S.; Hugonnet J. E.; Mengin-Lecreulx D.; Patin D.; Etheve-Quelquejeu M.; Fonvielle M.; Arthur M. (2018) Critical Impact of Peptidoglycan Precursor Amidation on the Activity of l,d-Transpeptidases from Enterococcus faecium and Mycobacterium tuberculosis. Chem. - Eur. J. 24, 5743–5747. 10.1002/chem.201706082. [DOI] [PubMed] [Google Scholar]
- Zapun A.; Philippe J.; Abrahams K. A.; Signor L.; Roper D. I.; Breukink E.; Vernet T. (2013) In vitro reconstitution of peptidoglycan assembly from the Gram-positive pathogen Streptococcus pneumoniae. ACS Chem. Biol. 8, 2688–2696. 10.1021/cb400575t. [DOI] [PubMed] [Google Scholar]
- Sacco E.; Cortes M.; Josseaume N.; Bouchier C.; Dubee V.; Hugonnet J. E.; Mainardi J. L.; Rice L. B.; Arthur M. (2015) Mutation landscape of acquired cross-resistance to glycopeptide and beta-lactam antibiotics in Enterococcus faecium. Antimicrob. Agents Chemother. 59, 5306–5315. 10.1128/AAC.00634-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubee V.; Arthur M.; Fief H.; Triboulet S.; Mainardi J. L.; Gutmann L.; Sollogoub M.; Rice L. B.; Etheve-Quelquejeu M.; Hugonnet J. E. (2012) Kinetic analysis of Enterococcus faecium L,D-transpeptidase inactivation by carbapenems. Antimicrob. Agents Chemother. 56, 3409–3412. 10.1128/AAC.06398-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainardi J. L.; Hugonnet J. E.; Rusconi F.; Fourgeaud M.; Dubost L.; Moumi A. N.; Delfosse V.; Mayer C.; Gutmann L.; Rice L. B.; Arthur M. (2007) Unexpected inhibition of peptidoglycan LD-transpeptidase from Enterococcus faecium by the beta-lactam imipenem. J. Biol. Chem. 282, 30414–30422. 10.1074/jbc.M704286200. [DOI] [PubMed] [Google Scholar]
- Ford M.; Perry J. D.; Gould F. K. (1994) Use of cephalexin-aztreonam-arabinose agar for selective isolation of Enterococcus faecium. J. Clin. Microbiol. 32, 2999–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hujer A. M.; Kania M.; Gerken T.; Anderson V. E.; Buynak J. D.; Ge X.; Caspers P.; Page M. G.; Rice L. B.; Bonomo R. A. (2005) Structure-activity relationships of different beta-lactam antibiotics against a soluble form of Enterococcus faecium PBP5, a type II bacterial transpeptidase. Antimicrob. Agents Chemother. 49, 612–618. 10.1128/AAC.49.2.612-618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lara J.; Needham A. J.; Foster S. J. (2005) Invertebrates as animal models for Staphylococcus aureus pathogenesis: a window into host-pathogen interaction. FEMS Immunol. Med. Microbiol. 43, 311–323. 10.1016/j.femsim.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Sifri C. D.; Begun J.; Ausubel F. M.; Calderwood S. B. (2003) Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun. 71, 2208–2217. 10.1128/IAI.71.4.2208-2217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin D. A.; Villanueva J. M.; Begun J.; Kim D. H.; Sifri C. D.; Calderwood S. B.; Ruvkun G.; Ausubel F. M. (2003) Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300, 1921. 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- Pidgeon S. E.; Pires M. M. (2017) Cell Wall Remodeling of Staphylococcus aureus in Live Caenorhabditis elegans. Bioconjugate Chem. 28, 2310–2315. 10.1021/acs.bioconjchem.7b00363. [DOI] [PubMed] [Google Scholar]
- Sabulski M. J.; Pidgeon S. E.; Pires M. M. (2017) Immuno-targeting of Staphylococcus aureus via surface remodeling complexes. Chem. Sci. 8, 6804–6809. 10.1039/C7SC02721D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranowski C.; Welsh M. A.; Sham L. T.; Eskandarian H. A.; Lim H. C.; Kieser K. J.; Wagner J. C.; McKinney J. D.; Fantner G. E.; Ioerger T. R.; Walker S.; Bernhardt T. G.; Rubin E. J.; Rego E. H. (2018) Maturing Mycobacterium smegmatis peptidoglycan requires non-canonical crosslinks to maintain shape. eLife 7, e37516. 10.7554/eLife.37516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Heredia A.; Pohane A. A.; Melzer E. S.; Carr C. R.; Fiolek T. J.; Rundell S. R.; Chuin Lim H.; Wagner J. C.; Morita Y. S.; Swarts B. M.; Siegrist M. S. (2018) Peptidoglycan precursor synthesis along the sidewall of pole-growing mycobacteria. eLife 7, e37243. 10.7554/eLife.37243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges H. L.; Brown R. A.; Crooks J. A.; Weibel D. B.; Kiessling L. L. (2018) Imaging mycobacterial growth and division with a fluorogenic probe. Proc. Natl. Acad. Sci. U. S. A. 115, 5271–5276. 10.1073/pnas.1720996115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella H.; Yang G.; Ouerfelli O.; Ehrt S.; Nathan C. F.; Vaubourgeix J. (2017) Distinct Spatiotemporal Dynamics of Peptidoglycan Synthesis between Mycobacterium smegmatis and Mycobacterium tuberculosis. mBio 8, e01183-17. 10.1128/mBio.01183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo J. T.; Adams S. R.; Deerinck T. J.; Boassa D.; Rodriguez-Rivera F.; Palida S. F.; Bertozzi C. R.; Ellisman M. H.; Tsien R. Y. (2016) Click-EM for imaging metabolically tagged nonprotein biomolecules. Nat. Chem. Biol. 12, 459–465. 10.1038/nchembio.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlot C.; Straume D.; Peters K.; Hegnar O. A.; Simon N.; Villard A. M.; Contreras-Martel C.; Leisico F.; Breukink E.; Gravier-Pelletier C.; Le Corre L.; Vollmer W.; Pietrancosta N.; Havarstein L. S.; Zapun A. (2018) Structure of the essential peptidoglycan amidotransferase MurT/GatD complex from Streptococcus pneumoniae. Nat. Commun. 9, 3180. 10.1038/s41467-018-05602-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T.; Pazos M.; Lara-Tejero M.; Vollmer W.; Galan J. E. (2018) Peptidoglycan editing by a specific LD-transpeptidase controls the muramidase-dependent secretion of typhoid toxin. Nat. Microbiol. 3, 1243–1254. 10.1038/s41564-018-0248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Gallay C.; Kjos M.; Domenech A.; Slager J.; van Kessel S. P.; Knoops K.; Sorg R. A.; Zhang J. R.; Veening J. W. (2017) High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol. Syst. Biol. 13, 931. 10.15252/msb.20167449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair J. M.; Webber M. A.; Baylay A. J.; Ogbolu D. O.; Piddock L. J. (2015) Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- Miller W. R.; Munita J. M.; Arias C. A. (2014) Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti-Infect. Ther. 12, 1221–1236. 10.1586/14787210.2014.956092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucault M. L.; Depardieu F.; Courvalin P.; Grillot-Courvalin C. (2010) Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc. Natl. Acad. Sci. U. S. A. 107, 16964–16969. 10.1073/pnas.1006855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C.; Massier S.; Le Breton Y.; Rince A. (2018) The role of the CroR response regulator in resistance of Enterococcus faecalis to D-cycloserine is defined using an inducible receiver domain. Mol. Microbiol. 107, 416–427. 10.1111/mmi.13891. [DOI] [PubMed] [Google Scholar]
- Kellogg S. L.; Little J. L.; Hoff J. S.; Kristich C. J. (2017) Requirement of the CroRS Two-Component System for Resistance to Cell Wall-Targeting Antimicrobials in Enterococcus faecium. Antimicrob. Agents Chemother. 61, e02461-16. 10.1128/AAC.02461-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbonnet C.; Tait-Kamradt A.; Garcia-Solache M.; Dunman P.; Coleman J.; Arthur M.; Rice L. B. (2016) Involvement of the Eukaryote-Like Kinase-Phosphatase System and a Protein That Interacts with Penicillin-Binding Protein 5 in Emergence of Cephalosporin Resistance in Cephalosporin-Sensitive Class A Penicillin-Binding Protein Mutants in Enterococcus faecium. mBio 7, e02188-15. 10.1128/mBio.02188-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.