Abstract
Background
Oxalobacter formigenes are bacteria that colonize the human gut and degrade oxalate, a component of most kidney stones. Findings of clinical and epidemiological studies suggest that O. formigenes colonization reduces the risk for kidney stones. We sought to develop murine models to allow investigating O. formigenes in the context of its native human microbiome.
Methods
For humanization, we transplanted pooled feces from healthy, noncolonized human donors supplemented with a human O. formigenes strain into recipient mice. We transplanted microbiota into mice that were treated with broad-spectrum antibiotics to suppress their native microbiome, were germ free, or received humanization without pretreatment or received sham gavage (controls).
Results
All humanized mice were stably colonized with O. formigenes through 8 weeks after gavage, whereas mice receiving sham gavage remained uncolonized (P < .001). Humanization significantly changed the murine intestinal microbial community structure (P < .001), with humanized germ-free and antibiotic-treated groups overlapping in β-diversity. Both germ-free and antibiotic-treated mice had significantly increased numbers of human species compared with sham-gavaged mice (P < .001).
Conclusions
Transplanting mice with human feces and O. formigenes introduced new microbial populations resembling the human microbiome, with stable O. formigenes colonization; such models can define optimal O. formigenes strains to facilitate clinical trials.
Keywords: Commensal bacteria, nephrolithiasis, oxalate, metabolism, microbiome, xenobiosis, antibiotics
Because the human intestinal bacterium Oxalobacter formigenes may provide protection against kidney stones, improved mouse models are needed to assess both pathophysiology and treatment options. Humanizing antibiotic-treated or germ-free mice before introducing O. formigenes led to comparable and stable colonization.
The prevalence of nephrolithiasis has risen in recent decades [1], affecting approximately 1 in 11 persons in the United States [2]. Nephrolithiasis also causes substantial financial burden [3], and can lead to chronic kidney disease [4]. Most kidney stones (approximately 80%) are composed of calcium oxalate, and urinary oxalate excretion is a major risk factor [5]. Oxalate is a byproduct of amino acid catabolism and also is present in the diet [6]. The intestinal microbiota includes bacteria that degrade oxalate, which can potentially lower the host’s oxalate load. Oxalobacter formigenes is the most efficient oxalate degrader in the human intestine, using oxalate as its exclusive carbon and energy source [7, 8]. O. formigenes also may secrete bioactive factors that induce oxalate transport across intestinal epithelial cells into the gut lumen [9, 10].
In case-control studies, O. formigenes colonization was associated with a 70% reduced risk of recurrent oxalate stones [11]. However, in contrast to those living under premodern conditions in which O. formigenes is ubiquitous, only a minority of persons in the United States, United Kingdom, and other industrialized countries carry O. formigenes [11–15]. Exposure to commonly used oral antibiotics can eliminate O. formigenes [16], and a 2018 epidemiological study provides evidence that antibiotic use is associated with higher nephrolithiasis risk [17]. Strategies to introduce O. formigenes into patients with a high risk of kidney stones are being considered [18, 19].
With the potential of O. formigenes to lower nephrolithiasis risk, study of its intestinal habitat is needed. However, there have been few prospective human studies, because it is challenging to control dietary calcium and oxalate levels and other clinically relevant variables [20, 21]. Therefore, to elucidate the biology of O. formigenes in animal hosts, and because it is not native to laboratory mice, we introduced it together with a humanized microbiota [22]. We hypothesized that by introducing feces from human donors into germ-free mice, we could create a humanized model for O. formigenes carriage superior to that in conventional or antibiotic pretreated mice. We now report that we could successfully colonize both germ-free and antibiotic pretreated mice with human microbiota and O. formigenes, creating a stable humanized mouse with O. formigenes persistence for ≥8 weeks.
METHODS
Mice
Groups consisted of 7–11-week-old C57BL/6 mice. Conventional mice were purchased from Jackson Laboratories, and germ-free mice obtained from the National Gnotobiotic Rodent Resource Center at the University of North Carolina. Mice initially received normal chow until their first gavage, when an irradiated, high-oxalate diet (1% oxalate, 0.5% calcium; D16080401, Research Diets) was begun, and continued for the duration of the experiment. Mice were maintained on 12-hour light-dark cycles with ad libitum access to food and water. All mouse experiments were approved by the New York University School of Medicine Institutional Animal Care and Use Committee (protocol 160724) and complied with federal and institutional regulations.
O. formigenes Culture Preparation
For this study, we selected O. formigenes strain OxCC13, a well-characterized group 1 strain that colonizes germ-free mice [22] and has been used in human challenge studies. As such, we believed it appropriate to test in mouse models. Pure cultures were grown in Schaedler broth containing 100 mmol/L oxalate and 10 mmol/L sodium acetate [23] at 37°C in an anaerobic chamber. Optical density at 595 nm was measured performed using a SpectraMas iD3 plate reader, with readings of 0.09–0.1 for cultures used for gavage, approximately 108 colony-forming units/mL.
Preparation of Human Donor Feces
Human fecal samples were selected from 3 O. formigenes–negative donors after samples were negative by (1) polymerase chain reaction (PCR), (2) quantitative PCR (qPCR), and (3) oxalate degradation assay (4 samples each, obtained over a 6-month period). Fecal samples were stored at −80°C and then resuspended in anaerobic transport medium (Anaerobe Systems) to create an 80-mg/mL pooled suspension for humanization.
Humanization Protocol
Pooled human donor feces and O. formigenes cultures were combined under anaerobic conditions and administered to mice via gastric gavage. Mice receiving gavage (Figure 1) were either (1) conventional, (2) germ free; or (3) pretreated with vancomycin (0.5 g/L), ampicillin (1 g/L), neomycin sulfate (1 g/L), and metronidazole (1 g/L) (all from Sigma) [24, 25] to suppress their native microbiome. Gavage was performed with 10 µL/g of donor inoculum or sham (anaerobic transport medium alone), 3 times at 48-hour intervals. The timing in all groups was coordinated to gavage germ-free mice immediately on arrival to our facility, minimizing microbial exposures before humanization.
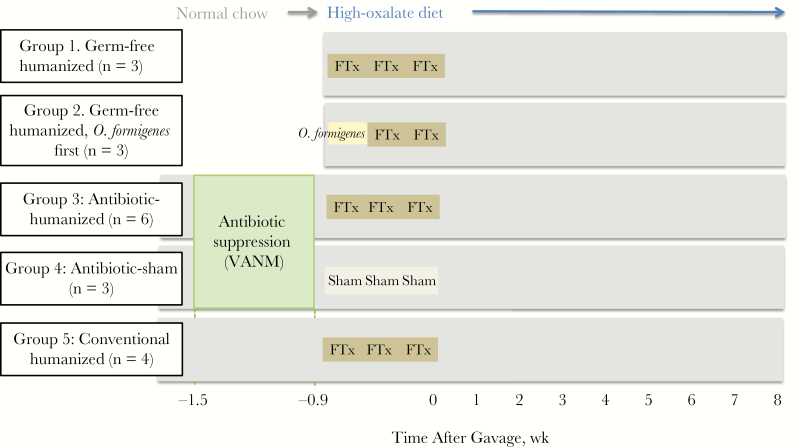
Figure 1.
Design of fecal humanization experiment in adult mice. Studies were conducted with germ-free (groups 1 and 2) or conventional (groups 3–5) C57BL/6J mice. Conventional mice either received antibiotics (0.5 g/L vancomycin, 1 g/L ampicillin, 1 g/L neomycin, 1 g/L metronidazole [VANM]) [24, 25] in their drinking water for 6 days or did not. Within 36 hours after ending the antibiotic course (or at the same time point in those who did not receive VANM), all mice were gavaged with either pooled human feces to which Oxalobacter formigenes was added (FTx), O. formigenes alone, or transport medium (sham) alone. All mice were followed up for 8 weeks after gavage.
Later Antibiotic Exposure
To attempt to eliminate O. formigenes from stably colonized mice, we used a pulsed antibiotic treatment (PAT) known to affect the murine microbiome [26–28]. Tylosin tartrate (Sigma) was dissolved (333 mg/L) in nonacidified water to achieve a therapeutic dose [29] of approximately 50 mg/kg body weight/d in recipient mice, as described elsewhere [26–28].
DNA Isolation
Murine fecal pellets and the human fecal gavage inoculum were collected and stored at −80°C until DNA was extracted, using the MoBio 96-well extraction kit, following the manufacturer’s instructions.
PCR Protocol
PCR to detect the O. formigenes oxc gene was performed using Taq DNA Polymerase and ×10 CoralLoad PCR buffer (Qiagen), with oligonucleotide primers (forward, 5’- ATGTAGAGTTGACTGATGGC-3’; reverse, 5’-TTGATG CTGTTGATACG-3’). PCR conditions included an initial 95°C denaturation for 3 minutes and 39 cycles at 95°C for 30 seconds, annealing at 54°C for 30 seconds, and extension at 72°C for 1 minute. PCR products were analyzed by electrophoresis using 2% agarose gels.
qPCR Protocol
qPCR to quantitate O. formigenes oxc copies was performed using LightCycler 480 SYBR Green І master mix and the LightCycler 480 system (Roche) (primers: forward, 5’-GTGTTGTCGGCATTCCTATC-3’; reverse 5’-GAAGCAGTTGGTGGTTGC-3’). Samples were prepared under a laminar flow hood, with each reaction well containing 16 µL of master mix, 0.5 µmol/L of each primer, and 2 µL of DNA template. Cycling conditions included an initial 95°C incubation for 10 minutes, followed by 40 cycles at 95°C for 23 seconds, annealing at 63°C for 20 seconds, and extension at 70°C for 40 seconds. Melting peak analysis was performed from 65°C to 95°C to confirm amplicon specificity. A positive result was defined by amplification >1.0E2, with melting peak between 86°C and 87°C. Levels of oxc were normalized to total 16S DNA by performing qPCR with universal 16S primers (forward 5′-GGM TTA GAT ACC CBG GTA GTC C-3′, reverse 5′-CCG TCA ATT CMT TTG AGT TT-3′) as follows: 95°C for 10 minutes, followed by 40 cycles of 95°C for 10 seconds, 60°C for 20 seconds, 72°C for 20 seconds, and a final 30 seconds at 40°C.
Oxalate Degradation Assay
Donor feces were inoculated into O. formigenes–specific growth medium B [8] supplemented with sodium oxalate (20 mmol/l) and then incubated at 37°C for 10 days. Degradation of oxalate by O. formigenes was assessed by adding 100 nmol/L calcium chloride to culture supernatant.
Sequencing
For amplicon library generation, the 16S ribosomal RNA (rRNA) gene V4 region was amplified with gene-specific primers, as described elsewhere [30]. The reverse amplification primers contained a 12–base pair (bp) Golay barcode for multiplexed sequencing runs. Amplicons were prepared in triplicate, and DNA concentrations were measured using the QuantiT PicoGreen double-stranded DNA Assay kit (Invitrogen) and pooled at equal DNA quantities. After samples were combined in subpools of approximately 96 samples, excess primers were removed with the Qiaquick PCR purification kit (Qiagen). DNA concentrations in these subpools were quantified with the Qubit high-sensitivity double-stranded DNA Assay (Invitrogen) and combined at equal concentrations. The 254-bp V4 region was sequenced using the Ilumina MiSeq 2 × 150–bp platform. The data sets generated and/or analyzed in the current study are available in the National Center for Biotechnology Information's Sequence Read Archive repository (under no. SUB4058073).
Data Analysis
Raw data processing was performed with QIIME-1 software [31]. Operational taxonomical units (OTUs) were picked against reference database GreenGenes 13_8 [32], using the open reference picking strategy [31]. For quality filtering, OTUs identified as chimerae (ChimeraSlayer method [33]) were excluded, as were OTUs with only a single read.
Statistics
An unpaired, 2-tailed t test was used to compare oxc/16S levels. For examination of the ß-diversity differences in microbial communities, Adonis tests with 999 permutations were used, and multiple-comparison P values were corrected using Bonferroni methods. Wilcoxon rank sum tests with multiple comparisons corrected by false discovery rate methods were used in the remaining statistical analyses, unless noted.
RESULTS
Study Design
We included 5 study groups to assess O. formigenes colonization efficacy and effects of both the humanizing gavage and antibiotics on the microbiota (Figure 1). In germ-free mice, group 1 (germ-free humanized) received 3 successive gavages of a pooled human fecal (O. formigenes–negative) suspension, to which O. formigenes was added. In group 2 (germ-free humanized, O. formigenes first), germ-free mice received a first gavage consisting only of O. formigenes to provide a potential colonization advantage, and with 2 subsequent gavages with the standard pooled feces together with O. formigenes. Two other groups received antibiotic treatment to suppress the normal microbiota; mice in group 3 (antibiotic-humanized) were given the same humanization regimen as in group 1, and those in group 4 (antibiotic-sham) were not humanized at all but were given a sham (control) gavage of transport medium. Mice in group 5 (conventional humanized) were gavaged with the human fecal suspension with O. formigenes, but did not receive any antibiotic before treatment. At the time of the first gavage, all mice were begun on the high-oxalate diet.
Oxalobacter Colonization
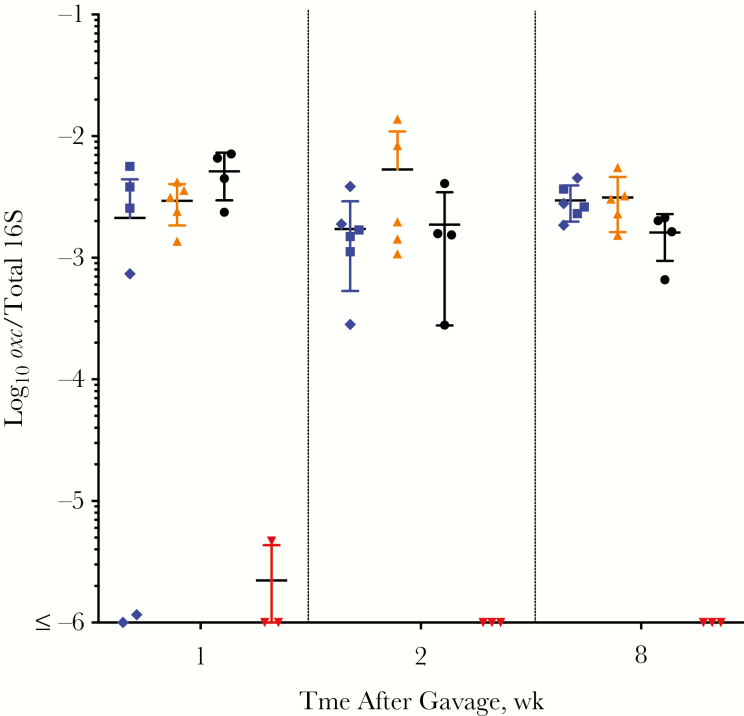
First, we asked whether these humanization methods permitted O. formigenes to be successfully introduced into the mouse intestine. Using an O. formigenes oxc-specific qPCR assay, we found that all of the humanization strategies were successful, with every challenged mouse colonized with O. formigenes, at levels significantly higher than sham (Figure 2). One week after gavage, the (group 2 [germ-free humanized, O. formigenes first]) germ-free mice that received O. formigenes alone in the first gavage initially had approximately 16-fold greater colonization than the mice (group 1 [germ-free humanized, O. formigenes first]) that did not receive the initial O. formigenes–alone inoculum, but the differences gradually diminished. All humanized mice remained colonized with O. formigenes for ≥8 weeks, stabilizing at approximately 2.7 × 10−3oxc/16S. These levels are comparable to the relative O. formigenes abundance in the human microbiome, as detected by means of a 16S rRNA sequencing approach using American Gut Project data [14]; that work showed that the O. formigenes relative abundance ranged from 10−6 to 10−3; the relative abundance we achieved in our mouse model parallels that finding. Similar results were found in the Human Microbiome Project, where the O. formigenes relative abundance was normally distributed around a median of 0.015% [13]. In total, this experiment demonstrated the feasibility of introducing O. formigenes to humanized mice, leading to stable levels with relative abundances that resemble those in naturally colonized humans [14].
Figure 2.
Detection of Oxalobacter formigenes in mice after humanization or sham gavage. Fecal samples from germ-free humanized (group 1, blue diamonds; group 2, blue squares), antibiotic-treated humanized (group 3, orange), conventional humanized (group 5, black), and antibiotic-treated sham-humanized (group 4, red) mice were assessed using O. formigenes–specific (oxc) quantitative polymerase chain reaction and normalized to total 16S DNA copies in each sample. Mice were followed up for 8 weeks after gavage.
Effect of Humanization on Intestinal Community Structure
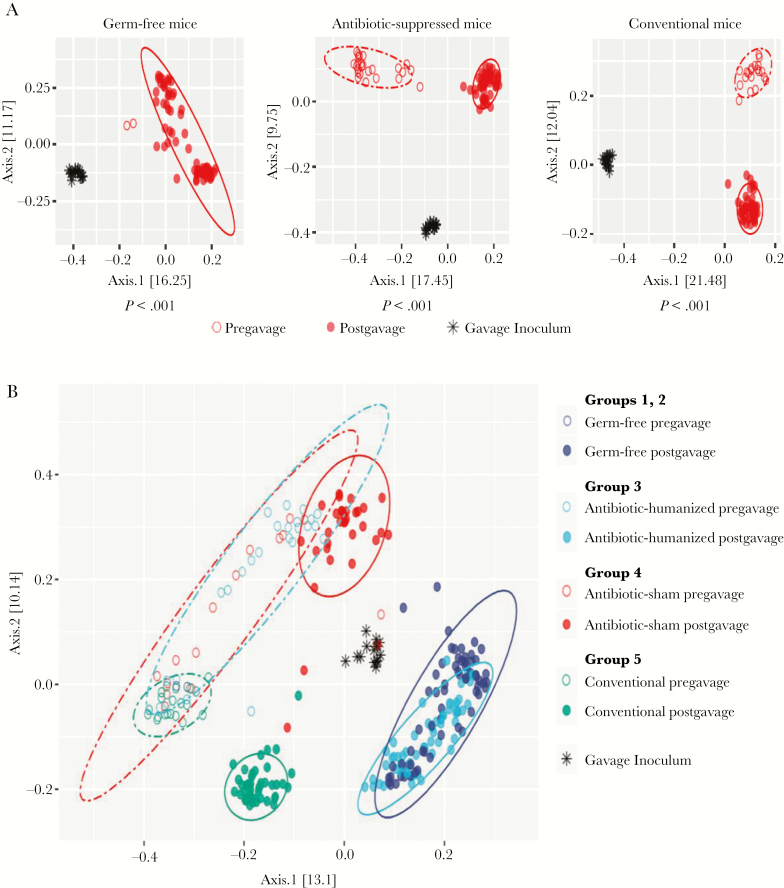
Our second objective was to evaluate murine O. formigenes colonization in which interactions were with the human microbiota rather than the endogenous mouse microbiota. To address this issue, we examined the community structure of the fecal microbiota using analysis of 16S rRNA sequences. Such analyses of β-diversity provide a way to detect differences in the aggregated composition between discrete groups. As expected, the pregavage mouse microbiota, whether conventional or antibiotic perturbed, differed significantly from the human microbiota (Figure 3A) (P < .05; Adonis test). Subsequently, in both the antibiotic-suppressed (group 3 [antibiotic-humanized]) and the conventional (group 5 [conventional-humanized]) mice, the human fecal gavage significantly shifted the community from the pregavage status (P < .001).
Figure 3.
β-Diversity before and after humanization or sham gavage. A, Pregavage and postgavage fecal samples from humanized mice. Mice were germ free (groups 1 and 2; left), were given broad-spectrum antibiotic treatment to suppress their own microbiota before gavage (groups 3 and 4; middle), or were untreated conventional mice (group 5; right). Principal coordinates analysis plots depict the unweighted UniFrac analysis of gavage inocula and fecal samples before and up to 8 weeks after gavage. The statistics below each panel reflect Adonis tests with 999 permutations to determine whether the microbiome community structures of inocula, before and after gavage, differed significantly (P < .05). B, Principal coordinates analysis plot depicts unweighted UniFrac analysis of the gavage inocula and fecal samples. Pregavage samples were collected from conventional mice and mice receiving high-dose, broad-spectrum antibiotics over 6 days (open circles). Postgavage samples were collected through 8 weeks (closed circles); 1 group of antibiotic-treated mice received sham gavage (red), and 3 other groups received humanization: untreated conventional mice (green); antibiotic-treated conventional mice (light blue), and germ-free mice (dark blue). By Adonis testing, the conventional humanized group differed significantly (P < .001) from both the antibiotic-treated humanized mice and the germ-free humanized mice. The antibiotic-treated humanized group also differed significantly (P < .001) from the antibiotic-treated sham-gavage group. Statistics were generated using Adonis tests with 999 permutations after Bonferroni correction for multiple testing.
Examining the communities together on a single plot further illustrated several relationships (Figure 3B). First, at baseline, the populations in the conventional mice (group 5; before gavage), formed a tight cluster. In contrast, in the 2 groups that received antibiotics (groups 3 and 4), there was marked heterogeneity compared with group 5 (P < .05 for each); however, the 2 groups largely overlapped (difference not significant), indicating the conserved antibiotic selection. In the conventional mice that were not humanized (group 4), antibiotic treatment led to a long-term shift in community structure (P < .05). In conventional mice that did not receive antibiotics (group 5), humanization significantly shifted the community (P < .05) (Figure 3B), indicating that introducing the heterologous microbiota had ecological impact.
The combined effect of antibiotic treatment and gavage (group 3) markedly shifted the community structure compared with before gavage (P < .05). Comparing the 2 groups that received antibiotics, there was a significant difference after gavage between the mice that received the sham (group 4 [antibiotic-sham]) or the human microbiota (group 3 [antibiotic-humanized]) (P < .001). After antibiotic treatment and humanization (group 3), the population structure almost completely overlapped with that of the humanized germ-free mice (groups 1 and 2), and these differed significantly (P < .001) from the postgavage conventional mice (group 5). In total, these studies show the significant ecological effects of both antibiotic treatment and humanization; humanization significantly altered the murine intestinal microbial composition, regardless of the method used. Notably, 8 weeks after gavage, the humanized samples remained distinct from the baseline conventional mouse samples, without evidence of regression toward a murine microbial phenotype.
Microbial Community Ecology
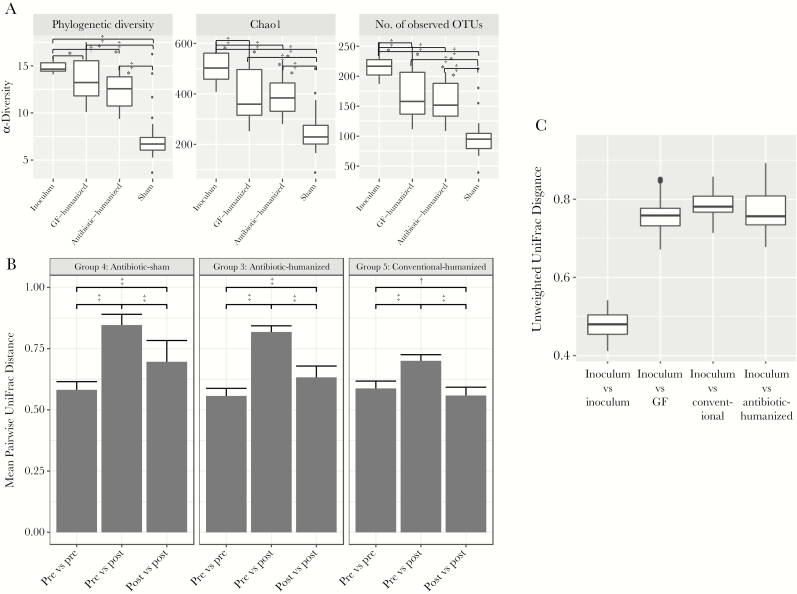
Next we assessed the extent to which humanization changed the gut microbial communities. After humanization, the initially germ-free (groups 1 and 2 [germ-free humanized]) and antibiotic-treated mice (group 3 [antibiotic-humanized]) differed significantly in α-diversity from sham-treated mice (group 4 [antibiotic-sham]) (P < .001) (Figure 4A). (α-Diversity compares the microbial richness of the specified communities.) The initially germ-free and antibiotic-treated mice did not differ significantly after humanization. We then assessed the changes in community structure (β-diversity) resulting from humanization, using UniFrac analysis (Figure 4B). With all 3 humanization protocols, the mean pairwise unweighted intergroup distances before and after gavage were significantly greater than the intragroup distances, indicating that the humanization significantly altered population structure. Weighted UniFrac analyses showed parallel results (data not shown). The β-diversity in all of the humanized mice differed significantly from the inoculum (Figure 4C), without significant differences between them. In total, these findings demonstrate that humanization significantly changed the community richness and structure in the recipient mice in relation to both their pregavage state and the inoculum.
Figure 4.
Microbial community ecology. A, α-Diversity in the gavage inoculum and in the treatment groups. The α-diversity metrics are represented using phylogenetic diversity (left), Chao1 index (middle), or number of observed operational taxonomical units (OTUs) (right). All comparisons were significant, except the comparison between germ-free (GF)–humanized and antibiotic-humanized groups using all 3 α-diversity measurements, based on false discovery rate–adjusted Wilcoxon rank sum tests. *P < .05; †P < .01; ‡P < .001 (for group comparisons). B, Analysis of β-diversity in samples before (pre) and after (post) gavage. Results show mean pairwise unweighted UniFrac distances with intragroup and intergroup (pre and post) comparisons. Groups were compared using Wilcoxon rank sum tests, corrected for multiple comparisons. *P < .01; †P < .001. C, β-Diversity (community structure) of samples from the treatment groups in relation to the gavage inoculum. Mean pairwise unweighted UniFrac distances are shown in relation to the intragroup variation for the inoculum. All comparisons were significant by false discovery rate–adjusted Wilcoxon rank sum tests (P < .001).
Effects of Humanization at the Taxonomic Level
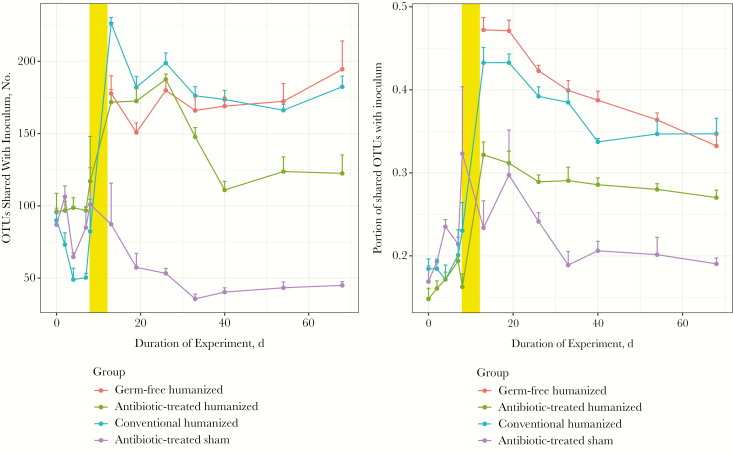
To examine the efficiency of gavage in transferring specific human taxa, we monitored the OTUs shared between the inoculum and the mouse intestinal contents after gavage (Figure 5). At baseline, the numbers of OTUs shared between the inoculum and the pregavage mice were approximately the same for the 3 groups of conventional mice, as expected, but the subsequent antibiotic treatment (groups 3 and 4 [antibiotic-humanized and antibiotic-sham]) reduced the number of shared taxa. After the humanizing gavage, the number of OTUs matching the human inoculum increased. However, in the mice that received sham gavage after antibiotic treatment (group 4), the number of shared OTUs declined below baseline.
Figure 5.
Operational taxonomical units (OTUs) shared between the human inoculum and the treatment groups. The number of OTUs shared between mouse fecal pellets and the human inoculum (donor feces supplemented with Oxalobacter formigenes) were compared before and through 8 weeks after gavage. Pregavage samples collected during antibiotic treatment are from day 5 to day 10. After gavage (yellow bar), all 3 humanized groups show a significantly greater number of shared OTUs with the inoculum, compared with the sham group (P < .001).
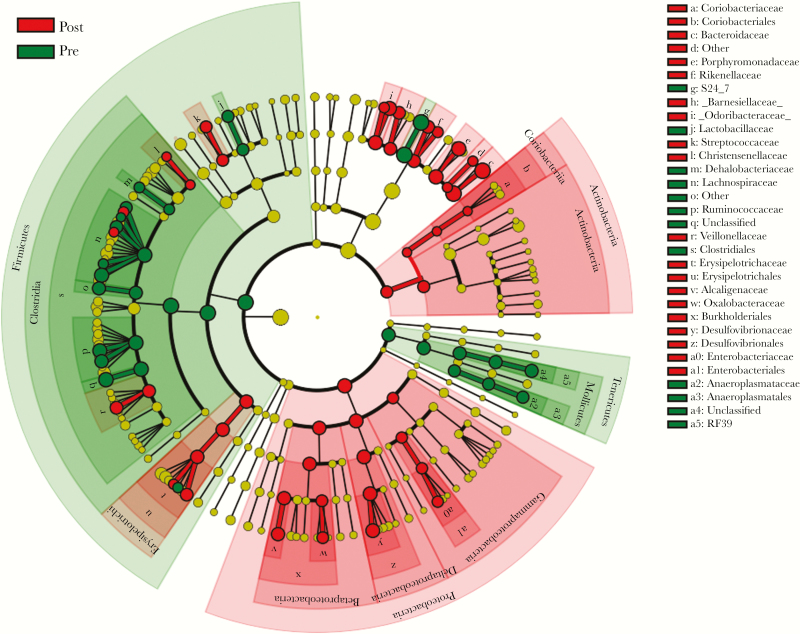
Over the 8-week study, significantly more OTUs were shared with inoculum in each of the 3 groups of humanized mice than in the sham-gavaged mice (P < .001). In conventional mice (group 5 [conventional-humanized), the modest increase in OTUs shared with inoculum declined by 2 weeks after gavage, with final levels similar to those at baseline, indicating that the humanization was transient. After humanization of the germ-free and antibiotic-suppressed recipients, the number of OTUs shared with donor inoculum increased in parallel, persisting throughout the study. Analyses at the taxonomic species level yielded similar results (Supplementary Figure 1), but because of intraspecies replacements, the magnitude of the observed effects was smaller than at the OTU level. In parallel, we determined which bacterial species were differentially abundant at baseline (before humanization) compared with postgavage samples. We found that 14 taxa, including S24_7, Lachnospiracaeae, and Ruminococcaceae, were depleted after gavage and that 29 taxa, including O. formigenes, Bacteroides fragilis, Bilophila spp., and Butyricimonas spp. were significantly increased (Figure 6 and Supplementary Figure 2).
Figure 6.
Differentially abundant taxa in feces of conventional mice before and after antibiotic-mediated humanization. Linear discriminant effect size analysis was used to identify bacterial taxa that were significantly enriched in pregavage (n = 12) or postgavage (n = 55) samples from 9 mice. Samples included represent baseline fecal samples from conventional mice and after high-dose antibiotics and humanization. Cladogram depicts significantly enriched taxa.
Next, we asked whether the method of humanization (initially germ free (groups 1 and 2) or antibiotic before treatment (group 3) resulted in different species composition after gavage. In parallel with the β-diversity similarities (Figure 3B), the 2 groups of mice had highly similar taxa present (Supplementary Figure 3), with only 3 significant differences.
Effect of Gavage Timing
An interval between antibiotic treatment and humanization carries the risk for rebound of murine microbiota or the acquisition of exogenous species in the setting of affected microbiota. Assessment the bacterial load (total 16S DNA) with qPCR showed continued suppression in the antibiotic-treated mice until immediately before gavage (Supplementary Figure 4). We also evaluated the microbiome community structure over time (Supplementary Figure 5); before treatment, all of the bacterial populations formed a tight cluster. Although the controls did not subsequently change, in the 9 mice receiving vancomycin, ampicillin, neomycin, and metronidazole, populations shifted significantly, forming 2 distinct clusters but never normalizing. Thus, the microbiota remained perturbed until the time of gavage.
Effect of Antibiotic Treatment on O. formigenes Colonization
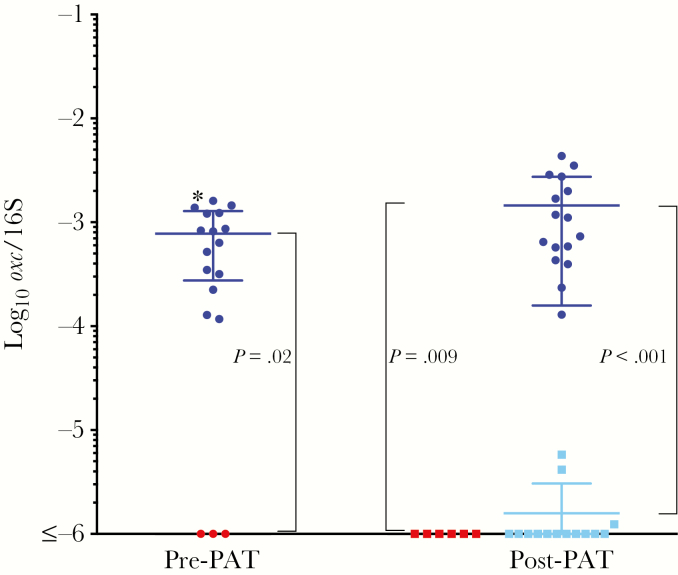
Finally, we asked whether the introduced O. formigenes could survive a conventional antibiotic challenge. To address this question, we used a single 6-day PAT with the macrolide tylosin, because we have observed its substantial effects on the intestinal microbiota [26–28]. By qPCR 9 weeks after gavage (before PAT), all humanized mice had significantly higher O. formigenes levels than did the sham-treated mice (Figure 7). For the humanized mice exposed to PAT, O. formigenes levels decreased to near the sham levels within 1 week, but there was no diminution in the untreated mice. Six weeks later, O. formigenes levels remained suppressed in the PAT-exposed mice compared with the unexposed mice (mean, 6.2 × 10−6 vs 3.4 × 10−3) (P <.001) These data indicate that the stable O. formigenes colonization in this model is nonetheless susceptible to a standard therapeutic antibiotic course.
Figure 7.
Intestinal Oxalobacter formigenes levels after a single pulsed antibiotic treatment (PAT). The number of oxc copies was normalized to the total 16S ribosomal RNA (rRNA) copies in each sample. Left, O. formigenes levels in mice that were humanized and received O. formigenes (group 3; blue circles) or that received sham gavage (group 4; red circles), before antibiotic exposure. Right, O. formigenes levels after half of the humanized mice (light blue squares) and sham mice (red squares) received a 6-day tylosin course (PAT). There was no change in levels in the humanized mice not exposed to PAT, which remained significantly elevated in relation to sham. Humanized mice exposed to PAT had significantly lower O. formigenes levels than unexposed humanized mice (blue circles), near the background (sham) level.
DISCUSSION
With increased nephrolithiasis incidence [1, 2], developing new ways to prevent calcium oxalate stones is urgently needed. Modifying the intestinal microbiota by increasing oxalate-degrading bacteria is attractive, but fundamental work is needed to assess both feasibility and utility. In conventionally raised mice and rats, O. formigenes colonization significantly reduces urinary oxalate levels [34–36]. However, treating patients with primary hyperoxaluria with the selected O. formigenes strain was not effective [19, 37]. Optimizing the strains and conditions is one way to improve the utility of administering O. formigenes, but with the difficulty of such studies in humans, relevant animal models must be developed. In particular, humanized murine models can permit comparisons among candidate O. formigenes strains before eventual human clinical trials.
Coadministering O. formigenes with altered Shaedler flora, a defined mixture of 8 murine bacterial species, to germ-free mice reduced urine oxalate more than using O. formigenes or altered Shaedler flora alone [38]. This synergistic effect suggests the importance of additional species that may degrade oxalate or creating an improved ecology for O. formigenes. However, more complex microbial networks are needed to study more human host-adapted O. formigenes strains, as well as the dynamics and effects of colonization on the host microbiome, and defining optimal conditions for colonizing the human gut.
Humanized mice have been increasingly used to study microbiome-related diseases [39–41] and, despite their limitations, may provide the best available models to examine the role of microbiome alterations on host oxalate metabolism and risk for stone disease. Although germ-free and antibiotic-treated mice can both be used for humanization, there are few data comparing these methods [42], and none in relation to O. formigenes. We found that both approaches were successful for stable and persistent colonization of both the human fecal microbiota and O. formigenes strain OXCC13, at relative abundances similar to those found in humans [13, 14]. Antibiotic treatment (with vancomycin, ampicillin, neomycin, and metronidazole) substantially suppressed the murine microbiome, as expected [24, 25], allowing human microbial species to be established in the host with characteristics similar to those seen in the humanized germ-free mice. Antibiotic-mediated humanization provides several advantages over germ-free methods. In addition to being more costly, the disturbed gastrointestinal and immunological physiology of germ-free mice also limits their utility [43]. Furthermore, null mouse lines that might be useful for oxalate studies, including Agxt, CFTR, SLC26A6, and SLC26A1, are not currently available as gnotobiotes.
Although we expected at least a temporary suppression of O. formigenes after administration of a macrolide [44], a single antibiotic pulse was completely suppressive for ≥6 weeks which is when the study ended. This supports the clinical observations that antibiotic use has a lasting effect on O. formigenes colonization [16], and potentially increases the risk for nephrolithiasis [17]. Whether humans can be recolonized after such loss requires investigation.
Our study had several limitations, including small sample sizes. Although we found significant effects of the treatment groups, future studies with larger samples may permit better understanding of different humanization methods. Useful information could have been obtained with additional control groups, including untreated conventional mice to control for variation over the course of the study or conventional mice gavaged with O. formigenes alone. Shortening the interval from ceasing antibiotic exposure to humanization also might minimize regrowth of the indigenous microbiota, although residual antibiotic effects may adversely affect humanization performed within 24 hours. The current study did not include any measurements of oxalate levels, since our aim was to optimize colonization; future studies with larger study groups will include in-depth analyses of oxalate metabolism.
In conclusion, we have created a humanized mouse model for O. formigenes colonization, in which both humanization and O. formigenes colonization were stable for several weeks. With persistent O. formigenes colonization at relevant abundances, and its removal with antibiotics, we can model host physiological effects in ways that are generalizable to humans. The model permits future research into differences in oxalate metabolism among O. formigenes strains and diets and the potential benefits of microbes interacting with O. formigenes and affecting host metabolism.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Zhan Gao for developing and sharing 16S quantitative polymerase chain reaction primers and protocol. We thank the National Gnotobiotic Rodent Resource Center, University of North Carolina, Chapel Hill for supplying mice for this study.
Financial support. This work was supported by: National Institute of Allergy and Infectious Diseases (grant U01 AI22245), the National Institute of Diabetes and Digestive and Kidney Diseases (grant U54 DK083908), the Oxalosis and Hyperoxaluria Foundation–American Society of Nephrology (career development Award to L. N.), and the C&D, Zlinkoff, and Diane Belfer funds. New York University School of Medicine’s Genome Technology Center (supported by Cancer Center Support Grant P30CA016087 at the Laura and Isaac Perlmutter Cancer Center).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: American Society of Nephrology annual conference, New Orleans, Louisiana, November 4, 2017.
References
- 1. Kittanamongkolchai W, Vaughan LE, Enders FT, et al. The changing incidence and presentation of urinary stones over 3 decades. Mayo Clin Proc 2018; 93:291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scales CD Jr, Smith AC, Hanley JM, Saigal CS; Urologic Diseases in America Project Prevalence of kidney stones in the United States. Eur Urol 2012; 62:160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Antonelli JA, Maalouf NM, Pearle MS, Lotan Y. Use of the national health and nutrition examination survey to calculate the impact of obesity and diabetes on cost and prevalence of urolithiasis in 2030. Eur Urol 2014; 66:724–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alexander RT, Hemmelgarn BR, Wiebe N, et al. ; Alberta Kidney Disease Network Kidney stones and kidney function loss: a cohort study. BMJ 2012; 345:e5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coe FL, Evan A, Worcester E. Kidney stone disease. J Clin Invest 2005; 115:2598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holmes RP, Goodman HO, Assimos DG. Contribution of dietary oxalate to urinary oxalate excretion. Kidney Int 2001; 59:270–6. [DOI] [PubMed] [Google Scholar]
- 7. Knight J, Deora R, Assimos DG, Holmes RP. The genetic composition of Oxalobacter formigenes and its relationship to colonization and calcium oxalate stone disease. Urolithiasis 2013; 41:187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Allison MJ, Dawson KA, Mayberry WR, Foss JG. Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch Microbiol 1985; 141:1–7. [DOI] [PubMed] [Google Scholar]
- 9. Arvans D, Jung YC, Antonopoulos D, et al. Oxalobacter formigenes-derived bioactive factors stimulate oxalate transport by intestinal epithelial cells. J Am Soc Nephrol 2017; 28:876–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hatch M, Gjymishka A, Salido EC, Allison MJ, Freel RW. Enteric oxalate elimination is induced and oxalate is normalized in a mouse model of primary hyperoxaluria following intestinal colonization with Oxalobacter. Am J Physiol Gastrointest Liver Physiol 2011; 300:G461–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaufman DW, Kelly JP, Curhan GC, et al. Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. J Am Soc Nephrol 2008; 19:1197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clemente JC, Pehrsson EC, Blaser MJ, et al. The microbiome of uncontacted Amerindians. Sci Adv 2015; 1:e1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barnett C, Nazzal L, Goldfarb DS, Blaser MJ. The presence of Oxalobacter formigenes in the microbiome of healthy young adults. J Urol 2016; 195:499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu M, Koh H, Kurtz ZD, et al. Oxalobacter formigenes-associated host features and microbial community structures examined using the American gut project. Microbiome 2017; 5:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. PeBenito A, Nazzal L, Wang C, et al. Comparative prevalence of Oxalobacter formigenes in three human populations. Sci Rep 2018; 9:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kharlamb V, Schelker J, Francois F, Jiang J, Holmes RP, Goldfarb DS. Oral antibiotic treatment of Helicobacter pylori leads to persistently reduced intestinal colonization rates with Oxalobacter formigenes. J Endourol 2011; 25:1781–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tasian GE, Jemielita T, Goldfarb DS, et al. Oral antibiotic exposure and kidney stone disease. J Am Soc Nephrol 2018; 29:1731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoppe B, Dittlich K, Fehrenbach H, Plum G, Beck BB. Reduction of plasma oxalate levels by oral application of Oxalobacter formigenes in 2 patients with infantile oxalosis. Am J Kidney Dis 2011; 58:453–5. [DOI] [PubMed] [Google Scholar]
- 19. Milliner D, Hoppe B, Groothoff J. A randomised phase II/III study to evaluate the efficacy and safety of orally administered Oxalobacter formigenes to treat primary hyperoxaluria. Urolithiasis 2018; 46:313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang J, Knight J, Easter LH, Neiberg R, Holmes RP, Assimos DG. Impact of dietary calcium and oxalate, and Oxalobacter formigenes colonization on urinary oxalate excretion. J Urol 2011; 186:135–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Duncan SH, Richardson AJ, Kaul P, Holmes RP, Allison MJ, Stewart CS. Oxalobacter formigenes and its potential role in human health. Appl Environ Microbiol 2002; 68:3841–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li X, Ellis ML, Knight J. Oxalobacter formigenes colonization and oxalate dynamics in a mouse model. Appl Environ Microbiol 2015; 81:5048–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ellis ML, Shaw KJ, Jackson SB, Daniel SL, Knight J. Analysis of commercial kidney stone probiotic supplements. Urology 2015; 85:517–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Diehl GE, Longman RS, Zhang JX, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature 2013; 494:116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 2004; 118:229–41. [DOI] [PubMed] [Google Scholar]
- 26. Livanos AE, Greiner TU, Vangay P, et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat Microbiol 2016; 1:16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ruiz VE, Battaglia T, Kurtz ZD, et al. A single early-in-life macrolide course has lasting effects on murine microbial network topology and immunity. Nat Commun 2017; 8:518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nobel YR, Cox LM, Kirigin FF, et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun 2015; 6:7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lewicki J. Tylosin: a review of pharmacokinetics, residues in food animals and analytical methods. Food and Agriculture Organization of the United Nations, 2006. http://www.fao.org/tempref/AG/agn/food/tylosin_2006.pdf. Accessed November 1, 2018. [Google Scholar]
- 30. Cox LM, Yamanishi S, Sohn J, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 2014; 158:705–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Haas BJ, Gevers D, Earl AM, et al. ; Human Microbiome Consortium Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 2011; 21:494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hatch M, Freel RW. A human strain of Oxalobacter (HC-1) promotes enteric oxalate secretion in the small intestine of mice and reduces urinary oxalate excretion. Urolithiasis 2013; 41:379–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hatch M, Cornelius J, Allison M, Sidhu H, Peck A, Freel RW. Oxalobacter sp. reduces urinary oxalate excretion by promoting enteric oxalate secretion. Kidney Int 2006; 69:691–8. [DOI] [PubMed] [Google Scholar]
- 36. Sidhu H, Allison MJ, Chow JM, Clark A, Peck AB. Rapid reversal of hyperoxaluria in a rat model after probiotic administration of Oxalobacter formigenes. J Urol 2001; 166:1487–91. [PubMed] [Google Scholar]
- 37. Hoppe B, Groothoff JW, Hulton SA, et al. Efficacy and safety of Oxalobacter formigenes to reduce urinary oxalate in primary hyperoxaluria. Nephrol Dial Transplant 2011; 26:3609–15. [DOI] [PubMed] [Google Scholar]
- 38. Li X, Ellis ML, Dowell AE, et al. Response of germ-free mice to colonization with O. formigenes and altered Schaedler flora. Appl Environ Microbiol 2016; 82:6952–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 2009; 1:6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arrieta MC, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 2015; 7:307ra152. [DOI] [PubMed] [Google Scholar]
- 41. Ridaura VK, Faith JJ, Rey FE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013; 341:1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lundberg R, Toft MF, August B, Hansen AK, Hansen CH. Antibiotic-treated versus germ-free rodents for microbiota transplantation studies. Gut Microbes 2016; 7:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009; 9:313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kelly JP, Curhan GC, Cave DR, Anderson TE, Kaufman DW. Factors related to colonization with Oxalobacter formigenes in U.S. adults. J Endourol 2011; 25:673–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.