Abstract
The Horn of Africa harbors the largest reservoir of Plasmodium vivax in the continent. Most of sub-Saharan Africa has remained relatively vivax-free due to a high prevalence of the human Duffy-negative trait, but the emergence of strains able to invade Duffy-negative reticulocytes poses a major public health threat. We undertook the first population genomic investigation of P. vivax from the region, comparing the genomes of 24 Ethiopian isolates against data from Southeast Asia to identify important local adaptions. The prevalence of the Duffy binding protein amplification in Ethiopia was 79%, potentially reflecting adaptation to Duffy negativity. There was also evidence of selection in a region upstream of the chloroquine resistance transporter, a putative chloroquine-resistance determinant. Strong signals of selection were observed in genes involved in immune evasion and regulation of gene expression, highlighting the need for a multifaceted intervention approach to combat P. vivax in the region.
Keywords: malaria, Plasmodium, vivax, genomics, Ethiopia, Duffy
Genome-wide signatures of selection were investigated in an Ethiopian Plasmodium vivax population. Antimalarial drug pressure appeared lower than in Asia, but there was a high prevalence (80%) of Duffy binding protein amplifications, potentially reflecting adaptation to Duffy-negative red blood cells.
(See the Editorial commentary by Sibley, on pages 1716–8.)
Reports of drug resistance and severe disease have refocused efforts to contain Plasmodium vivax, with almost 3 billion people living at risk of infection [1]. However, this species has proven to be highly resilient, with a high capacity for resurgence, and extensive genetic diversity providing a rich baseline for adaptation to local selective pressures [2–5].
Although the largest burden of P. vivax is in South and Southeast Asia, the Horn of Africa harbors 10%–20% of all cases, presenting an important reservoir of infection in Africa [1, 6]. Ethiopia suffers the largest number of clinical cases in the region [6], partly reflecting the large percentage of Duffy-positive individuals [7]. For several decades, the central dogma has been that the host Duffy antigen is essential for P. vivax to invade host red blood cells (RBCs) and thus establish a blood-stage infection [8]. However, recent reports have demonstrated the emergence of strains able to invade Duffy-negative RBCs, raising concern over the potential spread of vivax into the large regions of sub-Saharan Africa, which were previously assumed to be protected by high proportions of Duffy negativity [9].
Microsatellite-based studies of P. vivax in Ethiopia have revealed extensive parasite diversity [10–12], underpinning a high capacity for local adaptation to host and environmental challenges such as Duffy negativity or antimalarial drug pressure. However, while several population genomic studies have been undertaken in vivax populations from Asia, the Pacific, and South America [3, 5, 13–17], none has explored the genomes of P. vivax from the horn of Africa.
We used whole genome sequencing data to investigate adaptive molecular changes in an Ethiopian P. vivax population. Comparisons were undertaken against existing genomic data from Southeast Asian P. vivax populations to identify Ethiopian-specific adaptive changes.
METHODS
Ethical Considerations
Ethical approval for patient sampling in Ethiopia was granted by the ethics boards of the Addis Ababa University College of Natural Sciences (RERC/002/05/2013), the Armauer Hansen Research Institute (AHRI-ALERT P011/10), the National Research Ethics Committee of Ethiopia (3.10/580/06), and the Human Research Ethics Committee of Northern Territory Department of Health and Families, Australia (HREC-13–1942). Written informed consent was obtained from all participants or a legal guardian where participants were ≤18 years of age.
Study Site
The Ethiopian samples were sourced within the framework of previously described surveys recruiting symptomatic patients with P. vivax–positive blood smears [10]. Samples were collected between May and November 2013 from 4 sites in southern Ethiopia; Arba Minch, Misrak Badowacho, Halaba, and Hawasa (Supplementary Figure 1). Details on the local malaria epidemiology are published elsewhere [10]. In brief, Plasmodium falciparum and P. vivax are the dominant malaria-causing species, with vivax proportions ranging from 28% to 55%. The annual P. vivax parasite incidence (API) in 2012 ranged from 20–82 cases per 1000 population across the sites. The national policy for treating uncomplicated P. vivax infection is chloroquine plus primaquine, whereas artemether-lumefantrine is used to treat mixed-species infections of P. falciparum and P. vivax.
For comparative analysis, previously published genomic data were derived from Thailand, Indonesia, and Malaysia [5, 13]. These samples were sourced from symptomatic patients with P. vivax infection attending outpatient clinics in Tak Province, Thailand (2006–2013); Mimika district, Papua Indonesia (2011–2014); and Sabah, Malaysia (2010–2015). Estimates of the P. vivax incidence (API per 1000 population) at these times were 15.5 in Tak Province, 69.2–148.7 in Mimika district, and 0.02–0.32 in Sabah (data from Malaria Atlas Project and Sabah Department of Health). The first-line policy for treating P. vivax infection at the time of the enrollments was chloroquine plus primaquine in Thailand and Malaysia, and dihydroartemisinin-piperaquine plus primaquine in Indonesia.
Sample Processing
Patient sampling involved collection of 2–5 mL venous blood, with leukocyte depleted by cellulose filtration [18]. DNA extraction was performed using commercial kits (Qiagen), and Plasmodium species was confirmed by polymerase chain reaction [19, 20].
Whole Genome Sequencing
Leukocyte-depleted samples with ≥50 ng total DNA comprising <90% human DNA were subject to whole genome sequencing, read alignment, and variant calling within the framework of a P. vivax community study in the Malaria Genomic Epidemiology Network (MalariaGEN) [21]. Details on the library preparation, sequencing, and variant calling procedures are provided elsewhere [5]. In brief, sequencing was undertaken on the Illumina Hi-Seq platform using the manufacturer’s protocol for paired-end 150-bp reads. Reads were aligned against the P. vivax P01 reference [22] using bwa-mem version 0.7.15 [23], and single-nucleotide polymorphism (SNP) discovery and genotype calling were undertaken using previously defined methods [5], with data derived from the MalariaGEN P. vivax Genome Variation Project release 3.0. A set of 827 902 high-quality bi-allelic SNPs with a variant quality score log-odds >0 and <5% missing calls in high-quality samples (samples with ≥95% calls) were derived from an initial set of 4 084 419 discovered variants. Positions with <5 reads were defined as missing calls. For haplotype-based analyses, either the major or reference allele (at positions with equal allele depths) was used at heterozygote positions. Large copy number variations (CNVs) >3 kbp were detected using a hidden Markov model in pysamstats (http://github.com/alimanfoo/pysamstats) as described previously [24]. As the accuracy of the hidden Markov model in predicting the breakpoint sequence is imperfect, closer inspection of CNV boundaries was undertaken using Artemis software. The genomic data on the previously published and the new Ethiopian isolates are available in the European Nucleotide Archive (see [5, 13] and Supplementary Table 1, respectively).
Data Analysis
Within-host infection complexity was assessed using the within-sample F statistic (FWS) [25, 26]. A threshold of FWS > 0.95 was used as a proxy to monoclonal infection. Ethiopian isolates with FWS < 0.95 were deconvoluted (phased) using DEploid identity by descent (IBD) [27] to determine the within-sample relatedness. DEploid-IBD was run with the default settings using a panel of 4 distinct Ethiopian isolates with FWS > 0.95 (QS0002-C, QS0018-C, QS0028-C, and QS0042-C). Downstream haplotype-based analyses were undertaken on the samples with FWS > 0.95, and additionally explored with the deconvoluted infections. Infection complexity was also assessed using the proportion of runs of homozygosity (RoH) in the isolates from all populations using previously described methods [5].
The number of nucleotide differences between pairs of DNA sequences was calculated at the SNP positions using VCFtools version 0.1.13 [28] and divided by the number of nucleotides in the core genome (21 310 118 bp) to derive an approximation of the genome-wide average nucleotide diversity (pi) for each population. Neighbor-joining (NJ) and principal coordinates analyses (PCoA) were conducted using a pairwise distance matrix calculated using R, and NJ plots were created using iTOL software [29].
Tajima’s D was calculated using the scikit-allel package (https://github.com/cggh/scikit-allel). Only genes with ≥10 SNPs across all populations were assessed. Pairwise measures of the genetic differentiation (fixation index [FST]) at individual SNPs were calculated using the Weir and Cockerham formula [30]. The PlasmoDB Gene Ontology (GO) analysis tool was used to investigate enrichment of biological processes and molecular functions among the genes within the 1st and 99th percentile of the Tajima’s D distributions in Ethiopia and highly differentiated variants between Ethiopia and Asia (http://plasmodb.org).
The Rsb measure of cross-population extended haplotype homozygosity and the integrated haplotype score (iHS) were measured using the R-based rehh package [31]. For iHS analysis, ancestral alleles were derived by mapping Plasmodium cynomolgi reads [32] against the P. vivax P01 reference. Only positions where P. cynomolgi calls were homozygote and matched either the reference or alternative P. vivax allele were included in analysis.
Owing to extensive population structure in Malaysia, iHS, Rsb, and Tajima D were not investigated in this population, and FST results between Ethiopia and Malaysia were explored secondary to results against Thailand and Indonesia.
RESULTS
Genomic Data Summary
A total of 293 samples exhibited <5% missing genotype calls at 827 902 high-quality SNPs with genotype failure rates <5%. These samples include 24 new isolates from Ethiopia (detailed in Supplementary Data 1), and previously described samples from Thailand (n = 104), Indonesia (n = 111), and Malaysia (n = 54) [5, 13].
Comparable Superinfection and Cotransmission Rates in Ethiopia, Indonesia, and Thailand
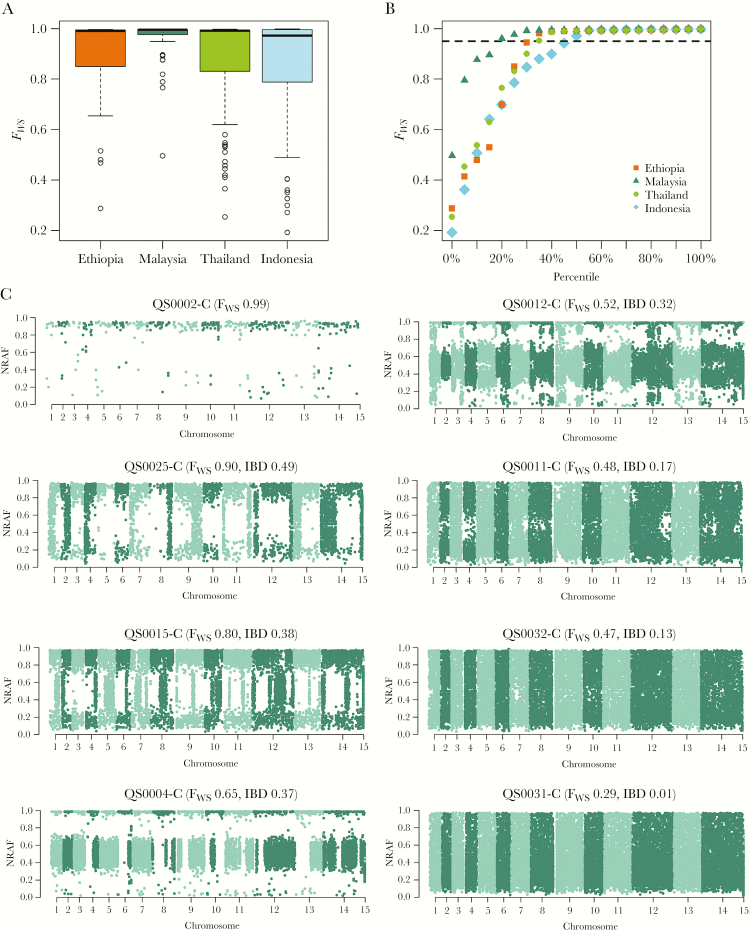
The proportion of monoclonal infections in Ethiopia (71% [17/24]) was similar to Indonesia (52% [58/111]; P = .151), Thailand (65% [68/104]; P = .787), and Malaysia (81% [44/54]; P = .451), although the latter displayed a skew toward higher FWS scores (Figure 1A and 1B). DEploid-IBD analysis of the 7 polyclonal Ethiopian samples revealed that 3 and 4 infections had 2 and 3 major clones (>10%), respectively. Four (57%) of the polyclonal infections displayed IBD >25% in pairwise comparisons of the clones, suggesting relatedness at the half-sibling level or greater, and likely reflecting cotransmission rather than superinfection (multiple inoculations either acquired within a few weeks of one another or over several months with hypnozoites; Supplementary Data 2). The remaining 3 infections exhibited IBD ranging from 0 to 17%, more consistent with superinfections (Figure 1C). There was no significant difference in the median RoH proportions in the polyclonal infections in Ethiopia (0.03 [interquartile range {IQR}, 0.01–0.51]) relative to Thailand (0.08 [IQR, 0.005–0.49]; P = .779) or Indonesia (0.28 [IQR, 0.03–0.54]; P = .519), suggesting similar rates of superinfection vs cotransmission in these populations (Supplementary Figure 2; Supplementary Data 2). However, median RoH was significantly higher in the preelimination setting of Malaysia (0.73 [IQR, 0.50–0.83]; P = .028), where high rates of inbreeding have been described in recent years [13, 33].
Figure 1.
Within-sample infection complexity in Ethiopia relative to the Southeast Asian populations and within polyclonal Ethiopian infections. The boxplots (A) and scatterplots (B) illustrate the distribution of within-sample F statistic (FWS) scores in Ethiopia relative to Thailand, Indonesia, and Malaysia. Data are presented on all 293 high-quality samples. B, Dashed line illustrates FWS = 0.95, above which infections are essentially monoclonal. Ethiopia exhibits an intermediate proportion of monoclonal infections relative to Malaysia and Indonesia, most comparable to Thailand. C, Manhattan plots of the nonreference allele frequency (NRAF) in 7 Ethiopian infections identified as polyclonal based on FWS < 0.95, and 1 monoclonal infection as a baseline reference (QS0002-C). Trends in the pairwise identity by descent (IBD) between clones in a given infection (as determined by DEploid-IBD) correlated positively with the FWS scores and approximated runs of homozygosity (RoH, not presented). DEploid-IBD found evidence of 2 major clones in QS0025-C, QS0015-C, and QS0004-C, and 3 clones in QS0012-C, QS0011-C, QS0032-C, and QS0031-C. The clones within QS0025-C demonstrated the highest pairwise IBD (0.49), indicative of siblings sharing approximately 50% of their genomes, as illustrated by the long stretches of homology (regions with NRAF approximating 0 or 1) on each chromosome. In striking contrast, the clones within QS0031-C exhibited the lowest pairwise IBD (<0.01%), with no evidence of recent common ancestry, rather reflecting a probable superinfection.
High Diversity and Frequent Outcrossing in Ethiopia
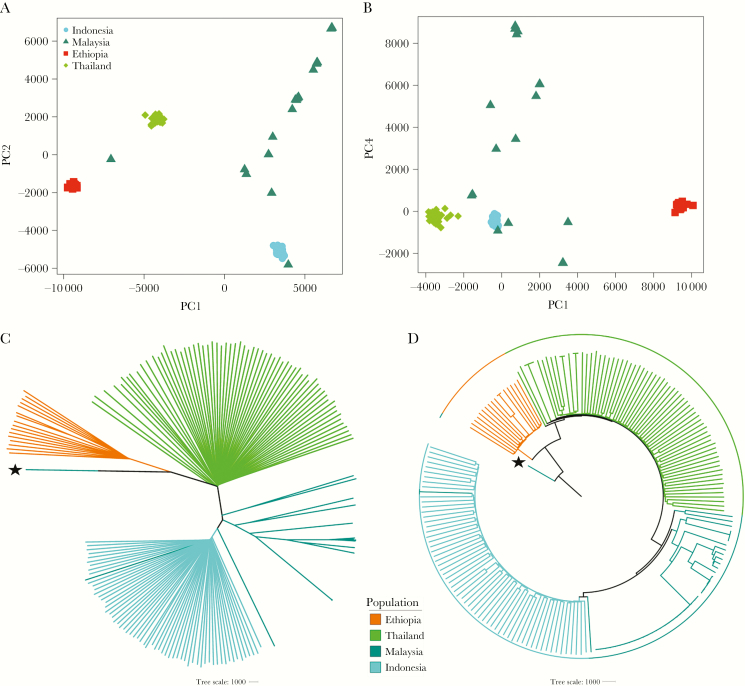
The average nucleotide diversity (pi) in Ethiopia (6.47 × 10-4) was lower than Thailand (7.69 × 10-4) and Indonesia (6.78 × 10-4) but higher than Malaysia (4.55 × 10-4). In contrast to the extensive substructure observed in Malaysia with PCoA and NJ analysis, higher levels of outcrossing were apparent in Ethiopia, Thailand, and Indonesia (Figure 2A–D). Similar results were observed with the deconvoluted Ethiopian haplotypes (Supplementary Figure 3A–D).
Figure 2.
Plasmodium vivax population structure and relatedness in Ethiopia relative to the Asian populations. All plots were generated using genomic data derived from 191 high-quality, monoclonal (within-sample F statistic [FWS] > 0.95) samples. A and B, Principal coordinates analysis plots illustrating the genetic differentiation within and between populations, respectively. Principal components (PC) 1–4 reflect 16.8%, 10.8%, 9%, and 2.9% of the variance, respectively. C and D, Unrooted and rooted neighbor-joining trees, respectively. The rooted tree is presented to illustrate similarity between infections in a given population rather than evolutionary patterns. The PY0120-C isolate from Malaysia, labeled with a star, was used as the ancestral sample; this sample is a suspected imported case that has been shown to have close identity with infections from India and Bangladesh (data not presented).
Varying Prevalence of Drug Resistance–Associated Mutations in Ethiopia
The prevalence of mutations associated with clinical or ex vivo antimalarial drug resistance was investigated in each population (Table 1). The multidrug resistance 1 (MDR1) Y976F variant, a minor modulator of CQ resistance [34], reached 32% (6/19) in Ethiopia, compared with 13% (14/104) in Thailand, 100% (111/111) in Indonesia, and 94% (51/54) in Malaysia. The prevalence of mutations in dihydrofolate reductase (DHFR) and dihydropteroate synthase (DHPS), which have been associated with antifolate resistance [34], was generally lower in Ethiopia than Asia. There were no DHFR triple or quadruple mutants (0% [0/18]) in Ethiopia, but 94% (17/18) of isolates had double mutations. Malaysia also exhibited a moderately low prevalence of triple and quadruple DHFR mutants (27% [14/51]), although these were prevalent in Thailand (99% [88/89]) and Indonesia (83% [77/93]). Ethiopia also exhibited a lower prevalence of DHPS A553G (0% [0/24]) and A383G (17% [4/24]) than the Asian populations (Table 1). None of the Ethiopian isolates had MDR1 CNVs. For reference purposes, the prevalence of nonsynonymous variants in orthologues of other genes implicated in drug resistance in P. falciparum is provided in Supplementary Data 3, including chloroquine resistance transporter (CRT), kelch-13, plasmepsin IV, multidrug resistance–associated proteins 1 and 2, and multidrug resistance protein 2.
Table 1.
Prevalence of Orthologous Drug Resistance Markers at Each Site
| Gene | Chr | Positions | Mutation | Drug | Frequency, % (no./No.)a | |||
|---|---|---|---|---|---|---|---|---|
| Ethiopia | Thailand | Indonesia | Malaysia | |||||
| MDR1 | 10 | 479 908 | F1076L | CQ | 100 (24/24) | 50 (54/104) | 100 (111/111) | 98 (53/54) |
| (PVP01_1010900) | 10 | 480 207 | Y976F | CQ, AQ + SP | 32 (6/19) | 13 (14/104) | 100 (111/111) | 94 (51/54) |
| 10 | Copy number variant | ≥2 copies | MQ | 0 (0/24) | 19 (20/104) | 0 (0/84) | 0 (0/49) | |
| DHFR-TS | 5 | 1 077 530; 1 077 532 | F57L/I | Antifolate, AQ + SP | 0 (0/24) | 91 (88/97) | 82 (77/94) | 94 (48/51) |
| (PVP01_0526600) | 5 | 1 077 533; 1 077 534; 1 077 535 | S58R | Antifolate, AQ + SP | 94 (17/18) | 100 (104/104) | 99 (104/105) | 36 (19/53) |
| 5 | 1 077 543 | T61M | Antifolate, AQ + SP | 0 (0/24) | 91 (90/99) | 82 (77/94) | 25 (13/51) | |
| 5 | 1 077 711 | S117N/T | Antifolate, AQ + SP | 100 (24/24) | 100 (102/102) | 99 (93/94) | 100 (53/53) | |
| … | … | Single mutant | Antifolate, AQ + SP | 6 (1/18) | 0 (0/89) | 1 (1/93) | 0 (0/51) | |
| … | … | Double mutant | Antifolate, AQ + SP | 94 (17/18) | 1 (1/89) | 16 (15/93) | 73 (37/51) | |
| … | … | Triple mutant | Antifolate, AQ + SP | 0 (0/18) | 0 (0/89) | 0 (0/93) | 2 (1/51) | |
| … | … | Quadruple mutant | Antifolate, AQ + SP | 0 (0/18) | 99 (88/89) | 83 (77/93) | 25 (13/51) | |
| DHPS | 14 | 1 270 401 | A553G | Antifolate | 0 (0/24) | 98 (98/100) | 16 (16/97) | 91 (48/53) |
| (PVP01_1429500) | 14 | 1 270 911 | A383G | Antifolate | 17 (4/24) | 100 (104/104) | 97 (106/109) | 94 (50/53) |
Mutation prevalence was calculated with homozygous calls only.
Abbreviations: AQ, amodiaquine; Chr, chromosome; CQ, chloroquine; MQ, mefloquine; SP, sulfadoxine-pyrimethamine.
aGenotype failures were <5% at all markers in all populations.
Evidence of Recent Directional Selection on an AP2 Domain Transcription Factor in Ethiopia Using iHS Analysis
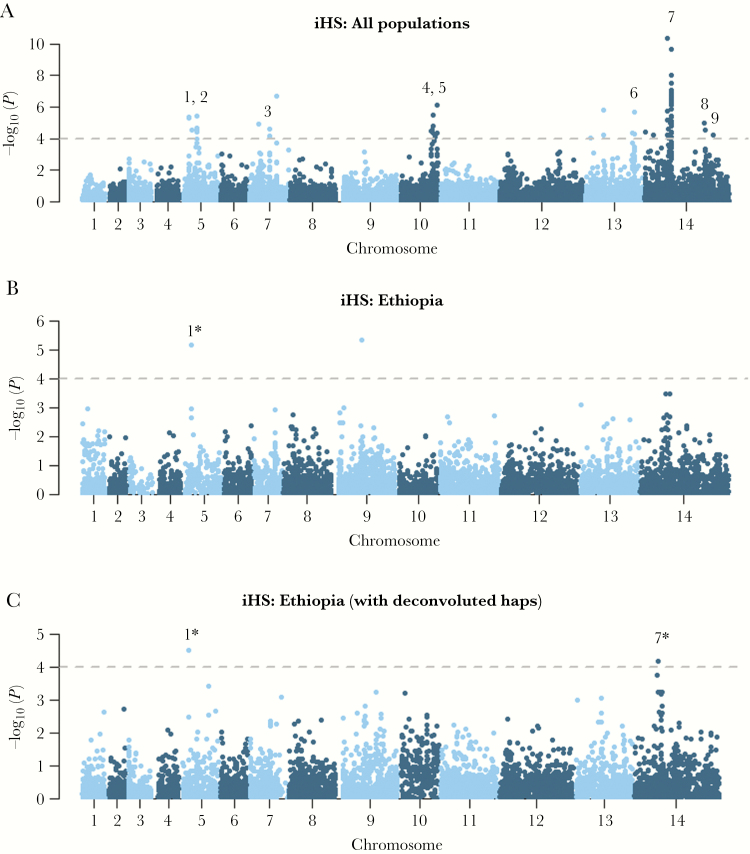
Analysis of other gene regions demonstrating evidence of recent directional selection was undertaken using the iHS score. Population-wide analyses revealed 9 regions with strong signals of selection, 2 of which demonstrated weak signals in Ethiopia (Figure 3; Supplementary Data 4). The largest signal in Ethiopia was a 220-kb region on chromosome 14 comprising 50 genes and overlapping with a previously described signal postulated to reflect selection at an AP2 domain transcription factor (PVP01_1418100) [17]. The other Ethiopian signal reflected a 1-kb region on chromosome 5 comprising a SPRY domain protein (PVP01_0508000).
Figure 3.
Genome-wide scans of extended haplotype homozygosity using the integrated haplotype score (iHS) illustrating regions under recent directional selection in Ethiopia and across populations. Manhattan plots of the iHS P value for the given populations: all monoclonal (within-sample F statistic [FWS] > 0.95) samples from Ethiopia, Thailand, and Indonesia (A), all monoclonal (FWS > 0.95) samples from Ethiopia (B), and all monoclonal (FWS > 0.95) samples from Ethiopia plus the 7 major haplotypes derived from polyclonal Ethiopian isolates that were deconvoluted (C) using DEploid identity by descent (IBD) software. Data are presented on 260 982 loci for which derived alleles could be confidently called. The dashed black lines demark the thresholds of –log10 (P value) > 4: signals supported by a minimum of 3 single-nucleotide polymorphisms (SNPs) above the threshold within 50 kb of one another and with an overall SNP density <10 kb per SNP are numbered. Details can be found in Supplementary Data 4. In brief, the putative genetic drivers include an SPRY domain protein (PVP01_0508000) (signal 1), 3 conserved proteins with unknown function (PVP01_0515000, PVP01_1029200, and PVP01_1455300) (signals 2, 4, and 9 respectively), gamma-glutamylcysteine synthetase (PVP01_0717300), an AP2 domain transcription factor (PVP01_1418100) or ferredoxin (PVP01_1419000) (signal 7), and an amino acid transporter (PVP01_1449600) (signal 8). The driver in the 92-kb region on chromosome 13 (signal 6) remains unclear. *Signals were supported by a minimum of 3 SNPs above the threshold in the population-wide data but only 1 SNP in Ethiopia.
Rsb-Based Signals of Differential Selection Between Ethiopia and Asia in Drug Resistance Candidates
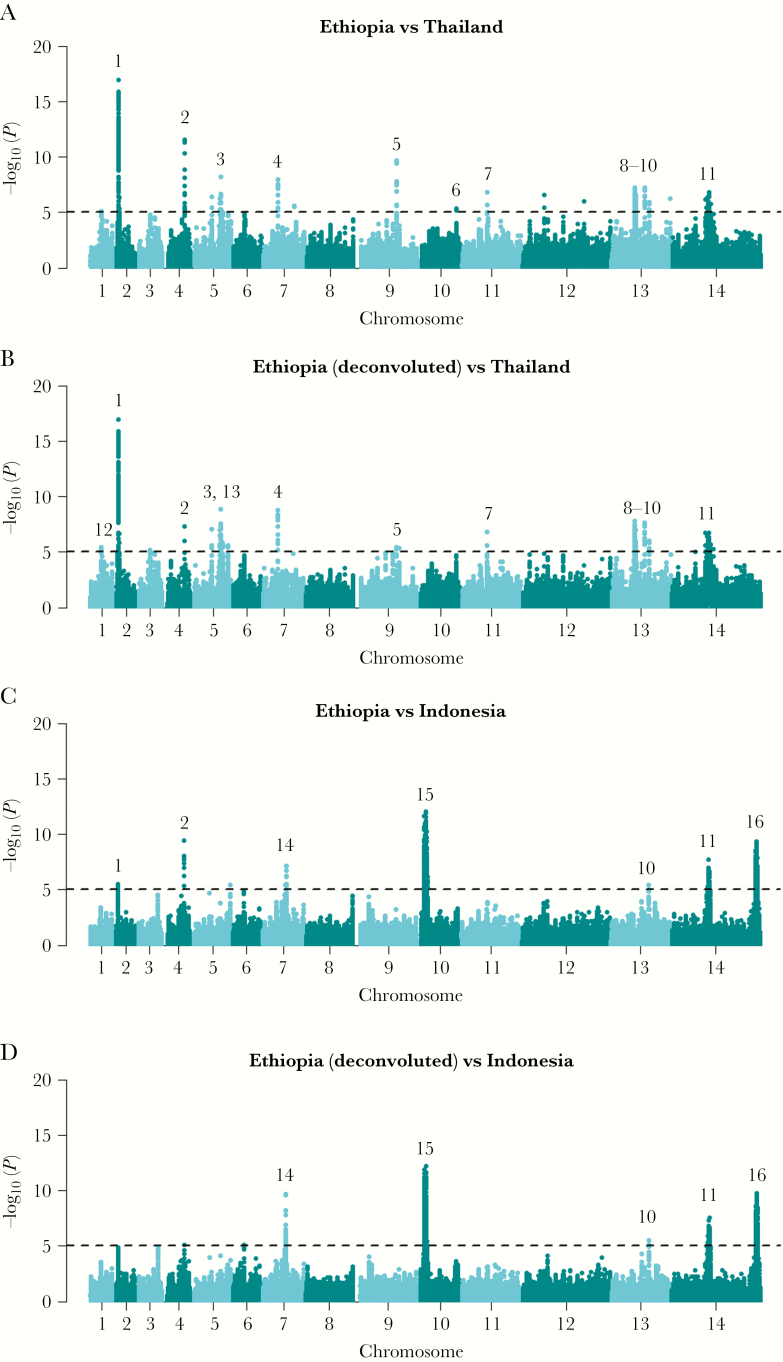
The Rsb metric was used to identify regions with recent directional selection in one population relative to another. Sixteen regions demonstrated strong differential selection between Ethiopia and Thailand or Indonesia, including 5 drug resistance candidates (Figure 4; Supplementary Data 4). The DHFR and DHPS regions demonstrated greater extended haplotype homozygosity indicative of stronger selection in Thailand and Indonesia relative to Ethiopia. Strong signals were also observed on chromosome 2, reflecting MRP1 selection in Thailand and Indonesia, and a region upstream of MDR1 on chromosome 10 in Indonesia. The only drug resistance candidate with evidence of recent selection in Ethiopia was a region upstream of CRT, with extended haplotypes relative to Thailand. Signals of selection were observed in Ethiopia in other gene functions including pathogenesis (merozoite surface protein 5 [MSP5], apical membrane antigen 1 [AMA1]) and regulation of gene expression (AP2 domain transcription factor [PVP01_0529800], tRNA [PVP01_0711200]).
Figure 4.
Genome-wide scans of Rsb-based cross-population extended haplotype homozygosity illustrating regions under divergent selection between Ethiopia and Thailand and Indonesia. A–D, Manhattan plots of the Rsb P value for the given populations. The dashed black lines demark the thresholds of –log10 (P value) > 5: signals supported by a minimum of 3 single-nucleotide polymorphisms (SNPs) above the threshold within 50 kb of one another and with an overall SNP density <10 kb per SNP are numbered. The multi-SNP signals are detailed in Supplementary Data 4. Several previously described signals in known or putative drug resistance candidates were identified including MRP1 (signal 1), DHFR (signal 3), and DHPS (signal 11). Signals were also observed in regions upstream of CRT (signal 12) and MDR1 (signal 15). The putative genetic drivers in other regions include MSP5 (signal 2), a tRNA (signal 4), AMA1 (signal 5), a merozoite surface protein 3 family cluster (signal 6), a voltage-dependent anion-selective channel (signal 8), liver-specific protein 1 (signal 10), and an AP2 domain transcription factor (signal 13). The putative genetic drivers in regions 14 and 16 are conserved Plasmodium proteins with unknown function, and those in regions 7 and 9 remain unclear.
F ST-Based Differentiation of Nonsynonymous Variants Between Ethiopia and Asia in Genes Involved in Regulation of Gene Expression
F ST analysis was undertaken to identify recent as well as older signals of selection that might not be detected with haplotype-based methods. A total of 306 and 1162 SNPs exhibited FST > 0.8 in Ethiopia vs Thailand and Indonesia, respectively; among these variants, 126 and 461 conferred nonsynonymous changes (Supplementary Data 5). After Bonferroni correction, there was no significant representation of any GO terms; however, the most enriched class of genes with highly differentiated nonsynonymous variants were those involved in the regulation of gene expression (11/68 genes with GO ID 0010468). Among the known drug resistance–associated determinants, high differentiation was observed between Ethiopia and Thailand in the DHPS A383G mutation.
Tajima D Analysis in Ethiopia
Tajima’s D analysis was conducted to identify regions under balancing as well as directional selection in Ethiopia (Supplementary Data 6). There was no significant enrichment of any GO term in either the 1st or 99th percentile of D scores in Ethiopia after Bonferroni correction. CRT and a proximal gene on chromosome 1 involved in ion transport (SCO1, PVP01_0109200) were among the 1st percentile in Ethiopia, suggesting evidence of positive selection. However, among 43 CRT and 15 SCO1 SNPs detected across the 3 populations, only 3 and 4 SNPs, respectively, appeared to be segregating in Ethiopia. Furthermore, neither CRT nor SCO1 were among the 1st percentile of genes in Thailand or Indonesia. A range of genes with various functions was observed among the 99th percentile in Ethiopia, reflecting genes putatively under balancing selection, including AMA1.
High Prevalence of Duffy Binding Protein 1 Copy Number Amplifications in Ethiopia
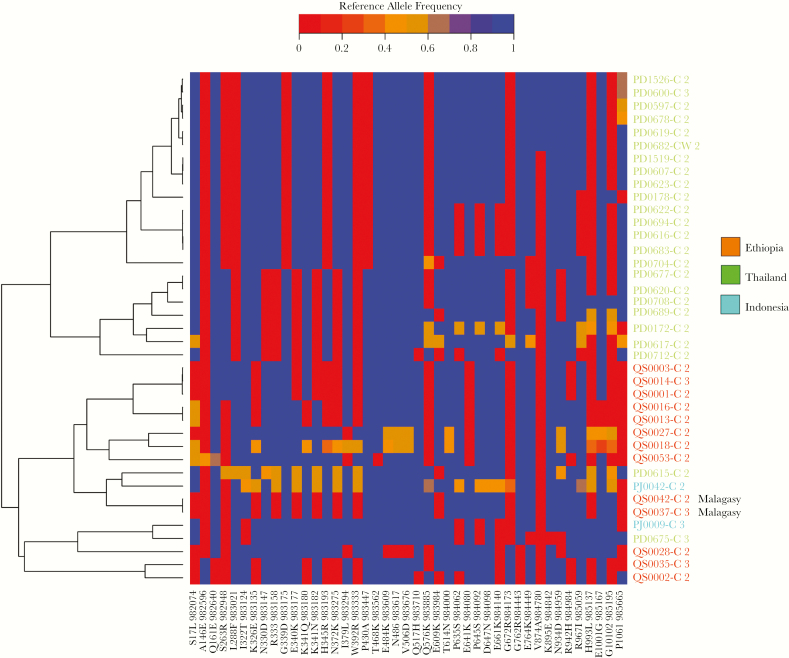
Copy number amplification was observed in 3 gene regions in Ethiopia; a 28S ribosomal RNA gene (PVP01_0504500) with amplification in 100% infections, an exported Plasmodium protein (PVP01_1470400) in 50% infections, and Duffy binding protein 1 (DBP1, PVP01_0623800) in 79% infections (Table 2; Supplementary Data 7). Deletions were also observed in 2 regions encompassing exported Plasmodium proteins: PVP01_0523500 present in 63% (15/24) of Ethiopian isolates and PVP01_0524900 + PVP01_0525000 in 4% (1/24). While the deletions were only observed in Ethiopia, the amplifications were also observed in Asia. The genetic architecture of the DBP1 amplification, which is putatively involved in P. vivax invasion of Duffy-negative RBCs [35], was explored further. DBP1 copy number ranged from 2 to 5 in Ethiopia, and 2 to 3 in the Asian populations. Artemis-based inspection of the amplification revealed a single 5′ and two 3′ breakpoints reflecting the previously described Malagasy and Cambodian type breakpoints [36, 37] in Ethiopia (Supplementary Figure 4). The most common breakpoint was the Cambodian type (63% [16/19]), also common in Indonesia (100% [5/5]) and Thailand (96% [30/31]). Inspection of the monoclonal infections with amplifications revealed a large range of DBP1 haplotypes in Ethiopia (minimum 8 haplotypes based on differences at nonheterozygous positions) (Figure 5). Furthermore, 38% (5/13) of isolates exhibited haplotype differences (heterozygote positions) between the copies within a given infection. The Thai parasites formed a largely separate cluster from Ethiopia, but with similar patterns of DBP1 diversity within and between infections (minimum 12 haplotypes, 35% [7/20] infections with heterozygotes). The sequence flanking DBP1 also displayed moderate diversity in Ethiopia and Asia, with no evidence of selective sweep dynamics (Supplementary Figure 5).
Table 2.
Summary of Copy Number Variants
| Gene | Description | Gene Coordinates | Variant Type | Frequency, % (no./No.) | |||
|---|---|---|---|---|---|---|---|
| Ethiopia | Thailand | Indonesia | Malaysia | ||||
| PVP01_0504500 | 28S ribosomal RNA | 5: 194 522-1 949 297 | Amplification | 100 (24/24) | 92 (96/104) | 95 (80/84) | 89 (41/46) |
| PVP01_0523500 | Plasmodium exported protein | 5: 951 202–952 041 | Deletion | 63 (15/24) | 0 (0/104) | 0 (0/84) | 0 (0/46) |
| PVP01_0524900, PVP01_0525000 | Plasmodium exported proteins | 5: 1 010 759-1 011 581 5: 1 013 733-1 014 550 |
Deletion | 4 (1/24) | 0 (0/104) | 0 (0/84) | 0 (0/46) |
| PVP01_0623800 | Duffy binding protein 1 | 6: 982 015–985 813 | Amplification | 79 (19/24) | 30 (31/104) | 6 (5/84) | 4 (2/46) |
| PVP01_1470400 | Plasmodium exported protein | 14: 3 009 446-3 010 524 | Amplification | 33 (8/24) | 49 (51/104) | 46 (39/84) | 89 (41/46) |
Twenty-two samples with an excess of copy number variants (≥18) were excluded from analysis.
Figure 5.
Heatplot illustrating multiple Duffy binding protein 1 (DBP1) haplotypes between samples and divergence between DBP1 copies within samples in monoclonal infections with copy number amplifications. The heat plot presents color-coded genotype calls at single-nucleotide polymorphisms in the DBP1 gene with minor allele frequency ≥1%. Genotypes are presented as reference allele frequencies ranging from 0 in red (homozygote alternative allele) to 1 in blue (homozygote reference allele). Samples are ordered on the y-axis according to their genetic relatedness as per the left-hand phylogram. Sample labels are color-coded according to country. Only monoclonal (within-sample F statistic > 0.95) infections with ≥2 DBP1 copies were included in the analyses. Therefore, heterozygous positions (in orange) reflect differences between the DBP1 copies within a given infection. Two Malagasy-type DBP1 amplifications are labeled; all other amplifications were Cambodian type.
DISCUSSION
This study presents the first population genomic investigation of P. vivax in Ethiopia, where the largest reservoir of this species in Africa persists. Our results reveal patterns of polyclonality, within-infection relatedness, population diversity, and structure comparable to Thailand and Indonesia, where transmission was stable at the time of sampling. We see evidence of genetic adaptations in various parasite mechanisms, which may facilitate the ongoing persistence of the population. The different forms of selection and adaptive responses that appear to be operating in the population are discussed further.
The striking prevalence of DBP1 amplification implies an important adaptive role in the host blood stage of infection in Ethiopia. The amplification has been found at low frequency in Malaysia (4%) and Indonesia (6%), and at higher frequencies in Cambodia (29%), Thailand (31%), and Madagascar (53%) [5, 13, 36, 37]. The prevalence in Ethiopia reached almost 80%, higher than in any population examined to date [5, 13, 36, 37]. The high haplotype diversity in the flanking regions of the DBP1 amplifications implies that it has most likely arisen independently on multiple occasions without being purged from the population. In a previous study in Thailand, a highly prevalent MDR1 amplification that conferred resistance to mefloquine was rapidly purged from the population after the drug was discontinued [24],. The fitness consequence of the DBP1 amplification is less clear. A genetic epidemiology study of DBP1 amplification in Cambodia speculated that it may have an immune-related role by enhancing antigenic diversity on the RBC membrane [36]. Our assessments in Ethiopia and Thailand also revealed moderately high DBP1 haplotype diversity, with haplotypic differences between copies enriching the diversity in >35% of monoclonal infections. However, the results of a recent functional study imply that the amplification may enhance parasite binding to Duffy-negative reticulocytes using a Duffy-independent invasion pathway [35]. Whether functioning in immune evasion or Duffy-negative reticulocyte invasion, the high prevalence of the DBP1 amplification in Ethiopia warrants close monitoring of this variant across the continent.
Antimalarial drug resistance presents another challenge to the containment of vivax in the Horn of Africa. Chloroquine (CQ) remains the first-line treatment for P. vivax blood-stage infection in Ethiopia, but recent surveys have demonstrated declining CQ efficacy against P. vivax, with recurrence ranging from 4% to 22% by day 28 [38–41]. Although the prevalence of the MDR1 Y976F mutation in Ethiopia (32%) was not as high as that documented in Papua Indonesia or Malaysia, where high-grade CQ resistance (CQR) has been reported [42, 43], ongoing surveillance is needed. Furthermore, since MDR1 Y976F mutation is not a prerequisite for high-grade CQR [34], additional markers are needed to facilitate these efforts. Several studies have explored the role of P. vivax CRT, the orthologue of PF3D7_0709000, the primary determinant of CQR in P. falciparum [44]; however, results are inconsistent [45–49]. Interestingly, we found evidence of extended haplotype homozygosity in Ethiopia relative to Thailand in a region upstream of CRT, potentially reflecting selection on a regulator of CRT expression in Ethiopia. While studies in Brazil have reported association between CRT expression and CQR in P. vivax using clinical phenotypes [46, 47], the lack of association using ex vivo phenotypes in isolates from Papua Indonesia [45], the epicenter of CQR for this species [50], is not consistent with a pivotal role. However, it is possible that different variants are operating in different populations. Further studies are warranted.
The absence of MDR1 amplification in Ethiopia suggests that mefloquine may be an appropriate treatment alternative for vivax, should CQR continue to rise. However, the high prevalence of DHFR double mutants (94%) raises concerns for pyrimethamine in regimens such as intermittent preventive treatment for pregnant women or infants. Interestingly, in contrast to the Asian populations, there were no triple or quadruple DHFR mutants and <20% prevalence of DHPS resistance-conferring mutants in Ethiopia, reflecting lower selective pressure. Accordingly, the haplotype-based signals of differential selection at the DHFR and DHPS loci reflected directional selection patterns in Thailand and Indonesia, not Ethiopia. It has been postulated that drug resistance variants may emerge more readily in populations with high rates of inbreeding. We did not find evidence of any significant difference in within-infection relatedness between Ethiopia and the Thai or Indonesian populations, implying comparable levels of inbreeding between these populations. Alternatively, differences in drug history or drug usage, which is generally postulated to be higher in Asia than Africa, might have contributed to lower selective pressure in Ethiopia.
Some of the strongest signals of selection in Ethiopia were observed in MSP5 and AMA1. Tajima D analysis demonstrated evidence of balancing selection in AMA1, consistent with high antigenic variation facilitating evasion of host immune pressure on the RBC stage of the parasite. Both AMA1 and MSP5 also exhibited evidence of differential selection between populations, reflecting complex haplotype structures of different strains circulating in different populations.
We also found evidence of adaptive variation in genes involved in regulation of gene expression, including several AP2 domain transcription factors. One of the strongest signals reflected an AP2 transcription factor on chromosome 14 (PVP01_1418100) under directional selection in Ethiopia and Asia. A previous study in Cambodia also reported strong selection on PVP01_1418100 and other transcription factors, highlighting the importance of these regulatory determinants in modulating mechanisms critical to the transmission and maintenance of vivax populations such as gametocytogenesis and hypnozoite dormancy and activation [17].
Although pressure from antimalarial drugs appears to be weaker in Ethiopia than Southeast Asia, the evidence of selection in genes involved in a variety of processes at various parasite life-cycle stages calls for multifaceted intervention approaches to contain P. vivax in the region. Ongoing genetic and genomic surveillance will aid in the detection of new forms of adaptation as they arise in the parasite population.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the patients who contributed their samples to the study; the health workers and field teams who assisted with the sample collections; the staff of the Wellcome Sanger Institute Sample Logistics, Sequencing, and Informatics facilities for their contributions; and the Director General of Health, Malaysia, for permission to publish this study.
Financial support. The sample and metadata collection in Ethiopian and Indonesia were supported by a Wellcome Trust Senior Research Fellowship in Clinical Science (grant number 200909 to R. N. P.); the National Health and Medical Research Council of Australia (Improving Health Outcomes in the Tropical North: A Multidisciplinary Collaboration “Hot North” Career Development Fellowship , grant number 1131932 to S. A.); and the Bill & Melinda Gates Foundation (award number OPP1164105). The patient sampling and metadata collection for Malaysia were funded by the Asia-Pacific Malaria Elimination Network (award number 108-07), the Malaysian Ministry of Health (award number BP00500420), and the National Health and Medical Research Council of Australia (award numbers 1037304 and 1045156; fellowships to N. M. A. [award numbers 1042072 and 1135820], B. E. B. [award number 1088738] and M. J. G. [award number 1074795]). M. J. G. was also supported by a National Health and Medical Research Council of Australia “Hot North” Early Career Fellowship (award number 1131932). The whole genome sequencing component of the study was supported by grants from the Medical Research Council and UK Department for International Development (award number M006212) and the Wellcome Trust (award numbers 206194 and 204911) to D. P. K. In addition, S. J. Z. and G. M. were funded by the Wellcome Trust (grant number 100956/Z/13/Z to G. M.). This work was supported by the Australian Centre for Research Excellence on Malaria Elimination (ACREME), funded by the National Health and Medical Research Council of Australia (APP 1134989).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Guerra CA, Howes RE, Patil AP, et al. The international limits and population at risk of Plasmodium vivax transmission in 2009. PLoS Negl Trop Dis 2010; 4:e774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sattabongkot J, Tsuboi T, Zollner GE, Sirichaisinthop J, Cui L. Plasmodium vivax transmission: chances for control? Trends Parasitol 2004; 20:192–8. [DOI] [PubMed] [Google Scholar]
- 3. Hupalo DN, Luo Z, Melnikov A, et al. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat Genet 2016; 48:953–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neafsey DE, Galinsky K, Jiang RH, et al. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat Genet 2012; 44:1046–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pearson RD, Amato R, Auburn S, et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet 2016; 48:959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. World malaria report 2016. Geneva, Switzerland: WHO, 2016. [Google Scholar]
- 7. Mathews HM, Armstrong JC. Duffy blood types and vivax malaria in Ethiopia. Am J Trop Med Hyg 1981; 30:299–303. [DOI] [PubMed] [Google Scholar]
- 8. Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med 1976; 295:302–4. [DOI] [PubMed] [Google Scholar]
- 9. Ménard D, Barnadas C, Bouchier C, et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci U S A 2010; 107:5967–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Getachew S, To S, Trimarsanto H, et al. Variation in complexity of infection and transmission stability between neighbouring populations of Plasmodium vivax in southern Ethiopia. PLoS One 2015; 10:e0140780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gunawardena S, Karunaweera ND, Ferreira MU, et al. Geographic structure of Plasmodium vivax: microsatellite analysis of parasite populations from Sri Lanka, Myanmar, and Ethiopia. Am J Trop Med Hyg 2010; 82:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lo E, Hemming-Schroeder E, Yewhalaw D, et al. Transmission dynamics of co-endemic Plasmodium vivax and P. falciparum in Ethiopia and prevalence of antimalarial resistant genotypes. PLoS Negl Trop Dis 2017; 11:e0005806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Auburn S, Benavente ED, Miotto O, et al. Genomic analysis of a pre-elimination Malaysian Plasmodium vivax population reveals selective pressures and changing transmission dynamics. Nat Commun 2018; 9:2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flannery EL, Wang T, Akbari A, et al. Next-generation sequencing of Plasmodium vivax patient samples shows evidence of direct evolution in drug-resistance genes. ACS Infect Dis 2015; 1:367–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Winter DJ, Pacheco MA, Vallejo AF, et al. Whole genome sequencing of field isolates reveals extensive genetic diversity in Plasmodium vivax from Colombia. PLoS Negl Trop Dis 2015; 9:e0004252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cowell AN, Valdivia HO, Bishop DK, Winzeler EA. Exploration of Plasmodium vivax transmission dynamics and recurrent infections in the Peruvian Amazon using whole genome sequencing. Genome Med 2018; 10:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parobek CM, Lin JT, Saunders DL, et al. Selective sweep suggests transcriptional regulation may underlie Plasmodium vivax resilience to malaria control measures in Cambodia. Proc Natl Acad Sci U S A 2016; 113:E8096–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Auburn S, Marfurt J, Maslen G, et al. Effective preparation of Plasmodium vivax field isolates for high-throughput whole genome sequencing. PLoS One 2013; 8:e53160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Padley D, Moody AH, Chiodini PL, Saldanha J. Use of a rapid, single-round, multiplex PCR to detect malarial parasites and identify the species present. Ann Trop Med Parasitol 2003; 97:131–7. [DOI] [PubMed] [Google Scholar]
- 20. Imwong M, Tanomsing N, Pukrittayakamee S, Day NP, White NJ, Snounou G. Spurious amplification of a Plasmodium vivax small-subunit RNA gene by use of primers currently used to detect P. knowlesi. J Clin Microbiol 2009; 47:4173–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sharrock WW, Suwanarusk R, Lek-Uthai U, et al. Plasmodium vivax trophozoites insensitive to chloroquine. Malar J 2008; 7:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Auburn S, Böhme U, Steinbiss S, et al. A new Plasmodium vivax reference sequence with improved assembly of the subtelomeres reveals an abundance of pir genes. Wellcome Open Res 2016; 1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Auburn S, Serre D, Pearson RD, et al. Genomic analysis reveals a common breakpoint in amplifications of the Plasmodium vivax multidrug resistance 1 locus in Thailand. J Infect Dis 2016; 214:1235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Auburn S, Campino S, Miotto O, et al. Characterization of within-host Plasmodium falciparum diversity using next-generation sequence data. PLoS One 2012; 7:e32891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Manske M, Miotto O, Campino S, et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 2012; 487:375–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu SJ, Hendry JA, Almagro-Garcia J, et al. The origins and relatedness structure of mixed infections vary with local prevalence of P. falciparum malaria. bioRxiv 2018. doi:10.1101/387266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Danecek P, Auton A, Abecasis G, et al. 1000 Genomes Project Analysis Group. The variant call format and VCFtools. Bioinformatics 2011; 27:2156–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 2016; 44:W242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting F(ST). Nat Rev Genet 2009; 10:639–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gautier M, Vitalis R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics 2012; 28:1176–7. [DOI] [PubMed] [Google Scholar]
- 32. Pasini EM, Böhme U, Rutledge GG, et al. An improved Plasmodium cynomolgi genome assembly reveals an unexpected methyltransferase gene expansion. Wellcome Open Res 2017; 2:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abdullah NR, Barber BE, William T, et al. Plasmodium vivax population structure and transmission dynamics in Sabah, Malaysia. PLoS One 2013; 8:e82553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Price RN, Auburn S, Marfurt J, Cheng Q. Phenotypic and genotypic characterisation of drug-resistant Plasmodium vivax. Trends Parasitol 2012; 28:522–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gunalan K, Lo E, Hostetler JB, et al. Role of Plasmodium vivax Duffy-binding protein 1 in invasion of Duffy-null Africans. Proc Natl Acad Sci U S A 2016; 113:6271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hostetler JB, Lo E, Kanjee U, et al. Independent origin and global distribution of distinct Plasmodium vivax Duffy binding protein gene duplications. PLoS Negl Trop Dis 2016; 10:e0005091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Menard D, Chan ER, Benedet C, et al. Whole genome sequencing of field isolates reveals a common duplication of the Duffy binding protein gene in Malagasy Plasmodium vivax strains. PLoS Negl Trop Dis 2013; 7:e2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Getachew S, Thriemer K, Auburn S, et al. Chloroquine efficacy for Plasmodium vivax malaria treatment in southern Ethiopia. Malar J 2015; 14:525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mekonnen SK, Aseffa A, Berhe N, et al. Return of chloroquine-sensitive Plasmodium falciparum parasites and emergence of chloroquine-resistant Plasmodium vivax in Ethiopia. Malar J 2014; 13:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Abreha T, Hwang J, Thriemer K, et al. Comparison of artemether-lumefantrine and chloroquine with and without primaquine for the treatment of Plasmodium vivax infection in Ethiopia: a randomized controlled trial. PLoS Med 2017; 14:e1002299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ketema T, Getahun K, Bacha K. Therapeutic efficacy of chloroquine for treatment of Plasmodium vivax malaria cases in Halaba district, South Ethiopia. Parasit Vectors 2011; 4:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ratcliff A, Siswantoro H, Kenangalem E, et al. Therapeutic response of multidrug-resistant Plasmodium falciparum and P. vivax to chloroquine and sulfadoxine-pyrimethamine in Southern Papua, Indonesia. Trans R Soc Trop Med Hyg 2007; 101:351–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grigg MJ, William T, Menon J, et al. Efficacy of artesunate-mefloquine for chloroquine-resistant Plasmodium vivax malaria in Malaysia: an open-label, randomized, controlled trial. Clin Infect Dis 2016; 62:1403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Djimdé A, Doumbo OK, Cortese JF, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med 2001; 344:257–63. [DOI] [PubMed] [Google Scholar]
- 45. Pava Z, Handayuni I, Wirjanata G, et al. Expression of Plasmodium vivax crt-o is related to parasite stage but not ex vivo chloroquine susceptibility. Antimicrob Agents Chemother 2016; 60:361–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Melo GC, Monteiro WM, Siqueira AM, et al. Expression levels of pvcrt-o and pvmdr-1 are associated with chloroquine resistance and severe Plasmodium vivax malaria in patients of the Brazilian Amazon. PLoS One 2014; 9:e105922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Silva SR, Almeida ACG, da Silva GAV, et al. Chloroquine resistance is associated to multi-copy pvcrt-o gene in Plasmodium vivax malaria in the Brazilian Amazon. Malar J 2018; 17:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nomura T, Carlton JM, Baird JK, et al. Evidence for different mechanisms of chloroquine resistance in 2 Plasmodium species that cause human malaria. J Infect Dis 2001; 183:1653–61. [DOI] [PubMed] [Google Scholar]
- 49. Sá JM, Yamamoto MM, Fernandez-Becerra C, et al. Expression and function of pvcrt-o, a Plasmodium vivax ortholog of pfcrt, in Plasmodium falciparum and Dictyostelium discoideum. Mol Biochem Parasitol 2006; 150:219–28. [DOI] [PubMed] [Google Scholar]
- 50. Price RN, von Seidlein L, Valecha N, Nosten F, Baird JK, White NJ. Global extent of chloroquine-resistant Plasmodium vivax: a systematic review and meta-analysis. Lancet Infect Dis 2014; 14:982–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.