Summary
Plant virus genome replication and movement is dependent on host resources and factors. However, plants respond to virus infection through several mechanisms, such as autophagy, ubiquitination, mRNA decay and gene silencing, that target viral components. Viral factors work in synchrony with pro‐viral host factors during the infection cycle and are targeted by antiviral responses. Accordingly, establishment of virus infection is genetically determined by the availability of the pro‐viral factors necessary for genome replication and movement, and by the balance between plant defence and viral suppression of defence responses. Sequential requirement of pro‐viral factors and the antagonistic activity of antiviral factors suggest a two‐step model to explain plant–virus interactions. At each step of the infection process, host factors with antiviral activity have been identified. Here we review our current understanding of host factors with antiviral activity against plant viruses.
Keywords: antiviral defence, host factors, virus–host interactions, virus resistance
Introduction
Infection of a plant by a virus initiates in a single cell. Viral proteins are synthesized by the host cell before genome replication, virion formation and movement to a neighbouring cell. The cycle is repeated at every newly infected cell (Nelson and Citovsky, 2005). Using the vascular system, plant viruses move long distances to infect tissues away from the initial site of infection, such as roots and young leaves (Heinlein, 2015; Wan et al., 2015). Multiple genetic analyses have shown that the entire infection cycle, including virus replication and movement, is genetically determined by viral and cellular factors that synchronize their activities in time and space (Fig. 1A) (Diaz et al., 2015; Hofius et al., 2007; Laliberte and Zheng, 2014; Li et al., 2016; Sasvari et al., 2018; Zhang et al., 2018).
Figure 1.
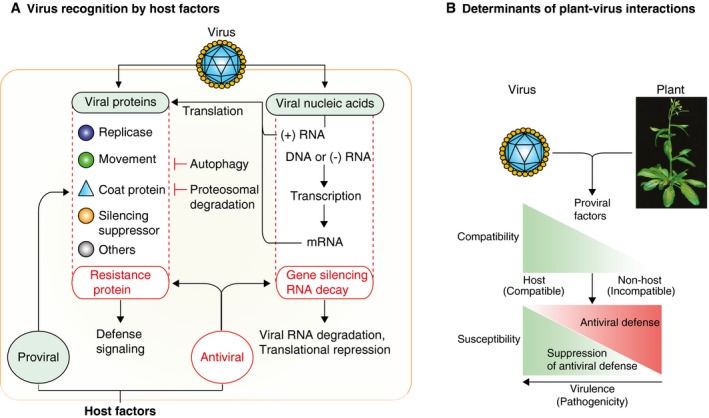
Genetic determinants of plant–virus interactions. (A) Viruses encode proteins to execute all parts of the infection cycle. Their expression is dependent on the host RNA translation machinery. Their activity requires host factors (pro‐viral) and resources. Antiviral immunity consists of host factors that target viral proteins or nucleic acids to restrict virus infection. (B) A two‐step model in plant–virus interactions. Compatibility is determined by the availability of pro‐viral host factors. Susceptibility is determined by the balance between antiviral defence and suppression of antiviral defence.
Plant–virus combinations could result in an incompatible or compatible interaction. Incompatible interactions occur between a virus and a non‐host plant, are characterized by the absence of virus infection and may be explained by the lack of cellular factors essential for the virus to replicate or move, antiviral defence or a combination (Fig. 1B) (Jaubert et al., 2011; Lellis et al., 2002). In contrast, compatible interactions occur between a virus and a susceptible host, are characterized by the establishment of virus infection and indicate the presence of pro‐viral cellular factors and resources necessary for virus infection and movement. Infection may spread through the entire plant, parts of the plant or be limited to the vascular system or the initially infected organ (Calvo et al., 2014a, 2014b; Lv et al., 2017; Otulak‐Koziel et al., 2018).
In susceptible hosts the absence of critical pro‐viral host factors results in the absence of infection and reduced virus replication, movement or both (Hofius et al., 2007; Lellis et al., 2002; Wang and Nagy, 2008). Accordingly, the absence of pro‐viral factors may turn a susceptible host into a non‐host, as is the case with resistant cultivars, landraces or ecotypes within a susceptible species (Hashimoto et al., 2016; Lellis et al., 2002). Because their presence conditions susceptibility, while their absence results in immunity or resistance, several terms have been used to describe host genes with pro‐viral activity, such as loss of susceptibility, recessive resistance or susceptibility genes (Garcia‐Ruiz, 2018; Hashimoto et al., 2016).
Interestingly, susceptible hosts harbour factors with antiviral activity (Diaz‐Pendon et al., 2007; Kushner et al., 2003; Panavas et al., 2005; Scholthof et al., 2011). To establish infection, viruses escape from or suppress antiviral defence activated by viral proteins or nucleic acids, particularly RNA (Fig. 1A) (Garcia and Pallas, 2015; Gorovits et al., 2016). With or without a hypersensitive reaction, the defence response restricts essential parts of the infection cycle, such as viral RNA translation, virus replication or movement, resulting in reduced virus accumulation and/or a delay in the establishment of systemic infection. Symptoms may or may not develop (Donze et al., 2014; Garcia‐Ruiz et al., 2018; Korner et al., 2018).
Multiple genetic analyses have shown that the outcome of plant–virus interactions is genetically determined by viral factors, host factors and their interaction (Fig. 1A) (Chisholm et al., 2001; Jaubert et al., 2011; Kushner et al., 2003; Lellis et al., 2002; Panavas et al., 2005). Consistent with this model, for all parts of the infection cycle, at least one host gene with antiviral activity has been identified (Table 1). Here I review our current understanding of host factors with antiviral activity against plant viruses. Their antagonistic activity is presented following sequential parts of the infection cycle.
Table 1.
Representative host factors with antiviral activity against plant viruses.
| Host factor | Cellular function | Virus | Viral factor | Host | Technique | Reference |
|---|---|---|---|---|---|---|
| Viral RNA translation | ||||||
| APUM5 | mRNA binding | CMV, TuMV | mRNA | Arabidopsis thaliana | T‐DNA mutant screen | Huh et al. (2013) |
| NIK1 | Receptor‐like kinase | CaLCuV | NSP | A. thaliana | Genetic analysis | Zorzatto et al. (2015) |
| Virus replication complex formation | ||||||
| PAH1 | Phospholipid biosynthesis | BMV, TBSB | 1a, p33 | Yeast and Nicotiana benthamiana | Genetic analysis | Chuang et al. (2014); Zhang et al. (2018) |
| Accumulation or activity of the replication proteins | ||||||
| Beclin1 (ATG6) | Autophagy | TuMV | NIb | N. benthamiana A. thaliana | Autophagosome marker, yeast two‐hybrid | Li et al. (2018) |
| Tm‐1 | NA | ToMV | 130K | Solanum lycopersicum | Cell fractionation and mass spectrometry | Ishibashi et al. (2007) |
| TARF | Ubiquitination | TMV | 126K | Nicotiana tabacum | Yeast two‐hybrid, VIGS | Yamaji et al. (2010) |
| Ubiquiting‐proteosome system | Protein degradation | TYMV | RdRp | A. thaliana | Pulse‐chase labelling | Camborde et al. (2010) |
| Rsp5p | Ubiquitination | TBSV | P92 | Yeast | Proteomics | Barajas et al. (2009) |
| PVR4 | NA | PepMV, PVY | NIb | Capsicum annum | Transient expression | Kim et al. (2015) |
| mRNA stability | ||||||
| DCP1, DCP2, XRN4, PARN | mRNA decay | TuMV | mRNA | N. benthamiana, A. thaliana | Genetic analysis | Li and Wang (2018) |
| XRN4 | mRNA decay | TBSV | mRNA | Yeast and N. benthamiana | Genetic mutation, VIGS | Jaag and Nagy (2009) |
| XRN4 | mRNA decay | TMV | mRNA | N. benthamiana | VIGS | Peng et al. (2011) |
| DCP1 | mRNA decay | TRV | mRNA | A. thaliana | Genetic mutation | Ma et al. (2015) |
| Virus movement | ||||||
| ESC1 (AtPiezo) | Mechanosensitive ion channel | CMV, TuMV | NA | A. thaliana | EMS mutagenesis | Zhang et al. (2019) |
| RTM1, RTM2, RTM3 | Protein binding | TEV | CP | A. thaliana | GUS or GFP‐fusion constructs | Chisholm et al. (2001); Decroocq et al. (2009) |
| KELP | Transcription coactivator | ToMV | p30 | N. benthamiana | Transient expression | Sasaki et al. (2009) |
| BTR1 | mRNA binding | ToMV | Genomic RNA | A. thaliana | Immunoprecipitation and mass spectrometry | Fujisaki and Ishikawa (2008) |
| Rsv3 | NA | SMV | CI | Glycine max | Genetic analysis | Zhang et al. (2009) |
| Rsv4 | NA | SMV | NA | G. max | Genetic analysis | Ma et al. (1995) |
| Ny‐1 | NA | PVY | NA | Solanum tuberosum | Hybrids between resistant and susceptible cultivars | Lukan et al. (2018); Szajko et al. (2008) |
| Antiviral gene silencing | ||||||
| DCL, AGO, RDR, SGS, DRB | gene silencing | CaMV, CymRSV, MNSV, PMMoV, ORMV, TuMV, SCMV, MCMV, CMV, PVA, TCV, TBSV, TSWV, PVX, ToRSV, RSV, TRV, TYLCV, WMV | RNA | A. thaliana, N. benthamiana Zea mays, Oryza sativa Cucumis melo, S. lycopersicum | Genetic analysis | Blevins et al., (2006); Brosseau and Moffett (2015); Diaz‐Pendon et al. (2007); Donaire et al. (2009); Garcia‐Ruiz et al. (2010, 2015); Jaubert et al. (2011); Karran and Sanfacon (2014); Ludman et al. (2017); Qu et al. (2008); Raja et al. (2014); Scholthof et al. (2011); Wu et al. (2015); Xia et al. (2016)) |
| Ty‐1 | RNA‐dependent RNA polymerase | ToYLCV | Genomic DNA | S. lycopersicum | Genetic analysis | Butterbach et al. (2014) |
| rgs‐Cam | Regulator of gene silencing | CMV | 2b | N. tabacum | Yeast two‐hybrid, transgenic overexpression | Anandalakshmi et al. (2000); Jeon et al. (2017) |
| PhOBF1 | Transcription factor | TRV | NA | Petunia hybrida | VIGS | Sun et al. (2017) |
| Accumulation or activity of viral proteins | ||||||
| NBR1 | Autophagy cargo receptor | TuMV | HC‐Pro | A. thaliana | Genetic analysis | Hafren et al. (2018) |
| ATG7, ATG8 | Autophagy | BSMV | NA | N. benthamiana | Yeast two‐hybrid, VIGS | Yang et al. (2018) |
| ATG8 | Autophagy | CLCuMuV | ßC1 | N. benthamiana | Yeast two‐hybrid, VIGS | Haxim et al. (2017) |
| rgs‐CaM | Immune receptor | CMV, TEV and TuMV | 2b, HC‐Pro | N. benthamiana | Surface plasmon resonance | Jeon et al. (2017); Nakahara et al. (2012) |
| RNA replication | ||||||
| GAPDH | Glycolysis | BaMV | 3ʹ UTR | N. benthamiana | UV‐crosslinking to RdRp preparations | Prasanth et al. (2011) |
| Virion formation | ||||||
| NBR1 | Autophagy cargo receptor | CaMV | CP, virions | A. thaliana | Genetic analysis | Hafren et al. (2017) |
| PUS4 | Pseudouridina synthase | BMV | Genomic RNA | N. benthamiana | Proteome array | Zhu et al. (2007) |
| Virus accumulation | ||||||
| CYR1 | NA | MYMIV | CP | Vigna mungo | Natural variation | Maiti et al. (2012) |
| NBR1 | Autophagy cargo receptor | TuMV and WMV | HC‐Pro | A. thaliana | Genetic analysis | Hafren et al. (2018) |
| RFP1 | Ubiquitination | TYLCCV | BC1 | N. tabacum | Yeast two‐hybrid | Shen et al. (2016) |
| PSBP | Kinase | AMV | CP | N. benthamiana | Yeast two‐hybrid | Balasubramaniam et al. (2014) |
| Cell death | ||||||
| N | Protein phosphatase | TMV | Helicase | N. tabacum 'Xanthi' | Transient expression | Abbink et al. (1998; Padgett et al. (1997) |
| RCY1 | NA | CMV strain Y | CP | A. thaliana | Genetic mapping | Takahashi et al. (2001) |
| Rx1, Rx2 | NA | PVX | CP | S. tuberosum | Transient expression | Bendahmane et al. (2000) |
| Tm‐2 | NA | TMV | MP | S. lycopersicum | Genetic analysis | Meshi et al. (1989) |
| Tm‐22 | NA | ToMV | MP | S. lycopersicum | Cloning, transgenic expression, localization | Chen et al. (2017); Lanfermeijer et al. (2003) |
| RPP8 | Protein binding | TCV | CP | A. thaliana | Cloning, transgenic expression | Cooley et al. (2000) |
| Rsv1 | NA | SMV | P3 and HC‐Pro | G. max | Virus mutagenesis | Eggenberger et al. (2008) |
| Tsw | NA | TSWV | NSs | Capsicum chinense | Transient expression | de Ronde et al. (2013) |
| Sw5b | NA | TSWV | NSm | S. tuberosum | Transient and transgene expression | Mariana et al. (2014) |
Virus names: alfalfa mosaic virus (AMV), bamboo mosaic virus (BaMV), barley stripe mosaic virus (BSMV), brome mosaic virus (BMV), cabbage leaf curl virus (CaLCuV), cauliflower mosaic virus (CaMV), cotton leaf curl multan virus (CLCuMuV), cymbidium ringspot virus (CymRSV), cucumber mosaic virus (CMV), cucumber necrosis virus (CNV), maize chlorotic mottle virus (MCMV), melon necrotic spot virus (MNSV), mungbean yellow mosaic india virus (MYMIV), oilseed rape mosaic virus (ORMV), pepper mild mottle virus (PMMoV), pepper mottle virus (PepMV), potato virus A (PVA), potato virus X (PVY), potato virus Y (PVY), rice stripe virus (RSV), soybean mosaic virus (SMV), sugarcane mosaic virus (SCMV), tobacco etch virus (TEV), tobacco mosaic virus (TMV), tobacco rattle virus (TRV), tomato bushy stunt virus (TBSV), tomato mosaic virus (ToMV), tomato ringspot virus (ToRSV), tomato yellow leaf curl virus (TYLCV), tomato spotted wilt virus (TSWV), tomato yellow leaf curl virus (ToYLCV), turnip crinkle virus (TCV), turnip mosaic virus (TuMV), turnip yellow mosaic virus (TYMV), watermelon mosaic virus (WMV).
Yeast: Saccharomyces cerevisiae.
Viral Determinants of Infection
RNA translation, genome replication, and virion formation and movement are core parts of the infection cycle of a plant by a virus (Ahlquist, 2006; Nelson and Citovsky, 2005). To accomplish these tasks, plant viruses encode replication, capsid, movement and auxiliary proteins. Additionally, to condition a cellular environment that is conducive to virus replication and movement, viral proteins target key components of antiviral immunity. Viral factors that determine the extent of infection and disease severity are considered pathogenicity determinants. Gene silencing suppressors are remarkable pathogenicity determinants of plant viruses. Virus‐encoded suppressors interfere with antiviral defence mechanisms mediated by gene silencing (Anandalakshmi et al., 1998; Diaz‐Pendon et al., 2007; Garcia‐Ruiz et al., 2015; Jaubert et al., 2011). Interestingly, the activity of several gene‐silencing suppressors promotes infection of heterologous viruses when expressed in cis‐, trans‐ or synergistic co‐infections (Garcia‐Ruiz et al., 2018; Gupta et al., 2018; Maliogka et al., 2012) (Fig. 2).
Figure 2.
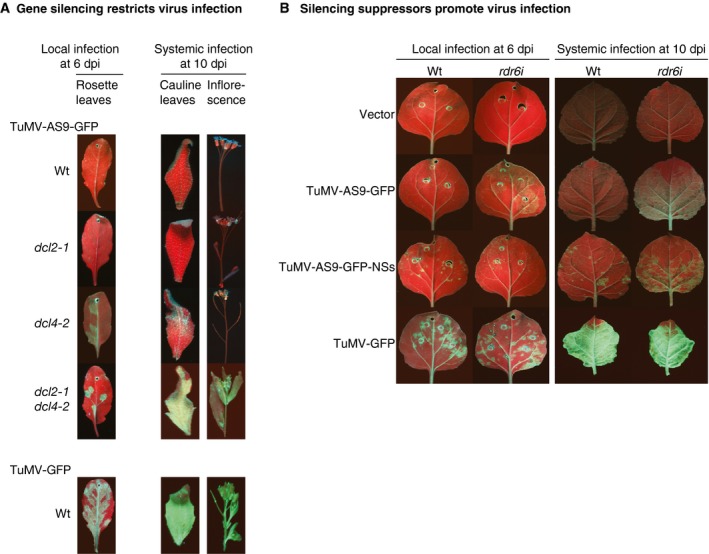
The balance between gene silencing and silencing suppression determines infection progression. Arabidopsis thaliana plants were mechanically inoculated with suppressor‐deficient turnip mosaic virus (TuMV)‐GFP or TuMV‐GFP. In Nicotiana benthamiana plants, infection was initiated by agroinfiltration. Pictures were taken under UV light. (A) In A. thaliana, Dicer‐like proteins 2 and 4 (DCL2 and DCL4) are core components of antiviral gene silencing and restrict virus infection in a tissue‐specific manner. In leaves, DCL4 is sufficient and DCL2 is dispensable. In the inflorescence, both DCL2 and DCL4 are necessary to restrict virus infection. TuMV‐encoded silencing suppressor (HC‐Pro) overcomes the antiviral effect of gene silencing and promotes the establishment of infection in leaves and the inflorescence. (B) In N. bethamiana RDR6 is an essential component of gene silencing. Suppressor‐deficient TuMV‐AS9‐GFP cannot infect wild‐type N. benthamiana. Local and systemic infection occurred by knocking down RDR6 in rdr6i plants, or by providing in cis the silencing suppressor from tomato spotted wilt virus. In normal and rd6i plants, local and systemic infection occur and the virus accumulates to high levels. Pathogenicity is determined by TuMV HC‐Pro.
Host Determinants of Infection
Genome‐wide screens of Saccharomyces cerevisiae (yeast) replicating brome mosaic virus (BMV) (Kushner et al., 2003) or tomato bushy stunt virus (TBSV) (Panavas et al., 2005) showed that a compatible host contains both pro‐viral and antiviral factors that affect virus replication at the cellular level. Mutagenesis and genetic analyses in Arabidopsis thaliana (Arabidopsis) allowed identification of pro‐viral and antiviral factors that affect virus replication at the cellular level, cell‐to‐cell and systemic movement (Diaz‐Pendon et al., 2007; Garcia‐Ruiz et al., 2010; Guo et al., 2017; Lellis et al., 2002; Zhang et al., 2019). Accordingly, viruses need pro‐viral host factors and are targeted by antiviral host factors. Pro‐viral host factors are necessary for essential steps of the infection cycle and work in synchrony with viral factors. In contrast, antiviral defence is mediated by host factors that target viral nucleic acids or proteins by multiple mechanisms such as autophagy, proteasome degradation, RNA decay and gene silencing (Fig. 1A).
Compatibility and Susceptibility in Plant–Virus Interactions
The role of host and viral factors, and their availability in the cell during the establishment of infection, suggests a two‐step model to explain plant–virus interactions (Fig. 1B). Compatibility is determined by the availability of pro‐viral host factors. The absence of one or more pro‐viral factors results in incompatibility in a host plant (Lellis et al., 2002). Viral proteins must be translated by the host translational machinery before replication can occur (Ahlquist, 2006; Machado et al., 2017; Miller et al., 2016). This feature makes viral RNA translation a critical determinant of the outcome in plant–virus interactions. An example is translation initiation factor eIF(iso)4E and potyviruses. Arabidopsis is susceptible to tobacco etch virus (TEV) and turnip mosaic virus (TuMV). However, mutant plants lacking eIF(iso)4E are immune to TEV and TuMV (Lellis et al., 2002). Similarly, down‐regulation of eIF(iso)4E in plum confers resistance to plum pox virus (PPV) (Wang et al., 2013). The mechanism is likely mediated by the lack of RNA translation to form potyviral polyproteins and possibly the lack of cell‐to‐cell movement (Contreras‐Paredes et al., 2013; Lellis et al., 2002). Similar effects have been detected for several plant species and their corresponding potyviruses (Sanfacon, 2015).
In compatible plant–virus combinations, susceptibility is determined by the balance between antiviral defence and suppression of antiviral defence (Fig. 1B). Defence responses may prevent infection from spreading to the entire plant, determining different levels of susceptibility to virus infection. If infection is stopped early, before the formation of local foci, the plant phenotype may be the same as that of a non‐host (Garcia‐Ruiz et al., 2015; Lellis et al., 2002; Qu et al., 2008).
A genetic analysis of infection of Arabidopsis by green‐fluorescence protein‐tagged TuMV (TuMV‐GFP) illustrates a two‐step model in plant–virus interactions (Fig. 2A). Suppressor‐deficient (mutant helper component proteinase HC‐Pro) TuMV‐AS9‐GFP cannot infect wild‐type plants or dcl2‐1 mutants. Infection is halted at the cellular level by gene silencing. However, suppressor‐deficient TuMV‐AS9‐GFP is able to infect dcl4‐2 mutants, which lack the contribution of Dicer‐like protein 4 (DCL4) to gene silencing. Visible infection foci form and the virus moves systemically into cauline leaves without reaching the inflorescence. Interestingly, suppressor‐deficient TuMV‐AS9‐GFP is able to establish local and systemic infection of cauline leaves and inflorescence in dcl2‐1 dcl4‐2 double mutants. Thus, in the absence of the HC‐Pro silencing suppression activity, gene silencing restricts infection in a tissue‐specific manner. In contrast, TuMV‐GFP establishes local and systemic infection, including the inflorescence, of Arabidopsis plants, wild‐type or mutants. Accordingly, the antiviral role of gene silencing is defeated by the TuMV‐encoded silencing suppressor HC‐Pro (Garcia‐Ruiz et al., 2010).
A genetic analysis of infection of Nicotiana benthamiana by TuMV‐GFP further supports the two‐step model (Fig. 2B). Suppressor‐deficient TuMV‐AS9‐GFP cannot infect wild‐type N. benthamiana. In Arabidopsis and in N. benthamiana, RNA‐dependent RNA polymerase 6 (RDR6) is a core component of antiviral gene silencing (Garcia‐Ruiz et al., 2010; Qu et al., 2005, 2008; Yang et al., 2004). In N. benthamiana, infection of the meristems by potato virus X (PVX) is prevented by RDR6 (Schwach et al., 2005), and infection of the Arabidopsis meristems by TuMV is restricted by argonaute (AGO) proteins 1, AGO2 and AGO10 (Garcia‐Ruiz et al., 2015).
Knockdown of RDR6 by RNA interference in N. benthamiana (rdr6i) (Schwach et al., 2005) rescued local and systemic infection by TuMV‐AS9‐GFP. In an alternative approach, expression in cis of the NSs protein from tomato spotted wilt virus (TSWV) (Garcia‐Ruiz et al., 2018) supported the establishment of local and systemic infection by TuMV‐AS9‐GFP (Fig. 2B). These observations show that tissue‐specific restriction of virus infection is determined by the balance between gene silencing and gene silencing suppression (Garcia‐Ruiz et al., 2015, 2018; Schwach et al., 2005).
Host Genes with Antiviral Activity
Host factors with antiviral activity (Fig. 2) limit virus accumulation, movement or both, resulting in a virus‐resistant or tolerant phenotype that normally displays symptoms less severe than susceptible plants (Diaz‐Pendon et al., 2007; Garcia‐Ruiz et al., 2010; Huh et al., 2013). For each part of the infection cycle, at least one host gene with antiviral activity has been identified (Table 1). Representative host factors are described below.
Viral RNA translation
Translation of viral proteins from genomic RNA, subgenomic RNA or mRNA is dependent on cellular factors and the protein translation machinery. Being a critical step that determines the availability of viral proteins, both host and viral factors regulate translation (Ahlquist, 2006; Miller et al., 2016; Sanfacon, 2015). In Arabidopsis, a leucine‐rich repeat receptor‐like kinase (NIK1) is a master regulator of translation. As a defence mechanism, using an NIK1‐dependent pathway, plants down‐regulate translation upon begomovirus infection. This effect results in a reduction in virus replication and accumulation. Remarkably, begomoviral nuclear shuttle protein (NSP) inactivates NIK1 to up‐regulate translation and promote susceptibility (Zorzatto et al., 2015).
Using nucleotide sequences as recognition signatures, Arabidopsis Pumilio RNA binding protein 5 (APUM5) binds cucumber mosaic virus (CMV) and TuMV mRNA to inhibit translation. Accordingly, mutant plants lacking APUM5 accumulate CMV and TuMV to higher levels than plants harbouring wild‐type APUM5 (Huh et al., 2013).
Virus replication complex formation
After reaching the nucleus of infected cells, DNA viruses form minichromosomes that are replicated by cellular DNA‐dependent DNA polymerases (Ceniceros‐Ojeda et al., 2016). In contrast, on cellular membranes, RNA viruses induce the formation of vesicles that contain RNA‐dependent RNA polymerases and genomic RNA, and are the sites of replication. Several cellular proteins that antagonize the formation of viral replication compartments have been identified and characterized (Table 1).
Phospholipids are crucial membrane components. Phosphatidic acid phosphohydrolase 1 (PAH1) limits phospholipid synthesis. Genetic analyses in yeast and N. benthamiana showed that PAH1 negatively regulates BMV and TBSV replication complex formation, resulting in reduced virus replication at the cellular level and reduced accumulation in plants (Chuang et al., 2014; Zhang et al., 2018).
Accumulation or activity of replication proteins
Virus‐encoded RNA‐dependent RNA polymerases replicate the genome of RNA viruses and, if present, transcribe subgenomic RNAs that are essential for gene expression. Viral RNA‐dependent RNA polymerases contain a conserved GDD motif (Li et al., 2018) and are targeted for degradation by autophagy protein 6 (ATG6 or Beclin1). Beclin1 is a core component of autophagy, interacting with and triggering degradation of the RNA‐dependent RNA polymerase (NIb) of several potyviruses, including TuMV, PPV, soybean mosaic virus (SMV) and TEV. Beclin1 also triggers degradation of the RNA‐dependent RNA polymerases of cucumber green mottle mosaic virus and pepino mosaic virus (Li et al., 2018). Additionally, in pepper (Capsicum annum), pathogenesis‐related protein 4c (Pvr4c) interacts with NIb and triggers cell death upon infection by pepper mottle virus or potato virus Y (PVY) (Kim et al., 2015).
In tomato (Solanum lycopersicum), tobacco mosaic virus resistance 1 (Tm‐1) confers resistance to tobacco mosaic virus (TMV) and to tomato mosaic virus (ToMV). Tm‐1 encodes a protein that binds ToMV replication protein 103K and prevents its normal activity (Ishibashi and Ishikawa, 2013; Ishibashi et al., 2007).
RNA replication
Glycolytic enzyme glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) binds the 3′ UTR of bamboo mosaic virus (BaMV) and satellite BaMV RNAs. This interaction down‐regulates BaMV negative‐strand RNA synthesis. Accordingly, GAPDH knockdown in N. benthamiana enhanced accumulation of BaMV. In contrast, GAPDH overexpression reduced BaMV accumulation (Prasanth et al., 2011). GAPDH has the opposite effect on TBSV replication. GAPDH preferentially binds to the 3′ end of negative‐strand TBSV RNA, retaining it in the replication complex to promote positive‐strand RNA synthesis. This activity results in asymmetric RNA replication characterized by higher synthesis and accumulation of positive‐ over negative‐strand genomic RNA, which is normal in TBSV replication (Wang and Nagy, 2008)
mRNA stability
Viruses express their genes through mRNA (Ahlquist, 2006). After translation, cellular and viral mRNAs are deadenylated, decapped and cleaved 5′ to 3′ by exoribonuclease 4 (XRN4) through the decapping‐dependent RNA decay pathway (Thran et al., 2012; Tsuzuki et al., 2017). Recent genetic analyses showed that RNA decay has an antiviral role that limits virus accumulation and may contribute to plant recovery from virus‐induced symptoms (Li and Wang, 2018; Ma et al., 2015; Moon and Wilusz, 2013; Tsuzuki et al., 2017). RNA decay and RNA silencing seem to act in coordination to suppress virus infection, and their activities partially overlap (Li and Wang, 2018; Peng et al., 2011).
Consistent with the antiviral role of RNA decay, potyviral HC‐Pro and genome‐linked protein (VPg) are silencing suppressor proteins that interfere with both gene silencing and mRNA decay. Interference with mRNA decay occurs through interactions with XRN4 and decapping protein 2 (DCP2), respectively, two core components of the 5′ to 3′ RNA decay pathway (Li and Wang, 2018).
Virus movement
Plant viruses move cell to cell as virions or nucleoprotein complexes through plasmodesmata. As a critical component of this process, plant viruses encode movement proteins that increase the plasmodesmata size exclusion limit and/or form microtubules (Taliansky et al., 2008). Several host factors that antagonize virus movement have been identified. They target viral proteins and RNA, or trigger cell death (Table 1).
Plants encode mechanosensitive ion channels that regulate ion movement across cells. In Arabidopsis, ESC1 encodes a piezo protein that functions as a mechanosensitive Ca2+ permeable channel and limits systemic infection of CMV and TuMV (Zhang et al., 2019).
Systemic movement of TEV, and some isolates of PPV and lettuce mosaic virus (Decroocq et al., 2009), is restricted by restricted‐TEV‐movement (RTM) genes RTM1, RTM2 and RTM3. These genes are expressed in phloem sieve elements and interact with the viral coat protein (Chisholm et al., 2001). Interestingly, resistance‐breaking isolates had mutations in the N‐terminus of the coat protein (Decroocq et al., 2009).
BTR1 is a ribonucleoprotein K‐homology RNA‐binding protein that binds ToMV genomic RNA and restricts cell‐to‐cell movement (Fujisaki and Ishikawa, 2008). In potato (S. tuberosum) plants the Ny‐1 gene confers resistance to PVY by triggering cell death at the infection sites, limiting cell‐to‐cell movement (Lukan et al., 2018).
Gene silencing
In plants, gene silencing is an essential mechanism of antiviral defence. Gene silencing targets viral RNA for degradation or translational repression. The result is restriction of virus replication and movement, and recovery from virus‐induced symptoms (Korner et al., 2018; Szittya and Burgyan, 2013). Gene silencing targets DNA and RNA viruses, satellite RNA viruses and viroids (Blevins et al., 2006; Diaz‐Pendon et al., 2007; Minoia et al., 2014; Shimura et al., 2011). All viruses express their genes and/or replicate their genome through an RNA intermediate (Ahlquist, 2006). This feature exposes viruses to gene silencing.
The core components of gene silencing include Dicer‐Like (DCL), Argonaute (AGO), double‐stranded RNA binding (DRB) and RNA‐dependent‐RNA‐polymerase (RDR) proteins. These proteins are conserved across plants (Incarbone and Dunoyer, 2013; Szittya and Burgyan, 2013; Zvereva and Pooggin, 2012). A signature feature of antiviral gene silencing is the accumulation of virus‐derived small interfering RNAs (siRNAs) in infected plants. Viral RNA is processed by DCL proteins into siRNAs that are 21 to 24 nucleotides long. Virus‐derived siRNAs are loaded into AGO proteins, programming them for specific slicing or translational repression of viral RNA (Garcia‐Ruiz et al., 2015; Karran and Sanfacon, 2014; Schuck et al., 2013). Accordingly, viral RNA is targeted by both DCL and AGO proteins.
Antiviral gene silencing might be triggered by viral RNA replication intermediates, self‐complementary sequences forming hairpin structures in viral single‐stranded RNA, and by products of overlapping transcription (Pantaleo et al., 2007; Szittya and Burgyan, 2013). DCL proteins process double‐stranded virus RNA into primary virus‐derived siRNAs that are necessary but not sufficient to prevent virus infection. Establishment of an antiviral state requires silencing amplification by plant RNA‐dependent RNA polymerases that synthesize double‐stranded RNA from single‐stranded viral RNA (Diaz‐Pendon et al., 2007; Garcia‐Ruiz et al., 2010).
Virus‐derived siRNA profiling has demonstrated that, in compatible plant–virus interactions, the entire genome of positive‐strand and negative‐strand RNA viruses is targeted by gene silencing in both monocot and dicot plants (Donaire et al., 2009; Garcia‐Ruiz et al., 2015; Margaria et al., 2015; Tatineni et al., 2014; Wang et al., 2011; Xia et al., 2016). However, gene silencing is not enough to restrict virus infection. That is due to the inhibitory activity of virus‐encoded gene silencing suppressors. Suppressors condition susceptibility, promote virus replication and movement, and promote symptom development by interfering with endogenous and antiviral gene silencing (Burgyan and Havelda, 2011; Garcia‐Ruiz et al., 2018; Kasschau et al., 2003). The mechanisms of silencing suppression include triggering the degradation of core components of gene silencing such as DCL, AGO, RDR6 and suppressor of gene silencing 3 (SGS3) proteins, and binding of both virus‐derived and cellular siRNAs including micro‐RNAs (miRNAs) (Burgyan and Havelda, 2011; Garcia‐Ruiz et al., 2015; Del Toro et al., 2017). These effects prevent the biogenesis and/or activity of virus‐derived and cellular siRNAs. Plant development and response to abiotic and biotic stress is in part regulated by miRNAs and other siRNAs. Accordingly, virus‐encoded gene silencing suppressors are determinants of symptom development (Garcia‐Ruiz et al., 2018; Kasschau et al., 2003). Furthermore, viral silencing suppressors impact virus accumulation and the spatial distribution of virus infection (Garcia‐Ruiz et al., 2018), and are frequently determinants of host range (Garcia‐Ruiz et al., 2015; Jaubert et al., 2011; Li et al., 2014).
The antiviral role of gene silencing was unambiguously demonstrated using viruses lacking gene silencing suppressors (Diaz‐Pendon et al., 2007; Garcia‐Ruiz et al., 2010, 2015; Pantaleo et al., 2007; Qu et al., 2008). Turnip crinkle virus (TCV), TBSV, CMV and TuMV accumulate to similar levels in wild‐type plants and in mutant plants lacking core components of the silencing machinery. However, suppressor‐deficient viruses cannot infect wild‐type plants. Instead, suppressor‐deficient viruses can only infect plants lacking core gene silencing components (Fig. 2). These genetic systems have been used to identify and characterize components of gene silencing (Diaz‐Pendon et al., 2007; Garcia‐Ruiz et al., 2010, 2015; Pantaleo et al., 2007; Qu et al., 2008).
As illustrated by genetic analyses using suppressor‐deficient TuMV‐AS9‐GFP, antiviral gene silencing restricts virus infection and movement in a tissue‐specific manner. In Arabidopsis plants lacking DCL4, AGO2, RDR1 or RDR6, TuMV‐AS9‐GFP established local infection and moved systemically into non‐inoculated leaves, without reaching the inflorescence. Systemic infection of the inflorescence only occurred in the absence of both DCL2 and DCL4, or RDR1 and RDR6, or AGO1, AGO2 and AGO10 (Garcia‐Ruiz et al., 2010, 2015).
To prevent their inhibitory effect on gene silencing, several plant factors target virus‐encoded silencing suppressors or regulate expression of gene silencing components (Table 1), as illustrated by the following examples. In Nicotiana tabacum, a calmodulin‐like protein (rgs‐CaM) binds to and, via autophagy directs degradation of, 2b, the silencing suppressor in CMV (Jeon et al., 2017). Autophagy cargo receptor NBR1 targets potyviral HC‐Pro for degradation, thus affecting silencing suppression and reducing accumulation of TuMV and watermelon mosaic virus (WMV). Interestingly, TuMV VPg and 6K2 prevent NBR1‐dependent degradation of HC‐Pro (Hafren et al., 2018).
In petunia (Petunia hybrida), PhOBF1, a leucine transcription factor, is up‐regulated by tobacco rattle virus (TRV) infection. PhOBF1 is a positive regulator of salicylic acid biosynthesis and of core components of gene silencing: DCL, AGO and RDRs. Thus, PhOBF1 enhances antiviral responses to TRV (Sun et al., 2017). In tomato, the Ty‐1 gene encodes an RNA‐dependent RNA polymerase that confers resistance to geminiviruses by enhancing transcriptional gene silencing (Butterbach et al., 2014).
Virion assembly and disassembly
In BMV, the negative‐strand RNA core promoter consists of a short stem with a three‐nucleotide loop that forms a clamp adenine motif. An array of 5000 yeast proteins was screened for proteins that bind the clamp adenine motif. Pseudouridine synthase 4 (PUS4) was identified. Functional characterization in N. benthamiana showed that PUS4 binding to BMV positive‐strand RNA prevented encapsidation, resulting in a slight reduction in viral RNA accumulation and a drastic reduction in BMV systemic movement (Zhu et al., 2007).
Host factors that condition virus resistance by undetermined mechanisms
For a growing number of plant–virus combinations, reduced virus accumulation has been observed in the presence of genes with antiviral activity although the part of the infection cycle that is affected has not been identified (Table 1). In these cases, virus infection triggers a hypersensitive response that results in the formation of necrotic lesions. Cell death might reduce virus movement and confine the virus to the infection sites and surrounding cells, but is not sufficient to prevent virus movement out of the cell death zone (Lukan et al., 2018).
The following plant–virus combinations are examples of host factors that condition virus resistance by undetermined mechanisms. The N resistance gene from Nicotiana glutinosa was introduced into N. tabacum and confers resistance to TMV (Levy et al., 2004). The N resistance protein is a receptor that contains three essential domains: a Toll‐interleukin‐1 (TIR), a nucleotide‐binding site (NBS) and a leucine‐rich repeat (LRR) (Dinesh‐Kumar et al., 2000). Transcription and alternative splicing of the N gene is stimulated by TMV infection (Levy et al., 2004), the protein coded by the N gene recognizes the helicase domain in TMV replication protein 126‐kD, and triggers a hypersensitive response visible as local necrotic lesions. As a result, TMV infection is restricted to cells surrounding the entry site (Abbink et al., 1998; Levy et al., 2004; Padgett et al., 1997).
The arginine‐rich cyclin 1 (RCY1) gene in Arabidopsis recognizes the coat protein in CMV strain Y and triggers local cell death (Takahashi et al., 2001). Similarly, TSWV infection triggers cell death in plants carrying the Tsw and Sw5b genes, which recognize the NSs or NSm proteins, respectively (Mariana et al., 2014). Likewise, the Tm‐2 2 gene in tomato encodes a leucine‐rich protein that interacts with the movement protein and confers resistance to tobamoviruses, including TMV. The response is mediated by a hypersensitive response and localized cell death (Chen et al., 2017). In soybean (Glycine max), the Rsv1 gene confers resistance to SMV strain N. SMV strain G7 is not affected. Both P3 and HC‐Pro mediate Rsv1‐dependent restriction of SMV strain N (Eggenberger et al., 2008). A component of the oxygen‐evolving complex pathosystem II, PSBP, interacts with alfalfa mosaic virus coat protein and delays activation of antiviral responses mediated by reactive oxygen species (Balasubramaniam et al., 2014).
Identification of Host Factors with Antiviral Activity
Host factors with antiviral activity have been identified and characterized using several experimental approaches (Table 1). For a small number of cases, natural genetic variation in plant populations has been used to identify, map and clone genes with antiviral activity (Lukan et al., 2018; Maiti et al., 2012; Szajko et al., 2008). However, experimental model systems based on yeast, Arabidopsis and N. benthamiana have contributed most of the genes with antiviral activity known to date (Table 1). These experimental systems allow systematic genetic analysis of virus–host interactions. Using yeast, genome‐wide screens have been conducted for BMV and TBSV. Based on the Arabidopsis mutant collection, multiple screens have been done for gene families such as DCL, AGO, RDRs, RNA decay or autophagy mutants (Blevins et al., 2006; Diaz‐Pendon et al., 2007; Garcia‐Ruiz et al., 2015; Jaubert et al., 2011; Qu et al., 2008).
Based on the concept that host and viral factors colocalize and may interact, yeast‐two hybrid assays and cell fractionation or immunoprecipitation followed by mass spectrometry has led to the identification of several genes with antiviral activity (Fujisaki and Ishikawa, 2008; Ishibashi et al., 2007). These assays involved a virus natural host or an experimental host (Table 1).
Functional characterization of the genes identified has been done using loss‐of‐function or gain‐of‐function mutants in Arabidopsis, N. benthamiana or natural hosts. Additionally, virus‐induced gene silencing (VIGS) has been widely used to down‐regulate genes in N. benthamiana. In both Arabidopsis and N. benthamiana, transient or transgenic expression has been used to validate the antiviral activity of a growing number of genes (Haxim et al., 2017; Jaag and Nagy, 2009; Sun et al., 2017).
Factors Essential and Non‐Essential for Host Survival
Genes with antiviral activity might be essential or non‐essential for plant survival. Non‐essential genes affect virus replication or movement without affecting the host (Table 1). However, essential genes cannot be removed from the host. AGO1 participates in antiviral defence (Garcia‐Ruiz et al., 2015; Qu et al., 2008; Wang et al., 2011) and is essential for miRNA‐dependent regulation of gene expression and development. Accordingly, ago1 null mutants show severe developmental phonotypes and are sterile. Hypomorphic ago1 mutant alleles retain part of their activity and have been used to genetically characterize the role of AGO1 in antiviral defence (Morel et al., 2002). In contrast, DCL2 and DCL4 are non‐essential, are redundant to each other, and single and double mutants show only mild leaf malformation (Diaz‐Pendon et al., 2007; Garcia‐Ruiz et al., 2010).
Conditional repression of expression or temperature‐sensitive expression were used to determine the role of yeast essential genes in BMV and TBSV replication (Gancarz et al., 2011; Nawaz‐ul‐Rehman et al., 2013). These genetic analyses identified 19 essential yeast genes that antagonized BMV (Gancarz et al., 2011) or TBSV replication (Nawaz‐ul‐Rehman et al., 2013).
Conclusions
The infection cycle of a plant by a virus is genetically determined by viral factors, cellular factors and their interaction. Viruses use cellular factors and resources to replicate and move. Viral protein or nucleic acids are targeted by antiviral immunity (Fig. 1A). A two‐step model for plant–virus interactions explains plant susceptibility to viruses (Fig. 1B). Initially, establishment of infection is determined by the level of plant–virus compatibility. Incompatibility might result from the lack of pro‐viral factors, while compatibility is determined by the availability of pro‐viral host factors. Subsequently, in compatible plant–virus combinations susceptibility is determined by the balance between antiviral defence and suppression of antiviral defence (Fig. 1B). Strong antiviral defence may stop infection at any point before spreading to the entire plant. This range results in plants with different levels of susceptibility. The lowest level of susceptibility, resulting from arrest of infection at the initially infected cell, is difficult to distinguish from an incompatible interaction that occurs in a non‐host (Fig. 1B). Susceptible hosts harbour both pro‐viral factors and factors with antiviral activity (Table 1). Their functional characterization has improved our understanding of the mechanisms of virus pathogenicity and antiviral defence in plant–virus interactions.
Future Directions
Host genes with antiviral activity provide an interesting option to develop genetic resistance to viruses in crops. However, viruses have a remarkable ability to mutate and are rapidly evolving (Duffy, 2018). Virus‐resistant plants select for variants capable of breaking genetic resistance. An example is the emergence of tomato brown rugose fruit virus (ToBRV), described in 2016. ToBRV originated from a recombination event between TMV and tomato mild mottle virus (ToMMV) (Salem et al., 2016). Interestingly, within a year, a ToBRV isolate that broke the Tm‐2 2‐dependent resistance was identified (Luria et al., 2017).
A complementary or alternative approach to the deployment of genes with antiviral activity is the identification, characterization and deployment of pro‐viral factors that determine susceptibility to plant viruses (Garcia‐Ruiz, 2018).
To date, genome‐wide screens and genetic analysis have been done mainly in model viruses using heterologous hosts and/or model plant systems (Table 1). Current advances in genome editing (Zhe et al., 2018) make it possible to implement genetic analysis in crop plants. Genome editing in combination with epitope‐tagging of viral proteins, either individually or in the context of an infectious clone, make it currently possible to identify and characterize host genes with antiviral activity and pro‐viral genes crop plants. Thus, it is safe to predict that viruses causing devastating diseases in staple crops will receive more attention in the near future. This includes orthotospoviruses, potyviruses, tobamoviruses and geminiviruses. Prime examples are the causal agents of maize lethal necrosis, maize chlorotic mottle virus and sugarcane mosaic virus (Wamaitha et al., 2018), and recently described ToBRV (Luria et al., 2017; Salem et al., 2016).
Conflicts of Interest
The author declares no conflict of interest. The founding sponsors had no role in the design of the study, in the collection, analyses or interpretation of data, in the writing of the manuscript or in the decision to publish the results.
Author Contributions
Hernan Garcia‐Ruiz conceived and wrote the paper.
Acknowledgements
This research was supported by NIH grant R01GM120108 to Hernan Garcia‐Ruiz and by the Nebraska Agricultural Experiment Station with funding from the Hatch Act (Accession Number 1007272) through the USDA National Institute of Food and Agriculture. Open access costs were provided by the same grant.
References
- Abbink, T.E.M. , Tjernberg, P.A. , Bol, J.F. and Linthorst, H.J.M. (1998) Tobacco mosaic virus helicase domain induces necrosis in N gene‐carrying tobacco in the absence of virus replication. Mol. Plant‐Microbe Interact. 11, 1242–1246. [Google Scholar]
- Ahlquist, P. (2006) Parallels among positive‐strand RNA viruses, reverse‐transcribing viruses and double‐stranded RNA viruses. Nat. Rev. Microbiol. 4, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandalakshmi, R. , Marathe, R. , Ge, X. , Herr, J.M. Jr. , Mau, C. , Mallory, A. , Pruss, G. , Bowman, L. and Vance, V.B. (2000) A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Sci. 290, 142–144. [DOI] [PubMed] [Google Scholar]
- Anandalakshmi, R. , Pruss, G.J. , Ge, X. , Marathe, R. , Mallory, A.C. , Smith, T.H. and Vance, V.B. (1998) A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA. 95, 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramaniam, M. , Kim, B.‐S. , Hutchens‐Williams, H.M. and Loesch‐Fries, L.S. (2014) The photosystem II oxygen‐evolving complex protein PsbP interacts with the coat protein of Alfalfa mosaic virus and inhibits virus replication. Mol. Plant‐Microbe Interact. 27, 1107–1118. [DOI] [PubMed] [Google Scholar]
- Barajas, D. , Li, Z. and Nagy, P.D. (2009) The Nedd4-type Rsp5p ubiquitin ligase inhibits tombusvirus replication by regulating degradation of the p92 replication protein and decreasing the activity of the tombusvirus replicase. J. Virol. 83, 11751–11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane, A. , Querci, M. , Kanyuka, K. and Baulcombe, D.C. (2000) Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J. 21, 73–81. [DOI] [PubMed] [Google Scholar]
- Blevins, T. , Rajeswaran, R. , Shivaprasad, P.V. , Beknazariants, D. , Si‐Ammour, A. , Park, H.S. , Vazquez, F. , Robertson, D. , Meins, F. , Hohn, T. and Pooggin, M.M. (2006) Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34, 6233–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosseau, C. and Moffett, P. (2015) Functional and genetic analysis identify a role for Arabidopsis ARGONAUTE5 in antiviral RNA silencing. Plant Cell. 27, 1742–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgyan, J. and Havelda, Z. (2011) Viral suppressors of RNA silencing. Trends Plant Sci. 16, 265–272. [DOI] [PubMed] [Google Scholar]
- Butterbach, P. , Verlaan, M.G. , Dullemans, A. , Lohuis, D. , Visser, R.G.F. , Bai, Y. and Kormelink, R. (2014) Tomato yellow leaf curl virus resistance by Ty‐1 involves increased cytosine methylation of viral genomes and is compromised by cucumber mosaic virus infection. Proc. Natl. Acad. Sci. 111, 12942–12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo, M. , Malinowski, T. and Garcia, J.A. (2014a) Single amino acid changes in the 6K1‐CI region can promote the alternative adaptation of Prunus‐ and Nicotiana‐propagated Plum pox virus C isolates to either host. Mol. Plant‐Microbe. Interact. 27, 136–149. [DOI] [PubMed] [Google Scholar]
- Calvo, M. , Martinez‐Turino, S. and Garcia, J.A. (2014b) Resistance to Plum pox virus strain C in Arabidopsis thaliana and Chenopodium foetidum involves genome‐linked viral protein and other viral determinants and might depend on compatibility with host translation initiation factors. Mol. Plant‐Microbe Interact. 27, 1291–1301. [DOI] [PubMed] [Google Scholar]
- Camborde, L. , Planchais, S. , Tournier, V. , Jakubiec, A. , Drugeon, G. , Lacassagne, E. , Pflieger, S. , Chenon, M. and Jupin, I. (2010) The ubiquitin-proteasome system regulates the accumulation of turnip yellow mosaic virus RNA-dependent RNA polymerase during viral infection. Plant Cell. 22, 3142–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceniceros‐Ojeda, E.A. , Rodriguez‐Negrete, E.A. and Rivera‐Bustamante, R.F. (2016) Two populations of viral minichromosomes are present in a geminivirus‐infected plant showing symptom remission (recovery). J. Virol. 90, 3828–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T. , Liu, D. , Niu, X. , Wang, J. , Qian, L. , Han, L. , Liu, N. , Zhao, J. , Hong, Y. and Liu, Y. (2017) Antiviral resistance protein Tm‐2(2) functions on the plasma membrane. Plant Physiol. 173, 2399–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T. , Parra, M.A. , Anderberg, R.J. and Carrington, J.C. (2001) Arabidopsis RTM1 and RTM2 genes function in phloem to restrict long‐distance movement of tobacco etch virus. Plant Physiol. 127, 1667–1675. [PMC free article] [PubMed] [Google Scholar]
- Chuang, C. , Barajas, D. , Qin, J. and Nagy, P.D. (2014) Inactivation of the host lipin gene accelerates RNA virus replication through viral exploitation of the expanded endoplasmic reticulum membrane. PLoS Pathog. 10, e1003944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras‐Paredes, C.A. , Silva‐Rosales, L. , Daros, J.A. , Alejandri‐Ramirez, N.D. and Dinkova, T.D. (2013) The absence of eukaryotic initiation factor eIF(iso)4E affects the systemic spread of a Tobacco etch virus isolate in Arabidopsis thaliana . Mol. Plant‐Microbe Interact. 26, 461–470. [DOI] [PubMed] [Google Scholar]
- Cooley, M.B. , Pathirana, S. , Wu, H.-J. , Kachroo, P. and Klessig, D.F. (2000) Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell. 12, 663–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ronde, D. , Butterbach, P. , Lohuis, D. , Hedil, M. , van Lent, J.W. and Kormelink, R. (2013) Tsw gene-based resistance is triggered by a functional RNA silencing suppressor protein of the Tomato spotted wilt virus . Mol. Plant Pathol. 14, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroocq, V. , Salvador, B. , Sicard, O. , Glasa, M. , Cosson, P. , Svanella‐Dumas, L. , Revers, F. , García, J.A. and Candresse, T. (2009) The determinant of potyvirus ability to overcome the RTM resistance of Arabidopsis thaliana maps to the N‐terminal region of the coat protein. Mol. Plant‐Microbe Interact. 22, 1302–1311. [DOI] [PubMed] [Google Scholar]
- Diaz, A. , Zhang, J. , Ollwerther, A. , Wang, X. and Ahlquist, P. (2015) Host ESCRT proteins are required for bromovirus RNA replication compartment assembly and function. PLoS Pathog. 11, e1004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz‐Pendon, J.A. , Li, F. , Li, W.X. and Ding, S.W. (2007) Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell, 19, 2053–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh‐Kumar, S.P. , Tham, W.H. and Baker, B.J. (2000) Structure–function analysis of the tobacco mosaic virus resistance gene N . Proc. Natl. Acad. Sci. 97, 14789–14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaire, L. , Wang, Y. , Gonzalez‐Ibeas, D. , Mayer, K.F. , Aranda, M.A. and Llave, C. (2009) Deep‐sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology, 392, 203–214. [DOI] [PubMed] [Google Scholar]
- Donze, T. , Qu, F. , Twigg, P. and Morris, T.J. (2014) Turnip crinkle virus coat protein inhibits the basal immune response to virus invasion in Arabidopsis by binding to the NAC transcription factor TIP. Virology, 449, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy, S. (2018) Why are RNA virus mutation rates so damn high? PLoS Biol. 16, e3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenberger, A.L. , Hajimorad, M.R. and Hill, J.H. (2008) Gain of virulence on Rsv1‐genotype soybean by an avirulent soybean mosaic virus requires concurrent mutations in both P3 and HC‐Pro. Mol. Plant‐Microbe Interact. 21, 931–936. [DOI] [PubMed] [Google Scholar]
- Fujisaki, K. and Ishikawa, M. (2008) Identification of an Arabidopsis thaliana protein that binds to tomato mosaic virus genomic RNA and inhibits its multiplication. Virology, 380, 402–411. [DOI] [PubMed] [Google Scholar]
- Gancarz, B.L. , Hao, L. , He, Q. , Newton, M.A. and Ahlquist, P. (2011) Systematic identification of novel, essential host genes affecting bromovirus RNA replication. PLoS One, 6, e23988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, J.A. and Pallas, V. (2015) Viral factors involved in plant pathogenesis. Curr. Opin. Virol. 11, 21–30. [DOI] [PubMed] [Google Scholar]
- Garcia‐Ruiz, H. (2018) Susceptibility genes to plant viruses. Viruses, 10, 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Ruiz, H. , Takeda, A. , Chapman, E.J. , Sullivan, C.M. , Fahlgren, N. , Brempelis, K.J. and Carrington, J.C . (2010) Arabidopsis RNA‐dependent RNA polymerases and dicer‐like proteins in antiviral defense and small interfering RNA biogenesis during Turnip mosaic virus infection. Plant Cell, 22, 481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Ruiz, H. , Carbonell, A. , Hoyer, J.S. , Fahlgren, N. , Gilbert, K.B. , Takeda, A. , Giampetruzzi, A. , Garcia Ruiz, M.T. , McGinn, M.G. , Lowery, N. , Martinez Baladejo, M.T. and Carrington, J.C. (2015) Roles and programming of Arabidopsis ARGONAUTE proteins during turnip mosaic virus infection. PLoS Pathog. 11, e1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Ruiz, H. , Gabriel Peralta, S.M. and Harte‐Maxwell, P.A. (2018) Tomato spotted wilt virus NSs protein supports infection and systemic movement of a potyvirus and is a symptom determinant. Viruses, 10, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovits, R. , Fridman, L. , Kolot, M. , Rotem, O. , Ghanim, M. , Shriki, O. and Czosnek, H. (2016) Tomato yellow leaf curl virus confronts host degradation by sheltering in small/midsized protein aggregates. Virus Res. 213, 304–313. [DOI] [PubMed] [Google Scholar]
- Guo, Z. , Lu, J. , Wang, X. , Zhan, B. , Li, W. and Ding, S.W. (2017) Lipid flippases promote antiviral silencing and the biogenesis of viral and host siRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA. 114, 1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, A.K. , Hein, G.L. , Graybosch, R.A. and Tatineni, S. (2018) Octapartite negative‐sense RNA genome of High Plains wheat mosaic virus encodes two suppressors of RNA silencing. Virology, 518, 152–162. [DOI] [PubMed] [Google Scholar]
- Hafren, A. , Macia, J.L. , Love, A.J. , Milner, J.J. , Drucker, M. and Hofius, D. (2017) Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA. 114, E2026–E2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafren, A. , Ustun, S. , Hochmuth, A. , Svenning, S. , Johansen, T. and Hofius, D. (2018) Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HCpro. Plant Physiol. 176, 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, M. , Neriya, Y. , Yamaji, Y. and Namba, S. (2016) Recessive resistance to plant viruses: potential resistance genes beyond translation initiation factors. Front. Microbiol. 7, 1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxim, Y. , Ismayil, A. , Jia, Q. , Wang, Y. , Zheng, X. , Chen, T. , Qian, L. , Liu, N. , Wang, Y. , Han, S. , Cheng, J. , Qi, Y. , Hong, Y. and Liu, Y. (2017) Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife. 6, e23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein, M. (2015) Plant virus replication and movement. Virology, 479–480, 657–671. [DOI] [PubMed] [Google Scholar]
- Hofius, D. , Maier, A.T. , Dietrich, C. , Jungkunz, I. , Bornke, F. , Maiss, E. and Sonnewald, U. (2007) Capsid protein‐mediated recruitment of host DnaJ‐like proteins is required for Potato virus Y infection in tobacco plants. J. Virol. 81, 11870–11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, S.U. , Kim, M.J. and Paek, K.H. (2013) Arabidopsis Pumilio protein APUM5 suppresses Cucumber mosaic virus infection via direct binding of viral RNAs. Proc. Natl. Acad. Sci. USA. 110, 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incarbone, M. and Dunoyer, P. (2013) RNA silencing and its suppression: novel insights from in planta analyses. Trends Plant Sci. 18, 382–392. [DOI] [PubMed] [Google Scholar]
- Ishibashi, K. and Ishikawa, M. (2013) The resistance protein Tm‐1 inhibits formation of a Tomato mosaic virus replication protein–host membrane protein complex. J. Virol. 87, 7933–7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi, K. , Masuda, K. , Naito, S. , Meshi, T. and Ishikawa, M. (2007) An inhibitor of viral RNA replication is encoded by a plant resistance gene. Proc. Natl. Acad. Sci. USA. 104, 13833–13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaag, H.M. and Nagy, P.D. (2009) Silencing of Nicotiana benthamiana Xrn4p exoribonuclease promotes tombusvirus RNA accumulation and recombination. Virology, 386, 344–352. [DOI] [PubMed] [Google Scholar]
- Jaubert, M. , Bhattacharjee, S. , Mello, A.F. , Perry, K.L. and Moffett, P. (2011) ARGONAUTE2 mediates RNA‐silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol. 156, 1556–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, E.J. , Tadamura, K. , Murakami, T. , Inaba, J.‐I. , Kim, B.M. , Sato, M. , Atsumi, G. , Kuchitsu, K. , Masuta, C. and Nakahara, K.S. (2017) rgs‐CaM detects and counteracts viral RNA silencing suppressors in plant immune priming. J. Virol. 91, e00761–00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran, R.A. and Sanfacon, H. (2014) Tomato ringspot virus coat protein binds to ARGONAUTE 1 and suppresses the translation repression of a reporter gene. Mol. Plant‐Microbe Interact. 27, 933–943. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D. , Xie, Z. , Allen, E. , Llave, C. , Chapman, E.J. , Krizan, K.A. and Carrington, J.C. (2003) P1/HC‐Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev. Cell, 4, 205–217. [DOI] [PubMed] [Google Scholar]
- Kim, S.B. , Lee, H.Y. , Seo, S. , Lee, J.H. and Choi, D. (2015) RNA‐dependent RNA polymerase (NIb) of the potyviruses is an avirulence factor for the broad‐spectrum resistance gene Pvr4 in Capsicum annuum cv. CM334. PLoS One. 10, e0119639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner, C.J. , Pitzalis, N. , Pena, E.J. , Erhardt, M. , Vazquez, F. and Heinlein, M. (2018) Crosstalk between PTGS and TGS pathways in natural antiviral immunity and disease recovery. Nat. Plants, 4, 157–164. [DOI] [PubMed] [Google Scholar]
- Kushner, D.B. , Lindenbach, B.D. , Grdzelishvili, V.Z. , Noueiry, A.O. , Paul, S.M. and Ahlquist, P. (2003) Systematic, genome‐wide identification of host genes affecting replication of a positive‐strand RNA virus. Proc. Natl. Acad. Sci. USA. 100, 15764–15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laliberte, J.F. and Zheng, H. (2014) Viral manipulation of plant host membranes. Annu. Rev. Virol. 1, 237–259. [DOI] [PubMed] [Google Scholar]
- Lanfermeijer, F.C. , Dijkhuis, J. , Sturre, M.J. , de Haan, P. and Hille, J. (2003) Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-2 2 from Lycopersicon esculentum . Plant Mol. Biol. 52, 1037–1049. [DOI] [PubMed] [Google Scholar]
- Lellis, A.D. , Kasschau, K.D. , Whitham, S.A. and Carrington, J.C. (2002) Loss‐of‐susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 12, 1046–1051. [DOI] [PubMed] [Google Scholar]
- Levy, M. , Edelbaum, O. and Sela, I. (2004) Tobacco mosaic virus regulates the expression of its own resistance gene N . Plant Physiol. 135, 2392–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. and Wang, A. (2018) RNA decay is an antiviral defense in plants that is counteracted by viral RNA silencing suppressors. PLoS Pathog. 14, e1007228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. , Huang, C. , Li, Z. and Zhou, X. (2014) Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin‐like protein to repress RDR6 expression. PLoS Pathog. 10, e1003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Xiong, R. , Bernards, M. and Wang, A. (2016) Recruitment of Arabidopsis RNA helicase AtRH9 to the viral replication complex by viral replicase to promote turnip mosaic virus replication. Sci. Rep. 6, 30297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. , Zhang, C. , Li, Y. , Wu, G. , Hou, X. , Zhou, X. and Wang, A. (2018) Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nat. Commun. 9, 1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludman, M. , Burgyan, J. and Fatyol, K. (2017) Crispr/Cas9 mediated inactivation of Argonaute 2 reveals its differential involvement in antiviral responses. Sci. Rep. 7, 1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukan, T. , Baebler, Š. , Pompe‐Novak, M. , Guček, K. , Zagorščak, M. , Coll, A. and Gruden, K. (2018) Cell death is not sufficient for the restriction of potato virus Y spread in hypersensitive response‐conferred resistance in potato. Front. Plant Sci. 9, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria, N. , Smith, E. , Reingold, V. , Bekelman, I. , Lapidot, M. , Levin, I. , Elad, N. , Tam, Y. , Sela, N. , Abu‐Ras, A. , Ezra, N. , Haberman, A. , Yitzhak, L. , Lachman, O. and Dombrovsky, A. (2017) A new Israeli tobamovirus isolate infects tomato plants harboring Tm‐22 resistance genes. PLoS One. 12, e0170429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, M.F. , Xie, L. , Song, X.J. , Hong, J. , Mao, Q.Z. , Wei, T.Y. , Chen, J.‐P. and Zhang, H.‐M . (2017) Phloem‐limited reoviruses universally induce sieve element hyperplasia and more flexible gateways, providing more channels for their movement in plants. Sci. Rep. 7, 16467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, G. , Chen, P. , Buss, G.R. and Tolin, S.A. (1995) Genetic characteristics of two genes for resistance to soybean mosaic virus in PI486355 soybean. Theor. Appl. Genet. 91, 907–914. [DOI] [PubMed] [Google Scholar]
- Ma, X. , Nicole, M.C. , Meteignier, L.V. , Hong, N. , Wang, G. and Moffett, P. (2015) Different roles for RNA silencing and RNA processing components in virus recovery and virus‐induced gene silencing in plants. J. Exp. Bot. 66, 919–932. [DOI] [PubMed] [Google Scholar]
- Machado, J.P.B. , Calil, I.P. , Santos, A.A. and Fontes, E.P.B. (2017) Translational control in plant antiviral immunity. Genet. Mol. Biol. 40, 292–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti, S. , Paul, S. and Pal, A. (2012) Isolation, characterization, and structure analysis of a non‐TIR‐NBS‐LRR encoding candidate gene from MYMIV‐resistant Vigna mungo . Mol. Biotechnol. 52, 217–233. [DOI] [PubMed] [Google Scholar]
- Maliogka, V.I. , Calvo, M. , Carbonell, A. , Garcia, J.A. and Valli, A. (2012) Heterologous RNA‐silencing suppressors from both plant‐ and animal‐infecting viruses support plum pox virus infection. J. Gen. Virol. 93, 1601–1611. [DOI] [PubMed] [Google Scholar]
- Margaria, P. , Miozzi, L. , Rosa, C. , Axtell, M.J. , Pappu, H.R. and Turina, M. (2015) Small RNA profiles of wild‐type and silencing suppressor‐deficient tomato spotted wilt virus infected Nicotiana benthamiana . Virus Res. 208, 30–38. [DOI] [PubMed] [Google Scholar]
- Mariana, H. , Silva, O.A. , Erico, C.D. , Dick, L. , Silva, B.L. , Kazuko, I.N.A. , Resende, R.O. and Kormelink, R. (2014) The Tomato spotted wilt virus cell‐to‐cell movement protein (NSM) triggers a hypersensitive response in Sw‐5‐containing resistant tomato lines and in Nicotiana benthamiana transformed with the functional Sw‐5b resistance gene copy. Mol. Plant Pathol. 15, 871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshi, T. , Motoyoshi, F. , Maeda, T. , Yoshiwoka, S. , Watanabe, H. and Okada, Y. (1989) Mutations in the tobacco mosaic virus 30-kD protein gene overcome Tm-2 resistance in tomato. Plant Cell. 1, 515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, W.A. , Shen, R. , Staplin, W. and Kanodia, P. (2016) Noncoding RNAs of plant viruses and viroids: sponges of host translation and RNA interference machinery. Mol. Plant‐Microbe Interact. 29, 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoia, S. , Carbonell, A. , Di Serio, F. , Gisel, A. , Carrington, J.C. , Navarro, B. and Flores, R. (2014) Specific argonautes selectively bind small RNAs derived from potato spindle tuber viroid and attenuate viroid accumulation in vivo. J. Virol. 88, 11933–11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, S.L. and Wilusz, J. (2013) Cytoplasmic viruses: rage against the (cellular RNA decay) machine. PLoS Pathog. 9, e1003762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, J.B. , Godon, C. , Mourrain, P. , Beclin, C. , Boutet, S. , Feuerbach, F. , Proux, F. and Vaucheret, H. (2002) Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post‐transcriptional gene silencing and virus resistance. Plant Cell, 14, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara, K.S. , Masuta, C. , Yamada, S. , Shimura, H. , Kashihara, Y. , Wada, T.S. , Meguro, A. , Goto, K. , Tadamura, K. , Sueda, K. , Sekiguchi, T. , Shao, J. , Itchoda, N. , Matsumura, T. , Igarashi, M. , Ito, K. , Carthew, R.W. and Uyeda, I. (2012) Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc. Natl. Acad. Sci. USA. 109, 10113–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz‐ul‐Rehman, M.S. , Reddisiva Prasanth, K. , Baker, J. and Nagy, P.D. (2013) Yeast screens for host factors in positive‐strand RNA virus replication based on a library of temperature‐sensitive mutants. Methods, 59, 207–216. [DOI] [PubMed] [Google Scholar]
- Nelson, R.S. and Citovsky, V. (2005) Plant viruses. Invaders of cells and pirates of cellular pathways. Plant Physiol. 138, 1809–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otulak‐Koziel, K. , Koziel, E. and Lockhart, B.E.L. (2018) Plant cell wall dynamics in compatible and incompatible potato response to infection caused by potato virus Y (PVYNTN). Int. J. Mol. Sci. 19, E862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett, H.S. , Watanabe, Y. and Beachy, R.N. (1997) Identification of the TMV replicase sequence that activates the N gene‐mediated hypersensitive response. Mol. Plant‐Microbe Interact. 10, 709–715. [Google Scholar]
- Panavas, T. , Serviene, E. , Brasher, J. and Nagy, P.D. (2005) Yeast genome‐wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc. Natl. Acad. Sci. USA. 102, 7326–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo, V. , Szittya, G. and Burgyan, J. (2007) Molecular bases of viral RNA targeting by viral small interfering RNA‐programmed RISC. J. Virol. 81, 3797–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J. , Yang, J. , Yan, F. , Lu, Y. , Jiang, S. , Lin, L. , Zheng, H. , Chen, H. and Chen, J. (2011) Silencing of NbXrn4 facilitates the systemic infection of Tobacco mosaic virus in Nicotiana benthamiana . Virus Res. 158, 268–270. [DOI] [PubMed] [Google Scholar]
- Prasanth, K.R. , Huang, Y.W. , Liou, M.R. , Wang, R.Y. , Hu, C.C. , Tsai, C.H. , Meng, M. , Lin, N‐S and Hsu, Y‐H . (2011) Glyceraldehyde 3‐phosphate dehydrogenase negatively regulates the replication of Bamboo mosaic virus and its associated satellite RNA. J. Virol. 85, 8829–8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, F. , Ye, X. , Hou, G. , Sato, S. , Clemente, T.E. and Morris, T.J. (2005) RDR6 has a broad‐spectrum but temperature‐dependent antiviral defense role in Nicotiana benthamiana . J. Virol. 79, 15209–15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, F. , Ye, X. and Morris, T.J. (2008) Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4‐initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. USA. 105, 14732–14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja, P. , Jackel, J.N. , Li, S. , Heard, I.M. and Bisaro, D.M. (2014) Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses. J. Virol. 88, 2611–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem, N. , Mansour, A. , Ciuffo, M. , Falk, B.W. and Turina, M. (2016) A new tobamovirus infecting tomato crops in Jordan. Arch. Virol. 161, 503–506. [DOI] [PubMed] [Google Scholar]
- Sanfacon, H. (2015) Plant translation factors and virus resistance. Viruses, 7, 3392–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, N. , Ogata, T. , Deguchi, M. , Nagai, S. , Tamai, A. , Meshi, T. , Kawakami, S. , Watanabe, Y. , Matsushita, Y. and Nyunoya, H. (2009) Over-expression of putative transcriptional coactivator KELP interferes with tomato mosaic virus cell-to-cell movement. Mol. Plant Pathol. 10, 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasvari, Z. , Kovalev, N. , Gonzalez, P.A. , Xu, K. and Nagy, P.D. (2018) Assembly‐hub function of ER‐localized SNARE proteins in biogenesis of tombusvirus replication compartment. PLoS Pathog. 14, e1007028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof, H.B. , Alvarado, V.Y. , Vega‐Arreguin, J.C. , Ciomperlik, J. , Odokonyero, D. , Brosseau, C. , Jaubert, M. , Zamora, A. and Moffett, P. (2011) Identification of an ARGONAUTE for antiviral RNA silencing in Nicotiana benthamiana . Plant Physiol. 156, 1548–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck, J. , Gursinsky, T. , Pantaleo, V. , Burgyan, J. and Behrens, S.E. (2013) AGO/RISC‐mediated antiviral RNA silencing in a plant in vitro system. Nucleic Acids Res. 41, 5090–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach, F. , Vaistij, F.E. , Jones, L. and Baulcombe, D.C. (2005) An RNA‐dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138, 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q. , Hu, T. , Bao, M. , Cao, L. , Zhang, H. , Song, F. , Xie, Q. and Zhou, X. (2016) Tobacco RING E3 ligase NtRFP1 mediates ubiquitination and proteasomal degradation of a geminivirus-encoded βC1. Mol. Plant. 9, 911–925. [DOI] [PubMed] [Google Scholar]
- Shimura, H. , Pantaleo, V. , Ishihara, T. , Myojo, N. , Inaba, J. , Sueda, K. , Burgyán, J. and Masuta, C. (2011) A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 7, e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, D. , Li, S. , Niu, L. , Reid, M.S. , Zhang, Y. and Jiang, C.‐Z. (2017) PhOBF1, a petunia ocs element binding factor, plays an important role in antiviral RNA silencing. J. Exp. Bot. 68, 915–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajko, K. , Chrzanowska, M. , Witek, K. , Strzelczyk‐Żyta, D. , Zagórska, H. , Gebhardt, C. , Hennig, J. and Marczewski, W. (2008) The novel gene Ny‐1 on potato chromosome IX confers hypersensitive resistance to Potato virus Y and is an alternative to Ry genes in potato breeding for PVY resistance. Theor. Appl. Genet. 116, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya, G. and Burgyan, J. (2013) RNA interference‐mediated intrinsic antiviral immunity in plants. Curr. Top. Microbiol. Immunol. 371, 153–181. [DOI] [PubMed] [Google Scholar]
- Takahashi, H. , Suzuki, M. , Natsuaki, K. , Shigyo, T. , Hino, K. , Teraoka, T. , Hosokawa, D. and Ehara, Y. (2001) Mapping the virus and host genes involved in the resistance response in cucumber mosaic virus‐infected Arabidopsis thaliana . Plant Cell Physiol. 42, 340–347. [DOI] [PubMed] [Google Scholar]
- Taliansky, M. , Torrance, L. and Kalinina, N.O. (2008) Role of plant virus movement proteins In: Plant Virology Protocols: from viral sequence to protein function, (Foster G.D., Johansen I.E., Hong Y. and Nagy P.D., eds), pp. 33–54. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Tatineni, S. , Riethoven, J.J. , Graybosch, R.A. , French, R. and Mitra, A. (2014) Dynamics of small RNA profiles of virus and host origin in wheat cultivars synergistically infected by Wheat streak mosaic virus and Triticum mosaic virus: virus infection caused a drastic shift in the endogenous small RNA profile. PLoS One, 9, e111577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thran, M. , Link, K. and Sonnewald, U. (2012) The Arabidopsis DCP2 gene is required for proper mRNA turnover and prevents transgene silencing in Arabidopsis. Plant J. 72, 368–377. [DOI] [PubMed] [Google Scholar]
- Del Toro, F.J. , Donaire, L. , Aguilar, E. , Chung, B.N. , Tenllado, F. and Canto, T. (2017) Potato virus Y HCPro suppression of antiviral silencing in Nicotiana benthamiana plants correlates with its ability to bind in vivo to 21‐ and 22‐nucleotide small RNAs of viral sequence. J. Virol. 91, e00367-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki, M. , Motomura, K. , Kumakura, N. and Takeda, A. (2017) Interconnections between mRNA degradation and RDR‐dependent siRNA production in mRNA turnover in plants. J. Plant. Res. 130, 211–226. [DOI] [PubMed] [Google Scholar]
- Wamaitha, M.J. , Nigam, D. , Maina, S. , Stomeo, F. , Wangai, A. , Njuguna, J.N. , Holton, T.A. , Wanjala, B.W. , Wamalwa, M. , Lucas, T. , Djikeng, A. and Garcia‐Ruiz, H. (2018) Metagenomic analysis of viruses associated with maize lethal necrosis in Kenya. Virol. J. 15, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, J. , Cabanillas, D.G. , Zheng, H. and Laliberte, J.F. (2015) Turnip mosaic virus moves systemically through both phloem and xylem as membrane‐associated complexes. Plant Physiol. 167, 1374–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R.Y. and Nagy, P.D. (2008) Tomato bushy stunt virus co‐opts the RNA‐binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe, 3, 178–187. [DOI] [PubMed] [Google Scholar]
- Wang, X.B. , Jovel, J. , Udomporn, P. , Wang, Y. , Wu, Q. , Li, W.X. , Gasciolli, V. , Vaucheret, H. and Ding, S.W. (2011) The 21‐nucleotide, but not 22‐nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana . Plant Cell, 23, 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Kohalmi, S.E. , Svircev, A. , Wang, A. , Sanfacon, H. and Tian, L. (2013) Silencing of the host factor eIF(iso)4E gene confers plum pox virus resistance in plum. PLoS One, 8, e50627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Yang, Z. , Wang, Y. , Zheng, L. , Ye, R. , Ji, Y. , Zhao, S. , Ji, S. , Liu, R. , Xu, L. , Zheng, H. , Zhou, Y. , Zhang, X. , Cao, X. , Xie, L. , Wu, Z. , Qi, Y. and Li, Y. (2015) Viral-inducible Argonaute18 confers broadspectrum virus resistance in rice by sequestering a host microRNA. eLife. 4, e05733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Z. , Zhao, Z. , Chen, L. , Li, M. , Zhou, T. , Deng, C. , Zhou, Q. and Fan, Z. (2016) Synergistic infection of two viruses MCMV and SCMV increases the accumulations of both MCMV and MCMV‐derived siRNAs in maize. Sci. Rep. 6, 20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji, Y. , Hamada, K. , Yoshinuma, T. , Sakurai, K. , Yoshii, A. , Shimizu, T. , Hashimoto, M. , Suzuki, M. , Namba, S. and Hibi, T. (2010) Inhibitory effect on the tobacco mosaic virus infection by a plant RING finger protein. Virus Res. 153, 50–57. [DOI] [PubMed] [Google Scholar]
- Yang, S.J. , Carter, S.A. , Cole, A.B. , Cheng, N.H. and Nelson, R.S. (2004) A natural variant of a host RNA‐dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana . Proc. Natl. Acad. Sci. USA. 101, 6297–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M. , Zhang, Y. , Xie, X. , Yue, N. , Li, J. , Wang, X.B. , Han, C. , Yu, J. , Liu, Y. and Li, D. (2018) Barley stripe mosaic virus γb protein subverts autophagy to promote viral infection by disrupting the ATG7–ATG8 interaction. Plant Cell. 30, 1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Hajimorad, M.R. , Eggenberger, A.L. , Tsang, S. , Whitham, S.A. and Hill, J.H. (2009) Cytoplasmic inclusion cistron of Soybean mosaic virus serves as a virulence determinant on Rsv3-genotype soybean and a symptom determinant. Virology. 391, 240–248. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , He, G. , Han, G.S. , Zhang, J. , Catanzaro, N. , Diaz, A. , Wu, Z. , Carman, G.M. , Xie, L. and Wang, X. (2018) Host Pah1p phosphatidate phosphatase limits viral replication by regulating phospholipid synthesis. PLoS Pathog. 14, e1006988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Tong, X. , Liu, S.‐Y. , Chai, L.‐X. , Zhu, F.‐F. , Zhang, X.‐P. , Zou, J.‐Z. and Wang, X.‐B. (2019) Genetic analysis of a Piezo‐like protein suppressing systemic movement of plant viruses in Arabidopsis thaliana . Sci. Rep. 9, 3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhe, N. , Qian, H. , Yong‐Qiang, H. and Sheng, H. (2018) Application of genome‐editing technology in crop improvement. Cereal Chem. 95, 35–48. [Google Scholar]
- Zhu, J. , Gopinath, K. , Murali, A. , Yi, G. , Hayward, S.D. , Zhu, H. , and Kao, C. (2007) RNA‐binding proteins that inhibit RNA virus infection. Proc. Natl. Acad. Sci. USA. 104, 3129–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorzatto, C. , Machado, J.P. , Lopes, K.V. , Nascimento, K.J. , Pereira, W.A. , Brustolini, O.J. , Reis, P.A.B. , Calil, I.P. , Deguchi, M. , Sachetto‐Martins, G. , Gouveia, B.C. , Loriato, V.A.P. , Silva, M.A.C. , Silva, F.F. , Santos, A.A. , Chory, J. and Fontes, E.P.B. (2015) NIK1‐mediated translation suppression functions as a plant antiviral immunity mechanism. Nature, 520, 679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvereva, A.S. and Pooggin, M.M. (2012) Silencing and innate immunity in plant defense against viral and non‐viral pathogens. Viruses, 4, 2578–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]


