Summary
Ralstonia solanacearum can metabolize ferulic acid (FA) and salicylic acid (SA), two representative phenolic acids, to protect it from toxicity of phenolic acids. Here, we genetically demonstrated a novel phenolic acid decarboxylase regulator (PadR)‐like regulator PrhP as a positive regulator on detoxification of SA and FA in R. solanacearum. Although the ability to degrade SA and FA enhances the infection process of R. solanacearum toward host plants, PrhP greatly contributes to the infection process besides degradation of SA and FA. Our results from the growth assay, promoter activity assay, RNA‐seq and qRT‐PCR revealed that PrhP plays multiple roles in the virulence of R. solanacearum: (1) positively regulates expression of genes for degradation of SA and FA; (2) positively regulates expression of genes encoding type III secretion system (T3SS) and type III effectors both in vitro and in planta; (3) positively regulates expression of many virulence‐related genes, such as the flagella, type IV pili and cell wall degradation enzymes; and (4) is important for the extensive proliferation in planta. The T3SS is one of the essential pathogenicity determinants in many pathogenic bacteria, and PrhP positively regulates its expression mediated with the key regulator HrpB but through some novel pathway to HrpB in R. solanacearum. This is the first report on PadR regulators to regulate the T3SS and it could improve our understanding of the various biological functions of PadR regulators and the complex regulatory pathway on T3SS in R. solanacearum.
Keywords: degradation of phenolic acid, PadR regulator, pathogenesis, Ralstonia solanacearum, type III secretion system
Introduction
Phenolic acids are abundant in the plant kingdom and are involved in the structure of plant cell walls. They belong to the class of secondary metabolites and bioactive compounds synthesized by plants (Verpoorte et al., 2002; Goleniowski et al., 2013). Phenolic compounds basically act as signalling molecules in the interaction between plants and microbes. In fact, they play a crucial role in initiating the symbiotic relationship between plants and useful microorganisms. Moreover, they usually disrupt membrane integrity and decouple the respiratory proton gradient, and hence are broadly antimicrobial (Fitzgerald et al., 2004; Harris et al., 2010). Hydroxybenzoic acids (typically as salicylic acid, SA) and hydroxycinnamic acids (HCAs, typically as ferulic acid, FA) are two important types of natural phenolic acids (Goleniowski et al., 2013; Heleno et al., 2015). Several lines of evidence suggest that phenolic acids are involved in the interaction between host plants and pathogens (Campos et al., 2014; Fry et al., 2000; Naoumkina et al., 2010). Many plants can release de novo synthesized HCAs into the rhizosphere in response to pathogen infection (Lanoue et al., 2010; Wallis and Chen, 2012). In order to protect themselves from the toxicity of phenolic acids, many bacterial species express phenolic acid decarboxylase (PADse, typically as padC gene product in Bacillus subtilis) to convert phenolic acids into less toxic 4‐vinyl and 4‐ethyl derivatives (Park et al., 2017; Tran et al., 2008). Phenolic acid decarboxylase regulators (PadRs) are a large group of transcriptional regulators that promote the PADse‐mediated detoxification of phenolic acids in many bacterial species. They can usually initiate the expression of PADse genes by release from their promoters in the presence of phenolic acids since they usually bind to promoters of PADse genes in the absence of phenolic acids and repress their expression (Florez et al., 2015; Huillet et al., 2006; Nguyen et al., 2011). The PadR regulators have been characterized to function in various survival processes of detoxification, antibiotic biosynthesis, multidrug resistance, toxin production and carbon catabolism (Florez et al., 2015; Huillet et al., 2006; Nguyen et al., 2011; Tran et al., 2008). Moreover, some bacteria, including Pseudomonas sp., Acinetobacter calcoaceticus and Ralstonia solanacearum, can protect themselves from the toxicity of phenolic acids by degrading them as carbon sources for metabolism at moderate concentrations (Gasson et al., 1998; Lowe‐Power et al., 2018; Overhage et al., 1999; Segura et al., 1999).
Ralstonia solanacearum is the causal agent of bacterial wilt disease in over 450 plant species of 50 botanical families worldwide (Jiang et al., 2017; Vasse et al., 1995). The ability to degrade SA and HCAs can facilitate its infection process in host plants to some extent (Lowe et al., 2015; Lowe‐Power et al., 2016). The bacterium usually remains invasive at moderate concentrations of these phenolic chemicals but just survives at higher concentrations. Different from the PADse‐mediated detoxification, degradation of the HCAs and SA in R. solanacearum is dependent on the fca‐fcs operon and nag operon, respectively. The fcs operon encodes a feruloyl‐CoA synthetase that converts HCAs into phenolic aldehydes, while the nag operon encodes dioxygenases that convert SA into double hydroxyl‐benzoic acid, which is unstable and causes the aromatic ring to open (Gasson et al., 1998; Lowe et al., 2015; Lowe‐Power et al., 2016). To date, a total of 9000 PadR members have been deposited to the Pfam database, while no PADse can be annotated in genomes of abundant R. solanacearum strains (https://iant.toulouse.inra.fr/bacteria/annotation/cgi/ralso.cgi). The regulation mechanism of degradation of phenolic acids remains to be further elucidated in R. solanacearum.
As a soil‐borne, vascular bacterium, R. solanacearum generally invades host plants through natural root openings or root wounds (Janse et al., 2004; Vasse et al., 1995). Once it invades the xylem vessels, it proliferates extensively and produces a large amount of exopolysaccharides (EPS) to block sap flow in the xylem vessels, causing plants to rapidly stunt and wilt (Denny, 1995; Roberts et al., 1988). In addition to the EPS, a syringe‐like type III secretion system (T3SS) is essential for the pathogenicity, and the bacteria use this to inject virulence factors (type III effectors, T3Es) into the host cytosol to subvert the host defence (Angot et al., 2006; Cunnac et al., 2004; Jones and Dangl, 2006). The T3SS is greatly conserved among numerous R. solanacearum strains, which is encoded by approximately 20 genes of the hypersensitive response and pathogenicity (hrp) regulon and is directly controlled by a master regulator HrpB, an AraC family of transcriptional regulator (Arlat et al., 1992; Coll and Valls, 2013; Mukaihara et al., 2010). Expression of the T3SS, hrpB and T3Es is not activated until the bacterium has contact with host signals or some mimic signals, such as those in nutrient‐limited medium that might mimic the plant apoplastic fluids (Marenda et al., 1998; Yoshimochi et al., 2009; Zhang et al., 2013). Expression of hrpB and T3SS has been well demonstrated to be globally regulated by a complex network including dozens of regulators, such as HrpG and PrhG, the PrhA‐PrhR/I‐PrhJ‐HrpG signalling cascade and some other well‐characterized regulators of PhcA, PrhN, PrhO and XpsR (Genin and Denny, 2012; Hikichi et al., 2017; Valls et al., 2006; Zhang et al., 2018). HrpG and PrhG are close paralogues of the OmpR/PhoB family of two‐component response regulators that respond to host signals by phosphorylation and activate the hrpB expression in a parallel way (Plener et al., 2010; Zhang et al., 2013). Host signals or mimic signals are presumed to be recognized by an outer membrane protein PrhA and transferred to HrpG via the PrhA‐HrpG signalling cascade, which begins to activate hrpB expression (Genin and Denny, 2012; Hikichi et al., 2017; Valls et al., 2006; Zhang et al., 2018). Moreover, R. solanacearum integrates numerous virulence factors, including the flagella, type IV pili and cell wall degradation enzymes (CWDEs) to promote its infection process toward host plants (Genin and Denny, 2012; Hikichi et al., 2007).
In order to further elucidate the global regulation of the T3SS in R. solanacearum, we generated a popA‐lacZYA fusion to monitor expression profiles of the T3SS in a Japanese R. solanacearum OE1‐1 and screened several T3SS‐regulating candidates with transposon mutagenesis. The popA gene is located on the left side of the hrp regulon that belongs to T3Es and is directly controlled by HrpB. The popA‐lacZYA exhibits an identical expression prolife to the hrp regulon under different conditions. Moreover, this fusion does not affect the infection process of OE1‐1 toward host plants. Among them is Rsp0309, which is annotated as a putative PadR‐like regulator (Zhang et al., 2013). The PadR regulators are known to be involved in various cellular processes, including virulence but not the T3SS. We thus focused on the Rsp0309, hereafter designated PrhP, to investigate its regulatory roles on the detoxification of phenolic acids, regulation of the T3SS and contribution to pathogenicity of R. solanacearum.
Results
PrhP is important for detoxification of SA and FA in R. solanacearum
The PadR regulators are well known to promote the detoxification of phenolic acids in many bacterial species (Gury et al., 2004, 2009). We thus generated a prhP mutant (RQ5649) to ascertain whether R. solanacearum PrhP is involved in the detoxification of SA and FA, two representative phenolic acids. Although growth of the wild‐type strain (RK5050) and RQ5649 was severely impaired with increasing concentration of supplementary SA and FA in both nutrient‐rich (Fig. 1a,b) and nutrient‐limited media (hrp‐inducing medium, data not shown), the prhP mutant was much more susceptible to SA and FA than the wild‐type strain, especially at low concentrations (Fig. 1a,b). Degradation of SA and FA is validated to be dependent on operons of nag and fcs in R. solanacearum, respectively (Lowe et al., 2015; Lowe‐Power et al., 2016), and we generated a nag mutant (RQ6058) and an fcs mutant (RQ5625) to compare their susceptibility with the prhP mutant. The nag mutant was substantially more susceptible than the prhP mutant to SA in rich medium (Fig. 1a), while the fcs mutant exhibited similar susceptibility as the prhP mutant to FA in rich medium (Fig. 1b). The complementary prhPOE1‐1 fully restored the impaired growth of the prhP mutant to that of the wild‐type strain in medium with supplementary SA and FA (Fig. 1a,b), confirming that PrhP is important for the detoxification of SA and FA in R. solanacearum.
Figure 1.
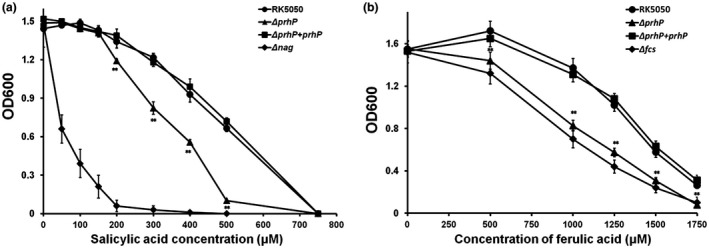
Growth of OE1‐1 derivates in nutrient‐rich medium with supplementary (a) salicylic acid (SA) and (b) ferulic acid (FA) at different concentrations. Strains were cultivated at 28 °C and OD600 was measured at about 15 h post‐inoculation (hpi). RK5050 refers to the wild‐type Ralstonia solanacearum strain, the ΔprhP refers to RQ5649 (RK5050, ΔprhP) and the ΔprhP + prhP refers to RQ5649 with complementary PrhPOE1‐1. The Δnag refers to RQ6058 (RK5050, ΔnagAL) and Δfcs refers to RQ5625 (RK5050, Δfcs). Mean values of four biological replicates were averaged and presented with SD (error bars). Statistical significance between RQ5649 and RK5050 was assessed using a post hoc Dunnett test following ANOVA. Significance level: ** indicates P < 0.01.
PrhP positively regulates expression of genes for SA degradation
The PadR regulators usually bind to promoters of PADse genes in the absence of phenolic acids to repress their expression, while they initiate the expression of PADse genes by release from their promoters in the presence of phenolic acids (Florez et al., 2015; Huillet et al., 2006; Nguyen et al., 2011). We generated a nagAa‐laZYA reporter fusion to monitor the expression of the nag operon and ascertained how PrhP regulates nag expression in conditions with or without supplementary SA. Consistent with a previous report (Lowe‐Power et al., 2016), supplementary SA greatly enhanced nag expression in RQC607 (OE1‐1, nagAa‐lacZYA), which reached to about 50‐ and 100‐fold higher levels in nutrient‐rich and nutrient‐limited media, respectively (Fig. 2a,b). Although nag expression remains at quite a low level in the absence of SA, it was substantially decreased in RQC609 (OE1‐1, ΔprhP, nagAa‐lacZYA) in both media, and the complementary prhP OE1‐1 fully restored the impaired nag expression to that of the parent strain (RQC607) (Fig. 2a). In the presence of SA, nag expression was significantly reduced with prhP deletion in nutrient‐limited medium (P = 0.0006), but there was no alteration in nutrient‐rich medium (P = 0.16) (Fig. 2b). It is worthwhile noting that the nagAa expression in the prhP mutant is slightly decreased to about 80% of levels of the parent strain (RQC607) in nutrient‐limited medium (Fig. 2b).
Figure 2.
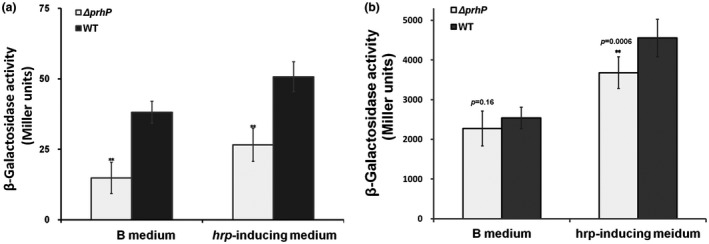
Expression of nagAa‐lacZYA in prhP mutants in nutrient‐rich and nutrient‐limited (hrp‐inducing) media (a) without salicylic acid (SA) and (b) with supplementary SA at a concentration of 250 μM. The WT refers to wild‐type Ralstonia solanacearum RQC607 (OE1‐1, nagAa‐lacZYA) and the ΔprhP refers to RQC609 (OE1‐1, ΔprhP, nagAa‐lacZYA). Cells were grown in medium to an OD600 of about 0.1 and subjected for the β‐galactosidase assay. Enzymatic activities are presented in Miller units (Miller, 1992). Mean values of four biological replicates were averaged and presented with SD (error bars). Statistical significance between RQC609 and RQC607 was assessed using a post hoc Dunnett test following ANOVA. Significance level: ** indicates P < 0.01.
PrhP positively regulates the T3SS expression both in vitro and in planta
The PrhP was originally screened as a T3SS‐regulating candidate by transposon mutagenesis, in which the popA‐lacZYA fusion was constructed to monitor the T3SS expression in R. solanacearum (Zhang et al., 2013). Consistent with those in transposon mutants, popA expression was significantly reduced in the prhP mutant (94 versus 256 Miller units of the wild‐type strain) in hrp‐inducing medium, and the complementary prhP OE1‐1 completely restored its reduced popA expression to that of the wild‐type strain (Fig. 3a).
Figure 3.
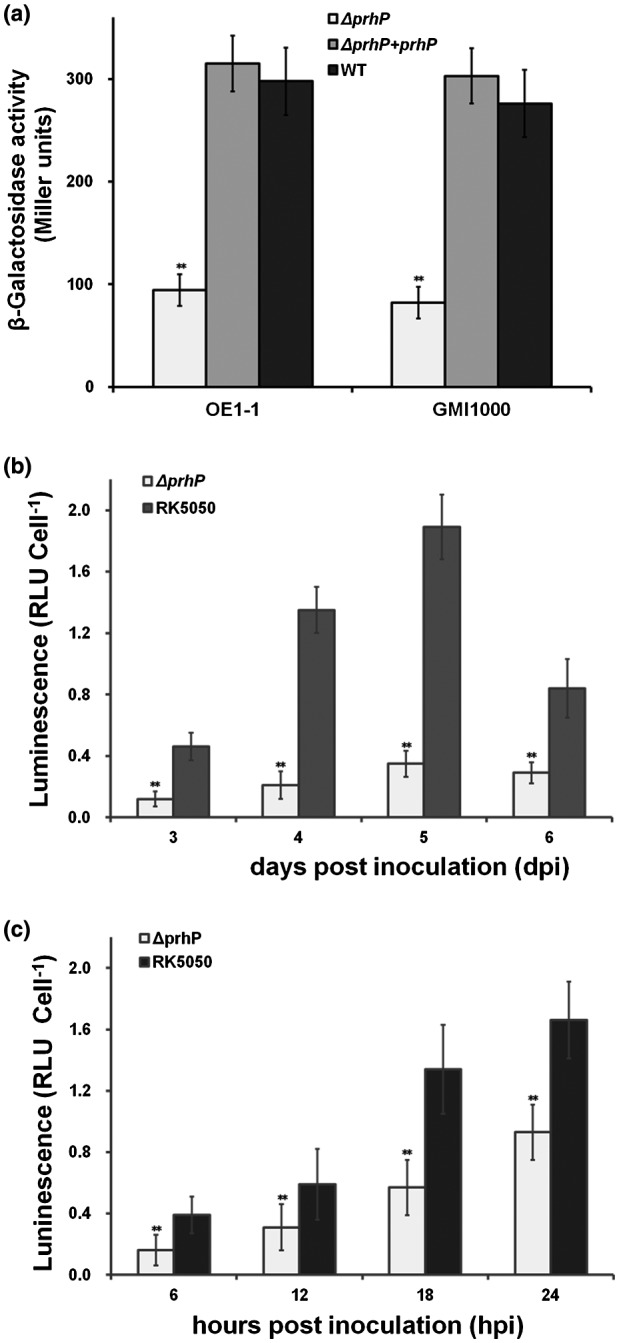
Expression of popA in different prhP mutants (a) prhP mutants from Ralstonia solancearum OE1‐1 and GMI1000 in hrp‐inducing medium, (b) prhP mutants from OE1‐1 in tomato stems, (c) prhP mutants from OE1‐1 in tobacco leaves. The WT refers to the wild‐type strain of RK5050 (OE1‐1, popA‐lacZYA) and GF0001(GMI1000, popA‐lacZYA). The ΔprhP from OE1‐1 refers to RQ5649 (RK5050, ΔprhP) and that from GMI1000 refers to GF0018 (GMI1000, popA‐lacZYA, ΔprhP). The ΔprhP + prhP from OE1‐1 refers to RQ5649 with complementary PrhPOE1‐1 and that from GMI1000 refers to GF0018 with complementary PrhPOE1‐1. (a) Cells were cultivated to an OD600 of approximately 0.1 and subjected for β‐galactosidase assay. Enzymatic activities are presented in Miller units. (b) Tomato plants were inoculated with petiole inoculation, which started wilting at about 3 days post‐inoculation (dpi) and died at about 6 dpi. Stem species were thus removed at 3–6 dpi and cells were harvested for the enzyme assay in planta with the Galacto‐Light Plus kit. (c) Tobacco leaves were infiltrated with cells suspension of about 0.1 OD600 and leaf disks were punched every 6 h for the enzyme assay in planta. Enzymatic activity was presented with luminescence normalized with cells numbers. Luminescence was evaluated using the GloMax20 luminometer (Promega) and cells numbers were quantified by dilution plating. Mean values of four biological replicates were averaged and presented with SD (error bars). Statistical significance between the ΔprhP and the parent strain was assessed using a post hoc Dunnett test following ANOVA. Significance level: ** indicates P < 0.01.
Ralstonia solanacearum is greatly heterogeneous and different strains usually exhibit different phenotypes even in the same host. For instance, the standard strain GMI1000 is avirulent in tobacco plants, while the Japanese strain OE1‐1 is virulent in tobacco plants. We therefore generated a prhP mutant (GF0018) from GF0001 (GMI1000, popA‐lacZYA) to ascertain whether PrhP regulates the T3SS expression in different R. solanacearum strains. Consistent with that from OE1‐1, popA expression was significantly impaired in the prhP mutant (GF0018), and the complementary PrhPOE1‐1 fully restored the reduced popA expression in GF0018 to that of the parent strain (GF0001) (Fig. 3a), confirming that the PrhP of OE1‐1 is functionally equivalent to that of GMI1000 and the regulation of PrhP on T3SS is conserved in different R. solanacearum strains.
The T3SS expression can be enhanced to a much higher level in planta than in hrp‐inducing media (Yoshimochi et al., 2009; Zhang et al., 2013). We thus recovered bacterial cells from tomato stems and tobacco leaves to ascertain whether PrhP is required for T3SS expression in planta. The wild‐type strain (RK5050) usually withers and kills petiole‐inoculated tomato plants at 3 and 7 days post‐inoculation (dpi), respectively. We thus recovered bacterial cells from petiole‐inoculated tomato stems at 3–6 dpi and subjected them for the enzyme assay. The prhP mutant (RQ5649) exhibited significantly impaired popA expression in tomato stems compared with the wild‐type strain at 3–6 dpi (Fig. 3b). Bacterial cells were also recovered from tobacco leaves, which were infiltrated with cell suspension at an OD600 of about 0.1, at 6–24 h post‐infiltration (hpi) and subjected for the enzyme assay. The prhP deletion significantly impaired the popA expression in tobacco leaves at 6–24 hpi (Fig. 3c). All these results confirmed that PrhP positively regulates T3SS expression both in vitro and in planta.
PrhP greatly promotes infection process of R. solanacearum besides degradation of SA and FA
The T3SS is essential for pathogenicity of R. solanacearum (Genin and Denny, 2012; Valls et al., 2006), and the ability to degrade SA and FA can facilitate its infection process toward host plants (Lowe et al., 2015; Lowe‐Power et al., 2016). We ascertained whether PrhP is required for the infection process of R. solanacearum towards host plants. When tomato plants were challenged with the petiole‐inoculation, the prhP mutant killed approximately 75% of test plants at 10 dpi, which is significantly less virulent than the wild‐type strain (Fig. 4a). With the soil‐soaking inoculation, the prhP mutant eventually killed about half the test tomato plants by 25 dpi, which is significantly less virulent than the wild‐type strain (Fig. 4b). It is worthwhile noting that the prhP mutant exhibited much less virulence in soil‐soaking inoculated tomato plants compared with those with petiole inoculation (Fig. 4a,b). Moreover, the prhP mutant exhibited significantly impaired virulence in tobacco plants with both soil‐soaking and leaf‐infiltration inoculation methods (Fig. 4c,d). The complementary PrhPOE1‐1 fully restored the impaired virulence of the prhP mutant to that of the wild‐type strain in both tomato and tobacco plants (Fig. 4a,b,c,d), confirming that PrhP can greatly promote the infection process of R. solanacearum toward host plants.
Figure 4.
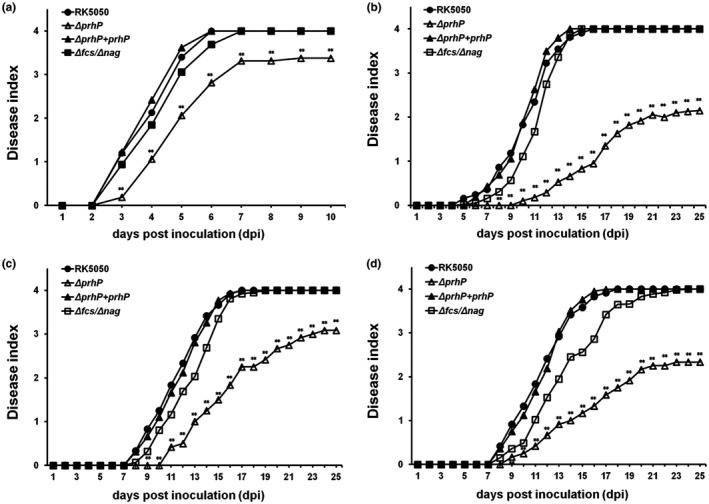
Virulence assay of Ralstonia solanacearum prhP mutants in (a) tomato plants with petiole inoculation, (b) tomato plants with soil‐soaking inoculation, (c) tobacco plants with leaf infiltration and (d) tobacco plants with soil‐soaking inoculation. The Δfcs/Δnag refers to RQ6207 (RK5050, Δfcs, Δnag). For the soil‐soaking inoculation, a bacterial suspension was poured into pot soil of plants at a final concentration of 107 cfu/g of soil. For the petiole inoculation, 3 µL of bacterial suspension at 108 cfu/mL was dropped onto the freshly cut surface of petioles. For the leaf infiltration, about 50 µL of bacterial suspension at 108 cfu/mL was infiltrated into tobacco leaves with a blunt‐end syringe. Wilt symptoms were inspected daily and scored on a disease index scale from 0 to 4 (0, no wilting; 1, 1–25% wilting; 2, 26–50% wilting; 3, 51–75% wilting; and 4, 76–100% wilted or dead). Each assay was repeated at least with four biological replicates and each trial contained at least 12 plants. Mean values of all results were averaged and presented with SD (error bars), but the SD is not presented in figures for aesthetic reasons. Statistical significance between the prhP mutant (RQ5649) and the wild‐type strain (RK5050) was assessed using a post hoc Dunnett test following ANOVA. Significance level: ** indicates P < 0.01.
The ability to degrade SA and FA facilitates the infection process of R. solanacearum toward host plants (Lowe et al., 2015; Lowe‐Power et al., 2016). We therefore generated a mutant with deletion of both fcs and nag operons (RQ6027) to ascertain whether the contribution of PrhP to pathogenicity depends on the degradation of SA and FA. The fcs and nag mutants (RQ6027) exhibited slightly less virulence than the wild‐type strain (RK5050), while the prhP mutant exhibited significantly less virulence than RQ6027 in tomato and tobacco plants regardless of inoculation method (Fig. 4a–d), indicating that PrhP greatly promotes the infection process of R. solanacearum as well as the degradation of SA and FA.
PrhP regulates the T3SS mediated with HrpB but through some novel pathway
In R. solanacearum, the T3SS and T3Es are directly controlled by the key regulator HrpB, and two close paralogues of HrpG and PrhG positively regulate hrpB expression in a parallel way (Mukaihara et al., 2010; Plener et al., 2010; Zhang et al., 2013). We deleted prhP from reporter strains of RK5046 (hrpB‐lacZYA), RK5120 (hrpG‐lacZAY) and RK5212 (prhG‐lacZAY) to ascertain how PrhP regulates the T3SS expression. The hrpB expression was significantly impaired with prhP deletion (53 versus 165 Miller units of RK5046) in hrp‐inducing medium, which was activated in hrp‐inducing medium, while expression of the hrpG and prhG was not altered with prhP deletion in either nutrient‐rich or nutrient‐limited (hrp‐inducing) media (Fig. 5), indicating that regulation of PrhP on the T3SS is mediated with the key regulator HrpB, but through some novel pathway to HrpB.
Figure 5.
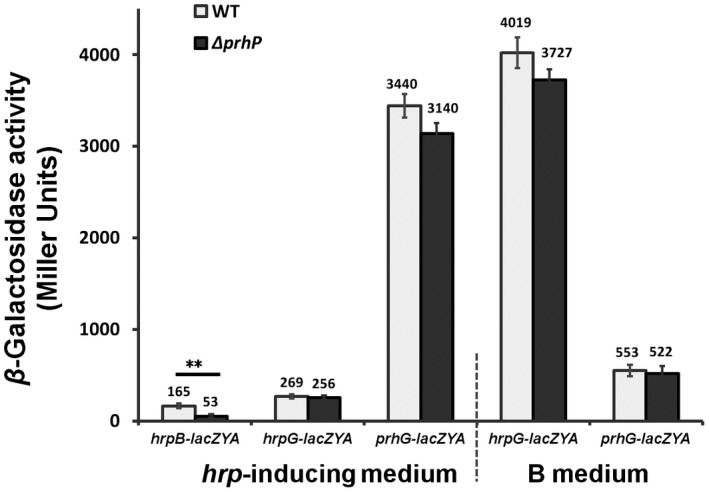
Expression of hrpB‐lacZYA, hrpG‐lacZYA and prhG‐lacZYA with prhP deletion. Ralstonia solanacearum cells were grown in nutrient‐rich or hrp‐inducing media to an OD600 of about 0.1 and subjected to enzyme assay. Mean values from all four biological replicates were averaged and presented with SD (error bars). Statistical significance between the prhP mutant (ΔprhP) and parent strains (WT) was assessed using a post hoc Dunnett test following ANOVA. Significance level: ** indicates P < 0.01.
PrhP is important for expression of a subset of T3Es but not for hypersensitive response (HR) elicitation
Deletion of prhP significantly impaired hrpB expression, which directly controls expression of abundant T3Es in R. solanacearum. We therefore ascertained whether expression of T3Es was impaired with prhP deletion. The wild‐type strain (RK5050) and the prhP mutant (RQ5649) were cultivated in hrp‐inducing medium to an OD600 of 0.1 and total RNA was extracted for subsequent qRT‐PCR. In this study, a total of 11 T3Es of RipAA, RipAR, RipB, RipD, RipE1, RipO, RipP2, RipR, RipTAL, RipW and RipX (PopA, positive control) were selected for quantification of mRNA levels with qRT‐PCR. The qRT‐PCR results showed that expression levels of all these T3Es were significantly or distinctly impaired with prhP deletion (Fig. S1), confirming that PrhP is important for expression of the T3SS and a subset of T3Es in R. solanacearum.
GMI1000 elicits HR in tobacco leaves, and several T3Es are experimentally validated to be responsible for the HR elicitation of GMI1000 in tobacco leaves (Peeters et al., 2013; Poueymiro et al., 2009). Expression of a subset of T3Es was significantly impaired with prhP deletion and we further ascertained whether PrhP affects the HR elicitation of GMI1000 in tobacco leaves. Tobacco leaves (Nicotiana tabacum 'Bright Yellow') were infiltrated with a cell suspension at a concentration of 0.1 OD600 and development of necrotic lesions was investigated periodically. The prhP mutant (GF0018) exhibited similar development of the necrotic lesions as GF0001 (GMI1000, popA‐lacZAY) in tobacco leaves (Fig. S2), indicating that PrhP is not essential for the HR elicitation of GMI1000 in tobacco leaves.
PrhP is important for the in planta growth of R. solanacearum
Plants accumulate phenolic acids to form a defence against the invasion of pathogens, which inhibits the in planta growth of pathogens (Bellés et al., 2006; Lowe‐Power et al., 2018), while R. solanacearum can overcome this inhibition and proliferate extensively in the xylem vessels (Lowe et al., 2015; Lowe‐Power et al., 2016). The extensive proliferation in planta is one of the most important pathogenicity determinants in R. solanacearum (Genin and Denny, 2012), and we ascertained whether the in planta growth of R. solanacearum was altered with the prhP deletion. Petiole‐inoculated tomato plants normally start wilting at about 3 dpi and die at about 6 dpi. Bacterial cells were thus recovered from stems of the petiole‐inoculated tomato plants from 2 to 6 dpi and subjected for quantification with dilution plating. The prhP mutant (RQ5649) exhibited significantly impaired growth compared to the wild‐type strain (RK5050) in tomato stems by approximately one order of magnitude (Fig. 6a). The prhP mutant caused some of the soil‐soaking inoculated tomato plants to wilt and die at 10 and 25 dpi, respectively (Fig. 4b), and we ascertained the growth of the prhP mutant in stems of the soil‐soaking inoculated tomato plants to 21 dpi. Growth of the prhP mutant was significantly impaired in stems of the soil‐soaking inoculated tomato plants, which remained at levels of about 103–104 cfu/g to 9 dpi, and increased slowly to a maximum of about 109 cfu/g at 21 dpi (Fig. 6b). It is worthwhile noting that growth of the prhP mutant in stems of the soil‐soaking inoculated tomato plants was significantly less than that in stems of the petiole‐inoculated tomato plants (Fig. 6a,b).
Figure 6.
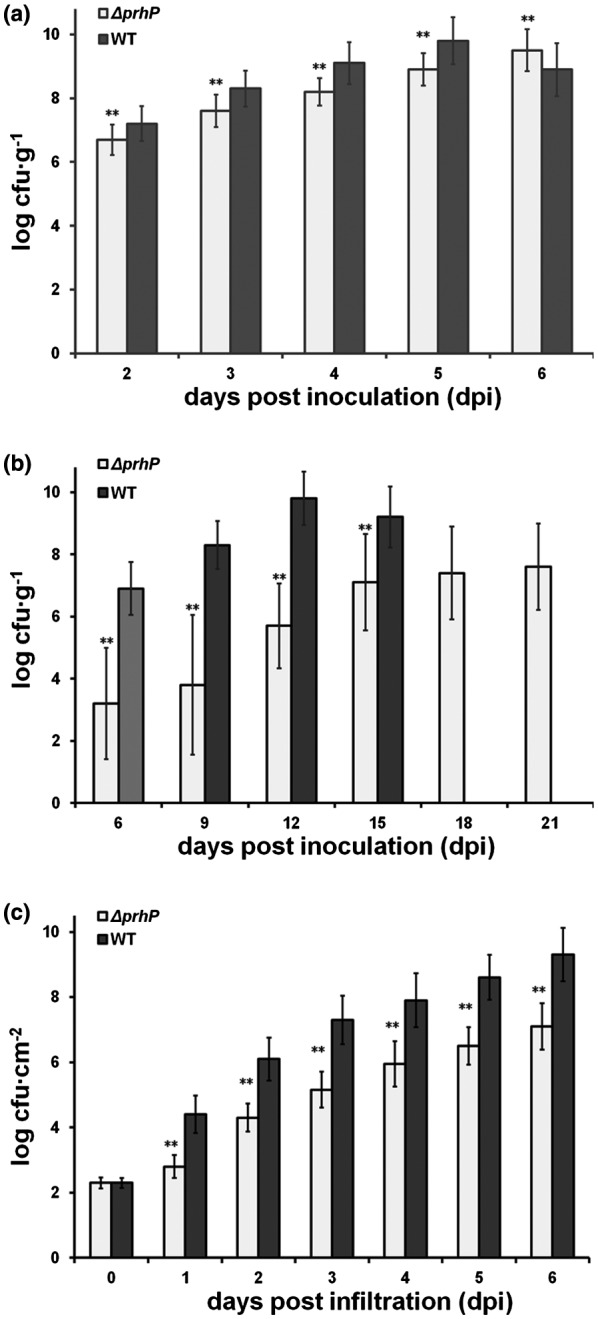
Bacterial growth in (a) tomato stems with petiole inoculation, (b) tomato stems with soil‐soaking inoculation and (c) tobacco leaves with leaf infiltration. With petiole inoculation, the wild‐type Ralstonia solanacearum strain (RK5050) normally causes tomato plants to wilt and die at about 3 and 6 days post‐inoculation (dpi), respectively. Bacterial cells were thus recovered from stems of petiole‐inoculated tomato plants at 2–6 dpi and subjected for quantification by dilution plating. With soil‐soaking inoculation, the wild‐type strain causes tomato plants to wilt and die at about 6 and 12 dpi, respectively. Cells were thus recovered from tomato stems at 6–12 dpi and subjected to quantification. The prhP mutant exhibited significantly less virulence on tomato plants, and bacterial cells were recovered from stems of the soil‐soaking inoculated tomato plants to 21 dpi. For leaf infiltration, about 50 µL of bacterial suspension at 104 cfu/mL was infiltrated into tobacco leaves with a blunt‐end syringe. Cells were recovered daily from tobacco leaf disks to 6 dpi, when tobacco leaves became withered and dried. Mean values of at least four biological replicates were averaged and presented with SD (error bars). Statistical significance between the prhP mutant (RQ5649) and the wild‐type strain (RK5050) was assessed using a post hoc Dunnett test following ANOVA. Significance level: ** indicates P < 0.01.
Tobacco plants exhibit different metabolic activities on SA from tomato plants (Bellés et al., 2006, 1999), and we ascertained whether growth of R. solanacearum in tobacco plants was altered with prhP deletion. Tobacco leaves were infiltrated with cell suspension at a concentration of 104 cfu/mL and the cell growth in tobacco leaves was quantified daily from 2 to 6 dpi, when tobacco leaves became withered and dried. Growth of the prhP mutant was also significantly impaired in tobacco leaves, but this was less than in the wild‐type strain by about one to two orders of magnitude (Fig. 6c). All these results confirm that PrhP is important for the in planta growth of R. solanacearum regardless of host plant species.
PrhP regulates expression of a large set of virulence‐related genes
Ralstonia solanacearum integrates numerous virulence factors to develop an infection process toward host plants, including the flagella, type IV pili, EPS and CWDEs (Genin and Denny, 2012; Hikichi et al., 2007). PrhP greatly promotes the infection process of R. solanacearum besides the degradation of SA and FA, and we profiled a transcriptome analysis to address its multiple roles in the pathogenicity. The transcriptomic analysis revealed that a total of 698 genes were differentially expressed by more than 2‐fold between the wild‐type strain (RK5050) and the prhP mutant (RQ5649), of these 20 and 678 genes were up‐ and down‐regulated, respectively (Fig. 7a).
Figure 7.
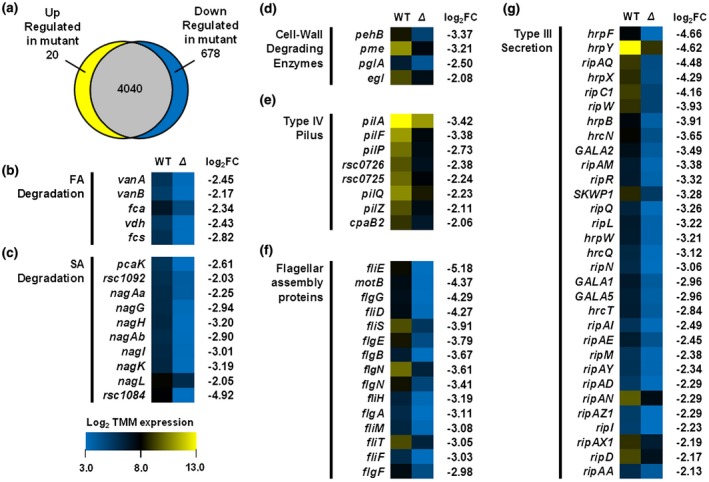
Gene expression in the Ralstonia solancearum prhP mutant with RNA‐seq. Briefly, total RNAs were isolated from three biological replicates according to the TRIzol reagent method (Life Technologies, Carlsbad, CA, USA) and subjected to RNA‐seq. Mean values of three biological replicates were averaged and subjected to statistical analysis. (a) A proportional Venn diagram of expression patterns created using BioVenn. Genes with relative expression levels greater than 2‐fold different between the prhP mutant and the wild‐type strain and adjusted P values of <0.05 were classified as differentially expressed genes (DEGs). (b)–(g) Heat maps show absolute expression of DEGs in different functional categories of (b) ferulic acid (FA) degradation, (c) salicylic acid (SA) degradation, (d) cell wall‐degrading enzymes, (e) type IV pilus, (f) flagellar assembly proteins and (g) T3SS and T3Es. Heat maps indicating low absolute expression (blue; 3.0) to high absolute expression (yellow; 13.0) are shown to the right of gene names. The fold change (prhP mutant versus RK5050) is shown to the right of each heat map.
Consistent with the above results from the promoter activity assay, the expression level of nagAa was significantly down‐regulated by 4.76‐fold in the prhP mutant (Fig. 7c). The nag operon contains 10 genes, which are responsible for SA degradation (Lowe‐Power et al., 2016). Expression of all these genes was significantly down‐regulated in the prhP mutant (Fig. 7c). Operons of fca‐vdh‐fcs and vanAB are responsible for FA degradation (Lowe et al., 2015). Expression of all these genes was significantly down‐regulated in the prhP mutant (Fig. 7b).
The T3SS is directly controlled by the key regulator HrpB and globally regulated with a complex network, including dozens of regulators (Hikichi et al., 2017; Vasse et al., 1995). Expression of hrpB was indeed significantly down‐regulated in the prhP mutant by 15.03‐fold, but no alteration in expression of prhA, prhI/R, hrpG, prhG, prhN, xpsR and phcA with more than 4‐fold in the prhP mutant (Fig. 7g). It is consistent with our above results from the promoter activity assay in respective reporter strains that PrhP positively regulates the T3SS mediated with the key regulator HrpB but through some novel pathway. The hrp gene cluster contains approximately 20 genes, and expression levels of 12 of these genes were significantly down‐regulated in the prhP mutant (Fig. 7g). Moreover, the HrpB directly controls expression of a great number of T3Es, and expression of most of the T3Es was significantly down‐regulated in the prhP mutant (Fig. 7g), which is consistent with the above results from the qRT‐PCR.
In addition, expression of many genes, encoding most of the components for the flagellum, was significantly down‐regulated in the prhP mutant (Fig. 7f), indicating that the flagellum assembly is impaired with prhP deletion. The flagella are known to be essential for bacterial swimming and we evaluated the impact of PrhP on swimming motility. Consistent with transcriptomic results, swimming halos produced by the prhP mutant were significantly smaller than those of the wild‐type strain on semi‐solid motility agar plates (Fig. S3). Expression of many genes related to type IV pili, which are known to be important for attachment to roots and twitching motility, and contribute to pathogenicity, was significantly down‐regulated with prhP deletion (Fig. 7e). Expression of most of the genes related to the type IV pili was significantly down‐regulated in the prhP mutant except three genes of cpaB, pilM and rsp1292, expression of which was up‐regulated by 2.1‐fold, 3.6‐fold and 2.7‐fold, respectively (Fig. 7e). Moreover, expression of several genes related to the CWDEs was significantly down‐regulated with prhP deletion (Fig. 7d), while expression of genes related to the EPS was slightly up‐regulated with prhP deletion (data not shown). All these results confirm that PrhP positively regulates expression of a large set of virulence‐related genes and promotes the infection process of R. solanacearum toward host plants.
Discussion
In the present study, we provided multiple lines of evidence to demonstrate that PrhP, a novel PadR regulator, plays a positive role in the detoxification of phenolic acids and greatly promotes the infection process of R. solanacearum toward host plants. Phenolic acids are broadly antimicrobial and many bacterial species express PADses to convert these chemicals into less toxic derivatives for detoxification (Park et al., 2017; Tran et al., 2008). The prhP mutant was much more sensitive to SA and FA (representative phenolic acids) than the wild‐type strain, confirming that the PrhP is important for R. solanacearum to promote the detoxification of phenolic acids. This is consistent with the fact that PadR regulators usually promote detoxification of phenolic acids in many bacterial species (Nguyen et al., 2011; Park et al., 2017). The PadR regulators usually repress the expression of PADse genes in the absence of phenolic acids by binding to their promoters but initiate the expression of PADse genes in the presence of phenolic acids by release from their promoters (Lowe‐Power et al., 2018; Nguyen et al., 2011). Different from the PADse‐based detoxification in many bacterial species, R. solanacearum detoxifies SA and FA by metabolizing them as carbon sources (Lowe et al., 2015; Lowe‐Power et al., 2016). Degradation of SA and FA in R. solanacearum is experimentally validated to be dependent on nag‐encoding dioxygenases and fcs‐encoding feruloyl‐CoA, respectively (Lowe et al., 2015; Lowe‐Power et al., 2016). It is not contradictory that PrhP was demonstrated to be a novel positive regulator of nag expression even in the absence of SA based our results from the RNA‐seq and promoter assay since the nag‐encoding dioxygenases and fcs‐encoding feruloyl‐CoA are not PADse. Note that no PADse is annotated in numerous genomes of R. solanacearum strains, indicating that degradation of SA and FA is mainly dependent on operons of the fcs and nag in R. solanacearum. PrhP displays about 37% of amino acid identity with several well‐characterized PadR regulators, such as the AphA in Vibrio cholerae and LadR in Lactobacillus plantarum (De Silva et al., 2005; Huillet et al., 2006). Moreover, no conserved palindromic sequences can be found for the PadR binding in promoter regions of fcs and nag operons. The regulation mechanism of PrhP on the expression of fcs and nag remains to be further elucidated.
The nag expression was greatly enhanced with supplementary SA (Fig. 2), which was consistent with the fact that most of the PADses are substrate inducible (Huillet et al., 2006, Lowe‐Power et al., 2018; Van Duy et al., 2007). Although the nagAa expression was significantly impaired with prhP deletion in the presence of SA, it remained at a level of about 80% of the wild‐type strain, indicating that the prhP mutant should be able to degrade SA to some extent. This could explain our result that the nag mutant was much more sensitive than the prhP mutant to supplementary SA (Fig. 1a). We supposed that some novel regulators should regulate the expression of fcs and nag operons besides the PrhP. This speculation is also demonstrated in some published papers that R. solanacearum might harbour some fcs‐independent pathway for specific degradation of HCAs (Lowe et al., 2015). We are currently screening dozens of candidates showing an impact on expression of nagAa‐lacZYA by transposon mutagenesis, and their transcriptional regulation on nag genes is under elucidation.
Phenolic acids play dual roles in the interaction between plants and plant pathogens to inhibit bacterial growth as antibiotics and also defend against pathogenic invasion by triggering an immune response, such as the SA‐triggered immune‐signalling pathways (Campos et al., 2014; Tsuda and Katagiri, 2010). The ability to degrade phenolic acids could thus protect R. solanacearum from the toxicity of these chemicals, and also impair the triggered plant immune response (Lowe‐Power et al., 2016, 2018). Supplementary SA and FA severely inhibited the growth of the prhP mutant in medium but at quite high concentrations, such as 500 μM of SA and 1500 μM of FA. Although it is complicated to measure concentrations of SA and FA in plants, several lines of evidence suggest that free SA can be accumulated to quite high concentrations in plant tissues that is capable to inhibit growth of R. solanacearum (Cameron and Zaton 2004; Huang et al., 2006; Smith‐Becker et al., 1998). Concentrations of some phenolic acids are also reported to be locally high in xylems of roots and stems where they are released by sentinel phenolic‐storing cells (Beckman, 2000; Wallis and Chen, 2012). In addition, bacterial growth is much more severely inhibited with the mixture of HCAs than those with limited compounds of HCAs (Harris et al., 2010). PrhP should be involved in the detoxification of phenolic acids more than SA and FA, which could explain the result that the fcs and nag mutants exhibit faintly impaired growth in host plants (Lowe et al., 2015; Lowe‐Power et al., 2016), while the prhP mutant exhibits significantly impaired growth in host plants. Extensive proliferation in xylem vessels is one of the most important pathogenicity determinants in R. solanacearum (Cunnac et al., 2004; Denny, 1995). The fcs and nag mutants exhibit faintly impaired virulence compared to the wild‐type strain (Lowe et al., 2015; Lowe‐Power et al., 2016), while the prhP mutant exhibits significantly less virulence compared to the wild‐type strain in host plants. The mutant lacking both fcs and nag exhibits slightly less virulence compared with the fcs and nag mutants, which is consistent with previous demonstration that the mixture of HCAs exhibits more a severe inhibitory effect on bacterial growth than the limited compounds of HCAs (Harris et al., 2010). The prhP mutant is significantly less virulent than the fcs and nag mutants in tomato and tobacco plants, indicating that PrhP might be involved in the detoxification of more phenolic acids than SA and FA, or is important for some other pathogenicity determinants.
The T3SS is another essential pathogenicity determinant in R. solanacearum (Genin, 2010; Hikichi et al., 2007) and is significantly impaired with prhP deletion both in vitro and in planta. To our knowledge, this is the first report on PadR regulators that positively regulates T3SS expression in pathogenic bacteria. The hrpB expression was also significantly impaired with prhP deletion, which is consistent with the fact that HrpB directly controls the entire T3SS and T3Es (Hikichi et al., 2007; Valls et al., 2006). PrhP positively regulates hrpB expression and in turn regulates the expression of T3SS and T3Es in R. solanacearum. hrpB expression is positively regulated by two close paralogues of HrpG and PrhG in a parallel way (Plener et al., 2010; Zhang et al., 2013), while no alteration was found in their expression with the prhP deletion, indicating that PrhP positively regulates hrpB expression through some novel pathway. As the OmpR/PhoB family of two‐component response regulators, HrpG and PrhG should respond to host signals by phosphorylation at some residues and then phosphorylated HrpG and PrhG begin to activate hrpB expression (Yoshimochi et al., 2009). It remains to be further elucidated whether PrhP is involved in host signal response with HrpG and PrhG. It is worthwhile noting that the HR elicitation of GMI1000 is not altered with prhP deletion in tobacco leaves, indicating that weakly expressed T3Es might be enough to trigger plant immunity since they are not completely diminished with prhP deletion. Moreover, our results from the RNA‐seq reveal that PrhP is involved in expression of a large set of virulence‐related genes, such as the flagella, type four pili and CWDEs. Expression of most of the genes for the flagellum assembly was significantly down‐regulated with prhP deletion, and the flagella‐mediated swimming motility was indeed impaired on semi‐solid motility agar plates with prhP deletion. The flagella, type 4 pili and CWDEs play important roles in adhesion to roots and cell wall destruction, which are especially important at the early stage of the infection process (Denny, 1995; Liu et al., 2005; Tans‐Kersten et al., 1998, 2004). Expression of many of these genes was significantly down‐regulated with prhP deletion, which could explain our results that the prhP mutants were much less virulent in soil‐soaking inoculated tomato plants than those with petiole inoculation. Tobacco plants exhibit different metabolic activities on phenolic acids, and different plants display different symptoms depending upon the infecting strains (Bellés et al., 1999, 2006). For instance, the secondary metabolites in xylem vessels are revealed to be quite different between these two plants, including phenolic acids, saccharides and amino acids (Lowe‐Power et al., 2018; Zuluaga et al., 2013). This might be a reason why the prhP mutants exhibit almost equally less virulence on tobacco plants regardless of inoculation method.
Taken together, our results from the growth assay, promoter assay, RNA‐seq and qRT‐PCR demonstrate that PrhP, a novel PadR regulator, plays an important regulatory role in multiple processes involved in the infection of R. solanacearum, such as the detoxification of phenolic acids, extensive proliferation in host plants and expression of a great number of virulence‐related genes. This is the first report on the PadR regulators that regulate the T3SS, which could improve our understanding of the various biological functions of PadR regulators and complex regulatory pathway on the T3SS in R. solanacearum.
Experimental Procedures
Bacterial strains and growth conditions
Escherichia coli strains of DH12S and S17‐1 were grown at 37 °C in LB medium for plasmid construction and conjugational transfer, respectively. Ralstonia solanacearum strains, listed in Table 1, were grown at 28 °C in nutrient‐rich medium (B medium) or nutrient‐limited medium (sucrose medium, hrp‐inducing medium) (Yoshimochi et al., 2009).
Table 1.
Bacterial strains used in this study
| Strain | Relative characteristics | References |
|---|---|---|
| OE1‐1 | Wild‐type, race 1, biovar 3 | Kanda et al. (2003) |
| RK5046 | OE1‐1, hrpB‐lacZYA | Yoshimochi et al. (2009) |
| RK5050 | OE1‐1, popA‐lacZYA | Yoshimochi et al. (2009) |
| RK5120 | OE1‐1, hrpG‐lacZYA | Yoshimochi et al. (2009) |
| RK5212 | OE1‐1, prhG‐lacZYA | Zhang et al. (2013) |
| RQ5649 | popA‐lacZYA, ΔprhP | This study |
| RQ5625 | RK5050, Δfcs | This study |
| RQ5651 | RK5046, ΔprhP | This study |
| RQ5657 | RK5212, ΔprhP | This study |
| RQ5660 | RK5120, ΔprhP | This study |
| RQ6207 | RQ5625, ΔnagAL | This study |
| RQ6058 | RK5050, ΔnagAL | This study |
| RQ6061 | OE1‐1, ΔprhP | This study |
| RQC380 | RQ5649, +prhPOE1‐1 | This study |
| RQC607 | OE1‐1, nagAa‐lacZYA | This study |
| RQC609 | RQ6061, nagAa‐lacZYA | This study |
| GMI1000 | Wild‐type, race 1, biovar 4 | Salanoubat et al. (2002) |
| GF0001 | GMI1000, popA‐lacZYA | Zhang et al. (2018) |
| GF0018 | GF0001, ΔprhP | This study |
| RQC089 | GF0018, +prhPOE1‐1 | This study |
Construction of prhp in‐frame deleted mutants
Mutants with in‐frame deletion of target genes were generated with pK18mobsacB‐based homologue recombination (Zhang et al., 2015). In general, two DNA fragments flanking the prhP were conjugated with joint PCR and subcloned into pK18mobsacB to generate pK18d0309. After validating the sequence, pK18d0309 was transferred into R. solanacearum by conjugation with S17‐1. The prhP mutants were generated (listed in Table 1) and confirmed by colony PCR with primer pairs of 0309A1B and 0309B2H. The primers used in this study are listed in Table S1.
Complementation analyses
Complementation analyses were performed in this study with the pUC18‐mini‐Tn7T‐Gm‐based site‐specific chromosome integration system (Choi et al., 2005; Zhang et al., 2015). In general, the prhP gene and its upstream region of about 600 bp, empirically harbouring the native promoter, was PCR amplified and finally cloned into pUC18‐mini‐Tn7T‐Gm to get pUCprhP. After validating the sequence, complementary prhP was integrated into R. solanacearum chromosome at a single attTn7 site (25 bp downstream of glmS) and confirmed by colony PCR with primers of glmsdown‐Tn7R (Zhang et al., 2011).
Construction of reporter strains with nagAa‐laZYA for promoter activity assay
The nagAa‐lacZYA reporter strains were generated with the pUC18‐mini‐Tn7T‐Gm‐based site chromosome integration system. In general, promoterless lacZYA was fused to nagAa at 54 bp after the start codon, in which 6 bp of nucleotide acids were replaced into KpnI by PCR for lacZYA insertion. A DNA fragment containing the promoter region and the KpnI site was first cloned into pUC18‐mini‐Tn7T‐Gm and then lacZYA was inserted to generate pUCnagAa‐lacZYA. After validating the sequence, this reporter fusion was integrated into the chromosome of the wild‐type strain and the prhP mutant to get the desired mutants (listed in Table 1).
β‐Galactosidase assay
The β‐galactosidase assay was performed to evaluate expression levels of lacZYA‐fused genes. The enzyme assay in vitro was expressed in Miller units (Miller, 1992), and that in planta was normalized with luminescence divided by cells number (Zhang et al., 2013). Each assay was repeated for four independent experiments with four replications per trial. The mean values of all the experiments were averaged with SD and the statistical significance was assessed using a post hoc Dunnett test following ANOVA.
Virulence assay and HR test
The virulence assay and HR test were performed as previously described (Yao and Allen, 2007). Wilt‐susceptible tomato plants (Solanum lycopersicum 'Moneymaker') and tobacco plants (N. tabacum 'Bright Yellow') were subjected to virulence assay with soil‐soaking inoculation method, which mimics natural invasion through roots, and petiole inoculation, which enables bacteria to directly invade xylems vessels. Each assay was repeated for four independent experiments with 12 plants per trial. Wilt symptoms of plants were inspected as 1–4 disease index and mean values of all experiments were averaged with SD. Whereas the SD was not presented in figures for virulence assay due to the consideration of aesthetic appearance. The statistical significance was assessed using a post hoc Dunnett test following ANOVA. The HR test was carried out on tobacco leaves of N. tabacum with leaf infiltration and the symptom development of HR was recorded periodically. Each test was repeated independently at least four times with six plants per trial and a representative result is presented.
Bacterial growth assay
Bacterial growth in planta was quantified by dilution plating (Zhang et al., 2013) and that in medium was assessed with OD600. Each assay was repeated for at least four independent experiments with three replications per trial. Mean values of all experiments were averaged with SD and statistical significance was assessed using a post hoc Dunnett test following ANOVA.
RNA extraction and deep sequencing
Ralstonia solanacearum strains were grown in hrp‐inducing medium to OD600 of about 0.1 and total RNA was isolated with TRIzol reagent method according to the manufacturer’s instructions (Life Technologies, Carlsbad, CA, USA). Total RNAs were isolated with three biological replicates and subjected to RNA‐seq. After validating the quality, RNA samples were entrusted to Shanghai Biozeron Biotechnology Co., Ltd (Shanghai, China) for deep sequencing. In general, contaminative genomic DNA was removed using RQ DNase I (Promega) and ribosomal RNA was removed using a Ribo‐Zero rRNA Removal Kit (Gram‐negative bacteria, Illumina, Madison, WI, USA). The cDNA synthesis, end repair and ligation of the Illumina adaptors were performed according to Illumina’s protocol, and subjected to RNA‐seq using Illumina HiSeq PE 2 × 151‐bp read length, resulting in c.10 million reads per sample.
Raw reads were trimmed and quality controlled by Trimmomatic software with default parameters (http://www.usadellab.org/cms/uploads/supplementary/Trimmomatic), and clean reads were separately aligned to the reference genome with orientation mode using hisat2 (https://ccb.jhu.edu/software/hisat2/index.shtml) software. The expression levels of each transcript were calculated using the fragments per kilobase of exon per million mapped reads to identify differential expression of genes between two different samples and cuffdiff (http://cufflinks.cbcb.umd.edu/) was used for differential expression analysis. The mean values of three biological replicates were averaged and subjected for statistical analysis.
qRT‐PCR analysis
cDNA was synthesized using the PrimeScript RT Reagent Kit with gDNA Eraser (Perfect for Real Time, Takara, Japan) according to the manufacturer’s instructions (contaminated genome DNA could be removed by the gDNA Eraser in this kit). The One Step SYBR PrimeScript PLUS RT‐PCR Kit (Takara, Dalian, China) was used for qRT‐PCRs with the Applied Biosystems 7500 Real‐Time PCR System. Primers used in this study were selected as previously described and serC gene was used as the reference gene for normalization of gene expression (Monteiro et al., 2012; Wu et al., 2015). Each assay was repeated from RNA isolation for at least three independent experiments with four replications per trial. The mean values of all experiments were averaged with SD, and the statistical significance between the wild‐type strain and the prhP mutant was assessed using a post hoc Dunnett test following ANOVA.
Supporting information
Fig. S1 Relative expression of T3Es genes in the prhP mutant. Strains were grown in hrp‐inducing medium to an OD600 of about 0.1 and total RNA was isolated. The cDNA was synthesized using the PrimeScript RT Reagent Kit with gDNA Eraser and mRNA levels of representative T3Es genes were determined by qRT‐PCR with reference gene as serC for normalization. Normalized values of the prhP mutant were divided with those of wild‐type (WT) strain and relative values (relative expression) were presented. Mean values of at least three biological replicates were averaged and presented with SD (error bars). Statistical significance was assessed between prhP mutants and WT strain. Significance level: * indicates P < 0.05 and ** indicates P < 0.01.
Fig. S2 HR test. Approximate 50 μL of bacterial suspension at 108 cfu/mL was infiltrated into tobacco leaves with a blunt‐end syringe: (A) GF001 (GMI1000, popA‐lacZYA), (B) GF0018 (GF0001, ΔprhP) and (C) distilled water. Development of necrotic lesions was observed periodically and pictures were taken. Each experiment was repeated at least four times and each treatment contained four plants. The results presented are from a representative experiment, and similar results were obtained in all experiments.
Fig. S3 Swimming motility of prhP mutants. Bacterial suspension (3 μL) at OD600 of 1.0 was dropped onto 0.3% agar plates and kept at 28 °C for 48 h. Swimming motility was quantified as halo diameters in millimetres. Mean values of three biological replicates with four replicates per trial were averaged and presented with SD (error bars). Statistical significance was assessed between RQ5649 and RK5050. Significance level: ** indicates P < 0.01.
Table S1 Primers used in this study.
Acknowledgements
We express thanks for funding support from the National Natural Science Foundation of China (31670082), the Chongqing Research Program of Basic Research and Frontier Technology (cstc2016jcjyA0470 to Y. Z and cstc2015jcyjA10011 to J. L) and the Graduate student scientific research innovation projects in Chongqing (CYS18105) to W.Z.
Contributor Information
Yong Zhang, Email: bioyongzhang@swu.edu.cn.
Kouhei Ohnishi, Email: kouheio@kochi-u.ac.jp.
References
- Angot, A. , Peeters, N. , Lechner, E. , Vailleau, F. , Baud, C. , Gentzbittel, L. , Sartorel, E. , Genschik, P. , Boucher, C. and Genin, S. (2006) Ralstonia solanacearum requires F‐box‐like domain‐containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. USA. 103, 14620–14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlat, M. , Gough, C.L. , Zischek, C. , Barberis, P.A. , Trigalet, A. and Boucher, C. (1992) Transcriptional organization and expression of the large hrp gene cluster of Pseudomonas solanacearum . Mol. Plant‐Microbe Interact. 5, 187–193. [DOI] [PubMed] [Google Scholar]
- Beckman, C.H. (2000) Phenolic‐storing cells, keys to programmed cell death and periderm formation in wilt disease resistance and in general defence responses in plants. Physiol. Mol. Plant Path. 57, 101–110. [Google Scholar]
- Bellés, J.M. , Garro, R. , Fayos, J. , Navarro, P. , Primo, J. and Conejero, V. (1999) Gentisic acid as a pathogen‐inducible signal, additional to salicylic acid for activation of plant defenses in tomato. Mol. Plant‐Microbe Interact. 12, 227–235. [Google Scholar]
- Bellés, J.M. , Garro, R. , Pallás, V. , Fayos, J. , Rodrigo, I. and Conejero, V. (2006) Accumulation of gentisic acid as associated with systemic infections but not with the hypersensitive response in plant‐pathogen interactions. Planta, 223, 500–511. [DOI] [PubMed] [Google Scholar]
- Cameron, R.K. and Zaton, K. (2004) Intercellular salicylic acid accumulation is important for age‐related resistance in Arabidopsis to Pseudomonas syringae . Physiol. Mol. Plant Path. 65, 197–209. [Google Scholar]
- Campos, L. , Lisón, P. , López‐Gresa, M.P. , Rodrigo, I. , Zacarés, L. , Conejero, V. and Bellés, J.M. (2014) Transgenic tomato plants overexpressing tyramine N‐hydroxycinnamoyl transferase exhibit elevated hydroxycinnamic acid amide levels and enhanced resistance to Pseudomonas syringae . Mol. Plant‐Microbe Interact. 27, 1159–1169. [DOI] [PubMed] [Google Scholar]
- Choi, K.H. , Gaynor, J.B. , White, K.G. , Lopez, C. , Bosio, C.M. , Karkhoff‐Chweizer, R.R. and Schweizer, H.P. (2005) A Tn7‐based broad range bacterial cloning and expression system. Nat. Methods. 2, 443–448. [DOI] [PubMed] [Google Scholar]
- Coll, N.S. and Valls, M. (2013) Current knowledge on the Ralstonia solanacearum type III secretion system. Microb. Biotechnol. 6, 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnac, S. , Occhialini, A. , Barberis, P. , Boucher, C. and Genin, S. (2004) Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53, 115–128. [DOI] [PubMed] [Google Scholar]
- De Silva, R.S. , Kovacikova, G. , Lin, W. , Taylor, R.K. , Skorupski, K. and Kull, F.J. (2005) Crystal structure of the virulence gene activator AphA from Vibrio cholerae reveals it is a novel member of the winged helix transcription factor superfamily. J. Biol. Chem. 280, 13779–13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny, T.P. (1995) Involvement of bacterial polysaccharides in plant pathogenesis. Ann. Rev. Phytopathol. 33, 173–197. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, D. , Stratford, M. , Gasson, M. , Ueckert, J. , Bos, A. and Narbad, A. (2004) Mode of antimicrobial action of vanillin against Escherichia coli, Lactobacillus plantarum and Listeria innocua . J. Appl. Microbiol. 97, 104–113. [DOI] [PubMed] [Google Scholar]
- Florez, A.B. , Alvarez, S. , Zabala, D. , Brana, A.F. , Salas, J.A. and Mendez, C. (2015) Transcriptional regulation of mithramycin biosynthesis in Streptomyces argillaceus: dual role as activator and repressor of the PadR‐like regulator MtrY. Microbiology, 161, 272–284. [DOI] [PubMed] [Google Scholar]
- Fry, S.C. , Willis, S.C. and Paterson, A.E. (2000) Intraprotoplasmic and wall‐localised formation of arabinoxylan‐bound diferulates and larger ferulate coupling‐products in maize cell‐suspension cultures. Planta, 211, 679–692. [DOI] [PubMed] [Google Scholar]
- Gasson, M.J. , Kitamura, Y. , McLauchlan, W.R. , Narbad, A. , Parr, A.J. , Parsons, E.L.H. , Payne, J. , Rhodes, M.J.C. and Walton, N.J. (1998) Metabolism of ferulic acid to vanillin. J. Biol. Chem. 273, 4163–4170. [DOI] [PubMed] [Google Scholar]
- Genin, S. (2010) Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum . New. Phytol. 187, 920–928. [DOI] [PubMed] [Google Scholar]
- Genin, S. and Denny, T.P. (2012) Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89. [DOI] [PubMed] [Google Scholar]
- Goleniowski, M. , Bonfill, M. , Cusido, R. and Palazón, J. (2013) Phenolic acids In: Natural Products (Amawat K. and Mérillon J.M., eds), Berlin, Heidelberg: Springer. [Google Scholar]
- Gury, J. , Barthelmebs, L. , Tran, N.P. , Divies, C. and Cavin, J.F. (2004) Cloning, deletion, and characterization of PadR, the transcriptional repressor of the phenolic acid decarboxylase‐encoding padA gene of Lactobacillus plantarum . Appl. Environ. Microbiol. 70, 2146–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gury, J. , Seraut, H. , Tran, N.P. , Barthelmebs, L. , Weidmann, S. , Gervais, P. and Cavin, J.F. (2009) Inactivation of PadR, the repressor of the phenolic acid stress response, by molecular interaction with Usp1, a universal stress protein from Lactobacillus plantarum, in Escherichia coli . Appl. Environ. Microbiol. 75, 5273–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, V. , Jiranek, V. , Ford, C.M. and Grbin, P.R. (2010) Inhibitory effect of hydroxycinnamic acids on Dekkera spp . Appl. Microbiol. Biotechnol. 86, 721–729. [DOI] [PubMed] [Google Scholar]
- Heleno, S.A. , Martins, A. , Queiroz, M.J.R.P. and Ferreira, I.C.F.R. (2015) Bioactivity of phenolic acids: metabolites versus parent compounds: a review. Food. Chem. 173, 501–513. [DOI] [PubMed] [Google Scholar]
- Hikichi, Y. , Yoshimochi, T. , Tsujimoto, S. , Shinohara, R. , Nakaho, K. , Kanda, A. , Kiba, A. and Ohnishi, K. (2007) Global regulation of pathogenicity mechanism of Ralstonia solanacearum . Plant Biotech. 24, 149–154. [Google Scholar]
- Hikichi, Y. , Mori, Y. , Ishikawa, S. , Hayashi, K. , Ohnishi, K. , Kiba, A. and Kai, K. (2017) Regulation involved in colonization of intercellular spaces of host plants in Ralstonia solanacearum . Front. Plant Sci. 8, 967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W.E. , Huang, L. , Preston, G.M. , Naylor, M. , Carr, J.P. , Li, Y. , Singer, A.C. , Whiteley, A.S. and Wang, H. (2006) Quantitative in situ assay of salicylic acid in tobacco leaves using a genetically modified biosensor strain of Acinetobacter sp. ADP1. Plant. J. 46, 1073–1083. [DOI] [PubMed] [Google Scholar]
- Huillet, E. , Velge, P. , Vallaeys, T. and Pardon, P. (2006) LadR, a new PadR‐related transcriptional regulator from Listeria monocytogenes, negatively regulates the expression of the multidrug efflux pump MdrL. FEMS Microbiol. Lett. 254, 87–94. [DOI] [PubMed] [Google Scholar]
- Janse, J.D. , Beld, H.E.V.D. , Elphinstone, J. , Simpkins, S. , Tjou‐Tam‐Sin, N.N.A. and Vaerenbergh, J.V. (2004) Introduction to Europe of Ralstonia solanacearum biovar 2 race 3 in Pelargonium zonale cuttings. J. Plant Pathol. 86, 147–155. [Google Scholar]
- Jiang, G. , Wei, Z. , Xu, J. , Chen, H. , Zhang, Y. and She, X. (2017) Bacterial wilt in china: history, current status, and future perspectives. Front. Plant Sci. 8, 1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kanda, A. , Ohnishi, S. , Tomiyama, H. , Hasegawa, H. , Yasukohchi, M. , Kiba, A. , Ohnishi, K. , Okuno, T. and Hikichi, Y. (2003) Type III secretion machinerydeficient mutants of Ralstonia solanacearum lose their ability to colonize resulting in loss of pathogenicity. J. Gen. Plant Pathol., 69, 250–257. [Google Scholar]
- Lanoue, A. , Burlat, V. , Henkes, G.J. , Koch, I. , Schurr, U. and Röse, U.S. (2010) De novo biosynthesis of defense root exudates in response to Fusarium attack in barley. New. Phytol. 185, 577–588. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Zhang, S. , Schell, M.A. and Denny, T.P. (2005) Pyramiding unmarked deletions in Ralstonia solanacearum shows that secreted proteins in addition to plant cell‐wall‐degrading enzymes contribute to virulence. Mol. Plant‐Microbe Interact. 18, 1296–1305. [DOI] [PubMed] [Google Scholar]
- Lowe, T.M. , Ailloud, F. and Allen, C. (2015) Hydroxycinnamic acid degradation, a broadly conserved trait, protects Ralstonia solanacearum from chemical plant defenses and contributes to root colonization and virulence. Mol. Plant‐Microbe Interact. 28, 286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe‐Power, T.M. , Jacobs, J.M. , Ailloud, F. , Fochs, B. , Prior, P. and Allen, C. (2016) Degradation of the plant defense signal salicylic acid protects Ralstonia solanacearum from toxicity and enhances virulence on tobacco, mBio. 7, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe‐Power, T.M. , Khokhani, D. and Allen, C. (2018) How Ralstonia solanacearum exploits and thrives in the flowing plant xylem environment. Trends Microbiol. 26, 929–942. [DOI] [PubMed] [Google Scholar]
- Marenda, M. , Brito, B. , Callard, D. , Genin, S. , Barberis, P. , Boucher, C. and Arlat, M. (1998) PrhA controls a novel regulatory pathway required for the specific induction of Ralstonia solanacearum hrp genes in the presence of plant cells. Mol. Microbiol. 27, 437–453. [DOI] [PubMed] [Google Scholar]
- Miller, J.H. (1992) The lac system In: A Short Course in Bacterial Genetics. A Laboratory Manual and Handbook for Escherichia Coli and Related Bacteria (Miller J.H., ed.), pp. 43–80, Plainview: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Monteiro, F. , Genin, S. , Van Dijk, I. and Valls, M. (2012) A luminescent reporter evidences active expression of Ralstonia solanacearum type III secretion system genes throughout plant infection. Microbiology, 158, 2107–2116. [DOI] [PubMed] [Google Scholar]
- Mukaihara, T. , Tamura, N. and Iwabuchi, M. (2010) Genome‐wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant‐Microbe Interact. 23, 251–262. [DOI] [PubMed] [Google Scholar]
- Naoumkina, M.A. , Zhao, Q. , Gallego‐Giraldo, L. , Dai, X. , Zhao, P.X. and Dixon, R.A. (2010) Genome‐wide analysis of phenylpropanoid defence pathways. Mol. Plant Pathol. 11, 829–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T.K. , Tran, N.P. and Cavin, J.F. (2011) Genetic and biochemical analysis of PadR‐padC promoter interactions during the phenolic acid stress response in Bacillus subtilis 168. J. Bacteriol. 193, 4180–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhage, J. , Prieffert, H. and Steinbüchel, A. (1999) Biochemical and genetic analyses of ferulic acid catabolism in Pseudomonas sp. strain HR199. Appl. Environ. Microbiol. 65, 4837–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.C. , Kwak, Y.M. , Song, W.S. , Hong, M. and Yoon, S. (2017) Structural basis of effector and operator recognition by the phenolic acid‐responsive transcriptional regulator PadR. Nucleic Acids. Res. 45, 13080–13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters, N. , Carrère, S. , Anisimova, M. , Plener, L. , Cazalé, A.C. and Genin, S. (2013) Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genom. 14, 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plener, L. , Manfredi, P. , Valls, M. and Genin, S. (2010) PrhG, a transcriptional regulator responding to growth conditions, is involved in the control of the type III secretion system regulon in Ralstonia solanacearum . J. Bacteriol. 192, 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymiro, M. , Cunnac, S. , Barberis, P. , Deslandes, L. , Peeters, N. , Cazale‐Noel, A.C. , Boucher, C. and Genin, S. (2009) Two type III secretion system effectors from Ralstonia solanacearum GMI1000 determine host‐range specificity on tobacco. Mol. Plant‐Microbe Interact. 22, 538–550. [DOI] [PubMed] [Google Scholar]
- Roberts, D.P. , Denny, T.P. and Schell, M.A. (1988) Cloning of the egl gene of Pseudomonas solanacearum and analysis of its role in phytopathogenicity. J. Bacteriol. 170, 1445–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanoubat, M. , Genin, S. , Artiguenave, F. , Gouzy, J. , Mangenot, S. , Arlat, M. , Billault, A. , Brottier, P. , Camus, J.C. , Cattolico, L. , Chandler, M. , Choisne, N. , Claudel‐Renard, C. , Cunnac, S. , Demange, N. , Gaspin, C. , Lavie, M. , Moisan, A. , Robert, C. , Saurin, W. , Schiex, T. , Siguier, P. , Thebault, P. , Whalen, M. , Wincker, P. , Levy, M. , Weissenbach, J. and Boucher, C.A. (2002) Genome sequence of the plant pathogen Ralstonia solanacearum . Nature. 415, 497–502. [DOI] [PubMed] [Google Scholar]
- Segura, A. , Bünz, P.V. , D’Argenio, D.A. and Ornston, L.N. (1999) Genetic analysis of a chromosomal region containing vanA and vanB, genes required for conversion of either ferulate or vanillate to protocatechuate in Acinetobacter. J. Bacteriol. 181, 3494–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith‐Becker, J. , Marois, E. , Huguet, E.J. , Midland, S.L. , Sims, J.J. and Keen, N.T. (1998) Accumulation of salicylic acid and 4‐hydroxybenzoic acid in phloem fluids of cucumber during systemic acquired resistance is preceded by a transient increase in phenylalanine ammonia‐lyase activity in petioles and stems. Plant Physiol. 116, 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tans‐Kersten, J. , Guan, Y. and Allen, C. (1998) Ralstonia solanacearum pectin methylesterase is required for growth on methylated pectin but not for bacterial wilt virulence. Appl. Environ. Microbiol. 64, 4918–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tans‐Kersten, J. , Brown, D. and Allen, C. (2004) Swimming motility, a virulence trait of Ralstonia solanacearum, is regulated by FlhDC and the plant host environment. Mol. Plant‐Microbe Interact. 17, 686–695. [DOI] [PubMed] [Google Scholar]
- Tran, N.P. , Gury, J. , Dartois, V. , Nguyen, T.K. , Seraut, H. , Barthelmebs, L. , Gervais, P. and Cavin, J.F. (2008) Phenolic acid‐mediated regulation of the padC gene, encoding the phenolic acid decarboxylase of Bacillus subtilis . J. Bacteriol. 190, 3213–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, K. and Katagiri, F. (2010) Comparing signaling mechanisms engaged in pattern triggered and effector‐triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. [DOI] [PubMed] [Google Scholar]
- Valls, M. , Genin, S. and Boucher, C. (2006) Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum . PLoS Pathog. 2, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duy, N. , Mäder, U. , Tran, N.P. , Cavin, J.F. , Tam, L.T. , Albrecht, D. , Hecker, M. and Antelmann, H. (2007) The proteome and transcriptome analysis of Bacillus subtilis in response to salicylic acid. Proteomics, 7, 698–710. [DOI] [PubMed] [Google Scholar]
- Vasse, J. , Frey, P. and Trigalet, A. (1995) Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum . Mol. Plant‐Microbe Interact. 8, 241–251. [Google Scholar]
- Verpoorte, R. , Contin, A. and Memelink, J. (2002) Biotechnology for the production of plant secondary metabolites. Phytochem. Rev. 1, 13. [Google Scholar]
- Wallis, C.M. and Chen, J. (2012) Grapevine phenolic compounds in xylem sap and tissues are significantly altered during infection by Xylella fastidiosa . Phytopathology, 102, 816–771. [DOI] [PubMed] [Google Scholar]
- Wu, D. , Ding, W. , Zhang, Y. , Liu, X. and Yang, L. (2015) Oleanolic acid induces the type III secretion system of Ralstonia solanacearum . Front. Microbiol. 6, 1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, J. and Allen, C. (2007) The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation and interactions with its tomato host. J. Bacteriol. 189, 6415–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimochi, T. , Zhang, Y. , Kiba, A. , Hikichi, Y. and Ohnishi, K. (2009) Expression of hrpG and activation of response regulator HrpG are controlled by distinct signal cascades in Ralstonia solanacearum . J. Gen. Plant. Pathol. 75, 196–204. [Google Scholar]
- Zhang, Y. , Kiba, A. , Hikichi, Y. and Ohnishi, K. (2011) prhKLM genes of Ralstonia solanacearum encode novel activators of hrp regulon and are required for pathogenesis in tomato. FEMS Microbiol. Lett. 317, 75–82. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Chen, L. , Takehi, Y. , Kiba, A. , Hikichi, Y. and Ohnishi, K. (2013) Functional analysis of Ralstonia solanacearum PrhG regulating the hrp regulon in host plants. Microbiology, 159, 1695–1704. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Luo, F. , Wu, D. , Hikichi, Y. , Kiba, A. , Igarashi, Y. , Ding, W. and Ohnishi, K. (2015) PrhN, a putative marR family transcriptional regulator, is involved in positive regulation of type III secretion system and full virulence of Ralstonia solanacearum . Front. Microbiol. 6, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Li, J. , Zhang, W. , Shi, H. , Luo, F. , Hikichi, Y. , Shi, X. and Ohnishi, K. (2018) A putative LysR‐type transcriptional regulator PrhO positively regulates the type III secretion system and contributes to the virulence of Ralstonia solanacearum . Mol. Plant Pathol. 19, 1808–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuluaga, A.P. , Puigvert, M. and Valls, M. (2013) Novel plant inputs influencing Ralstonia solanacearum during infection. Front. Microbiol., 20, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Relative expression of T3Es genes in the prhP mutant. Strains were grown in hrp‐inducing medium to an OD600 of about 0.1 and total RNA was isolated. The cDNA was synthesized using the PrimeScript RT Reagent Kit with gDNA Eraser and mRNA levels of representative T3Es genes were determined by qRT‐PCR with reference gene as serC for normalization. Normalized values of the prhP mutant were divided with those of wild‐type (WT) strain and relative values (relative expression) were presented. Mean values of at least three biological replicates were averaged and presented with SD (error bars). Statistical significance was assessed between prhP mutants and WT strain. Significance level: * indicates P < 0.05 and ** indicates P < 0.01.
Fig. S2 HR test. Approximate 50 μL of bacterial suspension at 108 cfu/mL was infiltrated into tobacco leaves with a blunt‐end syringe: (A) GF001 (GMI1000, popA‐lacZYA), (B) GF0018 (GF0001, ΔprhP) and (C) distilled water. Development of necrotic lesions was observed periodically and pictures were taken. Each experiment was repeated at least four times and each treatment contained four plants. The results presented are from a representative experiment, and similar results were obtained in all experiments.
Fig. S3 Swimming motility of prhP mutants. Bacterial suspension (3 μL) at OD600 of 1.0 was dropped onto 0.3% agar plates and kept at 28 °C for 48 h. Swimming motility was quantified as halo diameters in millimetres. Mean values of three biological replicates with four replicates per trial were averaged and presented with SD (error bars). Statistical significance was assessed between RQ5649 and RK5050. Significance level: ** indicates P < 0.01.
Table S1 Primers used in this study.


