Summary
Gene silencing exists in eukaryotic organisms as a conserved regulation of the gene expression mechanism. In general, small RNAs (sRNAs) are produced within the eukaryotic cells and incorporated into an RNA‐induced silencing complex (RISC) within cells. However, exogenous sRNAs, once delivered into cells, can also silence target genes via the same RISC. Here, we explored this concept by targeting the Cellulose synthase A3 (CesA3) gene of Hyaloperonospora arabidopsidis (Hpa), the downy mildew pathogen of Arabidopsis thaliana. Hpa spore suspensions were mixed with sense or antisense sRNAs and inoculated onto susceptible Arabidopsis seedlings. While sense sRNAs had no obvious effect on Hpa pathogenicity, antisense sRNAs inhibited spore germination and hence infection. Such inhibition of infection was not race‐specific, but dependent on the length and capping of sRNAs. Inhibition of infection by double stranded sRNA was more efficient than that observed with antisense sRNA. Thus, exogenous sRNA targeting conserved CesA3 could suppress Hpa infection in Arabidopsis, indicating the potential of this simple and efficient sRNA‐based approach for deciphering gene functions in obligate biotrophic pathogens as well as for R‐gene independent control of diseases in plants.
Keywords: Arabidopsis, downy mildew, oomycetes, spray‐induced gene silencing (SIGS), sRNA
Introduction
Noncoding 20–30 nucleotide (nt)‐long small RNAs (sRNAs) have been known to be involved in the regulation of gene expression and defence in eukaryotes (Chen et al., 2018; Qin et al., 2017; Zhang et al., 2019). Different types of RNAs, such as double‐stranded RNA (dsRNA) and small interfering RNA (siRNA), can trigger homologous RNA degradation or inhibit mRNA translation (Huang et al., 2016; Nejat and Mantri, 2018). This process is known as RNA silencing and plays a significant role in various biological processes, including innate immunity (Brant and Budak, 2018; Deng et al., 2018) and development (Li et al., 2017; Qin et al., 2017).
In plant–microbe interactions, plants and microbes can exchange RNA molecules, which then integrate into RNA silencing machinery in reciprocal recipient cells. Such cross‐kingdom RNA transfer was first demonstrated between fungus and plants (Weiberg et al., 2013). Botrytis cinerea, an ascomycete fungus infecting more than 200 plant species, transports its sRNAs that silence both Arabidopsis and tomato genes by hijacking plant cellular gene silencing machinery (Weiberg et al., 2013). On the other hand, a large number of cotton sRNAs were found in Verticillium dahliae hyphae recovered from V. dahliae-infected plant tissues and some of the cotton‐originated sRNAs can target essential fungal virulence genes (Zhang et al., 2016).
Movement of sRNAs from plant to pathogens has been explored using the host‐induced gene silencing (HIGS) technique where the sRNAs are generally made by producing dsRNA in transgenic plants using Agrobacterium or in viruses that replicate through dsRNA. HIGS has been successfully used to suppress essential pathogen genes in various plant–pathogen interaction systems including barley–Fusarium (Koch et al., 2013), Arabidopsis– and tomato–Verticillium (Song and Thomma, 2018), barley– and wheat–Blumeria (Nowara et al., 2010) and lettuce–Bremia (Govindarajulu et al., 2015). Similarly, HIGS has also been used against nematodes in Arabidopsis (Huang et al., 2006).
Several recent studies have shown that exogenously applied sRNAs can be taken up by fungal or plant cells and trigger RNA silencing. The exogenous application of RNA by spraying it directly onto plants has been referred to as spray‐induced gene silencing (SIGS) (Koch et al., 2016). The induction of gene silencing by spraying, or otherwise applying RNA, avoids the need to develop transgenic plants (Wang and Jin, 2017). This method has been tested against fungal pathogens, including Fusarium graminearum (Koch et al., 2016) and Fusarium culmorum (Koch et al., 2018). Several different methods have been used to deliver sRNAs onto plants. For example, high‐pressure spraying siRNAs was found to efficiently silence transgenic GFP gene expression in Nicotiana tabacum (Dalakouras et al., 2016). In a different approach, Mitter et al. (2017) used clay nanosheets to deliver dsRNA onto plants for silencing homologous viral RNA. For insect pests, an ingestion method seems to be another way to deliver RNAs. Insects were fed an artificial diet containing dsRNAs in order to induce RNA silencing. This strategy was successfully exploited to target against coleopteran species such as western corn rootworm Diabrotica virgifera virgifera (Baum et al., 2007).
The oomycetes include a unique group of biotrophic and hemibiotrophic plant pathogens and are distinct from fungi (Kamoun et al., 2014). The cell walls of oomycetes have been reported to be primarily β‐1,3‐glucans and cellulose with little or no chitin (Kamoun, 2003). Oomycete hyphae are coenocytic (multinucleate with no division by septa) and their vegetative nuclei are in a diploid state (Fugelstad, 2008; Coates and Beynon, 2010). Genetic manipulation has been developed for some oomycete pathogens. For example, stable transformations using protoplast uptake and regeneration have been reported for the culturable oomycetes including Phytophthora infestans, Phytophthora sojae and Phytophthora citricola (Kamoun, 2003; Mcleod et al., 2008). Using a DNA‐directed RNAi system, efficient gene silencing has been achieved in P. infestans (Abrahamian et al., 2016). Saraiva et al. (2014) used uptake of dsRNA into protoplasts of Saprolegnia parasitica and reported the efficient silencing of the tyrosinase gene. However, routine genetic transformations or gene silencing studies in obligate oomycete species have been hampered by the lack of efficient reliable methods.
Hyaloperonospora arabidopsidis (Hpa) is an obligate biotrophic oomycete pathogen that causes downy mildew disease on Arabidopsis thaliana. The Hpa–Arabidopsis system has been used as a model to investigate pathogen effectors (Woods‐Tör et al., 2018) and plant immunity (Holub, 2007). This pathogen has both sexual and asexual reproduction. For infection, an asexual conidiospore germinates on the surface of plant leaves, forms an appressorium, then a penetration hypha grows between the walls of neighbouring epidermal cells (Koch and Slusarenko, 1990). In susceptible plants, the hypha branches out into the intercellular space and forms haustoria in the epidermal and mesophyll cells. Within 1–2 weeks, conidiophores develop through stomata, carrying conidiospores that begin new rounds of infection. Sexual spores, called oospores, are produced within the cotyledons or on the leaves of the infected plant (Koch and Slusarenko, 1990).
To our knowledge, there is no reliable and efficient genetic transformation method for Hpa. Here, we used in vitro synthesized sRNAs targeting the Hpa‐CesA3 gene and report that antisense or double‐stranded capped sRNAs of 25 nt or longer inhibit spore germination and hence infection.
Results
Selection of target gene for sRNA‐mediated silencing in Hpa
The main cell wall components of oomycetes are β‐glucans and cellulose (Fugelstad, 2008; Raaymakers and Van den Ackerveken, 2016). We focused on a cellulose synthase gene as a target for sRNA‐mediated silencing. Using Pfam (Punta et al., 2012), we identified M4BU64 of Hpa belonging to the cellulose synthase gene family. In silico analysis revealed that this gene corresponds to HpaG810051 in the Emoy2 genome and exists as a single copy gene. EnsemblProtists gene annotation revealed HpaG810051 does not have an intron and the open reading frame encodes a predicted protein of 1144 amino acids (molecular mass 127.028 kDa). A BLASTX search against the database revealed that HpaG810051 has a high similarity to CesA3 proteins from other oomycetes, thus we designated HpaG810051 as Hpa‐CesA3. We then obtained the nucleotide and amino acid sequences of the CesA3 genes of Albugo candida, Albugo laibachii, Bremia lactucae, Phytopthora capsici, P. infestans and Plasmopara viticola and aligned them with those of Hpa‐CesA3. Alignment of amino acid sequences revealed a 93% identity of Hpa‐CesA3 with P. capsici‐CesA3, 92% with P. infestans‐CesA3 and P. viticola‐CesA3, 91% with B. lactucae‐CesA3, 77% with A. candida‐CesA3 and 76% with A. laibachii‐CesA3 (Fig. S1). Domain and motif searches of the Hpa‐CesA3 revealed a nucleotide‐diphospho‐sugar transferase domain (W545‐E891) and a cellulose synthase domain (F814‐Y1122), as well as 15 transmembrane domains (Fig. 1). Alignment of nucleotide sequences of these seven genes showed that Hpa‐CesA3 had 65–84% identity to its orthologues (Fig. S2). Interestingly, nucleotide alignment showed no region with 100% identity that would allow designing of a common sRNA for CesA3 gene silencing across these oomycete species.
Figure 1.

Domain structure of Hpa‐CesA3. The mature protein has 15 transmembrane domains, a nucleotide‐diphospho‐sugar transferase domain (amino acid residues 545‐891) and a cellulose synthase domain (amino acid residues 814‐1122). sRNA is the region where sRNAs were designed.
We then looked at the expression pattern of Hpa‐CesA3 using the available published transcriptome data in Arabidopsis Col‐0 inoculated with the avirulent or virulent Hpa isolates Emoy2 or Waco9, respectively (Asai et al., 2018). It is clear that Hpa‐CesA3 is expressed highly in the spores and the level of expression drops significantly in the mycelia during development (Fig. S3).
Hpa‐CesA3 antisense sRNA inhibits Hpa sporulation
Since Hpa is an obligate biotrophic pathogen and grows on Arabidopsis, we checked whether Hpa‐CesA3 had any homology to Arabidopsis CesA genes (Burn et al., 2002). BLASTN searches against the Arabidopsis database revealed no significant similarity. Subsequently, we designed 25‐nt sense and antisense RNA oligonucleotides from the 5ʹ region of the gene that does not have any homology in other genes in the Hpa genome. The sense or antisense sRNAs were mixed with Hpa spores at 5, 10 and 20 µM concentrations and 7‐day‐old Arabidopsis seedlings were drop inoculated. At 7 days post‐inoculation (dpi), Hpa sporulation was checked and no visible difference in sporulation was observed between control plants (Fig. 2a) and those inoculated with spore suspensions containing 5, 10 or 20 µM sense sRNA. However, sporulation was visibly reduced on plants inoculated with spore suspensions containing 5 and 10 µM antisense RNA (Fig. 2b,c). Interestingly, there was no sporulation on plants inoculated with a spore suspension containing 20 µM antisense RNA (Fig. 2d). Quantitative data analysis further demonstrated the significant reduction or no sporulation in plants inoculated with Hpa spores mixed with antisense sRNAs (Fig. 3). The experiment was repeated at least five times, each time with a minimum of three replicates, and similar results were obtained. We also designed 25‐nt sense and antisense DNA oligonucleotides from the same region of the gene and carried out similar inoculation experiments with 20 µM DNA oligonucleotides. There was no difference in the sporulation between control plants and those inoculated with sense or antisense DNA oligonucleotides (Fig. S4).
Figure 2.

Application of antisense sRNA targeting Hpa‐CesA3 inhibits sporulation. Hpa‐Emoy2 spores were mixed with antisense sRNA at different concentrations and 7‐day‐old Arabidopsis seedlings were drop inoculated. Seedlings were examined for sporulation at 7 days post‐inoculation. (a) Control (no antisense sRNA), (b) seedlings inoculated with spores mixed with 5 µM antisense sRNA, (c) seedlings inoculated with spores mixed with 10 µM antisense sRNA and (d) seedlings inoculated with spores mixed with 20 µM antisense sRNA. The inoculation experiments were repeated five times and similar observations were made. There was no sporulation in seedlings inoculated with spores mixed with 20 µM antisense sRNA.
Figure 3.
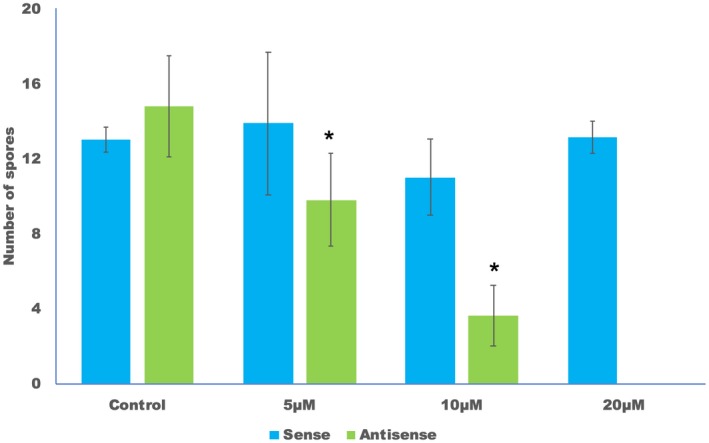
Antisense but not sense sRNA inhibits sporulation. Arabidopsis seedlings were drop inoculated with Hpa‐Emoy2 spores containing 0, 5, 10 and 20 µM sense or antisense sRNA. Ten inoculated seedlings from each sample were collected at 7 days post‐inoculation and placed in 250 µL H2O. The number of spores was counted using a haemocytometer. The average and standard error of three replicates are shown. The experiment was repeated five times with similar results. Asterisks (*) indicate significant difference to control inoculation at the corresponding sRNA (P < 0.05, paired Student’s t‐tests).
Hpa‐CesA3 antisense sRNA inhibits spore germination
To investigate how Hpa‐CesA3 antisense sRNA inhibits sporulation, we inoculated 7‐day‐old Arabidopsis seedlings with a spore suspension containing 20 µM antisense sRNA. Trypan blue staining of inoculated leaves was carried out at 7 dpi to reveal the extent of Hpa development in the tissues. Although normal pathogen development was observed in the control leaf tissues (Fig. 4a), there were no hyphae in, or pathogen spores on, the cotyledons inoculated with the spore suspension containing 20 µM sRNA (Fig. 4b,c). This indicates that antisense sRNA may have inhibited spore germination, thus preventing infection. Non‐germinating spores may have been washed away during trypan blue staining. To investigate this further, we set up germination assays using cellophane strips. After 48 h spores were examined for germination under a light microscope. Untreated, control spores were bright and produced germ tubes at various lengths within the 2 days (Fig. 5a,b). However, spores treated with antisense sRNA became dark brown and germination tubes were mainly absent or, in rare cases, were arrested (Fig. 5c,d). We repeated this assay five times and observed 100% inhibition of germination in all experiments (Table S1).
Figure 4.
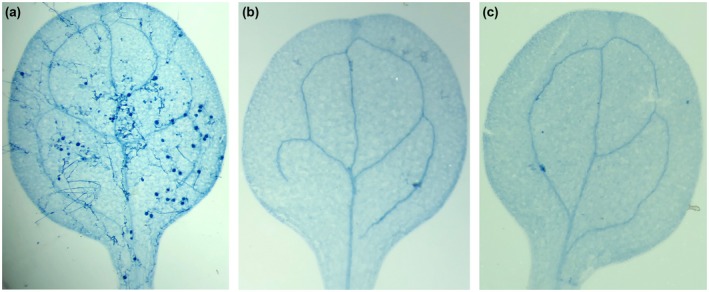
Pathogen inhibition at the infection stage. Arabidopsis seedlings were drop inoculated with Hpa‐Emoy2 spores containing 0 or 20 µM antisense sRNA and at 7 days post‐inoculation seedlings were stained with trypan blue. While there was normal infection with sporulation and oospore development was observed in the control (a), no infection or hyphal development were observed in seedlings inoculated with 20 µM antisense sRNA (b) and (c).
Figure 5.
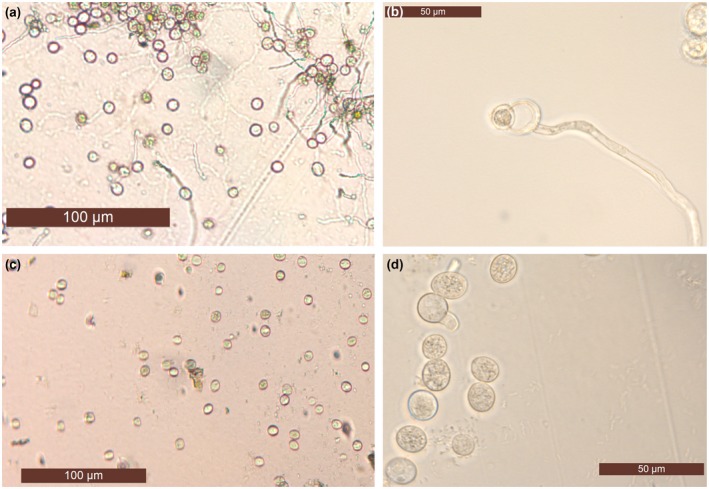
Germination is inhibited by antisense sRNA targeting the Hpa‐CesA3 gene. Hpa‐Emoy2 spores containing 0 or 20 µM antisense sRNA were placed on cellophane strips on MS medium and spore germination was examined using a Leica DM5500B light microscope after 48 h. Controls (a) and (b) produced long germination tubes, while spores incubated with 20 µM antisense sRNA (c) did not germinate or the germination tube was arrested (d) within the given time period.
Hpa‐CesA3 antisense sRNA‐mediated suppression of Hpa infection is not race specific
Hpa‐CesA3 antisense sRNA‐mediated suppression of Hpa infection was carried out using the Hpa‐Emoy2 isolate. To determine whether or not the suppression of infection we observed was isolate‐specific, we carried out a similar study using Hpa‐Cala2 isolate. Using BLASTN, we determined that the Emoy2 Hpa‐CesA3 gene had 99.97% identity to that of Cala2 and 100% identity at the sRNA target region (Fig. S5). Subsequently, we inoculated 7‐day‐old Arabidopsis seedlings with Hpa‐Cala2 spores mixed with and without 20 µM antisense sRNA. As expected, we observed normal sporulation in control seedlings while seedlings inoculated with spore suspension containing 20 µM antisense sRNA did not show any sporulation (data not shown), indicating Hpa‐CesA3 antisense sRNA‐mediated suppression of Hpa infection was not race‐specific.
Capping antisense sRNA is essential for suppression of Hpa infection
The experiments described above were carried out with 25 nt capped antisense sRNAs. To determine whether capping influenced the silencing of Hpa‐CesA3, we obtained an uncapped version of the same antisense sRNA and carried out similar inoculation studies. After 7 dpi, the control Arabidopsis seedlings and those seedlings inoculated with spores mixed with uncapped antisense 25 nt sRNA developed typical Hpa infection, resulting from normal sporulation and germination (Fig. 6a,b). In the same experiments, inoculations with spores mixed with capped antisense sRNA showed neither germination nor sporulation (Fig. 6c). These results reveal that sRNA capping is essential for sRNA biological activity in suppressing Hpa infection of plants.
Figure 6.

Capping sRNA has an effect on gene silencing. Arabidopsis seedlings were inoculated with Hpa‐Emoy2 spore suspension containing no sRNA (a), 20 µM uncapped antisense sRNA (b) and 20 µM capped antisense sRNA (c). Experiments were repeated five times with similar results. Samples were photographed at 7 days post‐inoculation.
sRNA length has an impact on Hpa‐CesA3 antisense sRNA‐mediated suppression of Hpa infection
The length of sRNA could influence the gene silencing (Vargason et al., 2003). In addition to the 25 nt sRNA, we tested the effect of 24 and 30 nt antisense Hpa‐CesA3 sRNAs on Hpa infection. Using 24 nt sRNAs, normal sporulation was observed on the seedlings inoculated with 20 µM sRNAs and plants became infected with Hpa. However, there was no sporulation on seedlings inoculated with spores containing 20 µM 30 nt antisense sRNAs and the inoculated Arabidopsis plants remained healthy (Fig. 7).
Figure 7.
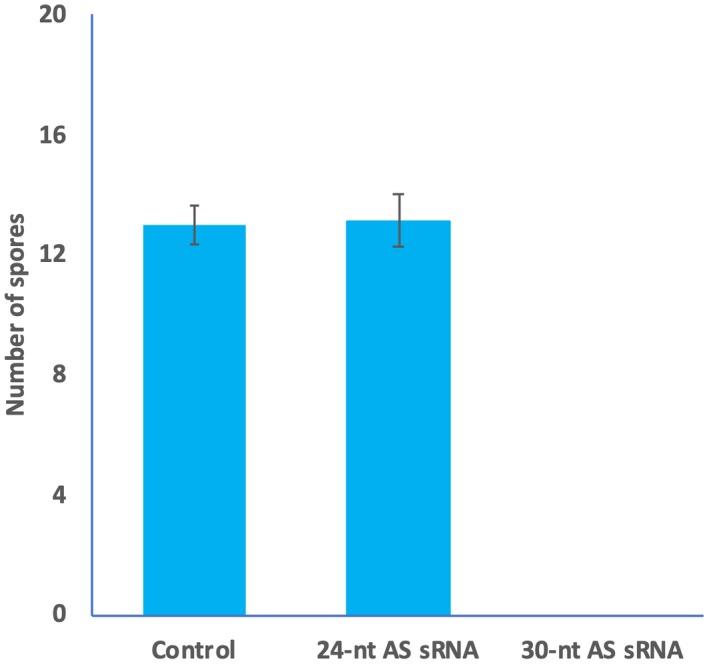
Length of sRNA has an effect on pathogen development. Arabidopsis seedlings were drop inoculated with Hpa‐Emoy2 spore suspension containing no sRNAs (control), 24 nt antisense (AS) sRNA and 30 nt antisense sRNAs. The inoculated seedlings from each sample were collected at 7 days post‐inoculation and placed in 250 µL H2O. The number of spores was counted using a haemocytometer. No spores were detected in samples inoculated with spore suspension containing 30 nt antisense sRNA. The average and standard error of three replicates are shown. The experiment was repeated five times with similar results.
Double‐stranded sRNA is more effective than single‐stranded sRNA in inhibiting infection
Since application of 5 and 10 µM antisense sRNA allowed reduced sporulation in infected plants (Fig. 2), we investigated whether double‐stranded (ds) sRNA would work better than the single‐stranded sRNA in inhibiting infection. We drop inoculated Arabidopsis seedlings with spores mixed with 0, 5, 10 and 20 µM 30‐nt dsRNA. At 7 dpi, Hpa sporulation was checked and while normal sporulation was observed in control seedlings, no sporulation was detected on seedlings inoculated with spore suspension containing 5, 10 and 20 µM 30‐nt dsRNA (Fig. 8). The experiment was repeated three times, each time with a minimum of three replicates, and similar results were obtained. Quantitative data analysis supported our observation (Table S2).
Figure 8.
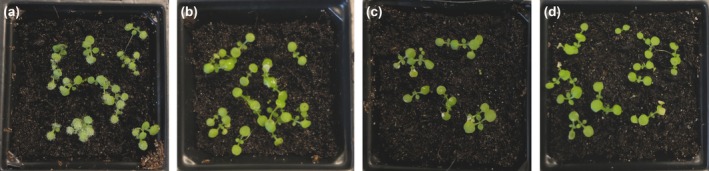
Double‐stranded (ds) sRNA is more effective in inhibiting infection than single‐stranded sRNA. Hpa‐Emoy2 spores were mixed with ds sRNA at 0, 5, 10 and 20 µM concentrations and 7‐day‐old Arabidopsis seedlings were drop inoculated. Seedlings were examined for sporulation at 7 days post‐inoculation. (a) Control (no sRNA), (b) seedlings inoculated with spores mixed with 5 µM ds sRNA, (c) seedlings inoculated with spores mixed with 10 µM ds sRNA and (d) seedlings inoculated with spores mixed with 20 µM ds sRNA. The inoculation experiments were repeated five times and similar observations were made. There was no sporulation in seedlings inoculated with spores mixed with 5, 10 or 20 µM ds sRNA.
Discussion
Using the Arabidopsis–Hyaloperonospora model system, we showed that targeting the Hpa‐CesA3 gene in Emoy2 and Cala2 isolates by exogenously applying gene‐specific sRNAs inhibits germination and hence infection of Arabidopsis.
We chose the Hpa‐CesA3 gene because cellulose is an important structural component of the cell wall of oomycetes (Raaymakers and Van den Ackerveken, 2016). Blum et al. (2012) investigated CesA3 genes in a total of 25 different oomycete species originating from the six ‘crown’ oomycete orders (Albuginales, Leptomitales, Peronosporales, Pythiales, Rhipidiales and Saprolegniales) for their sensitivity to the fungicide mandipropamid (MPD). Interestingly, those authors reported that only species belonging to the order of Peronosporales, of which Hpa is a member, were inhibited by this fungicide. Furthermore, Grenville‐Briggs et al. (2008) reported CesA3 to be the most strongly expressed gene during mycelial growth of Phytophthora, Saprolegnia and Pythium species. Working with P. infestans using a protoplast transfection strategy, Grenville‐Briggs et al. (2008) used in vitro‐generated long dsRNA to silence CesA genes. From their studies, they concluded that silencing these genes leads to disruption of cell walls surrounding appressoria and the inability to form functional appressoria. Interestingly, they reported that CesA3 is either less important or other CesA genes may be compensating for its loss of function (Grenville‐Briggs et al., 2008).
Being an obligate plant pathogen, Hpa is not amenable to the genetic transformation and manipulation reported for P. infestans (Zheng et al., 2014). Previously, we have tested several different methods, including electroporation, to obtain stable transgenic isolates and study gene functions. Although we observed marker gene expression in a few individual spores, we were unable to generate transgenic isolates or obtain uniformly transformed spore lines (N. Holton and M. Tör, unpublished data). Here, we used synthesized 25 nt sRNAs to silence the CesA3 gene in Hpa by simply mixing the sRNAs with spores and inoculating cotyledons of seedlings or applying to cellophane strips. In inoculation experiments with antisense sRNAs, we showed a dose‐dependent reduction in sporulation on the inoculated cotyledons. This clearly indicates that (a) exogenously applied single‐stranded and ds sRNAs can be taken up by the spores without electroporation or any other transfection method, (b) the single‐stranded sRNAs somehow bind to the native CesA3 RNA, forming a dsRNA and triggering the gene silencing machinery within the Hpa spores, (c) ds sRNA seems to be more effective in inhibiting infection than single‐stranded sRNA, (d) the silencing seems to be very effective, as Hpa is diploid and multinucleate, and (e) the CesA3 gene is essential for pathogenicity of Hpa. Use of sense and antisense DNA oligonucleotides served as controls.
Trypan blue staining of seedlings inoculated with a spore suspension containing 20 µM antisense sRNAs revealed a lack of infection, indicating the inhibition of pathogen development at the germination stage; this was confirmed by the germination assays, which showed 100% inhibition of germination. Our results are in contrast to the findings of Grenville‐Briggs et al. (2008) where silencing CesA genes in P. infestans resulted in abnormal appressorium development rather than the inhibition of germination. This may well have been due to the redundancy factor as P. infestans has a larger genome than Hpa (Haas et al., 2009).
Using the Hpa‐Cala2 genomic sequences (Woods‐Tör et al., 2018), we found that the CesA3 gene of Emoy2 has 99.97% identity to CesA3 from Cala2 and the designed sRNA was gene‐specific. In addition, the inhibition of sporulation of the Cala2 isolate by the sRNA confirmed that the method is effective and that this gene is necessary for infection.
Ideally, we would design an sRNA molecule that could inhibit infection by several oomycete species. However, gene silencing relies on a conserved nucleotide sequence and, although the domains of CesA3 are conserved at the amino acid level across some of the important oomycete species, this is not so at the nucleotide level; unfortunately, there is not a conserved region of the nucleotide sequence that is long enough to design a common sRNA. Nevertheless, the method we developed here can easily be adapted to other oomycete species using newly designed gene‐specific sRNAs.
It is well known that all eukaryotic mRNA contains a cap structure, an N7‐methylated guanosine linked to the first nucleotide of the RNA (Ramanathan et al., 2016). The cap has been reported to have several roles in cell viability, including promoting gene expression, mRNA stability and degradation, nuclear export of RNA and initiation of protein synthesis (Cowling, 2010). Recent studies also reported various cap structures in sRNAs in human cells (Abdelhamid et al., 2014), indicating that sRNAs can also go through a modification at their 5ʹ ends. Our inoculation studies clearly showed that the capping of sRNA was necessary for effective gene silencing. When Hpa spores are collected from infected seedlings, they are not derived from a sterile environment. Spore suspensions usually contain bacteria, tiny plant materials such as trichomes and other small substances. As the stability of exogenously applied sRNA in spore suspensions is very important, the cap structure of the sRNA may provide this required stability both outside and inside the spores.
In general, two classes of small non‐coding RNAs exist in plant cells: miRNAs (encoded by the genome) and siRNAs (derived from dsRNA produced by multiple sources) (Khraiwesh et al., 2012). In addition, the size of sRNAs in organisms can be different (Derbyshire et al., 2018). Several studies revealed that, in plants and animals, each sRNA (acting as a guide) binds to an Argonaute family protein and a sequence‐specific gene silencing ribonucleoprotein (RNP) is formed by base pairing between the sRNA and its target mRNA; this is known as the RNA‐induced silencing complex (RISC) in miRNA and siRNA pathways (Budak and Akpinar, 2015; Vargason et al., 2003; Wilson and Doudna, 2013).
In addition to the presence of a cap at the 5ʹ end of the sRNA, our results also showed the importance of the length of the sRNAs. We observed that 24 nt antisense sRNA did not inhibit infection, whereas it was completely inhibited by 25 or 30 nt antisense sRNAs. In a recent study, Åsman et al. (2016) co‐immunoprecipitated sRNAs with Argonaute proteins of the oomycete pathogen P. infestans and identified high enrichment of 24–26 nt sRNAs. In a similar study, Jia et al. (2017) sequenced sRNAs in another oomycete pathogen Phytophthora parasitica and reported that 25–26 nt sRNAs associate with efficient gene silencing in this pathogen. Although we do not know the exact reason of why 25 and 30 nt sRNAs silence Hpa‐CesA3 but not 24 nt, it is tempting to speculate that the 24 nt sRNAs may not be binding to the Hpa‐CesA3 transcript or may not be guiding the RISC complex to degrade CesA3 mRNA.
Application of sRNA is more advantageous than a transgenic approach. Using this system, it should be possible to study pathogen development and pathogenicity in obligate pathogens. Till now, effectors from obligate oomycetes such as Hpa have been studied either by bombardment or via a bacterial delivery system to plants (Bailey et al., 2011). An ideal method would be also to use reverse genetics to silence an effector gene within the pathogen and investigate whether this would alter pathogenicity. In addition, in some cases, effector genes in obligate pathogens are mapped to a locus where there are several genes within the interval. This system would allow rapid identification of the candidate gene. Using this simple method, we should now be able to study genes that are involved in pathogenicity and dissect different biological pathways of otherwise inaccessible obligate pathogens.
Experimental Procedures
Plant lines, pathogen isolates and propagation
Hyaloperonospora arabidopsidis isolates Emoy2 and Cala2 were maintained on A. thaliana Ws‐eds1 (Parker et al., 1996). Preparation of inoculum for experiments was performed as described previously (Tör et al., 2002). Sporulation was assessed 7 dpi, when the Hpa life cycle had been completed. To quantify sporulation, ten infected seedlings from each replicate were taken and placed into an Eppendorf tube containing 250 µl H2O. Samples were vortexed and conidiospores were counted using a haemocytometer.
sRNA and DNA oligonucleotide synthesis
The Hpa‐CesA3 gene (HpaG810051) was used as the target gene in this method. Sense and antisense sRNAs designed at various lengths were Hpa_CesA3_RNA_AS_24 5ʹ‐GCCGCAUCGCACGUACCUCAGUAC‐3', Hpa_CesA3_RNA_AS_25 5'‐GCCGCAUCGCACGUACCUCAGUACG‐3', Hpa_CesA3_RNA_S_25, 5'‐CGUACUGAGGUACGUGCGAUGCGGC‐3' and Hpa_CesA3_RNA_S_30 5ʹ‐GUCGUACUGAGGUACGUGCGAUGCGGCACU‐3ʹ. Hpa_CesA3_RNA_AS_30 5'‐AGUGCCGCAUCGCACGUACCUCAGUACGAC‐3' and double‐stranded sRNA were generated by mixing equal volume of Hpa_CesA3_RNA_S_30 and Hpa_CesA3_RNA_AS_30 oligos, heating at 95 ˚C for 5 min and allowing to anneal at room temperature for 20 min.
Similarly, sense and antisense DNA oligonucleotides were also designed to the same region. These were Hpa_CesA3_DNA_S_25 5ʹ‐GCCGCATCGCACGTACCTCAGTACG‐3ʹ and Hpa_CesA3_DNA_AS_25 5ʹ‐CGTACTGAGGTACGTGCGATGCGGC‐3ʹ. These were obtained as synthesized deoxyribonucleotides or ribonucleotides from Sigma (Gillingham, UK) or Eurofin (Ebersberg, Germany).
Application of sRNAs to pathogen spores and plant inoculations
Hpa spores were collected from infected A. thaliana Ws‐eds1 seedlings, washed twice in sterile distilled water and the spore concentration was adjusted to 5 × 104/mL using a haemocytometer. sRNAs were added to the spore suspension at a final concentration of 5, 10 and 20 µM and spores were subsequently drop inoculated onto 7‐day‐old seedlings. As a control, seedlings were also inoculated with spores in the same way with 20 µM DNA oligonucleotides or without sRNAs or DNA oligonucleotides. Seedlings were inspected from 3 dpi for sporulation. Sporulation was quantified as described above.
Spore germination assays
MS medium (Murashige and Skoog, 1962) was prepared with 4.3 g/L MS basal salt mixture powder (Sigma, M5524), agar (1.5%) (Sigma, A1296), sucrose (10 g/L) (Sigma, 84100), and distilled water. MS powder was dissolved in sterile distilled H2O, the pH was adjusted to 5.7 using 1 M NaOH/HCl, and agar and sugar were added. The medium was sterilized by autoclaving at 15 psi and 121 °C for 15 min. Approximately 20 mL of the medium was aliquoted into each sterile Petri dish in a laminar airflow unit.
Cellophane strips, 1.5 cm in length, were cut from plain transparent florists' cellophane and autoclaved in distilled water. After autoclaving, cellophane strips were placed onto the MS medium in Petri dishes under a laminar airflow and dishes were kept in the fridge for long‐term storage.
Hpa spores were collected, washed twice in sterile distilled water and the spore concentration was adjusted to 5 × 104 spores/mL using a haemocytometer. Approximately 10 µL spore suspension, with 0 or 20 µM antisense sRNA, was dropped onto each piece of cellophane. Plates were incubated with a 12 h light/12 h dark regime at 16 °C. Spores were examined under a light microscope 48 h after incubation and germinated spores were counted.
Staining plant tissues
Seedlings of infected and non‐inoculated controls were stained with a solution of phenol, lactic acid, glycerol and water (1:1:1:1) supplemented with 1 mg/mL trypan blue, decolorized in chloral hydrate and visualized under a compound microscope as described in Woods‐Tör et al. (2018).
Statistical analysis
For statistical analysis, paired Student’s t‐tests were performed on data obtained from plant infection assays.
Bioinformatics
IICB Genomics and Transcriptomics Resources (http://eumicrobedb.org) and the EnsemblProtist (http://protists.ensembl.org) database were used for information on Hpa. Web servers including InterPro (Quevillon et al., 2005) (http://www.ebi.ac.uk/interpro/) and Pfam (Punta et al., 2012) ((http://pfam.wustl.edu/) were used for the analysis of Hpa‐CesA3. BLAST (Altschul et al., 1997) was used to perform similarity‐search of nucleotide and amino acid sequences of Hpa‐CesA3 against oomycete and Arabidopsis sequences. Primer design was performed using Geneious v. 10.0 (Kearse et al., 2012).
Author Contributions
M.T. and Y.H. planned and designed the research. Ö.B., O.T., C.N. and M.T. conducted the laboratory work. M.T., Y.H. and H.B. analysed and interpreted the data and wrote the manuscript.
Conflict of Interest
The authors declare that there is no conflict of interest.
Supporting information
Fig. S1 Comparison of CesA3 amino acid sequences from different oomycete pathogens. Amino acid sequences of CesA3 proteins from Hyaloperonospora arabidopsidis (Hpa, M4BU64), Albugo candida (AFB77612), Albugo laibachii (CCA23182), Bremia lactucae (AFB20351), Phytophthora capsici (AFB20353), Phytophthora infestans (ABP9690), Plasmopara viticola (ADD84672) were aligned using Geneious v. 10. Black or dark grey boxes with white letters indicate identity or similarity to Hpa‐CesA3, respectively.
Fig. S2 Comparison of CesA3 nucleotide sequences from different oomycete pathogens. Nucleotide sequences of CesA3 gene from Hyaloperonospora arabidopsidis (Hpa), Albugo candida, Albugo laibachii, Bremia lactucae, Phytophthora capsici, Phytophthora infestans, Plasmopara viticola were aligned using Geneious v. 10. Black or dark grey boxes with white letters indicate identity or similarity to Hpa‐CesA3, respectively.
Fig. S3 Expression pattern of Hpa‐CesA3. Expression levels were represented as TPM (tags per million) of total reads mapped to Hyaloperonospora arabidopsidis genome. Data was acquired from Asai et al. (2018). Cs, conidiospore, dpi, days post‐inoculation.
Fig. S4 Sense and antisense DNA oligonucleotides do not inhibit sporulation. Arabidopsis seedlings were drop inoculated with Hpa‐Emoy2 spores containing 20 µM sense or antisense DNA oligonucleotides. Inoculated 10 seedlings from each sample were collected 7 days post‐inoculation and placed in 250 µL H2O. The number of spores was counted using a heamocytometer. Averages and standard errors of three replicates are shown. Experiment was repeated three times with similar results.
Fig. S5 Nucleotide sequence alignment of Hpa‐CesA3 from Emoy2 and Cala2 isolates. Sequences were aligned using Geneious v. 10. Black or dark grey boxes with white letters indicate identity or similarity to Hpa‐CesA3 from Emoy2, respectively. sRNA indicates the sequences where sRNAs were designed from.
Table S1 Spore germination assay using antisense sRNA.
Table S2 Number of spores detected in seedlings inoculated with dsRNA.
Acknowledgements
We are grateful to Dr Alison Woods‐Tör and Dr Volkan Çevik for critically reading the manuscript. Financial support from the Turkish Ministry of Agriculture to Ö.B. and Turkish Ministry of Education to O.T. is gratefully acknowledged. This work was also in part supported by grants from the Ministry of Science and Technology of the People's Republic of China (National Key R&D Program of China 2017YFE0110900) and the Ministry of Agriculture of the People’s Republic of China (2016ZX08009001‐004).
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.
References
- Abdelhamid, R.F. , Plessy, C. , Yamauchi, Y. , Taoka, M. , de Hoon, M. , Gingeras, T.R. , Isobe, T. and Carninci, P. (2014) Multiplicity of 5ʹ cap structures present on short RNAs Xing, Y., ed. PLoS One, 9, e102895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamian, M. , Ah‐Fong, A.M.V. , Davis, C. , Andreeva, K. and Judelson, H.S. (2016) Gene expression and silencing studies in Phytophthora infestans reveal infection‐specific nutrient transporters and a role for the nitrate reductase pathway in plant pathogenesis. PLoS Pathog. 12, e1006097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F. , Madden, T.L. , Schäffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, S. , Furzer, O.J. , Cevik, V. , Kim, D.S. , Ishaque, N. , Goritschnig, S. , Staskawicz, B.J. , Shirasu, K. and Jones, J.D.G. (2018) A downy mildew effector evades recognition by polymorphism of expression and subcellular localization. Nat. Commun. 9, 5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åsman, A.K.M. , Fogelqvist, J. , Vetukuri, R.R. and Dixelius, C. (2016) Phytophthora infestans Argonaute 1 binds microRNA and small RNAs from effector genes and transposable elements. New Phytol. 211, 993–1007. [DOI] [PubMed] [Google Scholar]
- Bailey, K. , Çevik, V. , Holton, N. , Byrne‐Richardson, J. , Sohn, K.H. , Coates, M. , Woods‐Tör, A. , Aksoy, H.M. , Hughes, L. , Baxter, L. and Jones, J.D. (2011) Molecular cloning of ATR5 Emoy2from Hyaloperonospora arabidopsidis, an avirulence determinant that triggers RPP5‐mediated defense in Arabidopsis . Mol. Plant-Microbe Interact. 24, 827–838. [DOI] [PubMed] [Google Scholar]
- Baum, J.A. , Bogaert, T. , Clinton, W. , Heck, G.R. , Feldmann, P. , Ilagan, O. , Johnson, S. , Plaetinck, G. , Munyikwa, T. , Pleau, M. , Vaughn, T. and Roberts, J. (2007) Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. [DOI] [PubMed] [Google Scholar]
- Blum, M. , Gamper, H.A. , Waldner, M. , Sierotzki, H. and Gisi, U. (2012) The cellulose synthase 3 (CesA3) gene of oomycetes: structure, phylogeny and influence on sensitivity to carboxylic acid amide (CAA) fungicides. Fungal Biol. 116, 529–542. [DOI] [PubMed] [Google Scholar]
- Brant, E.J. and Budak, H. (2018) Plant small non‐coding RNAs and their roles in biotic stresses. Front. Plant Sci. 9, 1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budak, H. and Akpinar, B.A. (2015) Plant miRNAs: biogenesis, organization and origins. Funct. Integr. Geno. 15, 523–531. [DOI] [PubMed] [Google Scholar]
- Burn, J.E. , Hocart, C.H. , Birch, R.J. , Cork, A.C. and Williamson, R.E. (2002) Functional analysis of the cellulose synthase genes CesA1, CesA2, and CesA3 in Arabidopsis. Plant Physiol. 129, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Zhang, X. , Fan, Y. , Li, B. , Ryabov, E. , Shi, N. , Zhao, M. , Yu, Z. , Qin, C. , Zheng, Q. and Zhang, P. (2018) A genetic network for systemic RNA silencing in plant. Plant Physiol. 176, 2700–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates, M.E. and Beynon, J.L. (2010) Hyaloperonospora arabidopsidis as a pathogen model. Annu. Rev. Phytopathol. 48, 329–345. [DOI] [PubMed] [Google Scholar]
- Cowling, V.H. (2010) Regulation of mRNA cap methylation. Biochem. J. 425, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalakouras, A. , Wassenegger, M. , McMillan, J.N. , Cardoza, V. , Maegele, I. , Dadami, E. , Runne, M. , Krczal, G. and Wassenegger, M. (2016) Induction of silencing in plants by high‐pressure spraying of in vitro‐synthesized small RNAs. Front. Plant Sci. 7, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y. , Wang, J. , Tung, J. , Liu, D. , Zhou, Y. , He, S. , Du, Y. , Baker, B. and Li, F. (2018) A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 14, e1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire, M.C. , Mbengue, M. , Barascud, M. , Navaud, O. and Raffaele, S. (2018) Small RNAs from the plant pathogenic fungus Sclerotinia sclerotiorum highlight candidate host target genes associated with quantitative disease resistance. bioRxiv. 354076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugelstad, J. (2008) Cellulose biosynthesis in oomycetes. Plant Cell, 20, 720–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajulu, M. , Epstein, L. , Wroblewski, T. and Michelmore, R.W. (2015) Host‐induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnol. J. 13, 875–883. [DOI] [PubMed] [Google Scholar]
- Grenville‐Briggs, L.J. , Anderson, V.L. , Fugelstad, J. , Avrova, A.O. , Bouzenzana, J. , Williams, A. , Wawra, S. , Whisson, S.C. , Birch, P.R. , Bulone, V. and van West, P. (2008) Cellulose synthesis in Phytophthora infestans is required for normal appressorium formation and successful infection of potato. Plant Cell, 20, 720–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H.Y. , Handsaker, R.E. , Cano, L.M. , Grabherr, M. , Kodira, C.D. , Raffaele, S. , Torto‐Alalibo, T. , Bozkurt, T.O. , Ah‐Fong, A.M.V. , Alvarado, L. , Anderson, V.L. , Armstrong, M.R. , Avrova, A. , Baxter, L. , Beynon, J. , Boevink, P.C. , Bollmann, S.R. , Bos, J.I.B. , Bulone, V. , Cai, G. , Cakir, C. , Carrington, J.C. , Chawner, M. , Conti, L. , Costanzo, S. , Ewan, R. , Fahlgren, N. , Fischbach, M.A. , Fugelstad, J. , Gilroy, E.M. , Gnerre, S. , Green, P.J. , Grenville‐Briggs, L.J. , Griffith, J. , Grünwald, N.J. , Horn, K. , Horner, N.R. , Hu, C.‐H. , Huitema, E. , Jeong, D.‐H. , Jones, A.M.E. , Jones, J.D.G. , Jones, R.W. , Karlsson, E.K. , Kunjeti, S.G. , Lamour, K. , Liu, Z. , Ma, L. , MacLean, D. , Chibucos, M.C. , McDonald, H. , McWalters, J. , Meijer, H.J.G. , Morgan, W. , Morris, P.F. , Munro, C.A. , O’Neill, K. , Ospina‐Giraldo, M. , Pinzón, A. , Pritchard, L. , Ramsahoye, B. , Ren, Q. , Restrepo, S. , Roy, S. , Sadanandom, A. , Savidor, A. , Schornack, S. , Schwartz, D.C. , Schumann, U.D. , Schwessinger, B. , Seyer, L. , Sharpe, T. , Silvar, C. , Song, J. , Studholme, D.J. , Sykes, S. , Thines, M. , van de Vondervoort, P.J.I. , Phuntumart, V. , Wawra, S. , Weide, R. , Win, J. , Young, C. , Zhou, S. , Fry, W. , Meyers, B.C. , van West, P. , Ristaino, J. , Govers, F. , Birch, P.R.J. , Whisson, S.C. , Judelson, H.S. and Nusbaum, C. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461, 393–398. [DOI] [PubMed] [Google Scholar]
- Holub, E.B. (2007) Natural variation in innate immunity of a pioneer species. Curr. Opin. Plant Biol. 10, 415–424. [DOI] [PubMed] [Google Scholar]
- Huang, G. , Allen, R. , Davis, E.L. , Baum, T.J. and Hussey, R.S. (2006) Engineering broad root‐knot resistance in transgenic plants by RNAi silencing of a conserved and essential root‐knot nematode parasitism gene. Proc. Natl. Acad. Sci. USA. 103, 14302–14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Yang, M. and Zhang, X. (2016) The function of small RNAs in plant biotic stress response. J. Integr. Plant Biol. 58, 312–327. [DOI] [PubMed] [Google Scholar]
- Jia, J. , Lu, W. , Zhong, C. , Zhou, R. , Xu, J. , Liu, W. , Gou, X. , Wang, Q. , Yin, J. , Xu, C. and Shan, W. (2017) The 25–26 nt small RNAs in Phytophthora parasitica are associated with efficient silencing of homologous endogenous genes. Front. Microbiol. 8, 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S. (2003) Molecular genetics of pathogenic oomycetes. Eukaryot. Cell, 2, 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S. , Furzer, O. , Jones, J.D.G. , Judelson, H.S. , Ali, G.S. , Dalio, R.J. , Roy, S.G. , Schena, L. , Zambounis, A. , Panabières, F. and Cahill, D. (2014) The top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse, M. , Moir, R. , Wilson, A. , Stones‐Havas, S. , Cheung, M. , Sturrock, S. , Buxton, S. , Cooper, A. , Markowitz, S. , Duran, C. , Thierer, T. , Ashton, B. , Meintjes, P. and Drummond, A. (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28, 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh, B. , Zhu, J.‐K. and Zhu, J. (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta. 1819, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, E. and Slusarenko, A. (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell, 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, A. , Kumar, N. , Weber, L. , Keller, H. , Imani, J. and Kogel, K.‐H. (2013) Host‐induced gene silencing of cytochrome P450 lanosterol C14α‐demethylase–encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. 110, 19324–19329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, A. , Biedenkopf, D. , Furch, A. , Weber, L. , Rossbach, O. , Abdellatef, E. , Linicus, L. , Johannsmeier, J. , Jelonek, L. , Goesmann, A. , Cardoza, V. , McMillan, J. , Mentzel, T. and Kogel, K.‐H. (2016) An RNAi‐based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 12, e1005901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, A. , Stein, E. and Kogel, K.‐H. (2018) RNA‐based disease control as a complementary measure to fight fusarium fungi through silencing of the azole target cytochrome P450 lanosterol C‐14 α‐demethylase. Eur. J. Plant Pathol. 303, 1–8. [Google Scholar]
- Li, S. , Castillo‐González, C. , Yu, B. and Zhang, X. (2017) The functions of plant small RNAs in development and in stress responses. Plant J. 90, 654–670. [DOI] [PubMed] [Google Scholar]
- Mcleod, A. , Fry, B.A. , Zuluaga, A.P. , Myers, K.L. and Fry, W.E. (2008) Toward improvements of oomycete transformation protocols. J. Eukaryot. Microbiol. 55, 103–109. [DOI] [PubMed] [Google Scholar]
- Mitter, N. , Worrall, E.A. , Robinson, K.E. , Li, P. , Jain, R.G. , Taochy, C. , Fletcher, S.J. , Carroll, B.J. , Lu, G.M. and Xu, Z.P. (2017) Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants, 3, 191. [DOI] [PubMed] [Google Scholar]
- Murashige, T. and Skoog, F. (1962) A Revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant, 15, 473–497. [Google Scholar]
- Nejat, N. and Mantri, N. (2018) Emerging roles of long non‐coding RNAs in plant response to biotic and abiotic stresses. Crit. Rev. Biotechnol. 38, 93–105. [DOI] [PubMed] [Google Scholar]
- Nowara, D. , Gay, A. , Lacomme, C. , Shaw, J. , Ridout, C. , Douchkov, D. , Hensel, G. , Kumlehn, J. and Schweizer, P. (2010) HIGS: host‐induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis . Plant Cell, 22, 3130–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E. , Holub, E.B. , Frost, L.N. , Falk, A. , Gunn, N.D. and Daniels, M.J. (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell, 8, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta, M. , Coggill, P.C. , Eberhardt, R.Y. , Mistry, J. , Tate, J. , Boursnell, C. , Pang, N. , Forslund, K. , Ceric, G. , Clements, J. , Heger, A. , Holm, L. , Sonnhammer, E.L.L , Eddy, S.R , Bateman, A. and Finn, R.D . (2012) The Pfam protein families database. Nucleic Acids Res. 40, D290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, C. , Li, B. , Fan, Y. , Zhang, X. , Yu, Z. , Ryabov, E. , Zhao, M. , Wang, H. , Shi, N. , Zhang, P. , Jackson, S. , Tör, M. , Cheng, Q. , Liu, Y. , Gallusci, P. and Hong, Y. (2017) Roles of dicer‐like proteins 2 and 4 in intra‐and intercellular antiviral silencing. Plant Physiol. 174, 1067–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon, E. , Silventoinen, V. , Pillai, S. , Harte, N. , Mulder, N. , Apweiler, R. and Lopez, R. (2005) InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaymakers, T.M. and Van den Ackerveken, G. (2016) Extracellular recognition of oomycetes during biotrophic infection of plants. Front. Plant Sci. 7, 5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan, A. , Robb, G.B. and Chan, S.‐H. (2016) mRNA capping: biological functions and applications. Nucleic Acids Res. 44, 7511–7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva, M. , de Bruijn, I. , Grenville‐Briggs, L. , McLaggan, D. , Willems, A. , Bulone, V. and van West, P. (2014) Functional characterization of a tyrosinase gene from the oomycete Saprolegnia parasitica by RNAi silencing. Fungal Biol. 118, 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y. and Thomma, B.P.H.J. (2018) Host‐induced gene silencing compromises Verticillium wilt in tomato and Arabidopsis. Mol. Plant Pathol. 19, 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tör, M. , Gordon, P. , Cuzick, A. , Eulgem, T. , Sinapidou, E. , Mert‐Türk, F. , Can, C. , Dangl, J.L. and Holub, E.B. (2002) Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell, 14, 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason, J.M. , Szittya, G. , Burgyán, J. and Hall, T.M.T. (2003) Size selective recognition of siRNA by an RNA silencing suppressor. Cell, 115, 799–811. [DOI] [PubMed] [Google Scholar]
- Wang, M. and Jin, H. (2017) Spray‐induced gene silencing: a powerful innovative strategy for crop protection. Trends Microbiol. 25, 4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg, A. , Wang, M. , Lin, F.‐M. , Zhao, H. , Zhang, Z. , Kaloshian, I. , Huang, H.‐D. and Jin, H. (2013) Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science, 342, 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R.C. and Doudna, J.A. (2013) Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 42, 217–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods‐Tör, A. , Studholme, D.J. , Çevik, V. , Telli, O. , Holub, E.B. and Tör, M. (2018) A suppressor/avirulence gene combination in Hyaloperonospora arabidopsidis determines race specificity in Arabidopsis thaliana . Front. Plant Sci. 9, 1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T. , Zhao, Y.‐L. , Zhao, J.‐H. , Wang, S. , Jin, Y. , Chen, Z.Q. , Fang, Y.Y. , Hua, C.L. , Ding, S.W. and Guo, H.S. (2016) Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants, 2, 1–6. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Lai, T. , Zhang, P. , Zhang, X. , Yuan, C. , Jin, Z. , Li, H. , Yu, Z. , Qin, C. , Tör, M. , Ma, P. , Cheng, Q. and Hong, Y. (2019) Mini review: revisiting mobile RNA silencing in plants. Plant Sci. 278, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X. , McLellan, H. , Fraiture, M. , Liu, X. , Boevink, P.C. , Gilroy, E.M. , Chen, Y. , Kandel, K. , Sessa, G. , Birch, P.R.J. and Brunner, F. (2014) Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22‐triggered immunity. PLoS Pathog. 10, e1004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Comparison of CesA3 amino acid sequences from different oomycete pathogens. Amino acid sequences of CesA3 proteins from Hyaloperonospora arabidopsidis (Hpa, M4BU64), Albugo candida (AFB77612), Albugo laibachii (CCA23182), Bremia lactucae (AFB20351), Phytophthora capsici (AFB20353), Phytophthora infestans (ABP9690), Plasmopara viticola (ADD84672) were aligned using Geneious v. 10. Black or dark grey boxes with white letters indicate identity or similarity to Hpa‐CesA3, respectively.
Fig. S2 Comparison of CesA3 nucleotide sequences from different oomycete pathogens. Nucleotide sequences of CesA3 gene from Hyaloperonospora arabidopsidis (Hpa), Albugo candida, Albugo laibachii, Bremia lactucae, Phytophthora capsici, Phytophthora infestans, Plasmopara viticola were aligned using Geneious v. 10. Black or dark grey boxes with white letters indicate identity or similarity to Hpa‐CesA3, respectively.
Fig. S3 Expression pattern of Hpa‐CesA3. Expression levels were represented as TPM (tags per million) of total reads mapped to Hyaloperonospora arabidopsidis genome. Data was acquired from Asai et al. (2018). Cs, conidiospore, dpi, days post‐inoculation.
Fig. S4 Sense and antisense DNA oligonucleotides do not inhibit sporulation. Arabidopsis seedlings were drop inoculated with Hpa‐Emoy2 spores containing 20 µM sense or antisense DNA oligonucleotides. Inoculated 10 seedlings from each sample were collected 7 days post‐inoculation and placed in 250 µL H2O. The number of spores was counted using a heamocytometer. Averages and standard errors of three replicates are shown. Experiment was repeated three times with similar results.
Fig. S5 Nucleotide sequence alignment of Hpa‐CesA3 from Emoy2 and Cala2 isolates. Sequences were aligned using Geneious v. 10. Black or dark grey boxes with white letters indicate identity or similarity to Hpa‐CesA3 from Emoy2, respectively. sRNA indicates the sequences where sRNAs were designed from.
Table S1 Spore germination assay using antisense sRNA.
Table S2 Number of spores detected in seedlings inoculated with dsRNA.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.


