Summary
The bacterial stringent response is a response to nutrition deprivation and other stress conditions. In Gram‐negative bacteria, this process is mediated by the small signal molecules guanosine pentaphosphate pppGpp and guanosine tetraphosphate ppGpp (collectively referred to as (p)ppGpp), and the RNA polymerase‐binding transcription factor DksA. The (p)ppGpp synthetase RelA and the bifunctional (p)ppGpp synthase/hydrolase SpoT are responsible for cellular (p)ppGpp levels. Here, we investigated the roles of DksA and (p)ppGpp in the virulence traits of Xanthomonas citri subsp. citri (Xcc), the causal agent of citrus canker. ΔdksA and (p)ppGpp‐deficient ΔspoTΔrelA strains caused reduced virulence and compromised growth in host plants, indicating that DksA and (p)ppGpp are required for full virulence of Xcc. To characterize the effect of stringent response regulators on gene expression, RNA‐seq was conducted using ΔdksA and ΔspoTΔrelA mutant strains grown in hrp‐inducing XVM2 medium. Transcriptome analyses showed that DksA and (p)ppGpp repressed the expression of genes encoding tRNAs, ribosome proteins, iron acquisition and flagellum assembly, and enhanced the expression of genes for histidine metabolism, type 3 secretion system (T3SS), type 2 secretion system (T2SS) and TonB‐dependent transporters. Phenotypically, the ΔdksA and ΔspoTΔrelA strains displayed altered motility, enhanced siderophore production and were unable to cause the hypersensitive response on non‐host plants. In conclusion, stringent response regulators DksA and (p)ppGpp play an important role in virulence, nutrition uptake and host adaptation of Xcc.
Keywords: citrus, host adaptation, RNA‐seq, stringent response, type 3 secretion system, Xanthomonas citri
Introduction
Xanthomonas spp. cause diseases in approximately 400 species of plant hosts, including many economically important crops (Ryan et al., 2011). Xanthomonas citri subsp. citri (Xcc) is the causal agent of citrus canker, one of the most destructive bacterial diseases in citrus (Vojnov et al., 2010). Xcc can be disseminated via wind‐driven rain and invades the host leaf mesophyll tissue through stomata or wounds (Brunings and Gabriel, 2003; Ference et al., 2017). To survive and multiply, Xcc needs to overcome the stress from both the non‐host and host environments such as nutrient limitations in the phyllosphere (Fatima and Senthil‐Kumar, 2015) and the host immunity response (Dodds and Rathjen, 2010).
Gram‐negative bacterial pathogens, including Xcc, deliver numerous effectors into the host cell via the type 3 secretion system (T3SS) to manipulate host signalling, suppress immune responses and/or induce plant susceptibility genes (Jacques et al., 2016; Tsuge et al., 2014; White et al., 2009). The T3SS is encoded by hrp (hypersensitive response and pathogenicity) genes and the regulation of hrp genes in Xanthomonas mainly depends on two key transcriptional regulators, HrpG and HrpX (Wengelnik and Bonas, 1996; Wengelnik et al., 1996). HrpG positively regulates the expression of HrpX, which in turn binds to a conserved motif (PIP box, plant‐inducible promoter) at promoter regions of some T3SS genes, effector genes and other virulence‐related genes (Guo et al., 2011; Koebnik et al., 2006). Several virulence regulators were found to act upstream of HrpG and/or HrpX in Xanthomonas, including post‐transcriptional regulator RsmA/CsrA (Andrade et al., 2014), Lon protease (Zhou et al., 2018), sensor kinase HpaS (Li et al., 2014), LysR‐type transcriptional activator GamR (Rashid et al., 2016) and small noncoding RNA sX13 (Schmidtke et al., 2013). In addition, the virulence of Xanthomonas is also controlled by two‐component systems, quorum sensing (QS) and cyclic di‐GMP (Büttner and Bonas, 2010). Beyond the T3SS induction during infection, little is known about how Xcc coordinates virulence‐associated regulatory networks.
The bacterial stringent response is a response to nutrition starvation and other stress conditions. During the stringent response, cellular resources are relocated from the synthesis of stable RNAs (tRNA and rRNA) and ribosomal proteins to promote the expression of components crucial for stress resistance (Dalebroux and Swanson, 2012; Potrykus and Cashel, 2008). This process is mainly regulated by the small signal molecules guanosine pentaphosphate pppGpp and guanosine tetraphosphate ppGpp (collectively referred to as (p)ppGpp for simplicity), and the RNA polymerase‐binding transcription factor DksA (Haugen et al., 2008). DksA, belonging to the DksA/TraR superfamily, binds to RNA polymerase (RNAP) through the secondary channel and augments the effect of (p)ppGpp on transcription initiation (Blankschien et al., 2009; Paul et al., 2004; Perederina et al., 2004). The cellular (p)ppGpp level is controlled by protein family RelA‐SpoT homologue (RSH), small alarmone synthetase (SAS) and small alarmone hydrolase (SAH) (Atkinson et al., 2011). Gram‐negative bacteria generally have two long‐RSH proteins: RelA (synthetase) and SpoT (synthase and hydrolase) (Irving and Corrigan, 2018). (p)ppGpp can be synthesized by both RelA and SpoT using GTP/GDP and ATP and degraded only by SpoT, but not by RelA, into GTP/GDP and pyrophosphate (PPi) due to the loss of hydrolytic activity in the RelA (p)ppGpp hydrolysis (HD) domain (Hauryliuk et al., 2015). The activity of RSH homologues in Escherichia coli could be regulated by small ligand, heterologous protein or at the transcriptional level (Irving and Corrigan, 2018).
Although the specific mechanism may vary between species, there are at least two paradigms for the broad effect of (p)ppGpp on cellular processes (Gourse et al., 2018). In E. coli, both (p)ppGpp and DksA can directly bind to RNAP and this interaction positively or negatively regulates the transcription initiation depending on the kinetics of promoter open complex (Haugen et al., 2008; Ross et al., 2016). In addition, (p)ppGpp also regulates the global cellular process in a RNAP‐independent manner by directly interacting with enzymes or transcription factors and therefore modulating their activities (Dalebroux and Swanson, 2012).
Transcriptome analysis showed that in E. coli the stringent response regulates more than 10% of all genes involved in synthesis of tRNA, ribosome proteins, flagella, fatty acids, amino acids, transporters and many other central cellular components (Aberg et al., 2009; Durfee et al., 2008; Traxler et al., 2008). Although the synergistic effect between DksA and (p)ppGpp exists in most cases, there is evidence showing that they may have divergent and even opposite effects on specific traits or gene expression (Lyzen et al., 2009, 2016; Magnusson et al., 2007). It is notable that even the basal level of (p)ppGpp still has a regulatory effect under balanced growth conditions (Gaca et al., 2013).
A stringent response not only helps bacteria adapt to a nutrient‐deprived environment, but also enhances bacterial virulence (Dalebroux et al., 2010). For plant pathogens, several reports showed the effect of stringent response regulators (p)ppGpp and/or DksA on virulence in plant pathogens like Erwinia amylovora (Ancona et al., 2015), Pectobacterium atrosepticum (Bowden et al., 2013) and Pseudomonas syringae (Chatnaparat et al., 2015a,b). The effect of stringent response regulators on virulence has yet to be studied in Xanthomonas genus. In this study, we examined stringent response regulators by generating ΔdksA and (p)ppGpp‐deficient ΔspoTΔrelA strains of Xcc. Using whole transcriptome analysis, virulence tests and phenotypic characterization, our study provides new insights into the functions of stringent response regulators as well as the positive and negative interplay between (p)ppGpp and DksA in Xanthomonas species.
Results
DksA and SpoT are required for the full pathogenicity of Xcc
A homology search showed that Xcc contains stringent response regulatory genes including dksA (locus tag XAC2358), relA (XAC3113) and spoT (XAC3393). BLASTP showed that DksA, RelA and SpoT from Xcc displayed 46.88%, 43.00% and 48.06% identity in amino acid sequence, respectively, with the homologous genes from E. coli K‐12 MG1655. DksA belongs to the DksA/TraR family and contains a zinc finger domain at the C‐terminal region, a typical feature of DksA/TraR family members (Blankschien et al., 2009) (Fig. S1). Both RelA and SpoT contain a (p)ppGpp synthesis (SYNTH) domain, a hydrolysis (HD) domain, a TGS (ThrRS, GTPase and SpoT) domain and an ACT (aspartokinase, chorismate mutase and TyrA) domain (Fig. S1).
Previously, a high‐throughput screen for genes of Xcc involved in citrus canker symptom development in our laboratory showed that a Tn5 transposon insertion mutant (dksA:Tn5) was unable to induce canker symptoms (Yan and Wang, 2012). To explore the effect of stringent response regulators on pathogenicity, three deletion mutants, ΔdksA, ΔrelA and double mutant ΔspoTΔrelA, were produced by double crossover homologous recombination. We were unable to obtain a spoT deletion mutant despite multiple attempts, which is consistent with previous studies in failing to generate the spoT mutant in E. coli. It was speculated that the hyper‐accumulation of (p)ppGpp due to disruption of spoT is lethal to bacteria (Xiao et al., 1991).
To test the pathogenicity, wild‐type strain Xcc306, ΔdksA, ΔrelA, double mutant ΔspoTΔrelA and complemented strains carrying recombinant plasmids were inoculated by syringe infiltration onto young Valencia sweet orange (Citrus sinensis) leaves. The ΔdksA strain with/without empty plasmid did not produce canker symptoms whereas the wild‐type strain and complemented strain caused typical canker symptoms characterized by necrosis with a corky appearance (Fig. 1A). While we did not observe any difference in symptoms between Xcc306 and ΔrelA, ΔspoTΔrelA induced much reduced canker symptoms (Fig. 1B). The complemented strain ΔspoTΔrelA (spoT) restored the canker symptoms (Fig. 1B). Consistent with reduced symptom development, bacterial growth of ΔdksA and ΔspoTΔrelA was substantially reduced in planta compared to that of the wild‐type Xcc306 and complemented strains (Fig. 1C).
Figure 1.
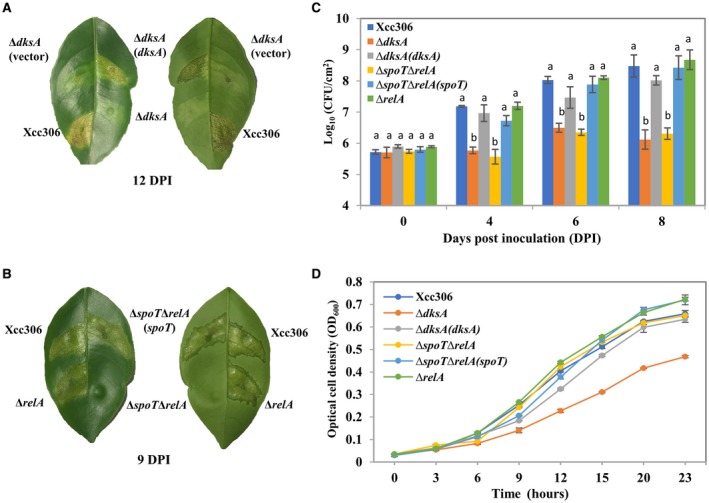
DksA and (p)ppGpp contribute to pathogenicity and bacterial growth in planta. (A, B) Bacterial cultures (108 CFU/mL) of the indicated strains were inoculated into sweet orange leaves. Pictures were taken at the indicated time points. (C) Bacterial cultures (107 CFU/mL) of the indicated strains were inoculated into sweet orange leaves and bacterial populations were determined at 0, 4, 6 and 8 days post‐inoculation. (D) Bacterial growth was monitored in the XVM2 medium. (C, D) Values represent means ± SD (n = 3). One‐way ANOVA with post hoc Tukey HSD test was applied to compare multiple strains. Statistical significance means P < 0.01. All the experiments were repeated three times independently with similar results.
Bacterial growth in vitro was also tested in the defined XVM2 medium. The XVM2 medium was reported to mimic the environment of plant intracellular spaces and induce hrp gene expression (Astua‐Monge et al., 2005; Wengelnik et al., 1996). ΔdksA growth rate was significantly lower than that of the wild‐type strain in the XVM2 medium (Fig. 1D), which might partially explain the reduced canker symptoms of the ΔdksA mutant. Complementation of ΔdksA through plasmid expression of DksA successfully recovered the growth of ΔdksA in the XVM2 medium (Fig. 1D). The ΔrelA and ΔspoTΔrelA mutants displayed similar or slightly enhanced growth in the XVM2 medium (Fig. 1D). To sum up, our data show that stringent response regulatory genes dksA and spoT are required for the pathogenicity of Xcc.
Transcriptome analysis of ΔdksA and ΔspoTΔrelA mutants compared to the wild‐type Xcc
To better understand the function of stringent response regulators in virulence and other cellular processes, RNA‐seq was conducted to determine the transcription profile of wild‐type Xcc306, ΔdksA and ΔspoTΔrelA strains. Bacteria were grown in the XVM2 medium and total RNA was extracted from the mid‐log phase (OD600 = 0.35) cultures. In total, nine RNA samples (three biological repeats for each strain) were sent for RNA‐seq (sequencing data information is provided in Table S1). To explore the similarities and dissimilarities between samples, principal component analysis (PCA) was performed. The three biological repeats of each strain clustered together into distinct groups from other strains, indicating that the major variation comes from the difference between each strain (Fig. 2A). Similarly, the heatmap shows that three biological repeats are clustered together (Fig. S2). In addition, RT‐qPCR was conducted to confirm the RNA‐seq data based on selected genes, which shows that log2‐transformed values derived from both methods are highly correlated (Fig. S3).
Figure 2.
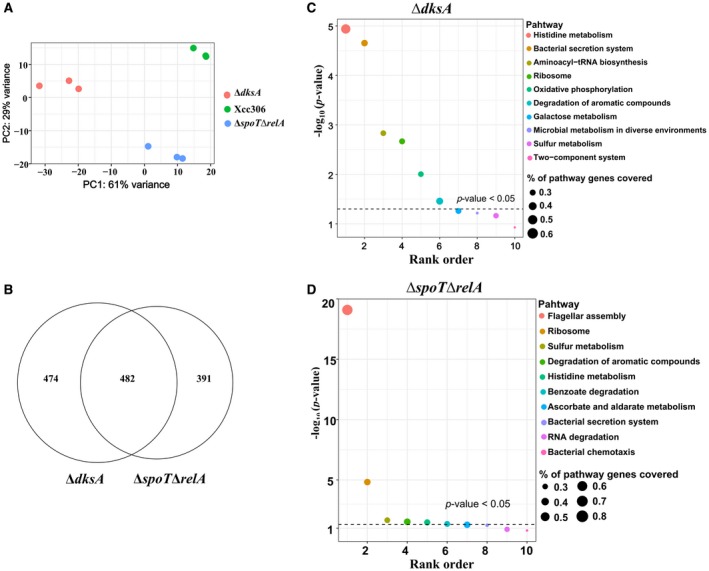
Functional enrichment analysis of differentially expressed genes (DEGs) in the ΔdksA and ΔspoTΔrelA strains compared to wild‐type Xanthomonas citri subsp. citri. (A) Principal component analysis (PCA) of nine bacterial samples. (B) Venn diagram of overlapping DEGs between ΔdksA and ΔspoTΔrelA. (C, D) Pathway enrichment analysis showing the over‐represented pathways for ΔdksA (C) and ΔspoTΔrelA (D). The dashed lines indicate the threshold of statistical significance (P = 0.05).
Differentially expressed genes (DEGs) were defined as genes with the absolute value of log2 fold change (FC) equal to or more than 2 and adjusted P‐value equal to or smaller than 0.01. The The differential gene expression test revealed that in the ΔspoTΔrelA mutant, 873 genes were identified as DEGs (Fig. 2B) and among them 179 genes were up‐regulated and 694 were down‐regulated (Table S2). In the ΔdksA mutant, 956 genes were identified as DEGs (Fig. 2B) and among them 196 genes were up‐regulated and 760 were down‐regulated (Table S3). There are approximately 4500 genes in the Xcc genome, of which 20% were regulated in the ΔdksA and ΔspoTΔrelA mutants. More than 50% of DEGs in both mutants (482 genes) are common, which supports the claim that DksA is the cofactor of (p)ppGpp in many cases (Fig. 2B).
To discover functionality‐related genes regulated by DksA and (p)ppGpp, DEGs were grouped into different functional categories based on the COG database (Fig. S4). The Fisher exact test was conducted to identify the over‐represented functional groups (P < 0.05) and showed that both DksA and (p)ppGpp are involved in cell motility, inorganic ion transport and metabolism, and intracellular trafficking and secretion (Table 1). To identify the specific pathways regulated by DksA and (p)ppGpp, pathway enrichment analysis was performed based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa and Goto, 2000). The results showed that the over‐represented pathways in both mutant strains included histidine metabolism, bacterial secretion systems, ribosome biosynthesis and degradation of aromatic compounds (P value < 0.05) (Fig. 2C,D). Flagellar assembly was the most over‐represented pathway in ΔspoTΔrelA rather than ΔdksA, which indicates the different effect on flagellar assembly between (p)ppGpp and DksA (Fig. 2C,D). To sum up, these results highlight the similarities in the regulons of DksA and (p)ppGpp, and reveal their potential roles.
Table 1.
Functional enrichment analysis based on COG data
| Functional classification (COGs) | Symbol | P‐value (ΔdksA) | P‐value(ΔspoTΔrelA) |
|---|---|---|---|
| RNA processing and modification | A | NS | NS |
| Chromatin structure and dynamics | B | NS | NS |
| Energy production and conversion | C | NS | NS |
| Cell cycle control, mitosis and meiosis | D | NS | NS |
| Amino acid transport and metabolism | E | NS | NS |
| Nucleotide transport and metabolism | F | NS | NS |
| Carbohydrate transport and metabolism | G | 0.0432 | NS |
| Coenzyme transport and metabolism | H | NS | NS |
| Lipid transport and metabolism | I | NS | NS |
| Translation, ribosomal structure and biogenesis | J | NS | NS |
| Transcription | K | NS | NS |
| Replication, recombination and repair | L | NS | NS |
| Cell wall/membrane biogenesis | M | NS | NS |
| Cell motility | N | 0.0209 | 5.5E‐05 |
| Post‐translational modification, protein turnover, chaperones | O | NS | NS |
| Inorganic ion transport and metabolism | P | 0.0004 | 0.0068 |
| Secondary metabolites biosynthesis, transport and catabolism | Q | NS | NS |
| General function prediction only | R | NS | NS |
| Function unknown | S | NS | NS |
| Signal transduction mechanisms | T | NS | NS |
| Intracellular trafficking and secretion | U | 0.0054 | 0.0056 |
| Defence mechanisms | V | NS | NS |
| Extracellular structures | W | NS | NS |
| Cytoskeleton | Z | NS | NS |
NS, no statistical significance.
DksA and (p)ppGpp repress tRNA and ribosome protein biosynthesis and activate histidine metabolism
A hallmark of the stringent response is the inhibition of stable RNA and ribosome protein biosynthesis (Lemke et al., 2011; Paul et al., 2004). KEGG database‐based enrichment analysis suggested that ribosome biosynthesis was over‐represented in the DksA and (p)ppGpp regulons. We analysed the transcript abundance of genes involved in tRNA and ribosome biosynthesis of Xcc. The relative gene expression levels of 54 tRNA‐encoding genes and 55 ribosome protein‐encoding genes in the ΔdksA and ΔspoTΔrelA mutants compared to the wild‐type Xcc were listed (Tables S4 and S5). Heatmap analysis displayed the relative gene expression profile for both ΔdksA and ΔspoTΔrelA strains compared to the wild‐type Xcc (Fig. 3A). Specifically, 24 of the 54 tRNA genes were up‐regulated DEGs (log2FC > 2) in the ΔdksA strain (Fig. 3B). Similarly, 10 of the 54 tRNA genes were up‐regulated DEGs (log2FC > 2) in the ΔspoTΔrelA strain (Fig. 3B). For ribosome protein‐encoding genes, 20 and 28 were up‐regulated DEGs (log2FC > 2) in ΔdksA and ΔspoTΔrelA, respectively (Fig. 3C).
Figure 3.
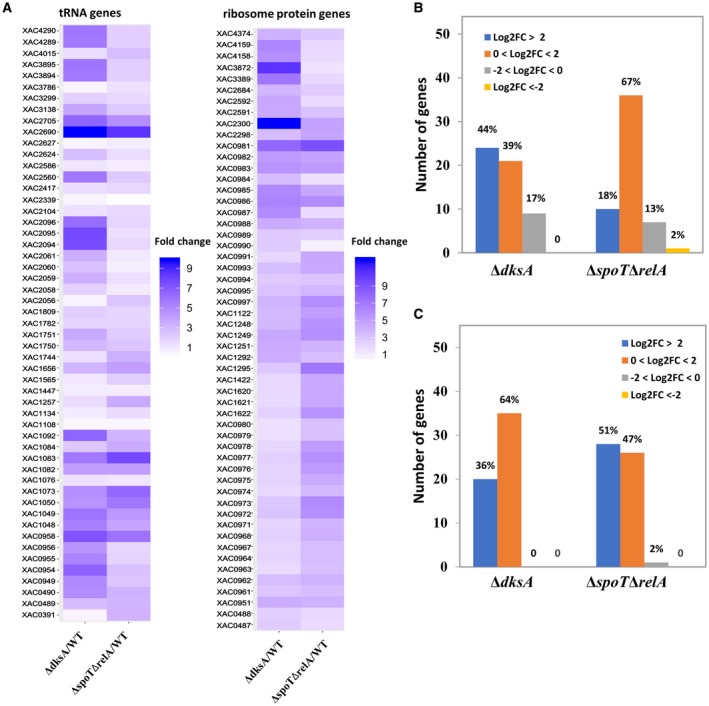
DksA and (p)ppGpp repress gene expression of tRNA and ribosome proteins. (A) Heatmap representing the gene expression profile for tRNA genes and ribosome protein genes. (B, C) Summary of gene expression levels of tRNA genes (B) and ribosome protein genes (C) in the ΔdksA and ΔspoTΔrelA strains. Log2FC refers to log2 fold change (Δ/WT; mutant/wild‐type). Data labels indicate the percentage of genes in each category.
Studies from E. coli have shown that DksA and (p)ppGpp can directly stimulate certain promoters for amino acid biosynthesis and transport (Paul et al., 2005). Based on pathway enrichment analysis, we found that many genes involved in histidine metabolism were down‐regulated in the ΔdksA and ΔspoTΔrelA strains (Fig. S5). Taken together, these results indicate that DksA and (p)ppGpp of Xcc negatively regulate tRNA and ribosome protein biosynthesis and positively regulate histidine metabolism.
DksA and (p)ppGpp positively regulate T3SS and T2SS
Xanthomonas possesses six protein secretion systems (type 1 to type 6). Type 2 secretion system (T2SS) and T3SS have been shown to be important to Xanthomonas virulence (Büttner and Bonas, 2010). Functional enrichment analysis showed that bacterial secretion systems are under the control of DksA and (p)ppGpp of Xcc (Table 1). Upon closer inspection, we identified that T3SS and the xcs T2SS genes are significantly down‐regulated in the ΔdksA and ΔspoTΔrelA mutants compared to the wild‐type (Table S6). The hrp/hrc cluster, which encodes the T3SS, is composed of 24 genes distributed in several operons. Twenty‐one (87.5%) and 17 (70.8%) T3SS genes were down‐regulated DEGs in the ΔdksA and ΔspoTΔrelA mutants compared to the wild‐type Xcc, respectively (Table 2). The xcs T2SS gene cluster contains 12 genes encoded in a single operon and 11 and 7 T2SS genes are down‐regulated DEGs in the ΔdksA and ΔspoTΔrelA mutants compared to wild‐type Xcc, respectively (Table S6). It should be noted that Xcc harbours a second T2SS (xps) that was reported to be more significant to bacterial virulence (Szczesny et al., 2010). No difference was observed in the expression of the xps genes in either ΔdksA or ΔspoTΔrelA compared to the wild‐type Xcc.
Table 2.
Down‐regulated type 3 secretion system (T3SS)‐related genes in mutants compared to wild‐type Xanthomonas citri subsp. citri
| Functional group | Genes | |||
|---|---|---|---|---|
| T3SS regulatory gene | hrpX ‡ | |||
| hrp/hrc gene cluster | hpaB | hrpD5 | hpaA | hrcS |
| hrcR | hrcQ | hpaP | hpa2 | |
| hrcU | hrcN | hrcJ | hrpB4 | |
| hrpB5 | hpa1 | hrpB7 | hrcT | |
| hrcC † | hrcV † | hrpB1 † | hrpB2 † | |
| hrpF † | hrpE ‡ | |||
| T3SS effector genes | xopI | xopK | xopN | xopZ |
| xopF | xopX † | xopM † | xopAP † | |
| xopE1 † | xopAU † | xopQ † | xopAV † | |
| xopL † | xopV † | xopP † | xopS ‡ | |
| avrBs2 ‡ | ||||
Down‐regulated DEGs only in ΔdksA.
Down‐regulated DEGs only in ΔspoTΔrelA.
The other genes are down‐regulated differentially expressed genes (DEGs) in both mutant strains.
In addition to the secretion apparatuses, we also observed significant differences in T3SS key regulatory gene hrpX, T3SS effector coding genes and T2SS hydrolase genes (Tables 2 and S6). Xcc encodes approximately 30 effector genes as described in the Xanthomonas Resource website (http://www.xanthomonas.org/t3e.html). Our data show that 15 (50.0%) and 7 (23.3%) effector genes are down‐regulated DEGs in the ΔdksA and ΔspoTΔrelA mutants compared to the wild‐type Xcc, respectively (Table 2).
To confirm the regulatory effect on the T3SS‐related genes, RT‐qPCR was used to measure the mRNA level of selected genes including two regulatory genes (hrpG, hrpX), one T3SS component gene (hrpF) and two effector genes (xopN, xopAU) in the ΔdksA and ΔspoTΔrelA mutants compared to the wild‐type Xcc. The transcript level of selected genes in ΔdksA and ΔspoTΔrelA were two to five times lower than that of wild‐type Xcc306 (Fig. 4A). Consistent with RT‐qPCR results, β‐glucuronidase (GUS) assays using translational fusion plasmids showed that the promoter activity of four selected genes (hrpG, hrpX, xopAU, hrpF) in either mutant strain was substantially lower than that of wild‐type Xcc306 (Fig. 4B). T3SS and effectors are known to be responsible for the hypersensitive response (HR) of Xcc on Nicotiana benthamiana (Adlung et al., 2016; Sankaranarayanan et al., 2014). To test whether the ΔdksA and ΔspoTΔrelA mutants were affected in the ability to trigger HR, wild‐type strain Xcc306, mutant strains including a T3SS‐disrupted mutant hrcV:Tn5 and complemented strains were inoculated by syringe infiltration on N. benthamiana. In contrast to wild‐type Xcc306 and complemented strains, hrcV:Tn5, ΔdksA and ΔspoTΔrelA were unable to cause HR (Fig. 4C). Taken together, these results indicate that DksA and (p)ppGpp positively regulate the T3SS system to promote virulence.
Figure 4.
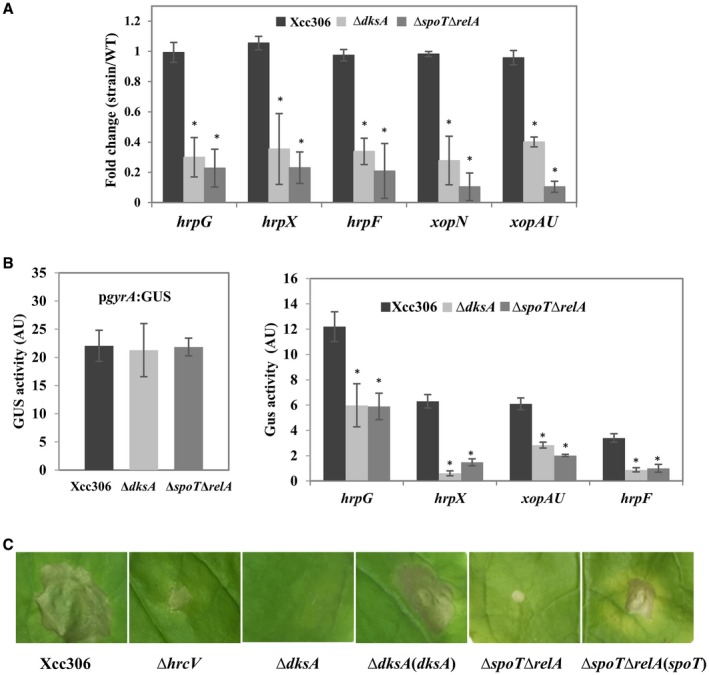
DksA and (p)ppGpp positively regulate expression of the type 3 secretion system (T3SS) genes. (A) mRNA abundance measurement of the selected T3SS genes by RT‐qPCR. For normalization, gyrA was used as an endogenous control. (B) Promoter activity of the indicated promoters was measured for Xcc306, ΔdksA and ΔspoTΔrelA strains harbouring translational fusion plasmids. (C) Bacterial cultures (2 × 108 CFU/mL) of the indicated strains were infiltrated into Nicotiana benthamiana leaves. Leaves were photographed at 8 days post‐inoculation. (A, B) Values represent means ± SD and asterisks indicate statistical significance using Student’s t‐test (P < 0.05, n = 3). All the experiments were repeated independently three times with similar results.
Motility‐related structural genes are negatively regulated by DksA and (p)ppGpp
Bacterial flagellum biosynthesis consumes considerable metabolic energy and has been considered as a virulence trait (Josenhans and Suerbaum, 2002). Both the flagellum‐dependent and type IV pilus‐dependent motility are present in Xcc and play important roles in adhesion, biofilm formation and virulence (Dunger et al., 2014; Malamud et al., 2011). COG‐based functional and KEGG‐based pathway enrichment analyses showed that motility‐related structural genes were affected in both ΔdksA and ΔspoTΔrelA strains. For flagellar assembly genes, 25 of 27 were up‐regulated DEGs in the ΔspoTΔrelA strain while only three were up‐regulated DEGs in the ΔdksA strain (Fig. 5A). Type IV pilus biogenesis‐related genes displayed different expression patterns: 22 of 28 type IV pilus biogenesis‐related genes, including the regulatory gene fimX (XAC2398), were up‐regulated DEGs in ΔdksA whereas no up‐regulated DEGs were found in ΔspoTΔrelA (Table S7).
Figure 5.
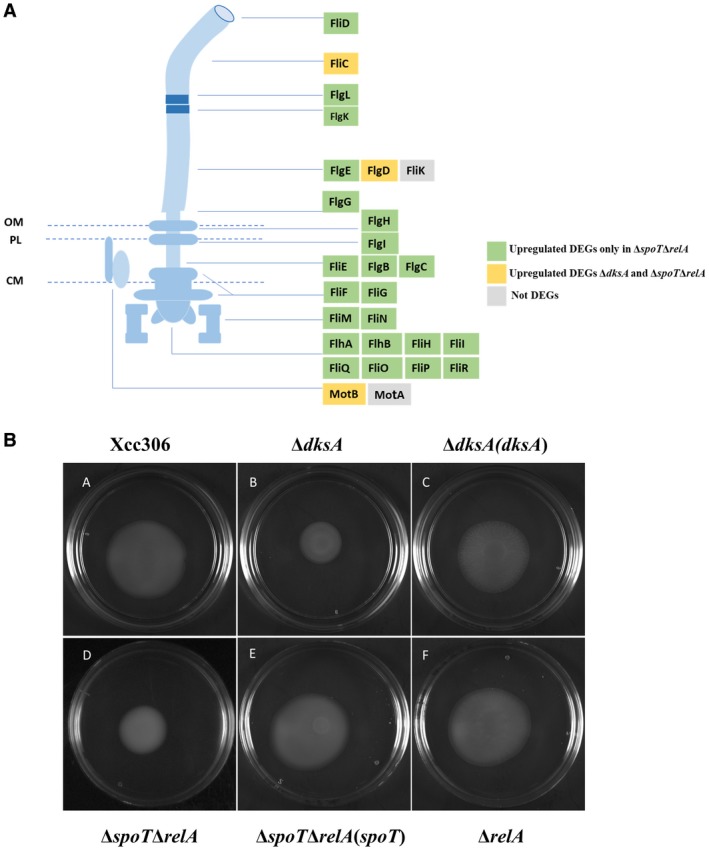
Negative regulation of flagellar assembly genes by (p)ppGpp. (A) Schematic diagram of bacterial flagellum. Gene expression changes in the ΔdksA and ΔspoTΔrelA strains compared to wild‐type Xanthomonas citri subsp. citri (Xcc306) are indicated by different colours. OM, outer membrane; PL, peptidoglycan layer; CM, cytoplasmic membrane. (B) Motility test of bacteria on 0.25% nutrient agar plate. Plates were incubated for 48 h before photographing. The experiment was repeated independently three times with similar results.
To test whether the gene expression changes affect flagellar morphology, wild‐type Xcc306, ΔdksA and ΔspoTΔrelA were grown on the XVM2 agar medium for observation under a transmission electron microscope (TEM). No obvious differences in flagellar morphology were observed (Fig. S6). Bacterial motility was tested on 0.25% semi‐solid nutrient agar plates. Unexpectedly, ΔdksA and ΔspoTΔrelA showed reduced motility compared to Xcc306, ΔrelA and complemented strains (Fig. 5B). To sum up, these results indicate that stringent response regulators DksA and (p)ppGpp inhibit the gene expression involved in flagellar and pili biosynthesis.
TonB‐dependent transporters genes are positively regulated by DksA and (p)ppGpp
TonB‐dependent transporters (TBDTs) are bacterial outer membrane proteins that are used for active uptake of nutrients such as siderophores, vitamin B12, nickel and carbohydrates in Gram‐negative bacteria (Noinaj et al., 2010; Schauer et al., 2008). Xcc306 encodes 46 TBDTs (da Silva et al., 2002), which are listed in Table S8. Heatmap analysis displayed the gene expression profile of TBDTs in the ΔdksA and ΔspoTΔrelA strains compared to the wild‐type Xcc (Fig. 6A). Among them, 23 (50%) TBDTs were down‐regulated DEGs (log2FC > −2) in the ΔdksA and ΔspoTΔrelA mutants compared to the wild‐type Xcc, respectively (Table S8).
Figure 6.
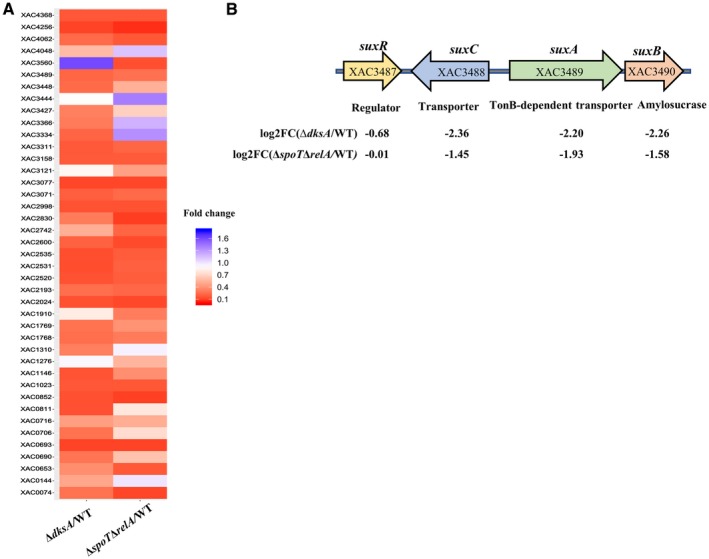
Positive regulation of TonB‐dependent transporter (TBDT) genes by DksA and (p)ppGpp. (A) Heatmap representing the gene expression profile of TBDT genes. (B) The organization of the sux locus and the gene expression changes in the ΔdksA and ΔspoTΔrelA strains compared to wild‐type Xanthomonas citri subsp. citri.
A previous study showed that TBDT‐coding genes were over‐represented in Xanthomonas spp. and Xanthomonas campestris pv. campestris employs TBDTs in the sucrose transport and metabolism as well as virulence through a conserved sux (sucrose utilization in Xanthomonas) locus (Blanvillain et al., 2007). The same sux locus was also found in Xcc and is composed of four genes encoding a regulatory protein (XAC3487), a sugar inner membrane transporter (XAC3488), a TonB‐dependent transporter (XAC3489) and an amylosucrase (XAC3490) (Fig. 6B). The suxA, a TonB‐dependent transporter, suxB and suxC genes were down‐regulated in both ΔdksA and ΔspoTΔrelA mutants compared to the wild‐type Xcc (Fig. 6B). To sum up, these results indicate that DksA and (p)ppGpp promote the expression of TBDTs, which probably enhance the uptake of nutrients, including sucrose.
Differential regulation of xss gene cluster by DksA and (p)ppGpp
Although DksA mostly acts cumulatively with (p)ppGpp, divergent and even opposite effects on gene expression and specific traits have been reported as well (Lyzen et al., 2009, 2016; Magnusson et al., 2007). We conducted cluster analysis and displayed the gene expression profile of 482 common DEGs in ΔdksA and ΔspoTΔrelA strains (Fig. S7). Among them, 426 genes were down‐regulated and 40 genes were up‐regulated in both mutant strains compared to the wild‐type Xcc. Divergent regulation between ΔdksA and ΔspoTΔrelA was observed for 16 genes (Table S9). Interestingly, we found that the xss gene cluster (composed of mhpE, xsuA and xssA‐E), which is involved in biosynthesis, export and utilization of siderophores (Pandey and Sonti, 2010; Pandey et al., 2017), was up‐regulated in ΔdksA and down‐regulated in ΔspoTΔrelA compared to the wild‐type Xcc (Fig. 7A).
Figure 7.
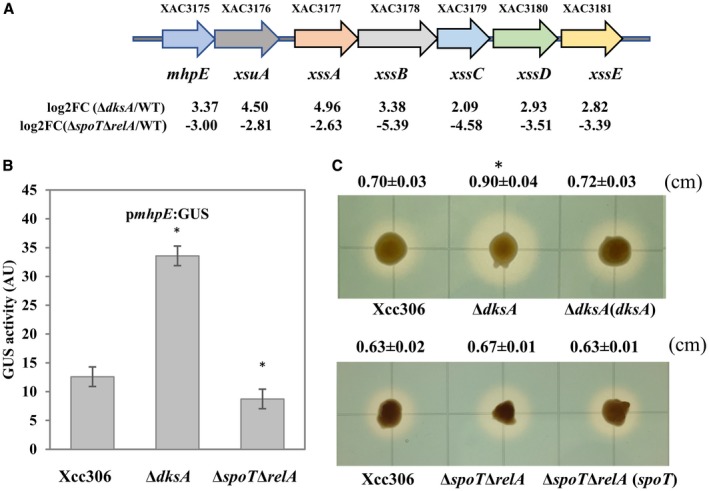
Differential regulation of the xss gene cluster by DksA and (p)ppGpp. (A) Organization of siderophore synthesis and utilization gene cluster (xss) in Xanthomonas citri subsp. citri (Xcc306). (B) Promoter activity of mphE in Xcc306, ΔdksA and ΔspoTΔrelA strains by β‐glucuronidase (GUS) assay. Values represent means ± SD and asterisks indicate statistical significance using Student’s t‐test (P < 0.05, n = 7). (C) Siderophore production by chrome azurol S (CAS) assay. Fresh bacteria were inoculated on nutrient agar plates supplemented with 200 µM 2,2'‐dipyridyl and photographed after 36 h. Values represent means ± SD of the halo diameter and the asterisk indicates statistical significance using Student’s t‐test (P < 0.05, n = 3). The experiments were repeated independently three times with similar results.
Consistent with the RNA‐seq data, the GUS assay showed that the promoter activity of mphE is significantly increased in ΔdksA and slightly decreased in ΔspoTΔrelA (Fig. 7B). The ability of these mutant strains to produce siderophores was assessed by a chrome azurol S (CAS) assay. The ΔdksA mutant produced a bigger halo around the colonies, indicating more siderophore production (Fig. 7C). However, no obvious difference was observed between wild‐type Xcc306, ΔspoTΔrelA and ΔspoTΔrelA (spoT) strains (Fig. 7C). Taken together, these results indicate that the xss gene cluster is differentially regulated by DksA and (p)ppGpp, and the DksA‐mediated repression of siderophore production might be due to the inhibition of xss gene cluster expression.
Discussion
To adapt to the host environment, plant pathogens need to rely on a sensory system to monitor external signals and subsequently regulate physiological processes and virulence traits. It has long been known that plant pathogens could activate T3SS in contact with host cells (Tang et al., 2006). However, beyond virulence induction, less is known about how plant pathogens prepare for infection regarding physiological changes. Our transcriptome analysis revealed that the stringent response regulator DksA and (p)ppGpp of Xcc have a profound effect on gene expression involved in biosynthesis of stable RNA, ribosome proteins, flagellum and type IV pilus, TBDTs, T2SS and T3SS. Further analysis showed changes in associated traits like pathogenicity, HR induction, motility and siderophore production. Our study further expands the understanding of interplay between the stringent response and virulence.
Deletion of dksA or double deletion of spoT and relA severely affects bacterial pathogenicity and growth in compatible host plants, indicating that stringent response regulators play an essential role in virulence regulation. Deletion of relA, which was reported as the main producer of (p)ppGpp in E. coli (Hauryliuk et al., 2015), had little to no effect on the virulence of Xcc. We speculate that since SpoT has weak (p)ppGpp synthetase activity, a certain amount of (p)ppGpp might be present in the ΔrelA strain, thus masking the effect of relA mutation.
The contribution of DksA and/or (p)ppGpp to virulence was reported in multiple animal and plant pathogenic bacteria such as Pseudomonas aeruginosa, Vibrio cholera, Streptococcus pneumonia, Salmonella enterica, enterohaemorrhagic E. coli (EHEC), E. amylovora and P. syringae (Ancona et al., 2015; Azriel et al., 2015; Bhadra et al., 2012; Chatnaparat et al., 2015a,b; Kazmierczak et al., 2009; Nakanishi et al., 2006; Xu et al., 2016). In EHEC, (p)ppGpp and DksA could directly activate transcription of two transcriptional regulators, Pch and Ler, which control the gene expression of T3SS and secreted proteins (Nakanishi et al., 2006). Consistent with previous study in other microorganisms, DksA and (p)ppGpp also regulate T3SS genes in Xcc, even though the detailed mechanism remains to be determined. Besides T3SS, our study also provides evidence indicating that T2SS is also under the control of DksA and (p)ppGpp, which further explains their contribution to Xcc virulence.
Under nutrient limitation, (p)ppGpp and DksA inhibit the biosynthesis of ribosome and cell surface organelles (flagella and pili), which is consistent with the high amount of energy needed in synthesizing those macromolecular complexes. For example, about 2% of biosynthetic energy was required for E. coli flagellar synthesis (Soutourina and Bertin, 2003). The negative regulation of ribosome and/or flagella biosynthesis was also observed in E. coli by DNA microarray (Durfee et al., 2008; Traxler et al., 2008). Another in vitro study showed that DksA and (p)ppGpp directly regulate the flagellar cascade of E. coli by inhibiting the transcription of two regulatory genes flhDC and fliA (Lemke et al., 2009). It is possible that Xcc may adopt a similar mechanism to regulate the flagellar transcriptional cascade. Interestingly, our RNA‐seq data indicate that in Xcc, (p)ppGpp has a strong negative effect on flagellar assembly whereas DksA mainly controls gene expression of type IV pili. Despite the higher expression of flagellar genes in the ΔdksA and ΔspoTΔrelA mutants compared to the wild‐type Xcc, reduced motility was observed for both mutants, which requires further investigation.
One striking feature identified in our analysis is the significant down‐regulation of TBDTs in the ΔdksA and ΔspoTΔrelA strains compared to the wild‐type Xcc. One community proteogenomics study revealed that various TBDTs were highly expressed in phyllosphere bacteria from soybean, clover and Arabidopsis thaliana plants, indicating the importance of TBDTs in microbial adaptation to the phyllosphere (Delmotte et al., 2009). Interestingly, TBDTs are significantly over‐represented in Xanthomonas genomes compared to other bacteria (Schauer et al., 2008). Although it is unclear whether over‐represented TBDTs are required for epiphytic fitness of Xanthomonas, the involvement of TBDTs in carbohydrate uptake and virulence was proposed (Blanvillain et al., 2007; Boulanger et al., 2010). Given that some effectors target plant SWEET genes to cause nutrient efflux for the benefit of some bacterial pathogens, one interesting question remaining to be addressed is whether effector‐mediated sucrose efflux could be acquired by bacterial TBDTs (Jacques et al., 2016).
Due to the low solubility and potential toxicity of iron, bacteria have evolved several iron acquisition mechanisms, including a siderophore‐based iron transport system, and iron homeostasis is tightly regulated in the cell (Andrews et al., 2003). The comparison of the DksA and (p)ppGpp regulons indicated that the xss gene cluster involved in siderophore production, transport and metabolism is differentially regulated. The GUS assay showed that the promoter activity of mphE was increased significantly in ΔdksA and slightly decreased in ΔspoTΔrelA compared to the wild‐type Xcc. Consistent with this result, more siderophore production was observed in ΔdksA. The relationship between iron uptake and stringent response regulators is not fully understood. It was reported that iron deprivation induces SpoT‐dependent accumulation of (p)ppGpp, which increases the expression of iron uptake genes (Vinella et al., 2005). Another study from S. enterica suggested that DksA represses the expression of some iron homeostasis‐related genes in response to nitrosative stress (Crawford et al., 2016). Our data show that DksA suppresses the expression of the iron‐uptake related genes such as the xss cluster under nutrient‐rich conditions whereas (p)ppGpp has an opposite regulation effect on those genes. It is possible that the interplay between these two factors enables bacteria to maintain iron homeostasis.
Based on our findings, a working model was proposed to demonstrate how DksA and (p)ppGpp contribute to Xcc regulation of different traits during colonization (Fig. 8). For survival and multiplication in the host apoplastic space, Xcc needs to conquer many barriers, including low nutrients, iron stress and plant immunity defence. These unfavourable stress conditions potentially activate a stringent response in the bacteria. In this case, accumulated (p)ppGpp together with DksA will relocate cellular resources and save biosynthetic energy by repressing the biosynthesis of stable RNA, ribosome proteins, flagella and type IV pili. Meanwhile, expression of genes for T3SS and TBDTs are enhanced to promote the bacterial virulence and nutrient uptake. The sophisticated control of different traits involving DksA and (p)ppGpp helps the pathogens achieve a balance between fitness and virulence, and contribute to host adaptation and colonization.
Figure 8.
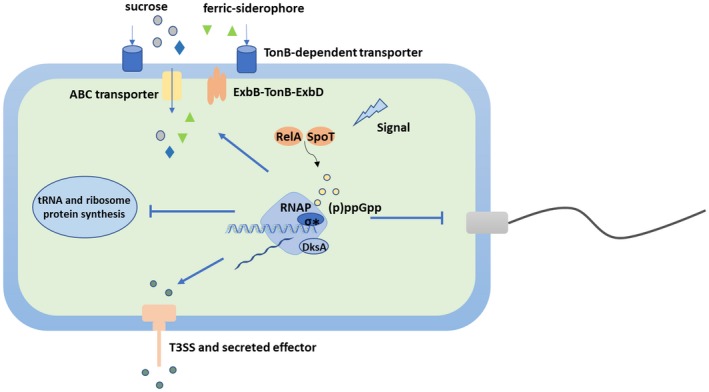
A working model of stringent response regulators of Xanthomonas citri subsp. citri during host colonization. ↓, positive regulation; ⊥, negative regulation. Regulatory steps in the model are mainly at the transcriptional level.
Experimental Procedures
Bacterial strains, growth conditions and plasmids
The strains, plasmids and primers used in this study are listed in Tables S10 and S11. Escherichia coli cells were cultured in lysogeny broth (LB) medium at 37 °C. Xanthomonas strains were cultured in nutrient broth (NB) medium and on nutrient agar (NA) plates at 28 °C. For induction of hrp genes, Xcc strains were grown in XVM2 medium (Wengelnik and Bonas, 1996). Concentrations of antibiotics were as follows: ampicillin (100 µg/mL), kanamycin (50 µg/mL), gentamycin (10 µg/mL) and spectinomycin (100 µg/mL).
Generation of mutant strains and complemented strains
The procedures for generating deletion mutants are described elsewhere and were used with slight modification (Murphy et al., 2000). Briefly, the genomic DNA template was extracted using a Genomic DNA Purification Kit (Promega, Madison, WI, USA). For each target gene (dksA, relA and spoT), the upstream and downstream flanking regions were amplified. A second overlap PCR was performed to connect two fragments using the forward primer of the upstream region and the reverse primer of the downstream region (Table S11). The whole fragment was sequenced and subsequently inserted into the multiple cloning sites of the pOK1 suicide vector (Huguet et al., 1998). The vector was transformed into Xcc by electroporation and markerless deletion mutants were produced using a two‐step sucrose counterselection procedure (Zhou et al., 2015). Note that the double mutant ΔspoTΔrelA was generated in the background of ΔrelA.
To construct the complemented strains, DNA fragments covering the entire coding region and the promoter region of target genes were amplified and inserted into the multiple cloning site of the plasmid pBBR1MCS‐2 (Kovach et al., 1995). Plasmids were introduced into corresponding mutant strains by electroporation.
RNA extraction, sequencing and data analysis
Fresh colonies of Xcc306, ΔdksA and ΔspoTΔrelA strains were picked up from NA plates and grown overnight in NB medium at 28 °C. Three biological repeats were used for each strain. Bacterial cells were harvested and washed before inoculation into the XVM2 medium. On reaching the exponential stage (OD600 = 0.35), bacterial cells were mixed with two volumes of RNA protect bacterial reagent (Qiagen, Valencia, CA, USA) and RNA was extracted following the instructions of the RNeasy Mini Kit (Qiagen). Residual genomic DNA was removed by a TURBO DNA‐free kit (Ambion, Austin, TX, USA). The mRNA enrichment and library construction were described elsewhere (Jalan et al., 2013). Briefly, the ribosomal RNA was removed using the Ribo‐Zero™rRNA Removal Kit for bacteria (Illumina, Madison, USA) according to the manufacturer’s instructions. The remaining transcripts were fragmented and cDNA was synthesized using mRNA templates via reverse transcription. cDNA libraries were constructed using the TrueSeq Stranded mRNA Sample Prep kit (Illumina). Paired end reads (150 bp) were generated for all the RNA samples using a Hiseq 3000 sequencer platform (Novo Gene, Beijing China). The RNA raw reads were deposited at the NCBI SRA database under the bio‐project accession no. PRJNA513356.
To determine the DEGs, mapping reads to the reference genome, transcript abundance quantification and differential expression analysis were performed using Rockhopper (McClure et al., 2013). The DEGs were selected based on an absolute value of log2 fold change (mutant strain/wild‐type strain) greater than or equal to 2 and an adjusted P‐value less than or equal to 0.01. The R package DESeq2 (v. 1.20.0) (Love et al., 2014) and Cluster 3.0 (de Hoon et al., 2004) were used for PCA analysis and cluster analysis. Based on the KEGG pathway database, clusterProfiler (v. 2.8.1) was used for pathway enrichment analysis (Yu et al., 2012). Enrichment analysis of all DEGs based on the Clusters of Orthologous Groups (COGs) of proteins database was performed using Fisher’s exact test (Abatangelo et al., 2009).
Pathogenicity tests, in planta bacterial growth and HR tests
Pathogenicity assays were performed in a quarantine greenhouse located at the Citrus Research and Education Center, Lake Alfred, FL, USA. The wild‐type strain Xcc306, ΔdksA, ΔrelA, ΔspoTΔrelA and complemented strains were cultured overnight in NB medium at 28 °C. After centrifugation, bacterial pellets were washed and resuspended in sterile water. The concentration of bacterial solution was adjusted to 108 (for monitoring symptoms in sweet orange), 2 × 108 (for monitoring HR) or 107 (for monitoring bacterial growth) CFU/mL (Andrade et al., 2014; Teper et al., 2019; Yan and Wang, 2012; Zhou et al., 2018). Bacteria were infiltrated into immature leaves of sweet orange (C. sinensis) and N. benthamiana.
To measure the bacterial population, three leaf disks were taken out from inoculation sites per strain per time point. Leaf disks with a diameter of 0.6 cm were put into a 1.5 mL Eppendorf tube containing sterile water and ground by a drill. The bacterial solution was serially diluted and spotted on NA plates for incubation at 28 °C. The bacterial population given as CFU/mL was calculated after 48 h.
In vitro bacterial growth
Fresh overnight bacterial culture was inoculated into a 50 mL centrifuge tube containing 10 mL NB or XVM2 media at an initial optical density of OD600 = 0.05. At each time point, 200 µL bacterial culture of each strain was taken out and measured using microplate spectrophotometer (Bio‐Rad Laboratories, Hercules, CA, USA). The experiments were repeated at least twice in triplicate with similar results.
RT‐qPCR
For two‐step quantitative reverse transcription PCR (RT‐qPCR), 1 µg RNA was used for cDNA synthesis using a qScript cDNA Synthesis Kit (Quantabio, Beverly, MA, USA). Primers were designed by online software Primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/). All primer sequences are listed and classified in Table S11. The reaction mixture was prepared following the instructions for the SYBR Green Master Mix kit (Clontech Laboratories, Mountain View, CA, USA) on a QuantStudio 3 Real‐Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA). The expression level of gyrA was used as an endogenous control. The fold change of target genes expression was calculated using the formula 2–ΔΔCt (Livak and Schmittgen, 2001). This experiment was repeated three times in triplicate with similar results.
Motility assay
Bacterial motility was tested using a semisolid NA plate containing 0.3% agar. Bacteria were grown in NB overnight with shaking at 200 rpm, and then centrifuged down, washed and diluted to OD600 = 0.5 in sterile water. A 5 µL suspension of each strain was spotted on the centre of the plates and incubated at 28 °C. Plates were photographed after 48 h. The assay was repeated three times independently in triplicate with similar results.
GUS activity assay
To generate GUS reporter plasmids the putative promoter region of the tested genes was amplified and cloned into pGUS vector (Teper et al., 2019) and transferred into wild‐type Xcc306 and mutant strains by electroporation.
GUS activity was quantified as described previously (Zhou et al., 2017) with slight modifications. Briefly, bacterial cells grown in the XVM2 medium were harvested and resuspended in phosphate‐buffered saline (PBS) and followed by sonication. A volume of 20 µL clear supernatant was added to 80 µL PBS buffer containing 1.25 µM p‐nitrophenyl β‐d‐glucopyranoside (PNPG) in a 96‐well plate. When a yellow colour developed during incubation at 37 °C, 100 µL stop buffer (0.4 M Na2CO3) was added to stop the reaction immediately. Meanwhile, the reaction time was recorded. The absorbance of the reaction solution was measured at 405 nm and normalized by a protein amount that was measured by the Bradford method at 595 nm using a Bio‐Rad Protein Assay Kit (Bio‐Rad). GUS activity was quantified by arbitrary units (AU) and determined as A405/(time in min × A595 × 0.02). The experiment was repeated at least twice in triplicate with similar results.
Siderophore production assay
The CAS assay was conducted as described elsewhere (Cordero et al., 2012). Briefly, 1 L of CAS‐Fe‐hexadecyl‐trimethyl‐ammonium bromide (HDTMA) dye was prepared as follows: 10 mL of a 10 mM ferric chloride (FeCl3) in 100 mM HCl solution was mixed with 590 mL of a 1 mM aqueous solution of CAS. The Fe‐CAS solution was then added to 400 mL of 2 mM aqueous solution of HDTMA. The resulting CAS‐Fe‐HDTMA solution was autoclaved for 20 min. The solution was stored at room temperature and covered with aluminium foil. For 100 mL of CAS‐agar, 10 mL of CAS‐Fe‐HDTMA dye was mixed with 90 mL of NB‐based media supplemented with 2,2ʹ‐dipyridyl (DP). To prepare samples, bacterial strains grown on NA plates were spotted onto the CAS agar plate and incubated at 28 °C for 36 h. Halo phenotype around colonies is indicative of siderophore production. The assay was repeated three times independently with similar results.
Supporting information
Fig. S1 Protein domain analyses of DksA, RelA and SpoT in Xcc. The domains of the DksA, SpoT and RelA are shown. Protein sequence analysis was performed using online software InterPro (https://www.ebi.ac.uk/interpro/).
Fig. S2 Heatmap analysis of the sample‐to‐sample distances. An overview over similarities and dissimilarities between samples. Sample‐to‐sample distances were calculated from a count matrix based on all chromosome genes. The heatmap was derived from the distance matrix using R package DESeq2 (v. 1.20.0).
Fig. S3 Comparison of gene expression by RT‐qPCR and RNA‐seq. At least 16 genes were randomly selected for ΔdksA and ΔspoTΔrelA strains. The mRNA abundance of these genes was tested by RT‐qPCR using gyrA as an endogenous control. The R‐squared value and the P‐value were calculated by R software (v. 3.5.1).
Fig. S4 Distribution of DEGs of ΔdksA and ΔspoTΔrelA compared to wild‐type Xcc according to COG categories. For Xcc306, the numbers mean the total genes in each category according to the Xanthomonas Genome Browser (http://xgb.leibniz‐fli.de/cgi/cog.pl?ssi=free).
Fig. S5 Down‐regulated DEGs involved in the histidine metabolism pathway. Genes in red represent down‐regulated DEGs in the ΔdksA and ΔspoTΔrelA mutants compared to wild‐type Xcc, genes in orange represent down‐regulated DEGs only in ΔdksA, genes in green represent down‐regulated DEGs only in ΔspoTΔrelA. This pathway is based on the KEGG pathway database (https://www.genome.jp/kegg‐bin/show_pathway?xac00340).
Fig. S6 Transmission electron microscopic observation of Xcc306, ΔdksA and ΔspoTΔrelA strains. Fresh bacteria of Xcc306, ΔdksA and ΔspoTΔrelA from XVM2 solid agar medium were transferred to formvar/carbon‐coated 400‐mesh copper grids and stained with 0.25% aqueous ammonium molybdate. The grids were allowed to dry for 1 h before observation with an FEI Morgagni 268 transmission electron microscope (FEI, OR, USA). The black arrows indicate bacterial polar flagella.
Fig. S7 Gene expression profile of common DEGs for ΔdksA and ΔspoTΔrelA. Hierarchical cluster analysis shows that all common DEGs were grouped into four clusters: I, II, III and IV. Forty genes (IV) were up‐regulated, and 426 genes (II) were down‐regulated in both mutants. Twelve genes (I) were up‐regulated in ΔdksA but down‐regulated in ΔspoTΔrelA. Four genes (III) were down‐regulated in ΔdksA but up‐regulated in ΔspoTΔrelA.
Table S1 Data quality summary.
Table S2 Differentially regulated genes in the ΔspoTΔrelA mutant compared to the wild‐type Xcc.
Table S3 Differentially regulated genes in the ΔdksA mutant compared to wild‐type (WT) Xcc.
Table S4 Gene expression of tRNA‐coding genes.
Table S5 Gene expression of ribosome protein genes.
Table S6 Gene expression profile of T3SS‐ and T2SS‐related genes in Xcc.
Table S7 Gene expression profile for putative type 4 pilus biogenesis and regulation genes of Xcc.
Table S8 Gene expression level of TBDT genes of Xcc.
Table S9 Differentially regulated genes by DksA and ppGpp.
Table S10 Strains and plasmids used in this study.
Table S11 Primer sequence used in this study.
Acknowledgement
This study was supported by the USDA‐NIFA Plant Biotic Interactions Program 2017‐67013‐26527 and a CSC Scholarship offered by China Scholarship Council.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.
References
- Abatangelo, L. , Maglietta, R. , Distaso, A. , D’Addabbo, A. , Creanza, T.M. , Mukherjee, S. and Ancona, N. (2009) Comparative study of gene set enrichment methods. BMC Bioinformatics, 10, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åberg, A. , Fernández‐Vázquez, J. , Cabrer‐Panes, J.D. , Sánchez, A. and Balsalobre, C. (2009) Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli . J. Bacteriol. 191, 3226–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlung, N. , Prochaska, H. , Thieme, S. , Banik, A. , Blüher, D. , John, P. , Nagel, O. , Schulze, S. , Gantner, J. , Delker, C. , Stuttmann, J. and Bonas, U. (2016) Non‐host resistance induced by the Xanthomonas effector XopQ is widespread within the genus Nicotiana and functionally depends on EDS1. Front. Plant Sci. 7, 1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancona, V. , Lee, J.H. , Chatnaparat, T. , Oh, J. , Hong, J.‐I. and Zhao, Y. (2015) The bacterial alarmone (p)ppGpp activates the type III secretion system in Erwinia amylovora . J. Bacteriol. 197, 1433–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade, M.O. , Farah, C.S. and Wang, N. (2014) The post‐transcriptional regulator rsmA/csrA activates T3SS by stabilizing the 5ʹ UTR of hrpG, the master regulator of hrp/hrc genes, in Xanthomonas . PLoS Pathog. 10, e1003945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S.C. , Robinson, A.K. and Rodríguez‐Quiñones, F. (2003) Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. [DOI] [PubMed] [Google Scholar]
- Astua‐Monge, G. , Freitas‐Astua, J. , Bacocina, G. , Roncoletta, J. , Carvalho, S.A. and Machado, M.A. (2005) Expression profiling of virulence and pathogenicity genes of Xanthomonas axonopodis pv. citri . J. Bacteriol. 187, 1201–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson, G.C. , Tenson, T. and Hauryliuk, V. (2011) The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS ONE, 6, e23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azriel, S. , Goren, A. , Rahav, G. and Gal‐Mor, O. (2015) The stringent response regulator DksA Is required for Salmonella enterica serovar typhimurium growth in minimal medium, motility, biofilm formation, and Intestinal colonization. Infect. Immun. 84, 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra, R.K. , Dasgupta, S. , Pal, R.R. , Bag, S. and Das, B. (2012) Functional characterization of the stringent response regulatory gene dksA of Vibrio cholerae and its role in modulation of virulence phenotypes. J. Bacteriol. 194, 5638–5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankschien, M.D. , Potrykus, K. , Grace, E. , Choudhary, A. , Vinella, D. , Cashel, M. and Herman, C. (2009) TraR, a homolog of a RNAP secondary channel interactor, modulates transcription. PLoS Genet. 5, e1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanvillain, S. , Meyer, D. , Boulanger, A. , Lautier, M. , Guynet, C. , Denancé, N. , Vasse, J. , Lauber, E. and Arlat, M. (2007) Plant carbohydrate scavenging through TonB‐dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One, 2, e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger, A. , Déjean, G. , Lautier, M. , Glories, M. , Zischek, C. , Arlat, M. and Lauber, E. (2010) Identification and regulation of the N‐acetylglucosamine utilization pathway of the plant pathogenic bacterium Xanthomonas campestris pv. campestris . J. Bacteriol. 192, 1487–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden, S.D. , Eyres, A. , Chung, J.C.S. , Monson, R.E. , Thompson, A. , Salmond, G.P.C. , Spring, D.R. and Welch, M. (2013) Virulence in Pectobacterium atrosepticum is regulated by a coincidence circuit involving quorum sensing and the stress alarmone, (p)ppGpp. Mol. Microbiol. 90, 457–471. [DOI] [PubMed] [Google Scholar]
- Brunings, A.M. and Gabriel, D.W. (2003) Xanthomonas citri: breaking the surface. Mol. Plant Pathol. 4, 141–157. [DOI] [PubMed] [Google Scholar]
- Büttner, D. and Bonas, U. (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev., 34, 107–33. [DOI] [PubMed] [Google Scholar]
- Chatnaparat, T. , Li, Z. , Korban, S.S. and Zhao, Y. (2015a) The stringent response mediated by (p)ppGpp is required for virulence of Pseudomonas syringae pv. tomato and its survival on tomato. Mol. Plant‐Microbe Interact. 28, 776–789. [DOI] [PubMed] [Google Scholar]
- Chatnaparat, T. , Li, Z. , Korban, S.S. and Zhao, Y. (2015b) The bacterial alarmone (p)ppGpp is required for virulence and controls cell size and survival of Pseudomonas syringae on plants. Environ. Microbiol. 17, 4253–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero, O.X. , Ventouras, L.‐A. , DeLong, E.F. and Polz, M.F. (2012) Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc. Natl. Acad. Sci. USA. 109, 20059–20064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, M.A. , Henard, C.A. , Tapscott, T. , Porwollik, S. , McClelland, M. and Vázquez‐Torres, A. (2016) DksA‐dependent transcriptional regulation in Salmonella experiencing nitrosative stress. Front. Microbiol. 7, 444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux, Z.D. and Swanson, M.S. (2012) ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10, 203–212. [DOI] [PubMed] [Google Scholar]
- Dalebroux, Z.D. , Svensson, S.L. , Gaynor, E.C. and Swanson, M.S. (2010) ppGpp conjures bacterial virulence. Microbiol. Mol. Biol. Rev. 74, 171–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmotte, N. , Knief, C. , Chaffron, S. , Innerebner, G. , Roschitzki, B. , Schlapbach, R. , von Mering, C. and Vorholt, J.A. (2009) Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. USA. 106, 16428–16433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Dunger, G. , Guzzo, C.R. , Andrade, M.O. , Jones, J.B. and Farah, C.S. (2014) Xanthomonas citri subsp. citri Type IV pilus is required for twitching motility, biofilm development, and adherence. Mol. Plant–Microbe Interact. 27, 1132–1147. [DOI] [PubMed] [Google Scholar]
- Durfee, T. , Hansen, A.‐M. , Zhi, H. , Blattner, F.R. and Jin, D.J. (2008) Transcription profiling of the stringent response in Escherichia coli . J. Bacteriol. 190, 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima, U. and Senthil‐Kumar, M. (2015) Plant and pathogen nutrient acquisition strategies. Front. Plant Sci. 6, 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ference, C.M. , Gochez, A.M. , Behlau, F. , Wang, N. , Graham, J.H. and Jones, J.B. (2017) Recent advances in the understanding of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management. Mol. Plant Pathol. 19, 1302–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaca, A.O. , Kajfasz, J.K. , Miller, J.H. , Liu, K. , Wang, J.D. , Abranches, J. and Lemos, J.A. (2013) Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond the stringent response. MBio, 4, e00646–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse, R.L. , Chen, A.Y. , Gopalkrishnan, S. , Sanchez‐Vazquez, P. , Myers, A. and Ross, W. (2018) Transcriptional responses to ppGpp and DksA. Annu. Rev. Microbiol. 72, 163–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y. , Figueiredo, F. , Jones, J. and Wang, N. (2011) HrpG and HrpX play global roles in coordinating different virulence traits of Xanthomonas axonopodis pv. citri . Mol. Plant–Microbe Interact. 24, 649–661. [DOI] [PubMed] [Google Scholar]
- Haugen, S.P. , Ross, W. and Gourse, R.L. (2008) Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 6, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauryliuk, V. , Atkinson, G.C. , Murakami, K.S. , Tenson, T. and Gerdes, K. (2015) Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon, M.J.L. , Imoto, S. , Nolan, J. and Miyano, S. (2004) Open source clustering software. Bioinformatics, 20, 1453–1454. [DOI] [PubMed] [Google Scholar]
- Huguet, E. , Hahn, K. , Wengelnik, K. and Bonas, U. (1998) hpaA mutants of Xanthomonas campestris pv. vesicatoria are affected in pathogenicity but retain the ability to induce host-specific hypersensitive reaction. Mol. Microbiol., 29, 1379–90. [DOI] [PubMed] [Google Scholar]
- Irving, S.E. and Corrigan, R.M. (2018) Triggering the stringent response: signals responsible for activating (p)ppGpp synthesis in bacteria. Microbiology, 164, 268–276. [DOI] [PubMed] [Google Scholar]
- Jacques, M.‐A. , Arlat, M. , Boulanger, A., Boureau, T., Carrère, S., Cesbron, S., Chen, N.W., Cociancich, S., Darrasse, A., Denancé, N., Fischer-Le Saux, M., Gagnevin, L., Koebnik, R., Lauber, E., Noël, L.D., Pieretti, I., Portier, P., Pruvost, O., Rieux, A., Robène, I., Royer, M., Szurek, B., Verdier, V. and Vernière, C. (2016) Using ecology, physiology, and genomics to understand host specificity in Xanthomonas . Annu. Rev. Phytopathol. 54, 163–187. [DOI] [PubMed] [Google Scholar]
- Jalan, N. , Kumar, D. , Andrade, M.O. , Yu, F. , Jones, J.B. , Graham, J.H. , White, F.F. , Setubal, J.C. and Wang, N. (2013) Comparative genomic and transcriptome analyses of pathotypes of Xanthomonas citri subsp. citri provide insights into mechanisms of bacterial virulence and host range. BMC Genom. 14, 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josenhans, C. and Suerbaum, S. (2002) The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291, 605–614. [DOI] [PubMed] [Google Scholar]
- Kanehisa, M. and Goto, S. (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak, K.M. , Wayne, K.J. , Rechtsteiner, A. and Winkler, M.E. (2009) Roles of relSpn in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol. Microbiol. 72, 590–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebnik, R. , Kruger, A. , Thieme, F. , Urban, A. and Bonas, U. (2006) Specific binding of the Xanthomonas campestris pv. vesicatoria AraC‐type transcriptional activator HrpX to plant‐inducible promoter boxes. J. Bacteriol. 188, 7652–7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach, M.E. , Elzer, P.H. , Steven Hill, D. , Robertson, G.T. , Farris, M.A. , Roop, R.M.M. and Peterson, K.M. (1995) Four new derivatives of the broad‐host‐range cloning vector pBBR1MCS, carrying different antibiotic‐resistance cassettes. Gene, 166, 175–176. [DOI] [PubMed] [Google Scholar]
- Lemke, J.J. , Durfee, T. and Gourse, R.L. (2009) DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol. Microbiol. 74, 1368–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke, J.J. , Sanchez‐Vazquez, P. , Burgos, H.L. , Hedberg, G. , Ross, W. and Gourse, R.L. (2011) Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc. Natl. Acad. Sci. USA. 108, 5712–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R.F. , Lu, G.T. , Li, L. , Su, H.Z. , Feng, G.F. , Chen, Y. , He, Y.Q. , Jiang, B.L. , Tang, D.J. and Tang, J.L. (2014) Identification of a putative cognate sensor kinase for the two‐component response regulator HrpG, a key regulator controlling the expression of the hrp genes in Xanthomonas campestris pv. campestris . Environ. Microbiol. 16, 2053–2071. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2− ΔΔ CT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzen, R. , Kochanowska, M. , Wegrzyn, G. and Szalewska‐Palasz, A. (2009) Transcription from bacteriophage pR promoter is regulated independently and antagonistically by DksA and ppGpp. Nucleic Acids Res. 37, 6655–6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzen, R. , Maitra, A. , Milewska, K. , Kochanowska‐Łyżeń, M. , Hernandez, V.J. and Szalewska‐Pałasz, A. (2016) The dual role of DksA protein in the regulation of Escherichia coli pArgX promoter. Nucleic Acids Res. 44, 10316–10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson, L.U. , Gummesson, B. , Joksimović, P. , Farewell, A. and Nyström, T. (2007) Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli . J. Bacteriol. 189, 5193–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamud, F. , Torres, P.S. , Roeschlin, R. , Rigano, L.A. , Enrique, R. , Bonomi, H.R. , Castagnaro, A.P. , Marano, M.R. and Vojnov, A.A. (2011) The Xanthomonas axonopodis pv. citri flagellum is required for mature biofilm and canker development. Microbiology, 157, 819–829. [DOI] [PubMed] [Google Scholar]
- McClure, R. , Balasubramanian, D. , Sun, Y. , Bobrovskyy, M. , Sumby, P. , Genco, C.A. , Vanderpool, C.K. and Tjaden, B. (2013) Computational analysis of bacterial RNA‐Seq data. Nucleic Acids Res. 41, e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, K.C. , Campellone, K.G. and Poteete, A.R. (2000) PCR‐mediated gene replacement in Escherichia coli . Gene, 246, 321–330. [DOI] [PubMed] [Google Scholar]
- Nakanishi, N. , Abe, H. , Ogura, Y. , Hayashi, T. , Tashiro, K. , Kuhara, S. , Sugimoto, N. and Tobe, T. (2006) ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol. Microbiol. 61, 194–205. [DOI] [PubMed] [Google Scholar]
- Noinaj, N. , Guillier, M. , Barnard, T.J. , Buchanan, S.K. , Barnard, T.J. and Buchanan, S.K. (2010) TonB‐dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, A. and Sonti, R.V. (2010) Role of the FeoB protein and siderophore in promoting virulence of Xanthomonas oryzae pv. oryzae on rice. J. Bacteriol. 192, 3187–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S.S. , Patnana, P.K. , Rai, R. and Chatterjee, S. (2017) Xanthoferrin, the α‐hydroxycarboxylate‐type siderophore of Xanthomonas campestris pv. campestris, is required for optimum virulence and growth inside cabbage. Mol. Plant Pathol. 18, 949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, B.J. , Barker, M.M. , Ross, W. , Schneider, D.A. , Webb, C. , Foster, J.W. and Gourse, R.L. (2004) DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell, 118, 311–322. [DOI] [PubMed] [Google Scholar]
- Paul, B.J. , Berkmen, M.B. and Gourse, R.L. (2005) DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. USA. 102, 7823–7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina, A. , Svetlov, V. , Vassylyeva, M.N. , Tahirov, T.H. , Yokoyama, S. , Artsimovitch, I. and Vassylyev, D.G. (2004) Regulation through the secondary channel–structural framework for ppGpp‐DksA synergism during transcription. Cell, 118, 297–309. [DOI] [PubMed] [Google Scholar]
- Potrykus, K. and Cashel, M. (2008) (p)ppGpp: Still magical? Annu. Rev. Microbiol. 62, 35–51. [DOI] [PubMed] [Google Scholar]
- Rashid, M.M. , Ikawa, Y. and Tsuge, S. (2016) GamR, the LysR‐type galactose metabolism regulator, regulates hrp gene expression via transcriptional activation of two key hrp regulators, HrpG and HrpX, in Xanthomonas oryzae pv. oryzae . Appl. Environ. Microbiol. 82, 3947–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, W. , Sanchez‐Vazquez, P. , Chen, A.Y. , Lee, J.‐H. , Burgos, H.L. and Gourse, R.L. (2016) ppGpp binding to a site at the RNAP‐DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol. Cell, 62, 811–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, R.P. , Vorhölter, F.‐J. , Potnis, N. , Jones, J.B. , Sluys, M.‐A. Van , Bogdanove, A.J. and Dow, J.M. (2011) Pathogenomics of Xanthomonas: understanding bacterium–plant interactions. Nat. Rev. Microbiol. 9, 344–355. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan, R. , Sonti, R.V. , Haque, A.S. , Gupta, M.K. , Nathawat, R. and Sinha, D. (2014) Mutations in the predicted active site of Xanthomonas oryzae pv. oryzae XopQ differentially affect virulence, suppression of host innate immunity, and induction of the HR in a nonhost plant. Mol. Plant–Microbe Interact. 28, 195–206. [DOI] [PubMed] [Google Scholar]
- Schauer, K. , Rodionov, D.A. and de Reuse, H. (2008) New substrates for TonB‐dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem. Sci. 33, 330–338. [DOI] [PubMed] [Google Scholar]
- Schmidtke, C. , Abendroth, U. , Brock, J. , Serrania, J. , Becker, A. and Bonas, U. (2013) Small RNA sX13: a multifaceted regulator of virulence in the plant pathogen Xanthomonas . PLoS Pathog. 9, e1003626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva, A.C.R. , Ferro, J.A. , Reinach, F.C. , Farah, C.S. , Furlan, L.R. , Quaggio, R.B. , Monteiro‐Vitorello, C.B. , Sluys, M.A.Van , Almeida, N.F. , Alves, L.M.C. , do Amaral, A.M. , Bertolini, M.C. , Camargo, L.E.A. , Camarotte, G. , Cannavan, F. , Cardozo, J. , Chambergo, F. , Ciapina, L.P. , Cicarelli, R.M.B. , Coutinho, L.L. , Cursino‐Santos, J.R. , El‐Dorry, H. , Faria, J.B. , Ferreira, A.J.S. , Ferreira, R.C.C. , Ferro, M.I.T. , Formighieri, E.F. , Franco, M.C. , Greggio, C.C. , Gruber, A. , Katsuyama, A.M. , Kishi, L.T. , Leite, R.P. , Lemos, E.G.M. , Lemos, M.V.F. , Locali, E.C. , Machado, M.A. , Madeira, A.M.B.N. , Martinez‐Rossi, N.M. , Martins, E.C. , Meidanis, J. , Menck, C.F.M. , Miyaki, C.Y. , Moon, D.H. , Moreira, L.M. , Novo, M.T.M. , Okura, V.K. , Oliveira, M.C. , Oliveira, V.R. , Pereira, H.A. , Rossi, A. , Sena, J.A.D. , Silva, C. , de Souza, R.F. , Spinola, L.A.F. , Takita, M.A. , Tamura, R.E. , Teixeira, E.C. , Tezza, R.I.D. , Trindade dos Santos, M. , Truffi, D. , Tsai, S.M. , White, F.F. , Setubal, J.C. and Kitajima, J.P . (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature, 417, 459–463. [DOI] [PubMed] [Google Scholar]
- Soutourina, O.A. and Bertin, P.N. (2003) Regulation cascade of flagellar expression in Gram‐negative bacteria. FEMS Microbiol. Rev. 27, 505–523. [DOI] [PubMed] [Google Scholar]
- Szczesny, R. , Jordan, M. , Schramm, C. , Schulz, S. , Cogez, V. , Bonas, U. and Büttner, D. (2010) Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria . New Phytol. 187, 983–1002. [DOI] [PubMed] [Google Scholar]
- Tang, X. , Xiao, Y. and Zhou, J.M. (2006) Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant–Microbe Interact., 19, 1159–1166. [DOI] [PubMed] [Google Scholar]
- Teper, D. , Zhang, Y. and Wang, N. (2019) TfmR, a novel TetR‐family transcriptional regulator, modulates the virulence of Xanthomonas citri in response to fatty acids. Mol. Plant Pathol. 20, 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler, M.F. , Summers, S.M. , Nguyen, H.T. , Zacharia, V.M. , Hightower, G.A. , Smith, J.T. and Conway, T. (2008) The global, ppGpp‐mediated stringent response to amino acid starvation in Escherichia coli . Mol. Microbiol. 68, 1128–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge, S. , Furutani, A. and Ikawa, Y. (2014) Regulatory network of hrp gene expression in Xanthomonas oryzae pv. oryzae . J. Gen. Plant Pathol. 80, 303–313. [Google Scholar]
- Vinella, D. , Albrecht, C. , Cashel, M. and D’Ari, R. (2005) Iron limitation induces SpoT‐dependent accumulation of ppGpp in Escherichia coli . Mol. Microbiol. 56, 958–970. [DOI] [PubMed] [Google Scholar]
- Vojnov, A.A. , Amaral, A.M.Do , Dow, J.M. , Castagnaro, A.P. and Marano, M.R. (2010) Bacteria causing important diseases of citrus utilise distinct modes of pathogenesis to attack a common host. Appl. Microbiol. Biotechnol. 87, 467–477. [DOI] [PubMed] [Google Scholar]
- Wengelnik, K. and Bonas, U. (1996) HrpXv, an AraC‐type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 178, 3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengelnik, K. , Van den Ackerveken, G. and Bonas, U. (1996) HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two‐component response regulators. Mol. Plant–Microbe Interact. 9, 704–712. [DOI] [PubMed] [Google Scholar]
- White, F.F. , Potnis, N. , Jones, J.B. and Koebnik, R. (2009) The type III effectors of Xanthomonas . Mol. Plant Pathol. 10, 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, H. , Kalman, M. , Ikehara, K. , Zemel, S. , Glaser, G. and Cashel, M. (1991) Residual guanosine 3’,5’‐bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266, 5980–5990. [PubMed] [Google Scholar]
- Xu, X. , Yu, H. , Zhang, D. , Xiong, J. , Qiu, J. , Xin, R. , He, X. , Sheng, H. , Cai, W. , Jiang, L. and Zhang, K. (2016) Role of ppGpp in Pseudomonas aeruginosa acute pulmonary infection and virulence regulation. Microbiol. Res. 192, 84–95. [DOI] [PubMed] [Google Scholar]
- Yan, Q. and Wang, N. (2012) High‐throughput screening and analysis of genes of Xanthomonas citri subsp. citri involved in citrus canker symptom development. Mol. Plant–Microbe Interact. 25, 69–84. [DOI] [PubMed] [Google Scholar]
- Yu, G. , Wang, L.-G. , Han, Y. and He, Q.-Y. (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS, 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , Hu, X. , Li, J. and Wang, N. (2015) A novel periplasmic protein, VrpA, contributes to efficient protein secretion by the type III secretion system in Xanthomonas spp. Mol. Plant–Microbe Interact. 28, 143–153. [DOI] [PubMed] [Google Scholar]
- Zhou, X. , Yan, Q. and Wang, N. (2017) Deciphering the regulon of a GntR family regulator via transcriptome and ChIP‐exo analyses and its contribution to virulence in Xanthomonas citri . Mol. Plant Pathol. 18, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , Teper, D. , Andrade, M.O.M.O. , Zhang, T. , Chen, S. , Song, W.‐Y.W.‐Y. and Wang, N. (2018) A phosphorylation switch on Lon protease regulates bacterial type III secretion system in host. MBio, 9, e02146–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Protein domain analyses of DksA, RelA and SpoT in Xcc. The domains of the DksA, SpoT and RelA are shown. Protein sequence analysis was performed using online software InterPro (https://www.ebi.ac.uk/interpro/).
Fig. S2 Heatmap analysis of the sample‐to‐sample distances. An overview over similarities and dissimilarities between samples. Sample‐to‐sample distances were calculated from a count matrix based on all chromosome genes. The heatmap was derived from the distance matrix using R package DESeq2 (v. 1.20.0).
Fig. S3 Comparison of gene expression by RT‐qPCR and RNA‐seq. At least 16 genes were randomly selected for ΔdksA and ΔspoTΔrelA strains. The mRNA abundance of these genes was tested by RT‐qPCR using gyrA as an endogenous control. The R‐squared value and the P‐value were calculated by R software (v. 3.5.1).
Fig. S4 Distribution of DEGs of ΔdksA and ΔspoTΔrelA compared to wild‐type Xcc according to COG categories. For Xcc306, the numbers mean the total genes in each category according to the Xanthomonas Genome Browser (http://xgb.leibniz‐fli.de/cgi/cog.pl?ssi=free).
Fig. S5 Down‐regulated DEGs involved in the histidine metabolism pathway. Genes in red represent down‐regulated DEGs in the ΔdksA and ΔspoTΔrelA mutants compared to wild‐type Xcc, genes in orange represent down‐regulated DEGs only in ΔdksA, genes in green represent down‐regulated DEGs only in ΔspoTΔrelA. This pathway is based on the KEGG pathway database (https://www.genome.jp/kegg‐bin/show_pathway?xac00340).
Fig. S6 Transmission electron microscopic observation of Xcc306, ΔdksA and ΔspoTΔrelA strains. Fresh bacteria of Xcc306, ΔdksA and ΔspoTΔrelA from XVM2 solid agar medium were transferred to formvar/carbon‐coated 400‐mesh copper grids and stained with 0.25% aqueous ammonium molybdate. The grids were allowed to dry for 1 h before observation with an FEI Morgagni 268 transmission electron microscope (FEI, OR, USA). The black arrows indicate bacterial polar flagella.
Fig. S7 Gene expression profile of common DEGs for ΔdksA and ΔspoTΔrelA. Hierarchical cluster analysis shows that all common DEGs were grouped into four clusters: I, II, III and IV. Forty genes (IV) were up‐regulated, and 426 genes (II) were down‐regulated in both mutants. Twelve genes (I) were up‐regulated in ΔdksA but down‐regulated in ΔspoTΔrelA. Four genes (III) were down‐regulated in ΔdksA but up‐regulated in ΔspoTΔrelA.
Table S1 Data quality summary.
Table S2 Differentially regulated genes in the ΔspoTΔrelA mutant compared to the wild‐type Xcc.
Table S3 Differentially regulated genes in the ΔdksA mutant compared to wild‐type (WT) Xcc.
Table S4 Gene expression of tRNA‐coding genes.
Table S5 Gene expression of ribosome protein genes.
Table S6 Gene expression profile of T3SS‐ and T2SS‐related genes in Xcc.
Table S7 Gene expression profile for putative type 4 pilus biogenesis and regulation genes of Xcc.
Table S8 Gene expression level of TBDT genes of Xcc.
Table S9 Differentially regulated genes by DksA and ppGpp.
Table S10 Strains and plasmids used in this study.
Table S11 Primer sequence used in this study.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.


