Abstract
RAS is one of the most well-known proto-oncogenes. Its gain-of-function mutations occur in approximately 30% of all human cancers. As the most frequently mutated RAS isoform, KRAS is intensively studied in the past years. Despite its well-recognized importance in cancer malignancy, continuous efforts in the past three decades failed to develop approved therapies for KRAS mutant cancer. KRAS has thus long been considered to be undruggable. Encouragingly, recent studies have aroused renewed interest in the development of KRAS inhibitors either directly towards mutant KRAS or against the crucial steps required for KRAS activation. This review summarizes the most recent progress in the exploration of KRAS-targeted anticancer strategies and hopefully provides useful insights for the field.
KEY WORDS: KRAS, Oncogene, Mutation, Cancer, Inhibitor, Targeted therapy
Graphical abstract
Continuous efforts in the past three decades failed to develop approved therapies for KRAS mutant cancer. Encouragingly, recent progress in the development of KRAS inhibitors either directly towards mutant KRAS or against the crucial steps required for KRAS activation may bring breakthrough for this long-pursued undruggable target.
1. Introduction
In the past decades, numerous oncogenes have been identified to be constitutively active in cancer due to genetic alterations, in the forms of mutations, amplification, rearrangement, etc. These mutant oncogenes often play a pivotal role in driving cancer progression, known as oncogene addiction. This insight has largely set the stage for the discovery of targeted anticancer therapies. The most successful example is protein kinase inhibitors, which have demonstrated the clinical benefits in a broad range of cancer types1. Among this growing list of oncogenes, KRAS, together with other RAS isoforms, represents the most prevalent oncogene in human cancers, yet decades long efforts in the discovery of RAS targeted therapies failed to obtain clinically approved drugs. Of note, recent years have witnessed the promising progress in exploring the therapeutic opportunities in RAS inhibition. As a wealth of excellent reviews has summarized the role of RAS signaling in driving tumorigenesis and possible direction of RAS-targeted anticancer therapies2., 3., 4., 5., 6., 7., 8., 9., we herein focuses on KRAS, the most frequently mutated RAS isoform, briefly summarizing the current knowledge of KRAS activating mutations in carcinogenesis and mainly revisiting the KRAS-targeted anticancer strategies in the recent years.
2. KRAS protein
KRAS (Kirsten rat sarcoma 2 viral oncogene homolog) gene is a proto-oncogene that encodes a small GTPase transductor protein called KRAS. KRAS belongs to a group of small guanosine triphosphate (GTP) binding proteins, known as RAS superfamily or RAS-like GTPases. Members of RAS superfamily are divided into families and subfamilies based on their structure, sequence and function. The five main families are RAS, RHO, RAN, RAB and ARF GTPases. The RAS family itself is further divided into 6 subfamilies (RAS, RAL, RAP, RHEB, RAD and RIT) and each subfamily shares the common core G domain, which provides essential GTPase and nucleotide exchange activity. RAS is the most frequently studied proteins in the RAS subfamily. In humans, three RAS genes encode highly homologous RAS proteins, HRAS, NRAS and KRAS. KRAS protein exists as two splice variants, KRAS4A and KRAS4B, in which KRAS4B is the dominant form in human cells.
KRAS protein contains four domains. The first domain at the N-terminus is identical in the three RAS forms, and the second domain exhibits relatively lower sequence identity. Both regions are important for the signaling function of the KRAS protein and jointly form the G-domain3. The G-domain of the KRAS protein includes the GTP-binding pocket, a region within which is essential for the interactions between the putative downstream effectors and GTPase-activating proteins (GAPs). KRAS protein also contains a hypervariable region at the C-terminus, which guides posttranslational modifications and determines plasma membrane anchoring. This region plays an important role in the regulation of the biological activity of RAS protein.
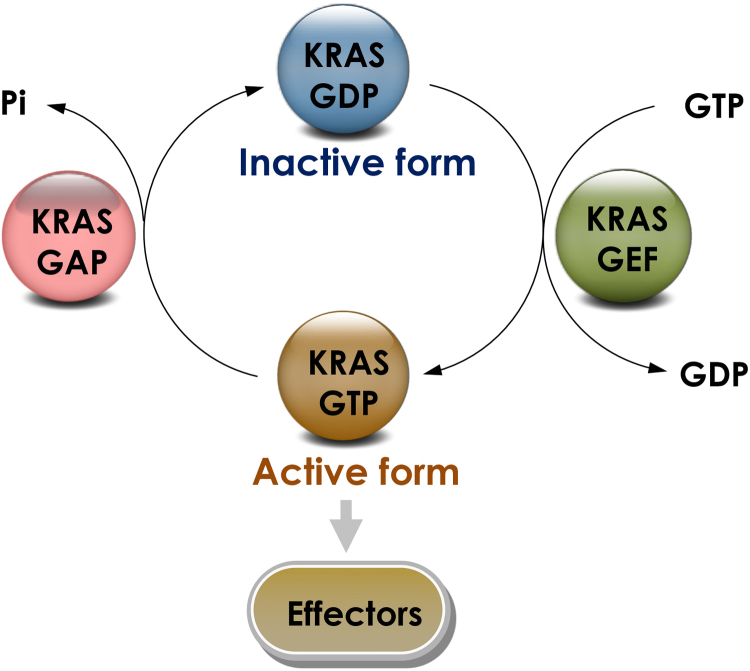
KRAS protein switches between an inactive to an active form via binding to GTP and guanosine diphosphate (GDP), respectively10. Under physiological conditions, the transition between these two states is regulated by guanine nucleotide exchange factors (GEFs), such as Son of Sevenless (SOS), or GAPs via different mechanisms that involve catalyzing the exchange of GDP for GTP, potentiating intrinsic GTPase activity or accelerating RAS-mediated GTP hydrolysis (Fig. 1). Under physiological conditions, KRAS is predominantly GDP-bound. Upon stimulation like growth factors, nucleotide binding of RAS-GEFs is disabled and releases the nucleotide. Upon binding to GTP, KRAS undergoes conformational changes known to result in two major consequences: 1) affecting KRAS interactions with GAPs, which amplify the GTPase activity of the RAS protein around 100,000-fold; 2) affecting the interactions with GEFs and promoting the release of GTP3., 11..
Figure 1.
KRAS GTPase cycle. KRAS regulation occurs through a GDP–GTP cycle that is controlled by the opposing activities of guanine nucleotide-exchange factors (GEFs), which catalyze the exchange of GDP for GTP, and GTPase-activating proteins (GAPs), which increase the rate of GTP hydrolysis to GDP. GTP bound KRAS interacts with various effector proteins, influencing the activity and/or localization of these effectors, which ultimately affects a wide spectrum of cellular pathways.
3. KRAS signaling
KRAS is one of front-line sensors that initiate the activation of an array of signaling molecules, allowing the transmission of transducing signals from the cell surface to the nucleus, and affecting a range of essential cellular processes such as cell differentiation, growth, chemotaxis and apoptosis. In addition to the aforementioned GTP/GDP binding, the activation of KRAS signaling is now known as a multi-step process that requires proper KRAS post-translation, plasma membrane-localization and interaction with effector proteins. These mechanistic insights pave the way for the exploration of KRAS signaling targeted therapies.
Newly synthesized KRAS is a cytosolic and inactive protein. A series of posttranslational modifications occur at the C-terminal CAAX (C, cysteine; A, aliphatic amino acid; X, terminal amino acid) tetrapeptide motif that allows KRAS membrane association12. Briefly, farnesyltransferase-catalyzed covalent addition of a farnesyl moiety to the cysteine residue starts this process. The following step is the proteolytic removal of the last three amino acids by RAS converting enzyme 1 (RCE1), which occurs at the cytosolic surface of the endoplasmic reticulum. Afterwards, isoprenylcysteine carboxyl methyltransferase (ICMT) catalyzes the methyl transfer to the C-terminal amino acid to negate the negative charge and prevent plasma membrane repulsion. In addition, the splice variant KRAS4A undergoes additional palmitoylation by palmitoyl transferase, resulting in proper targeting of KRAS4A to the membrane. However, there is no detectable palmitoylation of the predominant splice variant KRAS4B, which probably reaches the plasma membrane via a microtubule-dependent mechanism. The signal transduction of the KRAS protein does not exclusively occur at the plasma membrane. Activation of downstream signaling pathways by KRAS can also be triggered by signals from subcellular compartments, such as the endoplasmatic reticulum and the Golgi apparatus.
In response to extracellular stimuli, the conversion from inactive RAS-GDP to active RAS-GTP further promotes the activation of various signaling pathways, which includes mitogen-activated protein kinase (MAPK) pathway, phosphoinositide 3-kinase (PI3K) pathway and the Ral-GEFs pathway, among them the MAPK pathway is the best characterized. It is known that RAS-GTP directly binds to RAF protein, recruiting RAF kinase family from cytoplasm to membranes, where they dimerize and become active13. The activated RAF subsequently carries out a chain of phosphorylation reactions to its downstream substrates, namely MEK and ERK, and propagates the growth signal.
4. KRAS mutations in cancer
RAS is one of the most frequently mutated oncogenes in human cancer but the frequency and distribution of RAS gene mutations are not uniform4., 14.. KRAS is the isoform most frequently mutated, which constitutes 86% of RAS mutations. KRAS-4B is the dominant isoform in human cancers, and it is present in approximately 90% of pancreatic cancers, 30% to 40% of colon cancers, and 15% to 20% of lung cancers, mostly non-small-cell lung cancer (NSCLC). It is also present in biliary tract malignancies, endometrial cancer, cervical cancer, bladder cancer, liver cancer, myeloid leukemia and breast cancer (Table 1)3., 5..
Table 1.
Frequency of RAS isoform mutations in human cancers.
| Primary tissue | KRAS (%) | HRAS (%) | NRAS (%) | Total (%) |
|---|---|---|---|---|
| Pancreas | 90 | 0 | <1 | 90 |
| Colon | 30–50 | 1 | 6 | 42 |
| Small intestine | 35 | 0 | <1 | 35 |
| Biliary tract | 26 | 0 | 2 | 28 |
| Endometrium | 17 | <1 | 5 | 22 |
| Lung | 19 | <1 | 1 | 20 |
| Skin (melanoma) | 1 | 1 | 18 | 20 |
| Cervix | 8 | 9 | 2 | 19 |
| Urinary tract | 5 | 10 | 1 | 16 |
The mutations found most frequently in the KRAS gene are primarily at codons 12, 13, or 61. KRAS mutations also occur in codons 63, 117, 119, and 146 but with less frequency3. Structural studies have suggested that somatic missense mutations at these positions often enable perturbation of the intrinsic GTPase activity of the KRAS protein while the detailed the mechanisms could be variable between the specific mutations15. In details, mutation of glycine 12 (G12) causes RAS activation by interfering with GAP binding and GAP-stimulated GTP hydrolysis. Mutations at residue 13 sterically clash with the arginine and decrease GAP binding and hydrolysis. In contrast, glutamine 61 has a direct role in catalysis by positioning the attacking water molecule and helping to stabilize the transition state of the hydrolysis reaction16. As a consequence of diminished GTPase activity, the nucleotide state of KRAS becomes more dependent on relative nucleotide affinity and concentration. This gives GTP an advantage over GDP and increases the proportion of active GTP-bound RAS, causing the accumulation of the activated state. Moreover, mutations at residues 12, 13 and 61 were reported to decrease the affinity for the RAS-binding domain (RBD) of RAF as well but with different extent16.
Of interest, the extent to which specific mutations affect the biological behavior of RAS appears quite different as well3. Previous studies try to establish the link between particular amino acid substitutions and the transforming capabilities of RAS as well as the response to certain cancer therapies17., 18., 19.. For example, KRAS codon 12 valine-for-glycine (G12V) mutant has been associated with a worse prognosis than G12D mutation, from a glycine to an aspartic acid (D), in colorectal and lung cancers20., 21.. Consistently, HRAS-G12V exhibits weaker GTPase activity and stronger binding to GTP than HRAS-G12D, and it is also more potent in cell culture-based transformation assays. Regardless of these advancements, much remains to be learnt about the link between sequence permutations and functional alterations of oncogenic forms of RAS. Studies in patients in several cancer types failed to identify a correlation between the occurrence of specific RAS mutations and the aggressiveness of the disease22. Likewise, differences in KRAS mutations in the gastrointestinal tract appear failing to reflect variations in specific mutation-dependent disease characteristics.
Apart from mutation caused aberrant activation, the association between wild-type and mutant KRAS also plays an important role in mediating KRAS-driven cancer malignancy. It has been known for several years that wild-type KRAS exhibits a tumor-growth-restraining function in KRAS mutant cancer. Wild-type KRAS antagonizes oncogenic KRAS, resulting in inefficient cellular transformation and reduced tumor burden in several malignancies. This inhibitory effect is often overcome during tumor progression due to KRAS gene allele loss or copy number gain of the oncogenic form, resulting in the allelic imbalance23 and the enhanced tumor fitness24. Wild-type KRAS is believed to exert its growth-inhibitory function via the competition for proper membrane localization, shared regulators, downstream mediators, or activation of parallel signaling pathways25. A very recent study uncovered that the tumor-suppressive function of wildtype KRAS depends on its dimerization capacity with mutant KRAS. Impaired wild-type/mutant KRAS dimerization could abolish the growth inhibitory effects by wild-type KRAS; and the dimerization-deficient KRAS mutant retains critical biochemical properties but lacks the biological properties of mutant KRAS26. These insights suggest the importance of fully understanding the regulation of oncogenic KRAS activity and reveal the therapeutically exploitable role for KRAS dimerization in KRAS-driven cancer.
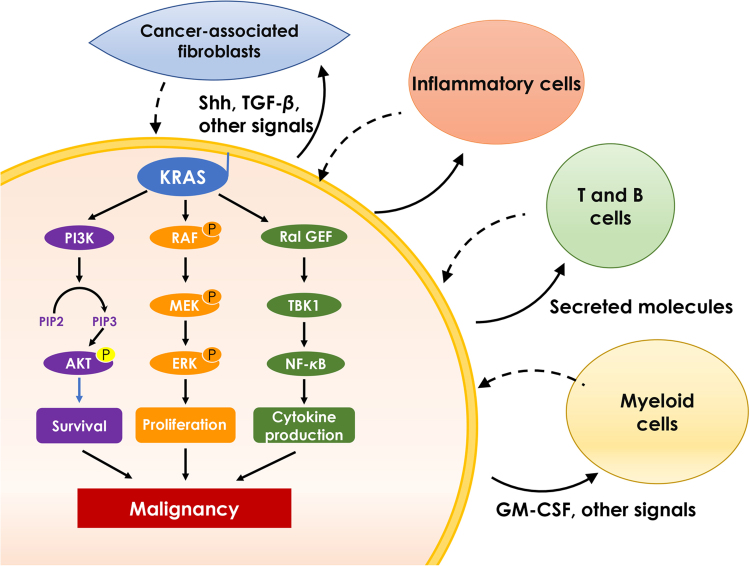
In addition to genetic alterations associated spontaneous tumor development, it has been increasingly appreciated that KRAS mutation also exhibits a broad impact on tumor microenvironment, which contributes to the promotion and maintenance of cancer malignancy. Tumor cells expressing oncogenic KRAS remodels surrounding stroma cells including fibroblasts, innate and adaptive immune cells via secreting molecules in a paracrine manner (Fig. 2)27. Indeed, oncogenic KRAS is a potent inducer of various chemokines, cytokines and growth factors, including interleukin-6 (IL-6), IL-8, chemokine (C-C motif) ligand 9 (CCL9), IL-23, hedgehog, etc, which function as the principal instructing signals for stromal reprogramming. For example, KRAS maintains the stromal inflammatory phenotype via producing IL-6 and IL-8 in pancreatic and lung cancer respectively28., 29.. Tumor cells derived granulocyte-macrophage colony stimulating factor (GM-CSF) could promote the infiltration of myeloid-derived suppressor cells and inhibit anti-tumor immunity30., 31.. It was also noted that MYC and RAS cooperate to program an immune suppressive stroma, which involves the CCL9 mediated recruitment of macrophages, programmed death-ligand 1 (PD-L1) dependent expulsion of T and B cells, as well as IL-23 orchestrated exclusion of adaptive T and B cells and innate immune natural killer (NK) cells28. These findings provide a glimpse of how KRAS is involved in remodeling the adjacent stroma. With the dramatically increased interest in the immune microenvironment, we may expect a better understanding of the role of KRAS in modulating anti-tumor immunity.
Figure 2.
The major KRAS effector pathways. Oncogenic KRAS activates intracellular PI3K, MAPK or RAL-GEF pathways to promote cell survival, proliferation and cytokine secretion. Oncogenic KRAS also induces secretion of molecules that affect surrounding components of the stroma, such as fibroblasts, innate and adaptive immune cells, in a paracrine manner. These stroma cells in turn promote cancer malignancy.
5. Targeting KRAS signaling in cancer therapy
Regardless of the tremendous attempts in the past decades that covered the multiple aspects of KRAS activation, KRAS mutant remains being considered as undruggable. As a result, much focus has been put on alternative approaches instead, such as inhibiting signaling cascades downstream of RAS, in particular the MAPK and PI3K pathways. Selective BRAF inhibitors (vemurafenib and dabrafenib) and dual-specificity MEK1/MEK2 inhibitors (trametinib and combimetinib) have been approved to treat BRAF-mutated melanoma alone or in combination. ERK1/2 kinases, as the exclusive downstream of MEK, have attracted intense efforts as well32. Moreover, inhibition of the downstream transcription factors, such as Fos-like antigen 1 (FOSL1), also showed therapeutic promise in KRAS mutant lung and pancreatic cancer33. All these inhibitors could potentially provide therapeutic solutions to a proportion of KRAS mutant cancer but will require proper patient stratification. For example, ERK1/2 inhibitors obtained about 40% response in KRAS mutant cancer based on cell-based assays in a large panel of cancer cell lines34 and this response rate is expected to largely decrease in clinical test.
Recently, innovative approaches demonstrated the previously unknown binding pockets on the surface of KRAS, which aroused the interests in this field. Several new strategies, such as covalently targeting mutant KRAS, inhibiting KRAS interaction with associated proteins required for membrane association, inhibiting KRAS-driven malignant phenotypes and KRAS synthetic lethal interactions, have showed some promise in cancer therapy and may open new window for KRAS targeted anticancer drugs4., 7., 16.. This review will mainly discuss the recent progress in these new directions. Of note, these strategies have catalyzed quite a few inhibitors that are currently under the development at various stages (Table 2).
Table 2.
Therapeutic strategies towards KRAS-driven cancera.
| Category | Target/mechanism | Compound | Stage |
|---|---|---|---|
| KRAS mutation | KRAS G12C | WW peptide | Preclinical |
| DC-032–759 (DC-040-466, DC-060-162) | Preclinical | ||
| PTD-RBD-VIF (PTD-RBD-Vif-C) | Preclinical | ||
| AU-8653 (AU-BEI-8653) | Preclinical | ||
| ARS-1620 | Preclinical | ||
| ADT-007 (DC-070-547) | Preclinical | ||
| KRAS G12D | KRAS_G12D_21 mer | Preclinical | |
| KRAS modifications | Farnesyltransferaseb | BMS-214662 | Phase II |
| EBP-994 (lonafarnib) | Phase II | ||
| NSC-702818 (R-115777, tipifarnib) | Phase II | ||
| AZD-3409 (EBP-921) | Phase I | ||
| A-228839 (ABT-839, A-228839.25) | Phase I | ||
| GGT | GGTI-2418 (PTX-001, PTX-100) | Phase I | |
| Membrane association | PDEδ | Deltasonamide 1/2 | Biological testing |
| KRAS expression | Oligonucleotide | AZD-4785 (IONIS-KRAS-2.5Rx) | Phase I |
| Oligonucleotide | KRAS-2(cRGD)2 | Preclinical | |
| siRNA | KRAS-siRNA NP | Preclinical | |
| siRNA | SGS6 siRNA | Preclinical | |
| Exosome siRNA | iExosomes | Preclinical | |
| DNA alkylating drugs | KR-12 | Preclinical | |
| KRAS degradation | Fused protein | PTD-RBD-VIF (PTD-RBD-Vif-C) | Preclinical |
| Anti-GTP-bound | Monoclonal antibody targeting the GTP-bound KRAS | RT-11 iMab (RT11 iMab, RT11-i) | Preclinical |
| KRAS-effector interaction | RAS-mimetic | Rigosertib | Preclinical |
| Metabolism | Glutaminase | CB-839 | Phase II |
| Synthetic lethality | BCL2 and MEK | Navitoclax (ABT-263) and trametinib | Phase I |
| TBK1 and MEK | Momelotinib and trametinib | Phase II | |
| CDK4 and MEK | Palbociclib and PD-0325901 | Phase I/II | |
| AKT and MEK | MK2206 and AZD6244 | Phase II | |
| SHP2 and MEK | SHP099 and AZD6244 | Preclinical | |
| Immunotherapy | PD-1 and MEK | Pembrolizumab and trametinib | Phase I |
According to Thomson Reuters Integrity database and www.clinicaltrails.gov.
Over 1000 inhibitors are reported. Listed here are those entered into clinical trials.
5.1. Directly targeting mutant KRAS
In principle, it should be possible to design small molecules that directly bind to GTP-binding site on KRAS and inhibit its interaction with GTP, similar to the approach that has been successfully used for the discovery of ATP-competitive inhibitors of protein kinases. However, this approach are currently considered as “mission impossible”, given the extremely high affinity of KRAS for GTP and the abundance of GTP in cell cytoplasm (~0.5 mmol/L)4. The GTP affinity of KRAS is extremely high, with a dissociation constant (Kd) at ~10−11 mol/L, which is in contrast to the growing list of targeted tyrosine kinases binding ATP at lower micromolar affinity. Thus far, the efforts targeting the GTP binding pocket fail to obtain effective compounds. These experiences, together with the lack of well-defined hydrophobic pockets on the surface of RAS protein, led to a perception that KRAS may be undruggable.
Recently, a new strategy which aimed to exploit a novel “Achilles heel” in KRAS, known as KRAS-G12C oncoprotein, was proposed15. KRAS-G12C is one of the three most common KRAS mutants in cancer, present in roughly 10%–20% of all KRAS G12 mutations and approximately 50% of KRAS-driven lung adenocarcinomas. The mutant cysteine 12 sits in close proximity to both the nucleotide pocket and the switch regions involved in effector interactions of KRAS protein. Small molecules that form covalent bond with the mutant cysteine demonstrated the possibility of directly and selectively targeting the mutant KRAS protein, with an apparent advantage of achieving specificity over the wild-type protein. These irreversible covalent compounds, which bind in an allosteric pocket beneath switch II of KRAS, allowed the identification of a new pocket beneath the effector binding region not apparent in previous structures of RAS. Biochemical analysis showed that these compounds preferentially bind to the GDP state of RAS, impair SOS-catalyzed nucleotide exchange and decrease the affinity of RAS for GTP relative to GDP. Moreover, they appear also blocking RAS–RAF association, which is probably resulted from conformation effects on switch I and switch II as well as other effects on nucleotide exchange and nucleotide affinities. A very recent study also suggests that the covalent inhibition towards G12C requires intact GTPase activity as drug-bound KRASG12C is insusceptible to nucleotide exchange factors and thus trapped in its inactive state. Drugs targeting the inactive or GDP-bound conformation are not expected to be effective. Indeed, mutants completely lacking GTPase activity and those promoting exchange reduced the potency of the drug. Suppressing nucleotide exchange activity enhanced KRAS G12C inhibition, whereas its potentiation had the opposite effect35., 36..
The first discovery of G12C allosteric regulatory site inspires increasing follow-up studies for G12C specific inhibitors. Among them, ARS-853 was a KRAS-G12C cell-specific inhibitor with improved efficacy in modifying KRAS-G12C and blocking exchange of GDP for GTP37. However, it did not show in vivo activity. Whether this KRAS-G12C strategy could translate to in vivo remained unclear. Recently, further optimization of ARS-series compounds led to the discovery of ARS-1620, which represents a new generation of KRASG12C-specific inhibitors with in vivo anticancer activity38.
Certainly, it is apparent that all these compounds will be limited to G12C mutant, as the covalent reaction occurs specifically with the thiol group of the cysteine residue. Recent study also reported the KRAS G12D-selective inhibitors. For example, a peptide generated by random peptide T7 phage display technology inhibited the enzyme activity of KRAS-G12D with IC50 at single-digit nanomolar level and significantly suppressed the downstream ERK-phosphorylation. This study suggested the possibility of targeting KRAS mutant beyond G12C, yet it remains at a very early stage as anticancer therapies39.
5.2. Targeting KRAS membrane association
RAS proteins require localization to the inner leaflet of the plasma membrane for the oncogenic activity, making this association a logical target for anti-RAS therapeutics8. Previous efforts in this direction primarily focused on targeting KRAS posttranslational modifications that modulate KRAS membrane association. The greatest drug discovery effort has gone into developing farnesyltransferase inhibitors (FTI) based on the knowledge that prenylation of the CAAX cysteine is required for oncogenic transformation40. Through a massive effort by many leading pharmaceutical companies, a large number of highly effective FTIs have been identified. But phase II and phase III trials of FTIs were disappointing. It was initially believed that blocking KRAS membrane association might be a flawed approach until further understanding of KRAS modification and trafficking has revealed that the root of the problem lies in the fact that KRAS4B can also be modified by geranylgeranyltransferase (GGT). Geranylgeranylation of KRAS, an alternative 20-carbon isoprenylation, could support the bioactivity of KRAS when farnesylation is impaired. Simultaneous genetic inactivation of FI and GGTI was shown to reduce KRAS-driven lung tumorigenesis in mice41. But the combined inhibition of these enzymes by small molecules has yet to show efficacy in KRAS-driven cancers and the high toxicity is concerned as well.
Recent findings regarding RAS isoform trafficking and the regulation of RAS subcellular localization have rekindled interest in efforts to target these processes. In particular, improved understanding of the palmitoylation/depalmitoylation cycle that regulates RAS interaction with the plasma membrane, endomembrane, and cytosol, and of the potential importance of RAS chaperones, have led to new approaches. Efforts to validate and target other enzymatically regulated posttranslational modifications are also ongoing. An important progress has recently been made by targeting prenyl-binding protein PDEδ, which is required for the correct localization and signaling of farnesylated RAS. PDEδ augments RAS signaling via enriching RAS at the plasma membrane. The GDI-like pocket of PDEδ binds and solubilizes farnesylated RAS proteins, thereby enhancing their diffusion in the cytoplasm. This mechanism allows more effective trapping of depalmitoylated RAS proteins at the Golgi and polycationic RAS proteins at the plasma membrane to counter the entropic tendency to distribute these proteins over all intracellular membranes42. This finding inspired the endeavor to disrupt the association of KRAS with PDEδ for the intervention of KRAS signaling. A work from Waldmann group reported the first small-molecule inhibitor deltarasin that interferes with binding of mammalian PDEδ to KRAS and impairs KRAS localization to endomembrane. In this study, biochemical screening and subsequent structure-based hit optimization yielded inhibitors of the KRAS-PDEδ interaction that selectively bind to the prenyl-binding pocket of PDEδ with nanomolar affinity, inhibit oncogenic KRAS signaling and suppress in vitro and in vivo proliferation of human pancreatic ductal adenocarcinoma cells dependent on oncogenic KRAS43. This work provided the proof-of-concept for this new strategy but the specificity of the compound raised concerns. A follow-up work from the same group reported a second series of inhibitors yielding an improved PDEδ inhibitor, designated as deltazinone 1, with high selectivity and less unspecific cytotoxicity than deltarasin and demonstrates a high correlation with the phenotypic effect of PDEδ knockdown in a set of human pancreatic cancer cell lines. However, the in vivo anticancer activity of this series of compounds was not assessed due to the stability issue44. Further chemical optimization will be required to fully evaluate this strategy.
5.3. Exploiting KRAS-regulated metabolic pathways
Recent advancement in cancer metabolism has brought the attention of KRAS-related studies to metabolic area. Oncogenic RAS promotes a metabolic reprogramming of tumor cells, leaning to an anabolic metabolism to produce biomass to support unrestricted proliferation. KRAS mutant cancers also use a diverse set of fuel sources to meet their metabolic needs and have developed a variety of mechanisms to obtain metabolic substrates from both extracellular and intracellular sources45., 46., 47.. These adaptations result in tumor specific metabolic vulnerabilities that tumor cells rely on particular pathways or rate-limiting metabolites. Inhibiting individual or combinations of these metabolic pathways open new therapeutic opportunities. Although targeting tumor metabolism is still in the early days of translation to patients, the continued advances in understanding critical metabolic adaptations in RAS-driven cancers, as well as the ability to study this altered metabolism in relevant tumor models, will accelerate the development of new therapeutic approaches. Because these dependencies are tumor selective, there is the opportunity for therapeutic intervention.
KRAS mutant human colon tumors are associated with the increased expression of glycolytic and glutamine metabolic proteins48. KRAS mutation in human pancreatic ductal adenocarcinoma drives the reprogramming of the glutamine metabolism. These cells rely on a distinct pathway in which glutamine-derived aspartate is transported into the cytoplasm and converted into oxaloacetate by aspartate transaminase (GOT1). This pathway is essential for maintaining the cellular redox state and supporting cancer cell growth49. Compared to KRAS wild-type cells, highly glycolytic KRAS mutant cells exhibit a metabolic vulnerability on glyceraldehyde 3-phosphate dehydrogenase (GAPDH), the inhibition of which leads to an energetic crisis and cell death50. In addition to glucose and glutamine metabolism, a recent study also reports that KRAS reprograms lipid homeostasis to support tumorigenesis. KRAS mutant lung cancer upregulates acyl-coenzyme A (CoA) synthetase long-chain family member 3 (ACSL3), which converts fatty acids into fatty acyl-CoA esters to supply lipid synthesis and β-oxidation51. Moreover, KRAS-G12D mutant drives a lipogenic gene-expression program to promote de novo lipogenesis52. All these findings have stimulated the attempts to treat KRAS mutant tumor with metabolic inhibitors, most of which have been tested in pre-clinical cancer models carrying KRAS mutations. Whether this may provide new opportunities for the therapy of KRAS mutant cancer still await clinical validation.
5.4. Synthetic lethality in KRAS mutant cancer
Synthetic lethal interactions have been widely exploited for cancer therapy, among which the most successful case might be PARP inhibitors treating cancers deficient in the homologous recombination repair pathway. Synthetic lethality defines the interaction between two co-essential genes that inhibiting both genes rather than either single gene could result in cell death53. Inspired by this idea, a variety of approaches including chemical, siRNA, shRNA, and CRISPR library screens have been implemented to identify synthetic lethal interactors with the KRAS oncogene. These efforts led to the identification of a wide array of pathways that are exquisitely required for the survival of KRAS mutant cells, including co-operating signaling, transcriptional regulation, maintenance of genomic stability, etc.4., 54.. These insights revealed the therapeutic promise of the combinational therapies for the treatment of KRAS mutant cancer55. For instance, it was lately reported that inhibition of SRC homology region 2-containing protein tyrosine phosphatase 2 (SHP2) provokes a senescence response in KRAS-mutant NSCLC, which is exacerbated by MEK inhibition. SHP2 inhibition gives rise to a vulnerability of KRAS-mutant NSCLC cells that could be exploited therapeutically. Thus far, several combination regimens are suggested for their potential therapeutic promise in treating KRAS mutant cancer and a couple of them, such as combing B-cell lymphoma 2 (BCL2) and MEK inhibitors, combined inhibition of MEK and AKT are undergoing clinical test56. However, at this moment, most of the interaction genes still lack effective therapeutics. For example, KRAS mutant NSCLC cells are shown to depend on the transcription factor GATA-binding factor 2 (GATA2) that is currently undruggable57.
Combinational therapy may represent the major solutions to eventually overcome this fatal disease. It should be noted that most of the synthetic lethality studies in KRAS mutant cancer are based on certain cancer cell lines or tumor models. As KRAS mutant cancers are indeed highly heterogeneous, a systematic study covering a spectrum of KRAS mutant cancer will be important for eventually coming up with feasible solutions that have translational value in clinic.
5.5. Immunotherapy for KRAS mutant cancer
Cancer immunotherapy is undoubtedly drawing the most attention in cancer treatment at the moment, especially for the immune checkpoint inhibitors that are aggressively tested in almost all cancer types. Recently, these efforts are also gradually expanding to KRAS mutant cancer. For example, anti-programmed cell death protein 1 (PD-1) antibody pembrolizumab has been tested in KRAS mutant NSCLC in combination with trametinib, a MEK inhibitor. While it is too early to conclude the clinical benefits, immunotherapies are expected to bring new hope for this type of cancer. In support of this notion, a multiple-dimensional analysis using genomic, transcriptomic, proteomic and clinical data in cohorts of lung adenocarcinoma immunotherapeutic patients observed that TP53/KRAS co-mutated subgroup manifested exclusive increased expression of PD-L1 and a highest proportion of PD-L1+/CD8+. KRAS mutant tumor showed prominently increased mutation burden as well. The prospective analysis in patients showed remarkable clinical benefit to PD-1 inhibitors in TP53 or KRAS mutant patients, especially those with co-occurring TP53/KRAS mutations58.
Moreover, it was recently revealed that the promotion and progression of KRAS-driven lung cancer was closely associated with the dysfunctional state of natural killer (NK) cells. The mechanism involves the aberrant expression of fructose-1,6-bisphosphatase (FBP1) in NK cells, which elicited this dysfunction by inhibiting glycolysis and impairing viability. This may suggest the potential directions for NK cell-based cancer immunotherapy involving FBP1 targeting59.
6. Perspectives
Recent progress in KRAS targeted anticancer drug discovery has aroused the interest to re-visit this long-pursued target. Nevertheless, we are still only near the beginning of a very long path for conquering the KRAS mutant cancer. Whether these compounds, with a new mechanism of action such as the G12C covalent inhibitors, would be advantageous compared with the previous attempts still await clinical validation. Nevertheless, these new directions bring the hope to the field and are stimulating more and more efforts to seek a better understanding of KRAS activation in cancer. In the meanwhile, it is important to mention even if these drugs are clinically effective, acquired resistance is expected to arise inevitably, given the strong selective pressure applied to the genetically unstable cancer cells. For example, mutations to cysteine at G12 site is expected to hinder the covalent binding at this residue.
KRAS itself apart, to target the KRAS-driven malignant phenotypes, such as the metabolic vulnerabilities of KRAS mutant cancer mentioned in this review, might represent another effective strategy. In this case, the challenge will be the compensatory effect that allows escaping the original dependency. To probe the proper drug combination could be an option. Also, the combined inhibition with KRAS mutant tumor and the programmed environment, such as the modulation of immune system, will be worthy to test.
Tumor stratification will be essential for the eventual success of KRAS targeted therapies. With increasingly revealed heterogeneous properties between KRAS-driven tumors, even between the same mutant forms, to precisely direct the therapies to selected patients are important to ensure the efficacy. Recently, the proteomic analysis of KRAS mutant cancer using tumor cells and patients samples has suggested the existence of multiple subtypes. These studies may potentially reveal the molecular markers for proper selection in the future.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
References
- 1.Huang M., Shen A., Ding J., Geng M. Molecularly targeted cancer therapy: some lessons from the past decade. Trends Pharmacol Sci. 2014;35:41–50. doi: 10.1016/j.tips.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Jancik S., Drabek J., Radzioch D., Hajduch M. Clinical relevance of KRAS in human cancers. J Biomed Biotechnol. 2010;2010:150960. doi: 10.1155/2010/150960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. Ras oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox A.D., Fesik S.W., Kimmelman A.C., Luo J., Der C.J. Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh H., Longo D.L., Chabner B.A. Improving prospects for targeting RAS. J Clin Oncol. 2015;33:3650–3659. doi: 10.1200/JCO.2015.62.1052. [DOI] [PubMed] [Google Scholar]
- 6.Stephen A.G., Esposito D., Bagni R.K., McCormick F. Dragging RAS back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 7.Papke B., Der C.J. Drugging RAS: know the enemy. Science. 2017;355:1158–1163. doi: 10.1126/science.aam7622. [DOI] [PubMed] [Google Scholar]
- 8.Cox A.D., Der C.J., Philips M.R. Targeting RAS membrane association: back to the future for anti-RAS drug discovery? Clin Cancer Res. 2015;21:1819–1827. doi: 10.1158/1078-0432.CCR-14-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konstantinopoulos P.A., Karamouzis M.V., Papavassiliou A.G. Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat Rev Drug Discov. 2007;6:541–555. doi: 10.1038/nrd2221. [DOI] [PubMed] [Google Scholar]
- 10.Bos J.L., Rehmann H., Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 11.Scheffzek K., Ahmadian M.R., Kabsch W., Wiesmuller L., Lautwein A., Schmitz F. The Ras–RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 12.Ahearn I.M., Haigis K., Bar-Sagi D., Philips M.R. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol. 2011;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavoie H., Therrien M. Regulation of RAF protein kinases in ERK signalling. Nat Rev Mol Cell Biol. 2015;16:281–298. doi: 10.1038/nrm3979. [DOI] [PubMed] [Google Scholar]
- 14.Prior I.A., Lewis P.D., Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostrem J.M., Peters U., Sos M.L., Wells J.A., Shokat K.M. K-Ras (G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostrem J.M., Shokat K.M. Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat Rev Drug Discov. 2016;15:771–785. doi: 10.1038/nrd.2016.139. [DOI] [PubMed] [Google Scholar]
- 17.Tyner J.W., Erickson H., Deininger M.W., Willis S.G., Eide C.A., Levine R.L. High-throughput sequencing screen reveals novel, transforming RAS mutations in myeloid leukemia patients. Blood. 2009;113:1749–1755. doi: 10.1182/blood-2008-04-152157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edkins S., O׳Meara S., Parker A., Stevens C., Reis M., Jones S. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006;5:928–932. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Roock W., Jonker D.J., Di Nicolantonio F., Sartore-Bianchi A., Tu D., Siena S. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 20.Andreyev H.J., Norman A.R., Cunningham D., Oates J.R., Clarke P.A. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 21.Keohavong P., DeMichele M.A., Melacrinos A.C., Landreneau R.J., Weyant R.J., Siegfried J.M. Detection of K-ras mutations in lung carcinomas: relationship to prognosis. Clin Cancer Res. 1996;2:411–418. [PubMed] [Google Scholar]
- 22.Kompier L.C., Lurkin I., van der Aa M.N., van Rhijn B.W., van der Kwast T.H., Zwarthoff E.C. FGFR3, HRAS, KRAS, NRAS and PIK3CA mutations in bladder cancer and their potential as biomarkers for surveillance and therapy. PLoS One. 2010;5:e13821. doi: 10.1371/journal.pone.0013821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westcott P.M., Halliwill K.D., To M.D., Rashid M., Rust A.G., Keane T.M. The mutational landscapes of genetic and chemical models of KRAS-driven lung cancer. Nature. 2015;517:489–492. doi: 10.1038/nature13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgess M.R., Hwang E., Mroue R., Bielski C.M., Wandler A.M., Huang B.J. KRAS allelic imbalance enhances fitness and modulates map kinase dependence in cancer. Cell. 2017;168:817–829. doi: 10.1016/j.cell.2017.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young A., Lou D., McCormick F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 2013;3:112–123. doi: 10.1158/2159-8290.CD-12-0231. [DOI] [PubMed] [Google Scholar]
- 26.Ambrogio C., Kohler J., Zhou Z.W., Wang H., Paranal R., Li J. KRAS dimerization impacts MEK inhibitor sensitivity and oncogenic activity of mutant KRAS. Cell. 2018;172 doi: 10.1016/j.cell.2017.12.020. [857-68 e15] [DOI] [PubMed] [Google Scholar]
- 27.di Magliano M.P., Logsdon C.D. Roles for KRAS in pancreatic tumor development and progression. Gastroenterology. 2013;144:1220–1229. doi: 10.1053/j.gastro.2013.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kortlever R.M., Sodir N.M., Wilson C.H., Burkhart D.L., Pellegrinet L., Brown Swigart L. Myc cooperates with Ras by programming inflammation and immune suppression. Cell. 2017;171:1301–1315. doi: 10.1016/j.cell.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukuda A., Wang S.C., Morris J.Pt, Folias A.E., Liou A., Kim G.E. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19:441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pylayeva-Gupta Y., Lee K.E., Hajdu C.H., Miller G., Bar-Sagi D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell. 2012;21:836–847. doi: 10.1016/j.ccr.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bayne L.J., Beatty G.L., Jhala N., Clark C.E., Rhim A.D., Stanger B.Z. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 2012;21:822–835. doi: 10.1016/j.ccr.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu F., Yang X., Geng M., Huang M. Targeting ERK, an Achilles׳ heel of the MAPK pathway, in cancer therapy. Acta Pharm Sin B. 2018;8:552–562. doi: 10.1016/j.apsb.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vallejo A., Perurena N., Guruceaga E., Mazur P.K., Martinez-Canarias S., Zandueta C. An integrative approach unveils FOSL1 as an oncogene vulnerability in KRAS-driven lung and pancreatic cancer. Nat Commun. 2017;8:14294. doi: 10.1038/ncomms14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris E.J., Jha S., Restaino C.R., Dayananth P., Zhu H., Cooper A. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 2013;3:742–750. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 35.Lito P., Solomon M., Li L.S., Hansen R., Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351:604–608. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shipman L. Signalling: putting the brakes on KRAS-G12C nucleotide cycling. Nat Rev Cancer. 2016;16:127. doi: 10.1038/nrc.2016.13. [DOI] [PubMed] [Google Scholar]
- 37.Patricelli M.P., Janes M.R., Li L.S., Hansen R., Peters U., Kessler L.V. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 2016;6:316–329. doi: 10.1158/2159-8290.CD-15-1105. [DOI] [PubMed] [Google Scholar]
- 38.Janes M.R., Zhang J., Li L.S., Hansen R., Peters U., Guo X. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell. 2018;172:578–589. doi: 10.1016/j.cell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 39.Sakamoto K., Kamada Y., Sameshima T., Yaguchi M., Niida A., Sasaki S. K-Ras (G12D)-selective inhibitory peptides generated by random peptide T7 phage display technology. Biochem Biophys Res Commun. 2017;484:605–611. doi: 10.1016/j.bbrc.2017.01.147. [DOI] [PubMed] [Google Scholar]
- 40.Berndt N., Hamilton A.D., Sebti S.M. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. 2011;11:775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu M., Sjogren A.K., Karlsson C., Ibrahim M.X., Andersson K.M., Olofsson F.J. Targeting the protein prenyltransferases efficiently reduces tumor development in mice with K-RAS-induced lung cancer. Proc Natl Acad Sci U S A. 2010;107:6471–6476. doi: 10.1073/pnas.0908396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandra A., Grecco H.E., Pisupati V., Perera D., Cassidy L., Skoulidis F. The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat Cell Biol. 2011;14:148–158. doi: 10.1038/ncb2394. [DOI] [PubMed] [Google Scholar]
- 43.Zimmermann G., Papke B., Ismail S., Vartak N., Chandra A., Hoffmann M. Small molecule inhibition of the KRAS—PDEδ interaction impairs oncogenic KRAS signalling. Nature. 2013;497:638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- 44.Papke B., Murarka S., Vogel H.A., Martin-Gago P., Kovacevic M., Truxius D.C. Identification of pyrazolopyridazinones as PDEδ inhibitors. Nat Commun. 2016;7:11360. doi: 10.1038/ncomms11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuneva M.O., Fan T.W., Allen T.D., Higashi R.M., Ferraris D.V., Tsukamoto T. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayers J.R., Torrence M.E., Danai L.V., Papagiannakopoulos T., Davidson S.M., Bauer M.R. Tissue of origin dictates branched-chain amino acid metabolism in mutant KRAS-driven cancers. Science. 2016;353:1161–1165. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bryant K.L., Mancias J.D., Kimmelman A.C., Der C.J. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39:91–100. doi: 10.1016/j.tibs.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hutton J.E., Wang X., Zimmerman L.J., Slebos R.J., Trenary I.A., Young J.D. Oncogenic KRAS and BRAF drive metabolic reprogramming in colorectal cancer. Mol Cell Proteom. 2016;15:2924–2938. doi: 10.1074/mcp.M116.058925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Son J., Lyssiotis C.A., Ying H., Wang X., Hua S., Ligorio M. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yun J., Mullarky E., Lu C., Bosch K.N., Kavalier A., Rivera K. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science. 2015;350:1391–1396. doi: 10.1126/science.aaa5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Padanad M.S., Konstantinidou G., Venkateswaran N., Melegari M., Rindhe S., Mitsche M. Fatty acid oxidation mediated by acyl-coA synthetase long chain 3 is required for mutant KRAS lung tumorigenesis. Cell Rep. 2016;16:1614–1628. doi: 10.1016/j.celrep.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh A., Ruiz C., Bhalla K., Haley J.A., Li Q.K., Acquaah-Mensah G. De novo lipogenesis represents a therapeutic target in mutant Kras non-small cell lung cancer. FASEB J. 2018;32:7018–7027. doi: 10.1096/fj.201800204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan D.A., Giaccia A.J. Harnessing synthetic lethal interactions in anticancer drug discovery. Nat Rev Drug Discov. 2011;10:351–364. doi: 10.1038/nrd3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aguirre A.J., Hahn W.C. Synthetic lethal vulnerabilities in KRAS-mutant cancers. Cold Spring Harb Perspect Med. 2018;8:a031518. doi: 10.1101/cshperspect.a031518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dietlein F., Kalb B., Jokic M., Noll E.M., Strong A., Tharun L. A synergistic interaction between Chk1- and MK2 inhibitors in KRAS-mutant cancer. Cell. 2015;162:146–159. doi: 10.1016/j.cell.2015.05.053. [DOI] [PubMed] [Google Scholar]
- 56.Corcoran R.B., Cheng K.A., Hata A.N., Faber A.C., Ebi H., Coffee E.M. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell. 2013;23:121–128. doi: 10.1016/j.ccr.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar M.S., Hancock D.C., Molina-Arcas M., Steckel M., East P., Diefenbacher M. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149:642–655. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
- 58.Dong Z.Y., Zhong W.Z., Zhang X.C., Su J., Xie Z., Liu S.Y. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 59.Cong J., Wang X., Zheng X., Wang D., Fu B., Sun R. Dysfunction of natural killer cells by FBP1-induced inhibition of glycolysis during lung cancer progression. Cell Metab. 2018;28:243–255. doi: 10.1016/j.cmet.2018.06.021. [DOI] [PubMed] [Google Scholar]