Abstract
Autophagy is essential for cellular metabolism and plays pivotal roles in carcinogenesis, while excessive autophagy induces toxicity and cell death. Our previous studies have suggested that let-7a-5p/BCL-xL might regulate autophagy in lung cancer, but the regulatory mechanism is unclear. The central goal of the study was to figure out the role of let-7a-5p/BCL-xL in the initiation of autophagy and its effect on the migration, invasion, and proliferation of A549 cells as well as its therapeutic potential in lung cancer. Based on the genome-wide expression profiles of lung cancer, BCL-xL and let-7a-5p were found to be dysregulated and negatively correlated in lung adenocarcinoma, which was associated with the survival of lung cancer. The crosstalk between BCL-xL and let-7a-5p was then investigated using dual-luciferase reporter assay, and it was found to suppress the migration and invasion of A549 cells. Further, we found that the crosstalk between BCL-xL and let-7a-5p could lead to toxic autophagy and cell death through activating the PI3K-signaling pathway, which was independent of apoptosis or pyroptosis. These findings indicate that let-7a-5p is a sensitive initiator for toxic autophagy in A549 lung cancer cells and is an appealing target for lung cancer therapy.
Keywords: lung adenocarcinoma, toxic autophagy, apoptosis, BCL-xL, let-7a-5p
Introduction
Lung cancer, a global public health problem, has been identified as the leading cause of cancer-associated death both in men and women.1 According to a report depicted by Cancer Statistics, there will be 246,440 estimated new lung cancer cases in the United States in 2019, accounting for 13% of all newly diagnosed cancer patients.2 Meanwhile, the estimated number of lung cancer deaths is 142,670, which is much higher than the number of prostate cancer and breast cancer, making it the most life-threatening cancer in human beings.2 Based on cancer registry data in 2014, Chen and colleagues3 indicated that about 78.2 new lung cancer cases were diagnosed in every 1 million Chinese people, and a total of 626,000 patients died of lung cancer in 2014. The latest report from the GLOBOCAN concerning the incidence and mortality of lung cancer showed that the developing countries faced more severe conditions than the developed countries.4
For the etiology of lung cancer, epidemiological studies revealed that smoking, alcohol, genetic factors, environmental air pollution, diet, and obesity contributed to the pathogenesis of lung cancer.5, 6, 7 Available studies have confirmed the relationship between lung cancer and tobacco smoking.8, 9, 10 Specially, other types of smoking, such as secondhand smoking, cigar smoking, and pipe smoking, have also been associated with lung cancer.11, 12, 13, 14 Moreover, Yoshida and colleagues15 demonstrated that a family history of cancer increased the susceptibility to lung cancer by 1.72 after adjusting for age, gender, history of smoking, and dust exposure. Fortunately, immunotherapy has made great progress in lung cancer treatment.16 Thus, a deep investigation into the mechanism of lung cancer may provide new therapeutic targets for lung cancer.
MicroRNAs (miRNAs), a subset of endogenously initiated non-coding RNAs with roughly 19–24 nt in length, could post-transcriptionally repress gene expression through mRNA degradation, and it is generally believed that miRNAs play dominant roles in the maintenance of nearly all physiological activities, including growth and development, learning, memory, as well as tumorigenesis.17, 18 A growing number of studies have demonstrated that miRNAs were tightly associated with lung cancer, while dysregulation of miRNAs led to the proliferation, migration, and invasion of lung cancer cells.19 Profiling of miRNAs in lung cancer revealed that a high expression of hsa-mir-155 corresponded to poor survival of lung cancer, which was proposed as a potential diagnostic and prognostic biomarker of lung cancer.20 Moreover, the expression of a certain number of miRNAs, including miR-146a-5p, miR-324-5p, miR-223-3p, and miR-223-5p, was detected to be altered in the cytologically normal bronchial epithelium of smokers with lung cancer, which may serve as potential targets for the treatment of lung cancer.21
Let-7 families are a group of well-studied miRNAs in many diseases, among which downregulation of let-7a-2 was associated with the poor survival of lung cancer, and let-7 was also found to inhibit the growth of multiple human lung cancer cell lines in vitro, as well as the growth of lung cancer xenografts in vivo.22 Li et al.23 reported that hyperoside and let-7a-5p synergistically repressed lung cancer cell proliferation via inducing G1/S phase arrest. Alternatively, it is suggested that let-7a-5p, as exosomal cargo, could be transported intercellularly and contribute to the migration and invasion of pulmonary cells.24 Therefore, it is essential to deeply investigate the role of let-7a-5p in the pathogenesis of lung cancer. Our previous studies revealed that let-7a-5p could target BCL-xL, IGF1R, mitogen-activated protein kinase (MAPK), and FAS and repress their expression, which might affect the autophagy and apoptosis of lung cancer cells.25 However, the putative regulatory mechanism of let-7a-5p in lung cancer has not been validated.
The role of autophagy in lung cancer is drawing more and more attention, while the activation and regulation of autophagy in lung cancer is unclear. Autophagy is a lysosome-dependent cellular degradation pathway that is essential for cell survival, differentiation, development, and homeostasis, which serves an adaptive role in protecting organisms against various damages, including cancer, aging, and cardiovascular diseases.26 Nonetheless, there are also studies indicating the harmful effect of autophagy in metabolisms. It is reported that autophagy suppresses tumor growth at the early stage of lung cancer but protects cancer cells from the immune system defense mechanisms.27 Mechanistically, it is widely accepted that the pro-survival BCL2 families could block apoptosis and autophagy by directly binding to BECN1/Beclin1,28 and the complex of VDAC2-BECN1-BCL-xL is further proved to suppress autophagy in ovary development in mammals;29 but, the role of let-7a-5p in BCL-xL-mediated autophagy is seldom investigated.
In this study, we identified the regulatory mechanism of let-7a-5p in lung cancer and demonstrated the role of autophagy in the proliferation, migration, and invasion of A549 lung cancer cells. Utilizing the genome-wide expression profiles of lung cancer in The Cancer Genome Atlas (TCGA) database, we explored the effect of let-7a-5p and BCL-xL on the survival of lung cancer patients. The crosstalk between let-7a-5p and BCL-xL was also investigated using dual-luciferase reporter assay, and the survival of A549 lung cancer cells under different expression levels of let-7a-5p/BCL-xL was checked using flow cytometry. The target genes at the downstream of the BCL-xL-mediated PI3K-signaling pathway were examined, and the morphological characteristics of A549 lung cancer cells were observed by a transmission electron microscope. Our research provides the basis for the mechanism study of lung cancer, and it highlights an immensely important role of autophagy in the treatment of lung cancer.
Results
Characterization of BCL-xL and let-7a-5p in Lung Cancer
A total of 969 lung cancer patients was recruited in this study; 483 of 969 lung cancer patients were diagnosed with lung adenocarcinoma, and 486 of 969 lung cancer patients were diagnosed with lung squamous cell carcinoma, among whom 59 patients with lung adenocarcinoma and 50 patients with lung squamous cell carcinoma simultaneously provided samples for BCL-xL and let-7a-5p profiling as control. The demographic details could be referred to TCGA database available online at https://portal.gdc.cancer.gov/. The genome-wide expression data were transformed with the algorithm of log2(transcript per million [TPM] + 1) and then evaluated using boxplot for homogeneity. As shown in Figures 1A–1D, the expressions of BCL-xL and let-7a-5p were compared between lung cancer and healthy control. The expression of BCL-xL was found significantly lower in lung adenocarcinoma, but it was found to be similarly expressed in lung squamous cell carcinoma. The expression of let-7a-5p decreased in both lung adenocarcinoma and lung squamous cell carcinoma.
Figure 1.
Expression, Correlation, and Survival Effect of BCL-xL/let-7a-5p in Lung Cancer
(A–D) Expressions of BCL-xL and let-7a-5p in lung adenocarcinoma and lung squamous cell carcinoma. Comparing with the control group, BCL-xL was downregulated in lung adenocarcinoma, and let-7a-5p was downregulated in both lung adenocarcinoma and lung squmous carcinoma. All data were deposited in TCGA and normalized by the algorithm of log2(transcript per million + 1) before comparison. *p < 0.05 (pooled t test). (E–H) Kaplan-Meier survival analysis of BCL-xL and let-7a-5p in lung adenocarcinoma and lung squamous cell carcinoma, the hazard ratio between different expression levels of BCL-xL/let-7a-5p was calculated. (I and J) Correlation between BCL-xL and let-7a-5p in lung adenocarcinoma and lung squamous cell carcinoma. The prediction ellipses in red and blue included 70% and 80% samples, respectively. *p < 0.05 (Pearson correlation test).
Accompanying the follow-up data of the 969 candidates, we explored the association of BCL-xL and let-7a-5p with the survival of lung cancer. Using the Kaplan-Meier survival analysis, we compared two patient cohorts and calculated the hazard ratio (HR) with 95% confidence intervals (95% CIs) and log-rank p value, as shown in Figures 1E–1H. We found that the high expression of BCL-xL adversely affected the survival of lung adenocarcinoma (HR = 1.6, p = 0.024), but dysregulation of BCL-xL did not alter the survival of lung squamous cell carcinoma (HR = 1.4, p = 0.096). Interestingly, elevated expression of let-7a-5p was beneficial for patients with lung adenocarcinoma (HR = 0.8, p = 0.022), while dysregulation of let-7a-5p did not affect the survival of lung squamous cell carcinoma (HR = 0.9, p = 0.725).
Furthermore, we investigated the relationship between BCL-xL and let-7a-5p using the genome-wide expression profiles normalized by log2(TPM + 1) (Figures 1I and 1J). For lung adenocarcinoma, the expression of BCL-xL was negatively correlated with the expression of let-7a-5p (correlation coefficient value −0.103, p value 0.0269). However, the correlation coefficient (−0.001) between BCL-xL and let-7a-5p was of no statistical significance in lung squamous cell carcinoma (p < 0.001).
Dysregulation of BCL-xL/let-7a-5p Alters the Migration and Invasion of A549 Lung Cancer Cells
To functionally investigate the biological activity of BCL-xL and let-7a-5p in lung adenocarcinoma, we altered the expressions of them via transfecting small hairpin RNA (shRNA)/pcDNA-BCL-xL plasmids or let-7a-5p mimics/inhibitors into A549 lung cancer cells, respectively. The migration of A549 lung cancer cells was found to be significantly decreased after inducing let-7a-5p mimics but elevated when transfecting with let-7a-5p inhibitors. Similarly, the downregulation of BCL-xL using shRNAs repressed A549 cell migration, but transfection of pcDNA-BCL-xL reversed the suppression caused by shRNAs. In comparison with the control group, the migration caused by either miRNA mimics/inhibitors or shRNA/pcDNA was significantly altered. Further, the invasion was found non-significantly changed in A549 cells treated with neither let-7a-5p mimics nor shRNA-BCL-xL plasmids but enhanced when cells were transfected with let-7a-5p inhibitors or pcDNA-BCL-xL plasmids (Figure 2). This observation confirmed that BCL-xL and let-7a-5p conversely acted in regulating the migration and invasion of A549 lung cancer cells.
Figure 2.
Dysregulation of BCL-xL/let-7a-5p Alters the Migration and Invasion of A549 Lung Cancer Cells
(A and B) Invasion and migration of A549 lung cancer cells transfected with let-7a-5p mimics or inhibitors (40×). (C and D) Invasion and migration of A549 lung cancer cells transfected with BCL-xL shRNA or BCL-xL pcDNAs (40×). (E–H) Quantitative analysis of (A)–(D). The migration and invasion of A549 lung cancer cells were enhanced in let-7a-5p inhibitor treated-cells, while the invasion was attenuated in cells treated with let-7a-5p mimics. For cells treated with sh-BCL2L1 or pc-BCL2L1, the invasion and migration were enhanced in pc-BCL2L1 group, but the invasion was attenuated in sh-BCL2L1 group. *p < 0.05 (one-way ANOVA, Bonferroni post hoc), error bars (standard error of mean).
let-7a-5p Directly Suppresses BCL-xL by Binding to Its 3′ UTRs
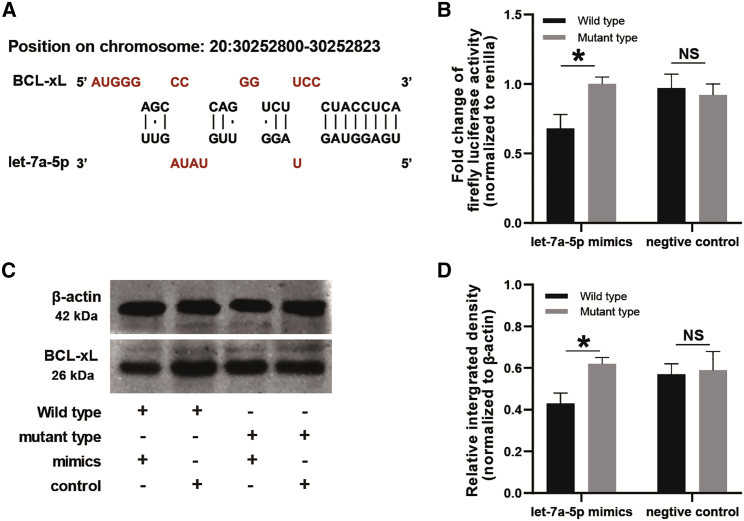
The interaction between BCL-xL and let-7a-5p was further investigated. Structurally, we showed the let-7a-5p binding site in the sequence of BCL-xL, in which the bases from the second to the seventh in the seed region of let-7a-5p could precisely bind to the sequence in the 3′ UTRs of BCL-xL (Figure 3A; Figure S1). Based on the base-pairing structure, we developed the pmirGLO dual reporter luciferase vector containing the putative binding site as well as the mutant binding site of let-7a-5p, and we transfected them into A549 lung cancer cells, respectively. It was observed that the luciferase activity significantly reduced in A549 lung cancer cells co-transfected with pmirGLO-BCL-xL-UTR vectors and let-7a-5p mimics as compared to cells co-transfected with mutant vectors and let-7a-5p mimics. However, alteration of luciferase activity was found non-significantly changed when cells were transfected with the let-7a-5p negative control (Figure 3B). Moreover, we detected the expression of BCL-xL in A549 lung cancer cells, and the expression values of BCL-xL were normalized to the values of β-actin. For cells co-transfected with let-7a-5p mimics and pmirGLO-BCL-xL-UTR vectors, the expression of BCL-xL was considerably lower in comparing with cells co-transfected with let-7a-5p mimics and mutant vectors (Figure 3D). However, for cells transfected with let-7a-5p negative control, the expression of BCL-xL was similar as compared to cells transfected with vectors of pmirGLO-BCL-xL-UTR and mutant type (Figure 3D).
Figure 3.
let-7a-5p Directly Targets and Inhibits BCL-xL
(A) Potential binding site targeted by let-7a-5 in BCL-xL. (B) Luciferase reporter assays display the fold change of firefly luciferase activity in A549 lung cancer cells. Cells were pre-transfected with pmirGLO reporter plasmid containing wild-type or mutant BCL-xL-3′ UTR and then treated with let-7a-5p mimics or negative control. *p < 0.05 (pooled t test), error bars (standard error of mean). (C) Western blot gels of BCL-xL and β-actin. (D) Quantitative analysis of (C). The relative integrated density of BCL2L1 was upreguated in A549 cells transfected with let-7a-5p mutant type mimics as compared to the wide type cells, while there was no statistical significance between groups in cells transfected with negtive controls. The relative expression of BCL-xL was normalized to β-actin. *p < 0.05 (pooled t test), error bars (standard error of mean).
let-7a-5p Promotes A549 Lung Cancer Cell Death
To investigate the effect of BCL-xL/let-7a-5p crosstalk on lung cancer cells, we examined the viability of A549 lung cancer cells. Using flow cytometry, we detected the cell death rate and apoptosis rate in A549 lung cancer cells (Figure 4A). It was found that the cell death rate increased in cells treated with let-7a-5p mimics but decreased in cells treated with let-7a-5p inhibitors, while the proportion of apoptotic A549 cells was found to be non-significantly altered among cells treated with let-7a-5p mimics or inhibitors as well as the untreated wild-type cells (Figures 4B and 4C). These data suggest that let-7a-5p induces apoptosis-independent cell death in A549 lung cancer cells.
Figure 4.
Crosstalk of BCL-xL and let-7a-5p Induces Apoptosis-Independent A549 Cell Death
(A) A549 cells treated with let-7a-5p mimics or inhibitors in flow cytometry. (B and C) Quantitative analysis of apoptotic and dead cells in (A). Comparing with the control group, the proportion of death cell in let-7a-5p mimic group or inhibitor group increased, and it was much higher in the inhibitor group. For apoptosis analysis of A549 cells with different treatments, there were no statistical significance found among different treatment groups. *p < 0.05 (pooled t test), error bars (standard error of mean). (D) Western blot gels of cleaved caspase-1/3, LC3-II, and β-actin. (E–G) Quantitative analysis of (D). The expression of cleaved caspase-1/3 was found to be of non-significant changes among different groups. The expression of LC-II increased in let-7a-5p mimic group, while it was downregulated as compared to let-7a-5p inhibitor group. The relative expression of cleaved caspase-1/3 and LC3-II was normalized to β-actin. *p < 0.05 (pooled t test), error bars (standard error of mean).
To identify the death pattern regulated by let-7a-5p, we detected the expressions of specific biomarkers related to different cell death categories. As has been suggested, caspase-1, caspase-3, and LC3-II were indicators for pyroptosis, apoptosis, and autophagy;30, 31 we therefore quantified the expressions of them in A549 lung cancer cells (Figures 4D–4G). Consistent with the proportion of apoptotic cells detected by flow cytometry, the expressions of caspase-3 and caspase-1 were found to be non-significantly altered among cells treated with let-7a-5p mimics or inhibitors as well as wild-type controls, while the expression of LC3-II was significantly elevated in cells treated with let-7a-5p mimics but downregulated when repressing let-7a-5p in A549 lung cancer cells.
let-7a-5p Induces Toxic Autophagy via Suppressing BCL-xL, and the Downstream Signaling Cascade of BCL-xL Entails PI3K Signaling
The morphological characteristics of A549 lung cancer cells were observed under the transmission electron microscope, and we found that cells treated with let-7a-5p mimics showed blurred cell contour and typical autophagosomes, in which undigested organelles were involved, but cells in the control group showed precise cell contour and fewer autophagosomes (Figure 5A). Furthermore, we investigated the mechanism of let-7a-5p promoting autophagy in A549 lung cancer cells. Given the crosstalk between let-7a-5p and BCL-xL and the putative mechanism reported in our previously published work,25 we detected the expression of genes at the downstream of BCL-xL in the PI3K-signaling pathway, including Beclin1, NRBF2, PIK3C3, and ATG5 (Figures 5B–5G). It was found that a high expression of let-7a-5p elevated the expressions of NRBF2, PIK3C3, and ATG5 as compared to the control group, while suppression of let-7a-5p inhibited the expressions of Beclin1, NRBF2, PIK3C3, and ATG5 compared with cells transfected with let-7a-5p mimics. These data suggested that autophagy in A549 lung cancer cells was induced by let-7a-5p and tightly associated with the PI3K-signaling pathway.
Figure 5.
Upregulation of let-7a-5p Induces Toxic Autophagy and Initiates PI3K-Signaling Pathway in A549 Cells
(A) Morphological characteristics of autophagosomes in A549 lung cancer cells under the transmission electron microscope. (B) Western blot gels of Beclin-1, NRBF2, PIK3C3, ATG5, and β-actin. (C–F) Quantitative analysis of western blot gels in (B). Comparing with the control group, the expressions of NRBF2, PIK3C3, and ATG5 in the mimic group were upregulated, while the expressions of Beclin-1, NRBF2, PIK3C3, and ATG5 in the inhibitor group was downregulated as compared to the mimic group. The relative expression of Beclin-1, NRBF2, PIK3C3, and ATG5 was normalized to β-actin. *p < 0.05 (pooled t test), error bars (standard error of mean). (G) Schematic representation of macroautophagy induced by the PI3K-signaling pathway.
Discussion
As the most critical component of non-small-cell lung cancer, lung adenocarcinoma has been widely investigated in most recent years; however, there have been no effective treatment strategies. For most of the current studies concerning the etiology of lung adenocarcinoma, the A549 cell line provides an excellent model for the investigation of lung cancer and, therefore, is widely used.32, 33, 34 In this study, we demonstrated that the suppression of BCL-xL by let-7a-5p enhanced autophagy but repressed migration and invasion in A549 lung cancer cells, which was tightly associated with the activation of the PI3K-signaling pathway.
The expression pattern of BCL-xL/let-7a-5p in A549 lung cancer cells provided emerging evidence for the etiological study of lung cancer. We found that the expressions of BCL-xL and let-7a-5p were significantly elevated in lung adenocarcinoma compared with the healthy adjacent control, while there were no significant alterations between lung squamous cell carcinoma and the corresponding control, indicating that dysregulation of BCL-xL and let-7a-5p might only affect the pathogenesis of lung adenocarcinoma, but not lung squamous cell carcinoma. In line with studies previously published, the expression of BCL-xL was not correlated with keratinization, a specific morphologic characteristic that was generally used to indicate the poor clinical outcome of lung squamous cell carcinoma.35 On the other hand, inhibition of the Bcl-2 family of anti-apoptotic proteins caused a dose-dependent decrease of viability and clonogenicity in A549 lung cancer cells,36 and downregulation of BCL-xL expression by 25% using an aptamer-targeted BCL-xL shRNA delivery system induced 14% late apoptosis in A549 lung cancer cells.37
In this study, we found that BCL-xL and let-7a-5p showed the opposite effect in deciding the survival of lung adenocarcinoma, while the survival of lung squamous cell carcinoma was not affected by them. Consistent with the expression patterns of BCL-xL and let-7a-5p, a high expression of BCL-xL was negatively correlated with the survival of lung adenocarcinoma, while a high expression of let-7a-5p prolonged the survival of the lung cancer. Maruoka et al.38 pointed out that the inhibition of BCL-xL using lemongrass essential oil resulted in the suppression of cancer cell proliferation and survival elevation in lung cancer. Pan-inhibition of BCL-xL protein family using sabutoclax was found to induce cell death and overcome drug resistance in breast cancer.39 Utilizing the expressions of miR-769-5p and let-7d-5p in lung cancer, Gasparini et al.40 generated a prognostic classifier that can predict overall survival of lung cancer patients. Besides, let-7 was also found collaborating with miR-34 to suppress non-small-cell lung cancer cell proliferation.41 However, the interaction between BCL-xL and let-7a-5p was seldom reported in lung cancer. Limited evidence suggests that let-7b inhibited the expression of BCL-xL in platelets42 and bioengineered let-7c inhibited orthotopic hepatocellular carcinoma via suppressing the expression of BCL-xL.43 Collectively, we may conclude that the expression of BCL-xL is negatively correlated with let-7a-5p and dysregulation of BCL-xL/let-7a-5p alters the survival of lung cancer, which is potentially associated with the pathogenesis of lung cancer.
Using the lung adenocarcinoma model constructed by the A549 cell line, we explored the role of BCL-xL and let-7a-5p on the migration and invasion of lung cancer cells in vitro. Interestingly, we found that the upregulation of BCL-xL or downregulation of let-7a-5p enhanced the migration and invasion of A549 lung cancer cells, which preliminarily confirmed the interaction between BCL-xL and let-7a-5p. Therefore, we adopted the dual-luciferase reporter assay to validate the hypothesis by inducing let-7a-5p putative binding sites or mutation in the 3′ UTR of BCL-xL in A549 lung cancer cells. As a result, the firefly luciferase activity decreased in cells with putative binding sites but was unchanged in cells with BCL-xL mutant phenotypes as compared to the wild-type control, and the expression of BCL-xL at the protein level further confirmed the hypothesis. Similarly, Liu et al.44 had reported that the upregulation of BCL-xL by TRIM59 might promote cell proliferation, invasion, migration, and cell cycle transition through the AKT-signaling pathway in breast cancer. Meanwhile, let-7 could act as a target of the competing endogenous RNA (ceRNA) HMGA2 in promoting lung cancer progression, where HMGA2 ceRNA activity promoted lung cancer growth, invasion, and dissemination,45 and let-7b/let-7c was found suppressing growth and invasion of lung cancer cells via repressing Cofilin-1 expression in vitro and in vivo.46 To the best of our knowledge, studies concerning the interaction between BCL-xL and let-7a-5p were limited, while data presented in this study provides evidence on the crosstalk between BCL-xL and let-7a-5p, and the biological activity of the crosstalk was further confirmed in aspects of migration and invasion in A549 lung cancer cells.
We also investigated the effect of BCL-xL/let-7a-5p crosstalk on the viability of A549 lung cancer cells, and we found that the dysregulation of let-7a-5p altered the cell death rate but did not change the cell apoptosis rate, which seemed to be that the cell death regulated by BCL-xL/let-7a-5p crosstalk was independent of the apoptosis-signaling pathway. To clarify the role of BCL-xL/let-7a-5p crosstalk in lung cancer cell death, we examined the expressions of caspase-1, caspase-3, and LC3-II, three specific biomarkers of pyroptosis, apoptosis, and autophagy, respectively. Consistent with the result of flow cytometry, the apoptosis represented by caspase-3 was not affected by let-7a-5p or BCL-xL, and a similar phenomenon was also found in pyroptosis detected using caspase-1. However, the expression of LC3-II changed with the alteration of let-7a-5p, indicating that BCL-xL/let-7a-5p crosstalk might induce lung cancer cell death through the autophagy-signaling pathway.
Recent studies also found the association between macroautophagy/autophagy and BCL-xL. For example, it was found that BCL-xL showed a high affinity toward the phosphorylated BECN1, a critical protein in the formation of autophagosomes, and the complex of BECN1-BCL-xL could block macroautophagy/autophagy.47, 48 Alternatively, dissociation of BCL-xL with the phosphorylated BECN1 also suppressed the autophagy and resulted in metabolic stress.49 Moreover, we examined the downstream cascade of BCL-xL in the PI3K-signaling pathway, including Beclin1, NRBF2, PIK3C3, and ATG5, and we found that the suppression of BCL-xL activated those proteins and resulted in macroautophagy in A549 lung cancer cells. However, autophagy is widely known as an essential factor for cell proliferation, migration, and invasion, yet it was seldom reported that autophagy contributed to cell death.
Chen et al.50 found that enhanced autophagy contributed to the proliferation and metastasis of pancreatic ductal adenocarcinoma and could be repressed by UBL4A via targeting LAMP1. Similar studies also demonstrated that a deficiency of basal autophagy induced thyroid follicular epithelial cell death,51 and autophagy played seminal roles for cells to adapt abdominal microenvironment in metastatic ovarian cancer.52 Different from findings in these studies, Emdad et al.53 systematically and comprehensively provided the conception of toxic autophagy, where excessive autophagy could promote type II cell death. Autophagy might also be toxic to cancer cells when facing impaired autophagic degradation conditions, as reported by Tang and colleagues.54 Despite the evidence mentioned above, studies concerning the toxicity of autophagy are limited, while the toxic property of autophagy may be potentially used as a critical therapeutic strategy for cancers. Interestingly, data in this study provided the basis for lung cancer treatment through the toxic autophagy-signaling pathway.
To sum up, it could be stated that the crosstalk between BCL-xL and let-7a-5p regulates the toxic autophagy as well as its therapeutic potential in lung cancer. BCL-xL and let-7a-5p were found to be dysregulated in lung adenocarcinoma, which was also associated with the survival of lung cancer. Inspired by the negative correlation between BCL-xL and let-a7-5p, we demonstrated their crosstalk and confirmed its effect on migration and invasion in A549 lung cancer cells. Further, we also found that the crosstalk between BCL-xL and let-7a-5p activated the PI3K-signaling pathway and induced toxic autophagy-dependent cell death. This study provides insight into the employment of let-7a-5p as an effective therapy targeting BCL-xL for the better management of lung cancer in clinical settings in the future.
Materials and Methods
Clinical Samples
We included 969 lung cancer patients registered at TCGA and collected their high-throughput sequencing datasets of mRNAs and miRNAs via the online platform at https://portal.gdc.cancer.gov/. 483 of 969 subjects were diagnosed with lung adenocarcinoma, and the adjacent healthy tissues of 59 of 483 subjects were recruited as the control for precise comparison with lung adenocarcinoma tissues. 486 of 969 subjects were diagnosed with lung squamous cell carcinoma, and the adjacent healthy tissues of 50 of 486 subjects were recruited as the control for comparison with lung squamous cell carcinoma tissues. The baseline characteristics of all subjects, including age, gender, smoking history, and cancer stages, were recorded and compared between lung cancer and the control. This study has been approved by the review board of Shandong Maternal and Child Health Care Hospital.
Cell Line
The lung adenocarcinoma cell line (A549) was purchased from the American Type Culture Collection (ATCC) and has been identified using short tandem polymorphism (STR) typing method. Cells were maintained in complete DMEM growth medium containing 10% fetal bovine serum, 100 μg/mL streptomycin, 100 U/mL penicillin, and 2 mM glutamine at 37°C in a humidified incubator with 5% CO2. Cells growing up to 80% confluence were rinsed using 0.25% (w/v) trypsin and passaged at the ratio of 1:3. Cell line experiments were carried out between 3 and 6 passages of revival.
Plasmid Design, Construction, and Transfection
Plasmids of shRNA and pcDNA using to silence or overexpress BCL-xL were designed and constructed, respectively (Hanbio Biotechnology, Shanghai, China). The full length of BCL-xL cDNA was chemically synthesized and cloned into the artificial-pcDNA promoter-reporter module, which was used to express BCL-xL, and the empty pcDNA was transfected as control. Meanwhile, an anti-BCL-xL element constructed using shRNA was transfected into A549 lung cancer cells.
Synthesis and Transfection of let-7a-5p Mimics and Inhibitors
The mature sequences of let-7a-5p mimic (5′-UGAGGUAGUAGGUUGUAUAGUU-3′), inhibitor (5′-AACUAUACAACCUACUACCUCA-3′), and their related controls (mimic, 5′-UUCUCCGAACGUGUCACGUTT-3′; inhibitor, 5′-AGCUCCCAAGAGCCUAACCCGU-3′) were chemically synthesized, respectively (Sangon Biotech, Shanghai, China). 2 × 105 A549 lung cancer cells were seeded in 12-well plates and maintained for 24 h before transfection. With the help of Lipo6000 (Beyotime, Shanghai, China), A549 lung cancer cells were transfected with the chemically synthesized mimics or inhibitors as well as their controls, respectively. Kept incubating for 18 h, cells were harvested, and the transfection efficiency was checked by detecting the relative expression of BCL-xL through PCR.
BCL-xL 3′ UTR Cloning and Dual-Luciferase Reporter Assay
The 3′ UTR of BCL-xL containing putative target sites of let-7a-5p was amplified via PCR and cloned into pmirGLO-dual-luciferase vector. Also, the related sequence in 3′ UTR of BCL-xL was then mutated by the overlap PCR-based site-directed mutagenesis method. 1 × 106 A549 lung cancer cells were seeded in a 6-well plate and starved for 2 h, and then cells were transfected with pmriGLO as well as control, respectively. After incubating for 48 h, A549 cells were harvested and subjected to the Dual-Luciferase Reporter Assay System for fluorescence detection and quantitative analysis.
Transwell Migration and Invasion Assay
For migration assay, 1 × 105 A549 cells were resuspended in 150 μL serum-free DMEM and plated in the upper compartment of the 24-well transwell; the lower compartment of the transwell was filled with 500 μL 10% FBS containing complete DMEM. For invasion assay, we added 50 μL Matrigel of 2 μg/μL concentration into the upper compartment and incubated them for 3 h at 37°C in a humidified incubator with 5% CO2 to solidify the Matrigel. A549 cells were then suspended with serum-free DMEM and plated in the upper compartment of the transwell, and they were maintained in a standard 5% CO2 incubator for 20 h. Cells in the lower compartment were collected via centrifugation, and the number was counted using a blood corpuscle counting meter under an optical microscope.
Western Blot
Protocols for western blot were as previously described.55 In brief, the total protein of A549 cells from different intervention groups was isolated and quantified. 20–40 μg total protein was resolved on SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes, followed by blocking the membrane for 2 h with fat-free milk, and then the membrane was incubated with the rabbit anti-human primary antibody of β-actin, BCL-xL, caspase-1, caspase-3, LC3-II, Beclin-1, NRBF2, PIK3C3, and ATG5 for 12 h at 4°C, respectively. Washing the protein-coated membrane with TBST 3 times, the membrane was then exposed to the secondary antibody for 2 h at room temperature. We repeated the washing procedure and subjected the membrane to Fluro Chem HD2Gel imaging system for signal detection.
Cell Flow Cytometry
A549 cells from different invention groups were digested and collected using 0.25% trypsin, followed by termination with complete medium, and then cells were centrifuged at 400 × g for 3 min, the supernatant was discarded, and the cell pellet at the bottom of the centrifuge tube was resuspended in PBS. 2 μL Annexin V Alexa Fluor488 was added following incubation for 15 min; 1 μL propidine iodide (PI) was then added to cells. Apoptotic cells and dead cells were measured using Accuri C6 flow cytometry (Becton Dickinson, Franklin Lakes, NJ, USA).
Transmission Electron Microscopy
According to protocols described above, A549 cells were harvested and fixed in the reagent composed of 2.5% glutaraldehyde and 0.1 M cacodylate buffer (pH 7.4) for 2 h. Then the pre-fixed cells were washed with PBS and post-fixed in 2% osmium tetroxide, followed by aspiration of the osmium tetroxide, and the cell pellet at the bottom of centrifuge tube was embedded in Epon resin and subjected to transmission electron microscopy at 80 kV voltage for ultrastructure observation.
Statistical Analysis
SAS version 9.2 was used for statistical assessment. Data were presented as mean ± SD for parametric data with normalized distribution and median (range) for non-parametric data. Pearson correlation analysis was adopted to determine the correlation between BCL-xL and let-7a-5p in lung adenocarcinoma. Kaplan-Meier survival analysis was used to assess the effect of BCL-Xl/let-7a-5p on the survival of lung cancer. Limma test was used to compare the genome-wide expression differences between lung cancer and the control. Pooled variance t test was applied to analyze differences between groups for parametric data. A p value less than 0.05 was considered statistically significant.
Author Contributions
L.Z., S.Y., and Y.W. designed the study. L.Z. and S.D. wrote the manuscript. S.D., J.L., J.T., and H.Y. performed the work. S.D. and J.L. analyzed the data and interpreted the results. All authors read and approved the final version of the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We express our sincere appreciation for members of the school of public health and experimental center, Weifang Medical University, for their help in providing facilities and reviewers who made valuable suggestions. This work was supported by Projects of Medical and Health Technology Development Program in Shandong Province (2018WS059).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2019.08.010.
Contributor Information
Yongjun Wu, Email: wuyongjun135@126.com.
Sanqiao Yao, Email: sanqiaoyao@126.com.
Lin Zhang, Email: zhanglin8901@163.com.
Supplemental Information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Chen W., Sun K., Zheng R., Zeng H., Zhang S., Xia C., Yang Z., Li H., Zou X., He J. Cancer incidence and mortality in China, 2014. Chin. J. Cancer Res. 2018;30:1–12. doi: 10.21147/j.issn.1000-9604.2018.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D.M., Piñeros M., Znaor A., Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 5.Dela Cruz C.S., Tanoue L.T., Matthay R.A. Lung cancer: epidemiology, etiology, and prevention. Clin. Chest Med. 2011;32:605–644. doi: 10.1016/j.ccm.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alberg A.J., Brock M.V., Samet J.M. Epidemiology of lung cancer: looking to the future. J. Clin. Oncol. 2005;23:3175–3185. doi: 10.1200/JCO.2005.10.462. [DOI] [PubMed] [Google Scholar]
- 7.Alberg A.J., Samet J.M. Epidemiology of lung cancer. Chest. 2003;123(1, Suppl):21S–49S. doi: 10.1378/chest.123.1_suppl.21s. [DOI] [PubMed] [Google Scholar]
- 8.O’Keeffe L.M., Taylor G., Huxley R.R., Mitchell P., Woodward M., Peters S.A.E. Smoking as a risk factor for lung cancer in women and men: a systematic review and meta-analysis. BMJ Open. 2018;8:e021611. doi: 10.1136/bmjopen-2018-021611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tremblay A., Taghizadeh N., Huang J., Kasowski D., MacEachern P., Burrowes P., Graham A.J., Dickinson J.A., Lam S.C., Yang H. A Randomized Controlled Study of Integrated Smoking Cessation in a Lung Cancer Screening Program. J. Thorac. Oncol. 2019;14:1528–1537. doi: 10.1016/j.jtho.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 10.Rosen R.J. Smoking and Lung Cancer Mortality in the United States From 2015 to 2065. Ann. Intern. Med. 2019;170:740. doi: 10.7326/L19-0065. [DOI] [PubMed] [Google Scholar]
- 11.de Groot P.M., Chung J.H., Ackman J.B., Berry M.F., Carter B.W., Colletti P.M., Hobbs S.B., McComb B.L., Movsas B., Tong B.C., Expert Panel on Thoracic Imaging ACR Appropriateness Criteria® Noninvasive Clinical Staging of Primary Lung Cancer. J. Am. Coll. Radiol. 2019;16(5S):S184–S195. doi: 10.1016/j.jacr.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 12.Kim C.H., Lee Y.C., Hung R.J., McNallan S.R., Cote M.L., Lim W.Y., Chang S.C., Kim J.H., Ugolini D., Chen Y. Exposure to secondhand tobacco smoke and lung cancer by histological type: a pooled analysis of the International Lung Cancer Consortium (ILCCO) Int. J. Cancer. 2014;135:1918–1930. doi: 10.1002/ijc.28835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pope C.A., 3rd, Burnett R.T., Turner M.C., Cohen A., Krewski D., Jerrett M., Gapstur S.M., Thun M.J. Lung cancer and cardiovascular disease mortality associated with ambient air pollution and cigarette smoke: shape of the exposure-response relationships. Environ. Health Perspect. 2011;119:1616–1621. doi: 10.1289/ehp.1103639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention (CDC) State-specific trends in lung cancer incidence and smoking--United States, 1999-2008. MMWR Morb. Mortal. Wkly. Rep. 2011;60:1243–1247. [PubMed] [Google Scholar]
- 15.Yoshida K., Takizawa Y., Nishino Y., Takahashi S., Kanemura S., Omori J., Kurosawa H., Maemondo M., Minami Y. Association between Family History of Cancer and Lung Cancer Risk among Japanese Men and Women. Tohoku J. Exp. Med. 2019;247:99–110. doi: 10.1620/tjem.247.99. [DOI] [PubMed] [Google Scholar]
- 16.Daly M.E., Monjazeb A.M., Kelly K. Clinical Trials Integrating Immunotherapy and Radiation for Non-Small-Cell Lung Cancer. J. Thorac. Oncol. 2015;10:1685–1693. doi: 10.1097/JTO.0000000000000686. [DOI] [PubMed] [Google Scholar]
- 17.Treiber T., Treiber N., Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019;20:5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- 18.Gebert L.F.R., MacRae I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu W., Zhang H., Niu Y., Wu Y., Sun W., Li H., Kong J., Ding K., Shen H.M., Wu H. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol. Cancer. 2017;16:118. doi: 10.1186/s12943-017-0685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R.M., Okamoto A., Yokota J., Tanaka T. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 21.Pavel A.B., Campbell J.D., Liu G., Elashoff D., Dubinett S., Smith K., Whitney D., Lenburg M.E., Spira A., AEGIS Study Team Alterations in Bronchial Airway miRNA Expression for Lung Cancer Detection. Cancer Prev. Res. (Phila.) 2017;10:651–659. doi: 10.1158/1940-6207.CAPR-17-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esquela-Kerscher A., Trang P., Wiggins J.F., Patrawala L., Cheng A., Ford L., Weidhaas J.B., Brown D., Bader A.G., Slack F.J. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 23.Li J.P., Liao X.H., Xiang Y., Yao A., Song R.H., Zhang Z.J., Huang F., Dai Z.T., Zhang T.C. Hyperoside and let-7a-5p synergistically inhibits lung cancer cell proliferation via inducing G1/S phase arrest. Gene. 2018;679:232–240. doi: 10.1016/j.gene.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L., Hao C., Yao S., Tang R., Guo W., Cong H., Li J., Bao L., Wang D., Li Y. Exosomal miRNA Profiling to Identify Nanoparticle Phagocytic Mechanisms. Small. 2018;14:e1704008. doi: 10.1002/smll.201704008. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L., Hao C., Zhai R., Wang D., Zhang J., Bao L., Li Y., Yao W. Downregulation of exosomal let-7a-5p in dust exposed- workers contributes to lung cancer development. Respir. Res. 2018;19:235. doi: 10.1186/s12931-018-0949-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saha S., Panigrahi D.P., Patil S., Bhutia S.K. Autophagy in health and disease: A comprehensive review. Biomed. Pharmacother. 2018;104:485–495. doi: 10.1016/j.biopha.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Lindqvist L.M., Vaux D.L. BCL2 and related prosurvival proteins require BAK1 and BAX to affect autophagy. Autophagy. 2014;10:1474–1475. doi: 10.4161/auto.29639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan J., Zhang Y., Sheng Y., Fu X., Cheng H., Zhou R. MYBL2 guides autophagy suppressor VDAC2 in the developing ovary to inhibit autophagy through a complex of VDAC2-BECN1-BCL2L1 in mammals. Autophagy. 2015;11:1081–1098. doi: 10.1080/15548627.2015.1040970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouyang L., Shi Z., Zhao S., Wang F.T., Zhou T.T., Liu B., Bao J.K. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45:487–498. doi: 10.1111/j.1365-2184.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Opdenbosch N., Lamkanfi M. Caspases in Cell Death, Inflammation, and Disease. Immunity. 2019;50:1352–1364. doi: 10.1016/j.immuni.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang C.H., Chen M.C., Chiu T.H., Li Y.H., Yu W.C., Liao W.L., Oner M., Yu C.R., Wu C.C., Yang T.Y. Arecoline Promotes Migration of A549 Lung Cancer Cells through Activating the EGFR/Src/FAK Pathway. Toxins (Basel) 2019;11:E185. doi: 10.3390/toxins11040185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z., Liu G., Jiang J. Profiling of apoptosis- and autophagy-associated molecules in human lung cancer A549 cells in response to cisplatin treatment using stable isotope labeling with amino acids in cell culture. Int. J. Oncol. 2019;54:1071–1085. doi: 10.3892/ijo.2019.4690. [DOI] [PubMed] [Google Scholar]
- 34.Duan S., Yu S., Yuan T., Yao S., Zhang L. Exogenous Let-7a-5p Induces A549 Lung Cancer Cell Death Through BCL2L1-Mediated PI3Kγ Signaling Pathway. Front. Oncol. 2019;9:808. doi: 10.3389/fonc.2019.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park H.J., Cha Y.J., Kim S.H., Kim A., Kim E.Y., Chang Y.S. Keratinization of Lung Squamous Cell Carcinoma Is Associated with Poor Clinical Outcome. Tuberc. Respir. Dis. (Seoul) 2017;80:179–186. doi: 10.4046/trd.2017.80.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H., Shao H., Golubovskaya V.M., Chen H., Cance W., Adjei A.A., Dy G.K. Efficacy of focal adhesion kinase inhibition in non-small cell lung cancer with oncogenically activated MAPK pathways. Br. J. Cancer. 2016;115:203–211. doi: 10.1038/bjc.2016.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayatollahi S., Salmasi Z., Hashemi M., Askarian S., Oskuee R.K., Abnous K., Ramezani M. Aptamer-targeted delivery of Bcl-xL shRNA using alkyl modified PAMAM dendrimers into lung cancer cells. Int. J. Biochem. Cell Biol. 2017;92:210–217. doi: 10.1016/j.biocel.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 38.Maruoka T., Kitanaka A., Kubota Y., Yamaoka G., Kameda T., Imataki O., Dobashi H., Bandoh S., Kadowaki N., Tanaka T. Lemongrass essential oil and citral inhibit Src/Stat3 activity and suppress the proliferation/survival of small-cell lung cancer cells, alone or in combination with chemotherapeutic agents. Int. J. Oncol. 2018;52:1738–1748. doi: 10.3892/ijo.2018.4314. [DOI] [PubMed] [Google Scholar]
- 39.Hu Y., Yagüe E., Zhao J., Wang L., Bai J., Yang Q., Pan T., Zhao H., Liu J., Zhang J. Sabutoclax, pan-active BCL-2 protein family antagonist, overcomes drug resistance and eliminates cancer stem cells in breast cancer. Cancer Lett. 2018;423:47–59. doi: 10.1016/j.canlet.2018.02.036. [DOI] [PubMed] [Google Scholar]
- 40.Gasparini P., Cascione L., Landi L., Carasi S., Lovat F., Tibaldi C., Alì G., D’Incecco A., Minuti G., Chella A. microRNA classifiers are powerful diagnostic/prognostic tools in ALK-, EGFR-, and KRAS-driven lung cancers. Proc. Natl. Acad. Sci. USA. 2015;112:14924–14929. doi: 10.1073/pnas.1520329112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stahlhut C., Slack F.J. Combinatorial Action of MicroRNAs let-7 and miR-34 Effectively Synergizes with Erlotinib to Suppress Non-small Cell Lung Cancer Cell Proliferation. Cell Cycle. 2015;14:2171–2180. doi: 10.1080/15384101.2014.1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan Y., Xie R., Zhang Q., Zhu X., Han J., Xia R. Bcl-xL/Bak interaction and regulation by miRNA let-7b in the intrinsic apoptotic pathway of stored platelets. Platelets. 2019;30:75–80. doi: 10.1080/09537104.2017.1371289. [DOI] [PubMed] [Google Scholar]
- 43.Jilek J.L., Zhang Q.Y., Tu M.J., Ho P.Y., Duan Z., Qiu J.X., Yu A.M. Bioengineered Let-7c Inhibits Orthotopic Hepatocellular Carcinoma and Improves Overall Survival with Minimal Immunogenicity. Mol. Ther. Nucleic Acids. 2019;14:498–508. doi: 10.1016/j.omtn.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y., Dong Y., Zhao L., Su L., Diao K., Mi X. TRIM59 overexpression correlates with poor prognosis and contributes to breast cancer progression through AKT signaling pathway. Mol. Carcinog. 2018;57:1792–1802. doi: 10.1002/mc.22897. [DOI] [PubMed] [Google Scholar]
- 45.Kumar M.S., Armenteros-Monterroso E., East P., Chakravorty P., Matthews N., Winslow M.M., Downward J. Retraction. Nature. 2015;523:370. doi: 10.1038/nature14551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai C.H., Lin L.T., Wang C.Y., Chiu Y.W., Chou Y.T., Chiu S.J., Wang H.E., Liu R.S., Wu C.Y., Chan P.C. Over-expression of cofilin-1 suppressed growth and invasion of cancer cells is associated with up-regulation of let-7 microRNA. Biochim. Biophys. Acta. 2015;1852:851–861. doi: 10.1016/j.bbadis.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 47.Sinha S., Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27(Suppl 1):S137–S148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee E.F., Smith N.A., Soares da Costa T.P., Meftahi N., Yao S., Harris T.J., Tran S., Pettikiriarachchi A., Perugini M.A., Keizer D.W. Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1. Autophagy. 2019;15:785–795. doi: 10.1080/15548627.2018.1564557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shiloh R., Gilad Y., Ber Y., Eisenstein M., Aweida D., Bialik S., Cohen S., Kimchi A. Non-canonical activation of DAPK2 by AMPK constitutes a new pathway linking metabolic stress to autophagy. Nat. Commun. 2018;9:1759. doi: 10.1038/s41467-018-03907-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen H., Li L., Hu J., Zhao Z., Ji L., Cheng C., Zhang G., Zhang T., Li Y., Chen H. UBL4A inhibits autophagy-mediated proliferation and metastasis of pancreatic ductal adenocarcinoma via targeting LAMP1. J. Exp. Clin. Cancer Res. 2019;38:297. doi: 10.1186/s13046-019-1278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurashige T., Nakajima Y., Shimamura M., Matsuyama M., Yamada M., Nakashima M., Nagayama Y. Basal Autophagy Deficiency Causes Thyroid Follicular Epithelial Cell Death in Mice. Endocrinology. 2019;160:2085–2092. doi: 10.1210/en.2019-00312. [DOI] [PubMed] [Google Scholar]
- 52.Kuo C.L., Jiang Z.Y., Wang Y.W., Lin T.Y., Huang W.L., Wu F.J., Luo C.W. In vivo selection reveals autophagy promotes adaptation of metastatic ovarian cancer cells to abdominal microenvironment. Cancer Sci. 2019 doi: 10.1111/cas.14162. Published online August 5, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emdad L., Bhoopathi P., Talukdar S., Pradhan A.K., Sarkar D., Wang X.Y., Das S.K., Fisher P.B. Recent insights into apoptosis and toxic autophagy: The roles of MDA-7/IL-24, a multidimensional anti-cancer therapeutic. Semin. Cancer Biol. 2019 doi: 10.1016/j.semcancer.2019.07.013. Published online July 26, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang Y., Hamed H.A., Cruickshanks N., Fisher P.B., Grant S., Dent P. Obatoclax and lapatinib interact to induce toxic autophagy through NOXA. Mol. Pharmacol. 2012;81:527–540. doi: 10.1124/mol.111.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang L., He Y.L., Li Q.Z., Hao X.H., Zhang Z.F., Yuan J.X., Bai Y.P., Jin Y.L., Liu N., Chen G. N-acetylcysteine alleviated silica-induced lung fibrosis in rats by down-regulation of ROS and mitochondrial apoptosis signaling. Toxicol. Mech. Methods. 2014;24:212–219. doi: 10.3109/15376516.2013.879974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.