Abstract
Malignant peripheral nerve sheath tumors (MPNSTs) are an aggressive soft-tissue sarcoma amenable only to surgical resection. Oncolytic herpes simplex viruses (oHSVs) are a promising experimental therapy. We previously showed that basal interferon (IFN) and nuclear factor κB (NFκB) signaling upregulate IFN-stimulated gene (ISG) expression and restrict efficient viral infection and cell-to-cell spread in ∼50% of tested MPNSTs. Stimulator of Interferon Genes (STING) integrates DNA sensor activity and mediates downstream IFN signaling in infected cells. We sought to identify STING’s role in oHSV resistance and contribution to basal ISG upregulation in MPNSTs. We show that the level of STING activity in human MPNST cell lines is predictive of oHSV sensitivity and that resistant cell lines have intact mechanisms for detection of cytosolic double-stranded DNA (dsDNA). Furthermore, we show that STING downregulation renders MPNSTs more permissive to oHSV infection and cell-to-cell spread. While next-generation viruses can exploit this loss of STING activity, first-generation viruses remain restricted. Finally, STING is not integral to the previously-observed basal ISG upregulation, indicating that other pathways contribute to basal IFN signaling in resistant MPNSTs. These data broaden our understanding of the intrinsic pathways in MPNSTs and their role in oHSV resistance and offer potential targets to potentiate oncolytic virus activity.
Keywords: STING, MPNST, ISG, oHSV, oncolytic virus, pattern recognition receptor, sarcoma, cGAS, Neurofibromatosis
Introduction
Malignant peripheral nerve sheath tumors (MPNSTs) are highly aggressive, treatment-refractory cancers of the peripheral nervous system with a 5-year overall survival rate of 44%. Conventional chemotherapies and radiotherapies have not improved patient survival in these cases, leaving surgical resection as the primary means of treatment.1 Oncolytic herpes simplex viruses (oHSVs) represent a maturing treatment modality and were effective in phase III trials for advanced melanoma.3 We previously showed that MPNST cell lines have varied levels of sensitivity to oHSV treatment and that sensitivity relates to the interferon (IFN) response. Specifically, we have shown that resistant cell lines constitutively express a set of IFN-stimulated genes (ISGs) and respond to oHSV infection with rapid phosphorylation of STAT1 and that NFκB-related signaling activity is involved in this basal ISG upregulation and oHSV resistance.2, 4
Among the hundreds of ISGs are a diverse set of genes that encode pattern recognition receptors (PRRs), which trigger IFN and pro-inflammatory signaling upon detection of conserved non-self molecular features called pathogen-associated molecular patterns (PAMPS). In addition to PAMPs, which are associated with fungal and bacterial pathogens, PRRs can also sense the nucleic acids that are associated with viral genomes and replication intermediates. Stimulator of Interferon Genes (STING) is a transmembrane endoplasmic reticulum (ER)-associated adaptor protein that integrates signals from numerous cytosolic and nuclear DNA and RNA-sensing PRRs, linking them to the downstream (IFN regulatory factor [IRF]3/NFκB) transcriptional machinery that mediates the antiviral IFN response and upregulation of ISGs.5
A growing body of work has shown that defects in STING signaling can enhance HSV replication and oncolytic activity in some tumors.6, 7, 8, 9, 10 Additionally, it has been shown that STING signaling, the IFN response, and ISG expression are related to radiotherapy resistance in a variety of cancers.11, 12, 13, 14 It is therefore logical to examine STING activity in radiation-resistant tumors that are known to have an upregulated IFN response, such as MPNSTs. While all MPNST cell lines have proven to be highly resistant to first-generation oHSVs, we have developed a chimeric oHSV (called C134) that contains a viral gene insertion that allows it to productively infect roughly 50% of tested cell lines.2 Although our previous work identified that C134-sensitive cell lines lack a rapid IFN response, the signaling pathways that drive MPNST resistance to C134 remain elusive.
In this work, we examine oHSV-sensitive and -resistant MPNST cell lines to identify potential differences in STING function. Since our previous work showed that increased basal JAK/STAT signaling and ISG expression (including PRRs) restricts oHSV infection in resistant MPNSTs,2 we sought to identify how PRRs upstream of STING contribute to this rapid antiviral response in MPNSTs. We hypothesized that oHSV-sensitive MPNSTs were capable of STING signaling, but that such signaling would be delayed as PRR expression ramps up in response to viral insult. To evaluate the contribution of STING signaling to C134 restriction, we screened our C134-sensitive and C134-resistant human MPNST cell lines for STING activation after viral infection. Further functional screening was conducted on the same set of cell lines with a small molecule inhibitor of STING (H-151) to evaluate the impact of STING inhibition upon C134 viral spread. Specific lines were also chosen from this group for genetic knockdown of STING and more in-depth studies of changes to STING signaling, activation, and impact upon both viral replication and spread. Finally, because signaling on the STING axis is only one potential stimulator of ISG expression, we evaluated the impact of STING knockdown on ISGs previously observed to be upregulated in resistant MPNST cell lines. In summary, our results show that there is greater STING-mediated activity in oHSV-resistant MPNST cell lines, this activity (while reduced) may still influence the antiviral response in certain sensitive lines (NMS-2PC), and this pathway, while important for the antiviral response, is dispensable for basal tonic ISG upregulation in MPNSTs.
Results
oHSV-Resistant MPNST Cell Lines Have Increased STING Activity
Our previous studies showed that ∼50% of MPNSTs resist viral infection. In an effort to simplify our analysis and move from our earlier flow-based approach, we examined the cell-to-cell spread of a GFP-expressing next-generation virus in six human MPNST cell lines using the Incucyte ZOOM live imaging platform. Human foreskin fibroblast (HFF) cells were included as a normal control. Based upon our earlier studies, we chose T265-luc (T265), STS26T-luc (STS26T), ST88-14-luc (8814), 90-8-luc (90-8), NMS-2PC (NMS2PC), and S462-luc (S462). Consistent with our earlier results, we found that oHSV-resistant lines T265, STS26T, and 8814 restricted next-generation (C134) viral spread more than sensitive lines 90-8, NMS2PC, and S462 (Figure 1A).2
Figure 1.
STING Function in Multiple Cell Lines
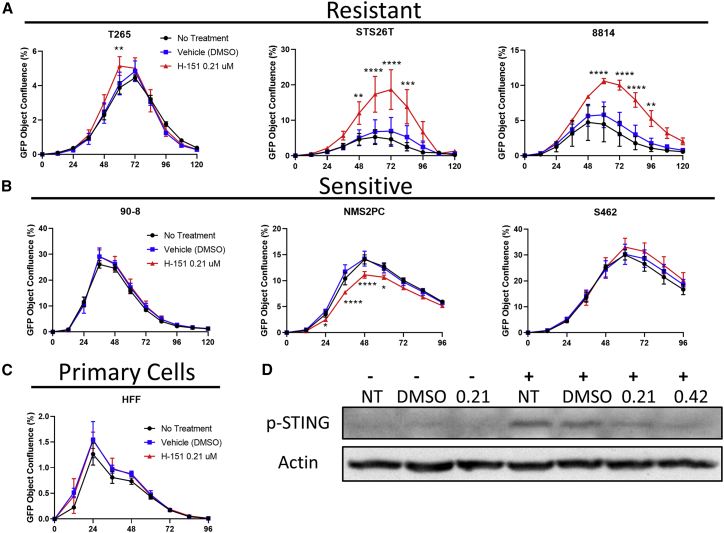
(A) Cell lines T265, STS26T, 8814, 90-8, NMS2PC, and S462 were infected with a GFP-expressing version of C134 (C154) at an MOI of 0.1 in triplicate. (As a control, HFF cells were similarly plated and treated.) The Incucyte Zoom platform was used to record viral spread as a function of GFP confluence. Measurements were taken at the indicated time points. (B) The same cell lines were infected either with mock (-) or C134 (+) at an MOI of 10. Lysate was analyzed by immunoblot for cGAS, p-STING, total STING, and actin. (C) STS26T, 8814, and NMS2PC were similarly infected. Lysate was collected at the indicated time points and analyzed by immunoblot with STING antibody. Densitometry shows normalized expression levels of STING relative to actin. Results are representative of 3 replicate experiments. (D) Confocal images (20×) of 8814 and NMS2PC were taken at 6 h post-infection with C134 at an MOI of 1. Coverslips were probed with anti-HSV glycoprotein D (gD) and anti-STING antibodies. Anti-gD is shown in red, anti-STING is shown in green, and merge appears as yellow. (Slides were also stained with DAPI.) Scale bar represents 50 μm; all error bars show SD.
Previous work has shown that phosphorylation of STING at the serine-366 site is consistent with its activation and downstream IFN signaling.15 Therefore, in order to detect potential differences in STING signaling in these cell lines, we conducted an immunoblot-based screen using an antibody that is specific to STING S366 phosphorylation. Our results show that our most resistant line (T265) demonstrates the highest level of STING phosphorylation in response to C134 infection. Resistant lines STS26T and 8814 also demonstrated clear phosphorylation of STING within 6 h of infection, similar to HFF control. In contrast, under the same conditions, STING remained unphosphorylated in all of the tested sensitive lines (Figure 1B).
In carcinomas, such loss of STING function is often related to changes in expression of cGAS (a STING-dependent dsDNA PRR).6, 7 Therefore, to complement the phosphorylation data, we assayed the total STING and cGAS present in these lines. Our results show that, in two of our sensitive lines (90-8 and S462), total STING expression was greatly diminished relative to all other lines. In the remaining sensitive line (NMS2PC), while STING expression was present at a level comparable to that of resistant lines, cGAS expression appeared to be non-existent (Figure 1B).
As a further check on STING function, we selected three cell lines with comparable levels of baseline STING expression—two resistant lines (STS26T and 8814) and one sensitive line (NMS2PC)—for analysis by STING degradation assay. Upon exposure to cytosolic dsDNA, cyclic dinucleotides, or viral infection, STING is activated; to prevent sustained immune signaling, activated cells often downregulate STING through lysosomal degradation.16, 17, 18, 19, 20, 21 We therefore measured STING expression levels in these lines at two time points after C134 infection. The results show that, while there are similar levels of STING expression prior to infection, STING is rapidly degraded in the oHSV-resistant MPNSTs post-infection, suggestive of increased activation (Figure 1C).
To visualize the previously observed differences in STING function, we conducted an immunofluorescent assay to determine the cellular localization of STING and HSV-1 glycoprotein D (gD) during C134 infection. In 8814, our confocal images recorded robust perinuclear aggregation of STING and gD in C134-infected cells in addition to co-localization of STING and gD in these regions (Figure 1D, top row). Neither similar aggregation of STING nor co-localization with gD was observed in NMS2PC (Figure 1D, bottom row). The results for 8814 are consistent with previously observed Golgi trafficking of STING and gD in response to viral infection.22, 23
In summary, these results show that, in oHSV-resistant MPNSTs, oHSV infection rapidly activates STING signaling, indicated by its phosphorylation, aggregation, and degradation. In contrast, oHSV infection in sensitive lines does not induce rapid phosphorylation of STING. Additionally, to the extent that STING is clearly observable in sensitive lines (such as in NMS2PC), the functional responses of aggregation and degradation also appear defunct. In these cell lines, lack of STING function may relate to deficient expression of either cGAS or STING.
STING Function Relates to oHSV Resistance
Although we have shown that STING function correlates with oHSV resistance in MPNST cell lines, it is possible that a causal relationship is lacking. For instance, oHSV resistance in these lines could be due primarily to the previously observed differences in STAT signaling and basal ISG expression.2 In such a case, differences in STING function might be a consequence of differential gene expression, but not a significant cause of increased viral resistance.
To rapidly screen multiple cell lines for evidence of a causal relationship, we chose to combine a small molecule inhibitor of STING (H-151) with a high-throughput viral spread assay on the Incucyte ZOOM. We hypothesized that, if STING function is integral to oHSV resistance in these lines, we would find that treatment with a STING inhibitor would cause an increase in viral spread.
Our results show that, in the three oHSV-resistant lines, STING inhibition significantly increased viral spread, particularly in the two lines with previously-demonstrated lower levels of total STING expression (Figure 2A). In contrast, STING inhibition did not significantly improve viral spread in the three oHSV-sensitive lines or primary HFF cells. The exception was for NMS2PC, where a significant decrease in viral spread was noted (Figures 2B and 2C).
Figure 2.
STING Interference with a Small Molecule (H-151) Increases Viral Spread in Resistant Cell Lines
(A) Resistant MPNST cell lines (T265, STS26T, 8814), (B) sensitive MPNST cell lines (90-8, NMS2PC, and S462), and (C) HFF primary cells were treated with 0.21 μM H-151 1 h prior to infection with a GFP-expressing version of C134 (C154) at an MOI of 0.1 in triplicate. The Incucyte Zoom platform was used to record viral spread as a function of GFP confluence. Measurements were taken at the indicated time points. (D) 8814 was similarly treated before infection with C134 at an MOI of 10. Lysates were collected at 6 h post-infection for analysis by immunoblot.-, Non-infected wells; +, infected wells with a plus; NT, no treatment; 0.21 or 0.42, μM H-151; DMSO, vehicle equivalent to 0.21 dose. All error bars show SD.
To verify that H-151 inhibited STING activation, we assayed STING phosphorylation in response to C134 infection in untreated media, vehicle-treated media, and media treated with two different doses of H-151 (0.21 and 0.42 μM final concentration). Based on image densitometry, we found that treatment with 0.21 μM H-151 yielded a 47% decrease in STING phosphorylation relative to vehicle (DMSO) control. Doubling the H-151 dose to 0.42 μM yielded only a further 5% decrease (Figure 2D). These results suggest that STING function in MPNST cell lines is a contributing factor to oHSV resistance.
STING Knockdown Reduces oHSV-induced Signaling in Resistant MPNST Cell Lines
Based upon the inhibitor studies, we sought to clarify STING’s role in oHSV restriction in MPNSTs. We hypothesized that STING was integral to oHSV restriction in resistant MPNSTs and that genetic knockdown of STING would reduce downstream IFN signaling in a resistant MPNST cell line. To test this, we created an 8814 STING knockdown cell line using a lentivirus expressing a short hairpin RNA (shRNA) targeting STING. As a control for our STING knockdown line (8814 shSTING) we also generated a cell line using a lentivirus expressing a non-target (scrambled) hairpin (8814 shSCR). To evaluate knockdown efficiency, we performed STING immunostaining and observed a 74% reduction in STING protein expression compared to scrambled hairpin control (SCR control) (Figure 3A). A similar knockdown was performed in the NMS2PC cell line with similar results (Figure S1A).
Figure 3.
Impact of STING Knockdown upon Signaling Pathway
We generated 8814 and NMS2PC cell lines, which stably express either a scrambled hairpin (shSCR) or a hairpin targeting STING (shSTING). (A) 8814, 8814 shSCR, and 8814 shSTING were analyzed by immunoblot using an anti-STING antibody to show knockdown efficiency. Densitometry shows normalized expression levels of STING relative to actin. (B) 8814 cell lines were plated under similar conditions and infected with C134. Lysates were collected at the indicated time points and analyzed by immunoblot for the indicated proteins of interest. (C) 8814 shSCR and 8814 shSTING were either infected at an MOI of 10 or mock infected in triplicate. Lysates were collected at 6 h post-infection and analyzed by qPCR for IFIT2. All error bars show SD.
To evaluate the impact of STING knockdown on downstream signaling, we next examined TBK1 and IRF3 phosphorylation after a high-multiplicity oHSV infection (MOI 10). The results show that, in the wild-type (WT) and shSCR cell lines, oHSV infection increases TBK1 and IRF3 phosphorylation rapidly (by 3 hours post-infection [hpi]). In contrast, in the 8814 shSTING cell line, while basal TBK1 phosphorylation was detectable, C134 infection did not increase TBK1 or IRF3 activity. Since STAT1 phosphorylation was our determinant of oHSV resistance in an earlier study,2 we also evaluated p-STAT1 and found that, consistent with our TBK1 and IRF3 results, oHSV infection induced STAT1 phosphorylation within 6 h in the WT and 8814 shSCR cells. In the 8814 shSTING line, however, oHSV did not similarly induce rapid STAT1 activation (Figure 3B).
Next, to determine if this reduction in activity also leads to decreased ISG expression, we examined IFN-induced protein with tetratricopeptide repeats 2 (IFIT2, also called ISG54) gene expression in the shSCR and shSTING cell lines before and after viral infection. IFIT2 is an ISG induced by viral dsDNA PRR activation and IFN induction24 and has been shown to disrupt STING-TBK1 signaling, thus acting as a STING negative-feedback regulator.25 We therefore hypothesized that, as its negative-feedback regulator, IFIT2 induction should be reduced concomitant with a reduction in STING signaling. To test this, we infected 8814 shSCR and 8814 shSTING with C134 at an MOI of 10 and collected total RNA at 0 and 6 hpi. A qPCR assay showed a 4-fold increase in IFIT2 mRNA level at 6 hpi in the control cell line relative to our STING knockdown cell line (Figure 3C). Taken together, these results suggest that STING knockdown downregulates oHSV-induced early IFN signaling in resistant MPNST cells.
Cytosolic dsDNA Detection Remains Intact and Is STING-Dependent in MPNST Cell Lines
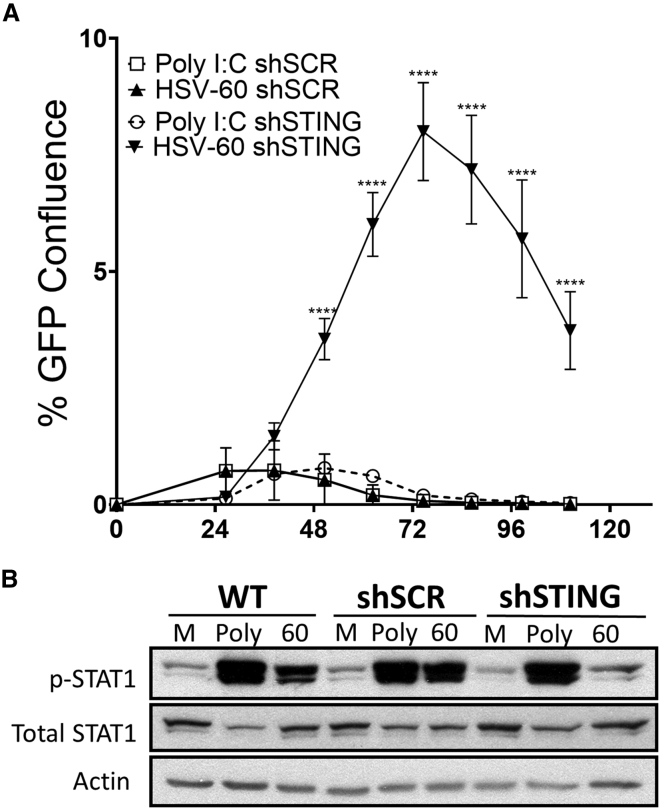
Upstream of STING, a PRR network detects cytosolic nucleic acids and initiates downstream antiviral signalin.5 Given that the STING-dependent dsDNA sensor (cyclic GMP-AMP synthase [cGAS]) was expressed abundantly in the more resistant cell lines and that HSV is a dsDNA virus, we next examined the fidelity and integration of dsDNA signaling in our knockdown line. We independently treated 8814 shSCR and 8814 shSTING cell lines with either HSV-60 (a dsDNA oligomer derived from the HSV-1 genome) or polyinosinic:polycytidylic acid (poly I:C; a chemical analog to dsRNA) 1 h prior to C134 infection and then measured viral spread (Incucyte ZOOM). Previous work has shown that poly I:C stimulates IFN signaling through a STING-independent mechanism,5 making it an ideal positive control. The results show that poly I:C pre-treatment was equally effective at limiting C134 spread (relative to vehicle) in both the shSCR (control) and STING knockdown cell lines. However, pre-treatment with HSV-60 was significantly less effective at limiting viral spread in our STING knockdown cell line (Figure 4A). These results show that shSTING knockdown disrupts dsDNA pathways but leaves the dsRNA-activated IFN pathways intact in our cell lines.
Figure 4.
STING Knockdown Selectively Impairs Cytosolic dsDNA Detection
(A) 8814 shSCR and 8814 shSTING were pre-treated for 1 h with poly I:C (5 μg/mL) or HSV-60 (5 μg/mL). Cells were infected with a GFP-expressing version of C134 (C154) at an MOI of 0.1 in triplicate. The Incucyte Zoom platform was used to record viral spread as a function of GFP confluence. (B) 8814, 8814 shSCR, and 8814 shSTING were transfected with vehicle (Mock), 5 μg/mL poly I:C (Poly), or 5 μg/mL HSV-60 (60). At 6 h post-transfection, cell lysates were collected and immunoblots of p-STAT1, total STAT1, and actin were performed. All error bars show SD.
Based upon this result and our previous data showing that oHSV induces STAT1 phosphorylation in resistant cell lines, we hypothesized that poly I:C would stimulate equivalent phosphorylation of STAT1 in our shSCR and shSTING cell lines but that HSV-60 would be less effective at stimulating p-STAT1 in our shSTING cell line. To test this, we transfected 8814, 8814 shSCR, and 8814 shSTING with vehicle only, poly I:C, or HSV-60 and collected cell lysate at 6 hpi. An immunoblot assay showed a clear deficit in dsDNA-stimulated signaling in our STING knockdown cell line relative to our control cell lines (Figure 4B). Taken together, these results show that dsDNA induces STING-dependent signaling in response to oHSV infections and rapid STAT1 activation in resistant MPNST cell lines.
STING Knockdown Enhances oHSV Replication and Spread in MPNST Cell Lines
Based upon our previous results with a small molecule STING inhibitor, we hypothesized that genetic depletion of STING in a resistant cell line would permit not only greater oHSV spread, but also enhanced levels of viral replication. To test this, we selected two representative cell lines with similar STING expression levels but with different oHSV infection responses (oHSV-resistant 8814, oHSV-sensitive NMS2PC) and reduced STING expression by shRNA. Our results show that STING knockdown significantly improved C134+EGFP spread in 8814-sh STING cells when compared to 8814 control and parental cell lines at 48 and 72 hpi (Figures 5A and S2A). Likewise, STING knockdown increased viral replication greater than 10 times at 48 and 72 h (Figure 5B). In independent studies, we found that STING knockdown also benefitted first-generation Δγ134.5 oHSV spread by nearly 6-fold (Figure S3A). However, overall first-generation viral spread was poor (maximum viral spread was 0.15% compared to 31% with C134). Even when infection was performed using 10-fold more virus, the maximum spread achieved by the first-generation virus was 0.39% (Figure S3B).
Figure 5.
STING Knockdown Improves Viral Replication and Spread
(A) 8814, 8814 shSCR, and 8814 shSTING were infected with a GFP-expressing version of C134 (C154) at an MOI of 0.1 in triplicate. The Incucyte Zoom platform was used to record viral spread as a function of GFP confluence. Measurements were taken at the indicated time points. (B) 8814 shSCR and 8814 shSTING were infected with C134 at an MOI of 0.1 in triplicate. Lysates were collected at the indicated times and analyzed by standard viral recovery assay. (C) An Incucyte Zoom viral spread assay was performed with NMS2PC, NMS2PC shSCR (control), and NMS2PC shSTING (STING knockdown) cell lines similar to (A). (D) A viral recovery assay was performed with NMS2PC shSCR and NMS2PC shSTING, similar to (B). All error bars show SD.
Our earlier studies showed that STING was present in oHSV-sensitive cell line NMS2PC but was less active than in resistant MPNSTs. Additionally, we noted that treatment with a small molecule inhibitor of STING appeared to decrease viral spread in this cell line. To gather more data about the effect of STING function in this cell line, we again used lentiviruses expressing the shSCR and shSTING RNA to generate control (NMS2PC shSCR) and knockdown (NMS2PC shSTING) cell lines and examined viral replication and spread. The results show that STING knockdown does improve viral cell-to-cell spread (Figures 5C and S2A) and replication (Figure 5D) in NMS2PC, but that this benefit was less pronounced than what was observed in resistant line 8814. These results suggest that some level of STING function remains present in NMS2PC, but that STING is more active in resistant cell lines, thus giving greater benefit upon knockdown.
STING Knockdown Has No Effect upon Basal ISG Upregulation in Resistant MPNST Cell Lines
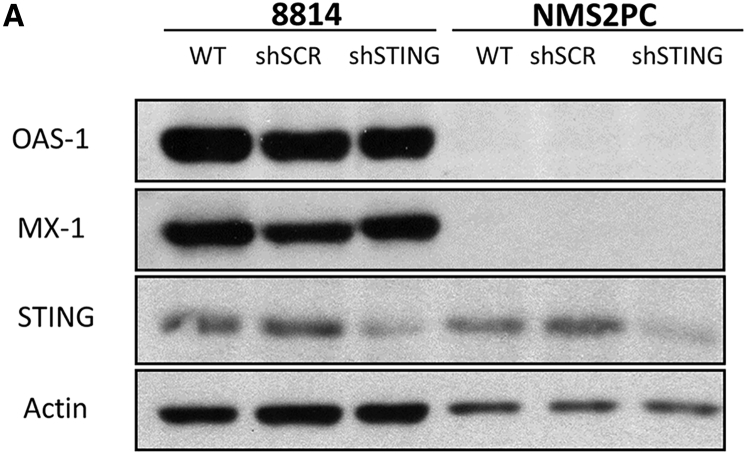
Our previous work identified that constitutive NFκB signaling in resistant MPNST cell lines upregulates ISG expression. This tonic NFκB and IFN stimulation contributes to oHSV resistance. Among these basally expressed genes are OAS-1 and MX-1.2 Although detection of viral DNA and signaling through STING is one mechanism for NFκB activation, other upstream pathways can also induce signaling on this pathway and cause ISG upregulation.26 To determine whether STING signaling contributes to tonic ISG expression in the oHSV resistant MPNSTs, we compared basal expression of our representative ISGs (OAS-1 and MX-1) in parental, control, and STING knockdown cell lines for both 8814 and NMS2PC. Our results show that, relative to our control cell line, STING knockdown does not negatively impact basal ISG expression in 8814 (Figure 6A). These results suggest that, while STING remains an important antiviral sensor in resistant MPNSTs and its activity is increased in the resistant MPNSTs, tonic ISG expression does not require STING, indicating that other pathways drive ISG upregulation in oHSV-resistant MPNSTs. This also suggests that these other pathways could be targeted to reduce ISG expression while leaving the STING antiviral signaling response intact for safe oHSV treatment.
Figure 6.
STING Knockdown Does Not Impact Basal ISG Expression
8814, 8814 shSCR, 8814 shSTING, NMS2PC, NMS2PC shSCR, and NMS2PC shSTING cell lines were analyzed by immunoblot using antibody against OAS-1, MX-1, STING, or actin.
Discussion
First-generation oHSVs contain dual deletions of the γ134.5 neurovirulence gene. Without an appropriate compensatory modification, this leaves first-generation viruses subject to PKR-mediated translational arrest, a cellular defense mechanism that the γ134.5 gene product would otherwise counter. C134 is a next-generation oHSV with improved intratumoral replication when compared to first-generation viruses. By inserting the human cytomegalovirus (HCMV) IRS1 gene, C134 compliments one γ134.5 gene function (late viral gene expression) but remains avirulent, similar to first-generation viruses. Our previous studies showed that even though C134 can evade a terminal ISG-mediated antiviral mechanism (PKR-mediated translational arrest), the virus remains restricted in cells with intact IFN signaling, similar to first-generation viruses.2, 27 Malignant transformation of cells often alters their IFN signaling, causing mutations that can complement the γ134.5 gene loss, allowing selective viral replication. However, many cancers are resistant to first-generation oHSVs, with MPNSTs being no exception.
We previously showed that 50% of MPNST cell lines restrict C134 infection and replication, thus prompting our investigation into the mechanisms mediating this resistance. Our work showed that MPNSTs contain sufficient entry receptors for viral entry but that ISG expression limits oHSV infection post-entry. These studies also showed that tonic NFκB and JAK/STAT signaling in resistant cells increases basal ISG expression, limiting efficient viral replication and spread.2
In this work, we have identified that dsDNA-activated STING signaling pathways are intact in C134-resistant MPNSTs and restrict viral replication. Specifically, we found that, while C134-resistant MPNST cell lines show expression of both cGAS and STING, sensitive lines show deficits in expression of at least one of the pair. Additionally, we found that C134-resistant cell lines are rendered more sensitive by interference with STING signaling (by either small molecule or genetic methods).
Studies involving the small-molecule STING-inhibitor (H-151) suggest that STING contributes to viral restriction in resistant MPNST cell lines, although the H-151 activity in T265 was less pronounced. Of the cell lines evaluated, T265 shows the highest level of basal STING expression, mirroring the levels seen in HFF cells. It is possible that with higher levels of STING expression, the dose of inhibitor used was less effective, since an excess of cellular STING might counteract the effects of a STING inhibitor.
Genetic knockdown of STING in resistant line 8814 improved C134 replication and spread to levels similar to what was observed for sensitive lines. However, under the same conditions, a first-generation virus remained restricted. This suggests that, while STING signaling is important to MPNST resistance in the case of C134, in the case of first-generation viruses, additional pathways remain a critical roadblock. Considered another way, the STING-mediated IFN response in MPNSTs becomes a critical factor for an invading oHSV only when PKR-mediated translational arrest can be successfully evaded. Such insights into the immune mechanisms of these tumors are critical if we are to devise improved treatments with oncolytic viruses.
In a previous work, we showed that a 48-h pre-treatment (but not co-treatment) of C134-resistant MPNSTs with JAK/STAT inhibitor, ruxolitinib, renders them C134-sensitive.2 We showed that this 48-h pre-treatment provided sufficient time to reduce ISG levels in the tumor cells. Importantly, we also identified that interrupting NFκB signaling also diminished this basal ISG overexpression.2 Whether or not this aberrant IFN signaling was STING-dependent was a critical question as we approached this work. As we began, it remained possible that the constitutive signaling through NFκB did not arise from aberrant STING signaling, but rather from an alternative pathway—for instance, as a result of constitutive MAPK signaling.26 Our current study suggests that, in C134-resistant MPNST cell lines, ISGs, which act as PRRs, are constitutively expressed, allowing these cells to mount a more immediate STING-dependent IFN response than occurs in the more sensitive MPNST lines. Interfering with STING impacts PRR downstream signaling but does not interrupt the underlying signaling pathways that drive this basal ISG upregulation in the tumor cells.
It is also possible that, in addition to enhancing STING-mediated resistance, these basally expressed ISGs restrict oHSV infection/replication through alternative STING-independent antiviral pathways. If so, this would indicate a multimodal resistance mechanism that could be separately targeted by future co-therapies. It is also interesting to consider the identity of the basally expressed upstream PRRs that allow the immediate STING-dependent response in these cell lines. While we identified in this work that dsDNA sensors such as cGAS likely contribute to oHSV resistance, many such DNA-sensing PRRs exist upstream of STING.28 Other work has suggested that certain dsRNA sensors (known to signal through STING) may also play an important role in the cellular response to HSV-I,29 so whether detection of dsRNA plays a role in oHSV resistance is also unknown. These questions remain to be addressed by future investigations.
Materials and Methods
Cell Lines and Viruses
MPNST cell lines have been previously described4 and were propagated in DMEM with 4.5 g/L glucose, L-glutamine, and sodium pyruvate (Corning, Corning, NY, USA) supplemented with 10% fetal bovine serum (FBS). Passages were kept under 12 for all experiments, and all cells were tested for mycoplasma contamination. Recombinant viruses C101 and C134 have been described previously.30, 31 Briefly, C101 and C134 were derived from the Δγ134.5 mutant HSV-1 R3616 by insertion, respectively, of the EGFP or HCMV IRS1 genes under the control of the cytomegalovirus (CMV) immediate early promoter in the UL3-UL4 intergenic region. C154 is derived from C134 by insertion of EGFP into the deletion loci of γ134.5.
Small Molecule Inhibition of STING
H-151, a small molecule inhibitor of STING, has been previously described.32 MPNST cell lines were plated into 48-well flat, clear-bottom polystyrene tissue culture-treated microplates (Corning, Corning, NY, USA) and allowed to adhere overnight. H-151 (Focus Biomolecules, Plymouth Meeting, PA, USA) was re-suspended in DMSO (Thermo Fisher Scientific, Rockford, IL, USA) and added to individual wells at a final concentration of 0.21 μM in culture media. Treatment was applied one hour prior to introduction of virus and subsequent viral spread assays on the IncuCyte ZOOM live cell imaging platform or by western blot (both described below).
shRNA, Lentivirus, and Transduction
Knockdown of STING was accomplished by transduction of cell lines with shRNA-expressing lentivirus. The STING knockdown shRNA used the target sequence: 5′-GTCCAGGACTTGACATCTTAA-3′. The control (SCR, scramble) shRNA used the target sequence 5′-CCTAAGGTTAAGTCGCCCTCG-3′, as previously described.33 Lentiviral production protocols have been previously described.34 Briefly, lentivirus was produced by transfecting HEK293T cells with a cocktail containing psPAX2 (a gift from Didier Trono, Addgene plasmid #12260), pCMV-vesicular stomatitis virus G (VSV-G; a gift from Bob Weinberg, Addgene plasmid #8454), and p.LKO1 (Addgene plasmid #24150) modified to express either the STING knockdown or control shRNA. HEK293T cultures were grown in T-25 flasks (Thermo Fisher Scientific, Rockford, IL, USA) until approximately 80% confluent and transfected with the plasmid cocktail using TransIT-LT1 (Mirus Bio, Madison, WI, USA) according to the manufacturer’s protocol. Lentiviral supernatants were collected at 48 and 72 hpi and filtered through 0.45 μm syringe filters (Advanced Microdevices, Ambala Cantt, India) and stored at −80°C until use. Stable cell lines were produced via lentiviral transduction. Hygromycin (Corning) selection (500 μg/mL for 8814, 300 μg/mL for NMS2PC) was applied at 48 hpi. Final cell lines were plated into 96-well flat, clear-bottom polystyrene tissue culture-treated microplates (Corning, NY, USA) and allowed to adhere overnight before subsequent viral spread assays on the IncuCyte ZOOM live cell imaging platform (described below). Cells were plated on 48- or 24-well plates (Corning, NY, USA) for western blot assays (also described below).
IncuCyte ZOOM Viral Spread Assay
The IncuCyte ZOOM viral spread assay has been previously described.35 GFP-expressing oHSV-1 (C154) was added at the indicated MOI and plates were transferred into the IncuCyte ZOOM live cell imaging system, which was housed inside a cell incubator at 37°C with 5% CO2, until the end of the assay. Four images per well from three technical replicates were taken every 3 h for the indicated duration of the assay with a 10× objective lens and then analyzed using the IncuCyte ZOOM Software. Green channel acquisition time was 400 ms in addition to phase contrast. Viral spread was quantified as a percentage of GFP confluence over time.
Viral Recovery
Assessment of viral replication was performed as previously described.30 In brief, triplicate samples of cells were infected in parallel with C134. Virus recovery was measured by limiting plaque dilution from infected cultures on days 2 and 3 post-infection. Average recovered virus and SD were calculated for the time points tested.
Transfection of HSV-60 and Poly I:C
HSV-60 has been previously described36 and was produced using cDNA oligomers (Thermo Scientific, Columbus, OH, USA) of the following sequences: 5′-TAAGACACGATGCGATAAAATCTGTTTGTAAAATTTATTA-AGGGTACAAATTGCCCTAGC-3′ 5′- CGATCCCGTTAAACATGGGATAATAAATTTTACAAACAGATTTTATCGCATCGTGTCTTA-3′. Oligomers were annealed in a stepwise (5°C steps) cool down from 95°C over a period of 2 h in 10 mM Tris, pH 8.0, 50 mM NaCl, 1 mM EDTA. Annealed oligomers were transfected into cell lines at a concentration of 5 μg/mL using 2 μL TransIT-LT1 (Mirus Bio, Madison, WI, USA) per microgram of DNA, according to the manufacturer’s instructions. poly I:C was transfected similarly. For Incucyte assays, cells were plated as described and given the indicated treatment 1 h before infection with virus.
Immunofluorescence
For immunofluorescent imaging, cells were plated on circular (12-mm diameter) borosilicate glass coverslips (Thermo Scientific, Columbus, OH, USA), with each coverslip placed in one well of a 24-well plate (Greiner Bio-One, Monroe, NC, USA). Cells were grown at 37°C with 5% CO2 and treated as described. Upon completion of the assay in cell culture, coverslips were washed in ice-cold PBS, soaked in 2.5% paraformaldehyde for 20 min, washed again in PBS, incubated with 0.1% Triton X-100 for 5 min, and then blocked with 10% goat serum for 1 h. Coverslips were incubated for 1 h at room temperature with primary antibody diluted in Tris-buffered saline with 0.1% Tween-20 (TBST). Coverslips were repeatedly washed with TBST, incubated in fluorescent conjugate secondary antibody (Thermo Scientific) diluted in TBST (1:500), and subsequently washed with TBST. Coverslips were mounted on glass slides and visualized on a Zeiss 710 confocal microscope.
Western Blotting
Cellular lysates were collected on ice in radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-Cl pH 8.0, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl) with protease inhibitor cocktail (Roche) and diluted in 4× sample buffer (240 mM Tris-Cl pH 6.8, 40% glycerol, 4% SDS, 20% β-mercaptoethanol, 0.04% bromophenol blue). Samples were denatured at 98°C for 5 min, chilled on ice, separated by PAGE, and transferred to a nitrocellulose membrane (Thermo Scientific) and blocked for 1 h at room temperature with 5% dry milk (S.T. Jerrell Co.) or BSA (Fisher). Membranes were incubated overnight at 4°C with primary antibody diluted in TBST. Membranes were repeatedly washed with TBST, incubated for 1 h with secondary antibody (Thermo Fisher) diluted in TBST (1:20,000) at room temperature, and subsequently washed with TBST. Membranes were developed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and exposed to X-ray film (Research Products International).
qPCR
Total RNA from 8814 cell lines was extracted with the Direct-Zol RNA MiniPrep Plus kit (Zymo Research, Irvine CA) and converted into cDNA by the SuperScript III First-Strand Synthesis System (Thermo Scientific), both according to the manufacturer’s instructions. Quantitation of IFIT2 gene expression was performed with SYBR Green I PCR Master Mix in the StepOnePlus Real-Time PCR System (both from Applied Biosystems) and expressed in relative copy numbers (RCN) as described earlier.37 The following sequences were used: human IFIT2, forward primer: 5′-AAGCACCTCAAAGGGCAAAAC-3′ and reverse primer: 5′-TCGGCCCATGTGATAGTAGAC-3′; GAPDH (control), forward primer: 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse primer: 5′-GGCTGTTGTCATACTTCTCATGG-3′.
Statistical Analysis
Statistical analysis was performed using Prism 8 (GraphPad Software).
For most analyses, statistical significance was established by two-way ANOVA with Bonferonni’s multiple comparisons test. In Figures 5B and 5D, statistical significance was established with an unpaired t test with Welch’s correction. For all analyses, the cutoff for statistical significance was set at p ≤ 0.05. The following notation was used: (NS) p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p < 0.0001. All error bars show SD.
Author Contributions
K.A.C., M.G., and J.M.L. conceived and designed the study. J.M.L., M.G., and K.A.C. developed the methodology. J.M.L. acquired the data. K.A.C., J.M.L., and M.G. analyzed and interpreted the data. J.M.L. wrote the manuscript. J.M.L and K.A.C. reviewed and revised the manuscript. K.A.C. supervised the study.
Conflicts of Interest
K.A.C., in full transparency, discloses a commercial interaction with Mustang Bio and Fortress Biotech for licensure of the C134 oncolytic virus.
Acknowledgments
This study was supported by Hyundai Hope on Wheels, Alex’s Lemonade Stand Foundation, CancerFree Kids, and Department of Defense NF170075. The authors wish to thank Dr. Uksha Saini for her assistance with editorial review of the manuscript.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2019.09.001.
Supplemental Information
References
- 1.Kolberg M., Høland M., Agesen T.H., Brekke H.R., Liestøl K., Hall K.S., Mertens F., Picci P., Smeland S., Lothe R.A. Survival meta-analyses for >1800 malignant peripheral nerve sheath tumor patients with and without neurofibromatosis type 1. Neuro-oncol. 2013;15:135–147. doi: 10.1093/neuonc/nos287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson J.D., Markert J.M., Li L., Carroll S.L., Cassady K.A. STAT1 and NF-κB Inhibitors Diminish Basal Interferon-Stimulated Gene Expression and Improve the Productive Infection of Oncolytic HSV in MPNST Cells. Mol. Cancer Res. 2016;14:482–492. doi: 10.1158/1541-7786.MCR-15-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andtbacka R.H., Kaufman H.L., Collichio F., Amatruda T., Senzer N., Chesney J., Delman K.A., Spitler L.E., Puzanov I., Agarwala S.S. Talimogene Laherparepvec Improves Durable Response Rate in Patients With Advanced Melanoma. J. Clin. Oncol. 2015;33:2780–2788. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 4.Jackson J.D., McMorris A.M., Roth J.C., Coleman J.M., Whitley R.J., Gillespie G.Y., Carroll S.L., Markert J.M., Cassady K.A. Assessment of oncolytic HSV efficacy following increased entry-receptor expression in malignant peripheral nerve sheath tumor cell lines. Gene Ther. 2014;21:984–990. doi: 10.1038/gt.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia T., Konno H., Ahn J., Barber G.N. Deregulation of STING Signaling in Colorectal Carcinoma Constrains DNA Damage Responses and Correlates With Tumorigenesis. Cell Rep. 2016;14:282–297. doi: 10.1016/j.celrep.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia T., Konno H., Barber G.N. Recurrent Loss of STING Signaling in Melanoma Correlates with Susceptibility to Viral Oncolysis. Cancer Res. 2016;76:6747–6759. doi: 10.1158/0008-5472.CAN-16-1404. [DOI] [PubMed] [Google Scholar]
- 8.de Queiroz N.M.G.P., Xia T., Konno H., Barber G.N. Ovarian Cancer Cells Commonly Exhibit Defective STING Signaling Which Affects Sensitivity to Viral Oncolysis. Mol. Cancer Res. 2019;17:974–986. doi: 10.1158/1541-7786.MCR-18-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bommareddy P.K., Zloza A., Rabkin S.D., Kaufman H.L. Oncolytic virus immunotherapy induces immunogenic cell death and overcomes STING deficiency in melanoma. OncoImmunology. 2019;8:1591875. doi: 10.1080/2162402X.2019.1591875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deschamps T., Kalamvoki M. Impaired STING Pathway in Human Osteosarcoma U2OS Cells Contributes to the Growth of ICP0-Null Mutant Herpes Simplex Virus. J. Virol. 2017;91:e00006-17. doi: 10.1128/JVI.00006-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang H., Deng L., Hou Y., Meng X., Huang X., Rao E., Zheng W., Mauceri H., Mack M., Xu M. Host STING-dependent MDSC mobilization drives extrinsic radiation resistance. Nat. Commun. 2017;8:1736. doi: 10.1038/s41467-017-01566-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Post A.E.M., Smid M., Nagelkerke A., Martens J.W.M., Bussink J., Sweep F.C.G.J., Span P.N. Interferon-Stimulated Genes Are Involved in Cross-resistance to Radiotherapy in Tamoxifen-Resistant Breast Cancer. Clin. Cancer Res. 2018;24:3397–3408. doi: 10.1158/1078-0432.CCR-17-2551. [DOI] [PubMed] [Google Scholar]
- 13.Weichselbaum R.R., Ishwaran H., Yoon T., Nuyten D.S., Baker S.W., Khodarev N., Su A.W., Shaikh A.Y., Roach P., Kreike B. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc. Natl. Acad. Sci. USA. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duarte C.W., Willey C.D., Zhi D., Cui X., Harris J.J., Vaughan L.K., Mehta T., McCubrey R.O., Khodarev N.N., Weichselbaum R.R., Gillespie G.Y. Expression signature of IFN/STAT1 signaling genes predicts poor survival outcome in glioblastoma multiforme in a subtype-specific manner. PLoS ONE. 2012;7:e29653. doi: 10.1371/journal.pone.0029653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.T., Grishin N.V., Chen Z.J. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 16.Kalamvoki M., Roizman B. HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc. Natl. Acad. Sci. USA. 2014;111:E611–E617. doi: 10.1073/pnas.1323414111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn J., Xia T., Rabasa Capote A., Betancourt D., barber G.N. Extrinsic Phagocyte-Dependent STING Signaling Dictates the Immunogenicity of Dying Cells. Cancer Cell. 2018;33:862–873.e5. doi: 10.1016/j.ccell.2018.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q., Lin L., Tong Y., Liu Y., Mou J., Wang X., Wang X., Gong Y., Zhao Y., Liu Y. TRIM29 negatively controls antiviral immune response through targeting STING for degradation. Cell Discov. 2018;4:13. doi: 10.1038/s41421-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong B., Zhang L., Lei C., Li Y., Mao A.P., Yang Y., Wang Y.Y., Zhang X.L., Shu H.B. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Konno H., Konno K., Barber G.N. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Lian Q., Yang B., Yan S., Zhou H., He L., Lin G., Lian Z., Jiang Z., Sun B. TRIM30α Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING. PLoS Pathog. 2015;11:e1005012. doi: 10.1371/journal.ppat.1005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukai K., Konno H., Akiba T., Uemura T., Waguri S., Kobayashi T., Barber G.N., Arai H., Taguchi T. Activation of STING requires palmitoylation at the Golgi. Nat. Commun. 2016;7:11932. doi: 10.1038/ncomms11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turcotte S., Letellier J., Lippé R. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J. Virol. 2005;79:8847–8860. doi: 10.1128/JVI.79.14.8847-8860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X., Michal J.J., Zhang L., Ding B., Lunney J.K., Liu B., Jiang Z. Interferon induced IFIT family genes in host antiviral defense. Int. J. Biol. Sci. 2013;9:200–208. doi: 10.7150/ijbs.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Li C., Xue P., Zhong B., Mao A.P., Ran Y., Chen H., Wang Y.Y., Yang F., Shu H.B. ISG56 is a negative-feedback regulator of virus-triggered signaling and cellular antiviral response. Proc. Natl. Acad. Sci. USA. 2009;106:7945–7950. doi: 10.1073/pnas.0900818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulze-Osthoff K., Ferrari D., Riehemann K., Wesselborg S. Regulation of NF-kappa B activation by MAP kinase cascades. Immunobiology. 1997;198:35–49. doi: 10.1016/s0171-2985(97)80025-3. [DOI] [PubMed] [Google Scholar]
- 27.Cassady K.A., Saunders U., Shimamura M. Delta gamma(1)34.5 Herpes Simplex Viruses Encoding Human Cytomegalovirus IRS1 or TRS1 Induce Interferon Regulatory Factor 3 Phosphorylation and an Interferon-Stimulated Gene Response. J. Virol. 2012;86:610–614. doi: 10.1128/JVI.05099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu J., Chen Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 29.Melchjorsen J., Rintahaka J., Søby S., Horan K.A., Poltajainen A., Østergaard L., Paludan S.R., Matikainen S. Early innate recognition of herpes simplex virus in human primary macrophages is mediated via the MDA5/MAVS-dependent and MDA5/MAVS/RNA polymerase III-independent pathways. J. Virol. 2010;84:11350–11358. doi: 10.1128/JVI.01106-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shah A.C., Parker J.N., Gillespie G.Y., Lakeman F.D., Meleth S., Markert J.M., Cassady K.A. Enhanced antiglioma activity of chimeric HCMV/HSV-1 oncolytic viruses. Gene Ther. 2007;14:1045–1054. doi: 10.1038/sj.gt.3302942. [DOI] [PubMed] [Google Scholar]
- 31.Cassady K.A. Human cytomegalovirus TRS1 and IRS1 gene products block the double-stranded-RNA-activated host protein shutoff response induced by herpes simplex virus type 1 infection. J. Virol. 2005;79:8707–8715. doi: 10.1128/JVI.79.14.8707-8715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haag S.M., Gulen M.F., Reymond L., Gibelin A., Abrami L., Decout A., Heymann M., van der Goot F.G., Turcatti G., Behrendt R., Ablasser A. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559:269–273. doi: 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- 33.Karthaus W.R., Iaquinta P.J., Drost J., Gracanin A., van Boxtel R., Wongvipat J., Dowling C.M., Gao D., Begthel H., Sachs N. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sprague L., Lee J.M., Hutzen B.J., Wang P.Y., Chen C.Y., Conner J., Braidwood L., Cassady K.A., Cripe T.P. High Mobility Group Box 1 Influences HSV1716 Spread and Acts as an Adjuvant to Chemotherapy. Viruses. 2018;10:E132. doi: 10.3390/v10030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghonime M.G., Jackson J., Shah A., Roth J., Li M., Saunders U., Coleman J., Gillespie G.Y., Markert J.M., Cassady K.A. Chimeric HCMV/HSV-1 and Δγ134.5 oncolytic herpes simplex virus elicit immune mediated antigliomal effect and antitumor memory. Transl. Oncol. 2018;11:86–93. doi: 10.1016/j.tranon.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S., Sirois C.M., Jin T., Latz E., Xiao T.S. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gavrilin M.A., Bouakl I.J., Knatz N.L., Duncan M.D., Hall M.W., Gunn J.S., Wewers M.D. Internalization and phagosome escape required for Francisella to induce human monocyte IL-1beta processing and release. Proc. Natl. Acad. Sci. USA. 2006;103:141–146. doi: 10.1073/pnas.0504271103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.