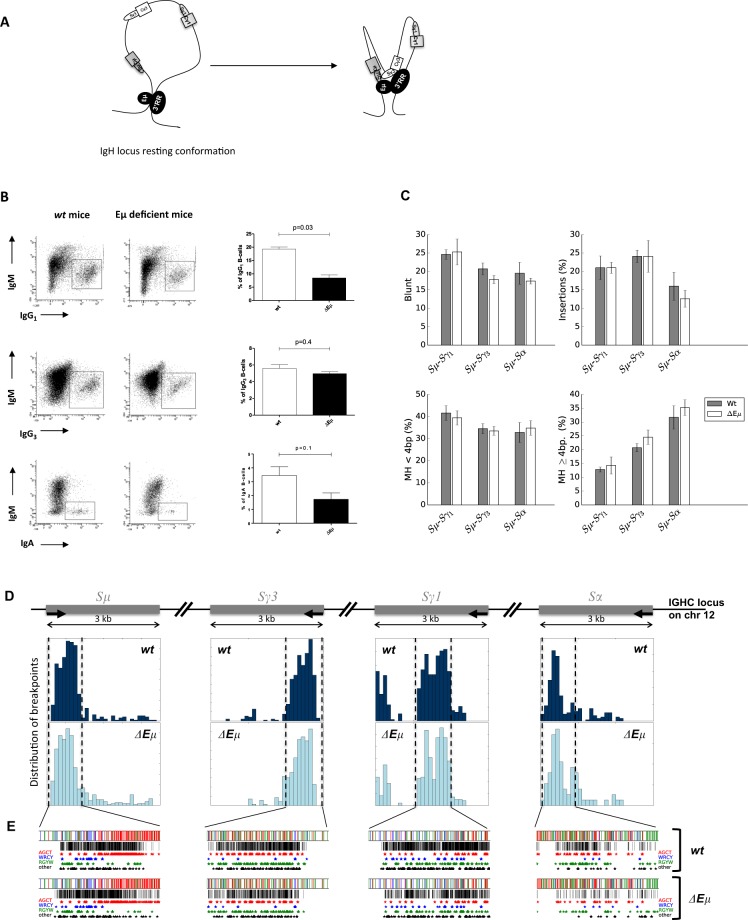
After encountering antigen, B-cells undergo class switch recombination (CSR) that substitutes the constant (C)μ gene with Cγ, Cε, or Cα, thereby generating IgG, IgE, and IgA antibodies with new effector functions but the same antigenic specificity.1 The DNA-editing enzyme activation-induced deaminase (AID) is required for CSR by targeting specific DNA switch (S) regions preceding the C region, except Cδ.2 Sμ is the donor region, while Sγ,ε,α are the acceptor regions. CSR is controlled in cis by the immunoglobulin heavy chain (IgH) 3’regulatory region (3’RR).3 The 3’RR is essential to poise AID on the S acceptor region. During CSR IgH, intrachromosomal interactions (schematized in Fig. 1a) are found between the 3’RR and the intronic Eμ enhancer.1,4 Looping allows transcriptional binding activators to enhancers to facilitate CSR. However, CSR is only modestly influenced by Eμ deletion.5–7 During CSR, two different DNA repair pathways take place: the classical nonhomologous end joining (c-NHEJ) and the alternative end joining (A-EJ) pathways. The c-NHEJ pathway (which uses components such as 53BP1, Ku70, ku80, XRCC4, and ligase 4) is preferentially implicated for blunt and microhomology structure junctions and the A-EJ pathway (which uses components such as CtIP, MRN, PARP1, DNA Pol θ) favors junctions with large homology.8,9 Data have reported that when an element of the DNA damage response pathway is defective, the molecular signature of CSR junctions is affected, suggesting that junctions are repaired through a different DNA repair pathway.10 We recently reported a new computational tool (CSReport) for the automatic analysis of CSR junctions sequenced by high-throughput sequencing,11 and used it to analyze the rare Sμ-Sδ junctions during IgD CSR,12,13 Sμ-Sγ,α junctions in wild-type (wt) mice14 and UNG-deficient mice.15 Even if Eμ-deficient mice exhibited a modest defect in IgH CSR, it is conceivable that its participation in CSR 3D loop interactions would be important for the DNA repair process. We used the CSReport and high-throughput sequencing to analyze the molecular signature of Sμ-Sγ3, Sμ-Sγ1, and Sμ-Sα junctions in Eμ-deficient mice to perform a more in depth search for a putative difference in their DNA repair pathways compared to those in wt mice.
Fig. 1.
Class switch recombination in Eμ-deficient mice. a Schematic representation of IgH intrachromosomal interactions between Eμ and 3’RR transcriptional enhancers with the S acceptor and S donor regions during CSR (according to references 1 and 4). b IgG1, IgG3, and IgA CSR in Eμ-deficient and wt mice. B-splenocytes were LPS-cultured for 4 days with (CSR toward IgG1) or without (CSR toward IgG3) the addition of IL-4 or TGFβ (CSR toward IgA). Cells gated on B220+ and/or CD138+ cells were investigated with anti-IgM, anti-IgG3, anti-IgG1, and anti-IgA antibodies. One representative experiment out of a total of 3–5 experiments for each isotype is shown. Mean ± SEM of three to five experiments for each isotype is reported. Significances were investigated using the Mann–Whitney U-test. c Structure profiles of Sμ-Sγ1, Sμ-Sγ3, and Sμ-Sα junctions in Eμ-deficient and wt mice. Junctions are classified in terms of junction with insertion, blunt junction, junction with microhomology (<4 bp) or large homology (≥4 bp). The junction profile was not significantly different in wt and Eμ-deficient mice. The results are pooled from three to seven mice. d Breakpoint localization in Sμ-Sγ1, Sμ-Sγ3, and Sμ-Sα junctions in wt and Eμ-deficient mice (same junctions as in Fig. 1c). e Motif targeting in Sμ-Sγ1, Sμ-Sγ3, and Sμ-Sα junctions. The stars identify breakpoint positions in AGCT, WRCY, and RGYW AID hotspots (same junctions as shown in Fig. 1c)
The mice were housed and the procedures were conducted in agreement with European Directive 2010/63/EU on animals used for scientific purposes applied in France as the « Décret n°2012–118 du 1er février 2013 relatif à la protection des animaux utilisés à des fins scientifiques ». Accordingly, the present project (APAFiS≠13855) was authorized by the « Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche ». Eμ-deficient and wt mice (housed in a conventional animal facility) were used. Single-cell suspensions of B-splenocytes were cultured for 4 days at 1 × 106 cells/ml in RPMI 1640 supplemented with 10% fetal calf serum and 5 μg/ml LPS, with (CSR toward IgG1) or without (CSR toward IgG3) the addition of 20 ng/ml IL-4 or 2 ng/ml TGFβ (PeproTech, Rocky Hill, NJ) (CSR toward IgA).14,16 Splenocytes were washed in PBS and stained with various antibodies: anti-B220-BV510 (BioLegends), anti-CD138-APC (Becton Dickinson Biosciences), anti-IgM PECy7 (ebiosciences), anti-IgG3-FITC (Becton Dickinson Biosciences), anti-IgG1-PE (Becton Dickinson Biosciences), and anti-IgA-FITC (Southern Biotech). The cells were analyzed on a Fortessa LSR2 (Beckman Coulter). In parallel experiments, stimulated B-cell splenocyte DNA was extracted for the investigation of Sμ-Sγ3, Sμ-Sγ1, and Sμ-Sα junctions. As previously described in detail,11 the junctions were PCR amplified. Libraries of 200 bp were prepared from the 1-2 kb PCR products of Sμ-Sγ3, Sμ-Sγ1, and Sμ-Sα amplification for Ion Proton sequencing (“GénoLim platform” of the Limoges University, France). The sequenced reads were then mapped to Sμ, Sγ1, Sγ3, and Sα regions using the BLAST algorithm. As previously reported,11 the computational tool developed for experiments performs junction assembly, identifies breakpoints in Sμ, Sγ1, Sγ3, and Sα, identifies junction structure (blunt, microhomology, large homology or insertion junctions) and outputs a statistical summary of the identified junctions.
Confirming previous studies,5–7 flow cytometry experiments indicated that compared to wt mice, the deletion of the Eμ enhancer had only a minor impact on CSR toward IgG3 (mean 5.0 vs 5.6%), IgG1 (mean 8.5 vs 19.3%), and IgA (mean 1.7 vs 3.5%) (Fig. 1b). We then extracted stimulated B-cell DNA to investigate the molecular signatures of these Sμ-Sγ3 (IgG3 CSR), Sμ-Sγ1 (IgG1 CSR), and Sμ-Sα (IgA CSR) junctions. When a few dozen CSR junctions are reported after cloning and subsequent Sanger sequencing, several hundred or thousand junctions can be documented using our computational tool of CSR junctions sequenced by high-throughput sequencing. The structural profiles (blunt, microhomology, large homology or junction with insertions) of 2443 Sμ-Sγ3, 8982 Sμ-Sγ1, and 617 Sμ-Sα Eμ-deficient junctions and 7209 Sμ-Sγ3, 14,700 Sμ-Sγ1, and 2639 Sμ-Sα wt junctions are reported in Fig. 1c. No significant difference (Mann–Whitney U-test) was found between Eμ-deficient mice and wt mice. In addition to the type of junctions, the distribution of IgG3, IgG1, and IgA junctions in terms of distance from the forward PCR primer in Sμ and from one of the reverse primers in Sγ3, Sγ1, and Sα was similar in Eμ-deficient and wt mice (Fig. 1d). The same results were found for the localization of the breakpoints within AID hotspots (AGCT, WRCY, RGYW) and other motifs (Fig. 1e) (displayed along specifically targeted segments within S regions).
As previously reported by others,5–7 the efficacy of CSR is, at most, modestly reduced by the deletion of the Eμ enhancer. Until recently, the transcriptional enhancers located in the IgH 3’RR are the sole cis-regulatory elements reported to affect conventional CSR.2,3,17 Compared with wt mice, the type and location of the CSR junctions in the absence of Eμ are unaffected. This observation is highlighted with three different isotypes (γ1, γ3, and α) by the analysis of a thousand independent junctions using our computational tool of CSR junctions sequenced by high-throughput sequencing. The Eμ enhancer is dispensable to poise AID to S donor/acceptor regions and for the generation of double strand breaks.18 Eμ IgH intrachromosomal interactions with S donor/acceptor regions and the 3’RR do not seem important to recruit/stabilize DNA repair factors and, thus, for the choice of the DNA repair pathway. In fact, do these Eμ IgH intrachromosomal interactions have a real relevance? Are these interactions only mechanical due to the IgH location of Eμ and to the 3D folding that brings S regions into contact with each other? These questions remain open. Deletion of the cis-regulatory IgH 3’RR affects CSR to all isotypes (except IgD).3,17 We currently investigate the molecular signature of the remaining CSR junctions in 3’RR-deficient mice to search for a potential role in the recruitment of specific DNA repair factors.
Acknowledgements
Authors are “Equipe Labellisée LIGUE 2018”. This work was supported by ANR (projet EpiSwitch-3’RR 2016). N.G. was supported by a grant from the “Société Française d’Hématologie”. H.I. and M.F. are supported by the University of Limoges and the “Région Nouvelle Aquitaine”. F.B. is supported by the Fondation Partenariale de l’Université de Limoges and ALURAD.
Author contributions
Y.D. designed the research. N.G., H.I., and M.F. performed the research. F.B. and Y.D. analyzed the data. N.G., H.I., M.F., F.B., and Y.D. wrote the paper.
Competing interests
The authors declare no competing interests.
References
- 1.Stavnezer J, Schrader CE. IgH chain class switch recombination: mechanism and regulation. J. Immunol. 2014;193:5370–5378. doi: 10.4049/jimmunol.1401849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rouaud P, et al. Elucidation of the enigmatic IgD class-switch recombination via germline deletion of the IgH 3’ regulatory region. J. Exp. Med. 2014;211:975–985. doi: 10.1084/jem.20131385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saintamand A, et al. Elucidation of IgH 3’ region regulatory role during class switch recombination via germline deletion. Nat. Commun. 2015;6:7084. doi: 10.1038/ncomms8084. [DOI] [PubMed] [Google Scholar]
- 4.Wuerffel R, et al. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27:711–722. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marquet M, et al. The Eµ enhancer region influences H chain expression and B cell fate without impacting IgVH repertoire and immune response in vivo. J. Immunol. 2014;193:1171–1183. doi: 10.4049/jimmunol.1302868. [DOI] [PubMed] [Google Scholar]
- 6.Li F, Yan Y, Pieretti J, Feldman DA, Eckhardt LA. Comparison of identical and functional Igh alleles reveals a nonessential role for Eμ in somatic hypermutation and class-switch recombination. J. Immunol. 2010;185:6049–6057. doi: 10.4049/jimmunol.0902992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perlot T, Alt FW, Bassing CH, Suh H, Pinaud E. Elucidation of IgH introninc enhancer functions via germ-line deletion. Proc. Natl Acad. Sci. USA. 2005;102:14362–14367. doi: 10.1073/pnas.0507090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 9.Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double break repair. Nat. Rev. Mol. Cell. Biol. 2017;18:495506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panchakshari RA, et al. DNA double-strand break response factors influence end-joining features of IgH class switch and general translocation junctions. Proc. Natl Acad. Sci. USA. 2018;115:762–767. doi: 10.1073/pnas.1719988115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyer F, et al. CSReport: a new computational tool designed for automatic analysis of class switch recombination junctions sequenced by high-throughput sequencing. J. Immunol. 2017;198:4148–4155. doi: 10.4049/jimmunol.1601924. [DOI] [PubMed] [Google Scholar]
- 12.Ghazzaui N, Issaoui H, Saintamand A, Boyer F, Denizot Y. Analysis of IgD CSR junctions by high-throughput sequencing. Immunol. Lett. 2017;188:86–88. doi: 10.1016/j.imlet.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Issaoui H, Ghazzaui N, Saintamand A, Denizot Y, Boyer F. IgD class switch recombination is not controlled through the immunoglobulin heavy chain 3’ regulatory region super-enhancer. Cell. Mol. Immunol. 2017;14:871–874. doi: 10.1038/cmi.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Issaoui H, Ghazzaui N, Saintamand A, Denizot Y, Boyer F. High throughput sequencing reveals similar molecular signatures for class switch recombination junctions for the γ and α isotypes. Cell. Mol. Immunol. 2019;16:90–92. doi: 10.1038/s41423-018-0025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghazzaui N, Issaoui H, Saintamand A, Denizot Y, Boyer F. Uracil-DNA glycosylase is not implicated in the choice of the DNA repair pathway during B-cell class switch recombination. Cell. Mol. Immunol. 2019;16:93–95. doi: 10.1038/s41423-018-0034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Issaoui H, et al. The IgH 3’ regulatory region super-enhancer does not control IgA class switch recombination in the B1 lineage. Cell. Mol. Immunol. 2018;15:289–291. doi: 10.1038/cmi.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent-Fabert C, et al. Genomic deletion of the whole IgH 3’ regulatory region (hs3a, hs1,2, hs3b, hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood. 2010;116:1895–1898. doi: 10.1182/blood-2010-01-264689. [DOI] [PubMed] [Google Scholar]
- 18.Saintamand A, et al. Eμ and 3’RR IgH enhancers show hierarchic unilateral dependence in mature B-cells. Sci. Rep. 2017;7:442. doi: 10.1038/s41598-017-00575-0. [DOI] [PMC free article] [PubMed] [Google Scholar]