Abstract
Duchenne muscular dystrophy is an inherited muscle wasting disease with severe symptoms and onset in early childhood. Duchenne muscular dystrophy is caused by loss-of-function mutations, most commonly deletions, within the DMD gene. Characterizing the junction points of large genomic deletions facilitates a more detailed model of the origins of these mutations and allows for a greater understanding of phenotypic variations associated with particular genotypes, potentially providing insights into the deletion mechanism. Here, we report sequencing of breakpoint junctions for seven patients with intragenic, whole-exon DMD deletions. Of the seven junction sequences identified, we found one instance of a “clean” break, three instances of microhomology (2–5 bp) at the junction site, and three complex rearrangements involving local sequences. Bioinformatics analysis of the upstream and downstream breakpoint regions revealed a possible role of short inverted repeats in the initiation of some of these deletion events.
Subject terms: Mutation, DNA sequencing, PCR-based techniques, Disease genetics
Duchenne muscular dystrophy: Understanding how the DMD gene breaks
Researchers in Australia have identified new examples of the genomic factors and mechanisms that lead to deletions linked with Duchenne muscular dystrophy (DMD). DMD is an inherited neuromuscular disease which causes progressive deterioration of muscles and, in some cases, intellectual impairment. Using samples from seven DMD patients, Niall Keegan of Murdoch University in Perth and colleagues sequenced the DNA left behind around the deletions in the DMD gene which cause the disease. They found one clean break, three sections with short repeated sequences, and three with more complex rearrangements. The diversity of these findings led them to suggest that the deletions resulted from a diversity of genomic factors and repair mechanisms. Future work could incorporate these findings into a model to predict where deletions will occur, expanding our understanding of DMD and its causes.
Introduction
Duchenne muscular dystrophy is an inherited neuromuscular disease arising from loss-of-function mutations in the DMD gene. The primary symptom of Duchenne muscular dystrophy is a progressive deterioration of all muscles in the body apart from the extraocular muscles1. Mild to moderate intellectual impairment is also observed in some cases, and there is evidence of an inverse relationship between the severity of cognitive effect and the age of physical onset2. Intellectual impairment can be exacerbated in cases where the causative genomic lesion extends into nearby genes such as NROB1 (ref. 3) and GKD4.
While a diverse array of mutation types can give rise to Duchenne muscular dystrophy or the symptomatically milder Becker muscular dystrophy, the majority of the pathological mutations of the DMD gene are deletions5, many of which encompass one or more entire exons.
The clearest predictor of the phenotype that will result from a large intragenic deletion is the set of exons lost. If the loss of these exons disrupts the gene’s open reading frame, the processed mRNA will either be translated into a truncated protein or be degraded by nonsense mediated decay before translation can be completed6. If, on the other hand, the deletion preserves the reading frame, it is far more likely that a functional protein isoform will be produced7, though this cannot be guaranteed8. While the DMD gene possesses a high degree of redundancy, especially in the central rod domain, not all DMD exons are equally dispensable9. An abridged dystrophin protein may retain much of its structural function, but loss of exons encoding the hinges10,11, actin-binding domain12, or dystroglycan-binding domain13 of the gene will necessarily impact the functions of those parts of the protein, leading to a phenotype distinct from that which may have arisen from a similarly sized in-frame deletion elsewhere in the gene.
The phenotype produced by a DMD whole-exon deletion is not predicted solely by the identities of the exons lost. Some intronic DMD sequences serve important regulatory roles both before14,15 and after mRNA splicing16, and deletions that affect functional regions such as these are likely to have negative consequences for the patient’s phenotype—consequences that can vary greatly even among patients with identical exon deletions16. In some cases, the unique conjunction of intronic sequences caused by a deletion can trigger the inclusion of a deleterious pseudoexon in the predominant transcript17,18. Given these factors and the role that intron secondary structure plays in pre-mRNA splicing19, it is plausible that the unique intronic span of a DMD deletion may also affect how readily the nearby exons will respond to antisense oligonucleotide-mediated exon-skipping therapy. For these reasons, it is of great value to both the patient and the researcher to fully map the intronic breakpoints in DMD whole-exon deletion cases, particularly those that manifest atypical phenotypes. Not only will this knowledge empower the patient to better understand their own disease, but comparison of symptoms between patients with fully mapped whole-exon deletions may lead to the discovery of new regulatory regions within the DMD introns and provide some insight into how these mutations occur.
In this study, successive rounds of PCR were used to delimit and amplify the genomic breakpoint junctions of seven Duchenne or Becker muscular dystrophy patients with whole-exon deletions. To economize on primers, we limited our study to deletions within the DMD deletion mutation hotspot, defined as spanning introns 44 to 55 (ref. 20). Bioinformatics web tools were then used to search the sequence around the breakpoints for features that may have contributed to each deletion event. Our analysis indicated that these deletions probably originated via multiple repair and recombination pathways.
Materials and methods
Selection of cell strains and DNA extraction
Seven patient myoblast or fibroblast cell strains were selected from our cell database (see Table 1). Each of the donor patients had been diagnosed with a deletion of at least one whole exon in the e45–e51 region of the DMD gene based on sequencing of their mRNA prior to the commencement of this study. We also selected a human myoblast strain not known to carry any disease-causing allele for use as a normal control. The eight cell strains were resurrected and cultured, and the DNA was extracted and purified using the PureLink Genomic DNA Kit from Invitrogen (ThermoFisher, Melbourne).
Table 1.
Details of the seven DMD exon deletion cell strains used in this study
| Patient ID | Exon deletion | ORF preserved? | Assigned phenotype | Cell type | Origin |
|---|---|---|---|---|---|
| d1 | 45–49 | Yes | Becker muscular dystrophy | Myoblast | Dubowitz Neuromuscular Centre, London, United Kingdom |
| d2 | 45–47 | Yes | Becker muscular dystrophy | Myoblast | Dubowitz Neuromuscular Centre, London, United Kingdom |
| d3 | 45–47 | Yes | Becker muscular dystrophy | Myoblast | Dubowitz Neuromuscular Centre, London, United Kingdom |
| d4 | 45–50 | No | Duchenne muscular dystrophy | Fibroblast | The Children’s Hospital at Westmead, Sydney, Australia |
| d5 | 46–51 | No | Duchenne muscular dystrophy | Myoblast | Hammersmith Hospital, London, United Kingdom |
| d6 | 48–50 | No | Duchenne muscular dystrophy | Myoblast | Dubowitz Neuromuscular Centre, London, United Kingdom |
| d7 | 51 | No | Duchenne muscular dystrophy | Fibroblast | Dystrophy Annihilation Research Trust Centre, Bengaluru, India |
PCR
PCRs were performed using AmpliTaq™ Gold DNA Polymerase from Invitrogen (ThermoFisher, Melbourne). Sets of up to seven primer pairs were designed at intervals across each intron expected to bear a deletion breakpoint. These primer sets were used to perform multiplex PCRs of the target genomic DNAs, and the reaction products were visualized on an agarose gel. The number of bands obtained for each reaction indicated the approximate extent of the deletion, and this information was used to inform successively more focused rounds of primer design and multiplex PCR. Once the breakpoints on either side of each deletion had been sufficiently delimited, a final PCR across the junction generated an amplicon containing the junction sequence (see Table 2 for primer sequences).
Table 2.
Sequences of primers used to amplify genomic deletion junctions (gene reference NG_012232.1(DMD_v001)) and corresponding amplicon regions
| Patient ID | Forward primer (5′-3′) | Reverse primer (5′-3′) | Amplicon |
|---|---|---|---|
| d1 | CATATGGTTTCTGGCCTTAG | TGCTGTGAACTACAAAGCAC | 1,171,882–1,518,667 |
| d2 | GGAACAGTATTCTAGGCAGG | CATCCCTCCCTTCTATGAAC | 1,265,936–1,449,576 |
| d3 | CACAAGGGTGTTAAGAACTACC | GATAGTTTCAATAATATGACCATG | 1,329,623–1,453,916 |
| d4 | CACCTCTTCTCATCTAATTCC | CGATCACAATCTTCTGTGAAG | 1,366,876–1,552,860 |
| d5 | CTATGAACAGGTATAAACCTG | CAGGACCAGCTTCTTGAACG | 1,377,531–1,608,719 |
| d6 | GCCTATGGTAAGATTGGTTTC | CCCTTGAGAAATATCTCCAAC | 1,427,297–1,563,083 |
| d7 | CTCCTATTTCAGCAAGTATC | GACCCTGGTAGGTACATCATG | 1,548,853–1,584,826 |
Sequencing
Junction amplicons were purified using Diffinity RapidTips™ (Chiral Technologies, Tokyo, Japan) and submitted to the Australian Genome Research Facility (Perth) for Sanger sequencing.
Bioinformatics
As per Verdin et al.21, we defined the breakpoint regions as the 150 bp surrounding each deletion breakpoint in the reference sequence (UCSC Genome Browser assembly ID hg38, Chromosome X). In cases of breakpoint junction microhomology, the 3′ end of the homologous sequence defined the center of the upstream and downstream sequence.
The web utility Non-B DB22 was used to search the 150 bp surrounding each breakpoint for non-B DNA features (i.e., A-phased repeats, direct repeats and slipped motifs, G-quadruplex forming repeats, inverted repeats and cruciform motifs, mirror repeats and triplex motifs, Z-DNA motifs, and short tandem repeats). Sequences were entered in FASTA format, and the results were saved as text files.
The “Repeating Elements” track of the UCSC Genome Browser23 was used to search the breakpoint regions for transposable elements and other repetitive sequences, and screen captures were taken of any relevant features. The gnomAD genome database24 was also searched for previously reported common polymorphisms at the junction sites, which, if present, may have contributed to the initiation of the observed mutation.
Results
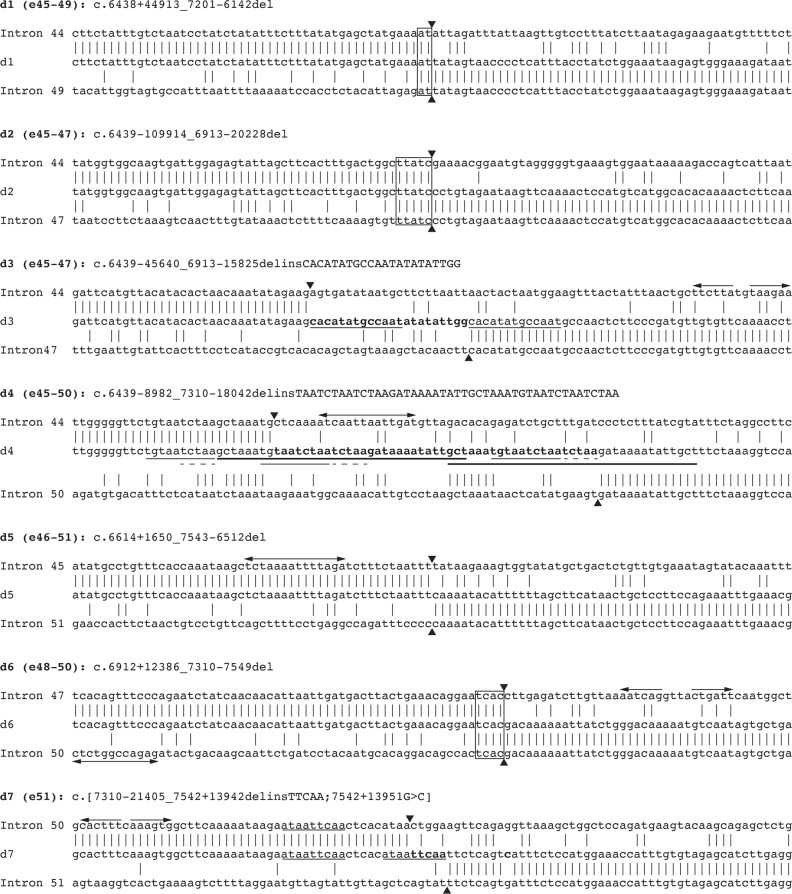
Sequences of the deletion breakpoint regions for seven DMD patient cell lines are shown in Fig. 1, formatted in the style of Esposito et al.25. Microhomologies and other sequence anomalies are indicated where they occur, as are inverted repeats discovered in the corresponding regions of the reference sequence (NG_012232.1(DMD_v001)). Aside from the inverted repeats, no other non-B DNA structures were detected.
Fig. 1. Junction sequences for seven patients with DMD whole-exon deletions, aligned to the corresponding regions of the reference sequence NG_012232.1(DMD_v001).
Regions of microhomology are enclosed in boxes. Arrowheads indicate inferred breakpoint sites. New nucleotides are in bold type. Nonconsecutive direct repeats are underlined, with dashed and thicker lines used where necessary to distinguish complex repeat arrangements (d4). Left- and right-pointing arrows indicate the 5′ and 3′ spans, respectively, of short inverted repeats
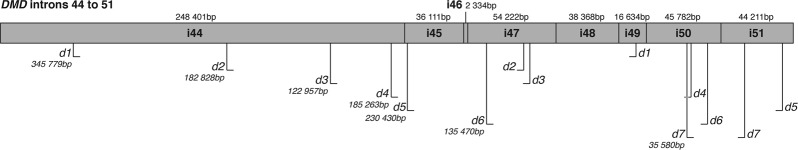
The relative positions of each breakpoint within introns 44 to 51 of the DMD gene are shown in Fig. 2. The deletion junctions of patients d1, d2, and d6 exhibited microhomologies of 2, 5, and 4 bp, respectively. Patients d3, d4, and d7 each had inserted tracts of sequence at their deletion junctions. For patient d3, this inserted tract consisted of a 13 bp copy of the intron sequence immediately 3′ of the junction, followed by a 9 bp novel sequence (22 bp total). Patient d7’s junction showed a similar feature, though in this case, the inserted sequence is only 5 bp and appears to be a partial copy of a local 9 bp tract. A G > C substitution was also noted 9 bp 3′ of patient d7’s junction, though this is a commonly reported variant (rs6527115, dbSNP build 151—see ref. 26). The deletion junction of patient d4 exhibited a 45 bp de novo insertion that appeared to be composed of disordered and partially nested copies of tracts of the surrounding sequence. Patient d5’s deletion junction appeared to be a “clean break”, with no microhomology or de novo sequence insertions.
Fig. 2. Relative positions of DMD gene deletion breakpoints for seven patients.
Patient IDs are indicated, as are the total sizes of the deletions (excluding any de novo sequence inserted at the junction). Due to the large size of many of the introns in this region of the gene, intervening exons are not visible at this scale
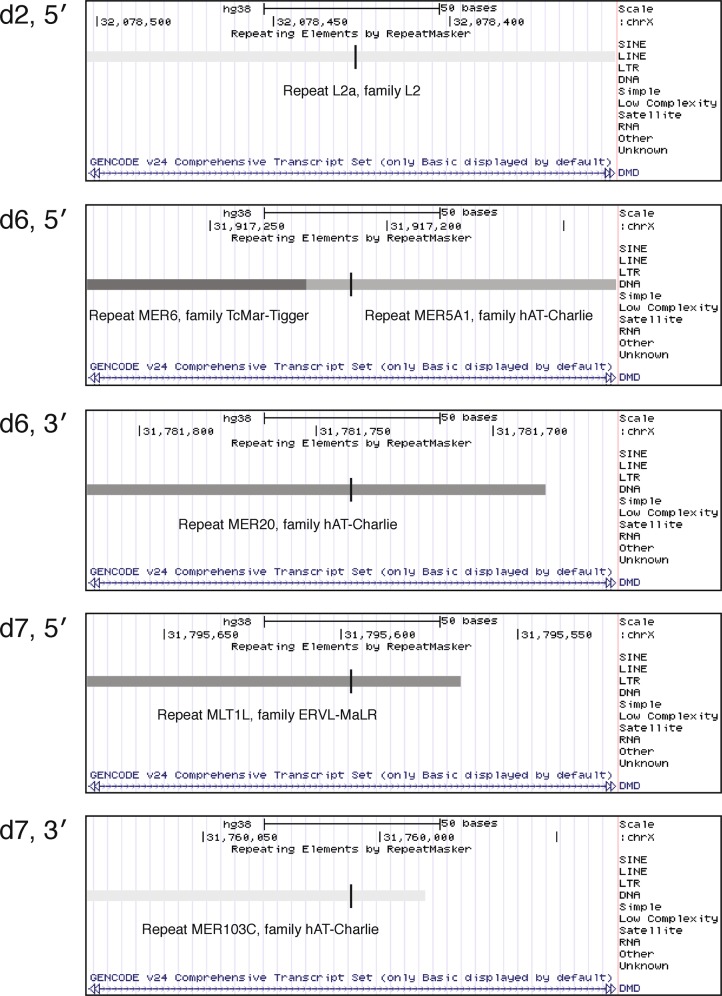
The “Repetitive Elements” track of the UCSC Genome Browser found transposons and retrotransposons at 5 of the 14 reference sequence breakpoint regions. Images of these elements are shown in Fig. 3.
Fig. 3. Repetitive elements detected within 150 bp deletion breakpoint regions of DMD exon deletion patients d2, d6, and d7.
Vertical dashes indicate the breakpoint location relative to the normal sequence. Darker shading on a repetitive element indicates a stronger match to that element’s consensus sequence
No previously reported common polymorphisms at the junction sites were found in the gnomAD database.
Discussion
Patients d1, d2, and d6—junction microhomologies
Deletion breakpoint junction microhomologies, such as those observed for patients d1, d2, and d6, are a feature common to many deletion mutations. In their 2013 study, Verdin et al.21 cataloged 22 microhomologies in the FOX2 genes of separate patients, ranging in size from 1 to 66 bp. They found that microhomologies occurred at a much higher rate than would be expected if the upstream and downstream breakpoints were completely random (p = 2.28 × 10–8) and noted that the regions around the breakpoints tend to be significantly enriched with repetitive elements.
Patient d2’s deletion could be a product of microhomology-mediated end joining (MMEJ), repairing a double-stranded break in the DMD27. Other research has implicated MMEJ in large deletions of other genes28, and the microhomology observed in this case (TTATC) was within the 5–25 bp size range required for this repair pathway.
The breakpoints of the deletions of patients d1 and d6 (Fig. 1) were too small to be attributed to MMEJ (2 and 4 bp, respectively). It is possible that they arose from nonhomologous end joining (NHEJ), as use of this pathway can be assisted by small microhomologies at the breakpoints29 even though it does not strictly require them. Microhomology-mediated break induced replication (MMBIR) is also a possibility, as this pathway does not always create new sequences. However, this hypothesis is less likely, as evidence from other mammals indicates that MMBIR is responsible for substantially fewer breakpoint junctions than NHEJ30.
Patients d3, d4, and d7—complex insertions at junctions
Patients d3, d4, and d7 all exhibited complex sequence insertions at their breakpoint junctions. A likely explanation for how these deletion junction sequences arose is MMBIR. This repair pathway is known to cause complex rearrangements of the DNA at the breakpoint junction, including duplications, inversions, and deletions31. MMBIR requires microhomologies at each breakpoint, and these were not observed for patients d3, d4, or d7. However, the microhomologies required for MMBIR need only be 1–3 bp long, and in cases where complex noncanonical junction sequences are created, such small sequence features could easily be obscured.
Patient d5 deletion junction—clean break
The clean break observed at patient d5’s deletion junction indicates that this deletion probably arose via NHEJ25, as this is the only repair or recombination pathway known to be capable of producing clean break junctions.
The role of transposable elements in facilitating large deletions
Transposable elements have been implicated as a causative factor in many large genomic deletions32,33, and it has been proposed that homology between two transposable elements facilitates non-allelic homologous recombination (NAHR)34. We detected a retrotransposon L2a site at the 5′ breakpoint region of patient d2, as well as transposons at both of patient d6’s breakpoints and both of patient d7’s breakpoints (Fig. 3). However, a BLAST-N analysis of each breakpoint pair for the deletions of patients d3, d4, and d7 could not detect any significant homology, and thus it does not appear that these sequence features contributed to the deletion events.
The role of non-B DNA in large deletions
Non-B DNA conformations may facilitate large genomic deletions by interacting with local microhomologies to errantly initiate the double-stranded break repair mechanism35,36. Our analysis detected six inverted repeats (size range 12–16 bp) across six of the deletion breakpoint regions but did not find any examples of the other searched-for features.
Short inverted repeats have previously been implicated in the occurrence of large genomic deletions37,38 and Lu et al.39 found that inverted repeats as small as 7–30 bp are positively associated with deletion breakpoint locations in human cancers. While it is not possible to replicate Lu et al.’s statistical analysis with a sample size of 14 breakpoints from seven nonrandomly selected patient cell lines, our findings are at least compatible with the hypothesis that short inverted repeats play a role in the initiation of some of the studied deletion events.
Conclusion
It has been suggested that large genomic deletion breakpoints do not occur randomly, but instead arise as a result of local features in the genome21. Indeed, there are a number of ways that asymmetrical chromosomal rearrangements can occur, producing deletions in tandem with other rearrangements, such as inversions or duplications40. Even with a sample size of just seven deletions from a single gene, we have observed a surprising diversity of junction phenomena and inferred a comparable diversity of contributing genomic factors and repair pathways. Developing a model that can incorporate all these factors and predict where and how deletions occur remains a daunting task—but it is a goal worth pursuing, both for its outcomes for human knowledge and for better informing genetic disease patients and their families.
Acknowledgements
The authors would like to thank the Enid and Arthur Home Scholarship and Murdoch University for the provision of funding and materials and their colleagues at the Centre for Molecular Medicine and Innovative Therapeutics for advice and constructive criticism.
Data availability
The reported nucleotide sequence data (d1–d7) are available in the GenBank database under accession numbers MK746139, MK829593, MK829594, MK829595, MK829596, MK829597, and MK829598.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Khurana TS, et al. Absence of extraocular muscle pathology in Duchenne’s muscular dystrophy: role for calcium homeostasis in extraocular muscle sparing. J. Exp. Med. 1995;182:467–475. doi: 10.1084/jem.182.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desguerre I, et al. Clinical heterogeneity of Duchenne muscular dystrophy (DMD): definition of sub-phenotypes and predictive criteria by long-term follow-up. PLoS ONE. 2009;4:e4347. doi: 10.1371/journal.pone.0004347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbaro M, et al. Multiplex ligation-dependent probe amplification analysis of the NR0B1(DAX1) locus enables explanation of phenotypic differences in patients with X-linked congenital adrenal hypoplasia. Horm. Res. Paediatr. 2012;77:100–107. doi: 10.1159/000336344. [DOI] [PubMed] [Google Scholar]
- 4.Lin L, et al. Analysis of DAX1 (NR0B1) and steroidogenic factor-1 (NR5A1) in children and adults with primary adrenal failure: ten years’ experience. J. Clin. Endocrinol. Metab. 2006;91:3048–3054. doi: 10.1210/jc.2006-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherratt TG, Vulliamy T, Dubowitz V, Sewry CA, Strong PN. Exon skipping and translation in patients with frameshift deletions in the dystrophin gene. Am. J. Hum. Genet. 1993;53:1007–1015. [PMC free article] [PubMed] [Google Scholar]
- 6.Hug N, Longman D, Caceres JF. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44:1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 8.Kesari A, et al. Integrated DNA, cDNA, and protein studies in Becker muscular dystrophy show high exception to the reading frame rule. Hum. Mutat. 2008;29:728–737. doi: 10.1002/humu.20722. [DOI] [PubMed] [Google Scholar]
- 9.van den Bergen JC, et al. Dystrophin levels and clinical severity in Becker muscular dystrophy patients. J. Neurol. Neurosurg. Psychiatry. 2014;85:747–753. doi: 10.1136/jnnp-2013-306350. [DOI] [PubMed] [Google Scholar]
- 10.Carsana A, et al. Analysis of dystrophin gene deletions indicates that the Hinge III region of the protein correlates with disease severity. Ann. Hum. Genet. 2005;69:253–259. doi: 10.1046/J.1469-1809.2005.00160.x. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura A, et al. Follow-up of three patients with a large in-frame deletion of exons 45-55 in the Duchenne muscular dystrophy (DMD) gene. J. Clin. Neurosci. 2008;15:757–763. doi: 10.1016/j.jocn.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Banks GB, Gregorevic P, Allen JM, Finn EE, Chamberlain JS. Functional capacity of dystrophins carrying deletions in the N-terminal actin-binding domain. Hum. Mol. Genet. 2007;16:2105–2113. doi: 10.1093/hmg/ddm158. [DOI] [PubMed] [Google Scholar]
- 13.Ishikawa-Sakurai M, Yoshida M, Imamura M, Davies KE, Ozawa E. ZZ domain is essentially required for the physiological binding of dystrophin and utrophin to beta-dystroglycan. Hum. Mol. Genet. 2004;13:693–702. doi: 10.1093/hmg/ddh087. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Chen D, Zhang J. Analysis of intron sequence features associated with transcriptional regulation in human genes. PLoS ONE. 2012;7:e46784. doi: 10.1371/journal.pone.0046784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rearick D, et al. Critical association of ncRNA with introns. Nucleic Acids Res. 2011;39:2357–2366. doi: 10.1093/nar/gkq1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/S1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 17.Khelifi MM, et al. Pure intronic rearrangements leading to aberrant pseudoexon inclusion in dystrophinopathy: a new class of mutations? Hum. Mutat. 2011;32:467–475. doi: 10.1002/humu.21471. [DOI] [PubMed] [Google Scholar]
- 18.Greer K, et al. Pseudoexon activation increases phenotype severity in a Becker muscular dystrophy patient. Mol. Genet. Genomic Med. 2015;3:320–326. doi: 10.1002/mgg3.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juan-Mateu J, et al. DMD mutations in 576 dystrophinopathy families: a step forward in genotype-phenotype correlations. PLoS ONE. 2015;10:e0135189. doi: 10.1371/journal.pone.0135189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verdin H, et al. Microhomology-mediated mechanisms underlie non-recurrent disease-causing microdeletions of the FOXL2 gene or its regulatory domain. PLoS Genet. 2013;9:e1003358. doi: 10.1371/journal.pgen.1003358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cer RZ, et al. Non-B DB: a database of predicted non-B DNA-forming motifs in mammalian genomes. Nucleic Acids Res. 2011;39(Database issue):D383–D391. doi: 10.1093/nar/gkq1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esposito G, et al. Precise mapping of 17 deletion breakpoints within the central hotspot deletion region (introns 50 and 51) of the DMD gene. J. Hum. Genet. 2017;62:1057–1063. doi: 10.1038/jhg.2017.84. [DOI] [PubMed] [Google Scholar]
- 26.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sfeir A, Symington LS. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem. Sci. 2015;40:701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas A, et al. Characterization of a novel large deletion caused by double-stranded breaks in 6-bp microhomologous sequences of intron 11 and 12 of the F13A1 gene. Hum. Genome Var. 2016;3:15059. doi: 10.1038/hgv.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lieber MR. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 30.Stewart C, et al. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 2011;7:e1002236. doi: 10.1371/journal.pgen.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang F, et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Zelm MC, et al. Gross deletions involving IGHM, BTK, or Artemis: a model for genomic lesions mediated by transposable elements. Am. J. Hum. Genet. 2008;82:320–332. doi: 10.1016/j.ajhg.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boone PM, et al. Alu-specific microhomology-mediated deletion of the final exon of SPAST in three unrelated subjects with hereditary spastic paraplegia. Genet. Med. 2011;13:582–592. doi: 10.1097/GIM.0b013e3182106775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robberecht C, Voet T, Esteki MZ, Nowakowska BA, Vermeesch JR. Nonallelic homologous recombination between retrotransposable elements is a driver of de novo unbalanced translocations. Genome Res. 2013;23:411–418. doi: 10.1101/gr.145631.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bacolla A, Wells RD. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 36.Inagaki H, et al. Chromosomal instability mediated by non-B DNA: cruciform conformation and not DNA sequence is responsible for recurrent translocation in humans. Genome Res. 2009;19:191–198. doi: 10.1101/gr.079244.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X, et al. Molecular analysis of a deletion hotspot in the NRXN1 region reveals the involvement of short inverted repeats in deletion CNVs. Am. J. Hum. Genet. 2013;92:375–386. doi: 10.1016/j.ajhg.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim C, et al. Size of gene specific inverted repeat–dependent gene deletion In Saccharomyces cerevisiae. PLoS ONE. 2013;8:e72137. doi: 10.1371/journal.pone.0072137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu S, et al. Short inverted repeats are hotspots for genetic instability: relevance to cancer genomes. Cell Rep. 2015;10:1674–1680. doi: 10.1016/j.celrep.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/S0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The reported nucleotide sequence data (d1–d7) are available in the GenBank database under accession numbers MK746139, MK829593, MK829594, MK829595, MK829596, MK829597, and MK829598.