Abstract
In human genome, there are approximately 1,500 RNA-binding proteins (RBPs). They can regulate mRNA stability or translational efficiency via ribosomes and these processes are known as ‘post-transcriptional regulation’. Accumulating evidences indicate that post-transcriptional regulation is the determinant of the accurate levels of cytokines mRNAs. While transcriptional regulation of cytokines mRNAs has been well studied and found to be important for the rapid induction of mRNA and regulation of the acute phase of inflammation, post-transcriptional regulation by RBPs is essential for resolving inflammation in the later phase, and their dysfunction may lead to severe autoimmune diseases such as rheumatoid arthritis or systemic lupus erythematosus. For post-transcriptional regulation, RBPs recognize and directly bind to cis-regulatory elements in 3′ untranslated region of mRNAs such as AU-rich or constitutive decay elements and play various roles. In this review, we summarize the recent findings regarding the role of RBPs in the regulation of inflammation.
Keywords: chronic inflammation, cis-regulatory element, post-transcriptional regulation, RNA-binding protein, translation
Aberrant expression of inflammatory cytokines causes severe autoimmune diseases, such as rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE). The transcriptional regulation mechanisms of inflammatory cytokines underlying these inflammatory diseases have been extensively studied for decades. Transcriptional regulation via the nuclear factor kappa B (NF-κB) and signal transducer and activator of transcription 3 (STAT3) pathways have been reported to be essential for the regulation of the immune system (1, 2).
However, recent studies have shown that the post-transcriptional regulation of inflammatory cytokines is also essential for their expression. In the acute phase of inflammation, the total amount of mRNA rapidly increases; while during the later phase of inflammation, post-transcriptional regulation determines the stability and translational efficiency of mRNA, thereby regulating the mRNA levels of cytokines (Fig. 1) (3–11). The role of RNA-binding proteins (RBPs) participate in post-transcriptional regulation is widely known and they perform multiple functions, including translational control and stabilization or degradation of mRNA. RBPs can recognize cis-elements or specific structures in the 5′ or 3′ untranslated regions (UTRs) of target mRNAs and determine mRNA fate by directly binding to these targets. There are approximately 1,500 RBPs in humans and some of them play important roles in the regulation of chronic inflammation (12). In this review, we summarize the post-transcriptional regulation of cytokines by RBPs that recognize cis-elements or specific structures of cytokines mRNAs.
Fig. 1.
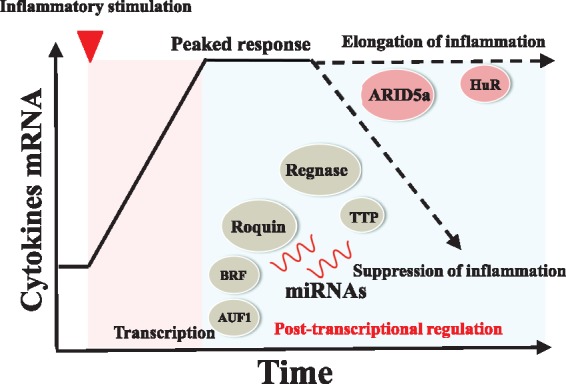
The post-transcriptional regulation of inflammatory cytokines is important for the regulation of chronic inflammation. In the acute phase of inflammation, mRNA levels of cytokines rapidly increase and reach the peak. In the chronic phase of inflammation, post-transcriptional regulation by RBPs or miRNA determines the prolongation or termination of inflammation (3–11).
Important cis-Elements and Structures of mRNA
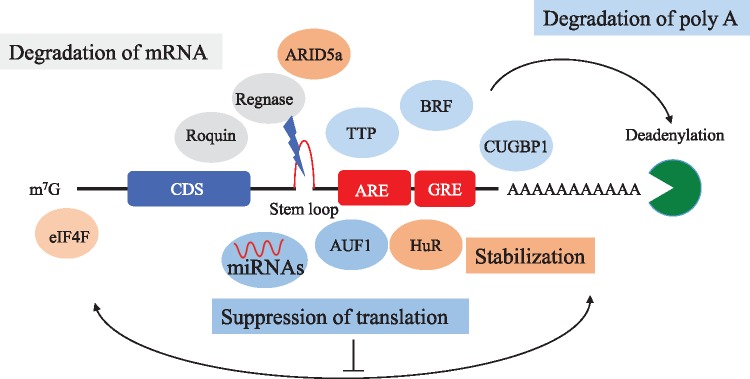
Most of RBPs recognize cis-elements or specific structures of cytokine mRNAs and elicit multiple functions, such as translational control, and destabilization of cytokine mRNAs. As shown in Fig. 2, these elements or specific structures are usually found in 3′ UTR of mRNA. In this section, we have introduced several vital cis-elements of mRNA (Fig. 2) (13–16).
Fig. 2.
Important RBPs and cis-elements for post-transcriptional regulation. Inflammatory cytokines contain several cis-elements, such as ARE. Many ARE-binding proteins bind to these cis-elements and regulate the levels of inflammatory cytokines (13–16).
Adenine and uridine-rich element (ARE) is one of the most important cis-elements of mRNA usually composed of AUUUA pentamers and are known to regulate the stability of inflammatory mRNAs (13). ARE was initially identified as a conserved AU sequence in the 3′ UTR, which mediated the selective degradation of colony stimulation factor 2 (CSF2) mRNA (17). AREs were also found in the 3′ UTRs of many inflammatory cytokines, such as IL-3, IL-6, IL-8, and tumour necrosis factor α (TNFα) (18–20). Furthermore, mice lacking AREs in the 3′ UTR of TNFα’s showed severe chronic inflammatory arthritis and Crohn’s like inflammatory bowel disease, suggesting an important physiological role of AREs (19). Approximately, 4,000 genes with ARE-mRNA exist and are classified into three classes (20–22). Class I (e.g. c-myc and c-fos) is characterized by having non-overlapping AUUUA pentamers, Class II (e.g. TNF alpha and IL-3) is characterized as having overlapping AUUUA pentamers and Class III AREs (e.g. c-jun) do not contain any AUUUA pentamer (21, 22). Though the mechanism of the regulation of ARE-containing mRNAs is still unknown, AREs are recognized and bound by many ARE-binding proteins (ARE-BPs). There are more than 20 ARE-BPs and they have multiple functions, such as degradation of mRNA and translational repression. The role of each ARE-BP is reviewed in the next section.
GU-rich element (GRE) is also an essential cis-element that was identified through computational analysis and is composed of GUUUG pentamers. GRE can be seen in mRNAs of inflammatory proteins, such as c-JUN, JUN-B and TNF-receptor 1B. The transcripts containing GRE tend to be short lived. CUGBP1 was the first reported GRE-binding protein and it strongly destabilizes GRE-containing mRNAs (14).
The constitutive decay element (CDE) is another crucial element for determining the stability of cytokine mRNA. CDE was firstly reported as an important cis-element of TNFα mRNA for preventing excessive induction of TNFα, and its mechanism of decay is distinct from that of ARE (15). The later research revealed CDE in more than 50 vertebrate mRNAs, many of them encoding essential factors for development and inflammation. The stem loop structure in the CDE is the most crucial structure for its regulation (16). Regnase-1 (ZC3H12A), Roquin1/2 (RC3H1/2) and Arid5a recognize and regulate the stem loop in CDE and strongly regulate the expression of inflammatory cytokines, as reviewed in the latter section.
ARE-Binding Proteins in the Immune System
As reviewed in the previous section, ARE is regulated by several ARE-binding proteins (ARE-BPs). In this section, we introduce several vital ARE-BPs involved in the regulation of inflammation.
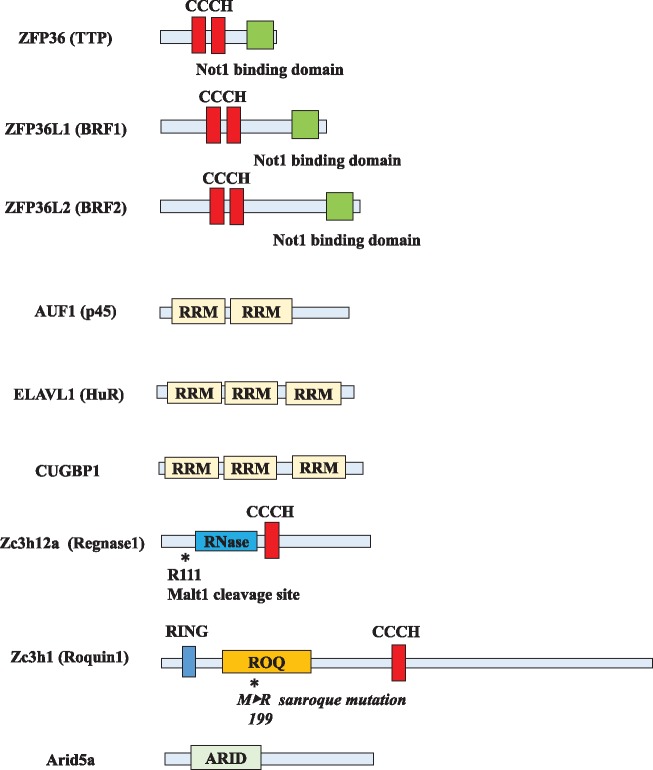
TTP (ZFP36, Tristetraproline) is one of the well-known ARE-BPs. It destabilizes the mRNAs of inflammatory cytokines, such as IL-2, IL-6, IL1-β and TNFα by directly binding to the ARE in their mRNAs. As shown in Fig. 3, TTP harbours an RNA-binding domain, CCCH (Cys-Cys-Cys-His) zinc finger (11). By directly binding to AREs in mRNA, it shortens the poly (A) tail of mRNAs and promotes mRNA decay through the recruitment of exosome, XRN1 and decapping enzymes (23, 24). Its expression is rapidly and transiently induced by stimulation with mitogens and growth factors, which indicates its roles in the negative feedback loop of inflammation (11). The expression of TTP is suppressed by phosphorylation caused by the mitogen-activated protein kinase (MAPK)-MAPKAK2 (MK2) kinase cascade and this suppression leads to the stabilization of cytokine mRNAs (25, 26). Mice lacking TTP showed severe autoimmune-like phenotypes such as arthritis and cachexia, suggesting the important role of TTP in the negative feedback of inflammatory stimulation (27). Latest reports have revealed the importance of TTP with respect to viral infection and tumour immunity; and is now considered to possess variable roles as a suppressor of the immune system owing to its functions (28, 29).
Fig. 3.
The structure of RBPs. The structure of RBPs. CCCH domain and RRM domain are important for RNA binding and function (8, 9, 35, 42, 55, 61, 66). ARID, AT-rich interactive domain; CCCH zinc finger, Cys-Cys-Cys-His zinc finger; RRM, RNA recognition motif; RING finger domain, Really interesting new gene finger domain; ROQ, ROQ RNA-binding domain.
BRF1 (ZFP36L1) and BRF2 (ZFP36L2) are members of the ZFP36 family, which share a highly conserved CCCH finger with TTP, and recognize AREs and accelerate mRNA decay (30). BRF1 KO mice were embryonic lethal because of chorioallantoic fusion, while mice lacking BRF2 survived only for a few days after birth possibly because of hematopoietic stem cell failure (31, 32). Both of BRF1 and BRF2 were expressed throughout the thymic development stage, and Zfp36l1fl/fl Zfp36l2fl/fl CD2-Cre dKO mice perturbed thymic development and T cell acute lymphoblastic leukemia (T-ALL) by aberrant expression of Notch1 (10). However, though their structures and regulatory mechanism seem to be similar, BRF1 and BRF2 play different roles in the immune system. BRF1 was reported to be essential for the maintenance of marginal-zone B cell compartment (33) and BRF2 was recently reported to prevent the chronic activation of memory T cells by blocking the recruitment (34).
AUF1 (hnRNP D) is another ARE-BP that downregulates ARE-containing mRNAs. There are four different alternatively spliced isoforms of AUF1 (p37, p40, p42 and p45) and all of them contain two RNA recognition motifs (35). The main role of AUF1 is the degradation of ARE-containing mRNAs and it plays an indispensable role in suppressing the immune system. Mice lacking AUF1 exhibited symptoms of severe endotoxin shock upon endotoxin challenge, due to the failure to degrade mRNAs of several critical inflammatory cytokines, such as IL1β and TNFα (6). Though the functions of AUF1 seem to be similar to that of TTP, AUF1-dependent decay of mRNA is different from the TTP-dependent destabilization of mRNA. AUF1 forms a protein assembly with the cap-dependent translation initiation factor, eIF4G and heat shock proteins such as, heat shock cognate 71 kDa protein (HSC70) and heat shock protein 70 (HSP70) (36, 37). AUF1-related mRNA decay is associated with displacement of eukaryotic initiation factor 4 G (eIF4G) from AUF1, and subsequent ubiquitination and degradation of AUF1 by proteasomes (38). Photoactivatable ribonucleotide enhanced crosslinking and immunoprecipitation (PAR-CLIP) analysis and ribosomal profiling showed that AUF1 preferentially recognizes U-rich sequences, and at times promotes the translation of numerous mRNAs under several conditions, which suggests the multiple and complex roles of AUF1 (39).
Contrary to these mRNA degradation factors, embryonic lethal abnormal vision (ELAV/Hu) family proteins are the only RBPs that stabilize mRNAs by recognizing AREs (7, 40, 41). Though most of Hu protein family members (Hu-B, C and D) are expressed only in the central nervous system, HuR (ELAVL1) is the only widely expressed factors in humans. HuR has three RNA recognition motif (RRM) domains that are important for RNA binding (42). Most of HuR proteins are localized in nucleus; however, HuR accumulates in the cytoplasm by inflammatory stimulation and delays the onset of decay of ARE-containing mRNAs (7, 41, 43, 44). These findings suggested that HuR competes with mRNA destabilizing ARE-BPs, such as TTP, and protect target mRNAs from decay. HuR targets mRNAs of several inflammatory cytokines, such as IL-4, IL-13, TNF-α and IL-17, and mice lacking HuR were embryonically lethal because of defects in placental branching morphogenesis and embryonic development (45–52). Further studies using conditional-knockout mice revealed that HuR deletion in CD4+ T cells impaired Th17 differentiation; these mice were resistant to experimental autoimmune encephalomyelitis (EAE) (46). Moreover, elucidation of HuR targets and functions in all transcriptomes using PAR-CLIP analysis showed most of the binding sites of HuR to be located in 3′ UTR of mRNAs with preferential bindings to the intronic sites or 3′ UTR of mRNA. Binding motifs are U-rich sequences such as UUUUUUU, UUUAUUU and UUUGUUU as well as common AU-rich motifs such as UAUUUAU (53). Furthermore, another report showed that HuR stabilized transcripts with both intronic and 3′ UTR biding sites rather than transcripts with only 3′ UTR sites or intronic-binding sites. HuR alters splicing pattern and also protects mRNA from microRNA (miRNA)-induced mRNA decay, suggesting that HuR regulates expression outcomes at multiple stages of RNA processing (54).
GRE-Regulating Proteins in the Immune System
CUGBP1 is a member of the CELF family of RBPs and is the only well-studied RBP to be reported as a GRE-regulating protein (14). CUGBP1 has two RRM domains at the N terminus and one RRM domain at its C terminus, which are essential for binding to mRNA (55). CUGBP1 is well-known for binding to the abnormally extended CUG mRNA repeats in type I myotonic dystrophy (56). However, it was found that CUGBP1 also had the ability to destabilize GRE-containing transcripts by recruiting the poly(A)-specific ribonuclease (PARN) deadenylase (14, 57). As GRE are present in the mRNAs of important inflammatory factors, such as JUN, JUNB or TNFRSF1B, CUGBP1 is thought to play a suppressive role in the immune system.
CDE-Regulating Proteins in the Immune System
The constitutive decay element is another essential structure for post-transcriptional regulation of the immune system. In this section, we focus on three characteristic CDE-regulating proteins, Roquin1/2, Regnase-1 and Arid5a.
Roquin1/2 (RC3H1 and RC3H2) are important factors for the destabilization of inflammatory cytokines mRNAs. Roquin has a CCCH zinc finger domain for RNA binding, a Really interesting new gene (RING) finger domain, a ROQ domain for recognizing stem loop structure and a proline-rich domain (16). The relationship between Roquin and inflammatory cytokines was studied in sanroque mice, generated by screening a mutated mouse genome strain (9). The sanroque mice carried a T to G substitution that resulted in a M199R codon change (Fig. 3) (9). The sanroque mutation increased the number of follicular helper T cells, and showed severe autoimmune phenotypes. In these mice, the mutations prevented the downregulation of the mRNA of inducible T-cell Co-stimulator (ICOS), which was an essential co-stimulatory receptor for follicular T cells, and resulted in aberrant production of the cytokine, IL-21 (9). Furthermore, a conserved 47-base pair minimal region in the 3′ UTR of ICOS mRNA was found to be essential for the regulation of ICOS by Roquin (58). However, though the mice lacking Roquin-1 exhibited perinatal lethality and conditional knockout of Roquin-1 in immune cells of mice that failed to maintain immune homeostasis, autoimmunity could be observed in these mice (59). Furthermore, even when Roquin-2 was ablated, a phenotype similar to that of Roquin-1-ablated mice was observed. T cells accumulated when both Roquin1/2 were ablated in CD4+ T cells, and these mice showed strong activation of effector and follicular helper T cells, resulting in severe autoimmune phenotype. Both Roquin1/2 shared target genes, such as ICOS or Ox40, which are co-stimulatory receptors and redundantly regulate the polarization into follicular helper T cells (60). These targeted genes consisted of stem-loop structure in CDE, and Roquin1/2 recognized and bound to it via their ROQ domain, and subsequently recruited the Ccr4-Caf1-Not deadenylase complex and causing the degradation of target genes. More than 50 vertebrate mRNAs harbour CDE and many of them are regulated by Roquin1/2 (16).
Regnase-1 (ZC3H12A, MCPIP1) is another important RBP that down-regulates inflammatory cytokines. As shown in Fig. 3, Regnase-1 has a CCCH zinc finger domain, a PilT N-terminus (PIN)-domain like RNase domain. The CCCH zinc finger domain is essential for its binding to target mRNA and PIN structure is especially important for Mg2+ binding and enzymatic activity. Regnase-1 knockout mice survived only for a few weeks, and they exhibited splenomegaly, pulmonary inflammation and larger lymph nodes, suggesting the essential roles of Regnase-1 in mRNA decay. Regnase-1 recognizes the stem loop in CDE of cytokine mRNAs and causes the mRNA decay with its RNase activity (8). The target genes of Regnase-1 are important inflammatory cytokines or immune-related genes such as c-Rel, Ox40 and Il-2, and T cell receptor (TCR) stimulation led to cleavage of Regnase-1 at R111 by Malt1, thereby decreasing the levels of Regnase-1 and upregulating inflammation (Fig. 3) (61). The Regnase family consists of ZC3H12B and ZC3H12D, and both repress the expression of inflammatory cytokines. The mechanisms of action of ZC3H12D seems to be similar to that of Regnase-1, whereas ZC3H12B downregulates mRNAs via distinct mechanisms, which are not yet fully understood (62, 63). Further studies would be needed to fully comprehend the regulatory mechanisms by ZC3H12 family genes.
As we have already reviewed, Regnase-1 and Roquin share their roles in the regulation of the immune system and the regulatory mechanism itself seems to be similar. The paracaspase, MALT1, cleaves both Roquin and Regnase-1 and repressed important targets for promoting Th17 differentiation (64). However, Regnase-1 preferentially cleaves translationally active mRNAs and requires the helicase activity of UPF1 for its action, while Roquin regulates translationally inactive mRNAs in stress granules or P-bodies. These observations suggest that Regnase-1 is involved in the acute phase of inflammation, while Roquin has a role in the chronic phase of inflammation (65). Therefore, Regnase-1 and Roquin play distinct roles in inflammation and their cooperation is an essential factor for regulating the expression of inflammatory cytokines.
The AT-rich interactive domain-containing protein 5a (Arid5a) is a counter factor for Regnase-1 and Roquin1-2. Arid5a was initially thought to be a transcriptional factor like other Arid family proteins (66). However, it was found to have important roles in post-transcriptional regulation as an RBP. It recognizes the stem loop structures in cytokines mRNAs and stabilizes them (5, 67). Arid5a can be induced by the stimulation of LPS, IL1-β and IL-6 (68). These inflammatory stimulations induce localization of Arid5a from the nucleus into the cytoplasm, thereby making it possible for Arid5a to upregulate cytokine genes (69). Arid5a KO mice showed resistance to endotoxin shock and EAE challenge, suggesting their important physiological roles in the immune system. Arid5a stabilizes mRNAs of Stat3, T-bet and Ox40, resulting in the upregulation of the polarization of each subset of helper T cells (67, 70, 71).
Conclusion
Post-transcriptional regulation by various RBPs through cis-elements is essential for the accurate regulation of inflammation. Though ARE and CDE are well-known cis-elements in mRNAs, there might still be other important regulatory elements. Furthermore, we speculated that similar to Arid5a, other factors that play the roles of both transcriptional factors and RBPs might exist. For example, approximately 800 zinc finger proteins are encoded by the human genomes and most of them have not been classified into transcriptional factors or RBPs yet; and some of them are thought to play both roles. For understanding the mechanism of inflammation, especially chronic inflammation, further elucidation of post-transcriptional regulation by RBPs is required. Recently, several ways for analysing the roles and target of RBPs have been proposed. One of the most important innovative techniques is the CLIP, which makes it possible for us to fully understand the target molecules of RBPs with high sensitivity (72, 73). These techniques for analysing the role of RBPs could clarify the whole scenario of chronic inflammation in the next decade. Furthermore, since many RBPs share the same cis-elements and play multiple roles, there should be some regulatory systems for the expression and subcellular localization of each RBP. However, the entire mechanism of such regulations is still unknown. Therefore, the technological innovation for analysing these interactions are needed to understand the complex roles of RBPs in inflammation.
Acknowledgement
We are grateful to all members of the Asahara Lab for their helpful discussion. This review article was partly supported by AMED-CREST from AMED (JP15gm0410001, JP18gm0810008) and grants from the National Institutes of Health [AR050631, AR065379 to H.A.].
Conflict of Interest
None declared.
Glossary
Abbreviations
- AUF1
AU-rich element polyU binding degradation factor1
- Arid5a
AT-rich interactive domain-containing protein 5a
- ARE
AU-rich element
- ARE-BP
AU-rich element-binding protein
- BRF
butyrate response factor
- CCCH zinc finger
Cys-Cys-Cys-His zinc finger
- CDE
constitutive decay element
- CELF
CUG-BP Elav-like family
- CLIP
crosslinking and immunoprecipitation
- CSF2
colony stimulating factor2
- CUGBP1
CUG triplet repeat RNA-binding protein1
- EAE
experimental autoimmune encephalomyelitis
- ELAVL1
ELAV-like protein 1
- GRE
GU rich element
- HSC70
heat shock cognate 71 kDa protein
- HSP70
heat shock protein 70
- ICOS
inducible T-cell co-stimulator
- LPS
lipopolysaccharide
- MAPK
mitogen-activated protein kinase
- MCPIP
monocyte chemotactic protein induced protein
- miRNA
micro RNA
- NF-κB
nuclear factor kappa B
- PAR-CLIP
photoactivatable ribonucleotide enhanced crosslinking and immunoprecipitation
- PARN
poly(A) specific ribonuclease
- PIN
PilT N-terminus
- RA
rheumatoid arthritis
- RBP
RNA-binding protein
- RING finger domain
really interesting new gene finger domain
- RRM
RNA recognition motif
- SLE
systemic lupus erythematosus
- STAT3
signal transducer and activator of transcription 3
- T-ALL
T cell acute lymphoblastic leukemia
- TNFα
tumor necrosis factor alpha
- TNFRSF1B
TNF receptor superfamily 1B
- TTP
tristetraproline
- UTR
untranslated region
- ZFP
zinc finger protein
References
- 1. Medzhitov R., Horng T. (2009) Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 9, 692–703 [DOI] [PubMed] [Google Scholar]
- 2. Yu H., Pardoll D., Jove R. (2009) STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer 9, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Elkon R., Zlotorynski E., Zeller K.I., Agami R. (2010) Major role for mRNA stability in shaping the kinetics of gene induction. BMC Genomics 11, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rabani M., Levin J.Z., Fan L., Adiconis X., Raychowdhury R., Garber M., Gnirke A., Nusbaum C., Hacohen N., Friedman N., Amit I., Regev A. (2011) Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 29, 436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Masuda K., Ripley B., Nishimura R., Mino T., Takeuchi O., Shioi G., Kiyonari H., Kishimoto T. (2013) Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc. Natl. Acad. Sci. U S A. 110, 9409–9414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lu J.Y., Sadri N., Schneider R.J. (2006) Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 20, 3174–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fan X.C., Steitz J.A. (1998) Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J 17, 3448–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Matsushita K., Takeuchi O., Standley D.M., Kumagai Y., Kawagoe T., Miyake T., Satoh T., Kato H., Tsujimura T., Nakamura H., Akira S. (2009) Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458, 1185–1190 [DOI] [PubMed] [Google Scholar]
- 9. Vinuesa C.G., Cook M.C., Angelucci C., Athanasopoulos V., Rui L., Hill K.M., Yu D., Domaschenz H., Whittle B., Lambe T., Roberts I.S., Copley R.R., Bell J.I., Cornall R.J., Goodnow C.C. (2005) A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature 435, 452–458 [DOI] [PubMed] [Google Scholar]
- 10. Hodson D.J., Janas M.L., Galloway A., Bell S.E., Andrews S., Li C.M., Pannell R., Siebel C.W., MacDonald H.R., De Keersmaecker K., Ferrando A.A., Grutz G., Turner M. (2010) Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat. Immunol. 11, 717–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carballo E., Lai W.S., Blackshear P.J. (1998) Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281, 1001–1005 [DOI] [PubMed] [Google Scholar]
- 12. Gerstberger S., Hafner M., Tuschl T. (2014) A census of human RNA-binding proteins. Nat. Rev. Genet. 15, 829–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zubiaga A.M., Belasco J.G., Greenberg M.E. (1995) The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol. 15, 2219–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vlasova I.A., Tahoe N.M., Fan D., Larsson O., Rattenbacher B., Sternjohn J.R., Vasdewani J., Karypis G., Reilly C.S., Bitterman P.B., Bohjanen P.R. (2008) Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell. 29, 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stoecklin G., Lu M., Rattenbacher B., Moroni C. (2003) A constitutive decay element promotes tumor necrosis factor alpha mRNA degradation via an AU-rich element-independent pathway. Mol. Cell Biol. 23, 3506–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leppek K., Schott J., Reitter S., Poetz F., Hammond M.C., Stoecklin G. (2013) Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell 153, 869–881 [DOI] [PubMed] [Google Scholar]
- 17. Shaw G., Kamen R. (1986) A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46, 659–667 [DOI] [PubMed] [Google Scholar]
- 18. Stoecklin G., Hahn S., Moroni C. (1994) Functional hierarchy of AUUUA motifs in mediating rapid interleukin-3 mRNA decay. J. Biol. Chem. 269, 28591–28597 [PubMed] [Google Scholar]
- 19. Kontoyiannis D., Pasparakis M., Pizarro T.T., Cominelli F., Kollias G. (1999) Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10, 387–398 [DOI] [PubMed] [Google Scholar]
- 20. Chen C.Y., Shyu A.B. (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20, 465–470 [DOI] [PubMed] [Google Scholar]
- 21. Chen C.Y., Xu N., Shyu A.B. (1995) mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 15, 5777–5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peng S.S., Chen C.Y., Shyu A.B. (1996) Functional characterization of a non-AUUUA AU-rich element from the c-jun proto-oncogene mRNA: evidence for a novel class of AU-rich elements. Mol. Cell. Biol. 16, 1490–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hau H.H., Walsh R.J., Ogilvie R.L., Williams D.A., Reilly C.S., Bohjanen P.R. (2007) Tristetraprolin recruits functional mRNA decay complexes to ARE sequences. J. Cell. Biochem. 100, 1477–1492 [DOI] [PubMed] [Google Scholar]
- 24. Lykke-Andersen J., Wagner E. (2005) Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 19, 351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bourcier C., Griseri P., Grepin R., Bertolotto C., Mazure N., Pages G. (2011) Constitutive ERK activity induces downregulation of tristetraprolin, a major protein controlling interleukin8/CXCL8 mRNA stability in melanoma cells. Am. J. Physiol. Cell Physiol. 301, C609–618 [DOI] [PubMed] [Google Scholar]
- 26. Deleault K.M., Skinner S.J., Brooks S.A. (2008) Tristetraprolin regulates TNF TNF-alpha mRNA stability via a proteasome dependent mechanism involving the combined action of the ERK and p38 pathways. Mol. Immunol. 45, 13–24 [DOI] [PubMed] [Google Scholar]
- 27. Taylor G.A., Carballo E., Lee D.M., Lai W.S., Thompson M.J., Patel D.D., Schenkman D.I., Gilkeson G.S., Broxmeyer H.E., Haynes B.F., Blackshear P.J. (1996) A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4 4, 445–454 [DOI] [PubMed] [Google Scholar]
- 28. Coelho M.A., de Carne Trecesson S., Rana S., Zecchin D., Moore C., Molina-Arcas M., East P., Spencer-Dene B., Nye E., Barnouin K., Snijders A.P., Lai W.S., Blackshear P.J., Downward J. (2017) Oncogenic RAS signaling promotes tumor immunoresistance by stabilizing PD-L1 mRNA. Immunity 47, 1083–1099.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moore M.J., Blachere N.E., Fak J.J., Park C.Y., Sawicka K., Parveen S., Zucker-Scharff I., Moltedo B., Rudensky A.Y., Darnell R.B. (2018) ZFP36 RNA-binding proteins restrain T cell activation and anti-viral immunity. Elife 7, e33057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stoecklin G., Colombi M., Raineri I., Leuenberger S., Mallaun M., Schmidlin M., Gross B., Lu M., Kitamura T., Moroni C. (2002) Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 21, 4709–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stumpo D.J., Byrd N.A., Phillips R.S., Ghosh S., Maronpot R.R., Castranio T., Meyers E.N., Mishina Y., Blackshear P.J. (2004) Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the Tristetraprolin family. Mol. Cell Biol. 24, 6445–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stumpo D.J., Broxmeyer H.E., Ward T., Cooper S., Hangoc G., Chung Y.J., Shelley W.C., Richfield E.K., Ray M.K., Yoder M.C., Aplan P.D., Blackshear P.J. (2009) Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood 114, 2401–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Newman R., Ahlfors H., Saveliev A., Galloway A., Hodson D.J., Williams R., Besra G.S., Cook C.N., Cunningham A.F., Bell S.E., Turner M. (2017) Maintenance of the marginal-zone B cell compartment specifically requires the RNA-binding protein ZFP36L1. Nat. Immunol. 18, 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Salerno F., Engels S., van den Biggelaar M., van Alphen F.P.J., Guislain A., Zhao W., Hodge D.L., Bell S.E., Medema J.P., von Lindern M., Turner M., Young H.A., Wolkers M.C. (2018) Translational repression of pre-formed cytokine-encoding mRNA prevents chronic activation of memory T cells. Nat. Immunol. 19, 828–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wagner B.J., DeMaria C.T., Sun Y., Wilson G.M., Brewer G. (1998) Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics 48, 195–202 [DOI] [PubMed] [Google Scholar]
- 36. Knapinska A.M., Gratacos F.M., Krause C.D., Hernandez K., Jensen A.G., Bradley J.J., Wu X., Pestka S., Brewer G. (2011) Chaperone Hsp27 modulates AUF1 proteolysis and AU-rich element-mediated mRNA degradation. Mol. Cell. Biol. 31, 1419–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sinsimer K.S., Gratacos F.M., Knapinska A.M., Lu J., Krause C.D., Wierzbowski A.V., Maher L.R., Scrudato S., Rivera Y.M., Gupta S., Turrin D.K., De La Cruz M.P., Pestka S., Brewer G. (2008) Chaperone Hsp27, a novel subunit of AUF1 protein complexes, functions in AU-rich element-mediated mRNA decay. Mol. Cell Biol. 28, 5223–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laroia G., Cuesta R., Brewer G.a., Schneider R.J. (1999) Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284, 499–502 [DOI] [PubMed] [Google Scholar]
- 39. Yoon J.H., De S., Srikantan S., Abdelmohsen K., Grammatikakis I., Kim J., Kim K.M., Noh J.H., White E.J., Martindale J.L., Yang X., Kang M.J., Wood W.H. 3rd, Noren Hooten N., Evans M.K., Becker K.G., Tripathi V., Prasanth K.V., Wilson G.M., Tuschl T., Ingolia N.T., Hafner M., Gorospe M. (2014) PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat. Commun. 5, 5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hinman M.N., Lou H. (2008) Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 65, 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peng S.S., Chen C.Y., Xu N., Shyu A.B. (1998) RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17, 3461–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ma W.J., Cheng S., Campbell C., Wright A., Furneaux H. (1996) Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271, 8144–8151 [DOI] [PubMed] [Google Scholar]
- 43. Lopez de Silanes I., Zhan M., Lal A., Yang X., Gorospe M. (2004) Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. U S A. 101, 2987–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tran H., Maurer F., Nagamine Y. (2003) Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2. Mol. Cell Biol. 23, 7177–7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cok S.J., Acton S.J., Morrison A.R. (2003) The proximal region of the 3′-untranslated region of cyclooxygenase-2 is recognized by a multimeric protein complex containing HuR, TIA-1, TIAR, and the heterogeneous nuclear ribonucleoprotein U. J. Biol. Chem. 278, 36157–36162 [DOI] [PubMed] [Google Scholar]
- 46. Chen J., Cascio J., Magee J.D., Techasintana P., Gubin M.M., Dahm G.M., Calaluce R., Yu S., Atasoy U. (2013) Posttranscriptional gene regulation of IL-17 by the RNA-binding protein HuR is required for initiation of experimental autoimmune encephalomyelitis. J. Immunol. 191, 5441–5450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Herjan T., Yao P., Qian W., Li X., Liu C., Bulek K., Sun D., Yang W.P., Zhu J., He A., Carman J.A., Erzurum S.C., Lipshitz H.D., Fox P.L., Hamilton T.A., Li X. (2013) HuR is required for IL-17-induced Act1-mediated CXCL1 and CXCL5 mRNA stabilization. J. Immunol. 191, 640–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Abdelmohsen K., Gorospe M. (2010) Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev. RNA 1, 214–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Casolaro V., Fang X., Tancowny B., Fan J., Wu F., Srikantan S., Asaki S.Y., De Fanis U., Huang S.K., Gorospe M., Atasoy U.X., Stellato C. (2008) Posttranscriptional regulation of IL-13 in T cells: role of the RNA-binding protein HuR. J. Allergy Clin. Immunol. 121, 853–859.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stellato C., Gubin M.M., Magee J.D., Fang X., Fan J., Tartar D.M., Chen J., Dahm G.M., Calaluce R., Mori F., Jackson G.A., Casolaro V., Franklin C.L., Atasoy U. (2011) Coordinate regulation of GATA-3 and Th2 cytokine gene expression by the RNA-binding protein HuR. J. Immunol. 187, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Atasoy U., Curry S.L., Lopez de Silanes I., Shyu A.B., Casolaro V., Gorospe M., Stellato C. (2003) Regulation of eotaxin gene expression by TNF-alpha and IL-4 through mRNA stabilization: involvement of the RNA-binding protein HuR. J. Immunol. 171, 4369–4378 [DOI] [PubMed] [Google Scholar]
- 52. Katsanou V., Milatos S., Yiakouvaki A., Sgantzis N., Kotsoni A., Alexiou M., Harokopos V., Aidinis V., Hemberger M., Kontoyiannis D.L. (2009) The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Mol. Cell Biol. 29, 2762–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kishore S., Jaskiewicz L., Burger L., Hausser J., Khorshid M., Zavolan M. (2011) A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods 8, 559–564 [DOI] [PubMed] [Google Scholar]
- 54. Mukherjee N., Corcoran D.L., Nusbaum J.D., Reid D.W., Georgiev S., Hafner M., Ascano M. Jr., Tuschl T., Ohler U., Keene J.D. (2011) Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell 43, 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tsuda K., Kuwasako K., Takahashi M., Someya T., Inoue M., Terada T., Kobayashi N., Shirouzu M., Kigawa T., Tanaka A., Sugano S., Guntert P., Muto Y., Yokoyama S. (2009) Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res 37, 5151–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Timchenko L.T., Miller J.W., Timchenko N.A., DeVore D.R., Datar K.V., Lin L., Roberts R., Caskey C.T., Swanson M.S. (1996) Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 24, 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moraes K.C., Wilusz C.J., Wilusz J. (2006) CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA 12, 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yu D., Tan A.H., Hu X., Athanasopoulos V., Simpson N., Silva D.G., Hutloff A., Giles K.M., Leedman P.J., Lam K.P., Goodnow C.C., Vinuesa C.G. (2007) Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 450, 299–303 [DOI] [PubMed] [Google Scholar]
- 59. Bertossi A., Aichinger M., Sansonetti P., Lech M., Neff F., Pal M., Wunderlich F.T., Anders H.J., Klein L., Schmidt-Supprian M. (2011) Loss of Roquin induces early death and immune deregulation but not autoimmunity. J. Exp. Med. 208, 1749–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pratama A., Ramiscal R.R., Silva D.G., Das S.K., Athanasopoulos V., Fitch J., Botelho N.K., Chang P.P., Hu X., Hogan J.J., Mana P., Bernal D., Korner H., Yu D., Goodnow C.C., Cook M.C., Vinuesa C.G. (2013) Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity 38, 669–680 [DOI] [PubMed] [Google Scholar]
- 61. Uehata T., Iwasaki H., Vandenbon A., Matsushita K., Hernandez-Cuellar E., Kuniyoshi K., Satoh T., Mino T., Suzuki Y., Standley D.M., Tsujimura T., Rakugi H., Isaka Y., Takeuchi O., Akira S. (2013) Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell 153, 1036–1049 [DOI] [PubMed] [Google Scholar]
- 62. Wawro M., Kochan J., Krzanik S., Jura J., Kasza A. (2017) Intact NYN/PIN-Like Domain is Crucial for the Degradation of Inflammation-Related Transcripts by ZC3H12D. J. Cell. Biochem. 118, 487–498 [DOI] [PubMed] [Google Scholar]
- 63. Wawro M., Wawro K., Kochan J., Solecka A., Sowinska W., Lichawska-Cieslar A., Jura J., Kasza A. (2019) ZC3H12B/MCPIP2, a new active member of the ZC3H12 family. RNA 25, 840–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jeltsch K.M., Hu D., Brenner S., Zoller J., Heinz G.A., Nagel D., Vogel K.U., Rehage N., Warth S.C., Edelmann S.L., Gloury R., Martin N., Lohs C., Lech M., Stehklein J.E., Geerlof A., Kremmer E., Weber A., Anders H.J., Schmitz I., Schmidt-Supprian M., Fu M., Holtmann H., Krappmann D., Ruland J., Kallies A., Heikenwalder M., Heissmeyer V. (2014) Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat. Immunol. 15, 1079–1089 [DOI] [PubMed] [Google Scholar]
- 65. Mino T., Murakawa Y., Fukao A., Vandenbon A., Wessels H.H., Ori D., Uehata T., Tartey S., Akira S., Suzuki Y., Vinuesa C.G., Ohler U., Standley D.M., Landthaler M., Fujiwara T., Takeuchi O. (2015) Regnase-1 and Roquin Regulate a Common Element in Inflammatory mRNAs by Spatiotemporally Distinct Mechanisms. Cell 161, 1058–1073 [DOI] [PubMed] [Google Scholar]
- 66. Amano K., Hata K., Muramatsu S., Wakabayashi M., Takigawa Y., Ono K., Nakanishi M., Takashima R., Kogo M., Matsuda A., Nishimura R., Yoneda T. (2011) Arid5a cooperates with Sox9 to stimulate chondrocyte-specific transcription. Mol. Biol. Cell 22, 1300–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hanieh H., Masuda K., Metwally H., Chalise J.P., Mohamed M., Nyati K.K., Standley D.M., Li S., Higa M., Zaman M.M., Kishimoto T. (2018) Arid5a stabilizes OX40 mRNA in murine CD4(+) T cells by recognizing a stem-loop structure in its 3′ UTR. Eur. J. Immunol. 48, 593–604 [DOI] [PubMed] [Google Scholar]
- 68. Nyati K.K., Masuda K., Zaman M.M., Dubey P.K., Millrine D., Chalise J.P., Higa M., Li S., Standley D.M., Saito K., Hanieh H., Kishimoto T. (2017) TLR4-induced NF-kappaB and MAPK signaling regulate the IL-6 mRNA stabilizing protein Arid5a. Nucleic Acids Res. 45, 2687–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Higa M., Oka M., Fujihara Y., Masuda K., Yoneda Y., Kishimoto T. (2018) Regulation of inflammatory responses by dynamic subcellular localization of RNA-binding protein Arid5a. Proc. Natl. Acad. Sci. U S A. 115, E1214–E1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zaman M.M., Masuda K., Nyati K.K., Dubey P.K., Ripley B., Wang K., Chalise J.P., Higa M., Hanieh H., Kishimoto T. (2016) Arid5a exacerbates IFN-gamma-mediated septic shock by stabilizing T-bet mRNA. Proc. Natl. Acad. Sci. U S A. 113, 11543–11548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Masuda K., Ripley B., Nyati K.K., Dubey P.K., Zaman M.M., Hanieh H., Higa M., Yamashita K., Standley D.M., Mashima T., Katahira M., Okamoto T., Matsuura Y., Takeuchi O., Kishimoto T. (2016) Arid5a regulates naive CD4+ T cell fate through selective stabilization of Stat3 mRNA. J. Exp. Med. 213, 605–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr., Jungkamp A.C., Munschauer M., Ulrich A., Wardle G.S., Dewell S., Zavolan M., Tuschl T. (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Licatalosi D.D., Mele A., Fak J.J., Ule J., Kayikci M., Chi S.W., Clark T.A., Schweitzer A.C., Blume J.E., Wang X., Darnell J.C., Darnell R.B. (2008) HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]