Abstract
Glomerella leaf spot (GLS) of apple (Malus×domestica Borkh.), caused by Glomerella cingulata, is an emerging fungal epidemic threatening the apple industry. Little is known about the molecular mechanism underlying resistance to this devastating fungus. In this study, high-throughput sequencing technology was used to identify microRNAs (miRNAs) involved in GLS resistance in apple. We focused on miRNAs that target genes related to disease and found that expression of a novel miRNA, Md-miRln20, was higher in susceptible apple varieties than in resistant ones. Furthermore, its target gene Md-TN1-GLS exhibited the opposite expression pattern, which suggested that the expression levels of Md-miRln20 and its target gene are closely related to apple resistance to GLS. Furthermore, downregulation of Md-miRln20 in susceptible apple leaves resulted in upregulation of Md-TN1-GLS and reduced the disease incidence. Conversely, overexpression of Md-miRln20 in resistant apple leaves suppressed Md-TN1-GLS expression, with increased disease incidence. We demonstrated that Md-miRln20 negatively regulates resistance to GLS by suppressing Md-TN1-GLS expression and showed, for the first time, a crucial role for miRNA in response to GLS in apple.
Subject terms: Plant immunity, Plant molecular biology
Introduction
Glomerella leaf spot (GLS) of apple (Malus×domestica Borkh.) is an emerging fungal epidemic that has caused great damage in China recently1. GLS, which is caused by Glomerella cingulata, was first reported in the US in 19702 but was not noticed in China until 20113. Today, GLS is an economically important disease that causes early severe defoliation, weakens tree vigor, and reduces apple production. As soon as 2 days after infection with GLS, reddish-purple spots appear on the leaves of infected plants. The spots rapidly expand and merge into necrotic lesions in ensuing days, after which the leaves often turn yellow and drop4. Fruits can also be infected, with small light-brown sunken lesions (1–5 mm in diameter), which do not increase in size over time5. In China, the varieties ‘Golden Delicious’, ‘Gala’, and ‘Qinguan’ are highly susceptible to GLS, whereas ‘Fuji’ and ‘Red Star’ are resistant. In apple, resistance to GLS is controlled by a single recessive gene6, and the location of a GLS resistance gene locus (Rgls) has been identified by bulked segregant analysis (BSA)7,8.
MicroRNAs (miRNAs) are small noncoding RNA molecules that exert regulatory functions by binding to complementary sequences in mRNAs to promote degradation or inhibit translation, resulting in silencing of the corresponding genes9. miRNAs play a crucial role in almost all biological processes in plants, including growth and development, hormone signaling, and stress responses10,11. Mounting evidence has revealed that miRNAs play a significant role in the response of plants to pathogens by directly and indirectly regulating expression of resistance (R) genes12,13. R genes encode a class of protein receptors, most of which contain a nucleotide-binding site (NBS) and one or more leucine-rich repeat (LRR) domains that recognize virulence effectors, leading to effector-triggered immunity (ETI)14,15. For example, members of the potato (Solanum tuberosum) miR482 superfamily target a class of disease resistance proteins with NBS and LRR motifs. Verticillium dahliae injection suppresses expression of miR482e and upregulates several NBS–LRR targets of miR482e, and transgenic potato plantlets overexpressing miR482e show hypersensitivity to V. dahliae infection16. Similarly, Soybean mosaic virus (SMV) counteracts soybean (Glycine max) defense responses through downregulation of several NBS–LRR family resistance genes by inducing accumulation of miR1507a and miR1507c17. In ‘Golden Delicious’ apple, suppression of five R genes, which are targeted by mdm-siR277-1 and mdm-siR277-2, results in susceptibility to Alternaria leaf spot fungus (ALT1)13. These findings highlight the fundamental role of miRNAs in plant immunity.
To date, research regarding GLS of apple has mainly focused on isolating and identifying the pathogen18, environmental conditions affecting infection and spread1, the disease mechanism19, control methods3, and genetic mapping of GLS resistance gene loci6–8. However, little is known about the molecular mechanism underlying the plant’s response to GLS infection.
In this study, we used high-throughput sequencing to identify miRNAs involved in apple resistance to GLS. We found that expression of a novel miRNA named Md-miRln20 was higher in susceptible than in resistant apple varieties, whereas its target gene, named Md-TN1-GLS, showed the opposite trend. These findings indicate that expression of Md-miRln20 and its target gene is closely related to GLS resistance in apple. Further analysis demonstrated that Md-miRln20 is able to regulate resistance to GLS by suppressing Md-TN1-GLS expression. These results illustrate for the first time the crucial role of miRNA in the response of apple to GLS.
Results
Resistance evaluation of apple germplasms to GLS
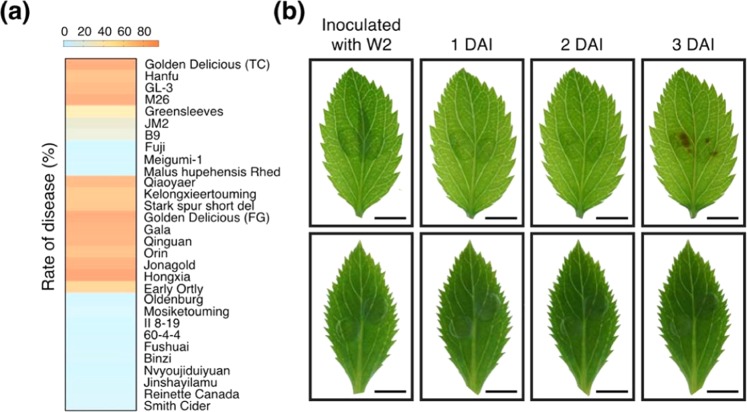
A spore suspension of G. cingulata strain W2 (W2) was inoculated onto detached leaves of 30 different apple germplasms, of which 10 were tissue cultured and 20 grown in the field, along with three F1 hybrid groups (Table S1; Figs. S1 and S2). On the inoculated leaves of susceptible apple germplasms, necrotic lesions became visible and expanded rapidly from 2 DAI; no changes were observed on the leaves of resistant germplasms (Fig. 1b). The incidence of GLS was calculated at 3 DAI. Four of the 10 tissue-cultured apple germplasms showed high disease rates, including ‘Golden Delicious’, ‘Hanfu’, ‘GL-3’, and ‘M26’. ‘Fuji’, ‘Meigumi-1’ (a self-crossed progeny of ‘Meigumi’), and ‘Malus hupehensis Rhed’ were almost completely immune to GLS (Figs. 1a, S1a, and S1b). We also used an SSR marker (S0405127)8 to further evaluate resistance, and a smaller band, 330 bp in size, was found only for susceptible plants. The results of the SSR analysis were essentially in agreement with the incidence of GLS (Figs. 1a and S1). The genotype of the resistant plants was rr, and the genotypes of the susceptible plants were RR and Rr6. ‘Nagafu 2’ and ‘Ryoka no kisetsu’ were resistant, with a genotype of rr. ‘Golden Delicious’, ‘62-45’, ‘Hanfu’, and ‘Yueshuai’ were susceptible, and each carried at least one R allele (Fig. S2). The separation ratios of susceptible-to-resistant plants in the three F1 hybrid groups we analyzed (‘Golden Delicious’ × ‘Nagafu 2’, ‘Hanfu’ × ‘Yueshuai’, and ‘62-45’ × ‘Ryoka no kisetsu’) were statistically consistent with the theoretical ratios of 1:1, 3:1, and 1:0 (Figs. S2 and S3; Table S2). These data verified that resistance to GLS is controlled by a single recessive gene in apple and indicated the genotypes ‘Golden Delicious’, ‘Hanfu’, ‘Yueshuai’, and ‘62-45’ to be Rr, Rr, Rr, and RR, respectively6–8.
Fig. 1. Resistance evaluation of apple germplasms to Glomerella leaf spot and small RNA sequencing of the leaves of susceptible ‘Golden Delicious’.
a The incidence index of apple germplasms. The first 10 were tissue-cultured (TC), and the last 20 were field-grown (FG). b Symptoms of the leaves of susceptible (upper) and resistant (lower) apple germplasms after inoculation with the W2 strain of G. cingulata. Bar = 0.5 cm
High-throughput sequencing of small RNAs and prediction of their targets in GD leaves infected with Glomerella cingulata
To identify miRNAs that respond to GLS in apple, we constructed a small RNA library from W2-inoculated leaves of ‘Golden Delicious’ (GD) collected at 3 DAI (Fig. 1b). A total of 10,096,436 raw reads were generated by Illumina high-throughput sequencing. After removing low-quality reads, 3′ and 5′ adaptors, and reads shorter than 18 nt and also filtering out other contaminant reads, 9,752,888 reads remained (Table S3). Unique sequences of 18–25 nt were mapped to the mature and precursor sequences of miRNAs from other plant species available in miRBase 21 (http://www.mirbase.org/)20. Based on this analysis, we identified 50 known and 101 novel miRNAs (Tables S4 and S5). To better understand the biological functions of the identified novel miRNAs, psRobot software was employed to predict their target genes21. According to target gene annotations in NCBI (https://www.ncbi.nlm.nih.gov), we found eight miRNAs likely to be involved in defense-related processes (Table 1).
Table 1.
Novel miRNAs and predicted targets chosen for expression analysis
| miRNA | Gene ID | Target description |
|---|---|---|
| Md-miRln2 | MDP0000277868 | TMV resistance protein N-like |
| Md-miRln8 | MDP0000935996 (MD11G1059400) | Probable WRKY transcription factor 26 |
| Md-miRln14 | MDP0000610304 (MD02G1052800) | Probable WRKY transcription factor 16 |
| Md-miRln20 | MDP0000234409 (MD02G1112700) | TMV resistance protein N-like |
| Md-miRln29 | MDP0000310092 (MD07G1205300) | Putative disease resistance protein At1g52660 |
| Md-miRln30 | MDP0000184270 (MD05G1025200) | Disease resistance protein RPM1-like |
| Md-miRln33 | MDP0000382588 (MD02G1165600) | Probable disease resistance protein At5g66910 |
| Md-miRln34 | MDP0000214891 | TMV resistance protein N-like |
Expression analysis of potential GLS-responsive novel miRNA-target pairs
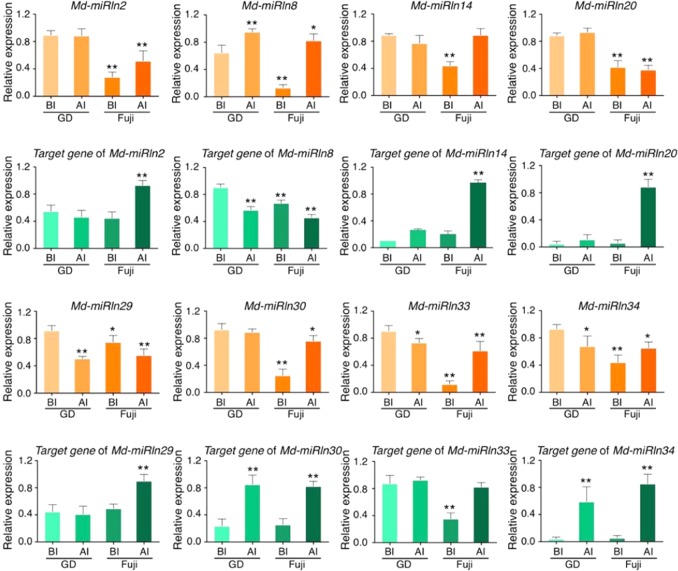
To uncover novel miRNAs that respond to GLS, we analyzed in GD (susceptible variety) and ‘Fuji’ (resistant variety) expression of eight miRNAs and their target genes (Table 1) related to defense according to annotations in NCBI. Notably, Md-miRln20 expression was approximately twice as high in GD as in ‘Fuji’, both before and after W2 inoculation. Moreover, expression of the corresponding target gene MDP0000234409 was significantly upregulated after inoculation in ‘Fuji’ but remained low in GD (Fig. 2). These results show that Md-miRln20 and its target gene likely play a role in the response of apple to GLS.
Fig. 2. Expression of novel miRNAs and their predicted targets in leaves of GD and ‘Fuji’.
RT-qPCR analysis of leaves before inoculation (BI) and 3 days after inoculation (AI) with W2. The orange bars indicate expression of miRNAs, and the green bars indicate that of their target genes. Error bars represent standard deviations calculated from three biological replicates. Student’s t-test: *P < 0.05 and **P < 0.01
Expression levels of Md-miRln20 and its target gene after inoculation are associated with GLS resistance in apple
Md-miRln20 was expressed at higher levels in the susceptible GD variety than in the resistant ‘Fuji’ variety, and its target gene was upregulated only in ‘Fuji’ after inoculation. Thus, we next examined whether these expression patterns extend to other resistant and susceptible varieties.
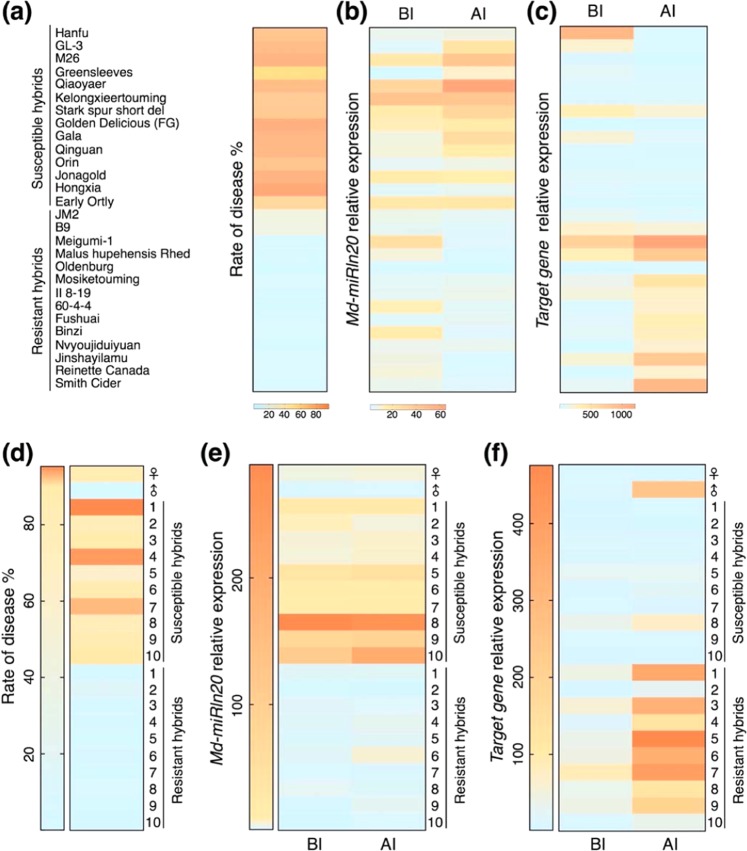
To determine whether Md-miRln20 expression and GLS resistance are associated in apple, expression of miRln20 and its target was analyzed in 29 varieties and 20 F1 hybrids (‘Golden Delicious’ × ‘Nagafu 2’), for which disease resistance had been previously evaluated (Fig. 3a, d). Md-miRln20 and its target gene showed different basal expression patterns in leaves of the 50 apple plants. Independent t-test analysis demonstrated that Md-miRln20 expression was significantly higher (P < 0.01) in the leaves of 24 susceptible apple plants than in those of 25 resistant ones at 3 days after inoculation with W2 (Fig. 3b, e). The target gene was induced only in W2-infected leaves of resistant apples (Fig. 3c, f).
Fig. 3. Expression pattern of Md-miRln20 and Target Gene among various apple germplasms with different resistance to GLS.
a The incidence index of apple germplasms. ‘Hanfu’, ‘GL-3’, ‘M26’, ‘Greensleeves’, ‘JM2’, ‘B9’, ‘Meigumi-1’, and ‘Malus hupehnensis Rhed’ were tissue-cultured plantlets, and others were grown in the field. b, c Expression level of Md-miRln20 (b) and Target Gene (c) in leaves before and after inoculation. d The incidence index of 10 susceptible and 10 resistant F1 hybrids of ‘Golden Delicious’ × ‘Nagafu 2’. e, f The expression level of Md-miRln20 (e) and Target Gene (f) in leaves of hybrids before and after inoculation
Thus, Md-miRln20 expression was higher in susceptible than in resistant apple varieties, whereas the opposite was true for its target gene. Additionally, Pearson correlation coefficients between disease rates and expression levels of Md-miRln20 and its target gene before and after inoculation were 0.28, 0.63, 0.00, and −0.63, respectively. These results suggest that the expression levels of Md-miRln20 and its target gene after inoculation are associated with resistance to GLS in apple leaves.
Validation of a predicted target of Md-miRln20 by a transient GUS expression assay
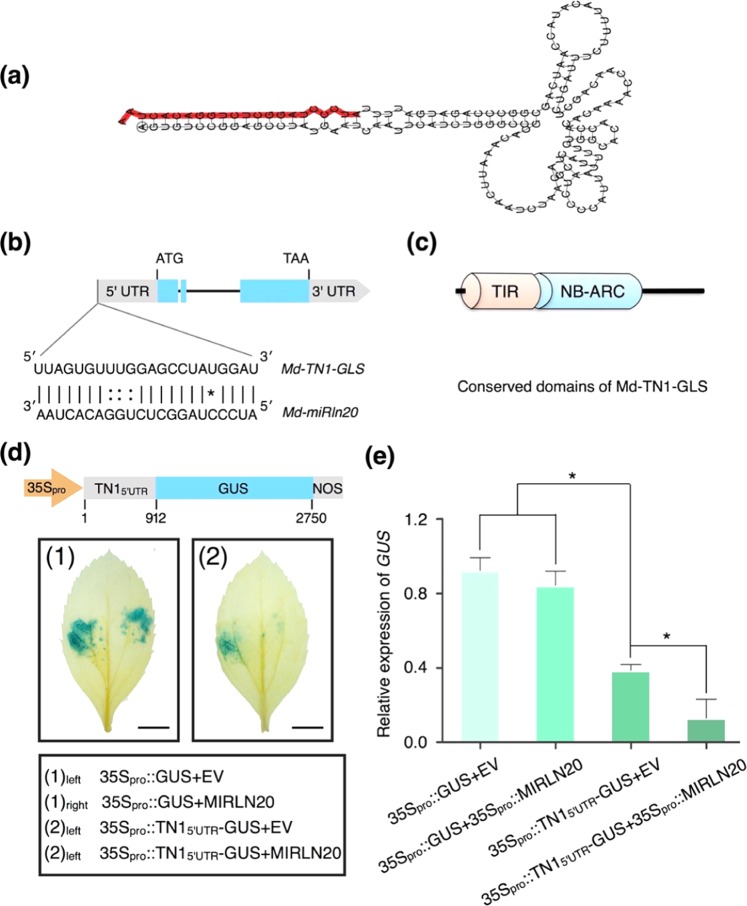
We next obtained the Md-miRln20 precursor sequence by BLAST searching the apple genome22 and EST libraries. Figure 4a displays the stem-loop structure of the precursor and the location of Md-miRln20, which is predicted to target the 5′ UTR of MDP0000234409 (Fig. 4b). Conserved domain analysis using Pfam (http://pfam.xfam.org)23 showed that the protein encoded by MDP0000234409 contains a Toll-like/interleukin-1 receptor (TIR) domain and a NBS, characteristics of the NBS gene family. We thus named the gene Md-TN1-GLS (Fig. 4c). To confirm that Md-TN1-GLS is regulated by Md-miRln20, the 5′ UTR region of Md-TN1-GLS containing the complementarity site was fused to a GUS reporter gene under control of the CaMV 35S promoter. Insertion of the 5′ UTR sequence caused downregulation of GUS in apple leaves, but when TN15′UTR-GUS was transiently coexpressed with MIRLN20, expression of GUS was much lower than that in controls (Fig. 4d, e). Together with the opposite expression patterns of Md-miRln20 and Md-TN1-GLS in apple leaves, these data suggest that Md-miRln20 negatively regulates Md-TN1-GLS at the post-transcriptional level.
Fig. 4. Validation of a predicted target of Md-miRln20.
a Secondary structure of the miRNA precursor and the location of Md-miRln20 (red). b Md-miRln20 targets the 5′ UTR of Md-TN1-GLS. Blue boxes indicate exons. Lines between bases indicate Watson–Crick base pairs, dots indicate wobble base pairs, and the asterisk indicates a mismatched pair. c Conserved domains of Md-TN1-GLS predicted by Pfam. d The 5′ UTR of Md-TN1-GLS containing the complementarity site was fused to the GUS reporter gene under control of the CaMV 35S promoter (35S::TN15′UTR-GUS). (1) and (2) Representative images of leaves expressing various constructs. EV, empty vector of pFGC5941; MIRLN20, overexpression of the precursor sequence of Md-miRln20 under control of the 35S CaMV promoter. Bar = 0.4 cm. e Quantification of relative GUS expression levels in transformed leaves was performed in three biological replicates. Statistical significance was determined by Student’s t-test: *P < 0.05
Md-miRln20 negatively regulates apple resistance to GLS by suppressing Md-TN1-GLS
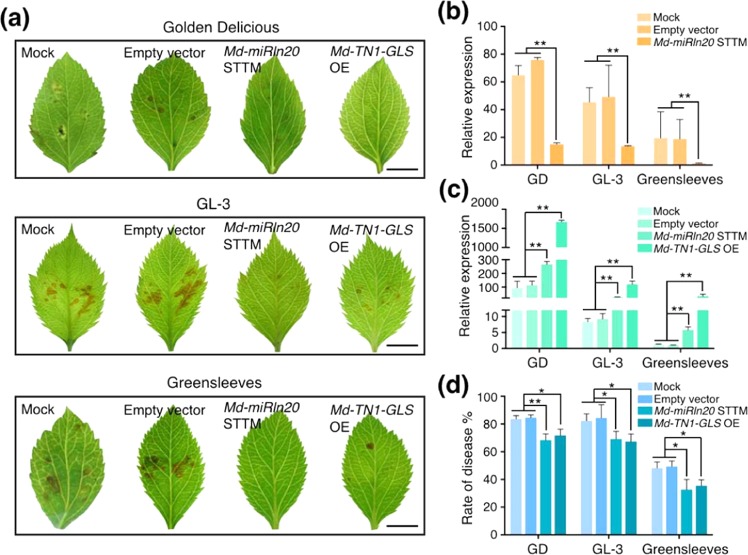
To investigate whether Md-miRln20 participates in resistance to GLS, a short tandem target mimic (STTM) of Md-miRln2024,25 to silence Md-miRln20 was cloned into the binary vector pFGC5941 and transiently expressed in the leaves of three susceptible tissue-cultured apple plantlets. RT-qPCR analysis indicated a significant decrease in the abundance of Md-miRln20 in the leaves of GD, ‘GL-3’, and ‘Greensleeves’ at 3 days after agro-infiltration (Fig. 5b).
Fig. 5. Transient expression of Md-miRln20 and Md-TN1-GLS in leaves from three susceptible tissue-cultured apple varieties.
a Symptoms of GD, ‘GL-3’, and ‘Greensleeves’ leaves at 3 DAI with W2. Bar = 0.5 cm. b, c Expression levels of Md-miRln20 (b) and Md-TN1-GLS (c) in leaves expressing the indicated constructs, as analyzed by RT-qPCR. d Disease incidence of leaves expressing the indicated constructs. EV, empty vector of pFGC5941; Md-miRln20 STTM, overexpression of the Md-miRln20 STTM sequence under control of the 35S CaMV promoter to silence Md-miRln20; Md-TN1-GLS OE, overexpression of Md-TN1-GLS under control of the 35S CaMV promoter. Expression values are means ± SD of three biological replicates, and statistical significance was determined by Student’s t-test: *P < 0.05 and **P < 0.01
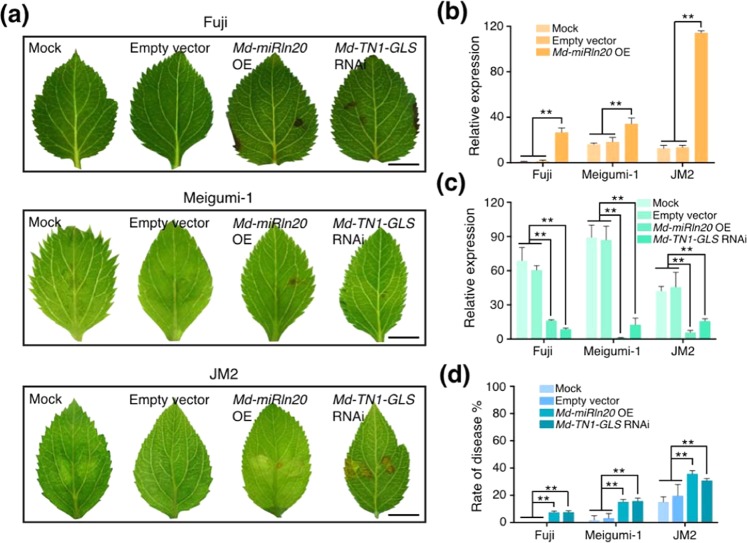
We then inoculated leaves harboring STTM-Md-miRln20 with W2 and found the mRNA level of Md-TN1-GLS to be increased at 3 DAI (Fig. 5c). In addition, expression of STTM-Md-miRln20 suppressed W2 infection symptoms (Fig. 5a) and significantly reduced the rate of disease (Fig. 5d), as did overexpression of Md-TN1-GLS (Fig. 5a, c, d). These results suggest that silencing of Md-miRln20 increased expression of Md-TN1-GLS, which contributed to enhanced resistance of susceptible apples to W2. In contrast, overexpression of MIRLN20 in ‘Fuji’, ‘Meigumi-1’, and ‘JM2’ (three resistant apple varieties) resulted in decreased expression of Md-TN1-GLS (Fig. 6b, c), and the leaves showed more necrotic lesions (Fig. 6a) as well as a higher disease incidence (Fig. 6d). Furthermore, silencing of Md-TN1-GLS rendered leaves more susceptible to W2 (Fig. 6a, c, d), as did overexpression of Md-MIRLN20. Therefore, Md-miRln20 appears to regulate the resistance of apple to GLS by suppressing expression of Md-TN1-GLS.
Fig. 6. Transient expression of Md-miRln20 and Md-TN1-GLS in leaves of three resistant tissue-cultured apple varieties.
a Symptoms of ‘Fuji’, ‘Meigumi-1’, and ‘JM2’ leaves at 3 DAI with W2. Bar = 0.5 cm. b, c Expression levels of Md-miRln20 (b) and Md-TN1-GLS (c) in leaves expressing the indicated constructs, as analyzed by RT-qPCR. d Disease incidence in leaves expressing the indicated constructs. Expression values are means ± SD of three biological replicates, and statistical significance was determined by Student’s t-test: *P < 0.05 and **P < 0.01
Discussion
miRNAs participate in plant disease
An increasing number of small RNAs (sRNAs) involved in plant immunity have been identified recently13,26,27. In this study, we used small RNA sequencing (sRNA-seq) to identify a novel miRNA, named Md-miRln20, as a negative regulator of resistance to GLS in apple.
Various sRNAs play crucial roles in plant defense processes, of which small interfering RNAs (siRNAs) and miRNAs are the two major groups. sRNAs actively regulate immunity in response to different pathogens, including bacteria28, fungi29, viruses30, and nematodes31. Upon infection, up- or downregulation of sRNAs leads to changes in target gene expression. Some sRNAs facilitate plant resistance. M. hupehensis plants transiently overexpressing miR168 exhibited reduced sensitivity to Botryosphaeria dothidea infection32. Similarly, transgenic rice (Oryza sativa) plants overexpressing miR160a and miR398b displayed enhanced resistance to Magnaporthe oryzae, as demonstrated by decreased fungal growth, increased hydrogen peroxide accumulation at the infection site, and upregulated expression of defense-related genes33.
Conversely, some sRNAs negatively regulate plant pathogen resistance. In rice, miRNA528 was upregulated and cleaved L-ASCORBATE OXIDASE (AO) messenger RNA upon viral infection, thereby reducing AO-mediated accumulation of reactive oxygen species and suppressing viral resistance34. Three Populous-specific miRNAs (Ptc-miR482, Ptc-miR482-1444, and Ptc-miR482-1448) promote cleavage of polyphenol oxidase (PPO) transcripts as well as the transcripts of R genes regulating biotic and abiotic stress tolerance. Downregulation of miR482 and miR1448 in Verticillium-infected Gossypium hirsutum resulted in accumulation of both PPO and disease resistance proteins35. In the present study, overexpression of a novel miRNA, Md-miRln20, resulted in susceptibility to GLS in apple leaves through suppression of the corresponding target gene Md-TN1-GLS (Fig. 6).
In addition, some sRNAs act as mobile signals that mediate cross-kingdom RNA interference (RNAi) during host–pathogen interactions36. In response to infection with V. dahliae, cotton plants showed increased production of miR166 and miR159 and exported both to the hyphae of this fungus to silence two essential fungal virulence genes37. Pst-milR1 is a novel miRNA-like RNA in Puccinia striiformis f. sp. tritici that acts as an effector that suppresses host immunity by binding to the transcripts of Triticum aestivum pathogenesis-related gene 2 (PR2)38. These findings suggest that cross-kingdom RNAi is likely to exist and that such a situation is worthy of exploration in the apple–G. cingulata interaction, the resistance mechanism of which remains unclear.
The contribution of Md-TN1-GLS to GLS resistance in apples is likely regulated by SA
Md-TN1-GLS is the target gene of Md-miRln20, and its expression promotes resistance to GLS in apple. Md-TN1-GLS contains a TIR domain and an NBS domain and belongs to the NBS gene family (Fig. 4c). NBS receptors are the most commonly known plant R proteins that detect effectors from pathogens. They are classified into two groups based on their N-terminal domains: Toll-like/interleukin-1 receptor (TIR)-type NBS proteins and coiled-coil (CC)-type NBS proteins39. TIR-NB (TN) proteins have also been identified in plants, containing TIR and NBS domains but lacking LRRs40. Md-TN1-GLS is a newly identified TN protein in apple that was upregulated in resistant apple leaves upon W2 infection. Overexpression of Md-TN1-GLS in susceptible apple varieties increased their resistance to GLS, whereas downregulation rendered apples more susceptible (Figs. 5 and 6). These results indicate that Md-TN1-GLS contributes to apple resistance to GLS.
NBS receptors trigger local resistance associated with programmed cell death as part of the hypersensitive response and also amplify basal defenses involving the signaling hormone salicylic acid (SA), leading to systemic resistance41. Arabidopsis lines stably overexpressing TX and TN genes display a variety of phenotypes associated with basal innate immune responses, which are associated with elevated levels of SA42. Pretreatment with SA strongly induces resistance to GLS in ‘Gala’ apple leaves, with SA-treated leaves showing a significant reduction in lesion number and disease index3. We analyzed the promoter sequences of Md-TN1-GLS in PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and identified several cis-acting elements involved in SA responsiveness (data not shown). Overall, upregulation of Md-TN1-GLS in resistant apple leaves is likely regulated by SA, though this remains to be confirmed.
Candidate genes influencing GLS resistance in apple
It has been reported that GLS resistance is controlled by a single recessive gene (Rgls) in apple6. The locus is on chromosome 15, sandwiched between the markers SNP4208 and SNP4257, with a recombination frequency of 0.97%6,7. Nine candidate genes are predicted in the 49-kb genetic interval between SNP4208 and SNP42577; however, the recessive major gene remains to be identified.
In this work, Md-miRln20 was found to negatively regulate resistance to GLS by suppressing expression of Md-TN1-GLS in apple. BLAST results showed that Md-TN1-GLS is located on chromosome 2 in a region that is homeologous to the central region of Chromosome 15 43. However, we found no Md-TN1-GLS-like gene in the region containing Rgls and tightly homeologous to Chromosome 2, where Md-TN1-GLS is located. These results indicate that resistance to GLS might be modified by several other genes, such as Md-TN1-GLS, in addition to the recessive major gene. Further molecular studies are underway to investigate this possibility.
In conclusion, we found that the expression levels of Md-miRln20 and its target gene correlate with resistance to GLS in apples. Furthermore, downregulating Md-miRln20 or overexpressing Md-TN1-GLS in susceptible apple leaves reduced the rate of disease, whereas overexpressing Md-miRln20 or silencing Md-TN1-GLS in resistant apple leaves increased the disease incidence. Our study demonstrates that Md-miRln20 negatively regulates resistance to GLS by suppressing expression of Md-TN1-GLS in apple.
Materials and methods
Plant materials and growth conditions
Tissue-cultured Malus×domestica Borkh. cv. ‘Golden Delicious’ and ‘Fuji’ plantlets were grown on Murashige and Skoog (MS) medium44 containing 0.6 mg L−1 6-benzylaminopurine (6-BA) and 0.15 mg L−1 1-naphthylacetic acid (NAA) in a climate-controlled culture room at 25 ± 1 °C with a 16/8-h light/dark photoperiod. The plants were transferred to fresh medium every 4 weeks.
Twenty apple varieties, 7–8 years old, were grown at the National Germplasm Repository of Apple (Institute of Pomology of Chinese Academy of Agricultural Sciences, CAAS, Xingcheng, Liaoning Province, China).
The crossing parents ‘Golden Delicious’, ‘Nagafu 2’, ‘Hanfu’, ‘Yueshuai’, ‘62-45’, and ‘Ryoka no kisetsu’ were 15-year-old trees, and the three hybrid groups were 6 years old and grown at the Liaoning Institute of Pomology.
Fungal growth, infection assay, and statistical analysis of disease incidence
The highly pathogenic fungal strain W2 (G. cingulata) was isolated by Professor Zongshan Zhou from the Institute of Pomology of CAAS. The W2 monospore culture was maintained on potato dextrose agar (PDA). Conidia were produced on PDA according to a method described previously3. A concentration of 1 × 105 conidia mL−1, which was assessed by microscopy (Olympus, CX31RTSF, Japan), was used to inoculate detached apple leaves by dropping 10 μL suspensions onto a unwounded abaxial surface.
For tissue-cultured plantlets, after 4 weeks of culture, expanded leaves of similar size were inoculated on the unwounded abaxial surface using 10 μL of conidia suspensions (1 × 105 conidia mL−1). For field-grown trees, 5-cm-long leaves were collected, disinfected with 75% (v/v) ethanol, placed on moist filter paper in a culture dish with the base of the petiole wrapped in a moist cotton ball, and incubated under the same conditions as the tissue culture plantlets (humidity, 75%; temperature, 25 °C). Necrotic lesions on each leaf were visually examined 3 days after inoculation (DAI), and disease incidence was recorded. The experiments were repeated three times, and t-test analysis was performed. Three biological replicates of approximately 50 leaves from different tissue-cultured plantlets were conducted to reduce experimental error.
RNA extraction, library construction, and sequencing
Mixed leaves of ‘Golden Delicious’ inoculated with W2 (G. cingulata (Stoneman) Spauld. & H. Schrenk) were collected at 3 DAI, and total RNA was isolated using the modified cetyltrimethyl ammonium bromide (CTAB) method45. RNA samples with high purity (OD 260/280 between 1.8 and 2.2) and integrity (RNA integrity number of 6.5 or higher) were used for small RNA library preparation and sequencing by Tianjin Biochip Corporation (Tianjin, China). Small RNA fragments (18–30 nucleotides) were purified on a 15% denaturing polyacrylamide gel and ligated with 3′ and 5′ adaptors. The ligated products were used for cDNA synthesis, followed by acrylamide gel purification and PCR amplification to generate small RNA libraries. An Agilent 2100 Bioanalyzer (Agilent, USA) was used for quantification and qualification of the sample libraries. The libraries were sequenced using the Illumina HiSeq 2500 sequencing platform (Illumina Inc., San Diego, CA, USA).
Sequence data analysis
The raw reads obtained by RNA sequencing (RNA-seq) were filtered by removing duplication sequences, low-quality reads, reads smaller than 18 nt, adaptor sequences, and contamination formed via adaptor–adaptor ligation. Other RNAs (rRNA, tRNA, snRNA, and snoRNA) were removed by BLAST searching against the GenBank database (http://blast.ncbi.nlm.nih.gov) and Rfam database (http://rfam.xfam.org/)21,46. The remaining clean reads were used for detecting conserved and novel miRNAs.
Identification of conserved and putative novel miRNAs and target prediction
To detect conserved and novel miRNAs, clean reads from the W2-inoculated library were searched against known plant miRNAs in miRBase 21.0 (www.mirbase.org/) with a maximum of three mismatches allowed to identify conserved miRNAs20. Sequences that did not align to any database entries were considered novel putative miRNAs. miREvo47 and mirdeep248 software programs were used to predict novel miRNAs that mapped to the apple genome v3.0.a122 (https://www.rosaceae.org/species/malus/all). The resulting sequences were screened for the presence of stem-loop secondary structures using mfold software49.
To predict miRNA target genes, miRNA sequences were aligned to the apple genome using Plant Small RNA Target Analysis Server (http://plantgrn.noble.org/psRNATarget/). The predicted target genes were evaluated based on complementarity and maximum expectation, as previously described50. The functions of the putative target genes were analyzed using Genome Database for Rosaceae22,43 (https://www.rosaceae.org) and the National Center for Biotechnology Information genomic database (http://www.ncbi.nlm.nih.gov).
Expression analysis of miRNAs and target genes in response to GLS by RT-qPCR
Leaves were obtained from plants at 3 DAI with W2 or water, frozen in liquid nitrogen, and stored at −80 °C until use. Total RNA was extracted using a modified CTAB method45. An ND-1000 NanoDrop spectrophotometer (Thermo Fisher Scientific, USA) was utilized to measure RNA concentrations. DNase-treated RNA (1 μg) was used for the reverse transcription reactions with miRcute Plus miRNA First-Strand cDNA Synthesis Kit (Tiangen Biotech Co., Ltd., China) or M-MLV Reverse Transcriptase (Promega, America). Reverse transcription-quantitative PCR (RT-qPCR) was performed using miRcute plus miRNA Premix and SuperReal PreMix Plus (Tiangen Biotech Co., Ltd., China). Thermocycling involved a program of 40 cycles of 95 °C for 10 s and 60 °C for 30 s with a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific, USA). Relative expression of the miRNAs and target genes was determined using the 2−ΔΔCt method46. U6 was used as the internal reference for miRNAs13, and MdActin was used as the reference for target genes46. All RT-qPCR analyses were performed in three biological replicates, each of which consisted of three technical replicates. Gene-specific primers are provided in Table S6.
Target validation by transient GUS expression assay
To generate the TN1s5′UTR-GUS construct, the 912-kb genomic sequence upstream of Md-TN1-GLS was cloned and ligated into pCAMBIA1305.1 (TransGen Biotech, China) between the CaMV 35S promoter and GUS gene. For expression of MdmiRln20, the precursor sequences obtained by PCR were ligated into the pFGC5941 vector. All of the constructs were transformed into Agrobacterium tumefaciens strain GV3101 by heat-shock transformation, and transient GUS expression assays were carried out by injecting Agrobacterium cells into the leaves of 4-week-old tissue-cultured ‘Golden Delicious’, as previously described13. The infiltrated plantlets were cultured on MS culture medium in a climate-controlled room at 25 ± 1 °C with a 16/8-h light/dark photoperiod. Leaves were collected for GUS staining and gene expression analysis at 3 days after Agrobacterium infiltration. GUS staining was performed as previously described13.
Functional analysis of MdmiRln20 and its target gene by transient expression in apple
STTMs technology-based vectors were used to block endogenous MdmiRln20 activity24. Briefly, the consensus trinucleotide bulge sequence CTA was inserted between the 11th and 12th position from the 3′ end of the reverse complementary sequences of mature miRNAs to generate small RNA-binding (SRB) sequences. Tandem repeats of these SRB sequences were interspersed by a 48-nt imperfect stem-loop linker (ISLL; 5′-GTTGTTGTTGTTATGGTCTAATTTAAATATGGTCTAAAGAAGAAGAAT-3′)46. The STTM sequences were synthesized and cloned into the pFGC5941 vector. To generate the Md-TN1-GLS overexpression vector, the full-length coding sequences were cloned into pFGC5941. For RNA interference of Md-TN1-GLS, a partial unique sequence (5′-AAAAGTTCAAAATTTGAAGACAACAAGGAGAAGATCCTCACATGGAGGAGTGCTCTTACAGATGCAGCAAGTTTGTCAGGATATACTTTCAAAGAGGGAGAGTATGAAGCTACATTTATCAGTAAGATCGTGGAGGAGATCTTCGT-3′) was ligated to the 5′ end of the intron in pFGC5941, and the reverse-complement sequence was ligated to the 3′ end of the intron to produce siRNAs. All of the constructs were transformed into Agrobacterium strain GV3101 by heat-shock transformation46. Transformed Agrobacterium cells were injected into the leaves of 4-week-old tissue-cultured ‘Golden Delicious’, ‘GL-3’, ‘Greensleeves’, ‘Fuji’, ‘Meigumi-1’, or ‘JM2’, as previously described46. The infiltrated plantlets were cultured on MS culture medium in a climate-controlled room at 25 ± 1 °C with a 16/8-h light/dark photoperiod. Leaves were inoculated with W2 at 2 days after Agrobacterium infiltration, and the disease incidence was recorded at 3 DAI. Three leaves from three different plantlets were collected as a sample for gene expression analysis.
DNA extraction and SSR analysis
Leaves from hybrids of ‘Golden Delicious’ × ‘Nagafu 2’, ‘Hanfu’ × ‘Yueshuai’, and ‘62-45’ × ‘Ryoka no kisetsu’ were ground for DNA extraction using a modified CTAB method13. Simple sequence repeat (SSR) analysis was carried out by PCR using a previously screened pair of primers (S0405127)8. The SSR PCR reaction was performed in a 15-μL volume, which consisted of 2 μL of genomic DNA (10 ng μL−1), 7.5 μL of 2× Master Mix (Cwbiotech, Beijing, China), and 0.8 μL of each primer (10 μM). The conditions for PCR amplification were as follows: 94 °C for 5 min, 34 cycles of 95 °C for 30 s, 54 °C for 30 s, and 72 °C for 20 s, followed by 72 °C for 10 min. The amplified products were separated on 2.5% agarose gels.
Supplementary information
Supplementary Fig. S1-S3 and supplementary Tables S1-S6.
Acknowledgements
The authors are grateful to Professor Zongshan Zhou (Institute of Pomology of CAAS, Liaoning, China) for providing the W2 strain of Glomerella cingulata. The authors also thank the Institute of Pomology of CAAS and Liaoning Institute of Pomology for providing the plant materials. This work was supported by funding from the National Natural Science Foundation of China (31872062) and the Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects (CEFF-PXM2019_014207_000032).
Author contributions
Yi Zhang and Tianzhong Li designed the study. Qiulei Zhang and Li Hao prepared the plant materials. Shengnan Wang and Shengyuan Wang performed the RT-qPCR experiments. Yi Zhang performed most of the experiments and wrote the article, and Wenna Zhang modified the language in the manuscript. Chaoran Xu and Yunfei Yu collected the samples.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-019-0175-x).
References
- 1.Wang B, Li B-H, Dong X-L, Wang C-X, Zhang Z-F. Effects of temperature, wetness duration, and moisture on the conidial germination, infection, and disease incubation period of Glomerella cingulata. Plant Dis. 2015;99:249–256. doi: 10.1094/PDIS-04-14-0361-RE. [DOI] [PubMed] [Google Scholar]
- 2.Vergutz L, et al. Thermodynamic characterization of humic acid–surfactant interaction: new insights into the characteristics and structure of humic acids. Rev. Bras. Cienc. Solo. 2015;39:1633–1642. doi: 10.1590/01000683rbcs20140258. [DOI] [Google Scholar]
- 3.Zhang Y, et al. Salicylic acid confers enhanced resistance to Glomerella leaf spot in apple. Plant Physiol. Biochem. 2016;106:64–72. doi: 10.1016/j.plaphy.2016.04.047. [DOI] [PubMed] [Google Scholar]
- 4.Velho AC, Alaniz S, Casanova L, Mondino P, Stadnik MJ. New insights into the characterization of Colletotrichum species associated with apple diseases in southern Brazil and Uruguay. Fungal Biol. 2015;119:229–244. doi: 10.1016/j.funbio.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Velho AC, Rockenbach MF, Mondino P, Stadnik MJ. Modulation of oxidative responses by a virulent isolate of Colletotrichum fructicola in apple leaves. Fungal Biol. 2016;120:1184–1193. doi: 10.1016/j.funbio.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, et al. Genetics and molecular marker identification of a resistance to Glomerella leaf spot in apple. Hortic. Plant J. 2016;2:121–125. doi: 10.1016/j.hpj.2016.06.002. [DOI] [Google Scholar]
- 7.Liu, Y. et al. Rapid location of Glomerella leaf spot resistance gene locus in apple by whole genome re-sequencing. Mol. Breed.37, 10.1007/s11032-017-0684-y (2017).
- 8.Liu Y, et al. Investigation and genetic mapping of a Glomerella leaf spot resistance locus in apple. Plant Breed. 2017;136:119–125. doi: 10.1111/pbr.12399. [DOI] [Google Scholar]
- 9.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 10.Bastias A, et al. Aging gene pathway of microRNAs 156/157 and 172 is altered in juvenile and adult plants from in vitro propagated Prunus sp. Cienc. Inv. Agr. 2016;43:429–441. doi: 10.4067/S0718-16202016000300009. [DOI] [Google Scholar]
- 11.Ding YH, et al. microRNAs involved in auxin signalling modulate male sterility under high-temperature stress in cotton (Gossypium hirsutum) Plant J. 2017;91:977–994. doi: 10.1111/tpj.13620. [DOI] [PubMed] [Google Scholar]
- 12.Ma C, et al. Cloning and characterization of miRNAs and their targets, including a novel miRNA-targeted NBS-LRR protein class gene in apple (Golden Delicious) Mol. Plant. 2014;7:218–230. doi: 10.1093/mp/sst101. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, et al. A single-nucleotide polymorphism in the promoter of a hairpin RNA contributes to Alternaria alternata leaf spot resistance in apple (Malus×domestica) Plant Cell. 2018;30:1924–1942. doi: 10.1105/tpc.18.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teixeira PJ, et al. High-resolution transcript profiling of the atypical biotrophic interaction between Theobroma cacao and the fungal pathogen Moniliophthora perniciosa. Plant Cell. 2014;26:4245–4269. doi: 10.1105/tpc.114.130807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinbrenner AD, Goritschnig S, Staskawicz BJ. Recognition and activation domains contribute to allele-specific responses of an Arabidopsis NLR receptor to an oomycete effector protein. PLoS Pathog. 2015;11:e1004665. doi: 10.1371/journal.ppat.1004665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang L, et al. Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J. Integr. Plant Biol. 2015;57:1078–1088. doi: 10.1111/jipb.12348. [DOI] [PubMed] [Google Scholar]
- 17.Bao D, et al. Down-regulation of genes coding for core RNAi components and disease resistance proteins via corresponding microRNAs might be correlated with successful Soybean mosaic virus infection in soybean. Mol. Plant Pathol. 2018;19:948–960. doi: 10.1111/mpp.12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreira Rafaele R., Peres Natalia A., May De Mio Louise L. Colletotrichum acutatum and C. gloeosporioides Species Complexes Associated with Apple in Brazil. Plant Disease. 2019;103(2):268–275. doi: 10.1094/PDIS-07-18-1187-RE. [DOI] [PubMed] [Google Scholar]
- 19.Liang X, et al. Transcriptomic analysis reveals candidate genes regulating development and host interactions of Colletotrichum fructicola. BMC Genomics. 2018;19:557. doi: 10.1186/s12864-018-4934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jian H, et al. Identification of rapeseed microRNAs Involved in early stage seed germination under salt and drought stresses. Front. Plant Sci. 2016;7:658. doi: 10.3389/fpls.2016.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velasco R, et al. The genome of the domesticated apple (Malus×domestica Borkh.) Nat. Genet. 2010;42:833. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 23.Finn RD, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang G, et al. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods. 2012;58:118–125. doi: 10.1016/j.ymeth.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reichel M, Li YJ, Li JY, Millar AA. Inhibiting plant microRNA activity: molecular SPONGEs, target MIMICs and STTMs all display variable efficacies against target microRNAs. Plant Biotechnol. J. 2015;13:915–926. doi: 10.1111/pbi.12327. [DOI] [PubMed] [Google Scholar]
- 26.Shivaprasad PV, et al. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 2012;24:859–874. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng Y, et al. A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 2018;14:e1006756. doi: 10.1371/journal.ppat.1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curaba J, Singh MB, Bhalla PL. miRNAs in the crosstalk between phytohormone signalling pathways. J. Exp. Bot. 2014;65:1425–1438. doi: 10.1093/jxb/eru002. [DOI] [PubMed] [Google Scholar]
- 29.Gupta OP, Sharma P, Gupta RK, Sharma I. Current status on role of miRNAs during plant–fungus interaction. Physiol. Mol. Plant Pathol. 2014;85:1–7. doi: 10.1016/j.pmpp.2013.10.002. [DOI] [Google Scholar]
- 30.Zhang C, et al. Suppression of jasmonic acid-mediated defense by viral-inducible microRNA319 facilitates virus infection in rice. Mol. Plant. 2016;9:1302–1314. doi: 10.1016/j.molp.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 31.Li X, et al. Identification of soybean microRNAs involved in soybean cyst nematode infection by deep sequencing. PLoS ONE. 2012;7:e39650. doi: 10.1371/journal.pone.0039650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu X, et al. Malus hupehensis miR168 targets to ARGONAUTE1 and contributes to the resistance against Botryosphaeria dothidea infection by altering defense responses. Plant Cell Physiol. 2017;58:1541–1557. doi: 10.1093/pcp/pcx080. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, et al. Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 2014;164:1077–1092. doi: 10.1104/pp.113.230052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, et al. ROS accumulation and antiviral defence control by microRNA528 in rice. Nat. Plants. 2017;3:16203. doi: 10.1038/nplants.2016.203. [DOI] [PubMed] [Google Scholar]
- 35.Lu S, Sun YH, Chiang VL. Stress-responsive microRNAs in Populus. Plant J. 2008;55:131–151. doi: 10.1111/j.1365-313X.2008.03497.x. [DOI] [PubMed] [Google Scholar]
- 36.Weiberg A, Jin HL. Small RNAs—the secret agents in the plant–pathogen interactions. Curr. Opin. Plant Biol. 2015;26:87–94. doi: 10.1016/j.pbi.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang T, et al. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants. 2016;2:16153. doi: 10.1038/nplants.2016.153. [DOI] [PubMed] [Google Scholar]
- 38.Wang B, et al. Puccinia striiformis f. sp tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol. 2017;215:338–350. doi: 10.1111/nph.14577. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Xia R, Kuang H, Meyers BC. The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them. Mol. Biol. Evol. 2016;33:2692–2705. doi: 10.1093/molbev/msw154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 42.Nandety RS, et al. The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol. 2013;162:1459–1472. doi: 10.1104/pp.113.219162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daccord N, et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017;49:1099. doi: 10.1038/ng.3886. [DOI] [PubMed] [Google Scholar]
- 44.Srinivasan C, Liu ZR, Scorza R. Ectopic expression of class 1 KNOX genes induce adventitious shoot regeneration and alter growth and development of tobacco (Nicotiana tabacum L) and European plum (Prunus domestica L) Plant Cell Rep. 2011;30:655–664. doi: 10.1007/s00299-010-0993-7. [DOI] [PubMed] [Google Scholar]
- 45.Gambino G, Perrone I, Gribaudo I. A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008;19:520–525. doi: 10.1002/pca.1078. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Q, et al. Md-miR156ab and Md-miR395 target WRKY transcription factors to influence apple resistance to leaf spot disease. Front. Plant Sci. 2017;8:526. doi: 10.3389/fpls.2017.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wen M, Shen Y, Shi S, Tang T. miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinformatics. 2012;13:140. doi: 10.1186/1471-2105-13-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang X, Li L. miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics. 2011;27:2614–2615. doi: 10.1093/bioinformatics/btr430. [DOI] [PubMed] [Google Scholar]
- 49.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1-S3 and supplementary Tables S1-S6.