Abstract
Circulating tumor cells (CTCs) are an important part in the field of “liquid biopsy.” However, major questions remain to be answered whether the mutations in the CTCs represent the mutations in primary tumor tissue and metastatic tumors. We compared the genetic mutations between CTCs and their matched tumors, and extracted data on the heterogeneity of the mutational status in CTCs and the change in mutations of CTCs before and during treatment. For mutations detected in single genes, we calculated the concordance of the mutations between the CTCs and primary tumor tissue. For mutations detected in multiple genes, we calculated the concordance of the mutations between the CTCs and primary/metastatic tumor tissue. The heterogeneity of the mutational status is clearly present in CTCs. For mutations detected in a single gene, the overall concordance of mutations is 53.05%. For mutations detected in multiple genes, the concordance of mutations is extremely different. The heterogeneity of the mutational status existed in single CTCs, and the mutational status of CTCs was discordant with that of tumor tissue.
Keywords: CTC, nutations, heterogeneity, clinical utility
Introduction
Cancer has been one of the leading causes of death worldwide.1 With precision medicine becoming more popular for treating cancers, non-invasive methods have gradually aroused everyone’s attention and interests2 for the diagnosis and treatment of tumors. It is unrealistic for traditional tissue biopsy to capture spatial and temporal heterogeneity during tumor evaluation, and this fact is being considered as one of the major reasons for the failure of systemic cancer treatments. First, it is difficult or impossible to obtain tumor tissues at multiple time points. Second, some tumor tissues in specific locations might be not accessible for biopsy, and the surgical procedure might increase the risk of cancer “seeding” to other sites.3, 4, 5 Third, extreme heterogeneity is discovered in primary and metastatic tumors, which indicates the bias of a single biopsy.6 Therefore, tissue biopsy is unable to meet the needs for diagnosis and treatment. “Liquid biopsy,” which is a new approach that has emerged in recent years, may be able to solve these issues.
With the rapid advances of technologies, liquid biopsy has started to enter the clinic. Circulating tumor cells (CTCs) are the main features in the field of “liquid biopsy.” CTCs are approved by the US Food and Drug Administration (FDA) to aid in monitoring the prognosis of patients. A number of CTCs have been applied for detecting metastatic breast cancer,7 prostate cancer,8 colon cancer,9 ovarian cancer,10 esophageal cancer,11 bladder cancer,12 and other cancers. Beyond cell counting, the genotypic characterization of CTCs has gained increasing attention. For example, a series of mutations on the TP53 gene was detected in CTCs from colorectal carcinoma patients in 2000.13 TP53 mutations were detected in CTCs from metastatic triple-negative breast cancer patients.14 Mutations on the epidermal growth factor receptor (EGFR) gene were detected in CTCs of non-small-cell lung cancer.15 In the meantime, with the rapid development of next-generation sequencing (NGS), there is a chance for using CTCs in high-throughput molecular diagnosis. Moreover, because intratumor heterogeneity has been found in many tumor tissues, including renal cell carcinoma, breast cancer, lung cancer, prostate cancer, leukemia, esophageal squamous cell carcinoma, and others,16, 17, 18, 19, 20 a subsequent question is whether CTCs could reveal the genetic information of tumors. To address this question, we analyzed the sequence profiles of CTCs and their corresponding primary/metastatic tumors, and refined some gene mutation heterogeneities in this study.
Results
The Comparison between CTCs and Matched Primary Tumors in Single Genes
In lung cancer, the concordance of EGFR mutation is 75.27% (79/93) and the concordance of KRAS mutations 73.77% (135/183). In colorectal cancer, the concordance of KRAS mutation is 56.10% (92/164), and the concordance of BRAF mutations 46.15% (6/13). In melanoma, the concordance of BRAF mutations is 80.00% (52/65). In breast cancer, the concordance of PIK3CA mutation is 13.73% (28/204). We collected data from the above studies and calculated the total concordance of single mutations between CTCs and matched primary tumor as 53.05% (383/722) (Table 1; Table S1).
Table 1.
Comparison of the Mutations in CTCs with Correspondent Primary Tumor Tissue in Single Genes in Different Subgroups
| No. of Patients | CTC+PTT+ | CTC+PTT− | CTC−PTT+ | Concordance | |
|---|---|---|---|---|---|
| Total | 722 | 383 | 117 | 222 | 0.530 |
| Disease | |||||
| Melanoma | 65 | 52 | 8 | 5 | 0.800 |
| Lung cancer | 276 | 205 | 27 | 44 | 0.743 |
| Colorectal cancer | 177 | 98 | 30 | 49 | 0.554 |
| Breast cancer | 204 | 28 | 52 | 124 | 0.137 |
| Mutation | |||||
| EGFR | 93 | 70 | 5 | 18 | 0.753 |
| BRAF | 78 | 58 | 8 | 12 | 0.744 |
| KRAS | 347 | 227 | 52 | 68 | 0.654 |
| PIK3CA | 204 | 28 | 52 | 124 | 0.137 |
| Country | |||||
| China | 192 | 155 | 15 | 22 | 0.807 |
| The Netherlands | 15 | 2 | 4 | 9 | 0.133 |
| Greece | 195 | 48 | 31 | 116 | 0.246 |
| United States | 55 | 33 | 9 | 13 | 0.60 |
| Singapore | 37 | 13 | 7 | 17 | 0.351 |
| France | 75 | 56 | 12 | 7 | 0.747 |
| Italy | 39 | 32 | 1 | 6 | 0.821 |
| Germany | 62 | 21 | 25 | 16 | 0.339 |
| UK | 28 | 10 | 9 | 9 | 0.357 |
| Japan | 7 | 6 | 1 | 0 | 0.857 |
| Brazil | 9 | 5 | 2 | 2 | 0.556 |
| Spain | 8 | 2 | 1 | 5 | 0.250 |
| Method | |||||
| Magnetically sensed antibody sandwich assays | 4 | 4 | 0 | 0 | 1.000 |
| Scorpion amplification refractory mutation system technology | 20 | 19 | 0 | 1 | 0.950 |
| Whole-genome amplification and Sanger sequencing | 10 | 9 | 1 | 0 | 0.900 |
| RELP-PCR | 34 | 29 | 3 | 2 | 0.853 |
| NGS | 37 | 31 | 0 | 6 | 0.838 |
| Membrane arrays | 190 | 153 | 15 | 22 | 0.805 |
| Immunohistochemistry | 59 | 46 | 8 | 5 | 0.780 |
| dd-PCR | 16 | 10 | 4 | 2 | 0.625 |
| The peptide nucleic acid (PNA)-mediated PCR clamp with TaqMan-MGB allelic discrimination assays | 13 | 5 | 2 | 6 | 0.385 |
| PCR sequencing | 157 | 51 | 52 | 54 | 0.325 |
| Quantitative real-time PCR | 14 | 3 | 5 | 6 | 0.214 |
| Melting analysis | 168 | 23 | 27 | 118 | 0.137 |
The concordance rate of mutations between CTCs and matched primary tumor was as follows: sum of patients with mutation existing in both CTCs and tumor tissue/sum of all patients. CTC, circulating tumor cell; dd-PCR, digital PCR; NGS, next-generation sequencing; PTT, primary tumor tissue; RELP-PCR, restriction fragment length polymorphism-PCR.
We next performed subgroup analyses of the data and concluded that melanoma (80%) and lung cancer (78.28%) had a relatively high concordance in CTCs and matched primary tumors. The mutations in EGFR, KRAS, and BRAF had high concordance values of 75.27%, 65.42%, and 74.36%, respectively. When analyzing a study from a different country, we reached a rough conclusion that the patient from a different country had extremely different concordance of mutations in CTCs and matched primary tumors. When comparing the different analysis methods, we observed that the concordances were all more than 90% by using magnetically sensed antibody sandwich assays, scorpion amplification refractory mutation system technology, whole-genome amplification (WGA), and Sanger sequencing (Table 1).
The Heterogeneity of the Mutational Status in Single Genes
In colorectal cancer, the KRAS (p.G13D) mutation and KRAS (p.G12D) mutation were observed from two CTCs samples isolated from the same patient.21 In a study, six of nine PIK3CA-mutated CTCs from one patient carried the PIK3CA (p. E545A) mutation, whereas the PIK3CA (p. E542K) mutation was present in three of nine CTCs.22 In breast cancer patients, heterogeneity of the PIK3CA mutational status was widely discovered. A study revealed one patient with three different PIK3CA mutations in single CTCs but wild-type PIK3CA status in pooled CTC samples.23 Different ESR1 mutations in a single CTC were also tested in breast cancer.24 In a similar study, six single CTCs were isolated from a metastatic triple-negative breast cancer patient, where one CTC exhibited the same TP53 (R110 delG) mutation and one revealed a TP53 (R110 delC) mutation; the remaining four single CTCs possessed the wild-type TP53 sequence.14 Moreover, mutational heterogeneity of the PIK3CA, TP53, ESR1, and KRAS genes was revealed in forty individual epithelial cell adhesion molecule (EpCAM)-positive CTCs from five patients with metastatic breast cancer.25 In addition, with some CTCs from inflammatory breast cancer patients harboring different combinations of mutated and wild-type genes, intra-patient CTC mutation heterogeneity was confirmed.26 In lung cancer, double or multiple EGFR mutations were observed in CTCs of patients with non-small-cell lung cancer, which indicated CTC mutation heterogeneity.27, 28 In pancreatic cancer, the presence of various KRAS mutations in codons 12 and 13 in CTCs indicated heterogeneity.29
The Heterogeneity of the Mutational Status in Multiple Genes
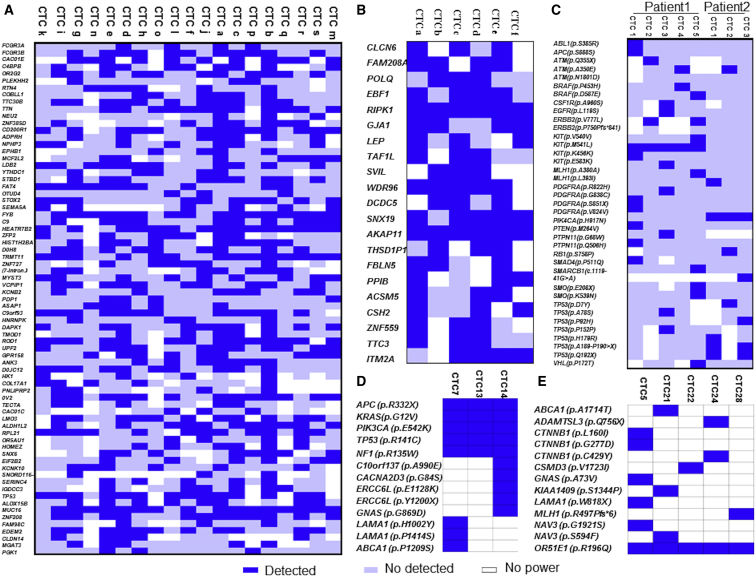
By collecting data from three experimental results and making a heatmap, we discovered that the mutational status in different CTCs had major differences (Figure 1).30, 31, 32 From the heatmap made from Lohr et al.’s32 research, we concluded that only two single-nucleotide variant (SNVs), RIPK1 and AKAP11, were ubiquitous among all six sequenced CTCs collected from patient 10, and that no SNV shared in all 19 sequenced CTCs (Figures 1A and 1B). Except for the KIT(p.M541L) mutation from the CTCs of patient 1 and the PDGFRA(p.V824V) mutation from the CTCs of patient 2, the other mutations of CTCs in metastatic breast cancer represented extreme uniformity (Figure 1C).30 Another study certified that the APC (p.R332X) mutation, KRAS (p.G12V) mutation, PIK3CA (p.E542K) mutation, TP53 (p.R141C) mutation, and NF1 (p.R135W) mutation were ubiquitous in all CTCs of patient 6, and the OR51E1 (p.R196Q) mutation was shared in all CTCs of patient 26; the other mutations demonstrated great heterogeneity (Figures 1D and 1E).31
Figure 1.
The Heatmap of the Mutations Detected in CTCs
(A) Detection of mutations in 19 CTCs of metastatic prostate cancer patient 36 by whole-exome sequencing in Lohr et al.’s32 research. (B) Detection of mutations in six CTCs of metastatic prostate cancer patient 10 by whole-exome sequencing in Lohr et al.’s32 research. (C) Detection of mutations in five CTCs of lung cancer patient 1 and mutations in three CTCs of lung cancer patient 2 by single-sell exome sequencing in De Luca et al.’s30 research. (D) Detection of mutations by massive parallel sequencing in three CTCs of colorectal cancer patient 6 in Heitzer et al.’s31 research. (E) Detection of mutations by massive parallel sequencing in five CTCs of colorectal cancer patient 26 in Heitzer et al.’s31 research. The areas shaded in dark blue represent the mutations were obtained; the blank areas represent underpowered. The mutated genes are listed in the left column.
The Concordance of Mutations between CTCs and Primary Tissues in Multiple Genes
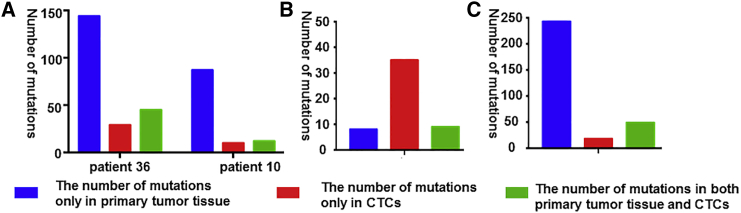
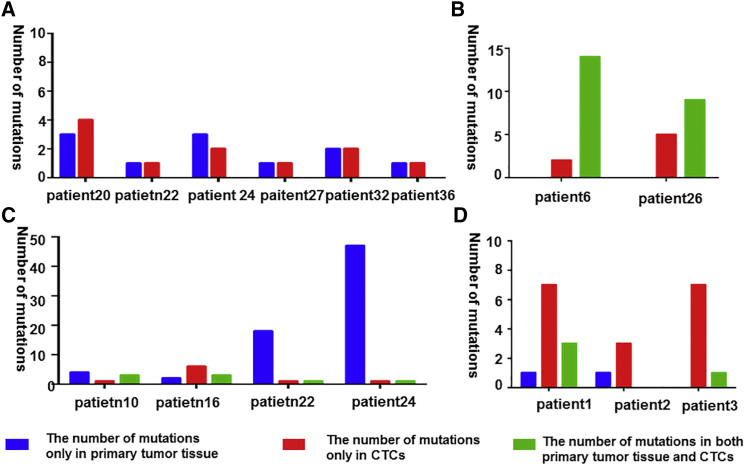
We analyzed data from studies of single CTCs by NGS and compared the mutations in CTCs and the primary tissues, and validated extreme discordance. After analyzing the research of Lohr et al.,32 who sequenced 9 primary tissues plus 19 CTCs in patient 36, and 11 primary tissues plus nine CTCs in patient 10, we observed that 144 mutations were unique to primary tissues and 29 mutations were shared only among CTCs. Only 45 mutations were shared in both CTCs and primary tissues in patient 36. In another patient, only 12 mutations were shared in both CTCs and primary tissues, with 87 mutations in tumor tissues and 10 mutations in CTCs alone (Figure 2A; Table S2).32 The other researchers sequenced one primary tissue and nine CTCs using whole-exome sequencing. They revealed relatively more unique mutations, with only 35 mutations in CTCs, 8 mutations in primary tissues alone, and 9 mutations in both of them (Figure 2B; Table S2).33 In research using target sequencing, one study concerned with mutations in the Catalogue of Somatic Mutations in Cancer (COSMIC) validated that there were no mutations that matched in both CTCs and primary tissues from six patients, which indicated great discordance (Figure 3A; Table S3).34 Four mutations were shared in both CTCs and primary tissues, with 16 mutations present in only primary tissues and 10 mutations present in only CTCs after combining all mutations from three patients (Figure 3B; Table S3).30 In another study, the mutations were extremely different in four patients. The majority of the mutations were found only in the primary tumor tissues in patients 22 and 24 (Figure 3C; Table S3).35 However, one study observed a high consistency of mutations between CTCs and primary tissues. When they performed ultra-deep sequencing of the primary tumors, the researchers found that some unique mutations only in CTCs were shared in primary tissues, with the SNVs qualified with the deep SNV algorithm (Figure 3D; Table S3).31 Except for the single-CTC sequencing, pooled CTC sequencing was recently performed by researchers. Lack et al.36 isolated EpCAM−/CD45+ cells, divided them into three equal pools of 500 cells, and then performed WGA through multiple displacement amplification (MDA), with sequencing performed on a HiSeq2000. However, authors compared mutations in pooled CTCs with mutations in matched treatment-naive tumor tissue and/or castration-resistant tumor tissue without clearing out the synonymous mutations. In this study, we removed the synonymous mutations and merely analyzed the non-synonymous and stop-gain mutations. After reprocessing the data, we concluded that 18 mutations were uniquely possessed in CTCs and 249 mutations were discovered in only matched treatment-naive tumor tissues and/or castrate-resistant tumor tissues, with 29 mutations sharing in both of them (Figure 2C; Table S2).36
Figure 2.
Comparison of the Mutations in Single/Pooled CTCs with Correspondent Primary Tumor Tissue by Whole-Exome Sequencing
(A) The results in patients 36 and 10 of comparing the mutations in CTCs with correspondent primary tumor tissue by whole-exome sequencing in Lohr et al.’s32 research. (B) The results of comparing the mutations in CTCs with correspondent primary tumor tissue by whole-exome sequencing in Ni et al.’s33 research. (C) The results of comparing the mutations in CTCs with correspondent primary tumor tissue by whole-exome sequencing in Kong et al.’s35 research.
Figure 3.
Comparison of the Mutations in CTCs with Correspondent Primary Tumor Tissue by Target Sequencing
(A) The results comparing the mutations in CTCs with corresponding primary tumor tissue by target sequencing in Yoo et al.’s34 research. (B) The results comparing the mutations in CTCs with corresponding primary tumor tissue by target sequencing in De Luca et al.’s30 research. (C) The results comparing the mutations in CTCs with corresponding primary tumor tissue by target sequencing in Heitzer et al.’s31 research. (D) The results comparing the mutations in CTCs with corresponding primary tumor tissue by target sequencing in Lack et al.’s36 research.
The Concordance of Mutations between CTCs and Metastatic Tissues
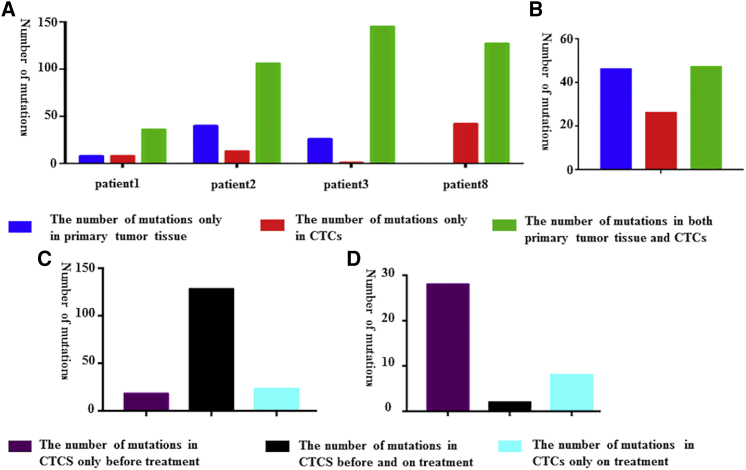
In metastatic prostate cancer, Lohr et al.32 performed whole-exome sequencing in 19 individual CTCs and compared the mutations with matching metastatic lymph nodes. We analyzed the data and came to the conclusion that 26 mutations were present in CTCs alone and 46 mutations were unique to metastatic tissues, with only 45 mutations in both CTCs and metastatic tumors tissues (Figure 4B; Table S4).32 In a similar lung cancer study, researchers sequenced mutations in individual CTC and metastatic tumors tissue in four patients, and observed that the majority of the mutations were shared between CTCs and metastatic tumor tissue (Figure 4A; Table S4).33
Figure 4.
Comparison of the Mutations in CTCs with Correspondent Metastatic Tumor Tissue and Comparison of the Mutations in CTCs before and during Treatment
(A) The results comparing the mutations in CTCs with corresponding metastatic tumor tissue in Ni et al.’s33 research. (B) The results comparing the mutations in CTCs with corresponding metastatic tumor tissue in Lohr et al.’s32 research. (C) The results comparing the mutations in CTCs before and during treatment in De Luca et al.’s30 research. (D) The results comparing the mutations in CTCs before and during treatment in Yoo et al.’s34 research.
The Genetic Shift of CTCs during Treatment
Given that repeat biopsy is undesirable, the ability to detect mutations in CTCs over time is important for monitoring the evolution of disease during the treatment. Independent of disease types, treatment methods, treatment time, and the detecting method, we analyzed data from two studies and reached opposite results. Two of the 38 mutations were always present in CTCs in one study, but the other study indicated that 128 out of 169 mutations were present all the time (Figures 4C and 4D; Table S5).30, 34
Discussion
The Different Definitions Caused by Amplification and Sequencing Brought Errors in Analyzing the Mutations Sequenced by NGS
For single-cell mutation analysis, it has been suggested that errors can be generated during whole-genome amplification and sequencing, which lead to high false discovery ratio (2–3 × 10−5) and high allele dropout ratio (11%).31 To eliminate the mutations introduced by amplification and sequencing errors, Ni et al.33 sequenced eight single CTCs from one patient, and only when an SNV in a CTC was detected in two other CTCs or in primary/metastatic tumors was it called a mutation. In another study, Lohr et al.32 sequenced 19 single CTCs from one patient and 6 single CTCs from another patient; they think that if a SNV in a CTC was detected in two other CTCs libraries, it will be called a mutation. On the other hand, Heitzer et al.31 do not agree with the view that a SNV should be defined as a true mutation only if it occurs in a certain number of cancer cells, because the available CTCs are often rare but of high quality. De Luca et al.30 trust high-depth and high-coverage sequencing; they sequenced CTCs with a mean depth of 1,500×, ranging from 1,046× to 2,478× depth of coverage for each amplicon per sample by Ion Torrent. Defining mutations after sequencing can lead to substantial controversies, and the methodological consensus must be unified.
To eliminate the artifacts caused by amplification and sequencing, it may be necessary for single-cell (not only in CTCs) analyses to sequence multiple single cells,37 such as at least three cells, to confirm the mutations in tumors.38 With the mutations correctly called, bioinformatics could play more important roles in the diagnosis and prognosis of cancer.39
The Possible Reasons for the Heterogeneity of Mutational Status
The limitations of the amplification and sequencing method might cause the heterogeneity. Besides, using different sequencing methods might yield different mutational results. Comparing the performance of high-resolution melt (HRM), allele-specific PCR (ASPCR), and pyrosequencing, Mohamed Suhaimi et al.40 observed that the mutation status sequenced by above methods were different.
In addition to the limitations of the amplification and sequencing methods, two other reasons may also explain the heterogeneity of the mutational statuses of single CTCs. There is an opinion that CTCs disseminated into the blood from primary tumors and/or metastatic tissues represent a surrogate for the tissue-based tumor, and analyses of CTCs might provide insight into tumor tissue heterogeneity. Therefore, the heterogeneity of the mutational status of single CTCs might be caused by the heterogeneity of the mutational status of single primary/metastatic tumor cells. Besides, some researchers first performed genome sequencing in which only the PIK3CA mutation was possessed in the cultured CTCs. When those cultured CTCs were sequenced, some newly acquired mutations in the ESR1 gene, PIK3CA gene, and FGFR2b gene were observed.41 Therefore, we suggest that the change in CTCs and their microenvironment, including epithelial-to-mesenchymal transitions,42 mesenchymal-to-epithelial transitions,43 blood flow, and immune cell attack might alter the mutations.
Finally, research about acquired mutation between targeted treatments reveals another potential reason for the heterogeneity of mutational status. For example, Blakely et al.44 confirmed increasing heterogeneity of an EGFR mutant cancer across multiple lines of therapy. Similarly, Piotrowska et al.45 showed the heterogeneity and coexistence of T790M and T790 wild-type resistant subclones drive mixed response to third-generation EGFR inhibitors in lung cancer. Thus, the subclonal acquisition of mutation during therapy might cause the heterogeneity of mutational status and discordance of mutations between CTCs with primary and/or metastatic tumor tissue.46
The Probable Reasons for the Discordance of Mutations between CTCs with Primary and/or Metastatic Tumor Tissue
It is necessary to elucidate the reasons for the discordance observed between primary/metastatic tumor tissues and CTCs. Several potential reasons have been proposed. First, limitations of current isolating and sequencing techniques may hamper the identification of the entire spectrum of mutational statuses and causes of the discrepancies.41 Second, with subcolonies and heterogeneity within the entire tumor tissue considered, certain numbers of CTCs are unlikely to contain the entire spectrum of mutations. When the CTCs isolating and enriching method improves to obtain sufficient numbers of CTCs, people might confirm if the discrepancies in the mutation statuses between tumor/metastatic tissues and CTCs are caused by the subcolonies and heterogeneity in bulk tumor tissue.16, 47 Third, the CTCs isolation technologies, including Cell Search, which is based on EpCAM, may have substantial limitations.48, 49 Many studies conclude that primary tumor cells should undergo epithelial-to-mesenchymal transition (EMT) to invade the blood vessels, which accelerates the production of CTCs. However, some technology depends on the EpCAM marker for the isolation of CTCs, which might ignore the EMT-transformed CTC.48, 49 To solve these problems, it has been suggested that using other antibodies or different enrichment systems to deplete normal blood cells and enrich the CTCs may be a solution.49, 50 Finally, the opinion that CTCs can acquire private and unique mutations during the evolution of the tumor has been proposed by some researchers, which might cause the large heterogeneity in CTCs and the discrepancies in mutation statuses between the tumor or metastatic tissues and CTCs.41, 51 A high proportion of concordance has been reported by most studies with mutations between CTCs and tumor tissues or metastatic tissues, which suggests the potential usefulness of using CTCs to profile mutations of tumors.
Minimally invasive access to CTCs offers a unique opportunity to monitor disease progression and guide drug management.52 However, amplification and sequencing errors, the lack of a uniform definition of mutations in the sequenced single CTCs, the heterogeneity of the mutational status in single CTCs, and the discordance of mutations between CTCs with primary and/or metastatic tumor tissue must be explored before CTCs can be widely used in clinics.
Materials and Methods
The Concordance Compared between CTCs and Primary Tissues in Single Genes
We retrieved in PubMed the studies comparing certain single-gene mutations between CTCs and primary tissues in many patients. The following data were extracted: the number of patients with the mutation shared in the CTCs and matched primary tumor, the number of patients with the mutation discovered in CTCs, but not in the matched tumors, and the number of patients with the mutation carried in the matched tumors, but not in CTCs. Then, we calculated the concordance rate of mutations between the CTCs and matched tumors as follows: sum of patients with mutation matched in both CTCs and primary tissues/sum of all patients. The data in different subgroups were also compared for the concordance of mutations.
The Concordance Compared between CTCs and Primary Tissues in Multiple Genes
We searched PubMed and collected the articles pertaining to multiple mutations (a larger number of mutations variants in at least 10 genes) sequenced by NGS. Then, we collected data about the comparison of the mutations between CTCs and primary tissues using NGS. Because of the different definitions of mutations in different studies, we gathered the mutations that were exclusively in CTCs, exclusively in primary tissues, and in both CTCs and primary tissues. The concordance rate of mutations between CTCs and matched primary tissues was calculated as follows: sum of mutations in both CTCs and primary tissues/sum of all mutations.
The Heterogeneity of Mutational Status in Multiple Genes
We searched PubMed and collected data comparing the mutations in different CTCs of the same patient using NGS. Then, we extracted the data about heterogeneity of mutational status and created a heatmap.
The Discordance between CTCs and Metastatic Tissues on Genetic Mutations
Studies comparing the mutations between CTCs and metastatic tissues using NGS were searched and collected in PubMed. Because of the different definitions of mutations in different studies, we gathered the mutations present exclusively in CTCs, exclusively in metastatic tissues, and in both CTCs and metastatic tissues. Then the concordance rate of mutations between CTCs and matched metastatic tissues was calculated as follows: sum of mutations in both CTCs and metastatic tissues/sum of all mutations.
The Genetic Shift of CTCs during Treatment
We searched PubMed and collected the studies comparing the mutations in CTCs before and during treatment using NGS. Because of the different definitions of mutations in different studies, we gathered information about the mutations that were confirmed exclusively in CTCs before treatment, exclusively in CTCs during treatment, and in CTCs both before and during treatment.
Author Contributions
Q.S. and Q.L. designed and configured this study. Qi Wang and L.Z. performed the majority of the experiments. L.H., X.T., S.M., and Y.W. prepared the figures. Qi Wang wrote the manuscript. X.F., D.L., C.S., and Qing Wang helped to revise the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81472823), the Fundamental Research Funds for Xi’an Jiaotong University (xjj2015093), and the Major Basic Research Project of Natural Science of Shaanxi Provincial Science and Technology Department (2017ZDJC-11). This study was also supported by the NIH (grants R21HG006173, R43HG007621, and HL117929). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2019.08.006.
Contributor Information
Qing Song, Email: qsong@msm.edu.
Qiling Li, Email: liqiling@mail.xjtu.edu.cn.
Supplemental Information
References
- 1.David A.R., Zimmerman M.R. Cancer: an old disease, a new disease or something in between? Nat. Rev. Cancer. 2010;10:728–733. doi: 10.1038/nrc2914. [DOI] [PubMed] [Google Scholar]
- 2.Mirnezami R., Nicholson J., Darzi A. Preparing for precision medicine. N. Engl. J. Med. 2012;366:489–491. doi: 10.1056/NEJMp1114866. [DOI] [PubMed] [Google Scholar]
- 3.Chen T., You Y., Jiang H., Wang Z.Z. Epithelial-mesenchymal transition (EMT): A biological process in the development, stem cell differentiation, and tumorigenesis. J. Cell. Physiol. 2017;232:3261–3272. doi: 10.1002/jcp.25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pittenger M.F., Martin B.J. Mesenchymal stem cells and their potential as cardiac therapeutics. Circ. Res. 2004;95:9–20. doi: 10.1161/01.RES.0000135902.99383.6f. [DOI] [PubMed] [Google Scholar]
- 5.Robertson E.G., Baxter G. Tumour seeding following percutaneous needle biopsy: the real story! Clin. Radiol. 2011;66:1007–1014. doi: 10.1016/j.crad.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Matkovic V., Fontana D., Tominac C., Goel P., Chesnut C.H., 3rd Factors that influence peak bone mass formation: a study of calcium balance and the inheritance of bone mass in adolescent females. Am. J. Clin. Nutr. 1990;52:878–888. doi: 10.1093/ajcn/52.5.878. [DOI] [PubMed] [Google Scholar]
- 7.Mu Z., Wang C., Ye Z., Austin L., Civan J., Hyslop T., Palazzo J.P., Jaslow R., Li B., Myers R.E. Prospective assessment of the prognostic value of circulating tumor cells and their clusters in patients with advanced-stage breast cancer. Breast Cancer Res. Treat. 2015;154:563–571. doi: 10.1007/s10549-015-3636-4. [DOI] [PubMed] [Google Scholar]
- 8.Zheng Y., Zhang C., Wu J., Cheng G., Yang H., Hua L., Wang Z. Prognostic Value of Circulating Tumor Cells in Castration Resistant Prostate Cancer: A Meta-analysis. Urol. J. 2016;13:2881–2888. [PubMed] [Google Scholar]
- 9.Thorsteinsson M., Söletormos G., Jess P. Low number of detectable circulating tumor cells in non-metastatic colon cancer. Anticancer Res. 2011;31:613–617. [PubMed] [Google Scholar]
- 10.Zhou Y., Bian B., Yuan X., Xie G., Ma Y., Shen L. Prognostic Value of Circulating Tumor Cells in Ovarian Cancer: A Meta-Analysis. PLoS ONE. 2015;10:e0130873. doi: 10.1371/journal.pone.0130873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S., Du H., Li G. Significant prognostic value of circulating tumor cells in esophageal cancer patients: A meta-analysis. Oncotarget. 2017;8:15815–15826. doi: 10.18632/oncotarget.15012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gazzaniga P., Gradilone A., de Berardinis E., Busetto G.M., Raimondi C., Gandini O., Nicolazzo C., Petracca A., Vincenzi B., Farcomeni A. Prognostic value of circulating tumor cells in nonmuscle invasive bladder cancer: a CellSearch analysis. Ann. Oncol. 2012;23:2352–2356. doi: 10.1093/annonc/mdr619. [DOI] [PubMed] [Google Scholar]
- 13.Khan Z.A., Jonas S.K., Le-Marer N., Patel H., Wharton R.Q., Tarragona A., Ivison A., Allen-Mersh T.G. P53 mutations in primary and metastatic tumors and circulating tumor cells from colorectal carcinoma patients. Clin. Cancer Res. 2000;6:3499–3504. [PubMed] [Google Scholar]
- 14.Fernandez S.V., Bingham C., Fittipaldi P., Austin L., Palazzo J., Palmer G., Alpaugh K., Cristofanilli M. TP53 mutations detected in circulating tumor cells present in the blood of metastatic triple negative breast cancer patients. Breast Cancer Res. 2014;16:445. doi: 10.1186/s13058-014-0445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pailler E., Faugeroux V., Oulhen M., Catelain C., Farace F. Routine clinical use of circulating tumor cells for diagnosis of mutations and chromosomal rearrangements in non-small cell lung cancer-ready for prime-time? Transl. Lung Cancer Res. 2017;6:444–453. doi: 10.21037/tlcr.2017.07.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerlinger M., Horswell S., Larkin J., Rowan A.J., Salm M.P., Varela I., Fisher R., McGranahan N., Matthews N., Santos C.R. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casasent A.K., Schalck A., Gao R., Sei E., Long A., Pangburn W., Casasent T., Meric-Bernstam F., Edgerton M.E., Navin N.E. Multiclonal invasion in breast tumors identified by topographic single cell sequencing. Cell. 2018;172:205–217.e12. doi: 10.1016/j.cell.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J., Fujimoto J., Zhang J., Wedge D.C., Song X., Zhang J., Seth S., Chow C.W., Cao Y., Gumbs C. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landau D.A., Carter S.L., Stojanov P., McKenna A., Stevenson K., Lawrence M.S., Sougnez C., Stewart C., Sivachenko A., Wang L. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao J.J., Lin D.C., Dinh H.Q., Mayakonda A., Jiang Y.Y., Chang C., Jiang Y., Lu C.C., Shi Z.Z., Xu X. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat. Genet. 2016;48:1500–1507. doi: 10.1038/ng.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo Y., Hayashi K., Kawakami K., Miwa Y., Hayashi H., Yamamoto M. KRAS mutation analysis of single circulating tumor cells from patients with metastatic colorectal cancer. BMC Cancer. 2017;17:311. doi: 10.1186/s12885-017-3305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasch C., Bauernhofer T., Pichler M., Langer-Freitag S., Reeh M., Seifert A.M., Mauermann O., Izbicki J.R., Pantel K., Riethdorf S. Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clin. Chem. 2013;59:252–260. doi: 10.1373/clinchem.2012.188557. [DOI] [PubMed] [Google Scholar]
- 23.Pestrin M., Salvianti F., Galardi F., De Luca F., Turner N., Malorni L., Pazzagli M., Di Leo A., Pinzani P. Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol. Oncol. 2015;9:749–757. doi: 10.1016/j.molonc.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paolillo C., Mu Z., Rossi G., Schiewer M.J., Nguyen T., Austin L., Capoluongo E., Knudsen K., Cristofanilli M., Fortina P. Detection of Activating Estrogen Receptor Gene (ESR1) mutations in single circulating tumor cells. Clin. Cancer Res. 2017;23:6086–6093. doi: 10.1158/1078-0432.CCR-17-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw J.A., Guttery D.S., Hills A., Fernandez-Garcia D., Page K., Rosales B.M., Goddard K.S., Hastings R.K., Luo J., Ogle O. Mutation Analysis of Cell-Free DNA and Single Circulating Tumor Cells in Metastatic Breast Cancer Patients with High Circulating Tumor Cell Counts. Clin. Cancer Res. 2017;23:88–96. doi: 10.1158/1078-0432.CCR-16-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bingham C., Fernandez S.V., Fittipaldi P., Dempsey P.W., Ruth K.J., Cristofanilli M., Katherine Alpaugh R. Mutational studies on single circulating tumor cells isolated from the blood of inflammatory breast cancer patients. Breast Cancer Res. Treat. 2017;163:219–230. doi: 10.1007/s10549-017-4176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maheswaran S., Sequist L.V., Nagrath S., Ulkus L., Brannigan B., Collura C.V., Inserra E., Diederichs S., Iafrate A.J., Bell D.W. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchetti A., Del Grammastro M., Felicioni L., Malatesta S., Filice G., Centi I., De Pas T., Santoro A., Chella A., Brandes A.A. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS ONE. 2014;9:e103883. doi: 10.1371/journal.pone.0103883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulemann B., Rösch S., Seifert S., Timme S., Bronsert P., Seifert G., Martini V., Kuvendjiska J., Glatz T., Hussung S. Pancreatic cancer: Circulating Tumor Cells and Primary Tumors show Heterogeneous KRAS Mutations. Sci. Rep. 2017;7:4510. doi: 10.1038/s41598-017-04601-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Luca F., Rotunno G., Salvianti F., Galardi F., Pestrin M., Gabellini S., Simi L., Mancini I., Vannucchi A.M., Pazzagli M. Mutational analysis of single circulating tumor cells by next generation sequencing in metastatic breast cancer. Oncotarget. 2016;7:26107–26119. doi: 10.18632/oncotarget.8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heitzer E., Auer M., Gasch C., Pichler M., Ulz P., Hoffmann E.M., Lax S., Waldispuehl-Geigl J., Mauermann O., Lackner C. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013;73:2965–2975. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- 32.Lohr J.G., Adalsteinsson V.A., Cibulskis K., Choudhury A.D., Rosenberg M., Cruz-Gordillo P., Francis J.M., Zhang C.Z., Shalek A.K., Satija R. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol. 2014;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ni X., Zhuo M., Su Z., Duan J., Gao Y., Wang Z., Zong C., Bai H., Chapman A.R., Zhao J. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc. Natl. Acad. Sci. USA. 2013;110:21083–21088. doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoo C.E., Park J.M., Moon H.S., Joung J.G., Son D.S., Jeon H.J., Kim Y.J., Han K.Y., Sun J.M., Park K. Vertical Magnetic Separation of Circulating Tumor Cells for Somatic Genomic-Alteration Analysis in Lung Cancer Patients. Sci. Rep. 2016;6:37392. doi: 10.1038/srep37392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong S.L., Liu X., Suhaimi N.M., Koh K.J.H., Hu M., Lee D.Y.S., Cima I., Phyo W.M., Lee E.X.W., Tai J.A. Molecular characterization of circulating colorectal tumor cells defines genetic signatures for individualized cancer care. Oncotarget. 2017;8:68026–68037. doi: 10.18632/oncotarget.19138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lack J., Gillard M., Cam M., Paner G.P., VanderWeele D.J. Circulating tumor cells capture disease evolution in advanced prostate cancer. J. Transl. Med. 2017;15:44. doi: 10.1186/s12967-017-1138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y., Xu X., Song L., Hou Y., Li Z., Tsang S., Li F., Im K.M., Wu K., Wu H. Single-cell sequencing analysis characterizes common and cell-lineage-specific mutations in a muscle-invasive bladder cancer. Gigascience. 2012;1:12. doi: 10.1186/2047-217X-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zong C., Lu S., Chapman A.R., Xie X.S. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou Y., Song L., Zhu P., Zhang B., Tao Y., Xu X., Li F., Wu K., Liang J., Shao D. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 40.Mohamed Suhaimi N.A., Foong Y.M., Lee D.Y., Phyo W.M., Cima I., Lee E.X., Goh W.L., Lim W.Y., Chia K.S., Kong S.L. Non-invasive sensitive detection of KRAS and BRAF mutation in circulating tumor cells of colorectal cancer patients. Mol. Oncol. 2015;9:850–860. doi: 10.1016/j.molonc.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C., Guan Y., Sun Y., Ai D., Guo Q. Tumor heterogeneity and circulating tumor cells. Cancer Lett. 2016;374:216–223. doi: 10.1016/j.canlet.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 42.Micalizzi D.S., Farabaugh S.M., Ford H.L. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J. Mammary Gland Biol. Neoplasia. 2010;15:117–134. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J., Du X., Wang G., Sun Y., Chen K., Zhu X., Lazar A.J., Hunt K.K., Pollock R.E., Zhang W. Mesenchymal to epithelial transition in sarcomas. Eur. J. Cancer. 2014;50:593–601. doi: 10.1016/j.ejca.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Blakely C.M., Watkins T.B.K., Wu W., Gini B., Chabon J.J., McCoach C.E., McGranahan N., Wilson G.A., Birkbak N.J., Olivas V.R. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat. Genet. 2017;49:1693–1704. doi: 10.1038/ng.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piotrowska Z., Hazar-Rethinam M., Rizzo C., Nadres B., Van Seventer E.E., Shahzade H.A., Lennes I.T., Iafrate A.J., Dias-Santagata D., Leshchiner I. Heterogeneity and coexistence of T790M and T790 wild-type resistant subclones drive mixed response to third-generation epidermal growth factor receptor inhibitors in lung cancer. JCO Precis. Oncol. 2018;2:1–15. doi: 10.1200/PO.17.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sundaresan T.K., Sequist L.V., Heymach J.V., Riely G.J., Jänne P.A., Koch W.H., Sullivan J.P., Fox D.B., Maher R., Muzikansky A. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin. Cancer Res. 2016;22:1103–1110. doi: 10.1158/1078-0432.CCR-15-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tannock I.F. Words of wisdom. Re: Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. Eur. Urol. 2014;65:846–847. doi: 10.1016/j.eururo.2013.12.033. [DOI] [PubMed] [Google Scholar]
- 48.Satelli A., Batth I., Brownlee Z., Mitra A., Zhou S., Noh H., Rojas C.R., Li H., Meng Q.H., Li S. EMT circulating tumor cells detected by cell-surface vimentin are associated with prostate cancer progression. Oncotarget. 2017;8:49329–49337. doi: 10.18632/oncotarget.17632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang K., Kong Y.Y., Dai B., Ye D.W., Qu Y.Y., Wang Y., Jia Z.W., Li G.X. Combination of circulating tumor cell enumeration and tumor marker detection in predicting prognosis and treatment effect in metastatic castration-resistant prostate cancer. Oncotarget. 2015;6:41825–41836. doi: 10.18632/oncotarget.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin H.K., Zheng S., Williams A.J., Balic M., Groshen S., Scher H.I., Fleisher M., Stadler W., Datar R.H., Tai Y.C., Cote R.J. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin. Cancer Res. 2010;16:5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu M., Bardia A., Aceto N., Bersani F., Madden M.W., Donaldson M.C., Desai R., Zhu H., Comaills V., Zheng Z. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wendel M., Bazhenova L., Boshuizen R., Kolatkar A., Honnatti M., Cho E.H., Marrinucci D., Sandhu A., Perricone A., Thistlethwaite P. Fluid biopsy for circulating tumor cell identification in patients with early-and late-stage non-small cell lung cancer: a glimpse into lung cancer biology. Phys. Biol. 2012;9:016005. doi: 10.1088/1478-3967/9/1/016005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.