Highlights
-
•
A single biopsy may not accurately reflect the global hypoxia status of a tumor due to intratumoral heterogeneity.
-
•
Compared with individual genes, hypoxia gene expression signatures are generally more consistent across multiple biopsies from different regions of a tumor and may give a more reliable estimate of global hypoxia status.
-
•
Wherever possible, the use of multiple biopsies provides greater assurance in correctly classifying a tumor as more or less hypoxic.
Keywords: Hypoxia, Gene signature, Heterogeneity, Cervical cancer
Abstract
Background and Purpose
Gene expression signatures are often used to identify hypoxic tumors. However, intratumoral heterogeneity raises concern that multiple biopsies may be necessary to assess global hypoxia status. The objective of this study was to compare the impact of heterogeneity on the discriminative capacity of several previously described hypoxia gene signatures and determine if a single biopsy is sufficient to obtain a reliable estimate of hypoxia in cervical cancer.
Materials and Methods
Multiple biopsies (33) were obtained from 11 locally advanced (FIGO IB to IVB) cervical cancers prior to treatment. Ten hypoxia gene signatures were analyzed. Variance component analysis was used to determine the ratio of within-tumor variability to total-tumor variability when one to five biopsies are available for analysis (W/T1–5). The mean standardized error in the signature scores was estimated by comparing the score using one biopsy randomly selected from each tumor to the ‘global’ score using all available biopsies.
Results
The ten hypoxia signatures were comprised of 6–99 genes each. The W/T1 ratios for individual genes commonly found in the signatures ranged from 0.17 to 0.73. W/T1 ratios for the signatures were generally lower (0.21–0.45), implying greater capacity to discriminate among tumors. With additional biopsies, the signature W/T ratios (ie W/T2-5) decreased further. The mean error in the signature scores varied from 0.27 to 0.40 of one standard deviation, suggesting high capacity to discriminate among tumors with different global hypoxia scores.
Conclusions
Compared with individual probes, hypoxia gene expression signatures are generally more consistent across multiple biopsies from different regions of a tumor and more tolerant of intratumoral heterogeneity.
1. Introduction
Tumor hypoxia is a poor prognostic factor in a wide range of malignancies [1], [2], [3], [4]. Several methods have been used to measure hypoxia, including: direct needle electrode measurements; endogenous biomarkers such as carbonic anhydrase IX (CAIX), glucose transporter 1 (GLUT-1) and vascular endothelial growth factor (VEGF); exogenous markers such as pimonidazole and EF5; and imaging techniques including positron emission tomographic (PET) evaluation of nitroimidazole uptake and binding, magnetic resonance (MR) blood-oxygen level dependent (BOLD) imaging and MR intravoxel incoherent motion (IVIM) imaging [5], [6], [7]. Oxygen electrodes, which have classically been considered the gold standard measurement of hypoxia in human cancers, can sense oxygen tensions from 0 to 100 mmHg and are strongly correlated with clinical outcome in patients with head and neck, cervical and prostate cancers [5], [6]. This technique, however, is invasive, only suitable for accessible tumors, and highly operator dependent [5]. The ideal method of measuring hypoxia in human tumors would be minimally invasive with low risk of morbidity and reproducible with high spatial and temporal resolution. Measurement approaches that meet these criteria and utilize only the diagnostic tumor biopsy are particularly attractive from the perspective of identifying patients with hypoxic tumors to participate in prospective clinical studies.
In recent years, gene expression profiling has been used to study tumor hypoxia. A number of genes are upregulated and play crucial roles in the adaptive responses of tumor cells to changes in oxygen. These include the family of hypoxia-inducible factors (HIFs) that regulate numerous hypoxia-dependent genes, many of which have been investigated as possible intrinsic biomarkers of tumor hypoxia [7], [8]. In cervical cancer, where hypoxia is known to portend a poor prognosis in terms of both local disease recurrence and the development of metastases, there are conflicting reports about the predictive value of HIF and downstream genes [9], [10], [11], [12]. Upregulation of HIF-dependent genes such as GLUT1, CAIX, and VEGF has been associated with poor treatment outcomes in some studies but not in others [13], [14], [15], [16], [17].
The variability in the published literature may partially be attributable to spatial and temporal heterogeneity of hypoxia and the resultant changes in gene expression. Several studies that used oxygen electrodes to measure hypoxia at multiple locations within tumors confirmed that oxygenation status varies both between tumors and within tumors [18], [19]. Unlike surgical specimens where an entire tumor is available for analysis, patients undergoing radiotherapy generally have only a single biopsy available for testing. The heterogeneity in tumor hypoxia raises concern that a single biopsy may not provide an accurate portrayal of global hypoxia status and that multiple biopsies may be necessary to correctly stratify patients in this context.
Gene signatures, which represent unique patterns of gene expression, may have improved ability to identify hypoxic tumors and predict clinical outcomes when compared with single genes. Multiple genes that together reflect the complex molecular adaptive response to hypoxia may be an effective way to personalize care by identifying tumors most likely to respond to hypoxia targeted treatment. We propose that gene signatures, in addition to providing more reliable predictions of hypoxia-dependent changes in tumor biology and clinical outcome than individual genes, may also be more robust to spatial and temporal differences in hypoxia and more suitable for use when only a single tumor biopsy is available. The effect of intratumoral heterogeneity on the utility of two head/neck hypoxia gene signatures has been reported previously [20], [21]. It is not known if the findings translate to other tumor types. Furthermore, there has not been a comparative analysis of the relative performance of different gene expression signatures in the face of heterogeneity.
The objective of this study was to examine the between and within tumor variability of several published hypoxia gene signatures using data obtained previously from multiple tumor biopsies in patients with cervical cancer [22], and the capacity of these signatures to identify patients with hypoxic tumors when only a single biopsy is available for analysis.
2. Material and methods
This study was initiated following institutional Research Ethics Board approval. Written consent was obtained from all participating patients.
2.1. Specimen collection and processing
Punch biopsies were obtained from cervical cancer patients who underwent examination under anesthesia as part of routine clinical staging, as previously described [22]. Four to seven biopsies were obtained trans-vaginally from different regions of the visible cervical tumor in each patient. Biopsy location in the tumor was not systematically recorded because of the wide variability in tumor size and configuration from patient to patient. The details of the pathologic analysis, RNA extraction and purification, microarray hybridization, filtering process, and clustering have been previously described [22]. The final cohort was comprised of 33 biopsies from 11 patients with cervical cancer, as outlined in Table 1. Two patients had five biopsies, five had three biopsies and four had two biopsies. The tumor cell fraction in the individual biopsies was between 60% and 100%.
Table 1.
Patient and tumor characteristics.
| Variable | Patients (Total n = 11) |
Biopsies Total (n = 33) |
|---|---|---|
| Age | ||
| Median (Range) | 47 (31–70) years | |
| Number of biopsies | ||
| 2 | 4 (36%) | |
| 3 | 5 (45%) | |
| 5 | 2 (18%) | |
| FIGO Stage | ||
| IB | 2 (18%) | 6 (18%) |
| IIB | 4 (36%) | 15 (46%) |
| IIIB | 4 (36%) | 10 (30%) |
| IVB | 1 (9%) | 2 (6%) |
| Tumor size (cm) | ||
| ≤5 | 4 (36%) | 13 (39%) |
| >5 | 7 (64%) | 20 (61%) |
| Histology | ||
| Squamous | 9 (82%) | 29 (88) |
| Adenosquamous | 2 (18%) | 4 (12) |
| Pelvic lymph node status at diagnosis | ||
| Positive | 2 (18%) | 4 (12%) |
| Equivocal | 3 (27%) | 10 (30%) |
| Negative | 6 (55%) | 19 (58%) |
2.2. Gene signature selection
A literature review of hypoxia gene expression signatures was published in 2015 and identified 32 signatures [23]. A supplementary review of the literature identified additional signatures published prior to 2018; the National Center for Biotechnology Information (PubMed) database was searched with combinations: of ‘hypox*’, ‘gene’, and ‘signature’ for articles published in English. Seven additional gene signatures were identified for a total of 39 signatures published between 2000 and 2018. These signatures were comprised of four to 759 genes and were derived from cell lines or clinical samples across a range of tumor types. We selected gene signatures that were derived from or validated in clinical samples (n = 10) as they may be more strongly correlated with clinical outcome [23], including two signatures that were derived from patients with cervical cancer. The gene signatures are summarized in Table 2.
Table 2.
Summary of the ten gene signatures selected for this analysis, in rank order from lowest to highest W/T.
| Signature | Development Site(s) | Number of Genes* | Number of Probe Sets† | Signature Score (Median, range) | W/T by Number of Biopsies |
||||
|---|---|---|---|---|---|---|---|---|---|
| W/T1 | W/T2 | W/T3 | W/T4 | W/T5 | |||||
| Fjeldbo [31] | Cervix | 6 (4) | 10 (6) | 7.87 (6.73–10.17) | 0.21 | 0.12 | 0.08 | 0.06 | 0.05 |
| Hu [32] | Breast | 15 (13) | 26 (20) | 7.44 (6.51–8.67) | 0.26 | 0.15 | 0.10 | 0.08 | 0.06 |
| Toustrup [24] | HN | 15 (15) | 37 (25) | 8.68 (7.19–9.63) | 0.28 | 0.17 | 0.12 | 0.09 | 0.07 |
| Halle [30] | Cervix | 31 (28) | 66 (43) | 0.01 (-0.52–0.49) | 0.29 | 0.17 | 0.12 | 0.09 | 0.08 |
| Buffa [26] | HN Breast |
51 (51) | 51 (51) | 8.77 (8.14–9.39) | 0.31 | 0.18 | 0.13 | 0.10 | 0.08 |
| Ghazoui [28] | Breast | 70 (68) | 136 (113) | 8.77 (8.25–9.15) | 0.33 | 0.20 | 0.14 | 0.11 | 0.09 |
| Betts [20], [29] | HN | 26 (25) | 58 (50) | 8.41 (7.54–8.84) | 0.40 | 0.25 | 0.18 | 0.14 | 0.12 |
| Winter [27] | HN | 99 (92) | 126 (119) | 7.96 (7.42–8.76) | 0.41 | 0.26 | 0.19 | 0.15 | 0.12 |
| Ragnum [33] | Prostate | 32 (28) | 61 (49) | 0.02 (-0.61–0.52) | 0.44 | 0.28 | 0.21 | 0.17 | 0.14 |
| Yang [34] | Prostate | 28 (26) | 66 (50) | 0.19 (-1.41–1.71) | 0.45 | 0.29 | 0.21 | 0.17 | 0.14 |
W/T1–5 Within-tumor variance divided by the Total sample variance when one to five biopsies are available. Lower W/T1–5 implies greater capacity of one or multiple biopsies to discriminate among tumors in the face of heterogeneity.
Number of genes in the original signatures vs. number of genes found in the Affymetrix array and used in this analysis.
Total number of probe sets vs. number of probe sets found in the Affymetrix array and used in this analysis.
2.3. Statistics
Hypoxia signature scores were calculated for each individual biopsy by replicating the methods outlined by the authors in their publications as closely as possible. All of the genes were available for only two of the ten signatures (Toustrup [24] and Buffa [26]); between one and seven genes were missing from the Affymetrix array for the other eight signatures (Table 2). For nine signatures, the scores were based on the mean or median of the available probe set values, with higher signature scores indicating higher levels of hypoxia. The Toustrup signature was designed to classify tumors as more or less hypoxic by comparing gene expression levels to those in a predefined reference cohort [21], [24]. This method could not be replicated without the reference cohort and the median of the probe set values was used instead. The global hypoxia score for each patient was calculated as the mean of the signature scores for all of the biopsies belonging to that patient.
Variance component analysis was used to calculate the within-tumor and between-tumor signature variances for the entire cohort [25]. Heterogeneity was expressed as the ratio of the within-tumor variance to the total-tumor variance with a single biopsy as the unit of measure (W/T1). A W/T1 ratio of zero implies that the signature score is spatially uniform within individual tumors, and that all of the variation is between tumors. In contrast, a W/T1 ratio of one implies that the signature score is highly variable within individual tumors and uniform among tumors. Therefore, the lower the W/T1 ratio, the higher the likelihood that a single biopsy will reliably discriminate among tumors on the basis of hypoxia. In addition, variance component analysis was modified to estimate W/T ratios for the scenario where multiple biopsies (two to five) are available (designated W/T2–5) [25]. The W/T1 and W/T2 ratios were also calculated for selected individual genes involved in four or more of the signatures using the same approach.
To further address the question of whether a single biopsy is sufficient to reliably determine the hypoxia status of a tumor, the error in the signature scores was estimated by comparing the score using one biopsy randomly selected from each tumor to the ‘global’ score calculated by using all available biopsies for that tumor. To facilitate comparison among the signatures, the scores were first standardized using the mean and standard deviation of the scores from the 33 individual biopsies. The absolute value of the difference between the single-biopsy score and the global score was calculated for each tumor. The mean error for the 11 tumors was then estimated. This process was repeated 1000 times, randomly selecting single biopsies from each tumor in each iteration, to yield a distribution of mean standardized error values.
3. Results
3.1. Variance analysis of individual hypoxia-related genes
Several hypoxia-related genes were consistently upregulated and included in multiple gene signatures. P4HA1 was the most common gene included in these signature (n = 6). Six genes were included in five signatures: ALDOA, KCTD11, NDRG1, PGAM1, SLC2A1/GLUT and VEGF. Ten other genes were included in four signatures: AK3, BNIP3, ADM, DDIT4, C20ORF20, LDHA, Lrp2bp, MRPS17, PGK1 and TPI1. The W/T1 ratio ranged from 0.17 to 0.73 for these 17 genes (Table 3).
Table 3.
The W/T1 and W/T2 ratios for selected genes commonly found in the ten gene signatures used in this analysis, in rank order from lowest to highest. CAIX is not found in any of the gene signatures, but included for comparison.
| Gene | Number of Signatures with the Gene |
W/T1 (1 Biopsy) |
W/T2 (2 Biopsies) |
|---|---|---|---|
| BNIP3 | 4 | 0.17 | 0.09 |
| AK3 | 4 | 0.19 | 0.10 |
| LDHA | 4 | 0.22 | 0.12 |
| KCTD11 | 5 | 0.25 | 0.14 |
| SLC2A1 | 5 | 0.26 | 0.15 |
| ADM | 4 | 0.28 | 0.16 |
| MRPS17 | 4 | 0.28 | 0.17 |
| NDRG1 | 5 | 0.30 | 0.17 |
| Lrp2bp | 4 | 0.32 | 0.19 |
| TPI1 | 4 | 0.33 | 0.20 |
| CAIX | 0 | 0.38 | 0.24 |
| DDIT4 | 4 | 0.56 | 0.39 |
| PGAM1 | 5 | 0.59 | 0.41 |
| C20ORF20 | 4 | 0.62 | 0.45 |
| ALDOA | 5 | 0.65 | 0.48 |
| PGK1 | 4 | 0.66 | 0.50 |
| VEGF | 5 | 0.70 | 0.54 |
| P4HA1 | 6 | 0.73 | 0.57 |
W/T1 – Within-tumor variance divided by the Total sample variance. Lower W/T1 implies greater capacity of a single biopsy to discriminate among tumors in the face of heterogeneity.
W/T2 – Within-tumor variance divided by the Total sample variance when two biopsies are available. Lower W/T2 implies greater capacity of two biopsies to discriminate among tumors in the face of heterogeneity.
CAIX, which is often promoted as an intrinsic biomarker of tumor hypoxia [15], [16] but not included in any of the signatures, is also included in Table 3 for comparison. These data suggest that there are significant differences in within-tumor heterogeneity among the genes that comprise the signatures. For example, the spatial distribution of BNIP3 (W/T1 = 0.17) and AK3 (W/T1 = 0.19) expression was relatively uniformly within individual tumors, while VEGF (W/T1 = 0.70) and P4HA1 (W/T1 = 0.73) varied from region to region. The W/T2 ratios estimated using two tumor biopsies were lower (0.09–0.57, Table 3) indicating greater capacity to discriminate among tumors with different gene expression levels.
3.2. Variance analysis of gene signatures
The median of the hypoxia signature scores for the 11 tumors taking all available biopsies into account is summarized in Table 2. There was no relationship between signature score and biopsy cellularity (data not shown). The signatures were correlated with each other – among the 45 Spearman correlation coefficients, 30 (67%) were greater than 0.7 (data not shown). It is interesting to note that all of the Spearman correlation coefficients for the Yang gene signature were less than 0.7; this may reflect the fact that although the signature includes genes associated with hypoxia, it was ultimately developed and validated as a prognostic tool for biochemical recurrence free survival in prostate cancer.
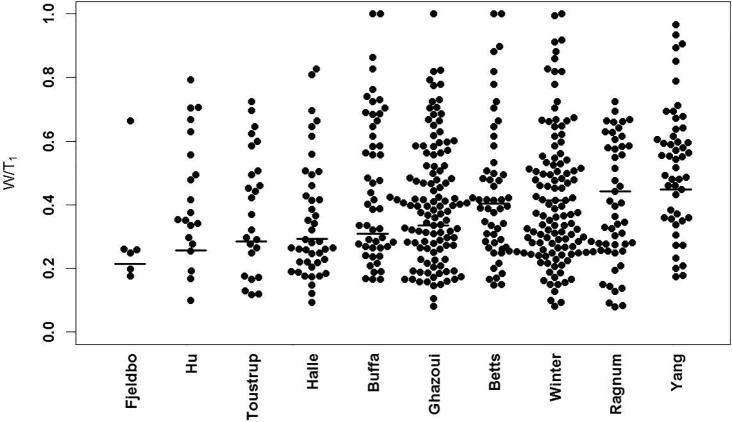
The W/T ratios for the signatures and the individual genes that comprise the signatures are shown in Fig. 1. The signature W/T1 scores were: Fjeldbo 0.21, Hu 0.26, Tostrup 0.28, Halle 0.29, Buffa 0.31, Ghazoui 0.33, Betts 0.40 and Winter 0.41, Ragnum 0.44, and Yang 0.45. Therefore, 21–45% of the total sample variation in hypoxia gene signature score was within tumors. When two biopsies were used, the W/T2 ratios ranged from 0.12 to 0.29. As more biopsies were added to the model, the W/T ratios continued to decrease: W/T3 (range: 0.08–0.21), W/T4 (range: 0.06–0.17), and W/T5 (range: 0.05–0.14). Regardless of the signature or the number of biopsies, the variation in signature scores was consistently less within tumors than between tumors. This suggests that gene signatures are more useful than individual genes in sorting tumors based on hypoxia status.
Fig. 1.
Within-tumor variability divided by total variability (W/T1) in gene expression and signature score for a single biopsy. The dots represent the W/T1 ratios for the individual genes included in the signatures. The horizontal lines represent the W/T1 ratios for each hypoxia gene signature.
3.3. Single vs. multiple biopsies to characterize tumor hypoxia
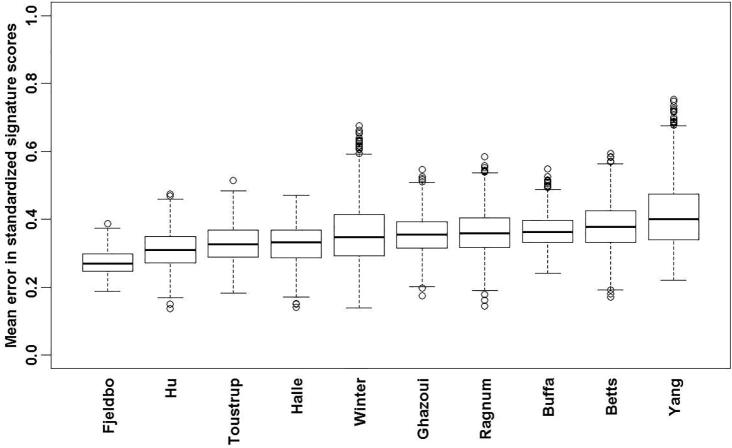
The mean standardized error in each of the signature scores with the use of one biopsy compared to all available biopsies was simulated to further address the question of whether a single biopsy is sufficient to reliably determine the hypoxia status of a tumor. The error calculated for the standardized signatures (means of zero and variances of 1) varied on average (over the 1000 repetitions) from a low of 0.27 for the Fjelbo signature to a high of 0.40 for the Yang signature (Fig. 2). Thus, for Fjeldbo, one can say that the standardized signature score based on one biopsy could differ by +/−0.27 from the global signature score using all biopsies. By dichotomizing the signature scores at their median values, with lower scores indicative of less hypoxic tumors and higher scores of more hypoxic tumors, it is expected that 20% of patients on average would be misclassified having oxic or hypoxic tumors using one biopsy compared to all available biopsies.
Fig. 2.
The mean standardized error between the hypoxia genes signature scores calculated using one biopsy selected at random from each tumor and the scores calculated using all available biopsies, repeated 1000 times. For each signature, the horizontal line represents the global mean error from the 1000 iterations, the edges of the rectangles represent the quartiles and the dots represent the numbers beyond the 1.5 interquartile range.
4. Discussion
This study examines the variability of hypoxia gene signatures and demonstrates that when only a single biopsy is available, the use of a hypoxia gene signature may give a more reliable estimate of global hypoxia status compared to individual genes, although the standardized error remains high. Defining tumor hypoxia status may help to stratify patients in terms of prognosis and allow for personalization of care by selecting those who may benefit most from intensification of standard treatment or the addition of hypoxia-targeted drugs.
Numerous hypoxic modification strategies have been investigated in clinical trials. In a large meta-analysis of 4805 patients with squamous cell carcinoma of the head/neck, the addition of hypoxic modification to radiotherapy led to improved loco-regional control, disease-specific survival and overall survival [35]. However, a phase III trial of standard radiotherapy and concurrent cisplatin chemotherapy, with or without the hypoxia cytotoxic drug tirapazamine failed to show a local control or survival benefit in over 850 patients with head/neck cancer [36]. Similarly, in cervical cancer, a phase III trial in 387 women with locally advanced cervical cancer showed no benefit of adding tirapazamine to standard-of-care radiotherapy and concurrent chemotherapy [37]. Patients in these studies were not stratified by the pre-treatment hypoxic status of their tumors. It is possible that the negative results were in part due to the inclusion of patients with tumors that had low or non-existent levels of hypoxia, who would not benefit from hypoxia-targeted treatment.
The utility of a gene signature to correctly identify patients for hypoxia-targeted treatment depends primarily on the integrated hypoxia dependency of the individual genes in the signature and their relationship to clinically-relevant biology that drives malignant progression, metastases and treatment resistance. It may also be influenced by intratumoral heterogeneity and the location of the biopsy in the tumor, although a previous cervical cancer study found no systematic difference in hypoxia as a function of where electrode measurements were made around the circumference of the visible tumor or at the periphery or in the centre of the tumor [19]. Overall, these factors have the potential to diminish the capacity of gene signatures to discriminate between tumors with different average signature scores and have important implications for future clinical trials where only one or two biopsies are available to evaluate hypoxia [25]. As intratumoral heterogeneity in the signature score increases, the effective statistical power to reliably predict hypoxia-dependent differences in clinical outcome decreases [25]. The design of future trials that rely on hypoxia gene signatures to select patients for hypoxia-targeted treatment should consider heterogeneity to ensure an adequate sample size and maximize the likelihood of achieving the study objectives.
Our study evaluated the effect of intratumoral spatial variation on hypoxia-dependent gene expression. We identified differences in the W/T1 ratios of individual genes common to many of the signatures included in our study, with values ranging from 0.17 to 0.73 (Table 3). Previous work has shown that a single biopsy is sufficient to reliably characterize the global expression level of individual genes with W/T1 < 0.15. However, two or three biopsies are needed for genes with W/T1 in the range of 0.16–0.3 [22]. The value of a second biopsy is illustrated by the W/T2 ratios in Table 3; a second biopsy substantially reduced the within-tumor uncertainty in estimating gene expression and additional biopsies would be expected to reduce the uncertainty even further.
For comparison, Wong et al., evaluated hypoxia in cervical cancer using Eppendorf polarographic electrodes and found much greater within than between-tumor variability (W/T 0.67–0.76) [19]. They demonstrated that five linear electrode tracks through the tumor with 20–30 measurements per track were needed to obtain a reliable estimate of global hypoxia status [19]. All of the ten signatures that we evaluated had W/T1 ratios ≤0.45 and four of the ten had W/T1 ratios <0.3 (Table 2). With two biopsies, the signature WT2 ratios ranged from 0.12 to 0.29; additional biopsies further reduced the W/T ratio. This implies that hypoxia gene signatures, in addition to identifying hypoxic tumors and predicting clinical outcomes better than single genes, may also be more reliable in the face of intratumoral heterogeneity. This may be due to the multi-parametric nature of gene signatures compared to individual genes or other methods for measuring hypoxia, with multiple complementary markers having different hypoxia sensitivities, biological associations and spatial-temporal dependencies.
In general, it is not known if hypoxia gene signatures derived for one type of cancer can reliably be applied to other cancers. Although some signatures appear broadly applicable [26], [27], [32], others may not translate as well [20], [29]. Even less is known about how the performance of gene signatures is influenced by spatial and temporal differences in hypoxia and gene expression among tumors of different types. Our findings, while based on cervical cancer, are in general agreement with previous studies in head/neck cancer, which also have shown that a single biopsy can reliably identify tumors with different levels of hypoxia. Betts et al. evaluated intra-tumoral variability in 13 tumors with three to four biopsies each [20]. Using their 26 gene signature (W/T1 0.4 in our analysis), the intra-tumoral variability was low with a median coefficient of variation of 23%. Similarly, in the DAHANCA clinical trial, a subset of 20 patients had two to four biopsies each and the Toustrup signature (W/T1 0.28 in our analysis) was used to classify the tumors as more or less hypoxic, considering one biopsy at a time [21], [24]. There was complete agreement in classifying 14 of the 20 tumors regardless of which of the available biopsies was used [21].
The small sample size is a limitation of our study; the results need to be confirmed using an independent cohort of patients. Not all of the genes in the ten signatures were available for analysis and there are now more refined and comprehensive methods for evaluating gene expression compared to the Affymetrix array we used; the implications for our findings are not known. We focused on the ability of hypoxic gene signatures to mitigate concerns about tumor heterogeneity when only a single biopsy is available for analysis. However, there are numerous other factors that could influence the overall performance of a gene signature as a predictor of tumor hypoxia, response to hypoxia-targeted treatment and/or patient outcome. The study design did not allow for more comprehensive comparison of the ten gene signatures. Lastly, the study focused on cervical cancer and we are unable to draw conclusions about the reliability of a single biopsy to evaluate hypoxia in other tumor types.
5. Conclusion
Compared with individual genes, hypoxia gene signatures are generally more consistent across multiple biopsies from different regions of a tumor and more tolerant of intratumoral heterogeneity. In settings where only a single biopsy is available, hypoxia gene signatures may give a more reliable estimate of global hypoxia status than individual genes. While multiple biopsies provide greater assurance in correctly classifying a tumor as more or less hypoxic, they often are not available. This underscores the importance of fully understanding the strengths and limitations of a single-biopsy approach to aid in applying hypoxia signatures appropriately in future clinical trials and routine practice.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The authors thank Dr. Barbara Bachtiary, Dr. Paul Boutros and Dr. Fei-Fei Liu for providing us with the gene expression data for this study.
Grant Support
This work was supported by program project grants from the Terry Fox Research Institute (Terry Fox Foundation New Frontiers Program Project Grants #20005 and #1036) and by the Princess Margaret Cancer Foundation.
Contributor Information
Jelena Lukovic, Email: Jelena.Lukovic@rmp.uhn.ca.
Kathy Han, Email: Kathy.Han@rmp.uhn.ca.
Melania Pintilie, Email: Melania.Pintilie@uhnresearch.ca.
Naz Chaudary, Email: Naz.Chaudary@uhnresearch.ca.
Richard P. Hill, Email: Richard.Hill@uhnresearch.ca.
Anthony Fyles, Email: Anthony.Fyles@rmp.uhn.ca.
Michael Milosevic, Email: Michael.Milosevic@rmp.uhn.ca.
References
- 1.Bristow R.G., Hill R.P. Hypoxia and metabolism: hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;9:180–182. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 2.Chaudary N., Hill R.P. Hypoxia and metastases. Clin Cancer Res. 2007;13:1947–1949. doi: 10.1158/1078-0432.CCR-06-2971. [DOI] [PubMed] [Google Scholar]
- 3.Vaupel P., Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26:225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 4.Dhani N., Fyles A., Hedley D., Milosevic M. The clinical significance of hypoxia in human cancers. Semin Nucl Med. 2015;45(2):110–121. doi: 10.1053/j.semnuclmed.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Dhani N., Milosevic M. Targeting tumoral hypoxia: finding opportunity in complexity. Future Oncol. 2012;8(9):1065–1068. doi: 10.2217/fon.12.100. [DOI] [PubMed] [Google Scholar]
- 6.Lima M., Le Bihan D. Clinical intravoxel incoherent motion and diffusion MR imaging: past, present, and future. Radiology. 2015;278(1):13–32. doi: 10.1148/radiol.2015150244. [DOI] [PubMed] [Google Scholar]
- 7.Semenza G.L. Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Curr Opin Genet Dev. 1998;8:588–594. doi: 10.1016/s0959-437x(98)80016-6. [DOI] [PubMed] [Google Scholar]
- 8.Höckel M., Knoop C., Schlenger K., Vorndran B., Baussmann E., Mitze M. Intratumoral pO2 predicts survival in advanced cancer of the uterine cervix. Radiother Oncol. 1993;26:45–50. doi: 10.1016/0167-8140(93)90025-4. [DOI] [PubMed] [Google Scholar]
- 9.Fyles A.W., Milosevic M., Wong R., Kavanagh M.C., Pintilie M., Sun A. Oxygenation predicts radiation response and survival in patients with cervix cancer. Radiother Oncol. 1998;48:149–156. doi: 10.1016/s0167-8140(98)00044-9. [DOI] [PubMed] [Google Scholar]
- 10.Fyles A., Milosevic M., Hedley D., Pintilie M., Levin W., Manchul L. Tumor hypoxia has independent predictor impact only in patients with node-negative cervix cancer. J Clin Oncol. 2002;20(3):680–687. doi: 10.1200/JCO.2002.20.3.680. [DOI] [PubMed] [Google Scholar]
- 11.Fyles A., Milosevic M., Pintilie M., Syed A., Levin W., Manchul L. Long-term performance of interstitial fluid pressure and hypoxia as prognostic factors in cervix cancer. Radiother Oncol. 2006;80(2):132–137. doi: 10.1016/j.radonc.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 12.Dellas K., Bache M., Pigorsch S., Taubert H., Kappler M., Holzapfel D. Prognostic impact of HIF-1α expression in patients with definitive radiotherapy for cervical cancer. Strahlenther Onkol. 2008;184:169–174. doi: 10.1007/s00066-008-1764-z. [DOI] [PubMed] [Google Scholar]
- 13.Haugland H.K., Vukovic V., Pintilie M., Fyles A.W., Milosevic M., Hill R.P. Expression of hypoxia-inducible factor-1alpha in cervical carcinomas: correlation with tumor oxygenation. Int J Radiat Oncol Biol Phys. 2002;53(4):854–951. doi: 10.1016/s0360-3016(02)02815-8. [DOI] [PubMed] [Google Scholar]
- 14.Airley R., Loncaster J., Davidson S., Bromley M., Roberts S., Patterson A. Glucose transporter Glut-1 expression correlates with tumor hypoxia and predicts metastasis-free survival in advanced carcinoma of the cervix. Clin Cancer Res. 2001;7:928–934. [PubMed] [Google Scholar]
- 15.Loncaster J.A., Harris A.L., Davidson S.E., Logue J.P., Hunter R.D., Wycoff C.C. Carbonic anhydrase (CA IX) expression, a potential new intrinsic marker of hypoxia: correlations with tumor oxygen measurements and prognosis in locally advanced carcinoma of the cervix. Cancer Res. 2001;61:6394–6399. [PubMed] [Google Scholar]
- 16.Hedley D., Pintilie M., Woo J., Morrison A., Birle D., Fyles A. Carbonic anhydrase IX expression, hypoxia and prognosis in patients with uterine cervical carcinomas. Clin Cancer Res. 2003;9:5666–5674. [PubMed] [Google Scholar]
- 17.Loncaster J.A., Cooper R.A., Logue J.P., Davidson S.E., Hunter R.D., West C.M.L. Vascular endothelial growth factor (VEGF) expression is a prognostic factor for radiotherapy outcome in advanced carcinoma of the cervix. Br J Cancer. 2000;83:620–625. doi: 10.1054/bjoc.2000.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaupel P., Kallinowski F., Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 1989;49(23):6449–6465. [PubMed] [Google Scholar]
- 19.Wong R.K., Fyles A., Milosevic M., Pintilie M., Hill R.P. Heterogneity of polarographic oxygen tension measurements in cervix cancer: an evaluation of within and variability, probe position, and track depth. IJROBP. 1997;39(2):404–412. doi: 10.1016/s0360-3016(97)00328-3. [DOI] [PubMed] [Google Scholar]
- 20.Betts G.N.J., Eustace A., Patiar S. Prospective technical validation and assessment of intra-tumor heterogeneity of low density array hypoxia gene profile in head and neck squamous cell carcinoma. Eur J Cancer. 2013;49(1):156–165. doi: 10.1016/j.ejca.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 21.Toustrup K., Sorensen B.S., Metwally M.A. Validation of a 15-gene hypoxia classifier in head and neck cancer for prospective use in clinical trials. J Acta Oncol. 2016;55:1091–1098. doi: 10.3109/0284186X.2016.1167959. [DOI] [PubMed] [Google Scholar]
- 22.Bachtiary B., Boutros P.C., Pintilie M., Shi W., Bastianutto C., Li J.H. Gene expression profiling in cervical cancer: an exploration of intratumor heterogeneity. Clin Cancer Res. 2006;12(19):5632–5640. doi: 10.1158/1078-0432.CCR-06-0357. [DOI] [PubMed] [Google Scholar]
- 23.Harris B.H., Barberis A., West C.M., Buffa F.M. Gene expression signatures as biomarkers of a hypoxia. Clin Oncol. 2015;27(10):547–560. doi: 10.1016/j.clon.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Toustrup K., Sørensen B.S., Lassen P., Wiuf C., Alsner J., Overgaard J. Danish Head and Neck Cancer Group (DAHANCA). Gene expression classifier predicts for hypoxic modification of radiotherapy with nimorazole in squamous cell carcinomas of the head and neck. Radiother Oncol. 2012;102(1):122–129. doi: 10.1016/j.radonc.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Pintilie M., Iakovlev V., Fyles A., Hedley D., Milosevic M., Hill R.P. Heterogeneity and power in clinical biomarker studies. J Clin Oncol. 2009;27(9):1517–1521. doi: 10.1200/JCO.2008.18.7393. [DOI] [PubMed] [Google Scholar]
- 26.Buffa F.M., Harris A.L., West C.M., Miller C.J. Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic metagene. Br J Cancer. 2010;102(2):428–435. doi: 10.1038/sj.bjc.6605450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winter S.C., Buffa F.M., Silva P. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 2007;67(7):3441–3449. doi: 10.1158/0008-5472.CAN-06-3322. [DOI] [PubMed] [Google Scholar]
- 28.Ghazoui Z., Buffa F.M., SDunbier A.K. Close and stable relationship between proliferation and a hypoxia metagene in aromatase inhibitor-treated ER positive breast cancer. Clin Cancer Res. 2011;17(9):3005–3012. doi: 10.1158/1078-0432.CCR-10-1704. [DOI] [PubMed] [Google Scholar]
- 29.Eustace A., Mani N., Span P.N. A 26-gene hhypoxia signature predicts benefit from hypoxia-modifying therapy in laryngeal cancer but not bladder cancer. Clin Cancer Res. 2013;19(17):4879–4888. doi: 10.1158/1078-0432.CCR-13-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halle C., Andersen E., Lando M. Hypoxia-induced gene expression in chemoresistant cervical cancer revealed by dynamic contrast-enhanced MRI. Cancer Res. 2012;72(20):5285–5295. doi: 10.1158/0008-5472.CAN-12-1085. [DOI] [PubMed] [Google Scholar]
- 31.Fjeldbo C.S., Julin C.H., Lando M. Integrative analysis of DCE-MRI and gene expression profiles in construction of a gene classifier for assessment of hypoxia-related risk of chemoradiotherapy failure in cervical cancer. Clin Cancer Res. 2016;22(16):4067–4076. doi: 10.1158/1078-0432.CCR-15-2322. [DOI] [PubMed] [Google Scholar]
- 32.Hu Z., Fan C., Livasy C., He Z., Oh D.S., Ewend M.G. A compact VEGF signature associated with distant metastases and poor outcomes. BMC Med. 2009;7(9) doi: 10.1186/1741-7015-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ragnum H.B., Vlatkovic L., Lie A.K. The tumor hypoxia marker pimonidazole reflects a transcriptional programme associated with aggressive prostate cancer. Br J Cancer. 2014:1–9. doi: 10.1038/bjc.2014.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang L., Roberts D., Takhar M. Development and validation of a 28-gene hypoxia-related prognostic signature for localized prostate cancer. E Bio Med. 2018;31:182–189. doi: 10.1016/j.ebiom.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Overgaard J. Hypoxic modification of radiotherapy in squamous cell carcinoma of the head and neck – a systematic review and meta-analysis. Radiother Oncol. 2011;100(1):22–32. doi: 10.1016/j.radonc.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Rischin D., Peters L.J., O’Sullivan B., Giralt J., Fisher R., Yuen K. Tirapazamine, cisplatin, and radiation versus cisplatin and radiation for advanced squamous cell carcinoma of the head and neck (TROG 02.02, HeadSTART): a phase III trial of the Trans-Tasman Radiation Oncology Group. J Clin Oncol. 2010;28(18):2989–2995. doi: 10.1200/JCO.2009.27.4449. [DOI] [PubMed] [Google Scholar]
- 37.DiSilvestro P.A., Ali S., Craighead P.S., Lucci J.A., Lee Y.C., Cohn D.E. Phase III randomized trial of weekly cisplatin and irradiation versus cisplatin and tirapazamine and irradiation in stages IB2, IIA, IIIB, and IVA cervical carcinoma limited to the pelvis: a Gynecologic Oncology Group study. J Clin Oncol. 2014;32(5):458–464. doi: 10.1200/JCO.2013.51.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]