ABSTRACT
In plants dehydration imposed by salinity can invoke physical changes at the interface of the plasma membrane and cell wall. Changes in hydrostatic pressure activate ion channels and cause depolarization of the plasma membrane due to disturbance in ion transport. During the initial phases of salinity stress, the relatively high osmotic potential of the rhizosphere enforces the plant to use a diverse spectrum of strategies to optimize water and nutrient uptake. Signals of salt stress are recognized by specific root receptors that activate an osmosensing network. Plant response to hyperosmotic tension is closely linked to the calcium (Ca2+) channels and interacting proteins such as calmodulin. A rapid rise in cytosolic Ca2+ levels occurs within seconds of exposure to salt stress. Plants employ multiple sensors and signaling components to sense and respond to salinity stress, of which most are closely related to Ca2+ sensing and signaling. Several tolerance strategies such as osmoprotectant accumulation, antioxidant boosting, polyaminses and nitric oxide (NO) machineries are also coordinated by Ca2+ signaling. Substantial research has been done to discover the salt stress pathway and tolerance mechanism in plants, resulting in new insights into the perception of salt stress and the downstream signaling that happens in response. Nevertheless, the role of multifunctional components such as Ca2+ has not been sufficiently addressed in the context of salt stress. In this review, we elaborate that the salt tolerance signaling pathway converges with Ca2+ signaling in diverse pathways. We summarize knowledge related to different dimensions of salt stress signaling pathways in the cell by emphasizing the administrative role of Ca2+ signaling on salt perception, signaling, gene expression, ion homeostasis and adaptive responses.
KEYWORDS: Calcium, salinity, secondary messengers, osmoprotection, signaling pathway
Introduction
Soluble salts are abundantly available in the soil as sources of essential nutrients for plant growth and development. Nevertheless, when the amount of salts in the soil exceeds a particular threshold, plant health and productivity are impaired. Over-salinized soil is a global concern that threatens approximately 20% of irrigated land with significant crop reduction.1,2 Salt stress frequently inhibits growth and influences key developmental stages, triggering premature senescence and death during prolonged exposure.3,4 Salinization has an osmotic effect on plant cells, leading to their shrinkage by dehydration. This effect is transient, with cells regaining their original volume within few hours of their initial exposure to chronic salt concentrations. Despite this recovery, cell division and mostly cell expansion are negatively impacted, resulting in reduced leaf and root growth. Various plant developmental stages comprising germination, vegetative and reproductive growth are also affected by salt stress. Plants dynamically respond to salt exposure, which in extreme conditions their response is accompanied with programmed cell death.5 Osmotic stress caused by a high salt environment constrains water uptake from rhizosphere, resulting in nutrient deficiency and dehydration damage to the plant. Plant responses to these constrains range from regulating external ion uptake to dynamic ion hemostasis pattern. For instance, phosphorus (P) uptake is reduced under saline conditions because Ca2+ inhibits P ions uptake by the root system, while in plant cells an increase in sodium (Na+) influx is accompanied with a decrease in potassium (K+) uptake and transport.6–8 As K+ is a pivotal component for modulating ion hemostasis and osmotic pressure, plants should define an adaptable strategy to regulate cation transporters in the plasma membrane (PM) to fulfill hampered ion hemostasis during saline stress event.
Plants utilize multiple strategies to alleviate salt stress damage. Na+ influx is powered by voltage-insensitive monovalent-cation channels (VIC).9 These channels are regulated using bivalent cations such as Ca2+ to maintain ion hemostasis. Over the course of evolution, plants have developed the capacity to detoxify excessive Na+ levels that accompany high salt concentrations. Recent studies have identified two classes of Na+/H+ antiporters responsible for Na+ detoxification: Na+/H+ exchanger (NHX1) and salt overly sensitive (SOS1), respectively localized to the vacuole and PM.10,11 Alternatively, to maintain an optimal osmotic pressure and alleviate the salt stress damage, Na+ accumulation in the cell is mediated by compartmentalization in the vacuole.12 This regulatory mechanism is triggered when the cell senses salt stress. Although the molecular mechanism of hyperosmotic perception in plants is still unknown, in yeast these signals are perceived in the PM and communicated downstream by means of the mitogen-activated proteins kinase (MAPK) pathway.4 Chloride (Cl−) ions are also toxic when they accumulate in the cytoplasm. Plants that are capable of Cl− compartmentalization in both shoot and root systems are potentially tolerant to NaCl stress. Alternatively, regulation of Cl− transport has also been suggested for induction of salt tolerance.13 In plant cells, ion concentration is directly regulated by cation-chloride co-transporters (CCCs).14–17 CCCs function has been described in mammals in which K+ and/or Na+ with the anion Cl– transport in a 1:1 ratio and promote electroneutral transport.18
Under salinity stress, abscisic acid (ABA) biosynthesis is up-regulated in guard cells, resulting in stomatal closure. This reduces photosynthetic capacity and triggers photoinhibition and oxidative stress. The biosynthesis and signaling activity of other phytohormones such as ethylene are also influenced by salinity stress. Induction of ethylene synthesis under a variety of stresses including salinity stress is well known. Biosynthesis of ethylene or its direct precursor, ACC (1-aminocyclopropane-1-carboxylate) is induced to a remarkable degree by salt stress.19 Therefore, ethylene accumulates in plants encumbered with salt shock. Transgenic plants, with ethylene overexpression display enhanced stress tolerance. Nevertheless, inconsistent findings also suggest an inhibitive effect of ethylene in the induction of salt tolerance. These contradictions lead us to speculate a dynamic mode of action for ethylene based on its concentration. In fact, various levels of ethylene may play distinct roles in regulation of salt stress responses. However, the mechanism/s controlling ethylene levels under salt stress condition could be a major point of interest. Earlier reports showed that under saline stress conditions, Ca2+-dependent protein kinases (CDPKs) phosphorylate the ACC synthase (ACS) protein stabilizing its conformation, leading to an increased ethylene content.20 Ca2+ levels act as a secondary messenger, becoming elevated early on when plants encounter biotic and abiotic stresses.21,22 Points of convergence in the ethylene and Ca2+ signaling pathways have been well characterized.23–26 Ethylene compromises the crosstalk between CDPK and MAPK. In general, the perception of salinity stress at the PM triggers a plethora of responses ranging from Ca2+ oscillation, gene expression, changes in phytohormone homeostasis and alteration in osmotic pressure. Furthermore, activated signaling machinery will ultimately lead to the saline stress responses. However, among all, Ca2+ oscillation plays multifaceted roles to rule out detrimental effects of saline shock by building up multiple cross nods through generation of dynamic amplitudes.27 This makes it capable of adjusting a range of variable downstream signaling cascades and influences the plant compatibility.
In this review, recent findings about Ca2+ signaling-related salt stress responses are discussed. In light of these findings, Ca2+ involvement in salt stress responses, with emphasis on salt perception, signaling events and Ca2+-mediated metabolic blueprints, will be discussed. Taken as a whole, these results underscore the fact that Ca2+ signaling and salt stress signaling and responses are inseparable.
Mechanism of salt stress perception
In plants, strong growth inhibition in different organs has been reported as a result of exposure to salt stress.28 Na+ influx into the cell is mediated via the Na+ transporter HKT.29 However, due to the high hydrated ionic radii similarity between Na+ and K+, PM ion transporters are generally not capable of discriminating their influx. The result is Na+ toxicity during exposure to high salt concentrations. This unidirectional influx disturbs various enzymatic processes and due to lack of need for high level of Na+, it becomes parallel with osmotic pressure impairment. Growth retardation can be due to changes in cell wall structure and disturbed cell division and expansion.30 Hampered growth, particularly in the leaves, leads to a sharp decline in plant biomass since reduced leaf area is accompanied by lower photosynthetic efficiency. It has been shown that salt-tolerant maize plants exhibit higher cell extensibility under salt stress.31 By contrast, hypersensitive Arabidopsis mutants are defective in cell-wall organization.32 Although the mechanism underlying cell wall expansion under osmotic shock is not completely known, cell-wall integrity during salt stress has been associated with the FERONIA (FER) receptor kinase activity. FER, a PM localized receptor kinase, plays a critical role in maintaining cell wall structure in plants exposed to saline shock; this role is mainly related to the Ca2+ signaling cascade. Upon exposure to salt stress, cell wall structure is damaged by Na+; this damage is perceived by FER which in turn activates a Ca2+ channel via unknown pathway, resulting in a transient Ca2+ signaling in the root cells (Figure 1).28,33 Na+ is sensed by salt sensitive receptors comprising SOS1, Na+/H+ antiporters, histidine kinases, AHK1/ATHK134–36 and nonselective cation channels (NSCCs).29,37 Elevation of Na+ concentration in the surrounding environment leads to an influx of Na+ into the cells through NSCCs. The two main categories of NSCCs in A. thaliana are cyclic nucleotide gated channels (CNGCs) and glutamate-activated channels (GLRs). CNGCs contribute to the regulation of ion homeostasis, which positively correlates with salt tolerance in Arabidopsis. CNGCs have also been proposed to be involved in Ca2+ influx and signaling.38,39 CNGS interacts with calmodulin (CaM), a Ca2+-modulated protein, in IQ-motif and regulates downstream signaling (Figure 1).40 Notably polymorphisms in IQ-peptide has been reported which suggests that there are diverse modes of action for CaM signaling. This explains how Ca2+-CaM binding to the CNGS induces flexible machinery to fine tune induced Ca2+ signals.
Figure 1.
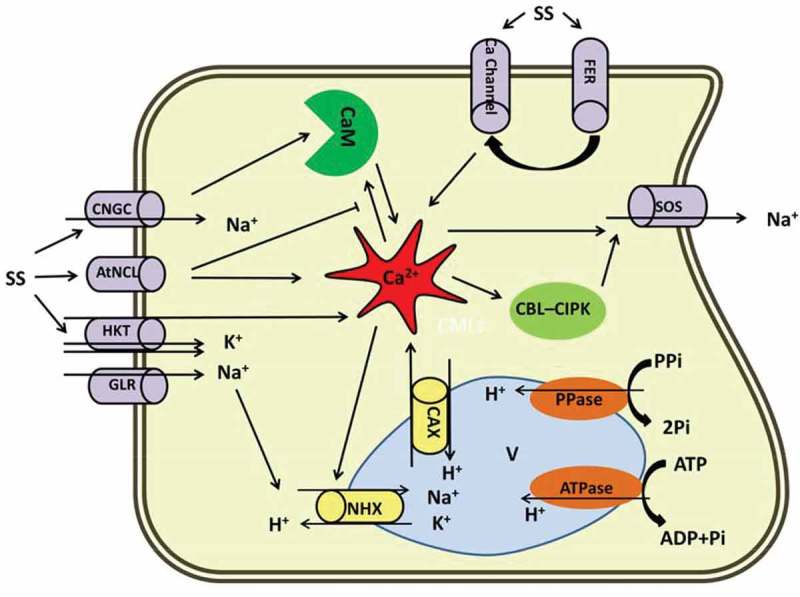
Diagrammatic representation of the Ca2+ role in salt stress responses in plants. Prominent role of Ca2+ under salt has been proposed in this scheme. Na+ influx to the cytosol is mediated by CNGC, HKT, GLR and AtNCl. Na+ efflux is derived by SOS. NHX and CAX are involved in Na+/H+ and Ca2+/H exchanges through vacuolar membrane, respectively. Ca2+ in the center implicates the Na+ current either by direct interaction with ion channels or indirectly through Ca2+ related modules (CBL-CIPK and CaM). FER activates Ca2+ channels via unknown pathway and bears a transient Ca2+ signaling. AtNCL: Na+/Ca2+ exchanger-like protein, CAX: Vacuolar H+/Ca2+ antiporter, CNGS: Cyclic nucleotide gated channels, FER: FERONIA, GLR: Glutamate-activated channels, HKT: Na+ transporter, NHX: Vacuolar located Na+/H+ antiporters, SOS: Salt overly sensitive, SS: Salt stress.
In animal cells, Na+/Ca2+ exchangers (NCXs) play important roles in Ca2+ homeostasis. Bioinformatic approaches in Arabidopsis have revealed a AtNCL gene encoding a PM-localized protein that has the capacity to bind Ca2+ and possesses NCX-like activity.41 It has been known that AtNCL is highly expressed under salt stress leading to Ca2+ ions being extruded (Figure 1). Consistently, plants defective in AtNCL functions exhibit elevated cytosolic Ca2+ levels and root surface Ca2+ fluxes.42,43 This suggests that AtNCL terminates Ca2+ signaling under salt stress, although because of availability of various transporters in response to Ca2+ signaling, a dynamic interaction between transporters across the PM is highly likely.
The potential functions of the SOS signaling pathway that are involved in the Na+ exclusion and ion homeostasis have been well described under salt stress.44,45 Previous studies reported that elevated Ca2+ levels can protect plants from Na+ toxicity. When Na+ is sensed by the root, the SOS signal transduction cascade is activated in the cell through Ca2+ oscillation and inhibits excessive ion accumulation generated by salt stress. Elevated concentrations of the Ca2+ generated by continuous Na+ influx into the cell activates the SOS proteins (a Ca2+ binding protein), in return SOS antiporters (such as SOS1) contribute to the Na+ exclusion and cellular ion (Na+/K+) homeostasis.46 This retro-controlling mechanism mediated by Ca2+ signaling balances the Na+ concentration in root cells under saline conditions. Salt stress signaling is regulated by ethylene biosynthesis and signaling in sos2 and sos3 mutants; changes in the expression of ethylene responsive genes (ERF) has been observed due to exposure to salt stress.47 Recent studies provide evidence that activation of SOS2 gene is responsive to phosphorylation and activation of ethylene insensitive 3 (EIN3), which leads to ethylene and salt-inducible ERF1 (ESE1) expression. This implies that the function of the SOS gene is closely linked to ethylene signaling during plant response to abiotic stresses.
Earlier reports pointed out that Ca2+ and Na+ can improve plant growth, photosynthesis, water and ion transport and these roles are mainly related to the direct interaction of Na+–Ca2+ at the surface of the PM, which is powered by Ca2+ signaling.48 Lynch and coworkers for the first time reported the involvement of Ca2+ in salt stress responses by discovering a rapid rise in intracellular Ca2+ in root protoplasts of maize (Zea mays L.) within seconds of being exposed to saline shock.49 Furthermore, induction of Ca2+ signals in response to osmotic and salt stresses are demonstrated by aequorin-based luminescence imaging of Ca2+ reporter protein.50 Ca2+ signaling is now regarded as a secondary messenger in stress signaling pathways, such as salt stress.51–54 It is noteworthy that, elevation in cytosolic Ca2+ in response to osmotic stress appeared to be a conserved signaling event in Plantae kingdom, which is manifested by the similar function across the higher plants to the basal-branching moss taxon Physcomitrella patens.55,56 The Ca2+ signaling pathway initiates with elevation in cytosolic Ca2+ content and is regulated by Ca2+ transporters such as cyclic CNGCs, doubled-pore Ca2+ channels (TPCs), Ca2+-ATPases and a vacuolar H+/Ca2+ antiporter (CAXs) (Figure 1).57 Moreover, it was postulated that an initial Ca2+-dependent signaling network involves in the salt stress responses comprising Ca2+ transport and downstream targets, such as CaM, CMLs (CaM-like protein), CDPKs, CBLs (calcineurin B-like protein) and CIPKs (CBL-interacting protein kinase).52,53
Both osmotic and ionic stresses have been implicated in triggering similar Ca2+ oscillations in Arabidopsis, suggesting a unique Ca2+ readout under both stress events.58,59 The amplitude and duration of the Ca2+ transient are determined by the activity of vacuolar membrane H+/Ca2+ antiporter (Vcx1p) and endomembrane localized Ca2+-ATPases.60,61 Ca2+ transients are also involved in activation of responsible channels for ion influx and energy dependent transport systems that contribute to the divalent cation compartmentalization. It has been reported that the salt stress-induced elevation in cytosolic Ca2+ and the new cytosolic Ca2+ status is regulated by CAX1, ECA (ATP-driven Ca2+ pump) and ACA Ca2+-ATPases (auto-inhibited Ca2+-ATPases).62 In general, two lines of evidence suggest that Ca2+ triggers salt stress responses: first, by direct inhibition of Na+ influx and second, as a downstream signaling component in response to salt stress.63
Ca+2 signaling and regulation of ion channels under salt stress conditions
NaCl is an immense challenge for salt-imposed plants; as the process of accumulation, exclusion, translocation and organelle sequestration of Na+ and Cl – are the major cause of salt-induced damages in plants.64,65 Na+ is the most abundant toxic ion in saline soils to enter the plant cells through Na+-permeable transporters, by way of an unknown molecular mechanism. It was assumed that Na+ can enter to the cell through nonselective cation transporters, such as K+ transporters. When plant roots encounter with a high dosage of Na+, three outcomes explain the exorbitant Na+ influx to the epidermal cells; (i) accumulation in the vacuoles, (ii) redirection to the soil or apoplastic area, and (iii) shift into the adjacent cells via plasmodesmata.66 In these ways, numerous transporters, ions, Ca2+ sensors, and their downstream interacting counterparts function in association to regulate the efflux of excess Na+ ions and to gain the ion hemostasis.67,68 Hyperosmolarity-gated Ca2+ permeable channel (OSCA1), a putative candidate for perception of salt stress,69 Na+/H+ antiporters (NHX), V-type H+-ATPase and H+-PPase are responsible for compartmentalization of Na+ into the vacuoles (Figure 1). These processes require the activation of a complex signaling network, which is initiated by Ca2+ signals in response to high levels of Na+70,71
Investigating NaCl-induced Ca2+ signaling in plants using ratiometric Ca2+ sensor, YC3.6, revealed that the localized application of NaCl results in a drastic and transient elevation in local Ca2+ and triggers a systemic feedback in distinct sites.72 However, it is not clarified whether physical (osmotic), chemical (ionic) or both features of Na+ trigger Ca2+ signaling responses.56 Under salt stress, cytosolic Ca2+ transiently activates CaM protein, which switches on the signaling networks through the calcineurin pathway so that salt-induced damage associated with high Na+ levels is mitigated.73 One of these networks consists of a Ca2+ sensor protein-kinase network like CBL-CIPK signaling complex that modulates protein transporters and serve the K+ and Na+ influx and transport to regulate ion hemostasis under salinity (Figure 1).74
An Na+ transporter was discovered to function in wheat as a ‘high-affinity K+-transporter’ and referred to as HKT according to its similarity to the high-affinity K+ transport in bacteria and fungi.75 In the absence of K+, this membrane protein is capable of Na+ transport with higher capacity (26 fold);76 hence HKT was further known as Na+–K+ symporter.77 Notably, it has been identified that a rice HKT protein encodes by OsHKT2;4 also functions as a Ca2+-permeable NSCC.75 It has also been reported that Na+ transport via NSCCs is sensitive to Ca2+, which may explain the inhibitory effect of Ca2+ on Na+ influx into the root epidermal cells. However, it remained controversial that whether the Ca2+ function is associated with the intracellular regulatory protein or may be regulated independently.
Different Na+ transport modes have also been suggested in plants by investigation of sos3-1 hkt1 double mutants in Arabidopsis. It was found that increased extracellular Ca2+ impedes the hkt1 knockout mutant to suppress Na+ sensitivity of sos3-1, leading researchers to postulate about the inhibitory effect of Ca2+ on Na+ influx in plants.63 Likewise, analysis of plant cell patches revealed the existence of both Ca2+ in/sensitive Na+ transport responses.63,78 According to the electrophysiological and in plants mutants dissection studies, two Na+ influx classifications have been defined in plants; one is directly inhibited by Ca2+, and the other is HKT.63 Given that Ca2+ functions as second messenger, the most probable scenario could be that the HKT links Ca2+ to the downstream signal transduction events.79 Consistently, a recent report has revealed that MtCML40 in MediCago truncatula triggers a salt stress response by targeting MtHKT-dependent Na+ accumulation.80 As Ca2+ has potentially displayed an array of physiological roles in both signaling and nutrient uptake, HKT could be considered as a Ca2+ mediating candidate in modulation of plant growth and development under salt stress.75
Na+/H+ (NHX) transporter has been suggested as a salt tolerance responsive transporter, since its overexpression is associated with increased tolerance to salt stress and conversely, plants with truncated NHX exhibit decreased tolerance.81–83 Moreover, NHX5/NH6X double mutants are hypersensitive to salt stress.84 Another NHX member localized to the PM, namely SOS1, has been suggested to contribute to Na+ loading into xylem vessels, though SOS1-related genes that are expressed in epidermal cells are rather low. These all can imply that NHX resembles to contribute in systemic retrieval and distribution of Na+ in plants.66
Salt susceptible mutants have shown K+ deficiency, which is consistent with the challenge of K+ and Na+ influx through similar transporter.85–87 Notably, independent influx of Ca2+ maintain its regulatory effect on K+/Na+ levels irrespective of K+/Na+ ratio. However, Ca2+ mainly contributes to K+ uptake, which results in a fine tuning of ion transport particularly when excessive levels of Na+ are present.88 Ca2+ may function in ion transport by interacting with NSCCs. Alternatively, as a second messenger, Ca2+ signaling triggers CBL–CIPK network with further effect on the SOS signal transduction pathway.63,66,85 In fact, under salinity, a causal link occurs between Na+ level regulated by NHX and SOS function modulated by CBL–CIPK signaling network, which is built up by Ca2+ signaling pathway.
To date several mutants of SOS genes have been empirically identified by a positional cloning approach.89 The SOS pathway consists of three major components including SOS3, a Ca2+ sensor (a myristoylated Ca2+-binding protein), SOS2 a serine/threonine protein kinase and SOS1 that functions as PM-Na+/H+ antiporter.89–91 According to previous studies, involvement of Ca2+ in salt stress signaling facilitates ion hemostasis.54,92,93 Salt perception by PM sensors induces the cytoplasmic Ca2+ oscillation. Furthermore, Ca2+ read out, sensed by SOS3 (also known as CBL4), is decoded by Ca2+-sensing proteins such as CBLs and their cooperative partners, CIPKs (Figure 2) 94 and ultimately brings SOS3 and SOS2 interaction. The myristoylation motif of SOS3 results in SOS3/SOS2 protein kinase complex, which further phosphorylates and activates SOS1. SOS1 facilitates excess of Na+ ions efflux and thus, contributes to Na+ ion homeostasis and long distance Na+ transport (Figure 2).95–97 SOS2 is also found to function in ion homeostasis by interacting with vacuolar Na+/H+ antiporter (NHX) and regulating Na+/H+ ratio, which results in sequestration of excess Na+ ions into the vacuole (Figure 2).98 Moreover, SOS2 is shown to regulate SOS1 and CAX1 independently of SOS3 resulting from external stimuli99 and contributes to Ca2+ homeostasis, demonstrating a metabolic link between Na+ and Ca2+ homeostasis in plants (Figure 2).100–103 This leads us to speculate that the salt stress responses, at least in part, are controlled by Ca2+ signal transduction pathway.
Figure 2.
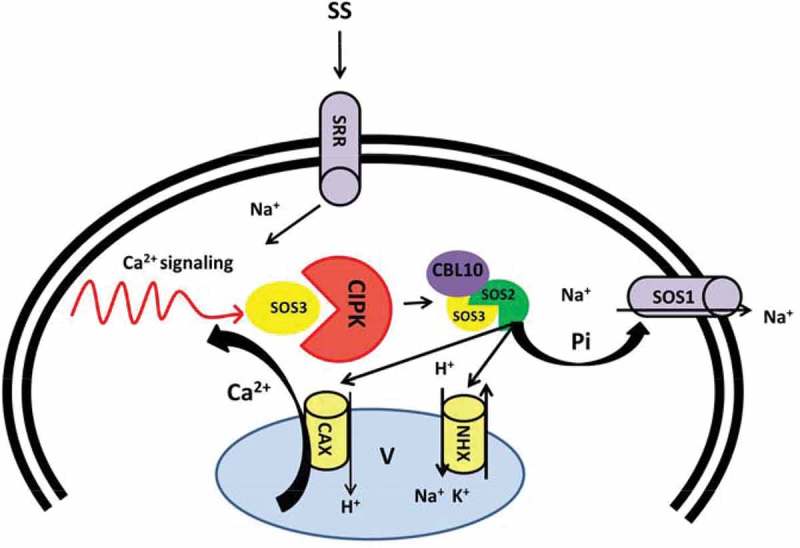
Schema model of SOS pathway regulated by Ca2+ under saline shock. Ca2+ signaling triggered by salt stress activate SOS3 causes interaction with SOS2. SOS3/SOS2 complex activates SOS1 by direct phosphorylation driven by SOS2 and result in Na+ efflux. Alternatively, SOS2 regulates vacuolar channels (CAX and NHX) and balance Na+ vacuolar sequestration. CAX: Vacuolar H+/Ca2+ antiporter, CBL: Calcineurin B-like protein, CIPK: CBL-interacting protein kinase, NHX: Vacuolar located Na+/H+ antiporters, SOS: Salt overly sensitive, SS: Salt stress. SRR: Sal responsive receptors.
As a second messenger, Ca2+ binds to the protein sensors and transmits stress cues to the downstream signaling routs. Several CBLs and their target kinases have been described as components of salt stress responses. Na+ efflux from the cytoplasm is mediated by an array of proteins comprising SOS3, its interacting kinase SOS2 and SOS1.101 The fact that sos1 mutant is hypersensitive to salt and contains more Na+ in comparison to the wild type, supports the postulation that the SOS1 protein may cause Na+ efflux from the cytoplasm. Mutations of SOS2 and SOS3 also produced a salt-hypersensitive phenotype; however, similar levels of Na+ in these mutants with wild type suggest distinct regulatory mechanisms for SOS2 and SOS3 compared to SOS1.104 CBL1 and its interacting kinase CIPK1, along with CBL9 with its kinase counterpart, CIPK3, were also found to contribute in salt stress tolerance signaling.105–107 CBL/CIPK signaling modules are involved in salt tolerance through modulation of PM processes. However, their metabolic mode of action in plants that are exposed to salt stress is not fully clarified.
For the first time, Kim and colleague (2007) reported lower Na+ content in a hyper salt-sensitive cbl10 mutant, which suggests a novel role for CBL10 in the regulation of a pathway for salt tolerance (Figure 2).104 In fact, plants with the capacity to mitigate excessive Na+ levels, either by limiting influx or by enhancing efflux, exhibit higher tolerance to salt stress. An alternative hypothesis posits that lower Na+ content in the cell is a function salt sequestration into the vacuoles. In this way defect/s in vacuolar Na+ sequestration inhibit Na+ transfer from cytoplasm to the vacuole which would be accompanied by salt susceptibility. This can be a plausible explanation for higher sensitivity of cb110 mutant plants when compare to the wild type, though they are more sensitive to salt shock.104 Consistently with this hypothesis, plants overexpressing AtNHX1 possess over-accumulation of Na+ while maintaining capability for salt tolerance.108
CBL10 interacts with the SOS2/SOS1 complex104,109,110 and localizes it to the PM where it mediates SOS1 induction and promotes Na+ efflux from aerial parts of the plants.109 The contribution of CBL10-SOS2 to salt stress responses by storing Na+ in the vacuole has also been documented; nevertheless, its vacuolar or endosomal target proteins have not been identified thus far.104 Alternatively, CBL10 triggers a stress response by interacting with AKT1 to hinder its accumulation at the PM and function as a negative regulator of K+ uptake and thus regulates the Na+ hemostasis.111 These finding reveal the related function of Ca2+ signaling/modules in salt tolerance mechanism in plants. According to the direct or indirect interactions of Ca2+ with salt responses queues, a generic role for Ca2+ signaling in the context of salt stress is conclusive.
Convergence of Ca2+ and salt stress signaling pathways
In response to salt stress, an array of genes including CAMTA (CaM binding transcription activator) and MYB have been shown to be up-regulated by Ca2+/CaM in Aegilops tauschii, implying the putative role of the Ca2+ signaling pathway in salt stress responses in plants.93,112,113 Transcriptomic analysis of the wild-type and myb59 mutant of A. thaliana using microarrays has revealed 45 differentially regulated transcripts with 33 and 12 genes up- and down-regulated, respectively. Twenty-five percent of up-regulated transcripts possess the expression of Ca2+-binding proteins and the rest are functionally involved in Ca2+ homeostasis, transport and signal transduction pathways.
Involvement of these genes in Ca2+ signaling has been confirmed according to the DAVID enrichment analysis.114 Comparison between wild-type, myb59 and overexpressed (OE) lines demonstrated that the genes involved in Ca2+ signaling (CML24, CML35, CML47, KIC and PILS4) have been constitutively expressed. Ca2+ signaling role in regulation of stomatal movement has been reportedly suggested.115–117 Likewise, Ca2+ has null effect on stomata of myb59 while in OE and wild type resulted in stomatal movement. This suggests that under salt stress, involvement of MYB59 in stomatal functioning through Ca2+ signaling is highly likely, as expressed MYB59 by salt stress can suppress CAX1 and trigger stomatal closure.54
It has been well documented that the expression of a Medicago truncatula CML, MtCML40, is up-regulated under salt stress. Surprisingly, overexpression of MtCML40 negatively affect the expression of MtHKT1;1 and MtHKT1;2, two genes responsible for encoding proteins with function in mitigation of exceed Na+ from shoots. This can indirectly connect MtCML40 regulatory role to the Ca2+ signaling through a MtHKT-dependent Na+ accumulation process when plants suffer from saline shock.
Gene duplication is defined as one of the major adaptation mechanisms developed by plants to cope with unexpected environmental stimuli. Gene duplication effect on plant salt tolerance has been investigated on the duplicated CBL10 genes of Eutrema salsugineum as a salt-tolerant plant. Results have indicated that, down-regulation of duplicated CBL10 genes (EsCBL10a and EsCBL10b) reduced the plant growth in response to salinity, while higher reduction was observed when expression level reduced in each gene individually, implying a distinct function for both genes in response to salt stress. Based on a cross-species complementation assay, EsCBL10b is a potential activator of SOS pathway while EsCBL10a exhibits a different function than EsCBL10b. Notably, N-terminus modification in homologues of EsCBL10s results in different functions of each protein. Regardless of different functions of EsCBL10a and EsCBL10b, their duplication increases Ca2+-mediated signaling capacity in Eutrema salsugineum.118 Moreover, it is believed that elevated number of active Ca2+ sensors causes salt tolerance and adaption. Eutrema penlandii comprises three Ca2+ sensors, EsCBL10a, EsCBL10b and EsSOS3, with both individual and combined functions. It has been revealed that co-expression of EsCBL10a and EsSOS3 in Atsos3 increases root growth, suggesting an additive effect of these genes on salt tolerance and thus root growth.118
It has been shown that salt tolerance is associated with higher K+/Na+ ratio119 and enhanced Na+ efflux.120 Transcriptome analysis in wheat genotypes revealed that NHX1 and the SOS pathway-related gene expression levels in salt-tolerant genotypes were higher in comparison with their expression in the other genotypes.121 Moreover, similar research in Aegilops tauschii during long-term exposure to saline stress demonstrated that genes encoding transporters and channels including SOS1, NHXs, HAKs, with contribution to K+ hemostasis have been up-regulated.93 In addition, it was shown that ENA1, which encodes the P-type ATPase required for Na+ efflux is expressed by elevation in cytosolic Ca2+ concentration.63,122
Transcriptomic analysis of Camellia sinensis revealed that expression levels of Ca2+-ATPases and CAX genes were upregulated in response to salt stress, this suggest the existence of overlapping expression patterns with Ca2+ signaling and salt stress responses.53 Moreover, three MAPK kinase and five MAPK kinase genes were also shown to be induced under salinity.53 As MAPK signaling pathway is mediated by Ca2+-dependent regulation pathway this suggests the involvement of Ca2+ in salt stress responses.123
WRKY gene expression under salt stress is also regulated by CaM-mediated Ca2+ signaling pathway.53,124–126 In this regard, it was reported that in Camellia sinensis multiple WRKY genes, including WRKY7, WRKY33, WRKY18 and WRKY40, were up-regulated under salinity.53 Among 74 WRKYs identified in Arabidopsis, expression of 18 genes drive salt stress responses in the root, whereas overexpression of AtWRKY25 and AtWRKY33 increased salt tolerance in the shoot.127,128
Studying the gene expression pattern in soybean under saline–alkaline and drought stress conditions revealed that genes associated with Ca2+ signaling are up-regulated in all stress treatments suggesting that Ca2+ signal transduction is casually linked to the external stimuli responses.129
In summary, physiological and metabolic analyses under salt stress condition demonstrated the involvement of several genes and transcription factors in salt stress responses that are linked to Ca2+-dependent signaling pathways. Future research on the role of Ca2+ signaling as stress signaling molecules on differential expression of genes can open a new avenue to reveal the controlling mechanism of salt stress in plants.
Polyamine/Ca2+-dependent salt stress responses
Polyamines (PAs), mainly known as putrescine (Put), spermidine (Spd) and spermine (Spm), are well recognized for their profound effects on plant growth, development and adaptation against various environmental cues.130 It has been well-established that PAs play indispensable roles in multiple cellular processes, such as cell division, differentiation, transcriptional regulation, translation, membrane and cell wall stabilization and programmed cell death.131–133 Transcriptome analysis of PAs in plants subjected to a wide array of environmental stressors has shown that modulation of PA levels is associated with tolerance and stress amelioration and is often accompanied by changes in expression level of PA biosynthesis-related genes.131,134 Studies concerning the regulation of PA concentrations in plants revealed that several genes involved in PA biosynthetic pathways are unregulated under salt stress.134,135 Activation and stimulation of arginine decarboxylase (ADC), which is involved in PA biosynthesis, in response to salinity has been regarded as a key regulator of adaptive responses in plants.136 An investigation on expression levels of PAs biosynthetic genes in 18 varieties of Oryza sativa demonstrated that ADC gene is induced under salt stress.137
Physiological studies have provided a wealth of information on the role of PAs degradation pathway driven by PAs oxidase enzyme in response to stress stimuli.138 Recent studies have indicated that the PAs metabolic pathway is in intricate crosstalk with other signaling molecules such as ABA, H2O2 and γ-aminobutyric acid (GABA).139,140 In this way, it has been shown that GABA enhances salt tolerance in some plant species such as lettuce.141 PA metabolism also stimulates NO production that may link PAs-mediated stress responses to the other stress mediators such as Ca2+ ions and protein kinases.
Under salinity conditions, PAs as positively charged molecules could substantially interact with negatively charged proteins including ion channels and affect their conductivity.142,143 Garufi et al. (2007) suggested that PAs may regulate the activity of numerous ion channels indirectly by affecting PM potential by activation of H+-ATPase through enhancement of interactions with 14-3-3 proteins, a family of highly conserved regulatory molecules (Figure 3).144 PAs directly inhibit both types (fast and slow acting) of NSCCs in vacuolar membrane with a high affinity.145,146 Consistently, it has been reported that PAs induce stomatal closure by regulating KAT-1 like voltage-dependent K+ channel. PAs also reduce Na+ influx through the NSCCs transporters, thus diminishing the Na+-induced membrane depolarization and inhibiting K+ losses.147,148 In pea leaves, the NSCC inhibition by external PAs, Put or Spm, incorporate to decreases in the salt-induced membrane depolarization and K+ efflux.146 Inhibitory effects of PAs in NSCC-mediated Na+ flow have been reported in leaf146 and root142 tissues. In this regard, PAs indirectly can be linked to the Ca2+ signaling as discussed transporters are modulated by Ca2+ or related modules.
Figure 3.
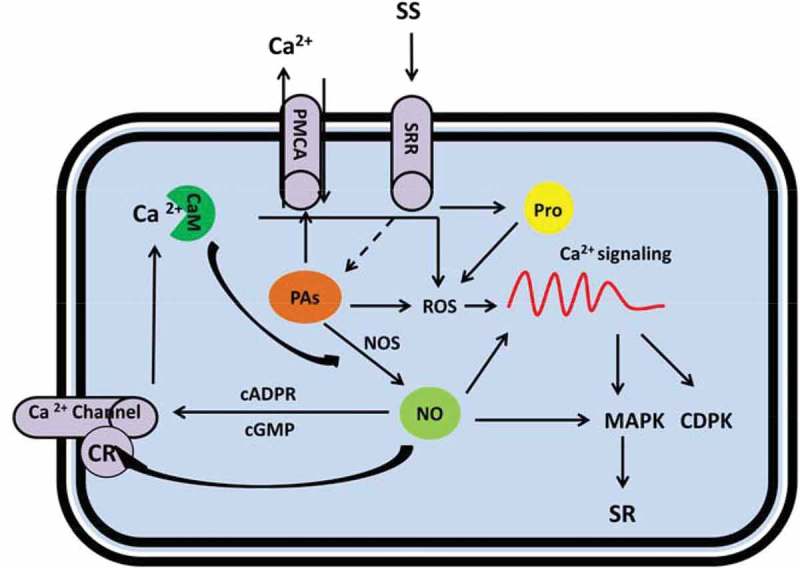
Metabolic scheme for regulation of salt stress responses by polyamines (PAs), nitric oxide (NO) and proline mediated by cytosolic Ca2+ signaling. Under salt stress, elevated PAs content affect Ca2+ level in the cell by either direct activation of PMCA and contributes to Ca2+ excursion (this will result in a steady state level of Ca2+ for normal metabolism in the cell); or indirectly by stimulation of NO and ROS production that results in downstream Ca2+ signaling. Similarly by salt shock, enhanced proline content influences the Ca2+ read out by ROS engagement; while NO is multiply associated with Ca2+ signaling comprising: direct manipulation of the Ca2+ signaling, effect on Ca2+ channels and regulation of MAPKs. CR: Cysteine residue, cGMP: Cyclic GMP, cADPR: Cyclic ADP ribose (cADPR), CaM: Calmodulin, CDPK: Ca2+ dependent protein kinases, MAPK: mitogen-activated proteins kinase, NO: Nitric oxide, PAs: Polyamines, PMCA: plasma membrane-Ca2+-ATPase, Pro: Proline, ROS: Reactive oxygen species, SR: Salt responses, SRR: Salt responsive receptors.
The H2O2 production in PAs-depleted cells, affect the PM ion transport in a ROS-dependent manner and activate Ca2+ signaling. Salt stress-induced decrease in the activity of the PM and vacuolar H+ pumps can be substantially retained by exogenous application of PAs (Figure 3).149 Spm-deficient mutant plants have constitutively active CAX channels implying negative role of PAs on Ca2+ current in vacuolar channels.150
Ca2+ influx across the PM plays a central role in ROS signaling. Free oxygen radicals like H2O2 and •OH can activate hyperpolarization of Ca2+-permeable channels. •OH is also able to activate both outward K+ current and voltage-dependent conductance (ROSIC). Unexpectedly, PAs potentiates •OH-induced K+ efflux, as well as ROSIC in isolated protoplasts. Thus, it is conceivable that adsorption of PAs may sensitize ROSIC for activation by •OH. ROS and PAs suppress the activity of some constitutively expressed K+ channels. In prolonged stress condition elevated cytosolic Ca2+ level may cause detrimental effect on the normal cell metabolism. Therefore, active Ca2+ efflux systems play a prominent role in maintaining basal Ca2+ content in the cell.150–152 PAs activate Ca2+ efflux systems, such as PM and endomembrane Ca2+-ATPase (PMCA) pumps and exchangers, thus balancing Ca2+ cytosolic levels (Figure 3).
These data are well in line with the idea that the PAs metabolic or catabolic pathways are joined to Ca2+ signaling with potential to provide salt stress responses in plants, although precise regulation mechanism remains elusive.
NO/Ca2+ regulated responses to salinity
NO has emerged as a potent signaling molecule in plant cells with various protective functions against abiotic stresses.153 This membrane-permeable free radical has also been associated with a wide range of physiological processes, such as root hair growth,154 germination155 stomatal movements,156 flowering,157 root nodulation158 and pollen tube growth.159 Dual effects of NO have been well defined: high cellular concentrations triggers cellular damage due to the generation of the nitro-oxidative pool while at low content it contributes to different redox-regulated gene expression and establishment of plant stress tolerance.160 An increasing number of studies report the modulation of NO metabolism during different abiotic stress conditions, such as high light intensity,161 low162 and high temperature,163 mechanical wounding164 and salinity.165,166 Arginine-dependent NO production by NO synthase (NOS) enzyme is the main pathway for NO synthesis in animals.167 A protein displaying NOS activity namely AtNOS1 has also been identified in A. thaliana (Figure 3).168 Considering the fact that NO is a molecule related to PAs by a common precursor of l–arginine, PAs like Spm and Spd have also been suggested to trigger NO production in plant (Figure 3).
Direct and indirect interactions with various signal transduction components, such as protein kinases, secondary messengers (e.g. Ca2+, cGMPs, phosphatidic acid and ROS), and phytohormones have also been proposed for NO.169–171 The function of NO in the signaling network might be mediated by mobilization of intracellular Ca2+ or interaction with Ca2+ channels,172 NO signaling has been shown to induce Ca2+ signaling in plants (Figure 3).173 This implies a feedback loop underlying Ca2+ and NO signaling (at least in part) are entwined. During salt stress, NO-induced elevated cytosolic Ca2+ levels modulate the activity of protein kinases, such as CDPKs and MAPKs.174,175 The primary targets of NO in plant cells might include MAPK. MAPKs can be activated in response to extracellular signals such as osmotic stress that cause the activation of signal transduction pathways resulting in altered gene expression (Figure 3).176 NO has been associated with ion transport and regulation of ion channels in mammalian tissues while in plant direct evidence of regulation of ion channel by NO comes mainly from electrophysiological studies in guard cells.156,177 ABA-induced stomatal closure via cytosolic Ca2+ has been implied to be associated with NO boost in the guard cells.115,178
Ca2+ channels are affected by NO through direct involvement with S-nitrosylation (binding of NO to the cysteine residue) or indirectly by effect on cyclic GMP (cGMP) and cyclic ADP ribose (cADPR) functionality to release Ca2+ from intracellular stores (Figure 3). Lamotte et al. (2006) emphasized the role of NO in activation of both PM and intracellular Ca2+-permeable channels via signaling cascades. In this way, PM depolarization, cADPR and protein kinases activity by NO was accompanied with Ca2+ reporter apoaequorin activation in Nicotiana plumbaginifolia subjected to hyperosmotic stress.179 Elevated levels of cytosolic Ca2+ elicits downstream physiological responses to a given signal and enhance/maintain NO generation. NO biosynthesis driven by NOS is regulated by plant-NOS activity on Ca2+ and CaM signaling basis (Figure 3).180,181
NO is known to be involved in plants tolerance to salt stress by improving antioxidative defense system, osmolyte accumulation and ionic homeostasis.182 The antioxidant role of NO during establishment of salinity tolerance is mainly due to the maintenance of cellular redox homeostasis and boosting of antioxidant metabolites, e.g. ascorbate, reduced glutathione, and osmolytes, e.g. proline and soluble sugar.183 Moreover, NO possesses antioxidant properties and might function as a signal to enhance the activity of H+-ATPase in PM and proton-pomp in tonoplast.184–186
An positive effect of exogenous NO on oxidative metabolism has been reported in NaCl-treated chickpea plants.187 Hasanuzzaman et al. (2011) demonstrated that ROS detoxification in Triticum aestivum under saline condition can be modulated by exogenous application of NO.188 Under salt stress NO maintain a high level of K+ ion, which is accompanied with low Na+ content in the cytosol by mediation of PM channels, such as H+-ATPase.189 Consistently, investigations conducted on salt-treated callus cell cultures showed that NO regulates PM-H+-ATPase activity, thus elevating the K+/Na+ ratio, leading to salt acclimation.190,191
These findings forged ahead our current understanding on the retro-controlling mechanism in response to salt stress powered by a system with three components involving salt perception and Ca2+/NO signaling. However, the metabolic route/s involved in this complex chain is still baffling and further studies are required to unravel the precise mechanism/s underlying salt-Ca2+-NO orchestration.
Osmoprotectant responses to salt and Ca2+ signaling
Various physiological and biochemical adaptive strategies have been developed in plants to minimize the detrimental effects of stress events, such as salinity. For instance, plants synthesize and accumulate compatible soluble substrates or osmoprotectants that are implicated to grow and develop under such conditions.192 Osmoprotectants are highly soluble organic compounds with low molecular weight and naturally safe at high concentrations.193 As evident from a series of in vivo and in vitro investigations on physiology, biochemistry, genetics and molecular biology of plants, osmoprotectants potentially provide adaptive steady status under various unfavorable conditions, such as drought,194 salinity,195 temperature196 and heavy metal stresses.197 Compatible solutes are integrated in adjustment of the plants turgor pressure against salinity stress. Reduction of the cytoplasm osmotic potential driven by compatible solutes permits the maintenance of the cell water content and retention of turgor pressure under stress conditions.198 In fact, osmotic adjustment as a fast acting adaptive response allows the plant to regulate ion homeostasis and maintain their turgor pressure and cell volumes under salinity shock199
The major osmolytes that are primarily synthesized and accumulated in the cytoplasm in response to abiotic stress can be characterized in three classes comprising (i) ammonium compounds such as PAs, glycinebetaine, b-alanine betaine, dimethyl-sulfonio propionate and choline-sulfate, (ii) sugars and sugar alcohols namely fructan, mannitol, D-ononitol and sorbitol and (iii) amino acids so-called proline and glycine betaine.200 Among them, proline, glycine betaine and mannitol are the most prominent osmoprotectants. Proline, an α-amino acid derivative compound, is the main and widely distributed compatible solutes with high capacity in maintaining osmotic balance in the cell under saline condition.201 Proline accumulates in response to saline stress and contributes to the alleviation of cytoplasmic acidosis and maintaining of the NADP/NADPH balance to conduct the natural metabolism.202 Additionally, the strong conformational rigidity of proline structure in retaining proteins stability and membrane structure when is paralleled with its integration to ROS scavenging makes it a vital key candidate for growth and survival under osmotic stress. Szabados and Savoure (2010) demonstrated that proline functions as a molecular chaperone capable of maintaining protein integrity with potential to enhance the activity of various enzymes attributed to the ability of this compound to form hydro-phil/phobic features dealing with various proteins.201 Investigations on the accumulation of proline in seven rice (Oryza sativa L.) cultivars showed that increased proline concentration progressively correlates with increased salt stress tolerance.203 Although, several lines of evidence indicate that under various stress circumstances, elevated levels of proline assume a protective role, excessive levels of proline are toxic due to its effect on increasing ROS production (Figure 3). ROS production is mediated by Ca2+/CaM signaling. Ca2+/CaM deals with the ROS in two divergent mechanisms. On one hand a feedback loop activates NAD(H) kinase and provides NADP(H) as primary substrate for ROS; on the other hand it induces the antioxidant enzymes activity and mitigates ROS accumulation.204 Ca2+ has also been proposed to stimulate antioxidant enzyme activity and maintain cellular redox potential in rice seedling under saline stress condition.205 This cross link between proline, Ca2+ and salt stress indicates that collapsed ion hemostasis derived by salt stress is partially ruled by Ca2+ signaling. However, knowledge about Ca2+ interrelation with diverse osmolytes is scant and requires further research to formulate the specific metabolic pathway.
Conclusion
To date, prevalent research has been conducted on salinity stress in plants and as a result diverse metabolic pathways have been elucidated, leading to the identification of important mechanisms for induction of salt tolerance in plants. Meanwhile, parallel investigations into Ca2+ signaling have revealed diverse roles for Ca2+ in the context of salt stress as well. Despite of these findings, the multifaceted role of Ca2+ in plant stress responses has been underestimated. In this review, the authors attempted to demonstrate the comprehensive effects of Ca2+, in terms of both quantity and signaling, in various aspects of plant responses to salt stress. Accordingly, the effect of Ca2+ in salinity begins from the initial stage of salinity perception, activating a signal transduction pathway to force Na+. Ca2+ dynamically affects diverse Na+ receptors and modulates salt-derived downstream signaling pathways. Consequently, Ca2+ can be considered as one of the most versatile and important molecules in the response to salinity. Due to the prominent role of Ca2+ in salinity stress, better understanding of Ca2+ signaling pathway can pave the way for release of salt-tolerant crops. This addresses the Ca2+ signaling as an imperative field for future research.
Funding Statement
Financial supports from Iran National Science Foundation (INSF) Grant number 97007583 to Dr. Maryam Seifikalhor and Grant number 96006991 to Dr. Sasan Aliniaeifard are gratefully acknowledged.
Acknowledgments
Authors want to sincerely thank Massimo Bosacchi from KWS Gateway Research Center for critical revising of the manuscript.
References
- 1.Negrão S, Schmöckel S, Tester M.. Evaluating physiological responses of plants to salinity stress. Ann. Bot. 2017;119:1–15. doi: 10.1093/aob/mcw191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qadir M, Quillérou E, Nangia V, Murtaza G, Singh M, Thomas RJ, Drechsel P, Noble AD. Economics of salt‐induced land degradation and restoration. Nat Resour Forum. 2014;38:282–295. doi:10.1111/1477-8947.12054. [Google Scholar]
- 3.Valenzuela CE, Acevedo-Acevedo O, Miranda GS, Vergara-Barros P, Holuigue L, Figueroa CR, Figueroa PM. Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. J Exp Bot. 2016;67:4209–4220. doi: 10.1093/jxb/erw202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J-K. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu P, Magwanga R, Lu H, Kirungu J, Wei Y, Dong Q, Wang X, Cai X, Zhou Z, Wang K, et al. A novel G-protein-coupled receptors gene from upland cotton enhances salt stress tolerance in transgenic Arabidopsis. Genes. 2018;9:209. doi: 10.3390/genes9040209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bano A, Fatima M. Salt tolerance in zea mays (L). following inoculation with rhizobium and pseudomonas. Biol Fertil Soils. 2009;45:405–413. doi: 10.1007/s00374-008-0344-9. [DOI] [Google Scholar]
- 7.Weimers K. Growth and phosphorus uptake of potato (Solanum tuberosum L.) in an alkaline soil as affected by mineral nitrogen forms and inoculation with phosphate-solubilizing bacteria and mycorrhizal fungi, SLU, Swedish University of Agricultural Sciences Faculty of Landscape Architecture, Horticulture and Crop Production Science, Department of Biosystems and Technology. 2017. [Google Scholar]
- 8.Keisham M, Mukherjee S, Bhatla S. Mechanisms of sodium transport in plants—progresses and challenges. Int J Mol Sci. 2018;19:647. doi: 10.3390/ijms19030647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pottosin I, Dobrovinskaya O. Non-selective cation channels in plasma and vacuolar membranes and their contribution to K+ transport. J Plant Physiol. 2014;171:732–742. doi: 10.1016/j.jplph.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Zhang W-D, Wang P, Bao Z, Ma Q, Duan L-J, Bao A-K, Zhang JL, Wang SM.. SOS1, HKT1; 5, and NHX1 synergistically modulate Na+ homeostasis in the halophytic grass Puccinellia tenuiflora. Front Plant Sci. 2017;8:576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Lu X, Shu N, Wang D, Wang S, Wang J, Guo L, Guo X, Fan W, Lin Z, et al. GhSOS1, a plasma membrane Na+/H+ antiporter gene from upland cotton, enhances salt tolerance in transgenic Arabidopsis thaliana. PLoS One. 2017;12:e0181450. doi: 10.1371/journal.pone.0181450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardo JM, Rubio F. Na+ and K+ transporters in plant signaling. Transporters and pumps in plant signaling. Berlin, Heidelberg: Springer; 2011. p. 65–98. doi: 10.1007/978-3-642-14369-4_3. [DOI] [Google Scholar]
- 13.Teakle NL, Tyerman SD. Mechanisms of Cl− transport contributing to salt tolerance. Plant Cell Environ. 2010;33:566–589. doi: 10.1111/j.1365-3040.2009.02060.x. [DOI] [PubMed] [Google Scholar]
- 14.Barbier-Brygoo H, De Angeli A, Filleur S, Frachisse J-M, Gambale F, Thomine S, Wege S. Anion channels/transporters in plants: from molecular bases to regulatory networks. Annu. Rev. Plant Biol. 2011;62:25–51. doi: 10.1146/annurev-arplant-042110-103741. [DOI] [PubMed] [Google Scholar]
- 15.Shabala S. Learning from halophytes: physiological basis and strategies to improve abiotic stress tolerance in crops. Ann. Bot. 2013;112:1209–1221. doi: 10.1093/aob/mct205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wegner LH. Root pressure and beyond: energetically uphill water transport into xylem vessels? J Exp Bot. 2013;65:381–393. doi: 10.1093/jxb/ert391. [DOI] [PubMed] [Google Scholar]
- 17.Fricke W. The significance of water co-transport for sustaining transpirational water flow in plants: a quantitative approach. J Exp Bot. 2015;66:731–739. doi: 10.1093/jxb/eru466. [DOI] [PubMed] [Google Scholar]
- 18.Henderson SW, Wege S, Qiu J, Blackmore DH, Walker AR, Tyerman SD, Walker RR, Gilliham M. Grapevine and Arabidopsis cation-chloride cotransporters localize to the golgi and trans-golgi network and indirectly influence long-distance ion transport and plant salt tolerance. Plant Physiol. 2015;169:2215–2229. doi: 10.1104/pp.15.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao -J-J, Chen H-W, Ma B, Zhang W-K, Chen S-Y, Zhang J-S. The role of ethylene in plants under salinity stress. Front Plant Sci. 2015;6:1059. doi: 10.3389/fpls.2015.01059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamiyoshihara Y, Iwata M, Fukaya T, Tatsuki M, Mori H. Turnover of LeACS2, a wound‐inducible 1‐aminocyclopropane‐1‐carboxylic acid synthase in tomato, is regulated by phosphorylation/dephosphorylation. Plant J. 2010;64:140–150. doi: 10.1111/j.1365-313X.2010.04316.x. [DOI] [PubMed] [Google Scholar]
- 21.Liang W, Ma X, Wan P, Liu L. Plant salt-tolerance mechanism: a review. Biochem Biophys Res Commun. 2018;495:286–291. doi: 10.1016/j.bbrc.2017.11.043. [DOI] [PubMed] [Google Scholar]
- 22.Aldon D, Mbengue M, Mazars C, Galaud J-P. Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 2018;19:665. doi: 10.3390/ijms19030665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang M, Smith JAC, Harberd NP, Jiang C. The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol Biol. 2016;91:651–659. doi: 10.1007/s11103-016-0488-1. [DOI] [PubMed] [Google Scholar]
- 24.Verma V, Ravindran P, Kumar PP. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016;16:86. doi: 10.1186/s12870-016-0796-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gravino M, Savatin DV, Macone A, De Lorenzo G. Ethylene production in B otrytis cinerea‐and oligogalacturonide‐induced immunity requires calcium‐dependent protein kinases. Plant J. 2015;84:1073–1086. doi: 10.1111/tpj.13057. [DOI] [PubMed] [Google Scholar]
- 26.Lee HY, Back K. Mitogen‐activated protein kinase pathways are required for melatonin‐mediated defense responses in plants. J Pineal Res. 2016;60:327–335. doi: 10.1111/jpi.12314. [DOI] [PubMed] [Google Scholar]
- 27.Schmöckel SM, Garcia AF, Berger B, Tester M, Webb AA, Roy SJ. Different NaCl-induced calcium signatures in the Arabidopsis thaliana ecotypes Col-0 and C24. PLoS One. 2015;10:e0117564. doi: 10.1371/journal.pone.0117564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q, Liu M-C, Maman J, Steinhorst L, Schmitz-Thom I, et al. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr Biol. 2018;28:666–75. e5. doi: 10.1016/j.cub.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu H. Plant salt tolerance and Na+ sensing and transport. Crop J. 2018;6:215–225. doi: 10.1016/j.cj.2018.01.003. [DOI] [Google Scholar]
- 30.Zörb C, Mühling KH, Kutschera U, Geilfus C-M. Salinity stiffens the epidermal cell walls of salt-stressed maize leaves: is the epidermis growth-restricting? PLoS One. 2015;10:e0118406. doi: 10.1371/journal.pone.0118406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geilfus C-M. Expansin expression and apoplastic pH in expanding leaves under NaCl stress. Christian-Albrechts Universität Kiel, Germany; 2011. [Google Scholar]
- 32.Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- 33.Okubo-Kurihara E, Ohtani M, Kurihara Y, Kakegawa K, Kobayashi M, Nagata N, Komatsu T, Kikuchi J, Cutler S, Demura T, et al. Modification of plant cell wall structure accompanied by enhancement of saccharification efficiency using a chemical, lasalocid sodium. Sci Rep. 2016;6:34602. doi: 10.1038/srep34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tran L-SP, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA 2007; 104:20623–20628. 10.1073/pnas.0706547105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu J-K. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6:441–445. [DOI] [PubMed] [Google Scholar]
- 36.Marin K, Suzuki I, Yamaguchi K, Ribbeck K, Yamamoto H, Kanesaki Y, Hagemann M, Murata N. Identification of histidine kinases that act as sensors in the perception of salt stress in synechocystis sp. PCC 6803. Proc Natl Acad Sci USA. 2003;100:9061–9066. https://www.scimagojr.com/journalsearch.php?q=21121&tip=sid. [DOI] [PMC free article] [PubMed]
- 37.Jin Y, Jing W, Zhang Q, Zhang W. Cyclic nucleotide gated channel 10 negatively regulates salt tolerance by mediating Na+ transport in Arabidopsis. J Plant Res. 2015;128:211–220. doi: 10.1007/s10265-014-0679-2. [DOI] [PubMed] [Google Scholar]
- 38.González-Fontes A, Navarro-Gochicoa MT, Ceacero CJ, Herrera-Rodríguez MB, Camacho-Cristóbal JJ, Rexach J. Understanding calcium transport and signaling, and its use efficiency in vascular plants. Plant Macronutrient Use Efficiency: Elsevier; 2017. p. 165–180. doi: 10.1016/B978-0-12-811308-0.00009-0 [DOI] [Google Scholar]
- 39.Finka A, Cuendet AFH, Maathuis FJ, Saidi Y, Goloubinoff P. Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell. 2012;24:3333–3348. doi: 10.1105/tpc.112.095844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer C, DeFalco TA, Karia P, Snedden WA, Moeder W, Yoshioka K, Dietrich P. Calmodulin as a Ca2+-sensing subunit of Arabidopsis cyclic nucleotide-gated channel complexes. Plant Cell Physiol. 2017;58:1208–1221. doi: 10.1093/pcp/pcx052. [DOI] [PubMed] [Google Scholar]
- 41.Wang P, Li Z, Wei J, Zhao Z, Sun D, Cui S. A Na+/Ca2+ exchanger-like protein (AtNCL) involved in salt stress in Arabidopsis. J Biol Chem. 2012;287:44062–44070. doi: 10.1074/jbc.M112.351643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shabala S, Wu H, Bose J. Salt stress sensing and early signalling events in plant roots: current knowledge and hypothesis. Plant Sci. 2015;241:109–119. doi: 10.1016/j.plantsci.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Li P, Zhang G, Gonzales N, Guo Y, Hu H, Park S, Zhao J. Ca2+‐regulated and diurnal rhythm‐regulated Na+/Ca2+ exchanger AtNCL affects flowering time and auxin signalling in Arabidopsis. Plant Cell Environ. 2016;39:377–392. doi: 10.1111/pce.v39.2. [DOI] [PubMed] [Google Scholar]
- 44.Huang WY, Alvarez S, Lee YK, Kondo Y, Chung JK, Lam HYM, Kuriyan J, Groves JT. Molecular timing of membrane signaling reactions. Biophys J. 2018;114:202a. doi: 10.1016/j.bpj.2017.11.1130. [DOI] [Google Scholar]
- 45.Ji H, Pardo JM, Batelli G, Van Oosten MJ, Bressan RA, Li X. The Salt Overly Sensitive (SOS) pathway: established and emerging roles. Mol Plant. 2013;6:275–286. doi: 10.1093/mp/sst017. [DOI] [PubMed] [Google Scholar]
- 46.Julkowska MM, Testerink C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 2015;20:586–594. doi: 10.1016/j.tplants.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 47.Quan R, Wang J, Yang D, Zhang H, Zhang Z, Huang R. EIN3 and SOS2 synergistically modulate plant salt tolerance. Sci Rep. 2017;7:44637. doi: 10.1038/srep44637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cramer GR. Sodium-calcium interactions under salinity stress. Salinity: environment-plants-molecules.Dordrecht: Springer; 2002. p. 205–227. doi:10.1007/0-306-48155-3_10. [Google Scholar]
- 49.Lynch J, Polito VS, Läuchli AJPP. Salinity stress increases cytoplasmic Ca activity in maize root protoplasts. Plant Physiol. 1989;90:1271–1274. doi: 10.1104/pp.90.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knight H, Trewavas AJ, Knight MRJTPC. Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant J. 1996;8:489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinhorst L, Kudla J. Signaling in cells and organisms—calcium holds the line. Curr Opin Plant Biol. 2014;22:14–21. doi: 10.1016/j.pbi.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 52.Boudsocq M, Sheen J. Stress signaling II: calcium sensing and signaling. Abiotic Stress Adaptation in Plants.Dordrecht: Springer; 2009. p. 75–90. doi:10.1007/978-90-481-3112-9_4. [Google Scholar]
- 53.Wan S, Wang W, Zhou T, Zhang Y, Chen J, Xiao B, Yang Y, Yu Y. Transcriptomic analysis reveals the molecular mechanisms of Camellia sinensis in response to salt stress. Plant Growth Regul. 2018;84:481–492. doi: 10.1007/s10725-017-0354-4. [DOI] [Google Scholar]
- 54.Fasani E, DalCorso G, Costa A, Zenoni S, Furini AJPMB. The Arabidopsis thaliana transcription factor MYB59 regulates calcium signalling during plant growth and stress response. Plant Mol Biol. 2019;99:517–534. [DOI] [PubMed] [Google Scholar]
- 55.Saidi Y, Finka A, Muriset M, Bromberg Z, Weiss YG, Maathuis FJ, Goloubinoff P. The heat shock response in moss plants is regulated by specific calcium-permeable channels in the plasma membrane. Plant Cell. 2009;21:2829–2843. doi: 10.1105/tpc.108.065318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stephan AB, Kunz -H-H, Yang E, Schroeder J. Rapid hyperosmotic-induced Ca2+ responses in Arabidopsis thaliana exhibit sensory potentiation and involvement of plastidial KEA transporters. Proc Natl Acad Sci U S A. 2016;113:E5242–E9. doi: 10.1073/pnas.1519555113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilkins KA, Matthus E, Swarbreck SM, Davies J. Calcium-mediated abiotic stress signaling in roots. Front Plant Sci. 2016;7:1296. doi: 10.3389/fpls.2016.01296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knight H, Trewavas AJ, Knight MRJTPJ. Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 1997;12:1067–1078. doi: 10.1046/j.1365-313x.1997.12051067.x. [DOI] [PubMed] [Google Scholar]
- 59.Tracy FE, Gilliham M, Dodd AN, Webb AA, Tester MJP. cell, environment. NaCl‐induced changes in cytosolic free Ca2+ in Arabidopsis thaliana are heterogeneous and modified by external ionic composition. Plant Cell Environ. 2008;31:1063–1073. doi: 10.1111/j.1365-3040.2008.01817.x. [DOI] [PubMed] [Google Scholar]
- 60.Miseta A, Kellermayer R, Aiello DP, Fu L, Bedwell D. The vacuolar Ca2+/H+ exchanger Vcx1p/Hum1p tightly controls cytosolic Ca2+ levels in S. cerevisiae. FEBS Lett. 1999;451:132–136. doi: 10.1016/s0014-5793(99)00519-0. [DOI] [PubMed] [Google Scholar]
- 61.Chakraborty K, Basak N, Bhaduri D, Ray S, Vijayan J, Chattopadhyay K, Sarkar RK.. Ionic basis of salt tolerance in plants: nutrient homeostasis and oxidative stress tolerance. Plant Nutrients and Abiotic Stress Tolerance.Singapore: Springer; 2018. p. 325–362. doi: 10.1007/978-981-10-9044-8_14. [DOI] [Google Scholar]
- 62.Sze H, Liang F, Hwang I, Curran AC, Harper J. Diversity and regulation of plant Ca2+ pumps: insights from expression in yeast. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:433–462. doi: 10.1146/annurev.arplant.51.1.433. [DOI] [PubMed] [Google Scholar]
- 63.Yokoi S, Bressan RA, Hasegawa P. Salt stress tolerance of plants. JIRCAS. 2002;23:25–33. [Google Scholar]
- 64.Munns R, James RA, Läuchli A. Approaches to increasing the salt tolerance of wheat and other cereals. J Exp Bot. 2006;57:1025–1043. doi: 10.1093/jxb/erj100. [DOI] [PubMed] [Google Scholar]
- 65.Flowers T, Koyama M, Flowers S, Sudhakar C, Singh K, Yeo A. QTL: their place in engineering tolerance of rice to salinity. J Exp Bot. 2000;51:99–106. [PubMed] [Google Scholar]
- 66.Luan S, Lan W, Lee S. Potassium nutrition, sodium toxicity, and calcium signaling: connections through the CBL–CIPK network. Curr Opin Cell Biol. 2009. Vol. 12 p. 339–346. [DOI] [PubMed] [Google Scholar]
- 67.Tuteja N. Mechanisms of high salinity tolerance in plants. Methods in enzymology. Elsevier; 2007. Vol.428. p. 419–438. doi:10.1016/S0076-6879(07)28024-3. [DOI] [PubMed] [Google Scholar]
- 68.Nikalje CG, Nikam DT, Suprasanna P. Looking at halophytic adaptation to high salinity through genomics landscape. Curr Genomics. 2017;18:542–552. doi: 10.2174/1389202918666170228143007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuan F, Yang H, Xue Y, Kong D, Ye R, Li C, Zhang J, Theprungsirikul L, Shrift T, Krichilsky B, Johnson DM.. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature. 2014;514:367–371. [DOI] [PubMed] [Google Scholar]
- 70.Chinnusamy V, Jagendorf A, Zhu J-KJCS. Understanding and improving salt tolerance in plants. Crop Sci. 2005;45:437–448. doi: 10.2135/cropsci2005.0437. [DOI] [Google Scholar]
- 71.Lv S, Jiang P, Tai F, Wang D, Feng J, Fan P, Bao H, Li Y. The V-ATPase subunit A is essential for salt tolerance through participating in vacuolar Na+ compartmentalization in Salicornia europaea. Planta. 2017;246:1177–1187. doi: 10.1007/s00425-017-2762-0. [DOI] [PubMed] [Google Scholar]
- 72.Choi W-G, Toyota M, Kim S-H, Hilleary R, Gilroy S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc Natl Acad Sci. 2014;111:6497–6502. doi: 10.1073/pnas.1319955111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ben-Johny M, Dick EI, Sang L, Limpitikul BW, Wei KP, Niu J, Banerjee R, Yang W, Babich JS, Issa JB, et al. Towards a unified theory of calmodulin regulation (calmodulation) of voltage-gated calcium and sodium channels. Curr Mol Pharmacol. 2015;8:188–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu Q, An L, Li W. The CBL–CIPK network mediates different signaling pathways in plants. Plant Cell Rep. 2014;33:203–214. doi: 10.1007/s00299-013-1507-1. [DOI] [PubMed] [Google Scholar]
- 75.Lan W-Z, Wang W, Wang S-M, Li L-G, Buchanan BB, Lin H-X, Gao JP, Luan S.. A rice high-affinity potassium transporter (HKT) conceals a calcium-permeable cation channel. Proc Natl Acad Sci USA. 2010;107 (15):7089–7094. doi: 10.1073/pnas.1000698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schachtman DP, Schroeder JIJN. Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature. 1994;370:655. doi: 10.1038/370655a0. [DOI] [PubMed] [Google Scholar]
- 77.Wang TT, Ren ZJ, Liu ZQ, Feng X, Guo RQ, Li BG, Li L-G, Jing H-C. SbHKT1; 4, a member of the high‐affinity potassium transporter gene family from Sorghum bicolor, functions to maintain optimal Na+/K+ balance under Na+ stress. J Integr Plant Biol. 2014;56:315–332. doi: 10.1111/jipb.12144. [DOI] [PubMed] [Google Scholar]
- 78.Amtmann A, Sanders D. Mechanisms of Na+ uptake by plant cells Advances in botanical research. 1998;29:75–112. doi: 10.1016/S0065-2296(08)60310-9. [DOI] [Google Scholar]
- 79.Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem WJTPC. Calmodulins and calcineurin B–like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell. 2002;14:S389–S400. doi: 10.1105/tpc.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang X, Wang T, Liu M, Sun W, W-HJE Z, Botany E. Calmodulin-like gene MtCML40 is involved in salt tolerance by regulating MtHKTs transporters in medicago truncatula. Environ Exp Bot. 2019;157:79–90. doi: 10.1016/j.envexpbot.2018.09.022. [DOI] [Google Scholar]
- 81.Huang Y, Guan C, Liu Y, Chen B, Yuan S, Cui X, Zhang Y, Yang F.. Enhanced growth performance and salinity tolerance in transgenic switchgrass via overexpressing vacuolar Na+ (K+)/H+ antiporter gene (PvNHX1). Front Plant Sci. 2017;8:458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li N, Wang X, Ma B, Du C, Zheng L, Wang Y. Expression of a Na+/H+ antiporter RtNHX1 from a recretohalophyte reaumuria trigyna improved salt tolerance of transgenic Arabidopsis thaliana. J Plant Physiol. 2017;218:109–120. doi: 10.1016/j.jplph.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 83.Khan MS, Ahmad D, Khan MA. Trends in genetic engineering of plants with (Na+/H+) antiporters for salt stress tolerance. Biotechnol Biotechnol Equip. 2015;29:815–825. doi: 10.1080/13102818.2015.1060868. [DOI] [Google Scholar]
- 84.Bassil E, Ohto M-A, Esumi T, Tajima H, Zhu Z, Cagnac O, Belmonte M, Peleg Z, Yamaguchi T, Blumwald E.. The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell. 2011, 23(1):224–239 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hasegawa PM, Bressan RA, Zhu J-K, Bohnert H. Plant cellular and molecular responses to high salinity. Plant Physiol Plant Mol Biol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- 86.Munns R, Tester MJARPB. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 87.Epstein EJPP. The essential role of calcium in selective cation transport by plant cells. Plant Physiol. 1961;36:437. doi: 10.1104/pp.36.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kramer G, Lynch J, Epstein EJPP. Influx of Na, K and Ca into roots of saltstressed cotton seedlings. Plant Physiol. 1987;83:510–516. doi: 10.1104/pp.83.3.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu J, Zhu J-KJS. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- 90.Liu J, Ishitani M, Halfter U, Kim C-S, Zhu J-K. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA. 2000. 97:3730–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shi H, Ishitani M, Kim C, Zhu J-K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matsumoto TK, Ellsmore AJ, Cessna SG, Low PS, Pardo JM, Bressan RA, Hasegawa PM. An osmotically induced cytosolic Ca2+ transient activates calcineurin signaling to mediate ion homeostasis and salt tolerance of Saccharomyces cerevisiae. J Biol Chem. 2002;277:33075–33080. doi: 10.1074/jbc.M205037200. [DOI] [PubMed] [Google Scholar]
- 93.Mansouri M, Naghavi MR, Alizadeh H, Mohammadi-Nejad G, Mousavi SA, Salekdeh GH, Tada Y. Transcriptomic analysis of aegilops tauschii during long-term salinity stress. Funct Integr Genomics. 2019;19:13–28. doi: 10.1007/s10142-018-0623-y. [DOI] [PubMed] [Google Scholar]
- 94.Batistic O, Kudla JJP. Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta. 2004;219:915–924. doi: 10.1007/s00425-004-1333-3. [DOI] [PubMed] [Google Scholar]
- 95.Zhao X, Wei P, Liu Z, Yu B, Shi H. Soybean Na+/H+ antiporter GmsSOS1 enhances antioxidant enzyme activity and reduces Na+ accumulation in Arabidopsis and yeast cells under salt stress. Acta Physiol Plant. 2017;39:19. doi: 10.1007/s11738-016-2323-3. [DOI] [Google Scholar]
- 96.Feki K, Tounsi S, Masmoudi K, Brini F. The durum wheat plasma membrane Na+/H+ antiporter SOS1 is involved in oxidative stress response. Protoplasma. 2017;254:1725–1734. doi: 10.1007/s00709-016-1066-8. [DOI] [PubMed] [Google Scholar]
- 97.Kumari PH, Kumar SA, Sivan P, Katam R, Suravajhala P, Rao K, Varshney RK, Kishor PBK. Overexpression of a plasma membrane bound Na+/H+ antiporter-like protein (SbNHXLP) confers salt tolerance and improves fruit yield in tomato by maintaining ion homeostasis. Front Plant Sci. 2017;7:2027. doi: 10.3389/fpls.2016.02027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qiu Q-S, Guo Y, Dietrich MA, Schumaker KS, Zhu J-K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci. 2002;99:8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cheng N-H, Pittman JK, Zhu J-K, Hirschi K. The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J Biol Chem. 2004;279:2922–2926. doi: 10.1074/jbc.M309084200. [DOI] [PubMed] [Google Scholar]
- 100.Zhu J-K. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu J-K. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6:441–445. [DOI] [PubMed] [Google Scholar]
- 102.Türkan I, Demiral TJE, Botany E. Recent developments in understanding salinity tolerance. Environmental and Experimental Botany 2009;67:2–9. [Google Scholar]
- 103.Hu DG, Ma QJ, Sun CH, Sun MH, You CX, Hao YJ. Overexpression of MdSOS2L1, a CIPK protein kinase, increases the antioxidant metabolites to enhance salt tolerance in apple and tomato. Physiol Plant. 2016;156:201–214. doi: 10.1111/ppl.12354. [DOI] [PubMed] [Google Scholar]
- 104.Kim BG, Waadt R, Cheong YH, Pandey GK, Dominguez‐Solis JR, Schültke S, SC Lee, Kudla J, Luan S.. The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 2007;52:473–484. doi: 10.1111/j.1365-313X.2007.03249.x. [DOI] [PubMed] [Google Scholar]
- 105.Albrecht V, Weinl S, Blazevic D, D’angelo C, Batistic O, Kolukisaoglu Ü, Bock R, Schulz B, Harter K, Kudla J. The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J. 2003;36:457–470. doi: 10.1046/j.1365-313x.2003.01892.x. [DOI] [PubMed] [Google Scholar]
- 106.Cheong YH, Kim K-N, Pandey GK, Gupta R, Grant JJ, Luan SJTPC. CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell. 2003;15:1833–1845. doi: 10.1105/tpc.012393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pandey GK, Cheong YH, Kim K-N, Grant JJ, Li L, Hung W, D’Angelo C, Weinl S, Kudla J, Luan S. The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell. 2004;16:1912–1924. doi: 10.1105/tpc.021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Apse MP, Aharon GS, Snedden WA, Blumwald EJS. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 109.Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell. 2007;19:1415–1431. doi: 10.1105/tpc.106.042291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lin H, Yang Y, Quan R, Mendoza I, Wu Y, Du W, Zhao S, Schumaker KS, Pardo JM, Guo Y. Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell. 2009;21:1607–1619. doi: 10.1105/tpc.109.066217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ren XL, Qi GN, Feng HQ, Zhao S, Zhao SS, Wang Y, Wu W-H. Calcineurin B‐like protein CBL 10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J. 2013;74:258–266. doi: 10.1111/tpj.12123. [DOI] [PubMed] [Google Scholar]
- 112.Pandey N, Ranjan A, Pant P, Tripathi RK, Ateek F, Pandey HP, Lo AC, D’Hooge R, Steer CJ, Thibodeau SN, et al. CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genomics. 2013;14:216. doi: 10.1186/1471-2164-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yoo JH, Park CY, Kim JC, Do Heo W, Cheong MS, Park HC, Kim MC, Moon BC, Choi MS, Kang YH, et al. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J Biol Chem. 2005;280:3697–3706. doi: 10.1074/jbc.M408237200. [DOI] [PubMed] [Google Scholar]
- 114.Huang DW, Sherman BT, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 115.Aliniaeifard S, van Meeteren U. Can prolonged exposure to low VPD disturb the ABA signalling in stomatal guard cells? J Exp Bot. 2013;64:3551–3566. doi: 10.1093/jxb/ert192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang W-H, Yi X-Q, Han A-D, Liu T-W, Chen J, Wu F-H, Dong X-J, He J-X, Pei Z-M, Zheng H-L. Calcium-sensing receptor regulates stomatal closure through hydrogen peroxide and nitric oxide in response to extracellular calcium in Arabidopsis. J Exp Bot. 2011;63:177–190. doi: 10.1093/jxb/err259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McAinsh MR, Evans NH, Montgomery LT, North KA. Calcium signalling in stomatal responses to pollutants. New Phytol. 2002;153:441–447. doi: 10.1046/j.0028-646X.2001.00336.x. [DOI] [PubMed] [Google Scholar]
- 118.Monihan SM, Ryu C-H, Magness CA, Schumaker K. Linking duplication of a calcium sensor to salt tolerance in Eutrema salsugineum. Plant Physiol. 2018;2019:01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Aliniaeifard S, Hajilou J, Tabatabaei SJ, Sifi-Kalhor M. Effects of ascorbic acid and reduced glutathione on the alleviation of salinity stress in olive plants. Int J Fruit Sci. 2016;16:395–409. doi: 10.1080/15538362.2015.1137533. [DOI] [Google Scholar]
- 120.Tester M, Davenport R. Na+ tolerance and Na+ transport in higher plants. Ann Bot. 2003;91:503–527. doi: 10.1093/aob/mcg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sathee L, Sairam RK, Chinnusamy V, Jha S. Differential transcript abundance of salt overly sensitive (SOS) pathway genes is a determinant of salinity stress tolerance of wheat. Acta Physiol Plant. 2015;37:169. doi: 10.1007/s11738-015-1910-z. [DOI] [Google Scholar]
- 122.Nakamura T, Liu Y, Hirata D, Namba H, Harada S-I, Hirokawa T, Miyakawa T. Protein phosphatase type 2B (calcineurin)‐mediated, FK506‐sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. Embo J. 1993;12:4063–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mehlmer N, Wurzinger B, Stael S, Hofmann‐Rodrigues D, Csaszar E, Pfister B, Bayer R, Teige M. The Ca(2+</sup>++) -dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 2010;63:484–498. doi: 10.1111/j.1365-313X.2010.04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jiang J, Ma S, Ye N, Jiang M, Cao J, Zhang J. WRKY transcription factors in plant responses to stresses. J Integr Plant Biol. 2017;59:86–101. doi: 10.1111/jipb.12513. [DOI] [PubMed] [Google Scholar]
- 125.Park CY, Lee JH, Yoo JH, Moon BC, Choi MS, Kang YH, Lee SM, Kim HS, Kang KY, Chung WS, et al. WRKY group IId transcription factors interact with calmodulin. FEBS Lett. 2005;579:1545–1550. doi: 10.1016/j.febslet.2005.01.057. [DOI] [PubMed] [Google Scholar]
- 126.Yan H, Jia H, Chen X, Hao L, An H, Guo XJP. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 2014;55:2060–2076. doi: 10.1093/pcp/pcu133. [DOI] [PubMed] [Google Scholar]
- 127.Jiang Y, Deyholos M. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol. 2009;69:91–105. doi: 10.1007/s11103-008-9408-3. [DOI] [PubMed] [Google Scholar]
- 128.Eulgem T, Somssich I. Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol. 2007;10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 129.Fan X-D, Wang J-Q, Yang N, Dong -Y-Y, Liu L, Wang F-W, Wang N, Chen H, Liu W-C, Sun Y-P, et al. Gene expression profiling of soybean leaves and roots under salt, saline–alkali and drought stress by high-throughput illumina sequencing. Gene. 2013;512:392–402. doi: 10.1016/j.gene.2012.09.100. [DOI] [PubMed] [Google Scholar]
- 130.Pathak MR, Teixeira Da Silva JA, Wani SH. Polyamines in response to abiotic stress tolerance through transgenic approaches. GM Crops Food. 2014;5:87–96. doi: 10.4161/gmcr.28774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gupta K, Dey A, Gupta B. Plant polyamines in abiotic stress responses. Acta Physiol Plant. 2013;35:2015–2036. doi: 10.1007/s11738-013-1239-4. [DOI] [Google Scholar]
- 132.Chen D, Shao Q, Yin L, Younis A, Zheng B. Polyamine function in plants: metabolism, regulation on development, and roles in abiotic stress responses. Front Plant Sci. 2018;9:1945. doi: 10.3389/fpls.2018.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gill SS, Tuteja N. Polyamines and abiotic stress tolerance in plants. Plant Signal Behav. 2010;5:26–33. doi: 10.4161/psb.5.1.10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shi H, Chan Z. Improvement of plant abiotic stress tolerance through modulation of the polyamine pathway. J Integr Plant Biol. 2014;56:114–121. doi: 10.1111/jipb.12128. [DOI] [PubMed] [Google Scholar]
- 135.Liu J, Nakajima I, Moriguchi T. Effects of salt and osmotic stresses on free polyamine content and expression of polyamine biosynthetic genes in Vitis vinifera. Biol. Plant. 2011;55:340–344. doi: 10.1007/s10535-011-0050-6. [DOI] [Google Scholar]
- 136.Liu J-H, Nada K, Honda C, Kitashiba H, Wen X-P, Pang X-M, Moriguchi T. Polyamine biosynthesis of apple callus under salt stress: importance of the arginine decarboxylase pathway in stress response. J Exp Bot. 2006;57:2589–2599. doi: 10.1093/jxb/erl018. [DOI] [PubMed] [Google Scholar]
- 137.Do PT, Drechsel O, Heyer AG, Hincha DK, Zuther E. Changes in free polyamine levels, expression of polyamine biosynthesis genes, and performance of rice cultivars under salt stress: a comparison with responses to drought. Front Plant Sci. 2014;5:182. doi: 10.3389/fpls.2014.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Eom SH, Lee JK, Kim D-H, Kim H, Jang K-I, Ryu H, Hyun TK. Identification and expression profiling of flax (Linum usitatissimum L.) polyamine oxidase genes in response to stimuli. Acta Bot Croat. 2018;77:97–101. doi: 10.1515/botcro-2017-0022. [DOI] [Google Scholar]
- 139.Seifikalhor M, Aliniaeifard S, Hassani B, Niknam V, Lastochkina O. Diverse role of γ-aminobutyric acid in dynamic plant cell responses. Plant Cell Rep. 2019;30:847–867. doi:10.1007/s00299-019-02396-z. [DOI] [PubMed] [Google Scholar]
- 140.Marco F, Alcazar R, Tiburcio AF, Carrasco P. Interactions between polyamines and abiotic stress pathway responses unraveled by transcriptome analysis of polyamine overproducers. OMICS. 2011;15:775–781. doi: 10.1089/omi.2011.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kalhor MS, Aliniaeifard S, Seif M, Asayesh EJ, Bernard F, Hassani B, Li T. Title: enhanced salt tolerance and photosynthetic performance: implication of ɤ-amino butyric acid application in salt-exposed lettuce (Lactuca sativa L.) plants. Plant Physiol Biochem. 2018;130:157–172. doi: 10.1016/j.plaphy.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 142.Zhao F, Song C-P, He J, Zhu H. Polyamines improve K+/Na+ homeostasis in barley seedlings by regulating root ion channel activities. Plant Physiol. 2007;145:1061–1072. doi: 10.1104/pp.107.105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Velarde-Buendía AM, Shabala S, Cvikrova M, Dobrovinskaya O, Pottosin I. Salt-sensitive and salt-tolerant barley varieties differ in the extent of potentiation of the ROS-induced K+ efflux by polyamines. Plant Physiol Biochem. 2012;61:18–23. doi: 10.1016/j.plaphy.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 144.Garufi A, Visconti S, Camoni L, Aducci P. Polyamines as physiological regulators of 14-3-3 interaction with the plant plasma membrane H+-ATPase. Plant Cell Physiol. 2007;48:434–440. doi: 10.1093/pcp/pcm010. [DOI] [PubMed] [Google Scholar]
- 145.Saha J, Brauer EK, Sengupta A, Popescu SC, Gupta K, Gupta B. Polyamines as redox homeostasis regulators during salt stress in plants. Front. Environ. Sci. 2015;3:21. doi: 10.3389/fenvs.2015.00021. [DOI] [Google Scholar]
- 146.Shabala S, Cuin TA, Pottosin I. Polyamines prevent NaCl‐induced K+ efflux from pea mesophyll by blocking non‐selective cation channels. FEBS Lett. 2007;581:1993–1999. doi: 10.1016/j.febslet.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 147.Alcázar R, Altabella T, Marco F, Bortolotti C, Reymond M, Koncz C, Carrasco P, Tiburcio AF. Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta. 2010;231:1237–1249. doi: 10.1007/s00425-010-1130-0. [DOI] [PubMed] [Google Scholar]
- 148.Zepeda-Jazo I, Velarde-Buendía AM, Enríquez-Figueroa R, Bose J, Shabala S, Muñiz-Murguía J, Pottosin II. Polyamines interact with hydroxyl radicals in activating Ca(2+++) and K(+) transport across the root epidermal plasma membranes. Plant Physiol. 2011;157:2167–2180. doi: 10.1104/pp.111.179671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pottosin I, Velarde-Buendía AM, Bose J, Fuglsang AT, Shabala S. Polyamines cause plasma membrane depolarization, activate Ca2+-, and modulate H+-ATPase pump activity in pea roots. J Exp Bot. 2014;65:2463–2472. doi: 10.1093/jxb/eru133. [DOI] [PubMed] [Google Scholar]
- 150.Pottosin I, Shabala S. Polyamines control of cation transport across plant membranes: implications for ion homeostasis and abiotic stress signaling. Front Plant Sci. 2014;5:154. doi: 10.3389/fpls.2014.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pottosin I, Velarde-Buendía A-M, Zepeda-Jazo I, Dobrovinskaya O, Shabala S. Synergism between polyamines and ROS in the induction of Ca2+ and K+ fluxes in roots. Plant Signal Behav. 2012;7:1084–1087. doi: 10.4161/psb.21185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pottosin I, Velarde-Buendía AM, Bose J, Zepeda-Jazo I, Shabala S, Dobrovinskaya O. Cross-talk between reactive oxygen species and polyamines in regulation of ion transport across the plasma membrane: implications for plant adaptive responses. J Exp Bot. 2014;65:1271–1283. doi: 10.1093/jxb/ert423. [DOI] [PubMed] [Google Scholar]
- 153.Sami F, Faizan M, Faraz A, Siddiqui H, Yusuf M, Hayat S. Nitric oxide-mediated integrative alterations in plant metabolism to confer abiotic stress tolerance, NO crosstalk with phytohormones and NO-mediated post translational modifications in modulating diverse plant stress. Nitric Oxide. 2018;73:22–38. doi: 10.1016/j.niox.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 154.Lombardo MC, Lamattina L. Abscisic acid and nitric oxide modulate cytoskeleton organization, root hair growth and ectopic hair formation in Arabidopsis. Nitric Oxide. 2018;80:89–97. doi: 10.1016/j.niox.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 155.He J, Ren Y, Chen X, Chen H. Protective roles of nitric oxide on seed germination and seedling growth of rice (Oryza sativa L.) under cadmium stress. Ecotoxicol Environ Saf. 2014;108:114–119. doi: 10.1016/j.ecoenv.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 156.Wang P, Du Y, Hou Y-J, Zhao Y, Hsu -C-C, Yuan F, Zhu X, Tao WA, Song C-P, Zhu J-K. Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc Natl Acad Sci U S A. 2015;112:613–618. doi: 10.1073/pnas.1423481112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Khurana A, Kumar R, Babbar SB. Nitric oxide is involved in salicylic acid-induced flowering of lemna aequinoctialis welw. Acta Physiol Plant. 2014;36:2827–2833. doi: 10.1007/s11738-014-1600-2. [DOI] [Google Scholar]
- 158.Hichri I, Boscari A, Castella C, Rovere M, Puppo A, Brouquisse R. Nitric oxide: a multifaceted regulator of the nitrogen-fixing symbiosis. J Exp Bot. 2015;66:2877–2887. doi: 10.1093/jxb/erv051. [DOI] [PubMed] [Google Scholar]
- 159.Wang W, Sheng X, Shu Z, Li D, Pan J, Ye X, Chang P, Li X, Wang Y.. Combined cytological and transcriptomic analysis reveals a nitric oxide signaling pathway involved in cold-inhibited camellia sinensis pollen tube growth. Front Plant Sci. 2016;7:456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Siddiqui MH, Al-Whaibi MH, Basalah MO. Role of nitric oxide in tolerance of plants to abiotic stress. Protoplasma. 2011;248:447–455. doi: 10.1007/s00709-010-0206-9. [DOI] [PubMed] [Google Scholar]
- 161.Xu Y, Sun X, Jin J, Zhou H. Protective effect of nitric oxide on light-induced oxidative damage in leaves of tall fescue. J Plant Physiol. 2010;167:512–518. doi: 10.1016/j.jplph.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 162.Puyaubert J, Baudouin E. New clues for a cold case: nitric oxide response to low temperature. Plant Cell Environ. 2014;37:2623–2630. doi: 10.1111/pce.12329. [DOI] [PubMed] [Google Scholar]
- 163.Hasanuzzaman M, Nahar K, Alam MM, Fujita M. Exogenous nitric oxide alleviates high temperature induced oxidative stress in wheat (Triticum aestivum L.) seedlings by modulating the antioxidant defense and glyoxalase system. Aust J Crop Sci. 2012;6:1314. [Google Scholar]
- 164.París R, Lamattina L, Casalongué CA. Nitric oxide promotes the wound-healing response of potato leaflets. Plant Physiolo Biochem. 2007;45:80–86. doi: 10.1016/j.plaphy.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 165.Ahmad P, Abdel Latef AA, Hashem A, Abd_Allah EF, Gucel S, Tran L-SP. Nitric oxide mitigates salt stress by regulating levels of osmolytes and antioxidant enzymes in chickpea. Front Plant Sci. 2016;7:347. doi: 10.3389/fpls.2016.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wimalasekera R, Tebartz F, Scherer GF. Polyamines, polyamine oxidases and nitric oxide in development, abiotic and biotic stresses. Plant Sci. 2011;181:593–603. doi: 10.1016/j.plantsci.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 167.Santolini J, André F, Jeandroz S, Wendehenne D. Nitric oxide synthase in plants: where do we stand? Nitric Oxide. 2017;63:30–38. doi: 10.1016/j.niox.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 168.Moreau M, Lee GI, Wang Y, Crane BR, Klessig DF. AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J Biol Chem. 2008;283:32957–32967. doi: 10.1074/jbc.M804838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Domingos P, Prado AM, Wong A, Gehring C, Feijo JA. Nitric oxide: a multitasked signaling gas in plants. Mol Plant. 2015;8:506–520. doi: 10.1016/j.molp.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 170.Weidinger A, Kozlov A. Biological activities of reactive oxygen and nitrogen species: oxidative stress versus signal transduction. Biomolecules. 2015;5:472–484. doi: 10.3390/biom5020472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sanz L, Albertos P, Mateos I, Sánchez-Vicente I, Lechón T, Fernández-Marcos M, Lorenzo O. Nitric oxide (NO) and phytohormones crosstalk during early plant development. J Exp Bot. 2015;66:2857–2868. doi: 10.1093/jxb/erv213. [DOI] [PubMed] [Google Scholar]
- 172.Khan MN, Mohammad F, Mobin M, Saqib MA. Tolerance of plants to abiotic stress: a role of nitric oxide and calcium. Nitric oxide in plants: metabolism and role in stress physiology.Cham: Springer; 2014. p. 225–242. doi:10.1007/978-3-319-06710-0_14. [Google Scholar]
- 173.Courtois C, Besson A, Dahan J, Bourque S, Dobrowolska G, Pugin A, Wendehenne D. Nitric oxide signalling in plants: interplays with Ca2+ and protein kinases. J Exp Bot. 2008;59:155–163. doi: 10.1093/jxb/erm197. [DOI] [PubMed] [Google Scholar]
- 174.Niu L, Yu J, Liao W, Yu J, Zhang M, Dawuda MM. Calcium and calmodulin are involved in nitric oxide-induced adventitious rooting of cucumber under simulated osmotic stress. Front Plant Sci. 2017;8:1684. doi: 10.3389/fpls.2017.01684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ma W, Smigel A, Tsai Y-C, Braam J, Berkowitz GA. Innate immunity signaling: cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol. 2008;148:818–828. doi: 10.1104/pp.108.125104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zaidi I, Ebel C, Belgaroui N, Ghorbel M, Amara I, Hanin M. The wheat MAP kinase phosphatase 1 alleviates salt stress and increases antioxidant activities in Arabidopsis. J Plant Physiol. 2016;193:12–21. doi: 10.1016/j.jplph.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 177.Chen ZH, Wang Y, Wang JW, Babla M, Zhao C, García‐Mata C, Sani E, Differ C, Mak M, Hills A, et al. Nitrate reductase mutation alters potassium nutrition as well as nitric oxide‐mediated control of guard cell ion channels in Arabidopsis. New Phytol. 2016;209:1456–1469. doi: 10.1111/nph.13714. [DOI] [PubMed] [Google Scholar]
- 178.Laxalt AM, García-Mata C, Lamattina L. The dual role of nitric oxide in guard cells: promoting and attenuating the ABA and phospholipid-derived signals leading to the stomatal closure. Front Plant Sci. 2016;7:476. doi: 10.3389/fpls.2016.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lamotte O, Courtois C, Dobrowolska G, Besson A, Pugin A, Wendehenne D. Mechanisms of nitric-oxide-induced increase of free cytosolic Ca2+ concentration in Nicotiana plumbaginifolia cells. Free Radic Biol Med. 2006;40:1369–1376. doi: 10.1016/j.freeradbiomed.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 180.Corpas FJ, Barroso JB, Carreras A, Quirós M, León AM, Romero-Puertas MC, Esteban FJ, Valderrama R, Palma JM, Sandalio LM, et al. Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant Physiol. 2004;136:2722–2733. doi: 10.1104/pp.104.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Corpas FJ, Barroso JB, Carreras A, Valderrama R, Palma JM, León AM, Sandalio LM, Del Río LA. Constitutive arginine-dependent nitric oxide synthase activity in different organs of pea seedlings during plant development. Planta. 2006;224:246–254. doi: 10.1007/s00425-005-0205-9. [DOI] [PubMed] [Google Scholar]
- 182.Khan MN, Siddiqui MH, Mohammad F, Naeem M. Interactive role of nitric oxide and calcium chloride in enhancing tolerance to salt stress. Nitric Oxide. 2012;27:210–218. doi: 10.1016/j.niox.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 183.Foyer CH, Noctor G. Redox signaling in plants. Antioxid Redox Signal. 2013;1;18(16):2087–2090. doi: 10.1089/ars.2013.5278. [DOI] [PubMed] [Google Scholar]
- 184.Liu S-L, Yang R-J, Ma M-D, Dan F, Zhao Y, Jiang P, Wang M-H. Effects of exogenous NO on the growth, mineral nutrient content, antioxidant system, and ATPase activities of Trifolium repens L. plants under cadmium stress. Acta Physiol Plant. 2015;37:1721. doi: 10.1007/s11738-014-1721-7. [DOI] [Google Scholar]
- 185.Zhang Y, Wang L, Liu Y, Zhang Q, Wei Q, Zhang W. Nitric oxide enhances salt tolerance in maize seedlings through increasing activities of proton-pump and Na+/H+ antiport in the tonoplast. Planta. 2006;224:545–555. doi: 10.1007/s00425-006-0242-z. [DOI] [PubMed] [Google Scholar]
- 186.Mazid M, Khan TA, Mohammad F. Role of Nitric oxide in regulation of H2O2 mediating tolerance of plants to abiotic stress: A synergistic signaling approach. J. Stress Physiol. Biochem. 2011;34–74. [Google Scholar]
- 187.Sheokand S, Kumari A, Sawhney V. Effect of nitric oxide and putrescine on antioxidative responses under NaCl stress in chickpea plants. Physiol Mol Biol Plants. 2008;14:355–362. doi: 10.1007/s12298-008-0034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hasanuzzaman M, Hossain MA, Fujita M. Nitric oxide modulates antioxidant defense and the methylglyoxal detoxification system and reduces salinity-induced damage of wheat seedlings. Plant Biotechnol Rep. 2011;5:353. doi: 10.1007/s11816-011-0189-9. [DOI] [PubMed] [Google Scholar]
- 189.Kovacic P, Somanathan R. Integrated approach to nitric oxide in animals and plants (mechanism and bioactivity): cell signaling and radicals. J Recept Signal Transduct Res. 2011;31:111–120. doi: 10.3109/10799893.2010.544317. [DOI] [PubMed] [Google Scholar]
- 190.Wang H, Liang X, Wan Q, Wang X, Bi Y. Ethylene and nitric oxide are involved in maintaining ion homeostasis in Arabidopsis callus under salt stress. Planta. 2009;230:293–307. doi: 10.1007/s00425-009-0946-y. [DOI] [PubMed] [Google Scholar]
- 191.Zhao L, Zhang F, Guo J, Yang Y, Li B, Zhang L. Nitric oxide functions as a signal in salt resistance in the calluses from two ecotypes of reed. Plant Physiol. 2004;134:849–857. doi: 10.1104/pp.103.030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Pereira A. Plant abiotic stress challenges from the changing environment. Front Plant Sci. 2016;7:1123. doi: 10.3389/fpls.2016.01123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Singh M, Kumar J, Singh S, Singh VP, Prasad SM. Roles of osmoprotectants in improving salinity and drought tolerance in plants: a review. Rev Environ Sci Bio/Technol. 2015;14:407–426. doi: 10.1007/s11157-015-9372-8. [DOI] [Google Scholar]
- 194.Nahar K, Hasanuzzaman M, Fujita M. Roles of osmolytes in plant adaptation to drought and salinity. Osmolytes and Plants Acclimation to Changing Environment: Emerging Omics Technologies.New Delhi: Springer; 2016. p. 37–68. doi:10.1007/978-81-322-2616-1_4. [Google Scholar]
- 195.Wutipraditkul N, Wongwean P, Buaboocha T. Alleviation of salt-induced oxidative stress in rice seedlings by proline and/or glycinebetaine. Biol. Plant. 2015;59:547–553. doi: 10.1007/s10535-015-0523-0. [DOI] [Google Scholar]
- 196.Awasthi R, Bhandari K, Nayyar H. Temperature stress and redox homeostasis in agricultural crops. Front Env Sci. 2015;3:11. doi: 10.3389/fenvs.2015.00011. [DOI] [Google Scholar]
- 197.Singh M, Kumar J, Singh S, Singh V, Prasad S, Singh M. Adaptation strategies of plants against heavy metal toxicity: a short review. Biochem Pharmacol (Los Angel). 2015;4:2167–2501. 1000161. [Google Scholar]
- 198.Chen H, Jiang J-G. Osmotic adjustment and plant adaptation to environmental changes related to drought and salinity. Environ Rev. 2010;18:309–319. doi: 10.1139/A10-014. [DOI] [Google Scholar]
- 199.Turan S, Cornish K, Kumar S. Salinity tolerance in plants: breeding and genetic engineering. Aust J Crop Sci. 2012;6:1337. [Google Scholar]
- 200.Slama I, Abdelly C, Bouchereau A, Flowers T, Savoure A. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann Bot. 2015;115:433–447. doi: 10.1093/aob/mcu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Szabados L, Savoure A. Proline: a multifunctional amino acid. Trends Plant Sci. 2010;15:89–97. doi: 10.1016/j.tplants.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 202.Rana V, Ram S, Nehra K. Review proline biosynthesis and its role in abiotic stress. IJAIR. 2017;6:2319–1473. [Google Scholar]
- 203.Joseph E, Radhakrishnan V, Mohanan K. A study on the accumulation of proline-an osmoprotectant amino acid under salt stress in some native rice cultivars of North Kerala, India. Universal J Agri Res. 2015;3:15–22. [Google Scholar]
- 204.Chen J, Zhang Y, Wang C, Lü W, Jin JB, Hua X. Proline induces calcium-mediated oxidative burst and salicylic acid signaling. Amino Acids. 2011;40:1473–1484. doi: 10.1007/s00726-010-0757-2. [DOI] [PubMed] [Google Scholar]
- 205.Rahman A, Nahar K, Hasanuzzaman M, Fujita M. Calcium supplementation improves Na+/K+ ratio, antioxidant defense and glyoxalase systems in salt-stressed rice seedlings. Front Plant Sci. 2016;7:609. doi: 10.3389/fpls.2016.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]


