Diabetes is a growing epidemic due to the systematic increase in overweight and obesity, and chronic diabetic foot ulcers (DFUs) are the most debilitating and costly complication of diabetes. Prevention is still essential for DFU care, but early detection is limited because we do not have a good DFU biomarker. Here, we report the preliminary results of a large longitudinal study to test the effectiveness of the use of T-cell phenotype and TCR repertoire diversity as DFU biomarkers. Our results show that the ratio of effector/naïve T-cells and the number of TCR expansions are valuable biomarkers of DFU prognosis.
Diabetes is the biggest epidemic in human history and is growing due to the systematic increase in overweight and obesity, and chronic diabetic foot ulcers (DFUs) are the most debilitating complications of diabetes.1 In DFUs, wound healing is compromised due to neuropathy, microangiopathy and impaired immune function.2
Since the most effective treatment strategies have limited results,3 prevention is essential for DFU care.1 Various genomic and proteomic biomarkers have been used for DFU prognosis in the past, with limited success.4 Among these, cytokines,5 nitric oxide,6 MMPs7 and especially microRNAs8 yielded the most promising results, but no biomarker is able to predict which diabetic wounds will fail to heal.
Our previous work has shown that diabetes significantly reduces naïve T-cell numbers and TCR repertoire diversity, mainly due to the accumulation of effector T-cells.9 These cells secrete large quantities of inflammatory cytokines that create a chronic inflammatory environment, thereby impairing wound healing.9 For these reasons, the accumulation of effector T-cells and the consequent reduction in TCR repertoire diversity are potential DFU biomarker candidates.
Here, we present the results of a longitudinal study and discuss the use of the effector/naïve blood T-cell ratio and number of TCR clonal expansions as biomarkers for the assessment of DFU prognosis.
We collected peripheral blood (PB) samples from 13 DFU patients (8F and 5M; 35–54 years; mean age = 43 years) into ethylenediamine tetraacetic acid (EDTA)-K3 tubes at the time of initial diagnosis after obtaining written informed consent. Patients were followed-up for 12 months, and relevant clinical information related to foot ulceration development and treatment was recorded. All diabetic patients enrolled in this study had no previous history of foot ulcerations, neoplastic malignancies or autoimmune disorders and had no clinically detectable ongoing infection at the time of sample collection, apart from foot infection. This study was approved by the Ethics Committee of Hospital de Santo António, Porto, Portugal.
DNA was extracted from the sorted populations using the Sigma GenElute Mammalian Genomic DNA Miniprep Kit (Sigma-Aldrich, St. Louis, MO, USA) as recommended by the manufacturer. Extracted DNA was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
PCR amplification was performed using 100 ng of DNA from each sample, as previously described.9 Amplification was performed using the Perkin Elmer model (PE) 9600 thermal cycler (Perkin Elmer Applied Biosystems, Inc., Foster City, CA, USA), and product sizes were detected with the Applied Biosystems ABI 310 capillary electrophoresis system (Thermo Fisher Scientific, Waltham, USA), as previously described.9 The resulting data were analyzed using Peak Scanner Software v1.0 (Thermo Fisher Scientific, Waltham, USA).
Analysis of surface antigen expression on PB T-cells was performed in all cases using whole blood direct immunofluorescence five-color staining, by flow cytometry as previously described.9 Flow cytometry experiments were performed on a FACSCANTO cytometer using Diva software for sample acquisition and data analysis (all from Becton-Dickinson, East Rutherford, NJ, USA).
Naïve (CD27+CD28+CD45RO-) and effector (CD27-CD28-) T-cells in both CD4 and CD8 subsets were quantified using anti-CD4 or anti-CD8 mAbs in conjunction with anti-CD27, anti-CD28 and/or anti-CD45RO mAbs as previously described.9
All statistical analyses were performed using one-way analysis of variance (ANOVA), followed by Tukey’s honest significant difference test with a 95% confidence interval using GraphPad Prism 6.0 software (GraphPad Software Inc., La Jolla, CA, USA). Differences were considered statistically significant at P < 0.05.
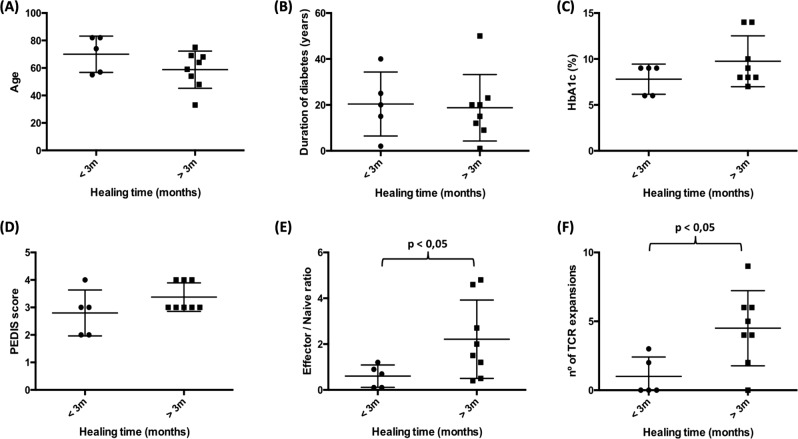
We divided the patients into two groups based on whether DFUs healed (n = 5) or not (n = 8) in less than 3 months. Both groups had a similar age, duration of diabetes (years from initial diagnosis), glycated hemoglobin (HbA1c) percentage and PEDIS score (Fig. 1a-d).
Fig. 1.
New biomarker candidates for DFUs. We collected blood samples from DFU patients at the time of initial diagnosis and followed them for 1 year. We divided these patients into two groups based on whether the ulcers healed or not in less than three months. Upon comparing patient age (a), duration of diabetes (from initial diagnosis to sample collection) (b), glycated hemoglobin HbA1c percentage (c) and PEDIS score (d), we found no significant differences between the groups. Both the effector/naïve T-cell ratio (e) and the number of TCR expansions (f) were significantly increased in diabetic patients
We quantified the percentage of blood effector (CD27-CD28-) and naïve (CD27+CD28+CD45RO-) T-cells and calculated the effector/naïve ratio. This ratio was significantly elevated in patients with non-healing DFUs compared to those with healed DFUs (2.5 ± 1.2 vs 0.6 ± 0.4; p < 0.05) (Fig. 1e).
The diversity in PCR product sizes directly correlates with the diversity in V(D)J recombination, which in turn correlates with T-cell clonal diversity, as previously described.9 For each sample, we counted the number of peaks that deviated more than two-fold from the Gaussian distribution, representing clonal expansions.9 TCR-αβ+ T-cells in blood from patients with non-healing DFUs underwent significantly more TCR expansions than those from patients with healed DFUs (4.8 ± 2.3 vs 1.0 ± 1.2; p < 0.05) (Fig. 1f).
Our results demonstrate that the most commonly used method for assessing DFU prognosis, the PEDIS score, is unable to predict which ulcers will or will not heal. By applying both an effector/naïve T-cell ratio above 1 and more than 2 TCR expansions as the cumulative criteria to assess DFU prognosis, we were able to identify which ulcers would fail to heal at the time of initial diagnosis, with a sensitivity of 75% and a specificity of 80%. These cumulative criteria yielded a positive predictive value of 86% and a negative predictive value of 67%, thus confirming the usefulness of simple blood analysis for the evaluation of DFU prognosis.
These are the first results of a large longitudinal study on the usefulness of T-cell phenotype and TCR repertoire diversity as prognostic biomarkers of DFUs; even with the currently small sample size, the results are already very promising, and this new biomarker has already demonstrated better prognostic potential than the current most effective and widely used prognosis system, the PEDIS score.
If confirmed by larger longitudinal studies, this new biomarker will empower clinicians to make early informed decisions regarding treatment and monitoring strategies, possibly avoiding millions of amputations worldwide, with enormous clinical and economic impacts.
Acknowledgements
We acknowledge support from the Sociedade Portuguesa de Diabetologia (SPD): Bolsa Almeida Ruas 2016, FEDER - strategic project COMPETE: POCI-01-0145-FEDER-007440, European Foundation for the Study of Diabetes (EFSD) and HealthyAging2020: CENTRO-01-0145-FEDER-000012N2323.
Competing interests
The authors declare no competing interests.
References
- 1.Lim JZ, Ng NS, Thomas C. Prevention and treatment of diabetic foot ulcers. J. R. Soc. Med. 2017;110:104–109. doi: 10.1177/0141076816688346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pereira SG, Moura J, Carvalho E, Empadinhas N. Microbiota of chronic diabetic wounds: ecology, impact, and potential for innovative treatment strategies. Front. Microbiol. 2017;8:1791. doi: 10.3389/fmicb.2017.01791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel S, Srivastava S, Singh MR, Singh D. Mechanistic insight into diabetic wounds: Pathogenesis, molecular targets and treatment strategies to pace wound healing. Biomed. Pharmacother. 2019;112:108615. doi: 10.1016/j.biopha.2019.108615. [DOI] [PubMed] [Google Scholar]
- 4.Pichu S, Patel BM, Apparsundaram S, Goyal RK. Role of biomarkers in predicting diabetes complications with special reference to diabetic foot ulcers. Biomark. Med. 2017;11:377–388. doi: 10.2217/bmm-2016-0205. [DOI] [PubMed] [Google Scholar]
- 5.Yager DR, Kulina RA, Gilman LA. Wound fluids: a window into the wound environment? Int. J. Low. Extrem. Wounds. 2007;6:262–272. doi: 10.1177/1534734607307035. [DOI] [PubMed] [Google Scholar]
- 6.Boykin JV., Jr. Wound nitric oxide bioactivity: a promising diagnostic indicator for diabetic foot ulcer management. J. Wound Ostomy Cont. Nurs. 2010;37:25–32. doi: 10.1097/WON.0b013e3181c68b61. [DOI] [PubMed] [Google Scholar]
- 7.Moura J, da Silva L, Cruz MT, Carvalho E. Molecular and cellular mechanisms of bone morphogenetic proteins and activins in the skin: potential benefits for wound healing. Arch. Dermatol. Res. 2013;305:557–569. doi: 10.1007/s00403-013-1381-2. [DOI] [PubMed] [Google Scholar]
- 8.Goodarzi G., Maniati M., Qujeq D. The role of microRNAs in the healing of diabetic ulcers. Int. Wound J. 2019. [DOI] [PMC free article] [PubMed]
- 9.Moura J. et al. Impaired T-cell differentiation in diabetic foot ulceration. Cell Mol. Immunol.14, 758–769 (2016). [DOI] [PMC free article] [PubMed]