Abstract
Purpose
The survival benefit with adjuvant chemotherapy for patients with resected stage II-III non–small-cell lung cancer (NSCLC) is modest. Efforts to develop prognostic or predictive biomarkers in these patients have not yielded clinically useful tests. We report findings from the Lung Adjuvant Cisplatin Evaluation (LACE)-Bio-II study, in which we analyzed next-generation sequencing and long-term outcomes data from > 900 patients with early-stage NSCLC treated prospectively in adjuvant landmark clinical trials. We used a targeted gene panel to assess the prognostic and predictive effect of mutations in individual genes, DNA repair pathways, and tumor mutation burden (TMB).
Methods
A total of 908 unmatched, formalin-fixed, paraffin-embedded, resected lung cancer tumor specimens were sequenced using a targeted panel of 1,538 genes. Stringent filtering criteria were applied to exclude germline variants and artifacts related to formalin fixation. Disease-free survival, overall survival, and lung cancer—specific survival (LCSS) were assessed in Cox models stratified by trial and adjusted for treatment, age, sex, performance score, histology, type of surgery, and stage.
Results
Nonsynonymous mutations were identified in 1,515 genes in 908 tumor samples. High nonsynonymous TMB (> 8 mutations/Mb) was prognostic for favorable outcomes (ie, overall survival, disease-free survival, and LCSS) in patients with resected NSCLC. LCSS benefit with adjuvant chemotherapy was more pronounced in patients with low nonsynonymous TMBs (≤ 4 mutations/Mb). Presence of mutations in DNA repair pathways, tumor-infiltrating lymphocytes, TP53 alteration subtype, and intratumor heterogeneity was neither prognostic nor predictive. Statistically significant effect of mutations in individual genes was difficult to determine due to high false-discovery rates.
Conclusion
High nonsynonymous TMB was associated with a better prognosis in patients with resected NSCLC. In addition, the benefit of adjuvant chemotherapy on LCSS was more pronounced in patients with low nonsynonymous TMBs. Studies are warranted to confirm these findings.
INTRODUCTION
Adjuvant chemotherapy in selected patients with resected stage IB non–small-cell lung cancer (NSCLC) and all patients with resected stages II and IIIA NSCLC is currently considered the standard of care and associated with an approximately 5% survival benefit at 5 years.1 Clinically useful prognostic and/or predictive biomarkers that can consistently identify patients most likely to benefit from adjuvant chemotherapy are yet to be developed. The Lung Adjuvant Cisplatin Evaluation (LACE)-Bio consortium includes investigators from the pivotal International Adjuvant Lung Trial, Cancer and Leukemia Group B–9633, National Cancer Institute of Canada Clinical Trials Group JBR.10, and Adjuvant Navelbine International Trialist Association (ANITA) trials, which have established the role of adjuvant chemotherapy in current-day clinical practice.1-5 Archival tissue specimens (ie, formalin fixed, paraffin embedded [FFPE]) collected from consenting patients enrolled in these trials by the consortium provide a valuable opportunity for developing prognostic and predictive biomarkers that could potentially inform clinical decision-making.6-10
The objective of LACE-Bio-II (LB2) was to comprehensively evaluate the prognostic value of copy number alterations and somatic mutations in resected NSCLC, and their predictive effect in guiding adjuvant therapy. Apart from serving as a valuable resource that can aid biomarker development, the comprehensive molecular characterization of patient samples from the LB2 trials using targeted next-generation sequencing has the potential to offer insights into the genomic underpinnings of NSCLC. We report here findings from targeted sequencing of 908 samples from the LB2 trials using a panel of 1,538 genes selected on the basis of The Cancer Genome Atlas (TCGA) Pan-Cancer analysis.11 Samples collected from patients enrolled in the ANITA trial were not included in these analyses, because the tissue from these samples was exhausted. Using these data, we explored the role of alterations in individual genes, various DNA repair pathways, type of TP53 mutation, intratumor heterogeneity, and nonsynonymous tumor mutation burden (TMB) in prognostication and predicting benefit from adjuvant chemotherapy. The prognostic and predictive effect of the presence of tumor infiltrating lymphocytes (TILs) and PDL1 expression, as measured by immunohistochemistry, was previously explored for LB2 samples, and these data were also incorporated into our analyses.8,9
METHODS
A total of 908 FFPE, resected lung cancer tumor specimens were sequenced on a targeted panel consisting of 1,538 genes. This gene set is larger than that typically assayed by currently available commercial assays, because genes frequently mutated across a wide variety of cancers sequenced by TCGA and implicated in lung cancer pathogenesis were included in our panel, regardless of their clinical actionability.11 Sequencing was performed using SeqCap custom capture probes (Roche NimbleGen, Madison, WI), designed for a curated set of putative cancer causing genes. Dual-indexed Illumina libraries (San Diego, CA) were combined in pools of 96 samples and were captured using the manufacturer’s protocol. Most (98.8%) of the targeted sequence was successfully covered, leading to a total coverage area of 4.88 Mb. The capture product was run on two lanes of the Illumina HiSeq 2000. A total of 908 samples passed a quality control cutoff threshold of 70% of the targets covered to a minimum depth of 20×. These included 414 squamous cell carcinoma (LUSC) and 375 lung adenocarcinoma (LUAD) samples. Given the lack of a matched normal to distinguish somatic from germline variants, single-nucleotide variants (SNVs) with a global minor allele frequency of > 1% in the exomes of the 1000 Genomes Project or National Heart, Lung, and Blood Institute, and those observed in five or more of the nearly 900 normal samples procured from patients with breast cancer, were considered germline and excluded from analysis. In addition, data from phase 3 of the 1000 Genomes Project, and The Exome Aggregation Consortium, consisting of data from 60,706 individuals from different populations, were also used for filtering germline variants from this data set (Data Supplement). The variant allelic frequency (VAF) cutoff for SNVs present in these data sets was set to a maximum of 0.0001%. To strike a balance between stringent filtering of putative germline mutations and sensitivity to detect potentially legitimate somatic variants, any discarded variant that was observed in TCGA LUAD and LUSC data sets was restored. Insertions and deletions (indels) were subjected to the same filtration steps as SNVs in both the initial pipeline and the downstream analysis. Given that indels present a greater challenge for accurate detection in comparison with SNVs, we also performed manual review on the most frequently occurring indels to eliminate common artifacts. A detailed description of methods used for variant calling, analyses, and filtering steps is available in the Data Supplement.
According to the predefined statistical analysis plan, the primary end point was disease-free survival (DFS), defined as the time from randomization to first recurrence (locoregional or distant) or death from any cause. Secondary end points were overall survival (OS), defined as the time from randomization to death from any cause, and lung cancer–specific survival (LCSS), defined as the time from randomization to death as a result of lung cancer. Because the functional consequence of synonymous mutations is uncertain, only nonsynonymous mutations were included in our analyses. Missense, nonsense, splice-site, and frameshift mutations were considered nonsynonymous. Genes altered by nonsynonymous mutations in < 20 samples were discarded from gene-specific prognostic analyses.
The presence of mutations in given genes was correlated to survival end points via Cox models stratified by trial and adjusted for treatment arm, patient age, sex, performance status, histology, and T and N stage. The relative hazard for a patient with at least one mutation in a gene as compared with none was expressed using hazard ratios. To evaluate the predictive role of mutations in a gene, a treatment-by-gene interaction was added to the Cox regression model stratified by study. P values were corrected to control for the false-discovery rate by computing q values.12 Alterations in the TP53 gene were also studied by considering disruptive and nondisruptive mutations separately (Data Supplement), on the basis of previous data that have shown this categorization to be prognostically important in lung and other cancers.13,14 In preplanned sensitivity analyses, we repeated these analyses in the histologic types: LUAD and LUSC. The frequencies of the mutations were compared between histologic subtypes using χ2 tests. To study the possible prognostic and predictive roles of mutations in various pathways, we used similar Cox models for (1) the presence versus absence of any mutation in a given pathway, and (2) the number of mutations in that pathway. These models were again adjusted for aforementioned clinicopathological factors and stratified by trial.
The association between TILs and TMB was assessed using a logistic model for TILs using TMB as the covariate. Likelihood-ratio tests were used to compare the models (adjusted for clinical variables) with versus without TMB. The prognostic and predictive roles of TMB and mutant allele tumor heterogeneity (MATH) score were assessed in Cox models, with TMB and MATH scores categorized into three groups according to tertiles.15 The estimated hazard ratios expressed the relative risk for low versus moderate and high versus moderate categories. To relax the assumption of linear effect and flexibly allow for nonlinear effects, we also modeled the effect of these risk factors using splines with three degrees of freedom (Data Supplement). Representativity analysis was performed to ensure the findings from the current analysis were applicable to all patients enrolled to the LACE-Bio trials, and no significant differences in DFS, OS, or LCSS were observed between patients with and without mutation data (Data Supplement).
RESULTS
Sample Availability, Demographics, and Clinical Characteristics
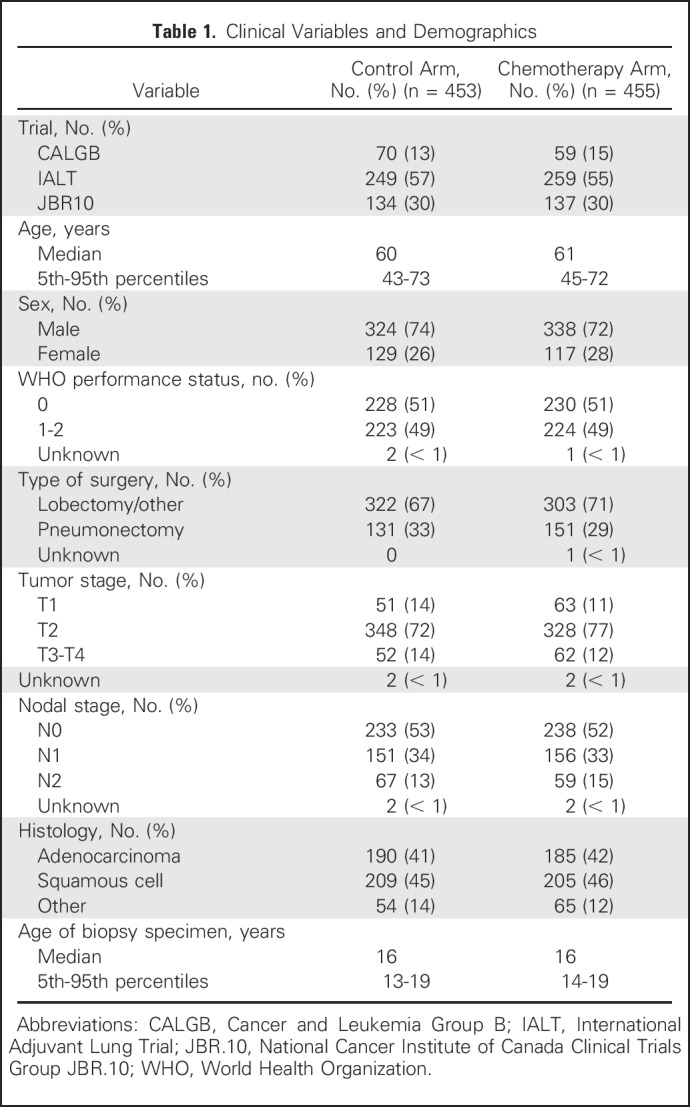
This study included 1,608 patients from three LB2 trials, of whom 1,008 had an FFPE tumor block available for assessment (Data Supplement). After excluding samples with tissue quantity or quality that was inadequate for sequencing, samples from 908 patients were sequenced on a targeted panel of 1,538 genes; 27% of the samples were obtained from female patients (Table 1). The median age at diagnosis was 60 years (range, 27 to 81 years). The majority of patients had stage II disease (n = 676; 75%), 12.5% had stage I disease (n = 114), and 12.5% had stage III/IV disease (n = 114). Smoking history was available for patients enrolled in only two of three LB2 trials.
Table 1.
Clinical Variables and Demographics
Mutation Burden, Signatures, and Frequently Mutated Genes
The mean depth of coverage across the 1,538 genes composing the sequence space was approximately 55×. The prevalence of C>T transitions was much higher in the LB2 cohort when compared with the previously published TCGA cohort, which was enriched for tobacco smoking–associated C>A transversions. Most C>T transitions were associated with a low VAF, suggesting that this mutational pattern was an artifact related to formalin fixation and specimen age (median, 16 years; range, 11 to 20 years).16 After applying additional filtering to exclude C>T transitions with a low VAF, the median nonsynonymous TMB was 5.7/Mb (range, 0.19 to 225/Mb; interquartile range, 3.2 to 9.7/Mb; Data Supplement). Missense variations (38%) were the most frequent mutation category. Other nonsynonymous mutations occurred at a lower rate, including nonsense (3.5%) and splice-site (4%) variations.
Trinucleotide mutation context of synonymous and nonsynonymous mutations was used to determine the predominant mutational signatures. This analysis showed enrichment for tobacco smoking (cosine similarity, 0.91), APOBEC-driven mutagenesis (cosine similarity, 0.75), and age-related spontaneous deamination mutation signatures (cosine similarity, 0.73; Data Supplement). We also identified the list of genes that were significantly mutated in the TCGA LUAD and LUSC data sets and compared the frequencies at which they were mutated in TCGA and LB2 samples. These frequencies were mostly comparable (paired Wilcoxon signed-rank test for LUAD, P = .706; for LUSC, P = .426; Fig 1; Data Supplement).17
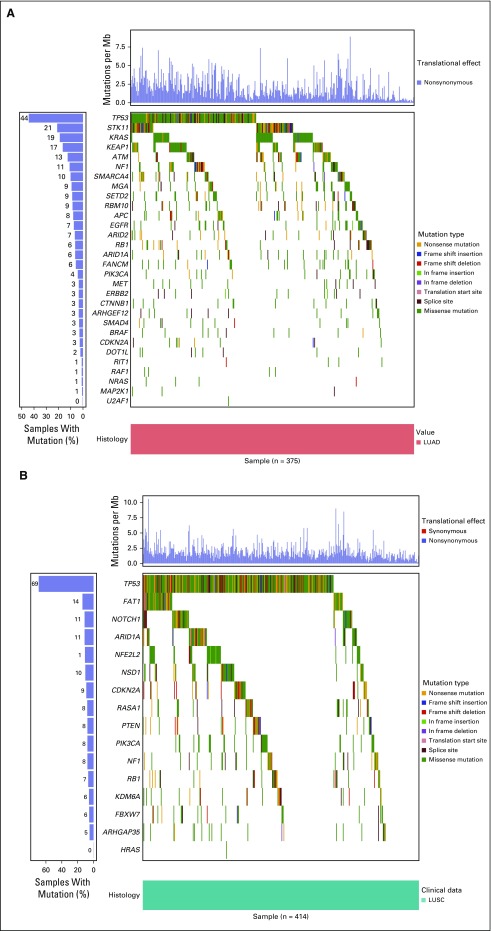
Fig 1.
Waterfall plots showing the frequency and types of mutations in genes mutated at a statistically significant level in The Cancer Genome Atlas cohort, across (A) LUAD (n = 375) and (B) LUSC (n = 414) samples from the LACE-Bio II cohort. LUAD, lung adenocarcinoma; LUSC, squamous cell carcinoma of the lung.
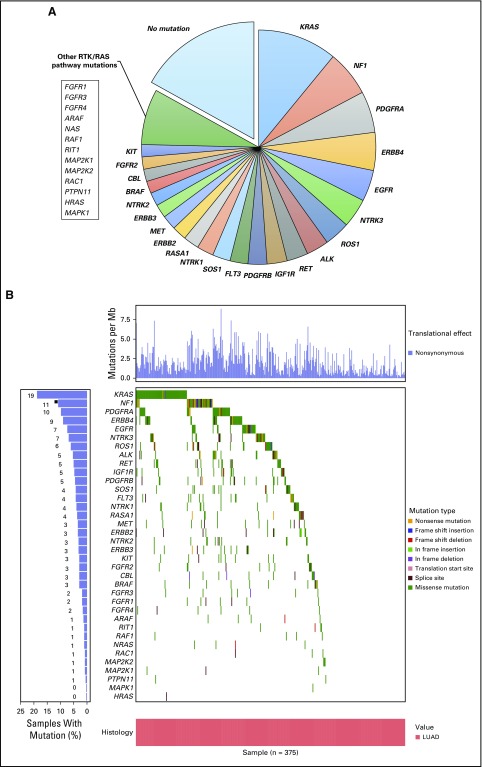
Within LB2 samples, the genes most differentially mutated between LUAD and LUSC were KRAS (19% v 2%), TP53 (44% v 69%), and STK11 (21% v 2%; q < 0.005; Data Supplement). Mutations in receptor tyrosine kinase/RAS signaling were identifiable in nearly 70% (n = 265) of LUAD samples (Fig 2). Among LUAD samples, KRAS was the most frequently mutated oncogene (19%; n = 72). As expected, activating mutations in KRAS, HRAS, NRAS, and EGFR were found in < 3% of LUSC samples. One LUSC sample in the LB2 cohort demonstrated a targetable L858R mutation in EGFR.
Fig 2.
(A) Approximately 70% of LUAD samples had at least one mutation in RTK/RAS/RAF pathway genes. (B) Distribution and type of mutation affecting these genes across all LUAD samples in the LACE-Bio II cohort. Percentages of samples with mutations listed for each gene are overlapping (and do not add up to 100%). LUAD, lung adenocarcinoma.
Prognostic and Predictive Analyses
Nonsynonymous mutations were identified in 1,515 of 1,538 genes in 908 LB2 specimens. Nonsynonymous TMB was estimated for all sequenced samples and analyses were performed to determine the effect of TMB on prognosis. Given that the TMB for LB2 samples was estimated using only a panel of 1,538 genes and nonsynonymous mutations, we assessed the ability of TMB calculated using this restricted gene panel to serve as a surrogate for TMB estimated through whole-exome sequencing. To this end, we used TCGA data to examine the correlation between the number of nonsynonymous mutations involving LB2 panel genes and mutations across the whole exome. We limited our results to 1,214 LB2 genes that could be mapped back to the TCGA data set, including all key oncogenes. Our analysis indicated a significant correlation between these parameters (R2 = 0.96; P < .001; Data Supplement).
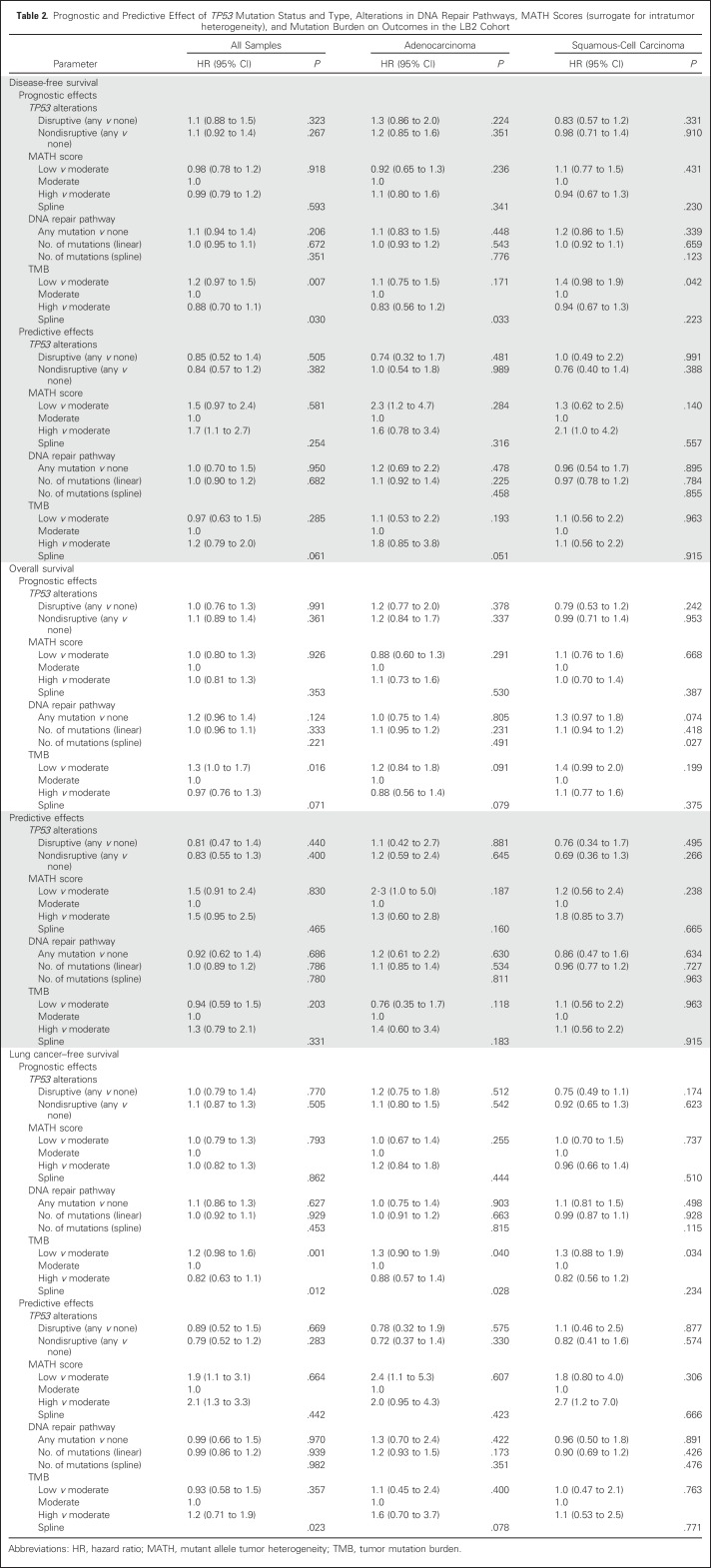
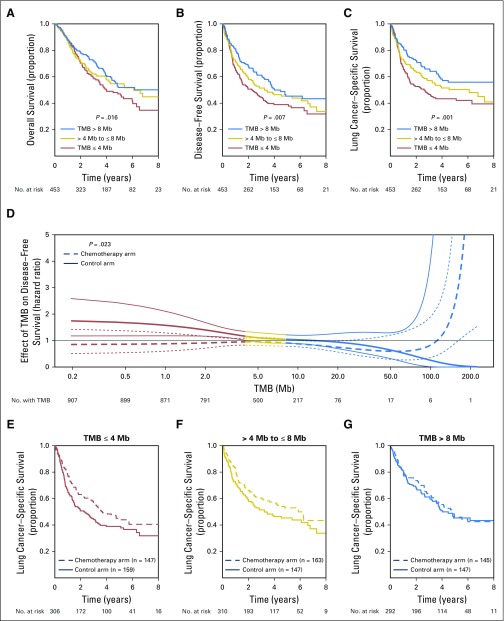
The nonsynonymous TMB was significantly prognostic for DFS, OS, and LCSS (Table 2). When tumors from all histologies were categorized into tertiles (low, ≤ 4 mutations/Mb; intermediate, > 4 and ≤ 8 mutations/Mb; high, > 8 mutations/Mb) on the basis of mutational burden, tumors with a high nonsynonymous TMB had favorable outcomes, whereas tumors with a low nonsynonymous TMB were associated with poor outcomes (DFS, OS, and LCSS P for trend = .007, .016, and .001, respectively). For DFS, the hazard ratio for patients with a TMB in the lower tertile was 1.2 (95% CI, 0.97 to 1.5) relative to the moderate category, whereas it was 0.88 (95% CI, 0.70 to 1.1) for patients with a high TMB. Nonsynonymous TMB was also significantly prognostic for LCSS in LUAD (P = .04) and LCSS and DFS in LUSC (P = .034 and .042, respectively), when these histologies were independently analyzed (Table 2).
Table 2.
Prognostic and Predictive Effect of TP53 Mutation Status and Type, Alterations in DNA Repair Pathways, MATH Scores (surrogate for intratumor heterogeneity), and Mutation Burden on Outcomes in the LB2 Cohort
Exploratory analyses suggested that nonsynonymous TMB might have a significant predictive effect on LCSS, when a flexible (ie, splines) model was used (P = .023; Table 2, Fig 3), but not for DFS (P = .06) or OS (P = .33). The spline model allowed us to model the effect of nonsynonymous TMB on outcomes in a flexible manner, without assuming a linear effect between these variables (Data Supplement).18 This effect was significant for samples from all histologies, but not when LUAD and LUSC samples were independently analyzed (Table 2). This model seemeded to suggest that the beneficial effect of adjuvant chemotherapy on LCSS was most pronounced in samples with lower nonsynonymous TMBs and decreased as nonsynonymous TMB increased. However, this effect may be driven by the small size of samples with high nonsynonymous TMB (Data Supplement) and the linear test for trend was not significant when samples were regrouped by tertiles of TMB (P = .357; Table 2).
Fig 3.
Kaplan-Meier plots showing prognostic effect of nonsynonymous TMB on (A) overall survival, (B) disease-free survival, and (C) lung cancer–specific survival (LCSS) across all tumor samples. (D) Effect of mutational burden on hazard ratio for LCSS benefit with adjuvant chemotherapy when a flexible “spline” model is used. (E–G) Predictive effect of nonsynonymous TMB on LCSS at different levels of TMB (P = .357). TMB, tumor mutation burden.
No significant prognostic or predictive association was observed between tumor MATH scores and outcomes among LB2 samples (Table 2). Hypothesizing that nonsynonymous TMB would serve as a marker of neo-epitope burden and antitumor immune response, we estimated the intensity of TILs in the biopsy specimens as described in our previous study, to determine if it correlated with the nonsynonymous TMB and MATH scores.8 The intensity of TILs, however, did not correlate with the nonsynonymous TMB or MATH scores at a statistically significant level (Data Supplement). In addition, 805 of 908 tumor samples were categorized on the basis of PDL1 expression scores in tumor and stroma using cutoff values of < 1%, < 25%, and < 50%, and < 1%, < 10%, < 25%, respectively, using the E1L3N antibody.9 High nonsynonymous TMB or MATH score did not correlate with PDL1 expression, although we found that stromal PDL1 expression correlated with intense TIL infiltration (Data Supplement).
As determined in the statistical analyses plan, 845 of 1,515 genes with nonsynonymous mutations were excluded from prognostic and predictive analyses, because they were mutated in < 20 specimens (Data Supplement). Although our analyses showed mutations in a few individual genes to be prognostic and predictive for outcomes, the q values for these associations were high, limiting our ability to interpret the role of these alterations as biomarkers (Data Supplement). A total of 571 (60%) samples showed nonsynonymous TP53 mutations. The presence of TP53 mutations, either disruptive or nondisruptive, was neither prognostic nor predictive (Table 2). Mutations in DNA repair, antigen presentation and processing, and developmental pathways such as WNT signaling possibly play an important role in determining prognosis and response to therapy in patients with cancers.19-23 As a result, LB2 samples were categorized on the basis of presence or absence of nonsynonymous alterations in genes participating in these pathways (Data Supplement). However, the presence of mutations in any of these pathways was neither prognostic nor predictive of chemotherapy benefit (Data Supplement).
DISCUSSION
Although cytotoxic chemotherapy improves outcomes in patients with resected NSCLC, the magnitude of this benefit is modest at best.1 The availability of targeted sequencing and associated long-term outcomes data for tumor specimens obtained from nearly 900 patients clearly makes LB2 a valuable resource for development of prognostic and predictive biomarkers in early-stage NSCLC. Results from this analysis showed high nonsynonymous TMB to be significantly associated with a favorable prognosis. In addition, in line with this observation, our results suggest a role for low nonsynonymous TMB in predicting for LCSS benefit with adjuvant chemotherapy. Nevertheless, this result should be interpreted with caution because it is likely driven by few samples with extremely high TMB and the linear tests for trend did not suggest significantly different effects when samples were regrouped by tertiles of TMB. Although this predictive effect was not significant for OS and DFS, LCSS may be a more meaningful end point to assess the efficacy of adjuvant chemotherapy in this cohort of patients who have a high risk of dying from competing causes.24-26 These results are supported by findings from other cancers. A prognostic and predictive role for mismatch repair deficiency, which is associated with high TMBs, has been well described in patients with colon cancer.27,28 In patients with resected stage II colon cancer, mismatch repair deficiency, is utilized as a biomarker that is prognostic for outcomes and predictive for lack of benefit from adjuvant chemotherapy.28,29
Lung cancer is characterized by a high TMB because of its association with smoking. Given that some mutations can result in the production of neo-epitopes, TMB in lung cancer is a predictor of response to immunotherapies.30 McGranahan et al31 observed an association between longer OS and high neo-epitope burden in patients with early-stage LUADs. Similarly, in a previous analysis using LB2 samples, intense TIL infiltration was associated with favorable outcomes.8 These data suggest that the prognostic effect of nonsynonymous TMB in lung cancer may, in part, be mediated by its role in shaping tumor-host immune interactions. Apart from high TMBs, lung cancers also show marked chromosomal instability. Preliminary findings from the TRACERx (Tracking Cancer Evolution Through Therapy) prospective analysis, which characterized the evolutionary patterns of NSCLC in 100 patients through whole-exome sequencing, showed that a high fraction of subclonal copy number alterations was associated with a short relapse-free survival in patients undergoing resection.32 Taken together, these results suggest that quantifying genomic instability at the nucleotide and/or chromosomal level is likely to prove useful in prognosticating disease outcome for patients with lung cancer and guiding treatment in the near future. These findings also emphasize a role for nonsynonymous TMB as a potential stratification variable in clinical trials.
We found TIL infiltration correlated with stromal PDL1 expression in LB2 samples, but neither TIL infiltration nor PDL1 expression was associated with TMB. These results are comparable to the Checkmate 026 trial, in which PDL1 expression was not associated with TMB.33 However, it is possible that these correlative analyses are limited by technical challenges associated with sequencing a limited number of genes, PDL1 antibody selection, and absence of multiregion tumor sequencing data, because the clonality of a mutation in a tumor specimen is known to influence antigen presentation, T-cell effector function, and migration.31
There are several important limitations to interpreting data presented in this study. First, many mutations encountered in sequenced samples were C>T transitions with a low VAF that required additional filtering.34 Although this step did not affect the frequency of known hotspot mutations in cancer-causing genes such as TP53, KRAS, and EGFR (Data Supplement), it is possible that this could have led to the inadvertent exclusion of other relevant but less-established variants. Second, the mean depth of coverage achieved was lower than planned, possibly due to DNA breakage from formalin fixation.34 Third, the nonavailability of matched normal tissue samples for tumor specimens in our cohort limited the ability to distinguish somatic from germline variants accurately. Fourth, using a targeted gene panel for sequencing precluded us from studying the prognostic or predictive effect of alterations in genes not included in the panel. Despite these challenges, except for the relatively lower frequency of EGFR mutations (7.5%; n = 28) in the LB2 sample set, which likely is explained by the underrepresentation of samples obtained from never-smokers, the frequency of alterations detected in our analysis was comparable with that of TCGA, and our findings are supported by corroborating evidence from other cancers.29 Finally, nonsynonymous TMB in our study was inferred from a limited gene panel. Although whole-exome or whole-genome sequencing data are likely to provide a more accurate assessment of nonsynonymous TMB, TMB estimated using gene panels has been shown to be representative of TMB inferred from whole-exome sequencing.35 In addition, we were able to demonstrate good concordance between whole-exome derived TMB and TMB estimated by our custom gene panel, by using TCGA data (R2 = 0.96; P < .001).
Overall, findings from the current study suggest that high nonsynonymous TMB represents a strong and favorable prognostic factor for outcomes in patients with resected NSCLC and could potentially be used to identify patients less likely to benefit from adjuvant chemotherapy. Studies are warranted to confirm these observations and explore the mechanisms underlying this association.
ACKNOWLEDGMENT
We acknowledge Ni Liu and Shakeel Virk for technical assistance.
Footnotes
Supported by National Institutes of Health Grant No. CA165958, Canadian Cancer Society Research Institute Grants No. 021039 and 704970, and La Ligue Nationale Contre le Cancer.
See accompanying Editorial on page 2978
AUTHOR CONTRIBUTIONS
Conception and design Siddhartha Devarakonda, Federico Rotolo, Ming-Sound Tsao, Irena Lanc, Ken A. Olaussen, Robert Fulton, Keyue Ding, Gwénaël Le Teuff, Frances A. Shepherd, Jean-Pierre Pignon, Stephen L. Graziano, Lesley Seymour, Ramaswamy Govindan, Stefan Michiels
Administrative support: Jean-Pierre Pignon, Lesley Seymour
Provision of study materials or patients: Lesley Seymour
Collection and assembly of data: Federico Rotolo, Ming-Sound Tsao, Elisabeth Brambilla, Robert Fulton, Shingo Sakashita, Anne McLeer-Florin, Keyue Ding, Gwénaël Le Teuff, Stephen L. Graziano, Jean-Charles Soria, Lesley Seymour, Ramaswamy Govindan, Stefan Michiels
Data analysis and interpretation: Siddhartha Devarakonda, Federico Rotolo, Ming-Sound Tsao, Irena Lanc, Elisabeth Brambilla, Ashiq Masood, Robert Fulton, Shingo Sakashita, Anne McLeer-Florin, Gwénaël Le Teuff, Frances A. Shepherd, Robert Kratzke, Jean-Charles Soria, Lesley Seymour, Ramaswamy Govindan, Stefan Michiels
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Tumor Mutation Burden as a Biomarker in Resected Non–Small-Cell Lung Cancer
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Siddhartha Devarakonda
No relationship to disclose
Federico Rotolo
Employment: Innate Pharma
Honoraria: Chugai Pharma
Ming-Sound Tsao
Consulting or Advisory Role: Merck Sharp & Dohme, Bristol-Myers Squibb, Pfizer, Ventana Medical Systems, AbbVie, Celgene, AstraZeneca/MedImmune
Research Funding: AstraZeneca (Inst), Merck Sharp & Dohme (Inst)
Irena Lanc
No relationship to disclose
Elisabeth Brambilla
No relationship to disclose
Ashiq Masood
Honoraria: Bristol-Myers Squibb, Boehringer Ingelheim
Speakers' Bureau: Bristol-Myers Squibb, Boehringer Ingelheim
Research Funding: Boston Biomedical
Ken A. Olaussen
No relationship to disclose
Robert Fulton
Honoraria: Roche
Patents, Royalties, Other Intellectual Property: Patent pending for a capture array for multiple myeloma
Shingo Sakashita
No relationship to disclose
Anne McLeer-Florin
No relationship to disclose
Keyue Ding
No relationship to disclose
Gwénaël Le Teuff
No relationship to disclose
Frances A. Shepherd
Stock or Other Ownership: Eli Lilly, AstraZeneca
Honoraria: Eli Lilly, AstraZeneca, Bristol-Myers Squibb, Roche, Merck Sharp & Dohme, Merck Serono, Boehringer Ingelheim
Consulting or Advisory Role: Eli Lilly, AstraZeneca, Boehringer Ingelheim, Merck Serono
Research Funding: Eli Lilly (Inst), Pfizer (Inst), Bristol-Myers Squibb (Inst), AstraZeneca/MedImmune (Inst), Roche Canada (Inst), Merrimack (Inst)
Jean-Pierre Pignon
No relationship to disclose
Stephen L. Graziano
No relationship to disclose
Robert Kratzke
No relationship to disclose
Jean-Charles Soria
Employment: MedImmune
Stock or Other Ownership: AstraZeneca/MedImmune
Honoraria: Roche, AstraZeneca, Sanofi, Servier
Lesley Seymour
Stock or Other Ownership: AstraZeneca
Consulting or Advisory Role: Boehringer Ingelheim, Hanmi
Research Funding: Pfizer (Inst), AstraZeneca (Inst), Innate Pharma (Inst), Oncolytics (Inst), Merck (Inst), Senwha (Inst)
Ramaswamy Govindan
Honoraria: AbbVie, Genentech
Consulting or Advisory Role: Pfizer, Celgene, AstraZeneca, AbbVie, Merck, Inivata, EMD Serono, Genentech, Bristol-Myers Squibb, Nektar, Merck Serono, Adaptimmune, Phillips Gilmore Oncology, GlaxoSmithKline
Research Funding: National Cancer Institute
Stefan Michiels
Consulting or Advisory Role: IDDI, Hexal, Johnson & Johnson, Genticel, Mabxience, Roche, QuintilesIMS
Patents, Royalties, Other Intellectual Property: Patent pending of a prognostic gene score in early breast cancer: WO2017EP66533
REFERENCES
- 1.Pignon JP, Tribodet H, Scagliotti GV, et al. : Lung adjuvant cisplatin evaluation: A pooled analysis by the LACE Collaborative Group. J Clin Oncol 26:3552-3559, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Douillard JY, Rosell R, De Lena M, et al. : Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): A randomised controlled trial. Lancet Oncol 7:719-727, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Butts CA, Ding K, Seymour L, et al. : Randomized phase III trial of vinorelbine plus cisplatin compared with observation in completely resected stage IB and II non-small-cell lung cancer: Updated survival analysis of JBR-10. J Clin Oncol 28:29-34, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strauss GM, Herndon JE, II, Maddaus MA, et al. : Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non-small-cell lung cancer: CALGB 9633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol 26:5043-5051, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arriagada R, Bergman B, Dunant A, et al. : Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med 350:351-360, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Ma X, Le Teuff G, Lacas B, et al. : Prognostic and predictive effect of TP53 mutations in patients with non-small cell lung cancer from adjuvant cisplatin-based therapy randomized trials: A LACE-bio pooled analysis. J Thorac Oncol 11:850-861, 2016 [DOI] [PubMed] [Google Scholar]
- 7.Tsao MS, Marguet S, Le Teuff G, et al. : Subtype classification of lung adenocarcinoma predicts benefit from adjuvant chemotherapy in patients undergoing complete resection. J Clin Oncol 33:3439-3446, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brambilla E, Le Teuff G, Marguet S, et al. : Prognostic effect of tumor lymphocytic infiltration in resectable non-small-cell lung cancer. J Clin Oncol 34:1223-1230, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsao MS, Le Teuff G, Shepherd FA, et al. : PD-L1 protein expression assessed by immunohistochemistry is neither prognostic nor predictive of benefit from adjuvant chemotherapy in resected non-small cell lung cancer. Ann Oncol 28:882-889, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shepherd FA, Lacas B, Le Teuff G, et al. : Pooled analysis of the prognostic and predictive effects of TP53 comutation status combined with KRAS or EGFR mutation in early-stage resected non–small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol 35:2018-2027, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kandoth C, McLellan MD, Vandin F, et al. : Mutational landscape and significance across 12 major cancer types. Nature 502:333-339, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Storey JD: A Direct Approach to False Discovery Rates. J R Stat Soc Series B Stat Methodol 64:479-498, 2002 [Google Scholar]
- 13.Poeta ML, Manola J, Goldwasser MA, et al. : TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med 357:2552-2561, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molina-Vila MA, Bertran-Alamillo J, Gasco A, et al. : Nondisruptive p53 mutations are associated with shorter survival in advanced non-small-cell lung cancer patients. Clin Cancer Res 20:4647-4659,2014 [DOI] [PubMed] [Google Scholar]
- 15.Mroz EA, Rocco JW: MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol 49:211-215, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong SQ, Li J, Tan AY, et al. : Sequence artefacts in a prospective series of formalin-fixed tumours tested for mutations in hotspot regions by massively parallel sequencing. BMC Med Genomics 7:23, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell JD, Alexandrov A, Kim J, et al. : Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 48:607-616, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durrleman S, Simon R: Flexible regression models with cubic splines. Stat Med 8:551-561, 1989 [DOI] [PubMed] [Google Scholar]
- 19.Van Allen EM, Mouw KW, Kim P, et al. : Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov 4:1140-1153, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fridman WH, Zitvogel L, Sautès-Fridman C, et al. : The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 14:717-734, 2017 [DOI] [PubMed] [Google Scholar]
- 21.Fong CY, Gilan O, Lam EY, et al. : BET inhibitor resistance emerges from leukaemia stem cells. Nature 525:538-542, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagaraj AB, Joseph P, Kovalenko O, et al. : Critical role of Wnt/β-catenin signaling in driving epithelial ovarian cancer platinum resistance. Oncotarget 6:23720-23734, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spranger S, Bao R, Gajewski TF: Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 523:231-235, 2015 [DOI] [PubMed] [Google Scholar]
- 24. Strauss G, Wang X, Maddaus M, et al: Adjuvant chemotherapy (AC) in stage IB non-small cell lung cancer (NSCLC): Long-term follow-up of Cancer and Leukemia Group B (CALGB) 9633. J Clin Oncol 29, 2011 (suppl; abstr 7015) [Google Scholar]
- 25.Mell LK, Jeong JH: Pitfalls of using composite primary end points in the presence of competing risks. J Clin Oncol 28:4297-4299, 2010 [DOI] [PubMed] [Google Scholar]
- 26.Eguchi T, Bains S, Lee MC, et al. : Impact of increasing age on cause-specific mortality and morbidity in patients with stage I non-small-cell lung cancer: A competing risks analysis. J Clin Oncol 35:281-290, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de la Chapelle A, Hampel H: Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol 28:3380-3387, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth AD, Delorenzi M, Tejpar S, et al. : Integrated analysis of molecular and clinical prognostic factors in stage II/III colon cancer. J Natl Cancer Inst 104:1635-1646, 2012 [DOI] [PubMed] [Google Scholar]
- 29.Sargent DJ, Marsoni S, Monges G, et al. : Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol 28:3219-3226, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizvi NA, Hellmann MD, Snyder A, et al. : Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348:124-128, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGranahan N, Furness AJ, Rosenthal R, et al. : Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351:1463-1469, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jamal-Hanjani M, Wilson GA, McGranahan N, et al. : Tracking the evolution of non-small-cell lung cancer. N Engl J Med 376:2109-2121, 2017 [DOI] [PubMed] [Google Scholar]
- 33.Carbone DP, Reck M, Paz-Ares L, et al. : First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med 376:2415-2426, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh E, Choi YL, Kwon MJ, et al. Comparison of accuracy of whole-exome sequencing with formalin-fixed paraffin-embedded and fresh frozen tissue samples PLoS One 10:e01441622015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zehir A, Benayed R, Shah RH, et al. : Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23:703-713, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]