Abstract
The tomato (Solanum lycopersicum) male sterile 32 (ms32) mutant has been used in hybrid seed breeding programs largely because it produces no pollen and has exserted stigmas. In this study, histological examination of anthers revealed dysfunctional pollen and tapetum development in the ms32 mutant. The ms32 locus was fine mapped to a 28.5 kb interval that encoded four putative genes. Solyc01g081100, a homolog of Arabidopsis bHLH10/89/90 and rice EAT1, was proposed to be the candidate gene of MS32 because it contained a single nucleotide polymorphism (SNP) that led to the formation of a premature stop codon. A codominant derived cleaved amplified polymorphic sequence (dCAPS) marker, MS32D, was developed based on the SNP. Real-time quantitative reverse-transcription PCR showed that most of the genes, which were proposed to be involved in pollen and tapetum development in tomato, were downregulated in the ms32 mutant. These findings may aid in marker-assisted selection of ms32 in hybrid breeding programs and facilitate studies on the regulatory mechanisms of pollen and tapetum development in tomato.
Subject terms: Pollen, Plant breeding
Introduction
Tomato (Solanum lycopersicum) is one of the most important vegetable crops in the world. Hybrid vigor has been extensively utilized in tomato production because F1-hybrid cultivars usually exhibit a greater yield and higher disease resistance than open-pollinated varieties1,2. Male sterility has proven to be a useful trait for hybrid seed production3. Male-sterile mutants are classified as structural, functional, and sporogenous types4. Structural male-sterile mutants of tomato usually bear extremely deformed stamens and do not produce pollen, such as stamenless (sl)5 and sl-26. Functional male-sterile mutants can produce normal pollen, but the pollen cannot reach the stigma because of indehiscent anthers or dehiscent anthers with morphological abnormalities, such as positional sterile (ps)7, ps-28, and exserted stigma (ex)9. Sporogenous male-sterile mutants exhibit relatively normal floral morphology but produce little or no fertile pollen. Most tomato male-sterile mutants belong to this type10. Thus, fine mapping and identifying the genes for sporogenous male-sterile mutants of tomato will not only help us understand the regulation of pollen development but also facilitate the use of these mutants in hybrid seed breeding programs.
Pollen development has been widely studied in Arabidopsis and rice (Oryza sativa)11. Several transcription factor genes have been identified as regulators involved in pollen development in Arabidopsis, such as DYSFUNCTIONAL TAPETUM1 (DYT1, referred to as AtDYT1 hereafter), DEFECTIVE TAPETUM DEVELOPMENT AND FUNCTION1 (TDF1, AtTDF1, syn. AtMYB35), ABORTED MICROSPORES (AMS, AtAMS), basic helix-loop-helix 10 (bHLH10, AtbHLH10), bHLH89 (AtbHLH89), bHLH91 (AtbHLH91), and MYB80 (AtMYB80, syn. AtMYB103)12–17. AtDYT1 encodes a bHLH transcription factor that directly regulates the expression of AtTDF118. AtTDF1 affects the expression of AtAMS19. AtAMS protein can interact with AtbHLH89/91 and regulate the expression of AtMYB8020,21. The interaction between AtDYT1 and AtbHLH10/89/91 is also important for the regulation of pollen development22. Therefore, they form a DYT1-TDF1-AMS-bHLH10/89/91-MYB80 transcriptional cascade to precisely regulate pollen and tapetum development in Arabidopsis17,23,24. A similar signaling pathway including the UDT1-TDR1-TIP2-EAT1 transcriptional cascade is also involved in the regulation of pollen and tapetum development in rice18. UDT1 (UNDEVELOPED TAPETUM1, OsUDT1) is a homolog of AtDYT125, TDR1 (TAPETUM DEGENERATION RETARDATION1, OsTDR1) is a homolog of AtAMS26, and TIP2 (TDR INTERACTING PROTEIN2, OsTIP2) and EAT1 (ETERNAL TAPETUM 1, OsEAT1) are homologs of AtbHLH10/89/9018,27. These findings suggest that the signaling pathway regulating pollen and tapetum development is highly conserved in higher plants23. In tomato, until now, only one regulator gene, SlMS10, a homolog of AtDYT1 and OsUDT1, has been found to be involved in pollen and tapetum development. A putative regulatory network for pollen and tapetum development in tomato, which is similar to the networks in Arabidopsis and rice, was presented based on the evolutionary relationships and conserved functions of the genes differentially expressed between the wild type (WT) and male sterile 1035 (ms1035)24. However, except for SlMS10, the regulators in the network need further investigation.
The tomato ms32 mutant is completely sterile because it does not produce pollen28. It has been used in hybrid seed breeding programs3. In this study, histological examination revealed dysfunctional microgametogenesis and tapetum degradation in the ms32 mutant. The ms32 locus was fine mapped to a 28.5 kb interval that encoded four putative genes. Solyc01g081100 was proposed to be the candidate gene of MS32 as it contained a single nucleotide polymorphism (SNP) that resulted in the formation of a premature stop codon. Solyc01g081100 is a homolog of AtbHLH10/89/90 and OsEAT1. Real-time quantitative reverse-transcription PCR (qPCR) showed that most of the regulator genes, which were proposed to be involved in pollen and tapetum development, were downregulated in the ms32 mutant. However, the expression of SlMS10 did not show a significant difference in the ms32 mutant compared to the WT. These findings may help in marker-assisted selection of ms32 in hybrid breeding programs and facilitate studies on the regulatory mechanisms of pollen and tapetum development in tomato.
Results
Phenotypic investigation of the ms32 mutant
The morphologies of the ms32 mutant and WT plants were the same until the flowering stage. At the flowering stage, the ms32 mutant had longer sepals and shorter petals than the WT (Fig. 1). In addition, the stamens in ms32 flowers were greatly reduced and shrunken and pale in color, usually with the stigma remaining exserted (Fig. 1). Pollen viability analysis detected no pollen in the flowers of the ms32 mutant (Fig. 1). The ms32 mutant could not set fruit after self-pollination with an electric vibrator, but it was able to produce fruit when manually pollinated with pollen from WT plants (Fig. S1). These findings were consistent with a previous study of ms3228.
Fig. 1. Flower phenotypes and pollen production of the ms32 mutant.
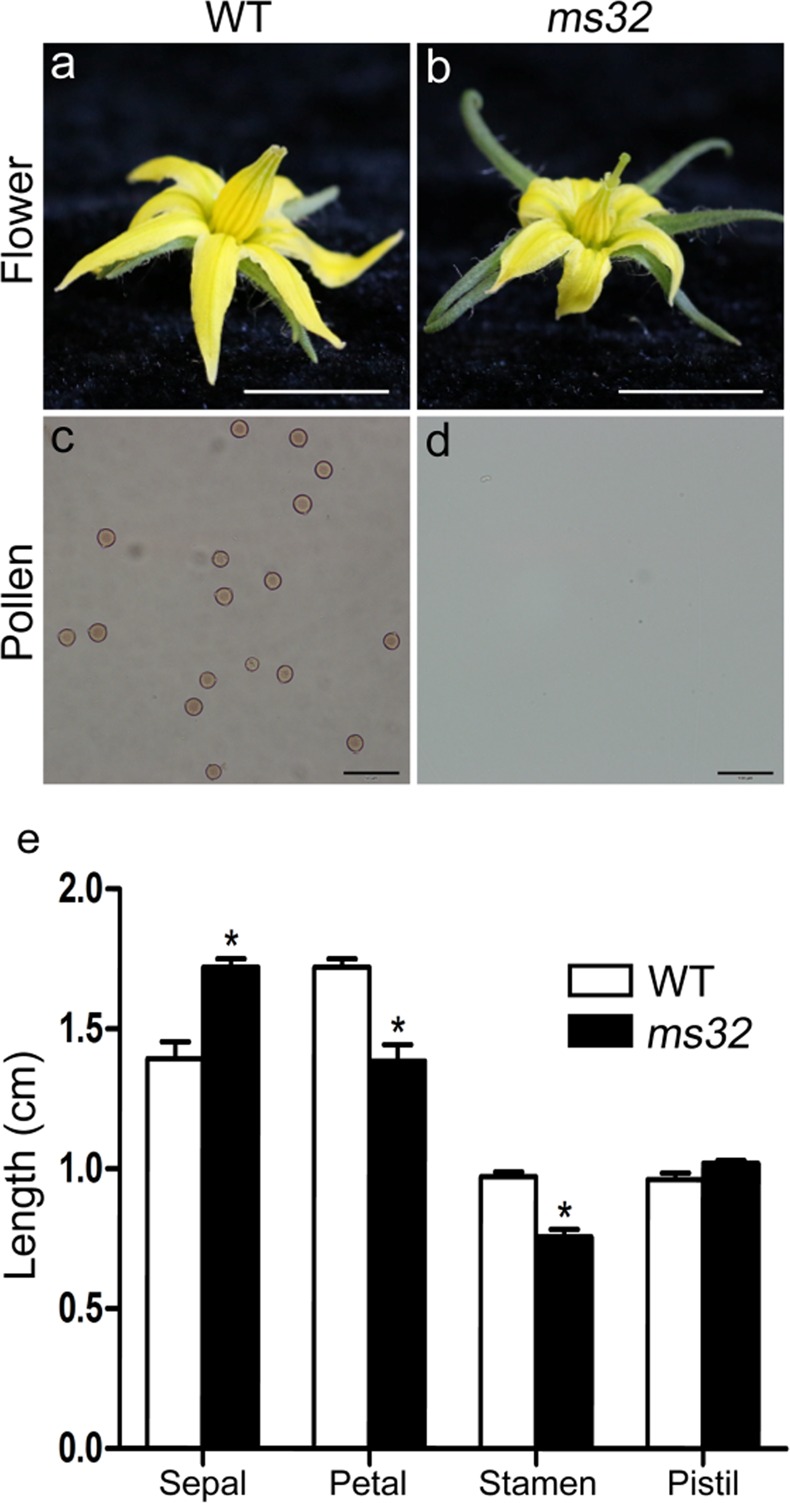
a, b Flower of WT (a) and ms32 plants (b). Scale bars, 1 cm. c, d Analysis of pollen viability by acetocarmine staining of pollen from anthers of WT (c) and ms32 plants (d). Scale bars, 50 μm. e Length of floral organs in WT and ms32 flowers at anthesis. Values represent the means ± SEs. Asterisks indicate a significant difference (p < 0.05) between the WT and ms32 mutant
Histological examination of ms32 anthers
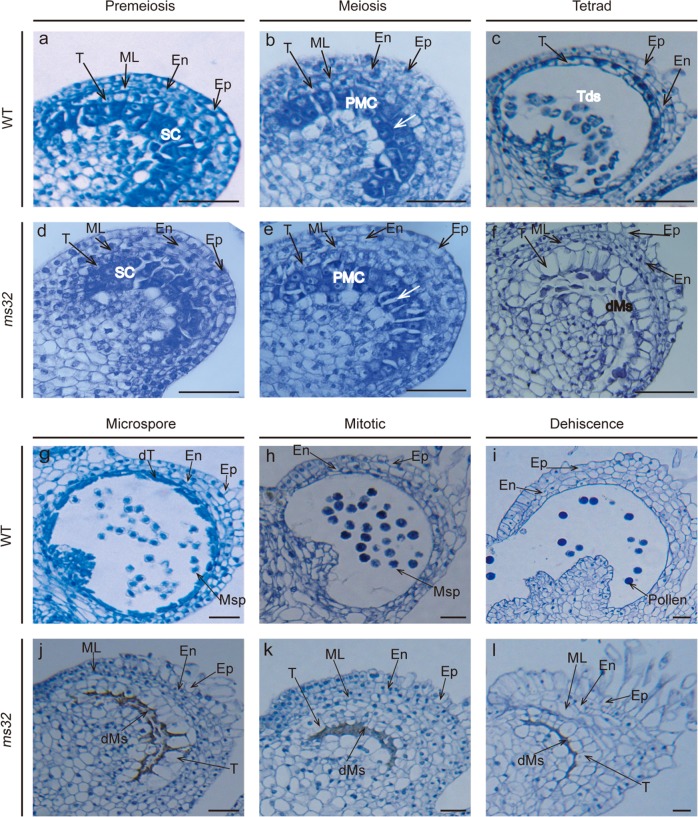
Histological examination of anthers at different developmental stages was performed to detect the spatial and temporal occurrence of defects in ms32 anthers. At the premeiotic stage, the cell layer differentiation in ms32 anthers seemed to be similar to that in WT anthers (Fig. 2a, d). At the meiotic stage of WT anthers, sporogenous cells developed in pollen mother cells (PMCs), and meiosis was initiated and completed (Fig. 2b). However, PMCs began to crush and separate from each other in ms32 anthers (Fig. 2e). At the tetrad, microspore, mitotic and dehiscence stages, dramatic changes in morphology were detected between WT and ms32 anthers. In WT anthers, PMCs successively developed into tetrads (Fig. 2c), microspores (Fig. 2g), vacuolated microspores (Fig. 2h), and pollen grains (Fig. 2i). In addition, the tapetal cells gradually degenerated and disappeared (Fig. 2c, g–i). In ms32 anthers, PMCs were crushed and could not produce tetrads, microspores and pollen grains successively. In contrast, tapetal cells were enlarged and vacuolated at the tetrad stage and remained until the dehiscence stage (Fig. 2f, j–l).
Fig. 2. Histological examination of anthers at different developmental stages in WT and ms32 plants.
Transverse sections of WT (a–c, g–i) and ms32 (d–f, j–l) anthers at different developmental stages. a, d Premeiotic stage; b, e Meiotic stage, with white arrows pointing to the crushed and separated PMCs; c, f Tetrad stage; g, j Microspore stage; h, k Mitotic stage; i, l Dehiscence stage. dMs, degenerated meiocytes; dT, degenerated tapetum; En, endothecium; Ep, epidermis; ML, middle cell layer; Msp, microspore; PMC, pollen mother cell; SC, sporogenous cell; T, tapetum; Tds, tetrads. Scale bars, 50 μm
Genetic analysis of the ms32 locus
To determine the inheritance of the ms32 locus, F1 plants were obtained from a cross between the ms32 mutant and LA1589. All F1 plants developed normal flowers and fruits. The F2 population derived from the abovementioned F1 plants segregated in an approximate 3:1 ratio (fertility:sterility = 500:165, χ2 = 0.27, less than χ20.05 = 3.84, χ2-test), which indicated that the sterility phenotype was caused by a single recessive mutation.
Fine mapping of the ms32 locus
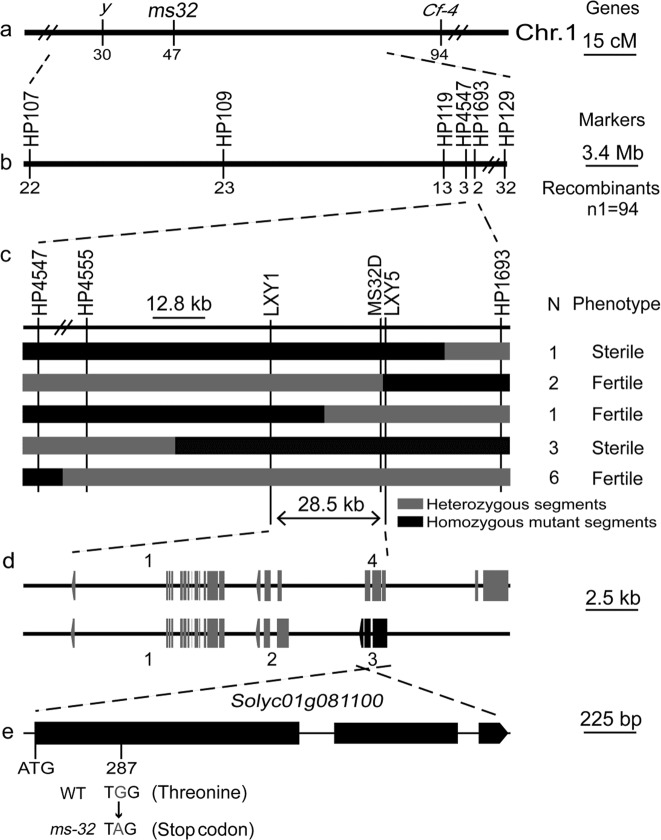
The ms32 locus has been determined to be located between the y locus and Cf-4 locus on chromosome 1 (Fig. 3a)29. The genes Y and CF-4 have been cloned30,31. Based on this information, six insertion/deletion (InDel) markers were developed in the region between the two genes (Table S1). For preliminary mapping of the ms32 locus, these InDel markers were used to genotype 94 sterile male plants in an F2 population. The genotype of the recombinant individuals, which was detected by these markers, indicated that the ms32 locus was located between markers HP4547 and HP1693 (Fig. 3b). For fine mapping of the ms32 locus, three InDel markers were developed within the region between markers HP4547 and HP1693. The markers HP4547 and HP1693 were utilized to genotype 665 individuals from the whole F2 population. A total of 13 recombinants, which were determined to be homozygous for the ms32 mutant and for just one of these two markers, were selected for further genotyping. Based on the genotype and phenotype of these recombinants, the ms32 locus was narrowed to an interval between markers LXY1 and LXY5, a 28.5 kb interval on chromosome 1 (Fig. 3c).
Fig. 3. Fine mapping of the ms32 locus.
a Genetic position of the ms32 locus on the genetic map constructed by Mutschler et al.29. b Preliminary mapping of the ms32 locus using 94 sterile male plants. c Fine mapping of the ms32 locus. N indicates the number of recombinants. d The annotated genes near the ms32 locus according to ITAG release 3.2. Rectangles and arrows indicate the exons and direction of transcription, respectively. e The exon-intron structure of the coding region of Solyc01g081100 and the sequence polymorphism in the ms32 mutant
Candidate gene analysis
Four putative genes were identified in the 28.5 kb region that contained the ms32 locus by searching the tomato genome annotation database (ITAG release 3.2) in the Sol Genomics Network (SGN, https://solgenomics.net/; Fig. 3d and Table 1). qPCR analysis revealed that Solyc01g081090 and Solyc01g081100 were highly expressed in stamens (Fig. S3). However, the expression of Solyc01g081080 was barely detected in any organs analyzed in this study (Fig. S3), and the expression of Solyc01g081110 in sepals and petals was higher than that in stamens and carpels (Fig. S3). The genes Solyc01g081090 and Solyc01g081100 encoded bHLH transcription factors. Phylogenic analysis showed that they were within the same clade, which also included AtbHLH10/89/91, OsETA1, and OsTIP2 (Fig. 4), which play important roles in pollen and tapetum development17,18,27. Therefore, Solyc01g081090 and Solyc01g081100 seemed to be candidate genes for ms32. Sequencing results showed that there was no sequence difference between the WT and ms32 mutant within Solyc01g081090. However, there was a SNP from G to A in the coding region of Solyc01g081100 (Fig. 3e), which resulted in a premature stop codon and protein truncation (Fig. 3e and S4). Therefore, Solyc01g081100 was the most likely candidate gene of MS32.
Table 1.
Predicted genes in the ms32 region
| ID | Gene namea | Position on SL3.0ch01 | Putative function |
|---|---|---|---|
| 1 | Solyc01g081080.2.1 | 80273359-80281965 (−) | Replication factor C subunit |
| 2 | Solyc01g081090.2.1 | 80284835-80287067 (−) | bHLH transcription factor 003 |
| 3 | Solyc01g081100.2.1 | 80290985-80292911 (−) | bHLH transcription factor 004 |
| 4 | Solyc01g081110.3.1 | 80284835-80300423 (−) | Pentatricopeptide repeat-containing protein |
aGenes were identified by using the tomato gene model (ITAG release 3.2, SL3.0) in the SGN (https://solgenomics.net/)
Fig. 4. Phylogenetic analysis of bHLH proteins related to pollen and tapetum development in several plant species.
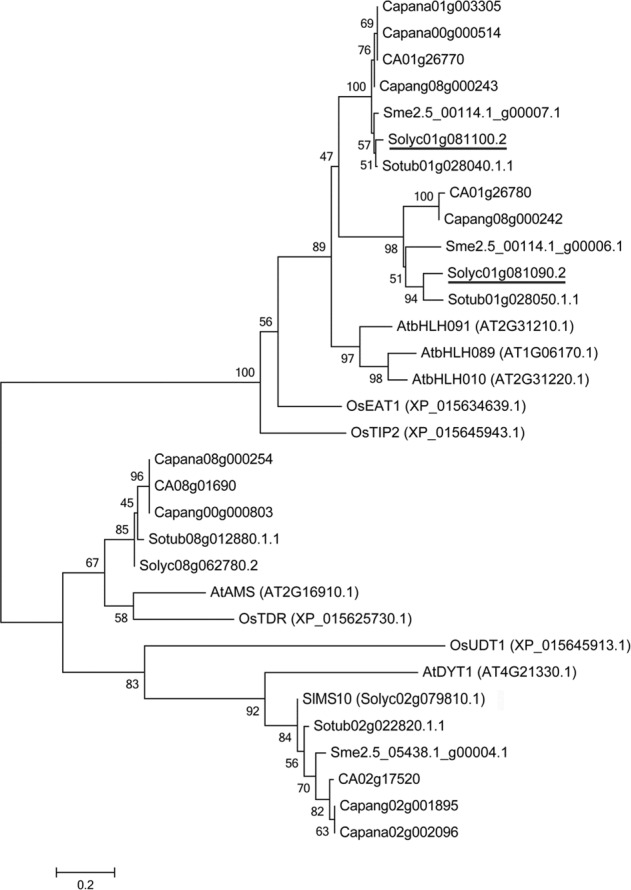
The phylogenetic tree was constructed with the neighbor-joining method by using MEGA6 (Tamura et al., 2013) with 1000 bootstrap replicates, which was based on the full-length deduced amino acid sequences. Solyc01g081090.2 and Solyc01g081100.2 are underlined. At, Arabidopsis thaliana; Ca, Capsicum annuum; Capana, Capsicum annuum var. zunla; Capang, Capsicum annuum var. glabriusculum; Os, Oryza sativa; Sl, Solanum lycopersicum; Sme, Solanum melongena; Solyc, Solanum lycopersicum; Sotub, Solanum tuberosum
Expression analysis of the genes involved in pollen and tapetum development
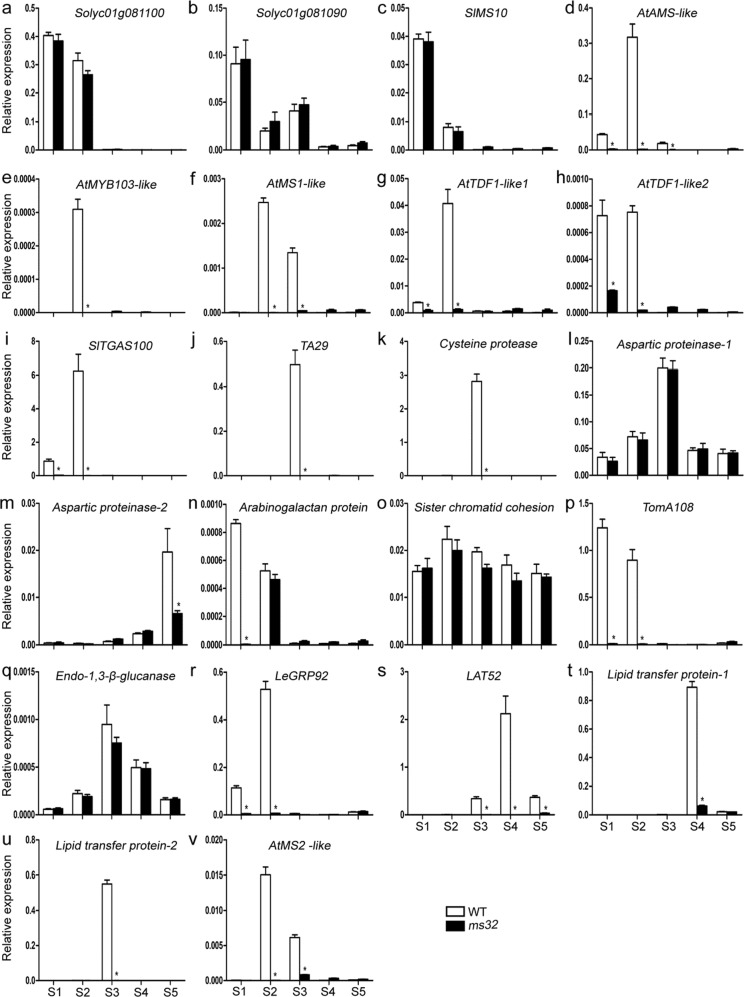
Given that the ms32 mutant exhibited abnormal pollen and tapetum development (Fig. 2) as well as contained a SNP that was predicted to cause a premature stop codon in the Solyc01g081100 gene (Fig. 3e and S4), which is a homolog of the pollen and tapetum development regulator genes AtbHLH10/89/91, OsETA1, and OsTIP2 (Fig. 4)17,18,27, the transcriptional expression of several genes previously proposed to be related to tomato pollen and tapetum development were detected in flowers of WT and ms32 plants24,32. For the genes encoding transcription factors, Solyc01g081090, Solyc01g081100, and SlMS10 (Solyc02g079810) were expressed highly and almost equally in the early stages of flower development in WT and ms32 plants (Fig. 5a–c). In contrast, AtAMS-like (Solyc08g062780), AtTDF1-like1 (Solyc03g113530), AtTDF1-like2 (Solyc03g059200), AtMS1-like (Solyc04g008420), and AtMYB103-like (Solyc10g005760) showed significant downregulation in the ms32 mutant (Fig. 5d–h).
Fig. 5. Transcription/expression of the genes related to pollen and tapetum development in flower buds of WT and ms32 plants.
qPCR of the Solyc01g081100 (a), Solyc01g081090 (b), SlMS10 (c), AtAMS-like (d), AtMYB103-like (e), AtMS1-like (f), AtTDF1-like1 (g), AtTDF1-like2 (h), SlGAS100 (i), TA29 (j), cysteine protease (k), aspartic protease-1 (l), aspartic protease-2 (m), arabinogalactan protein (n), sister chromatid cohesion (o), TomA108 (p), endo-1,3-β-glucanase (q), LeGRP92 (r), LAT52 (s), lipid transfer protein-1 (t), lipid transfer protein-1 (u), and AtMS2-like (v) genes. Values represent the means ± SEs. Asterisks indicate a significant difference (p < 0.05) between WT and ms32 plants. S1–S5, flower buds at five stages of anther development. S1, meiosis and tetrad stage; S2, microspore stage; S3, mitotic stage; S4, dehiscence stage; S5, opened flower stage
For the genes related to tapetum degradation, Cysteine protease (Solyc07g053460), TA29 (Solyc02g078370)33, and TGAS100 (Solyc06g064470)34 were downregulated in the ms32 mutant (Fig. 5i–k); Arabinogalactan protein (Solyc11g072780) and Aspartic proteinase-2 (Solyc08g068870) were only significantly downregulated at the earliest and latest stages of flower development in the ms32 mutant, respectively (Fig. 5m, n). However, Aspartic proteinase-1 (Solyc06g069220) was expressed almost equally between WT and ms32 flowers in all stages examined (Fig. 5l). For the genes associated with pollen formation and maturation, TomA108 (Solyc01g009590)35, Lipid transfer protein-1 (Solyc06g059790), Lipid transfer protein-2 (Solyc01g095780), AtMS2-like (Solyc03g051960), LAT52 (Solyc10g007270)36,37, and LeGRP92 (Solyc02g032910)38 showed downregulation in the ms32 mutant (Fig. 5p, r–v). In contrast, Sister chromatid cohesion (Solyc03g116930) and Endo-1,3-β-glucanase (Solyc03g046200) were expressed almost equally between WT and ms32 flowers in all stages examined (Fig. 5o, q).
Discussion
Solyc08g091100, a putative bHLH transcription factor gene, is a candidate gene for the ms32 locus controlling male sterility in tomato
The tomato ms32 mutant is considered useful in hybrid seed production, mainly due to the following advantages: (1) stably and completely sterile and (2) accessible for pollination without emasculation due to the exerted stigma3. However, ms32 is a recessive mutation, and its mutant phenotype is expressed at the flowering stage (Fig. 1); thus, it is inconvenient to distinguish fertile and sterile plants when the ms32 line is used in hybrid seed production and the ms32 locus is introduced into the inbred line. The availability of molecular markers tightly linked to the MS32 mutant gene would facilitate functional studies and breeding. In this study, the ms32 locus was fine mapped to a 28.5 kb interval that encoded four putative genes (Fig. 3). Solyc01g081100 contained a SNP in the coding region in the ms32 mutant, which resulted in a premature stop codon and protein truncation (Fig. 3e and S4). Solyc01g081100 is a homolog of AtbHLH10/89/91, OsETA1, and OsTIP2 (Fig. S4), whose loss-of-function mutants are also male sterile17,18,27. However, no sequence differences and no expression differences were detected between the WT and ms32 mutant for the genes Solyc01g081080, Solyc01g081090, and Solyc01g081110 (Fig. 5b and S5). Therefore, Solyc01g081100 was the most likely candidate gene of MS32. A MS32 candidate gene-specific marker, MS32D, was developed based on the SNP in the coding region of Solyc01g081100. MS32D successfully distinguished the homozygous WT, heterozygote, and homozygous ms32 mutant (Fig. S6). Therefore, this marker may facilitate MAS of ms32 in the breeding of tomato male-sterile female parents in hybrid seed production.
The MS32 gene regulates pollen and tapetum development
The tomato ms32 mutant did not release pollen from the flowers at anthesis (Fig. 1). Histological examination revealed defects in microgametogenesis and tapetum degradation resulting from the ms32 mutation (Fig. 2). At and after the tetrad stage, in ms32 anthers, PMCs were degenerated and failed to successively produce tetrads, microspores, and pollen grains. Additionally, tapetal cells were abnormally enlarged and vacuolated without degeneration (Fig. 2). Fine-mapping and sequencing results indicated that Solyc01g081100, a homolog of AtbHLH10/89/91 and OsEAT1 that regulates pollen and tapetum development, was the most likely candidate gene of MS32 (Figs. 3 and 4). qPCR also showed that most of the genes proposed to be involved in pollen and tapetum development in tomato were downregulated in the ms32 mutant (Fig. 5). These findings suggest that the MS32 gene is involved in the regulation of pollen and tapetum development.
Previous studies have suggested that Arabidopsis and rice share a highly conserved regulatory network of pollen and tapetum development23. Two pathways, namely, the DYT1-TDF1-AMS-bHLH10/89/91-MYB80 transcriptional cascade in Arabidopsis and the UDT1-TDR1-TIP2-EAT1 transcriptional cascade in rice, have been suggested to regulate pollen and tapetum development17,18,23,24. Recently, SlMS10 (Solyc02g079810), a homolog of AtDYT1 and OsUDT1, has been proposed to lead a similar transcriptional cascade regulating pollen and tapetum development in tomato24. Loss of function of the SlMS10 gene results in downregulation of the transcription factor genes AtTDF1-like, AtAMS-like, AtbHLH89/91-like (Solyc01g081100), AtMYB103-like, and AtMS1-like24. In this study, most of these genes were downregulated in the ms32 mutant, except the AtbHLH89/91-like genes (Solyc01g081090 and Solyc01g081100) and SlMS10 (Fig. 5a–h). Based on the fine-mapping and sequencing results in this study and transcription analysis results in this and previous studies24, Solyc01g081100 is proposed to be the candidate gene of MS32 and to be a regulator gene downstream of SlMS10. The results also suggested that there was no self-feedback regulation of Solyc01g081100 expression and no feedback regulation between the MS32 and SlMS10 genes. However, SlMS10 and MS32 might work together to regulate pollen and tapetum development in tomato. In Arabidopsis, AtDYT1 upregulates the expression of AtbHLH89 and forms a protein complex with AtbHLH89 protein to activate the expression of AtTDF122. Similarly, in this and previous studies24, mutations in SlMS10 and Solyc01g081100 were related to the downregulation of AtTDF1-like genes (Fig. 5g, h). It is necessary to detect whether SlMS10 interacts with Solyc01g081100 to promote the expression of AtTDF1-like genes.
The AtbHLH10, AtbHLH89, and AtbHLH91 genes resulted from duplication and are functionally redundant for normal pollen and tapetum development in Arabidopsis. Single mutants in each of the AtbHLH10, AtbHLH89, and AtbHLH91 genes are developmentally normal and without obviously defective pollen grains17. Two AtbHLH89/91-like genes, namely, Solyc01g081090 and Solyc01g081100, were identified in tomato. These two genes might be the result of tandem duplication as they are closely linked on the same chromosome. This tandem duplication might have occurred before the separation of Solanaceae species because potato, pepper, and eggplant all contain two AtbHLH89/91-like genes that are tightly linked on the same chromosome (Fig. 4). The tomato ms32 mutant was completely male sterile (Fig. 1 and S1). It contained a mutation in Solyc01g081100, while there was no sequence polymorphism or expression difference in Solyc01g081090 (Figs. 3 and 5b), which suggested functional divergence of these two genes. In rice, OsEAT1, a homolog of Solyc01g081090 and Solyc01g081100, activates tapetal cell death by regulating aspartic proteases27. An aspartic protease gene (Solyc06g069220) was also found to be downregulated in the tomato ms1035 mutant and the SlMED18 RNA interference line, which are both defective in tapetum development24,32. However, this aspartic protease gene did not show significantly different expression in the tomato ms32 mutant (Fig. 5l). Furthermore, Solyc01g081100, the candidate gene of MS32, is downregulated in the tomato ms1035 mutant but not in the SlMED18 RNA interference line. In contrast, SlMS10, putatively encoding a regulator upstream of Solyc01g081100 in the transcriptional cascade in tomato, is downregulated in both the tomato ms1035 mutant and the SlMED18 RNA interference line24,32. These findings suggest that the other bHLH transcription factor, in addition to Solyc01g081100, might also be involved in the regulation of tapetum development. Further studies on Solyc01g081090 and Solyc01g081100 may help us understand the details of pollen and tapetum development regulation in tomato.
In summary, in this study, the tomato ms32 mutant did not release pollen and exhibited abnormalities in pollen and tapetum development (Figs. 1 and 2). The ms32 locus was fine mapped to a 28.5 kb interval that encoded four putative genes. Solyc01g081100 was proposed to be the candidate gene of MS32 as it contained a SNP that resulted in the formation of a premature stop codon (Fig. 3 and S4). The candidate gene-specific marker MS32D (Fig. S6), which was developed according to the SNP in the gene Solyc01g081100, can be used for MAS of male-sterile inbred lines of tomato. Solyc01g081100 can also be used as the target for gene editing to quickly develop such inbred lines. qPCR analysis showed that the expression levels of several genes involved in pollen and tapetum development were changed in the ms32 mutant (Fig. 5). These findings will support studies on the regulatory mechanisms of pollen and tapetum development in tomato.
Materials and methods
Plant materials
Seeds of ms32 (TGRC accession number LA2714, background genotype: Montfavet-168) and LA1589, the wild tomato species Solanum pimpinellifolium, were obtained from the Tomato Genetics Resource Center (TGRC, Davis, CA, USA). The source of the other seeds used in this study was previously described39. The male-sterile plant and homozygous male-fertile plant derived from LA2714 were named the ms32 mutant and WT, respectively. A total of 665 plants of an F2 population derived from a cross between the ms32 mutant and the wild tomato species LA1589 were grown in an open field in Shunyi District (Beijing, China) in the spring and summer of 2016. For the phenotypic analysis and expression analysis, the WT and ms32 mutant plants were grown in a greenhouse in Haidian District (Beijing, China) in the spring and summer of 2018.
Phenotypic analysis and histological examination
The morphology of the WT and ms32 mutant flowers was observed at anthesis. The entire flower was photographed by a camera (Canon EOS 70D, Canon Inc., Japan). Pollen viability was tested using the method described by Sinha and Rajam40. Histological examination was performed using the method described by Brukhin et al.41. Slides were observed under a microscope (Olympus BX51TRF, Olympus Corporation, Tokyo, Japan).
Molecular marker development and genotyping
InDels were identified by comparing the sequences of chromosome 1 of tomato lines Heinz 1706 and LA1589, whose whole-genome sequence was published and released in the SGN (https://solgenomics.net/)42. PCR primers matching the flanking regions of these InDels were designed using the Primer-BLAST tool available from the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/tools/primer-blast/). These primers were used to test for polymorphisms between the ms32 mutant and LA1589. The MS32 candidate gene-specific marker MS32D was developed based on DNA polymorphism within the MS32 candidate gene. General information regarding the DNA markers used for fine mapping of ms32 is given in Table S1. The PCR conditions were as follows: 94 °C for 4 min; followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30–60 s; and 72 °C for 5 min. The PCRs were performed on a PCR instrument (Bio-Rad, Hercules, CA, USA), and the PCR products of these markers were separated on a 3.0% agarose gel. For the marker MS32D, the PCR products were digested with 2 U DdeI restriction enzyme for 2 h, after which the digested products were separated on a 3% agarose gel.
Genetic analysis and fine mapping
The segregation ratio of the ms32 locus was tested by chi-square analysis using 665 individuals of an F2 population. For preliminary mapping of the ms32 locus, 94 sterile plants in the F2 population were genotyped using six InDel markers in the interval between genes Y (SlMYB12) and Cf-4 on chromosome 1, as the ms32 locus is located in this interval in the genetic map reported in 198729–31. For fine mapping of the ms32 locus, the whole F2 population was genotyped using markers HP4547 and HP1693. A total of 13 recombinants, which were determined to be homozygous for the ms32 mutant for just one of these two markers, were selected for further genotyping using additional markers (see Table S1).
Gene prediction
The putative genes in the fine-mapping region harboring the ms32 locus were identified using the tomato gene model (ITAG release 3.2) in the SGN (https://solgenomics.net).
Phylogenetic tree construction
The protein sequences of the known bHLH transcription factors AtAMS, AtbHLH10, AtbHLH89, AtbHLH91, AtDYT1, OsEAT1, OsTDR1, OsTIP2, and OsUDT1, which are related to pollen and tapetum development in Arabidopsis and rice, were downloaded from The Arabidopsis Information Resource (https://www.arabidopsis.org/) and GenBank. Using the full-length protein sequences of these bHLH transcription factors as queries, the homologs of eggplant, pepper, potato and tomato were identified by BLASTP in the databases of eggplant genome protein sequences (release 2.5), Capsicum annuum cv CM334 genome protein sequences (release 1.55), Capsicum annuum var. glabriusculum genome protein sequences (v2.0), Capsicum annuum var. zunla genome protein sequences (v2.0), potato ITAG Release 1 predicted proteins (ST1.0), and tomato genome proteins (ITAG release 3.20) in the SGN with an E-value ≤ 1 × 10−50. Multiple alignment of the full-length sequences of these bHLH transcription factors was performed by Clustal X ver. 2.143 using the default settings. An unrooted phylogenetic tree was constructed by using MEGA644 with the neighbor-joining method, Jones-Taylor-Thornton (JTT) model, and 1000 replicates.
Sequence polymorphism analysis
The genomic fragments of the candidate genes of MS32, namely, Solyc01g081090 and Solyc01g081100, were obtained from the WT and ms32 mutant by overlapping PCR. The amplified fragments were sequenced at the Beijing Genomics Institute (Beijing, China). The partial cDNAs of the Solyc01g081100 gene were obtained by reverse-transcription PCR (RT-PCR), which was performed using Phusion High-Fidelity DNA Polymerase (Cat. No. M0530L; New England Biolabs, Ipswich, MA, USA) and the primers for cDNA cloning (Table S1). The amplified fragments were cloned using a pEASY-Blunt Zero Cloning Kit (Cat. No. CB501-02; TransGen Biotech, Beijing, China). The cDNA clones were sequenced at the Beijing Genomics Institute.
RNA extraction, cDNA synthesis, and qPCR analysis
Flower buds at different developmental stages were collected from WT and ms32 plants and rapidly frozen in liquid nitrogen. Each tissue sample comprised three biological replicates, and each replicate contained samples from at least three plants. Total RNA was isolated using the Quick RNA Isolation Kit (Cat. No. BC1803, Huayueyang Biotech Co. Ltd., Beijing, China), and cDNA was synthesized from 2 μg total RNA using GoScript Reverse Transcriptase (Cat. No. A5003; Promega, Madison, WI, USA). The qPCRs were conducted using GoScriptTM qPCR Master Mix (Cat. No. A6002; Promega) and a LightCycler 480 Detection System (Roche Diagnostics GmbH, Mannheim, Germany). The primers for qPCR are provided in Table S1. qPCR was performed using the methods described previously by Cao et al.45.
Supplementary information
Tomato ms32 supplementary figures and tables
Acknowledgements
We thank the Tomato Genetics Resource Center (Davis, CA, USA) for providing seed stocks. This work was supported by the National Natural Science Foundation of China (No. 31471889), the National Key Research and Development Program of China (No. 2016YFD0101703), the Central Public-interest Scientific Institution Basal Research Fund (No. Y2017PT52), and the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (No. CAAS-ASTIP-IVFCAAS).
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Yong Qin, Phone: +86 991 8762361, Email: 352167610@qq.com.
Zejun Huang, Phone: +86 10 82109530, Email: huangzejun@caas.cn.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-019-0170-2).
References
- 1.Bai YL, Lindhout P. Domestication and breeding of tomatoes: What have we gained and what can we gain in the future? Ann. Bot.-Lond. 2007;100:1085–1094. doi: 10.1093/aob/mcm150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tamta S, Singh JP. Heterosis in tomato for growth and yield traits. Int. J. Veg. Sci. 2018;24:169–179. doi: 10.1080/19315260.2017.1407857. [DOI] [Google Scholar]
- 3.Atanassova, B. Genic male sterility and its application in tomato (Lycopersicon esculentum mill.) hybrid breeding and hybrid seed production. Proceedings of the IIIrd Balkan Symposium on Vegetable and Potatoes, 45–51 (2007).
- 4.Kaul, M. L. H. Male sterility in higher plants (Springer, Berlin, 1988).
- 5.Gómez Pedro, Jamilena Manuel, Capel Juan, Zurita Sergio, Angosto Trinidad, Lozano Rafael. Stamenless , a tomato mutant with homeotic conversions in petals and stamens. Planta. 1999;209(2):172–179. doi: 10.1007/s004250050619. [DOI] [PubMed] [Google Scholar]
- 6.Sawhney VK, Greyson RI. Morphogenesis of the stamenless-2 mutant in tomato. I. Comparative description of the flowers and ontogeny of stamens in the normal and mutant plants. Am. J. Bot. 1973;60:514–523. doi: 10.1002/j.1537-2197.1973.tb05952.x. [DOI] [Google Scholar]
- 7.Larson, R. E. & Paur, S. The description and the inheritance of a functionally sterile flower mutant in tomato and its probable value in hybrid tomato seed production. Proc. Am. Soc. Hort. Sci. 52, 355–364 (1948).
- 8.Atanassova B. Functional male sterility (ps-2) in tomato (Lycopesicon esculentum Mill.) and its application in breeding and hybrid seed production. Euphytica. 1999;107:13–21. doi: 10.1023/A:1003527714805. [DOI] [Google Scholar]
- 9.Rick CM, Robinson J. Inherited defects of floral structure affecting fruitfulness in Lycopersicon esculentum. Am. J. Bot. 1951;38:639–652. doi: 10.1002/j.1537-2197.1951.tb14871.x. [DOI] [Google Scholar]
- 10.Gorman SW, McCormick S, Rick C. Male Sterility in Tomato. Crit. Rev. Plant Sci. 1997;16:31–53. doi: 10.1080/07352689709701945. [DOI] [Google Scholar]
- 11.Wilson ZA, Zhang DB. From Arabidopsis to rice: pathways in pollen development. J. Exp. Bot. 2009;60:1479–1492. doi: 10.1093/jxb/erp095. [DOI] [PubMed] [Google Scholar]
- 12.Phan HA, Iacuone S, Li SF, Parish RW. The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell. 2011;23:2209–2224. doi: 10.1105/tpc.110.082651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sorensen AM, et al. Plant J. 2003. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor; pp. 413–423. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W. Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development. 2006;133(16):3085–3095. doi: 10.1242/dev.02463. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Zai-Bao, Zhu Jun, Gao Ju-Fang, Wang Chen, Li Hui, Li Hong, Zhang Hui-Qi, Zhang Sen, Wang Dong-Mi, Wang Quan-Xi, Huang Hai, Xia Hui-Jun, Yang Zhong-Nan. Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. The Plant Journal. 2007;52(3):528–538. doi: 10.1111/j.1365-313X.2007.03254.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhu J, et al. Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 2008;55:266–277. doi: 10.1111/j.1365-313X.2008.03500.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhu E, et al. The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J. 2015;83:976–990. doi: 10.1111/tpj.12942. [DOI] [PubMed] [Google Scholar]
- 18.Fu Z, et al. The Rice Basic Helix-Loop-Helix Transcription Factor TDR INTERACTING PROTEIN2 Is a Central Switch in Early Anther Development. Plant Cell. 2014;26:1512–1524. doi: 10.1105/tpc.114.123745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu J, Lou Y, Xu X, Yang ZN. A genetic pathway for tapetum development and function in Arabidopsis. J. Integr. Plant Biol. 2011;53:892–900. doi: 10.1111/j.1744-7909.2011.01078.x. [DOI] [PubMed] [Google Scholar]
- 20.Xu, J. et al. The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell22, 91–107 (2010). [DOI] [PMC free article] [PubMed]
- 21.Ferguson AC, et al. Biphasic regulation of the transcription factor ABORTED MICROSPORES (AMS) is essential for tapetum and pollen development in Arabidopsis. New Phytol. 2017;213:778–790. doi: 10.1111/nph.14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui J, et al. Feedback Regulation of DYT1 by Interactions with Downstream bHLH Factors Promotes DYT1 Nuclear Localization and Anther Development. Plant Cell. 2016;28:1078–1093. doi: 10.1105/tpc.15.00986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomez JF, Talle B, Wilson ZA. Anther and pollen development: A conserved developmental pathway. J. Integr. Plant Biol. 2015;57:876–891. doi: 10.1111/jipb.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong HJ, et al. Tomato Male sterile 1035 is essential for pollen development and meiosis in anthers. J. Exp. Bot. 2014;65:6693–6709. doi: 10.1093/jxb/eru389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung KH, et al. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell. 2005;17:2705–2722. doi: 10.1105/tpc.105.034090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N, et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell. 2006;18:2999–3014. doi: 10.1105/tpc.106.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niu N, et al. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. commun. 2013;4:1445. doi: 10.1038/ncomms2396. [DOI] [PubMed] [Google Scholar]
- 28.Rick CM. New male-sterile mutants. Rep. Tomato Genet Coop. 1960;10:35–37. [Google Scholar]
- 29.Mutschler, M. A., Rick, C. M. & Tanksley, S. D. Linkage maps of the tomato (Lycopersicon esculentum). Rep. Tomato Genet Coop.37, 5–34 (1987).
- 30.Lin T, et al. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 2014;46:1220–1226. doi: 10.1038/ng.3117. [DOI] [PubMed] [Google Scholar]
- 31.Parniske Martin, Hammond-Kosack Kim E, Golstein Catherine, Thomas Colwyn M, Jones David A, Harrison Kate, Wulff Brande B.H, Jones Jonathan D.G. Novel Disease Resistance Specificities Result from Sequence Exchange between Tandemly Repeated Genes at the Cf-4/9 Locus of Tomato. Cell. 1997;91(6):821–832. doi: 10.1016/S0092-8674(00)80470-5. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Martin F, et al. Developmental role of the tomato Mediator complex subunit MED18 in pollen ontogeny. Plant J. 2018;96:300–315. doi: 10.1111/tpj.14031. [DOI] [PubMed] [Google Scholar]
- 33.Nawaz-ul-Rehman, M. S., Mansoor, S., Khan, A. A., Zafar, Y. & Briddon, R. W. RNAi-mediated male sterility of tobacco by silencing TA29. Mol. Biotechnol.36, 159–165 (2007). [DOI] [PubMed]
- 34.Van den Heuvel, K. J., Van Lipzig, R. H., Barendse, G. W. & Wullems, G. J. Regulation of expression of two novel flower-specific genes from tomato (Solanum lycopersicum) by gibberellin. J. Exp. Bot.53, 51–59 (2002). [DOI] [PubMed]
- 35.Chen RD, Zimmermann E, Xu SX, Liu GS, Smith AG. Characterization of an anther- and tapetum-specific gene and its highly specific promoter isolated from tomato. Plant cell rep. 2006;25:231–240. doi: 10.1007/s00299-005-0056-7. [DOI] [PubMed] [Google Scholar]
- 36.Twell D, Wing R, Yamaguchi J, McCormick S. Isolation and expression of an anther-specific gene from tomato. Mol. Gen. Genet. 1989;217:240–245. doi: 10.1007/BF02464887. [DOI] [PubMed] [Google Scholar]
- 37.Muschietti, J., Dircks, L., Vancanneyt, G. & McCormick, S. LAT52 protein is essential for tomato pollen development: pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J.6, 321–338 (1994). [DOI] [PubMed]
- 38.McNeil KJ, Smith AG. A glycine-rich protein that facilitates exine formation during tomato pollen development. Planta. 2010;231:793–808. doi: 10.1007/s00425-009-1089-x. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L, et al. Fine mapping and molecular marker development of anthocyanin absent, a seedling morphological marker for the selection of male sterile 10 in tomato. Mol. Breed. 2016;36:107. doi: 10.1007/s11032-016-0531-6. [DOI] [Google Scholar]
- 40.Sinha R, Rajam MV. RNAi silencing of three homologues of S-adenosylmethionine decarboxylase gene in tapetal tissue of tomato results in male sterility. Plant Mol. Biol. 2013;82:169–180. doi: 10.1007/s11103-013-0051-2. [DOI] [PubMed] [Google Scholar]
- 41.Brukhin, V., Hernould, M., Gonzalez, N., Chevalier, C. & Mouras, A. Flower development schedule in tomato Lycopersicon esculentum cv. sweet cherry. Sex. Plant Reprod.15, 311–320 (2003).
- 42.Sato S, et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 44.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao Xue, Qiu Zhengkun, Wang Xiaotian, Van Giang Tong, Liu Xiaolin, Wang Jing, Wang Xiaoxuan, Gao Jianchang, Guo Yanmei, Du Yongchen, Wang Guoping, Huang Zejun. A putative R3 MYB repressor is the candidate gene underlying atroviolacium, a locus for anthocyanin pigmentation in tomato fruit. Journal of Experimental Botany. 2017;68(21-22):5745–5758. doi: 10.1093/jxb/erx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tomato ms32 supplementary figures and tables