Abstract
Purpose
High-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements (HGBL-DH/TH) has a poor outcome after standard chemoimmunotherapy. We sought to understand the biologic underpinnings of HGBL-DH/TH with BCL2 rearrangements (HGBL-DH/TH-BCL2) and diffuse large B-cell lymphoma (DLBCL) morphology through examination of gene expression.
Patients and Methods
We analyzed RNA sequencing data from 157 de novo germinal center B-cell-like (GCB)-DLBCLs, including 25 with HGBL-DH/TH-BCL2, to define a gene expression signature that distinguishes HGBL-DH/TH-BCL2 from other GCB-DLBCLs. To assess the genetic, molecular, and phenotypic features associated with this signature, we analyzed targeted resequencing, whole-exome sequencing, RNA sequencing, and immunohistochemistry data.
Results
We developed a 104-gene double-hit signature (DHITsig) that assigned 27% of GCB-DLBCLs to the DHITsig-positive group, with only one half harboring MYC and BCL2 rearrangements (HGBL-DH/TH-BCL2). DHITsig-positive patients had inferior outcomes after rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone immunochemotherapy compared with DHITsig-negative patients (5-year time to progression rate, 57% and 81%, respectively; P < .001), irrespective of HGBL-DH/TH-BCL2 status. The prognostic value of DHITsig was confirmed in an independent validation cohort. DHITsig-positive tumors are biologically characterized by a putative non–light zone germinal center cell of origin and a distinct mutational landscape that comprises genes associated with chromatin modification. A new NanoString assay (DLBCL90) recapitulated the prognostic significance and RNA sequencing assignments. Validating the association with HGBL-DH/TH-BCL2, 11 of 25 DHITsig-positive–transformed follicular lymphomas were classified as HGBL-DH/TH-BCL2 compared with zero of 50 in the DHITsig-negative group. Furthermore, the DHITsig was shared with the majority of B-cell lymphomas with high-grade morphology tested.
Conclusion
We have defined a clinically and biologically distinct subgroup of tumors within GCB-DLBCL characterized by a gene expression signature of HGBL-DH/TH-BCL2. This knowledge has been translated into an assay applicable to routinely available biopsy samples, which enables exploration of its utility to guide patient management.
INTRODUCTION
Recognition of the biologic heterogeneity in diffuse large B-cell lymphoma (DLBCL) has prompted significant effort to define distinct molecular subgroups within the disease that have prognostic significance and, more importantly, harbor potentially targetable biology.1-3 Accordingly, the most recent revision of the WHO classification, which divides tumors with DLBCL morphology into cell-of-origin (COO) molecular subtypes activated B-cell-like (ABC) and germinal center B-cell-like (GCB), recognizes high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements (HGBL-DH/TH).4 This newly defined entity comprises tumors with either DLBCL or high-grade morphology. Approximately 8% of tumors with DLBCL morphology are HGBL-DH/TH, and all HGBL-DH/TH with BCL2 translocations (HGBL-DH/TH-BCL2) of DLBCL morphology belong to the GCB molecular subgroup.5,6 Clinically, despite the generally superior prognosis of GCB-DLBCLs, patients with HGBL-DH/TH-BCL2 have poor outcomes,7-12 which has prompted many institutions to treat these tumors with dose-intensive immunochemotherapy. GCB patients that do not respond to standard therapies and HGBL-DH/TH patients that remain refractory to intensified therapy represent a critical unmet need and would benefit from novel treatment strategies that better address their unique biology.
Owing to the relative rarity of HGBL-DH/TH-BCL2, a comprehensive biologic characterization of this entity is lacking, and there has been speculation that additional molecular features may refine the definition of these cases. Genomic studies in DLBCL have identified recurrent mutations and revealed the association of many with COO.13-16 Although the most recent genomic landscape studies defined genetic subgroups on the basis of somatic mutation and structural variants, they did not address the genetic background of HGBL-DH/TH-BCL2.17-19
We sought to identify gene expression features that distinguish HGBL-DH/TH-BCL2 from the remainder of GCB-DLBCLs. In doing so, we discovered a distinct molecular subgroup that comprises 27% of GCB-DLBCLs, only one half of which were HGBL-DH/TH-BCL2, with poor prognosis after rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) treatment, a finding validated in an independent cohort using a gene expression signature that robustly identifies these patients (double-hit signature [DHITsig]). Relative to the other GCB-DLBCLs, these tumors have significantly lower expression of genes associated with the light zone (LZ) of the germinal center and have mutations and gene expression features that imply potentially targetable vulnerabilities. The signature has been translated into a new assay on the NanoString Technologies (Seattle, WA) platform that allows application to routinely available formalin-fixed paraffin-embedded (FFPE) biopsy samples.
PATIENTS AND METHODS
Patient Cohorts
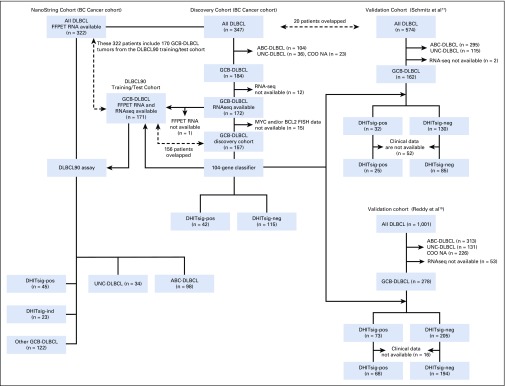
To enhance our understanding of HGBL-DH/TH-BCL2, we analyzed RNA sequencing (RNAseq) data from 157 de novo GCB DLBCLs, including 25 HGBL-DH/TH-BCL2, to define gene expression differences between HGBL-DH/TH-BCL2 and other GCB-DLBCLs (discovery cohort). These are GCB-DLBCLs with available MYC and BCL2 fluorescent in situ hybridization (FISH) results from our previously described cohort of 347 diagnostic biopsy samples of patients with de novo DLBCL treated with R-CHOP who were selected from the BC Cancer population-based registry6 (Appendix Fig A1, online only). Selection of the 347 patients was described previously.6 This study was reviewed and approved by the University of British Columbia-BC Cancer Research Ethics Board in accordance with the Declaration of Helsinki.
We used two external cohorts with RNAseq data available (Reddy et al,19 278 patients with GCB-DLBCL; Schmitz et al,17 162 patients with GCB-DLBCL) to explore the prognostic significance and molecular features associated with DHITsig DLBCL. FFPE biopsy samples of 322 of the 347 DLBCLs plus 88 transformed follicular lymphomas (tFLs)20 with DLBCL morphology and 26 HGBLs from patients treated in BC Cancer were analyzed for the validation of the novel NanoString assay. Patient characteristics and pathology data for the BC Cancer DLBCL cohort are listed in the Data Supplement. Methods for FISH and immunohistochemistry are described in the Data Supplement.
Gene Expression Profiling and Mutational Analysis
RNAseq was applied to RNA extracted from fresh frozen biopsy samples. We compiled mutations from targeted sequencing of the discovery cohort and existing exome data from two validation cohorts, each with matched RNAseq.17,19 Sample processing of RNA and DNA, library construction and detailed analytic procedures for RNAseq, targeted resequencing, and mutational analysis of exome data were either previously described6,21-23 or are included in the Data Supplement.
Digital Gene Expression Profiling
To translate the signature into an assay applicable to FFPE, we performed digital expression profiling on RNA derived from FFPE biopsy samples using the NanoString assay; methods are provided in the Data Supplement.
Statistical Analysis
The Kaplan-Meier method was used to estimate the time to progression (TTP; progression/relapse or death as a result of lymphoma or acute treatment toxicity), progression-free survival (PFS; progression/relapse or death as a result of any cause), disease-specific survival (DSS; death as a result of lymphoma or acute treatment toxicity), and overall survival (OS; death as a result of any cause), with log-rank test performed to compare groups. Univariable and multivariable Cox proportional hazard regression models were used to evaluate proposed prognostic factors (Data Supplement).
Fisher’s exact test was used for between-categorical data comparisons. For the comparison of two continuous variables, data were tested by Wilcoxon rank sum test, except where noted. Multiple testing correction was performed where necessary using the Benjamini-Hochberg procedure. All P values result from two-sided tests, and a threshold of .05 was used for significance, except where noted. All analyses were performed using R-3.4.1 software (https://cran.r-project.org/bin/windows/base/old/3.4.1).
RESULTS
Development of DHITsig
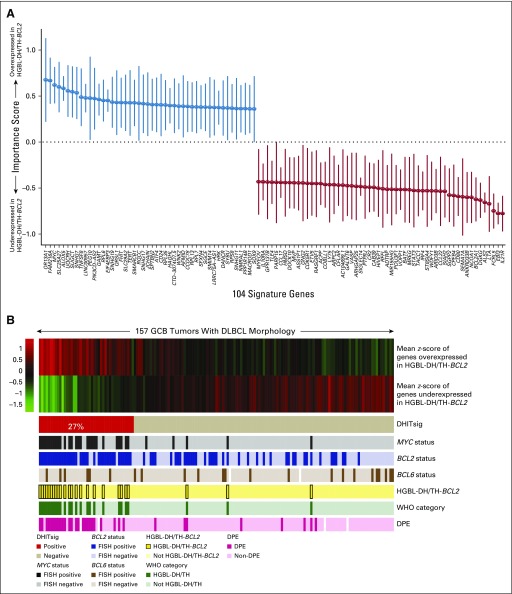
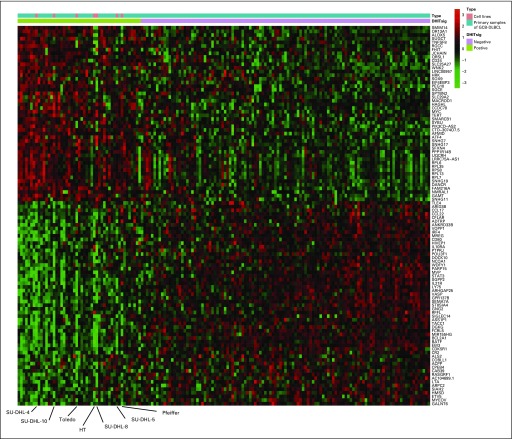
We identified 104 genes that were most significantly differentially expressed between HGBL-DH/TH-BCL2 and other GCB-DLBCLs (Fig 1A; Data Supplement). We devised a model score using the expression of these 104 genes (Data Supplement) that separates GCB-DLBCL into two groups. The smaller group, which comprised 42 tumors (27%), was termed DHITsig positive (DHITsig-pos) and included 22 of the 25 HGBL-DH/TH-BCL2 tumors as determined by FISH. The remaining 115 GCB tumors (73%) were considered DHITsig negative (DHITsig-neg), including three HGBL-DH/TH-BCL2 tumors (Fig 1B).
FIG 1.
The gene expression–based model of 104 genes on the basis of high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements with BCL2 translocations (HGBL-DH/TH-BCL2) status. (A) Importance score with 95% CI of the 104 most significantly differentially expressed genes between HGBL-DH/TH-BCL2 and germinal center B-cell-like (GCB) diffuse large B-cell lymphoma (DLBCL). Genes with blue and red bars are over- and underexpressed in HGBL-DH/TH-BCL2, respectively. (B) Mean z-score of genes over- or underexpressed in HGBL-DH/TH-BCL2 is shown in the form of a heat map, with the 157 patient biopsy samples shown as columns. Double-hit signature (DHITsig) groups identified by the signature are shown below the heat map. The status of MYC, BCL2, and BCL6 genetic alterations; HGBL-DH/TH-BCL2; WHO categories; and MYC/BCL2 dual protein expression (DPE) status are displayed beneath the heat map. FISH, fluorescent in situ hybridization.
Prognostic Value of DHITsig
Having developed the DHITsig while blinded to patient outcomes, we then explored the prognostic impact of the DHITsig within the 157 patients with de novo GCB-DLBCL uniformly treated with R-CHOP6,24 using assignments from the locked RNAseq model. DHITsig was not associated with clinical variables, including the factors of International Prognostic Index (IPI), IPI subgroups, B symptoms, and tumor volume. As expected, MYC and BCL2 translocations and protein expression of MYC and BCL2 were significantly more frequent in DHITsig-pos patients (all P < .001; Table 1).
TABLE 1.
Patient Characteristics According to DHITsig in Germinal Center B-Cell-Like Diffuse Large B-Cell Lymphoma

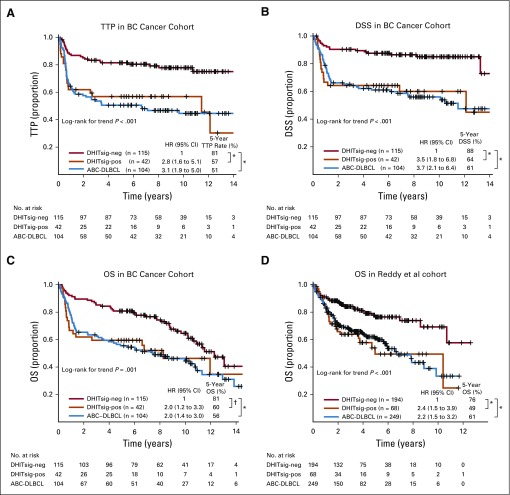
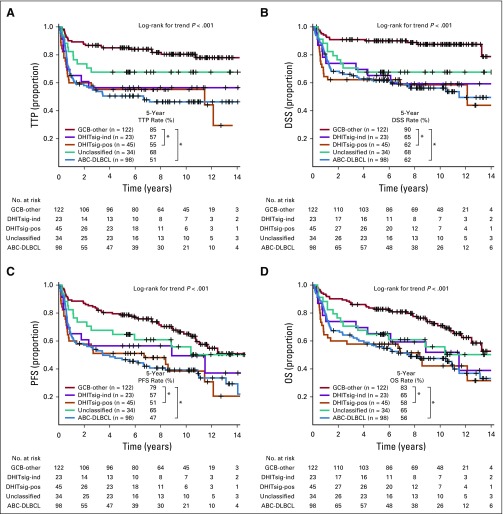
DHITsig-pos patients had significantly shorter TTP, DSS, and OS compared with those with DHITsig-neg GCB (log-rank P < .001, < .001, .012, respectively) and exhibited outcomes comparable to those of ABC-DLBCL from the cohort of 347 patients (Figs 2A to 2C). Of note, the DHITsig-pos patients without HGBL-DH/TH-BCL2 showed comparably poor prognosis to patients with HGBL-DH/TH-BCL2 (Appendix Fig A2, online only). Although IPI factors, IPI subgroups, and dual protein expression (DPE) of MYC and BCL2 also were associated with survival in GCB-DLBCL (Data Supplement), DHITsig remained prognostic in multivariable analyses that included these factors (Data Supplement). In particular, DPE did not provide a statistically significant risk stratification within either the DHITsig-pos or the DHIT-neg groups (Appendix Fig A3, online only), which indicated that the DHITsig designation subsumes the prognostic impact of DPE within GCB-DLBCL. Similarly, DHITsig designation subsumed the prognostic impact of the established pathology subgroups (ie, HGBL-DH/TH-BCL2). We then applied this gene expression model to GCB-DLBCL from an independent data set (Reddy et al,19 262 patients with GCB-DLBCL) in which the DHITsig-pos group also had significantly inferior OS compared with other patients with GCB-DLBCLs (P < .001; Fig 2D). Characteristics of the cohorts of Reddy et al and BC Cancer are compared in the Data Supplement
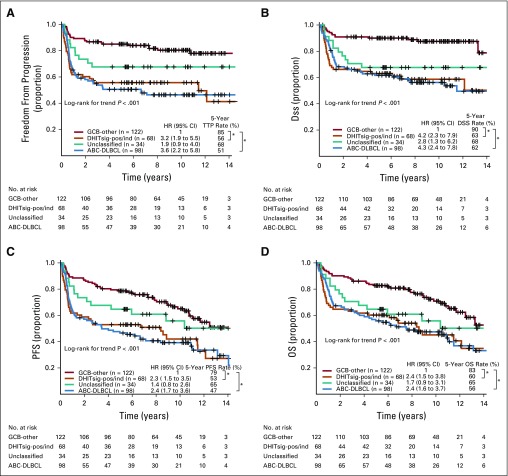
FIG 2.
Prognostic association of double-hit signature (DHITsig) in patients with diffuse large B-cell lymphoma (DLBCL) treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. Kaplan-Meier curves of the DHITsig-positive (DHITsig-pos) germinal center B-cell-like DLBCL v DHITsig-negative (DHITsig-neg) GCB-DLBCL v activated B-cell-like (ABC)-DLBCL for (A) time to progression (TTP), (B) disease-specific survival (DSS) and (C) overall survival (OS) in the BC Cancer cohort and (D) OS in the Reddy et al19 validation cohort. *P < .001, †P = .011. HR, hazard ratio; PFS, progression-free survival.
DHITsig Defines a Biologically Distinct Subgroup Within GCB-DLBCL
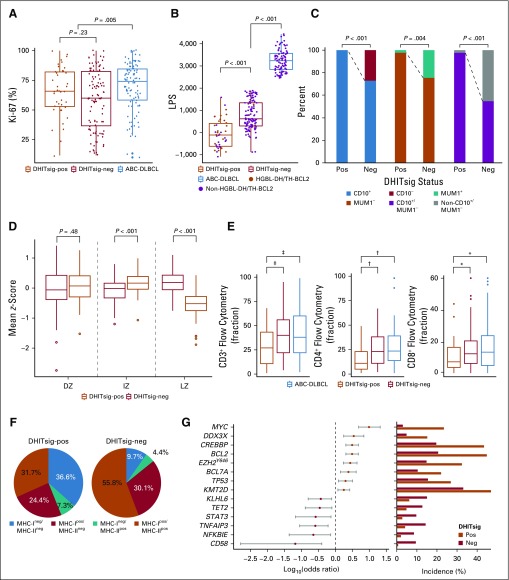
Exploration of the pathology and gene expression patterns demonstrated that DHITsig-pos tumors form a distinct biologic subgroup of GCB-DLBCL characterized by a COO from the intermediate zone (IZ)/dark zone (DZ) of the germinal center. In a first step, a pathology re-review of the entire cohort of 347 patients with DLBCL from BC Cancer was performed by a panel of expert hematopathologists (G.W.S., P.F., and K.T.) who confirmed that DHITsig-pos tumors were indeed of DLBCL morphology. No morphologic features were observed that distinguished these tumors from DHITsig-neg tumors. Furthermore, the proliferation index, assessed by immunohistochemistry for Ki67, was not significantly different between DHITsig groups (Fig 3A).
FIG 3.
Genetic, molecular, and phenotypic features of the double-hit signature (DHITsig). (A) Comparison of protein encoded by the MKI67 gene (Ki-67) staining by immunohistochemistry between DHITsig-positive (DHITsig-pos), DHITsig-negative (DHITsig-neg) germinal center B-cell-like (GCB) diffuse large B-cell lymphoma (DLBCL) and activated B-cell-like (ABC) DLBCL. (B) Comparison of linear predictor score (LPS) provided by the Lymph2Cx assay between DHITsig-pos, DHITsig-neg GCB-DLBCL and ABC-DLBCL. Dark orange dots represent the high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements with BCL2 translocations (HGBL-DH/TH-BCL2) tumors. (C) Comparison of immunohistochemistry staining pattern of CD10 (membrane metallo-endopeptidase) and MUM1 (IRF4) between DHITsig-pos and DHITsig-neg GCB-DLBCL tumors. (D) Comparison of mean z-scores of dark zone (DZ), intermediate zone (IZ), and light zone (LZ) signature genes (20 genes each) between DHITsig-pos and DHITsig-neg groups. (E) Comparison of fraction of tumor-infiltrating T cells (CD3+, CD4+, and CD8+ T cells) measured by flow cytometry between DHITsig-pos, DHITsig-neg GCB-DLBCL and ABC-DLBCL. (F) Frequencies of MHC class I (MHC-I) and MHC-II double-negative, isolated MHC-II negative, isolated MHC-I negative, and MHC-I and -II double-positive cases in DHITsig-pos and DHITsig-neg cases. (G) Forest plots summarize the results of Fisher’s exact tests that compare the frequency of mutations that affect individual genes in DHITsig-neg (dark orange dots) and DHITsig-pos (red dots) GCB-DLBCL tumors. Significantly enriched genes in either DHITsig-pos or DHITsig-neg cases (false discovery rate < .10) are represented. Log10 odds ratios and 95% CIs are shown. Bar plots represent the frequency of mutations in either DHITsig-pos or DHITsig-neg groups. *P < .10, †P < .05, ‡P <.01.
In the Lymph2Cx assay, low linear predictor scores (LPSs) result in an assignment to the GCB group, whereas high LPSs result in an ABC assignment. Among the GCB DLBCLs, DHITsig-pos tumors had significantly lower LPSs than DHITsig-neg tumors (P < .001; Fig 3B). Moreover, DHITsig-pos tumors were universally positive for CD10 (membrane metallo-endopeptidase) staining, and the majority were MUM1 (IRF4) negative. CD10+/MUM1− cases were significantly more frequent in DHITsig-pos tumors (P < .001; Fig 3C). Most GCB-DLBCLs have been demonstrated previously to have a COO consistent with B lymphocytes from the LZ of the germinal center.25 Given that the gene features in the Lymph2Cx and these immunohistochemistry markers are associated with B-cell differentiation states, we considered whether the two DHITsig groups had gene expression patterns that imply distinct putative COOs. Gene signatures associated with DZ, LZ, and the more recently described IZ, which represents the transition stage between DZ and LZ, were explored within the GCB-DLBCLs.26 Of note, DHITsig-pos tumors showed significantly lower expression of LZ genes compared with DHITsig-neg tumors (P < .001; Fig 3D). The expression of genes in the DZ cluster were not statistically different between the two groups, whereas genes associated with the IZ had higher expression within the DHITsig-pos tumors. Furthermore, genes characteristic of the IZ are part of the 104-gene DHITsig model. Collectively, these findings demonstrate that although DHITsig-neg tumors have an LZ COO, we postulate that the COO for DHITsig-pos tumors are IZ B cells transitioning from the LZ to the DZ.
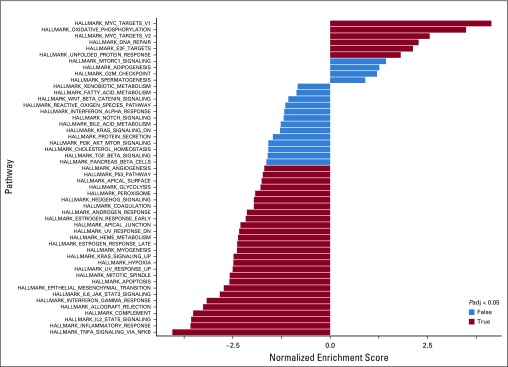
Gene set enrichment analysis was then used to uncover additional biologic differences between DHITsig-pos and DHITsig-neg tumors. We found that DHITsig-pos tumors demonstrated overexpression of MYC and E2F targets and genes associated with oxidative phosphorylation and MTORC1 signaling (Appendix Fig A4, online only). Conversely, DHITsig-pos tumors exhibit lower expression of genes associated with apoptosis, tumor necrosis factor-α signaling through nuclear factor-κB and decreased interleukin-6/Janus kinase/signal transducers and activators of transcription 3, processes upregulated in centrocytes. DHITsig-pos tumors also exhibited lower expression of immune and inflammation signatures. Consistently, tumor-infiltrating lymphocytes, especially CD4+ T cells, had significantly lower representation in DHITsig-pos tumors relative to other GCBs (Fig 3E). Loss of surface MHC class I (MHC-I) and MHC-II protein expression was also more frequent in DHITsig-pos tumors (Fisher’s exact test for MHC-I and MHC-II, 61% v 40% [P = .020] and 44% v 14% [P < .001], respectively; Fig 3F), with 68% of DHITsig-pos tumors having a loss of either MHC-I or MHC-II expression. Finally, we identified that all representative GCB-DLBCL cell lines tested belonged to the DHITsig-pos subgroup (Appendix Fig A5, online only), consistent with the notion that DHITsig-pos tumors harbor strong cell-autonomous survival and proliferation signals and reduced dependence on the microenvironment.
Mutational Landscape of DHITsig-pos GCB-DLBCL
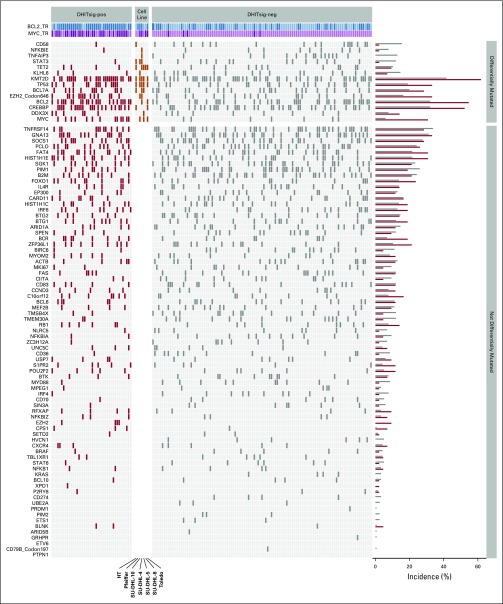
We next sought genetic features associated with DHITsig status within GCB-DLBCL. For this, we used the combined mutation data derived from 569 unique patients with GCB-DLBCL in three cohorts (BC Cancer, Reddy et al,19 and Schmitz et al17). Along with the expected enrichment of mutations in MYC and BCL2 (false discovery rate [FDR] < .001), mutations that affect CREBBP, EZH2Y646, DDX3X, TP53, and KMT2D were more frequent in DHITsig-pos tumors (FDR < .10). In contrast, the mutations of TNFAIP3, KLHL6, NFKBIE, TET2, CD58, and STAT3 were more common among DHITsig-neg GCB tumors (FDR < .10; Fig 3G; Appendix Fig A6, online only; Data Supplement). With regard to the recently genetically defined subgroups of DLBCL, exploration of the DHITsig groups in Schmitz et al showed a statistically significant association between DHITsig-pos and EZB (P = .001; Data Supplement). However, these groups showed only partial overlap, with 38% of the DHITsig-pos tumors falling outside the EZB group, and the majority (67%) of EZB were DHITsig-neg.
MYC rearrangements were not detected in 15 of the DHITsig-pos patients, despite the use of two break-apart probe sets and an MYC/IGH dual-fusion probe set (Data Supplement). One tumor had a FISH pattern consistent with amplification of MYC as double minutes. Other mechanisms of MYC dysregulation that are cryptic to FISH may be operative in the remaining 14 tumors.
Translation of DHITsig Into a Clinically Relevant Assay
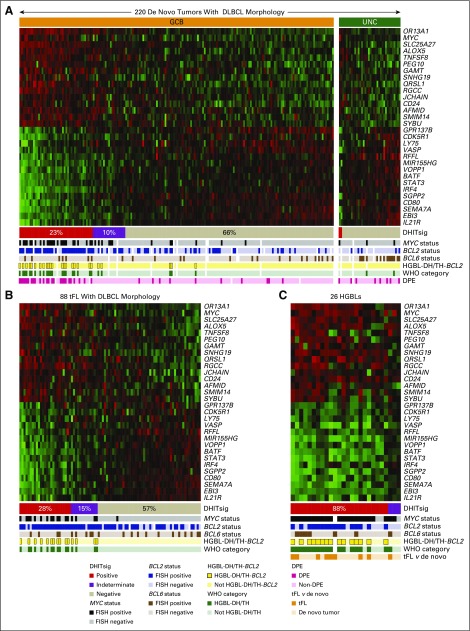
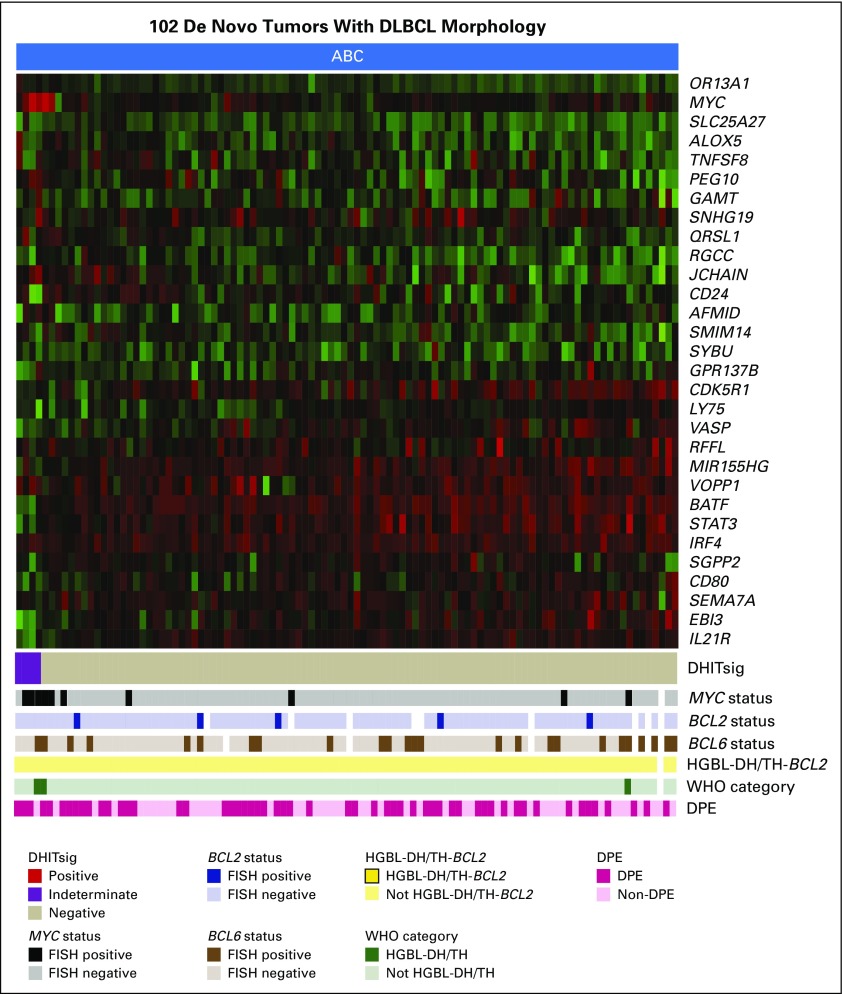
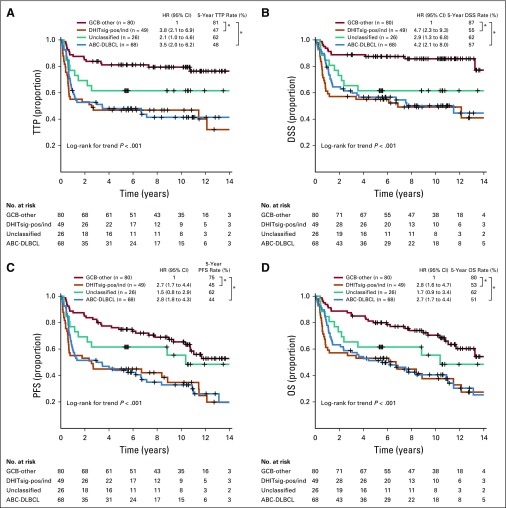
To provide an assay applicable to routinely available biopsy samples, the 104-gene RNAseq model was reduced to a 30-gene module. This module was added to the Lymph3Cx,27 which in turn is an extension of Lymph2Cx that contains a module to distinguish primary mediastinal B-cell lymphomas (PMBCL). This NanoString-based assay, named DLBCL90, assigns tumors into DHITsig-pos and DHITsig-neg groups using a Bayes rule with 20% and 80% probability thresholds and with an indeterminate group (DHITsig-ind) where the tumor could not be assigned with sufficient confidence (Appendix Figs A7 to A9, online only; Data Supplement). This was applied to 171 GCB-DLBCL tumors from the 347-patient cohort (including 156 from the discovery cohort), which gave 26% DHITsig-pos, 64% DHITsig-neg, and 10% DHITsig-ind with a frank misclassification rate of 3% against the RNAseq comparator (Data Supplement; Appendix Fig A10, online only). The integrity of the Lymph2Cx assay was maintained (Appendix Fig A11, online only). The assay was then applied to the remainder of the available 322 FFPE biopsy samples from the 347-patient de novo DLBCL cohort, which showed that the DHITsig was not seen in ABC-DLBCL, with four (4%) of 102 being DHITsig-ind (Fig 4A; Appendix Fig A12, online only). The prognostic significance for TTP, DSS, PFS, and OS of DHITsig was maintained (all P < .001). Because the DHITsig-ind group had similar outcomes to DHITsig-pos, these two groups are shown together in Figure 5 and separately in Appendix Figure A13 (online only). Of note, the assay identified a group with very good prognosis, with DHITsig-neg GCB-DLBCLs exhibiting a DSS rate of 90% at 5 years. In advanced-stage disease (defined in the Data Supplement), the DSS rate of the DHITsig-neg patients with GCB-DLBCL was 87% at 5 years (Appendix Figs A14 and A15, online only, show outcomes in advanced- and limited-stage disease, respectively). Characteristics of the rare DHITsig-neg HGBL-DH/TH-BCL2 tumors along with the outcomes of these patients are discussed in the Data Supplement.
FIG 4.
The gene expression–based model for the double-hit signature (DHITsig). The DLBCL90 assay is shown in the form of a heat map, with the 30 informative genes shown as rows, and the tumors shown as columns, separated into (A) 220 germinal center B-cell-like (GCB) and unclassified (UNC) diffuse large B-cell lymphomas (DLBCLs). (B) Eighty-eight transformed follicular lymphomas (tFLs) with DLBCL morphology. (C) Twenty-six high-grade B-cell lymphomas (HGBLs). The tumors are arrayed from highest DHITsig score on the left to lowest DHITsig score on the right. DHITsig groups identified by the signature are shown below the heat map. The status of MYC, BCL2, and BCL6 genetic alterations; HGBL with MYC and BCL2 and/or BCL6 rearrangements with BCL2 translocations (HGBL-DH/TH-BCL2) status; and WHO categories also are shown. White bars indicate data that are not available or assignments that could not be made on the basis of the available data. DPE, dual protein expression; FISH, fluorescent in situ hybridization.
FIG 5.
Prognostic association of DLBCL90 in patients with diffuse large B-cell lymphoma (DLBCL) treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. Kaplan-Meier curves of the germinal center B-cell-like (GCB) DLBCL v double-hit signature (DHITsig)-positive (DHITsig-pos) and DHITsig-indeterminate (DHITsig-ind) v unclassified v activated B-cell-like (ABC) DLBCL for (A) time to progression (TTP), (B) disease-free survival (DSS), (C) progression-free survival (PFS), and (D) overall survival (OS) in 322 patients with de novo tumors of DLBCL morphology treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. The same curves with separation of DHITsig-pos and DHITsig-ind are shown in Appendix Figure A13 (online only). *P < .001. HR, hazard ratio.
To validate the association between the DHITsig and HGBL-DH/TH-BCL2, DLBCL90 was applied to 88 patients with tFL with DLBCL morphology. Within these patients, 11 of the 25 DHITsig-pos tumors were HGBL-DH/TH-BCL2 compared with zero of 50 in the DHITsig-neg group. Within the DHITsig-ind group, four of 13 tumors were HGBL-DH/TH-BCL2 (Fig 4B). Finally, the DLBCL90 assay was applied to 26 HGBL tumors, including seven classified as HGBL not otherwise specified and 18 classified as HGBL-DH/TH with high-grade morphology; one tumor could not be assigned because of an unknown MYC rearrangement status. Among these tumors, the majority were assigned to the DHITsig-pos group (23 [88%]) with three (12%) being DHITsig-ind (Fig 4C).
DISCUSSION
We have defined a novel molecular subgroup within tumors with DLBCL morphology characterized by distinct biology and prognosis after standard immunochemotherapy. Twenty-seven percent of GCB-DLBCLs share this signature, with only one half of these harboring concurrent MYC and BCL2 rearrangements. Furthermore, the poor prognosis, comparable with ABC-DLBCL, was irrespective of HGBL-DH/TH-BCL2 status. The robustness of this prognostic gene expression signature was confirmed through validation in an independent cohort of R-CHOP–treated patients.
This signature was also evident in tumors with high-grade morphology, which currently fall into the HGBL not otherwise specified and HGBL-DH/TH with high-grade morphology groups in the revised 2017 WHO classification.4 We propose that this signature identifies tumors with high-grade molecular features, with FISH for MYC and BCL2 as surrogate features that represent a subset of these. As genomic testing gains adoption in clinical practice, this signature could form the basis of a more inclusive category that encompasses DLBCLs both with and without these rearrangements. This system would classify tumors with shared biology together while significantly expanding a group of patients with an established need for dose-intensive regimens or alternative therapies and may drive acceleration of clinical trials aimed at improving outcomes. Meanwhile, the removal of these tumors from GCB-DLBCL leaves a group with a 5-year DSS rate of 90%—strong evidence that R-CHOP is sufficient for these patients.
The DHITsig identifies tumors with distinct biology with potential targetable vulnerabilities. As opposed to other GCB-DLBCLs, which have a putative COO of the germinal center LZ, DHITsig-pos tumors display a pattern of gene expression consistent with an intermediate zone COO. The expression of MYC in normal germinal center biology is restricted to cells selected within the LZ for re-entry into the DZ,28,29 and we postulate that these recycling cells are the physiologic counterpart of DHITsig-pos tumor cells. They have a high incidence of mutations within chromatin-modifier genes and are cold tumors with a paucity of infiltrating T cells and a high incidence of low MHC-I and MHC-II expression.23,30 Furthermore, gene expression suggests that the tumor cells are highly metabolically charged with a high expression of genes associated with oxidative phosphorylation. This biology provides rationale for exploring targeted agents beyond the current focus on BCL2 inhibitors.31 Such strategies could target cell differentiation block (enhancer of zeste homolog 2 inhibitors), reducing immune evasion (histone deacetylase inhibitors), enhancing T-cell activation (newer-generation anti-CD20 antibodies), and targeting oxidative phosphorylation and the proteasome. Finally, high expression of certain genes raises the possibility that specific inhibitors may be useful (eg, inhibitors of arachidonate 5-lipoxygenase).
Two groups recently have described genetically defined subgroups within GCB-DLBCL that carry a poor prognosis, namely EZB and cluster 3 subgroups.17,18 Examination of RNAseq data from Schmitz et al17 shows that there is only partial overlap between DHITsig-pos tumors and the EZB category, which indicates that this new biologic group is not adequately identified by this genetics-based classification.
To allow the identification of DHITsig tumors in routinely available biopsy samples, we reduced this gene expression classifier to a module that was added to the Lymph3Cx COO/PMBCL assay. This assay faithfully reproduced the RNAseq-based model of DHITsig, while maintaining the accuracy of the base Lymph2Cx assay, and could identify HGBL-DH/TH within tumors of DLBCL morphology in a cohort of tFL. Recently, the Efficacy and Safety Study of Lenalidomide Plus R-CHOP Chemotherapy Versus Placebo Plus R-CHOP Chemotherapy in Untreated ABC Type Diffuse Large B-Cell Lymphoma (ROBUST) phase III clinical trial in DLBCL used a gene expression–based assay for real-time patient selection, which demonstrated the feasibility of such assays in prospective clinical trial designs.32 Furthermore, the retrospective application of this assay to completed clinical trials will allow exploration of the impact of therapeutic strategies on this newly described entity.
This new DHITsig-pos subgroup of GCB-DLBCL roughly doubles the number of DLBCL tumors that would be classified as HGBL on the basis of FISH testing. We envision that this will afford an immediate clinical impact because the signature identifies a large group of DHITsig-neg patients with GCB-DLBCL in whom R-CHOP is sufficient to effect cure, obviating the need for therapy escalation in this group. Finally, the translation of this biology to an assay facilitates clinical trials aiming to improve outcomes, assigning DLBCL COO, and identifying distinct biologic groups within GCBs that harbor potentially targetable biology.
ACKNOWLEDGMENT
We thank the biospecimen, library, sequencing, and bioinformatics teams of Canada’s Michael Smith Genome Sciences Centre for RNAseq data generation. We also thank Holly Eely for biopsy acquisition and Sylvia Lee for histology expertise in FFPE block processing and tissue microarray construction.
Appendix
FIG A1.
Patient flow for the discovery, two independent validation cohorts, and NanoString cohort. ABC, activated B-cell-like; COO, cell of origin; DHITsig, double-hit signature; DLBCL, diffuse large B-cell lymphoma; FFPET, formalin-fixed paraffin-embedded tissue; FISH, fluorescent in situ hybridization; GCB, germinal center B-cell-like; ind, indeterminate; neg, negative; pos, positive; RNAseq, RNA sequencing; UNC, unclassified.
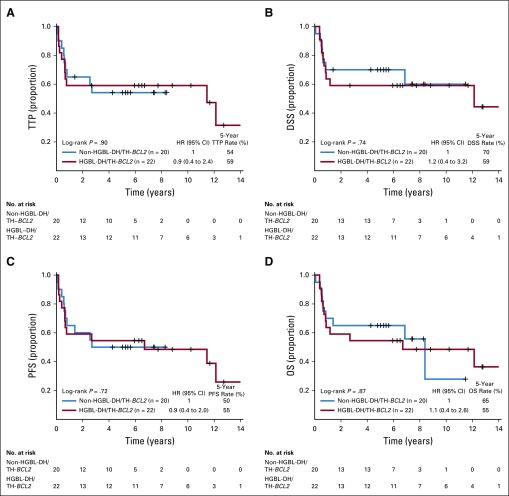
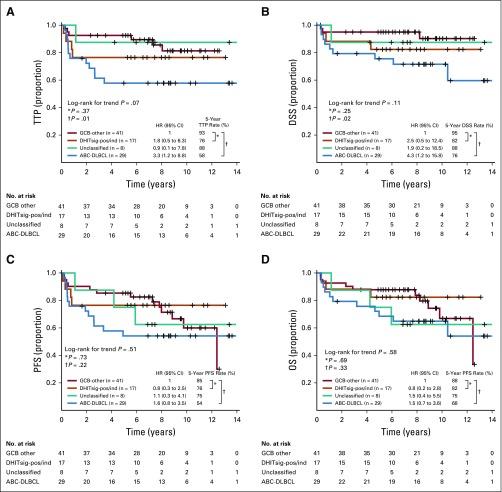
FIG A2.
Kaplan-Meier curves of the patients with high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements with BCL2 translocations (HGBL-DH/TH-BCL2) v non-HGBL-DH/TH-BCL2 within double-hit signature–positive (DHITsig-pos) germinal center B-cell-like (GCB) diffuse large B-cell lymphoma (DLBCL) for (A) time to progression (TTP), (B) disease-specific survival (DSS), (C) progression-free survival (PFS), and (D) overall survival (OS). HR, hazard ratio.
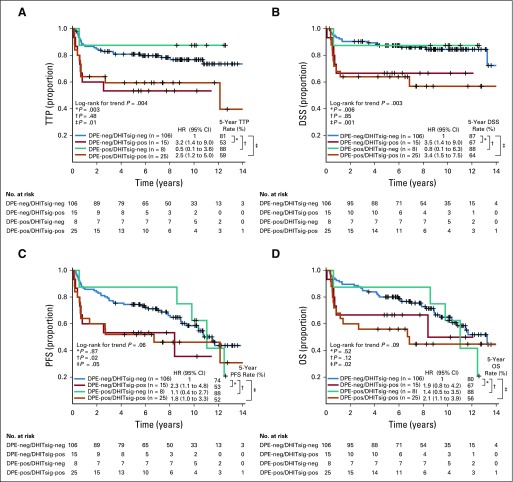
FIG A3.
Kaplan-Meier curves of patients stratified by double-hit signature (DHITsig) combined with dual protein expression (DPE) status in germinal center B-cell-like (GCB) diffuse large B-cell lymphoma (DLBCL) for (A) time to progression (TTP). (B) Disease-specific survival (DSS). (C) Progression-free survival (PFS). (D) Overall survival (OS). DPE-negative (DPE-neg)/DHITsig-positive (DHIT-pos) compared with DPE-positive (DPE-pos)/DHITsig-pos: TTP hazard ratio (HR), 1.4 (95% CI, 0.5 to 3.5; log-rank P = .51); DSS HR, 1.0 (95% CI, 0.4 to 2.8; log-rank P = .99); PFS HR, 1.2 (95% CI, 0.5 to 2.8; log-rank P = .67); and OS HR, 0.9 (95% CI, 0.4 to 2.2; log-rank P = .79).
FIG A4.
Bar plot of the gene set enrichment analysis. This analysis includes differential expression genes between double-hit signature–positive (DHITsig-pos) and DHITsig-negative (DHITsig-neg) groups with false discovery rate < 0.1 and log2 fold change greater than an absolute value of 0.5.
FIG A5.
Heat map of primary samples with germinal center B-cell-like (GCB) diffuse large B-cell lymphoma (DLBCL) along with seven GCB-DLBCL cell lines arrayed according to the double-hit signature (DHITsig) score.
FIG A6.
Mutations detected by targeted sequencing of the discovery cohort and seven diffuse large B-cell lymphoma (DLBCL) cell lines are shown with germinal center B-cell-like (GCB) cases classified as double-hit signature–positive (DHITsig-pos) shown on the left. Genes with significantly different mutation abundance between groups are at the top. The incidence of mutations in each of the genes for the two groups is shown on the right. ind, indeterminate; neg, negative.
FIG A7.
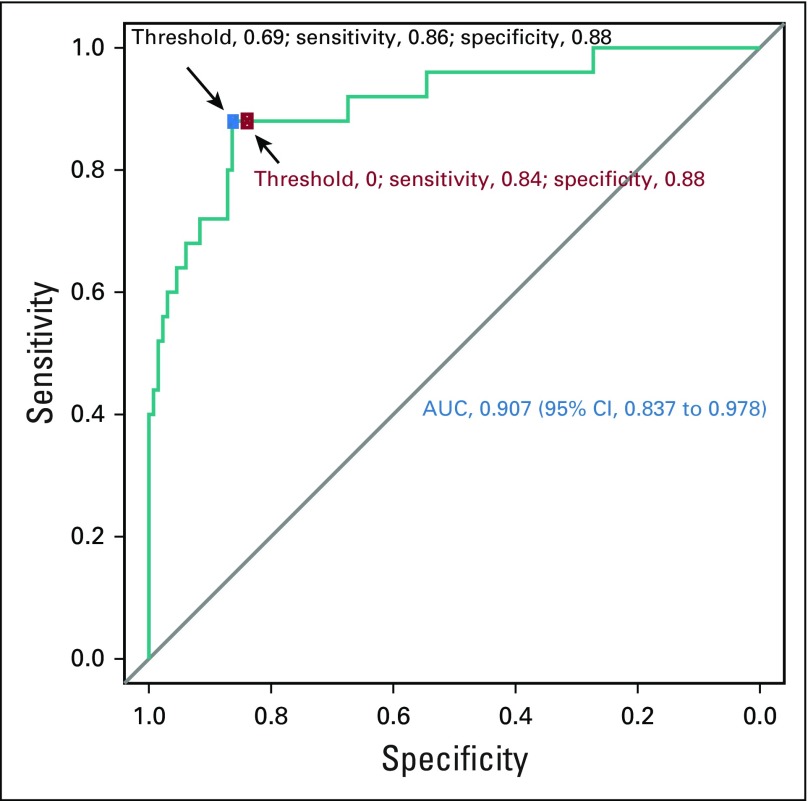
Receiver operating characteristic curve for the RNA sequencing double-hit signature model score for high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements with BCL2 translocations. The threshold of 0 is indicated by the red dot, whereas the blue dot represents the threshold selected by the Youden index. AUC, area under the curve.
FIG A8.
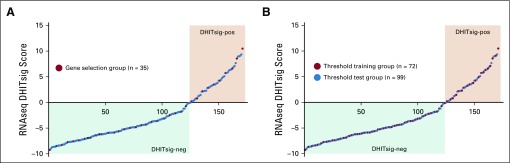
RNA sequencing (RNAseq) double-hit signature (DHITsig) scores from 171 germinal center B-cell-like (GCB) diffuse large B-cell lymphomas (DLBCLs) used to train and test the DLBCL90 assay. The tumors are arrayed from left to right with increasing DHITsig scores, with tumors with a score < 0 being designated DHITsig-negative (DHITsig-neg) and > 0 being DHITsig-positive (DHITsig-pos). (A) Tumors highlighted in red had digital expression performed using a code set that contained all 104 genes in the RNAseq model. (B) Tumors highlighted in red were used to train the threshold for the DLBCL90 assay. Note that all of the tumors highlighted in (A) are also highlighted in (B).
FIG A9.
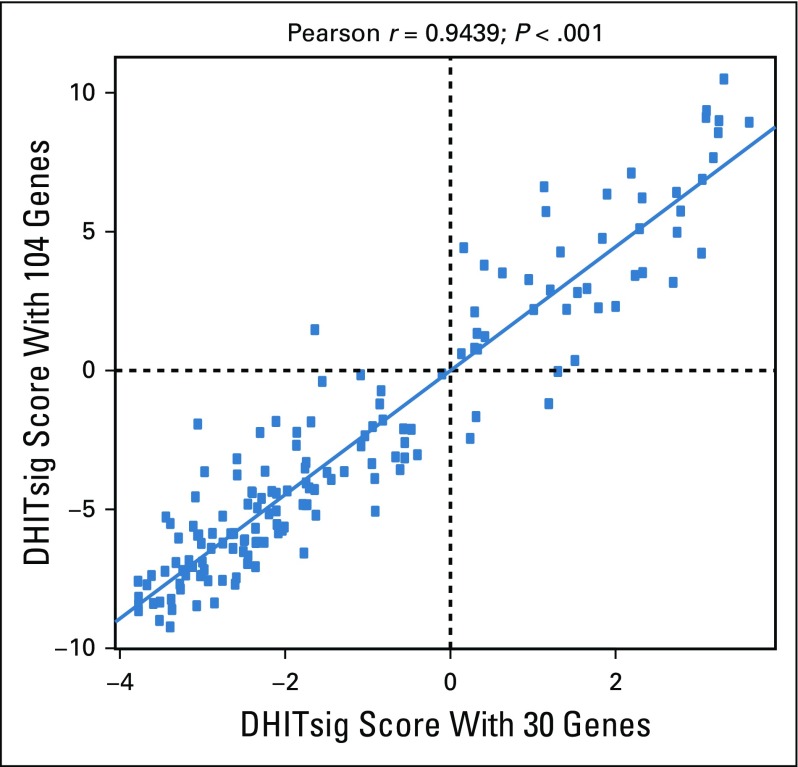
Plot that shows the association between the double-hit signature (DHITsig) scores from RNA sequencing using the full 104-gene model and the model using the 30 genes selected for the DLBCL90 model.
FIG A10.
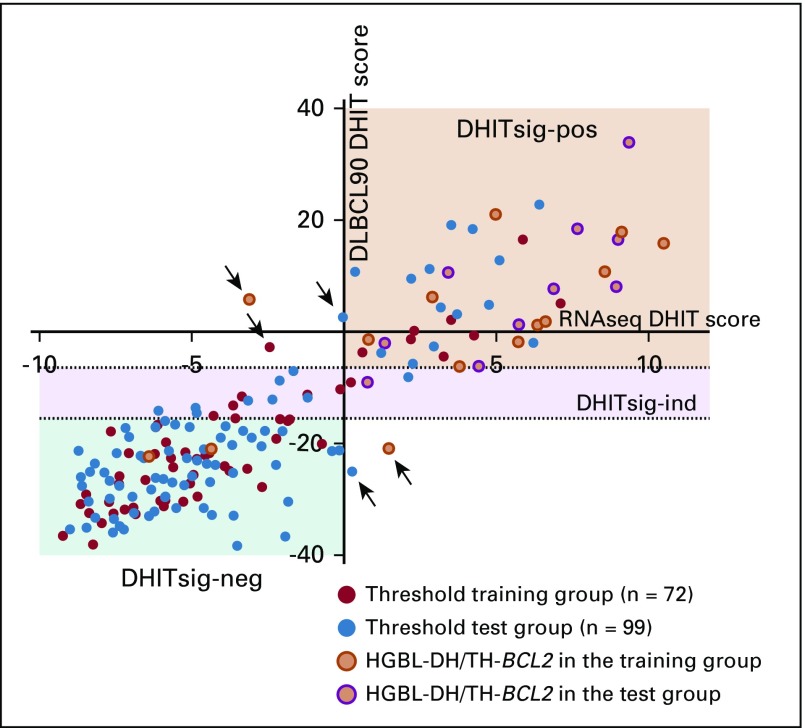
Plot of the double-hit signature (DHITsig) score from the RNA sequencing (RNAseq) model (x-axis) against the DHIT score from the DLBCL90 assay in 171 samples of germinal center B-cell-like (GCB) diffuse large B-cell lymphoma (DLBCL). The 72 biopsy samples highlighted in red were used to establish the thresholds for the assay. Arrows point to the five tumors (3%) that were frankly misclassified. HGBL-DH/TH-BCL2, high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements with BCL2 translocations; ind, indeterminate; neg, negative; pos, positive.
FIG A11.
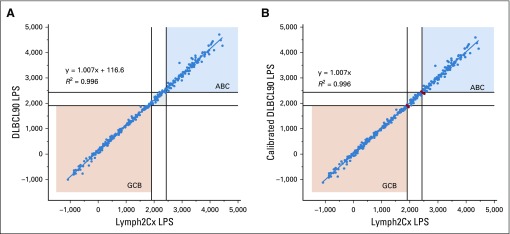
Comparisons between the linear predictor score (LPS) from the Lymph2Cx24 and the DLBCL90 assay. (A) The uncalibrated DLBCL90 LPS scores. (B) The calibrated DLBCL90 LPS scores where 116.6 points were removed from the uncalibrated scores. The six tumors (2%) that moved from a definitive category to unclassified (or vice versa) are highlighted in red. ABC, activated B-cell-like; GCB, germinal center B-cell-like.
FIG A12.
Heat map that shows the results of the DLBCL90 assignment of double-hit signature (DHITsig) in 102 activated B-cell-like (ABC) diffuse large B-cell lymphoma (DLBCL) tumors. The columns, which represent the tumors, are arrayed from highest scores on the left to lowest on the right. Arrayed below the heat map are pathology characteristics of the tumors. DPE, dual protein expression; FISH, fluorescent in situ hybridization; HGBL-DH/TH-BCL2, high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements with BCL2 translocations.
FIG A13.
Kaplan-Meier curves of the outcomes in 322 patients with de novo diffuse large B-cell lymphoma treated with curative intent with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone in the BC Cancer cohort grouped according to double-hit signature (DHITsig) status and cell of origin. (A) Time to progression (TTP). (B) Progression-free survival (PFS). (C) Disease-specific survival (DSS). (D) Overall survival (OS). *P < .001. ABC, activated B-cell-like; GCB, germinal center B-cell-like; ind, indeterminate; pos, positive.
FIG A14.
Kaplan-Meier curves of the outcomes in 223 patients with de novo diffuse large B-cell lymphoma (DLBCL) with advanced-stage disease treated with curative intent with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone in the BC Cancer cohort grouped according to double-hit signature (DHITsig) status and cell of origin. (A) Time to progression (TTP). (B) Disease-specific survival (DSS). (C) Progression-free survival (PFS). (D) Overall survival (OS). *P < .001. ABC, activated B-cell-like; GCB, germinal center B-cell-like; HR, hazard ratio; ind, indeterminate; pos, positive.
FIG A15.
Kaplan-Meier curves of the outcomes in 95 patients with de novo diffuse large B-cell lymphoma (DLBCL) with limited-stage disease treated with curative intent with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone in the BC Cancer cohort grouped according to double-hit signature (DHITsig) status and cell of origin. (A) Time to progression (TTP). (B) Disease-specific survival (DSS). (C) Progression-free survival (PFS). (D) Overall survival (OS). ABC, activated B-cell-like; GCB, germinal center B-cell-like; HR, hazard ratio; ind, indeterminate; pos, positive.
Footnotes
Presented at the 60th American Society of Hematology Annual Conference, San Diego, CA, December 1-4, 2018.
Supported by the Canadian Cancer Society Research Institute (704848, 705288), Genome Canada (4108), Genome British Columbia (171LYM), the Canadian Institutes of Health Research (GPH-129347, 300738), the Terry Fox Research Institute (1061, 1043), and the BC Cancer Foundation. C.S. and R.D.M are supported by a Michael Smith Foundation for Health Research career investigator award (5120) and Scholar Award (16805), respectively. D.W.S. is supported by the BC Cancer Foundation.
Processed as a Rapid Communication manuscript.
See accompanying editorial on page 175
See accompanying article on page 202
AUTHOR CONTRIBUTIONS
Conception and design: Daisuke Ennishi, Aixiang Jiang, Marco A. Marra, Christian Steidl, Ryan D. Morin, David W. Scott
Financial support: Ryan D. Morin
Administrative support: Joseph M. Connors, Ryan D. Morin
Provision of study material or patients: Merrill Boyle, Kerry J. Savage, Joseph M. Connors, Ryan D. Morin, David W. Scott
Collection and assembly of data: Daisuke Ennishi, Merrill Boyle, Brett Collinge, Susana Ben-Neriah, Graham W. Slack, Pedro Farinha, Anja Mottok, Barbara Meissner, Laurie H. Sehn, Robert Kridel, Andrew J. Mungall, Joseph M. Connors, Randy D. Gascoyne, Ryan D. Morin, David W. Scott
Data analysis and interpretation: Daisuke Ennishi, Aixiang Jiang, Brett Collinge, Bruno M. Grande, Christopher Rushton, Jeffrey Tang, Nicole Thomas, Katsuyoshi Takata, Tomoko Miyata-Takata, Saeed Saberi, Ali Bashashati, Jeffrey Craig, Diego Villa, Kerry J. Savage, Laurie H. Sehn, Robert Kridel, Sohrab P. Shah, Christian Steidl, Joseph M. Connors, Ryan D. Morin
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Double-Hit Gene Expression Signature Defines a Distinct Subgroup of Germinal Center B-Cell-Like Diffuse Large B-Cell Lymphoma
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Graham W. Slack
Consulting or Advisory Role: Seattle GeneticsAnja Mottok
Patents, Royalties, Other Intellectual Property: Patent pending “Methods to Determine Lymphoma Subtype” (Inst)
Travel, Accommodations, Expenses: Pfizer
Diego Villa
Honoraria: Roche Canada, Janssen Pharmaceuticals, Lundbeck Canada, Seattle Genetics, Gilead Science, Acerta Pharma, AstraZeneca, Celgene, Merck
Consulting or Advisory Role: Roche Canada, Janssen Pharmaceuticals, Lundbeck Canada, Seattle Genetics, Gilead Science, Acerta Pharma, AstraZeneca, Celgene
Research Funding: Roche (Inst)
Travel, Accommodations, Expenses: Roche Canada, Janssen Pharmaceuticals, Lundbeck Canada, Acerta Pharma
Kerry J. Savage
Honoraria: Seattle Genetics, Bristol-Myers Squibb, Merck, Novartis, Takeda Pharmaceuticals
Consulting or Advisory Role: Seattle Genetics, Bristol-Myers Squibb, Merck, Servier, Novartis, AbbVie, Verastem
Research Funding: Roche (Inst)
Laurie H. Sehn
Honoraria: Amgen, Apobiologix, AbbVie, Celgene, Gilead Sciences, Janssen Pharmaceuticals, Karyopharm Therapeutics, Kite Pharma, Lundbeck, Merck, Roche, Genentech, Seattle Genetics, Takeda Pharmaceuticals, TEVA Pharmaceuticals Industries, TG Therapeutics
Consulting or Advisory Role: Celgene, AbbVie, Seattle Genetics, TG Therapeutics, Janssen Pharmaceuticals, Amgen, Roche, Genentech, Gilead Sciences, Lundbeck, Amgen, Apobiologix, Karyopharm Therapeutics, Kite Pharma, Merck, Takeda Pharmaceuticals, TEVA Pharmaceuticals Industries
Research Funding: Roche (Inst), Genentech (Inst)
Robert Kridel
Travel, Accommodations, Expenses: Roche, Genentech
Marco A. Marra
Travel, Accommodations, Expenses: Roche Canada
Sohrab P. Shah
Employment: Contextual Genomics
Leadership: Contextual Genomics
Stock and Other Ownership Interests: Contextual Genomics
Consulting or Advisory Role: Contextual Genomics
Christian Steidl
Consulting or Advisory Role: Seattle Genetics, Roche
Research Funding: Tioma Therapeutics, Bristol-Myers Squibb
Patents, Royalties, Other Intellectual Property: Patent “Method for Determining Lymphoma Type” using the NanoString Technologies platform
Expert Testimony: Juno Therapeutics, Bayer AG
Joseph M. Connors
Research Funding: Seattle Genetics (Inst), Bristol-Myers Squibb (Inst), Amgen (Inst), Cephalon (Inst), Roche (Inst), Genentech (Inst), Janssen Pharmaceuticals (Inst), Eli Lilly (Inst), Merck (Inst), NanoString Technologies (Inst), Takeda Pharmaceuticals, Bayer AG
Patents, Royalties, Other Intellectual Property: Evaluation of mantle cell lymphoma and methods related thereof (Inst), methods for selecting and treating lymphoma types (Inst)
Randy D. Gascoyne
Consulting or Advisory Role: Roche, Genentech
Ryan D. Morin
Consulting or Advisory Role: Epizyme
David W. Scott
Consulting or Advisory Role: Celgene, Janssen Pharmaceuticals
Research Funding: Janssen Pharmaceuticals, Roche, Genentech, NanoString Technologies
Patents, Royalties, Other Intellectual Property: Named inventor on a patent pending that describes gene expression profiling in prognostication in classical Hodgkin lymphoma (Inst), potentially named inventor as a member of the Lymphoma/Leukemia Molecular Profiling Project on a patent pending on the use of gene expression profiling to assign cell of origin in diffuse large B-cell lymphoma (Inst), named inventor on a patent pending on the use of gene expression profiling to determine the proliferation signature in mantle cell lymphoma (Inst), named inventor on a patent pending that describes use of gene expression profiling to identify molecular subtypes of germinal center B-cell-like diffuse large B-cell lymphoma (Inst)
Travel, Accommodations, Expenses: Celgene
No other potential conflicts of interest were reported.
REFERENCES
- 1.Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shipp MA, Ross KN, Tamayo P, et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 3.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 4.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott DW, King RL, Staiger AM, et al. High-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements with diffuse large B-cell lymphoma morphology. Blood. 2018;131:2060–2064. doi: 10.1182/blood-2017-12-820605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ennishi D, Mottok A, Ben-Neriah S, et al. Genetic profiling of MYC and BCL2 in diffuse large B-cell lymphoma determines cell-of-origin-specific clinical impact. Blood. 2017;129:2760–2770. doi: 10.1182/blood-2016-11-747022. [DOI] [PubMed] [Google Scholar]
- 7.Ott G, Rosenwald A, Campo E. Understanding MYC-driven aggressive B-cell lymphomas: Pathogenesis and classification. Blood. 2013;122:3884–3891. doi: 10.1182/blood-2013-05-498329. [DOI] [PubMed] [Google Scholar]
- 8.Sarkozy C, Traverse-Glehen A, Coiffier B. Double-hit and double-protein-expression lymphomas: Aggressive and refractory lymphomas. Lancet Oncol. 2015;16:e555–e567. doi: 10.1016/S1470-2045(15)00005-4. [DOI] [PubMed] [Google Scholar]
- 9.Johnson NA, Slack GW, Savage KJ, et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30:3452–3459. doi: 10.1200/JCO.2011.41.0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green TM, Young KH, Visco C, et al. Immunohistochemical double-hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30:3460–3467. doi: 10.1200/JCO.2011.41.4342. [DOI] [PubMed] [Google Scholar]
- 11.Johnson NA, Savage KJ, Ludkovski O, et al. Lymphomas with concurrent BCL2 and MYC translocations: The critical factors associated with survival. Blood. 2009;114:2273–2279. doi: 10.1182/blood-2009-03-212191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savage KJ, Johnson NA, Ben-Neriah S, et al. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood. 2009;114:3533–3537. doi: 10.1182/blood-2009-05-220095. [DOI] [PubMed] [Google Scholar]
- 13.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morin RD, Mungall K, Pleasance E, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122:1256–1265. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohr JG, Stojanov P, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378:1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24:679–690. doi: 10.1038/s41591-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy A, Zhang J, Davis NS, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171:481–494.e15. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kridel R, Mottok A, Farinha P, et al. Cell of origin of transformed follicular lymphoma. Blood. 2015;126:2118–2127. doi: 10.1182/blood-2015-06-649905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. doi: 10.1038/s41467-018-06354-3. Arthur S, Jiang A, Grande B, et al: Genome-wide discovery of regulatory variants in diffuse large B-cell lymphoma. Nat Commun 9:4001, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortega-Molina A, Boss IW, Canela A, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21:1199–1208. doi: 10.1038/nm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y, Ortega-Molina A, Geng H, et al. CREBBP inactivation promotes the development of HDAC3-dependent lymphomas. Cancer Discov. 2017;7:38–53. doi: 10.1158/2159-8290.CD-16-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott DW, Mottok A, Ennishi D, et al. Prognostic significance of diffuse large B-cell lymphoma cell of origin determined by digital gene expression in formalin-fixed paraffin-embedded tissue biopsies. J Clin Oncol. 2015;33:2848–2856. doi: 10.1200/JCO.2014.60.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Victora GD, Dominguez-Sola D, Holmes AB, et al. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 2012;120:2240–2248. doi: 10.1182/blood-2012-03-415380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milpied P, Cervera-Marzal I, Mollichella M-L, et al. Human germinal center transcriptional programs are de-synchronized in B cell lymphoma. Nat Immunol. 2018;19:1013–1024. doi: 10.1038/s41590-018-0181-4. [DOI] [PubMed] [Google Scholar]
- 27. Mottok A, Wright G, Rosenwald A, et al: Molecular classification of primary mediastinal large B-cell lymphoma using routinely available tissue specimens. Blood 10.1182/blood-2018-05-851154 [Epub ahead of print on September 26, 2018] [DOI] [PMC free article] [PubMed]
- 28.Dominguez-Sola D, Victora GD, Ying CY, et al. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat Immunol. 2012;13:1083–1091. doi: 10.1038/ni.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calado DP, Sasaki Y, Godinho SA, et al. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol. 2012;13:1092–1100. doi: 10.1038/ni.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green MR, Kihira S, Liu CL, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci U S A. 2015;112:E1116–E1125. doi: 10.1073/pnas.1501199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davids MS, Roberts AW, Seymour JF, et al. Phase I first-in-human study of venetoclax in patients with relapsed or refractory non-Hodgkin lymphoma. J Clin Oncol. 2017;35:826–833. doi: 10.1200/JCO.2016.70.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowakowski GS, Chiappella A, Witzig TE, et al. ROBUST: Lenalidomide-R-CHOP versus pacebo-R-CHOP in previously untreated ABC-type diffuse large B-cell lymphoma. Future Oncol. 2016;12:1553–1563. doi: 10.2217/fon-2016-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]