Abstract
PURPOSE
The wide heterogeneity in multiple myeloma (MM) outcome is driven mainly by cytogenetic abnormalities. The current definition of high-risk profile is restrictive and oversimplified. To adapt MM treatment to risk, we need to better define a cytogenetic risk classification. To address this issue, we simultaneously examined the prognostic impact of del(17p); t(4;14); del(1p32); 1q21 gain; and trisomies 3, 5, and 21 in a cohort of newly diagnosed patients with MM.
METHODS
Data were obtained from 1,635 patients enrolled in four trials implemented by the Intergroupe Francophone du Myélome. The oldest collection of data were used for model development and internal validation. For external validation, one of the two independent data sets was used to assess the performance of the model in patients treated with more current regimens. Six cytogenetic abnormalities were identified as clinically relevant, and a prognostic index (PI) that was based on the parameter estimates of the multivariable Cox model was computed for all patients.
RESULTS
In all data sets, a higher PI was consistently associated with a poor survival outcome. Dependent on the validation cohorts used, hazard ratios for patients in the high-risk category for death were between six and 15 times higher than those of patients in the low-risk category. Among patients with t(4;14) or del(17p), we observed a worse survival in those classified in the high-risk category than in those in the intermediate-risk category. The PI showed good performance for discriminating between patients who died and those who survived (Harrell’s concordance index greater than 70%).
CONCLUSION
The cytogenetic PI improves the classification of newly diagnosed patients with MM in the high-risk group compared with current classifications. These findings may facilitate the development of risk-adapted treatment strategies.
INTRODUCTION
Multiple myeloma (MM) is characterized by a wide heterogeneity in outcome, with patients staying alive more than 10 years after diagnosis and others dying in a few months. Among all prognostic factors described in MM, chromosomal abnormalities present in tumor plasma cells and detected by interphase fluorescence in situ hybridization (FISH) or by single nucleotide polymorphism (SNP) array have a substantial impact.1 Indeed, the International Myeloma Working Group has recommended the incorporation of these factors along with serum lactate dehydrogenase in the revised International Staging System (ISS) for Multiple Myeloma (R-ISS).2 Among the high-risk chromosomal changes described in MM, those established and on which there is general consensus are del(17p) and t(4;14). These abnormalities negatively affect both progression-free survival and overall survival and, respectively, approximately 8% and 15% of patients with newly diagnosed MM (NDMM).3-7 A consensus also seems to exist on the protective role of hyperdiploidy.8-11 Among all trisomies involved in hyperdiploid MM, three have been shown to modulate overall survival of these high-risk patients. First, patients with trisomy of chromosome 3 or 5 have a significantly improved overall survival compared with those who lack these trisomies, whereas patients with trisomy 21 have worse outcomes than those who lack this trisomy.12 Second, a more recently described high-risk chromosomal change is the del(1p32), which affects 8% of patients with NDMM,13 the presence of which dramatically worsens the poor outcome value of t(4;14).14 Finally, although it displays a lower prognostic impact, the frequent gain of 1q21 is also significantly associated with reduced survival and affects approximately one third of patients with NDMM.15 All combinations among these abnormalities are theoretically possible, even if some of them are more frequent, such as the association among trisomies.16 Nevertheless, they rarely have been analyzed together.17-19 Today, the definition of cytogenetic risk profile in patients with MM, on the basis of two or three unfavorable prognostic markers, seems restrictive and oversimplified. We aimed to adapt MM treatment to risk by defining a cytogenetic risk classification that could be used systematically (hence routinely). We simultaneously examined the prognostic impact of del(17p); t(4;14); del(1p32); 1q21 gain; and trisomies 3, 5, and 21 in a large cohort of patients with NDMM. Our main objective was to develop and validate a prognostic model that is based on these seven cytogenetic abnormalities and to evaluate how successful this model is when used in a more recent series of patients with MM.
METHODS
Patients and Methods
Cytogenetics results and clinical data were obtained from patients enrolled in four different randomized clinical trials implemented by the Intergroupe Francophone du Myélome (IFM): IFM 99-02 (ClinicalTrials.gov identifier: NCT00222053), IFM 99-06 (ClinicalTrials.gov identifier: NCT00367185), IFM 2005-01 (ClinicalTrials.gov identifier: NCT00200681), and IFM 2009 (ClinicalTrials.gov identifier: NCT01191060). Complete details of these trials have been reported.20-23 The oldest collection of IFM cytogenetic data was from 960 patients, and these data were used as the primary data set for model development and internal validation. This data set was chosen not only because it already contained the common cytogenetic aberrations and trisomy abnormalities described in patients with MM (ie,1p12, 1p22, 1p32, 1q, 3q, 5p, 6p, 6q, 8p, 12p, 14q, 16q, 20p, 20q, 22q plus trisomies) but also because a long-term follow-up period over several years was available for all patients. Seventy-four percent of the patients received an intensive approach with either vincristine, doxorubicin, and dexamethasone or bortezomib and dexamethasone (VD) followed by high-dose melphalan (HDM) and autologous stem-cell transplantation (ASCT), whereas 26% received a nonintensive treatment because they were older than age 65 years.
After model development and internal validation, two other data sets were formed to include ISS disease stage, which was unavailable in the primary data set, and to test whether the prognostic model applies in patients treated with more current and effective treatment regimens. The first data set included 359 patients enrolled during the same period as the training set, and the second was derived from the IFM 2009 study and included 322 patients. For these two external validation cohorts, only conventional high-risk cytogenetic factors were available, and new cytogenetic analyses were performed to derive the prognostic index (PI). Hence, patients were selected if an excess of bone marrow plasma cells were still stored in the IFM myeloma biobank (DC-201-1654). None of these patients were included in the primary data set. Similar to the primary data set, the first external validation cohort comprised patients who received induction therapy with either vincristine, doxorubicin, and dexamethasone (n = 152) or VD (n = 148) followed by HDM and ASCT and 59 patients (16%) who received a nonintensive treatment because they were older than age 65 years. The second external validation cohort included 167 patients (52%) who received a lenalidomide and VD (RVD) induction followed by an RVD consolidation and 155 patients (48%) who received an RVD induction followed by HDM and ASCT and an RVD consolidation. All patients gave written informed consent before entering the source trials.
Bone marrow samples were obtained at diagnosis before treatment initiation and were shipped overnight to a central laboratory. Upon receipt, plasma cells were isolated from bone marrow using CD138+ magnetic-activated cell sorting (Miltenyi Biotec, Paris, France). Postsorting purity was checked by cytologic analysis of a spin from positive fraction. Only samples with 70% or more of plasma cells after sorting were kept, per protocol, for the cytogenetic analysis. Plasma cells from all samples were analyzed by FISH for t(4;14)(p16;q32) determination using specific probes from Abbott Molecular (Paris, France) and by SNP array (Affymetrix, Santa Clara, CA) using the Cytoscan HD Array Kit (Affymetrix) for the detection of the six other anomalies. In cases where del(17p) was detected, an additional FISH analysis was performed to evaluate the percent of involved plasma cells using specific probe TP53/CEP17 (Cytocell AmpliTech, Compiègne, France). Only del(17p) present in at least 60% of plasma cells were taken into account.24 This stringent definition has been confirmed by a large meta-analysis25 in patients who displayed the anomaly in less than 60% of the plasma cells presenting a non–high-risk profile. For the second external validation cohort, SNP array analyses were performed for only 32% of patients because of a lack of DNA. For the other 68% of patients, we performed FISH experiments using probes that targeted chromosomes 1p32, 1q21, 5p23, 17p13, and 21q21. These probes were provided by Abbott Molecular. The choice of the 5p23 probe was based on the analysis of more than 1,500 SNP arrays, which showed that the minimal gained region was 5p. All cytogenetic assessments were performed by biologists blinded to clinical data.
Statistical Analysis
MM-specific survival was defined as the time from diagnosis to death. Patients whose cause of death was definitely unrelated to MM were censored at the time of death. Death events excluded from analysis were a result of other malignancies, lethal toxicities from treatments under investigation, and violence (eg, suicide, road traffic accident). Alive patients were censored at the last date known alive. The primary data set, which comprised 960 patients, was split into two parts (3:1) according to bone marrow sampling date to create two sets of patients used for the development (n = 720) and the internal validation (n = 240) of the model. Because the split was in time, the internal validation set can be viewed as a temporal validation. In the training set, 73 of the 720 patients had a missing value for t(4;14), and multiple imputations that were based on 30 imputed data sets were applied to replace missing values by imputed values from a logit model. In the internal validation set, six patients had missing values for trisomy 21 or t(4;14), but no imputation was performed because model development was solely on the basis of data from the training set. A multivariable Cox proportional hazards regression model stratified on treatment was used to identify cytogenetic abnormalities [del(17p), t(4;14), del(1p32), and 1q21 gain and trisomies 3, 5, and 21] predictive of survival (Appendix Table A1, online only). All first-order interactions between abnormalities were tested, and none were significant. The proportionality assumption was checked with Cox-Snell residuals and log-log plots. There was evidence of a time-varying effect associated with the del(17p) abnormality, which was shown to diminish progressively with time. [The average relative hazard for patients with del(17p) during the first 5 years after diagnosis was reduced from 3.2 to 1.5 after 5 years.]
To avoid complex modeling of the cytogenetic PI and to facilitate its application in practice and research settings, the validation of the final model was thereafter limited to the prognosis of death within the first 5 years. A PI that was based on the parameter estimates of the multivariable Cox model was computed for all patients. To check the validity of the PI, measures of discrimination and calibration were performed in the validation cohorts where follow-up was censored at 5 years. Discrimination was assessed using Kaplan-Meier survival curves for the cytogenetic risk groups and by estimating hazard ratios along with their 95% CIs as well as using Harrell’s concordance index (C-index). The C-index estimates the proportion of all pairs of patients in whom prediction and outcome are concordant and takes values from 0.5 (no discrimination) to 1.0 (perfect discrimination). The model was recalibrated to account for different baseline hazard functions in the validation data sets, and calibration was checked by performing Cox proportional hazards regression modeling in the validation data sets with the PI as a single covariate. Calibration was deemed valid if the estimate parameter was not statistically different from 1. Calibration also was checked by plotting the observed survival probability versus the expected survival probability at 5 years for different values of the PI. Tests were two-sided, and P < .05 was considered significant. Statistical analyses were performed using SAS 9.4 (SAS Institute, Cary, NC) and R version 3.0.2 (R Foundation for Statistical Computing, Vienna, Austria) software.
RESULTS
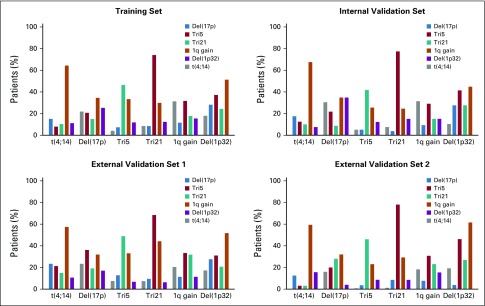
Altogether, 1,635 patients with MM diagnosed between 2000 and 2012 were analyzed. Patient characteristics, treatment, and cytogenetic abnormalities are listed in Table 1. The adverse cytogenetic lesions were slightly over-represented in the training set. The median number of cytogenetic abnormalities was two per patient (interquartile range, 1-3) in the training set and one (interquartile range, 0-2) in the other data sets. In the training and internal validation sets, no cytogenetic abnormality was found in 20% of patients, whereas in the external validation data sets, there were 25% without any cytogenetic abnormality. The complexity of the associations between cytogenetic abnormalities is shown in Appendix Figure A1 (online only). The most frequent combination was the one between trisomies 5 and 21 followed by 1q gain among patients with t(4;14) and del(1p32). The rarest combination was the one between trisomy 5 and t(4;14).
TABLE 1.
Patient Characteristics and Cytogenetic Abnormalities in the Four Data Sets
Because of a difference in cohort construction, the median follow-up time was 8.2, 6.0, 7.3, and 4.9 years in the training, internal validation, and external validation sets 1 and 2, respectively. The estimated 5-year survival probabilities were 58%, 62%, 54%, and 80%, respectively.
In the training set, six cytogenetic abnormalities were identified as statistically relevant (Table 2), and the PI was computed as follows: 0.4 × t(4;14) + 1.2 × del(17p) − 0.3 × trisomy 5 + 0.3 × trisomy 21 + 0.5 × 1q gain + 0.8 × del(1p32). On the basis of the distribution of the PI and the 5-year survival probability, three categories of cytogenetic risk were created. The low-risk group included all patients with a PI less than or equal to 0 in whom 5-year survival probability was greater than 75%, the high-risk group included all patients with a PI greater than 1 in whom 5-year survival probability was less than 50%, and the intermediate-risk group included patients with a PI of 0 to 1 in whom 5-year survival probability was between 50% and 75%.
TABLE 2.
Multivariable Cox Proportional Hazard Regression Models of Myeloma-Specific Survival Stratified by Treatment in the Training Set
The PI ranged from −0.3 to 2.9 in all data sets. A score of 0 or less was observed only in patients with no adverse cytogenetic abnormalities. In the training set, a score greater than 1 was observed in 94% of patients with del(17p), 29% with t(4;14), 51% with either del(17p) or t(4;14), and 4% with neither of these two adverse cytogenetic abnormalities as a result of the combination of del(1p32) with either or both 1q gain and trisomy 21. The distribution of the PIs and the repartition of risk categories according to del(17p), t(4;14), and ISS disease stages were similar across data sets (Appendix Figs A2 and A3, online only).
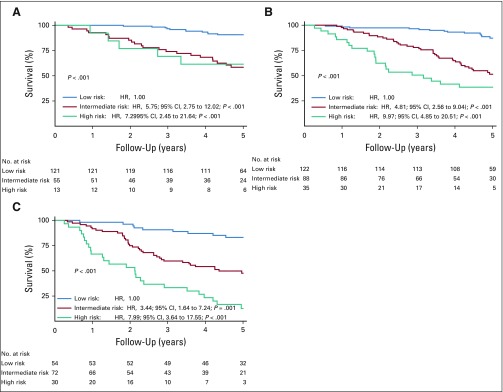
In all the data sets, a higher PI was consistently associated with a poor survival outcome, and patients classified in intermediate- and high-risk cytogenetic groups experienced shorter survival times than those in the low-risk group (Fig 1). Dependent on the validation cohorts used, patients in the high-risk cytogenetic category had hazard ratios for death that were six to 15 times higher than those of patients in the low-risk category. These results were confirmed in the two external validation data sets after adjustment for ISS disease stages, which indicates that the PI provided independent information over that obtained from the ISS disease stage to predict death events in the external validation data sets (Table 3). By pooling all the validation data sets to show the impact of the PI on patients who displayed or not t(4;14) and del(17p) and according to ISS disease stages at diagnosis, we always observed a worse survival in patients classified in the high-risk category than in those classified in the intermediate-risk category (Appendix Figs A4 to A6, online only).
FIG 1.
Kaplan-Meier survival curves for myeloma-specific survival according to the three categories of the cytogenetic prognostic index. (A) Training set (n = 647). (B) Internal validation set (n = 234). (C) External validation data set 1 (n = 359). (D) External validation data set 2 (n = 322). Cox proportional hazards regression models were stratified by treatment group. HR, hazard ratio.
TABLE 3.
Cox Proportional Hazard Regression Models for Myeloma-Specific Survival at Up to 5 Years, Including ISS Disease Stage Adjusted or Not on the PI
The prognostic significance carried by the six cytogenetic abnormalities demonstrated good performance for discriminating patients who died from those who survived in all the data sets. The C-indices for the PI that defined the three risk categories were 0.71 (95% CI, 0.66 to 0.77), 0.76 (95% CI, 0.72 to 0.79), and 0.71 (95% CI, 0.64 to 0.78) for the internal validation set and external validation data sets 1 and 2, respectively. Results were similar if the PI was used as a linear score.
Using external validation data sets 1 and 2, we also assessed the discriminatory ability of ISS disease stage alone or in combination with the PI and adverse cytogenetic profile defined by either del(17p) or t(4;14). The better discriminatory performance was observed for PI alone used as a continuous index or in three risk categories. Any other prognostic models that were based on ISS disease stage and/or adverse cytogenetic profile systematically had a lower ability to discriminate patients than a model that included only the PI. Moreover, when ISS disease stage was combined with the PI, the added prognostic value of ISS was not clinically relevant (Table 4). Using external validation data set 2, we also were able to derive the R-ISS. The C-index for the R-ISS disease stage alone was 0.55, which means that the PI had a better prognostic accuracy. Observed versus predicted probabilities, after model recalibration to account for differences in baseline survival function, showed that calibration was correct in the validation data sets (Appendix Fig A7, online only).
TABLE 4.
Cox Proportional Hazards Regression Models for Myeloma-Specific Survival, Including the Cytogenetic PI With or Without Other Known Prognostic Factors in the External Validation Data Sets
Finally, we graphically assessed the prognostic value of the PI relative to the study of time to progression, which is defined as the time from diagnosis to disease progression or death as a result of disease. In external validation data set 2, the risk of progression or death related to MM was higher for patients classified in the high- and intermediate-risk categories than in those in the low-risk category (Fig 2). Median time to progression was 3.6, 3.0, and 2.4 years in the low-, intermediate-, and high-risk groups, respectively.
FIG 2.
Kaplan-Meier survival curves for time to progression according to the three categories of the cytogenetic prognostic index in the external validation data set 2. Cox proportional hazards regression models were stratified by treatment group and adjusted by International Staging System disease stage. aHR, adjusted hazard ratio.
DISCUSSION
Specific chromosomal abnormalities play a major role in MM prognostication, and combinations of cytogenetic lesions are more often the norm rather than the exception.17,18,26,27 With improvement in patients’ survival, those who display del(17p) or t(4;14) were observed to have heterogeneous survival that depended on the co-occurrence of other lesions.11,12,14,17,18 Hence, it became obvious that the current definition of a high-risk cytogenetic group defined by the presence of del(17p), t(4;14), or t(14;16) was oversimplified and could lead to misclassification of patient prognosis because it was only based on a few cytogenetic abnormalities, which gave them the same weight in assigning patients in a high-risk group. The cytogenetic PI presented here is innovative because it is based on a weighted score, including several cytogenetic abnormalities, and because it also includes a good prognostic factor, the trisomy 5.12 According to our PI, patients were identified at high risk if they displayed the only del(17p) abnormality that induced a poor prognosis in itself or various combinations of adverse cytogenetic lesions. Consequently, less than 35% of patients who displayed t(4;14) were identified as high risk, and those patients had worse survival than patients with t(4;14) from the intermediate-risk category. This is important to take into account not only in the analysis of published prospective trials but also in the design of future risk-adapted trials.
This PI was developed from a large series of patients with sufficient follow-up to identify good prognostic factors (using a Cox proportional hazards regression model stratified by chemotherapy received during induction) predicted by cytogenetic markers only. Age was not considered during model development because none of the cytogenetic lesions studied were associated with age.28-30 We show that the PI performance for discriminating patients who died versus those who survived was 10 points higher compared with conventional stratification factors, which means that the PI alone was the better risk score for differentiating risk categories in patients with MM. Its discriminatory ability was also better than other combinations of traditional risk scores, including R-ISS.
The strength of this study lies in the external validations performed, especially using patients from validation data set 2 who differed from those of the training set because they received up-to-date induction and consolidation therapy and effective second-line therapies. Even though probability of survival was higher in this cohort, the PI retained good levels of prognostic and discriminative abilities. These findings suggest that the PI contains the main cytogenetic lesions and validate its transportability to patients with MM in other settings.
A limitation of this study is the development of a score in patients only included in clinical trials. This limits its application in daily clinical practice. Nevertheless, the score provides the advantage to require standardized methods because the six anomalies were assessable by widely available FISH and/or SNP array. Another limitation is the noninclusion of cytogenetic aberrations that are potentially important for the survival of patients, such as t(14;16), which is now part of the definition of the R-ISS, although its prognostic significance is not confirmed by all studies.2,8,31 Data were available for external validation data set 2, and we observed that patients who displayed a t(14;16) had the same median overall survival as those without it. In addition, Walker et al26 showed that the number of copies of 1q21 gain seems to be important to consider for correctly modeling its prognostic impact.
In conclusion, our data suggest that the cytogenetic PI improves the classification of patients in high-risk groups compared with classifications commonly used during the past few years. These findings may provide better prognostic stratification for patients with NDMM included in clinical trials and accelerate the development of trials on the basis of risk-adapted treatment strategies.
ACKNOWLEDGMENT
We thank IFM for providing patient samples and clinical data, with special thanks to Sandrine Rollet. We thank Abbott Molecular for providing probes for cytogenetic analysis.
APPENDIX
FIG A1.
Associations among the six cytogenetic anomalies in the four data sets. Tri, trisomy.
FIG A2.
Distribution of the linear prognostic index in the four data sets.
FIG A3.
Distribution of risk categories in function of t(4;14) and del(17p) in the four data sets.
FIG A4.
Kaplan-Meier survival curves for myeloma specific survival according to the risk categories of the cytogenetic prognostic index in patients displaying t(4;14) (A) and del17p (B). All validation datasets were pooled for this analysis. Cox proportional regression models were stratified on treatment groups. CI, confidence interval; HR, hazard ratio.
FIG A5.
Kaplan-Meier survival curves for myeloma specific survival according to the risk categories of the cytogenetic prognostic index in patients negative for both t(4;14) and del(17p)(A) and in patients displaying t(4;14) or del(17p) (B). External validation datasets were pooled for this analysis. Cox proportional regression models were stratified on treatment groups. CI, confidence interval; HR, hazard ratio.
FIG A6.
Kaplan-Meier survival curves for myeloma specific survival according to the risk categories of the cytogenetic prognostic index for patients with ISS disease stage I (A), stage II (B) and stage III (C) at diagnosis. External validation datasets were pooled for this analysis. Cox proportional regression models were stratified on treatment groups. CI, confidence interval; HR, hazard ratio.
FIG A7.
Comparison of empirical 5-year survival probability according to Kaplan-Meier method and 5-year survival probability predicted by Cox proportional hazards (CPH) regression modeling.
TABLE A1.
Multivariable Cox Proportional Hazard Regression Models of Myeloma Specific Survival Stratified by Treatment in the Training Set
Footnotes
Supported by National Cancer Institute grants P01-155258 and P50-100707 (H.A.-L.) and the Cancer Pharmacology of Toulouse and Region program. The Centre de Recherches en Cancérologie de Toulouse Team 13 is supported by la Fondation Association pour la Recherche sur la Cancer grant PGA1*20160203788.
AUTHOR CONTRIBUTIONS
Conception and design: Sabine Brechignac, Claudine Sohn, Mohamad Mohty, Xavier Leleu, Thierry Facon, Michel Attal, Hervé Avet-Loiseau, Jill Corre
Administrative support: Frédérique Orsini-Piocelle
Provision of study material or patients: Mamoun Dib, Laurent Voillat, Hélène Demarquette, Philippe Collet. Mohamad Mohty, Margaret Macro, Philippe Moreau, Xavier Leleu
Collection and assembly of data: Aurore Perrot, Cyrille Hulin, Marie-Lorraine Chretien, Mamoun Dib, Arnaud Jaccard, Karim Belhadj, Sabine Brechignac, Jean Fontan, Laurent Voillat, Hélène Demarquette, Philippe Collet, Philippe Rodon, Claudine Sohn, François Lifermann, Frédérique Orsini-Piocelle, Valentine Richez, Margaret Macro, Stéphane Minvielle, Xavier Leleu, Thierry Facon, Jill Corre
Data analysis and interpretation: Valérie Lauwers-Cances, Elodie Tournay, Cyrille Hulin, Bruno Royer, Olivier Decaux, Sabine Brechignac, Claudine Sohn, Philippe Moreau, Xavier Leleu, Thierry Facon, Michel Attal, Hervé Avet-Loiseau, Jill Corre
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Development and Validation of a Cytogenetic Prognostic Index Predicting Survival in Multiple Myeloma
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Aurore Perrot
Honoraria: Janssen Pharmaceuticals, Amgen, Celgene, Sanofi, Takeda Pharmaceuticals
Cyrille Hulin
Honoraria: Celgene, Janssen-Cilag, Amgen, AbbVie, Takeda Pharmaceuticals
Travel, Accommodations, Expenses: Celgene, Janssen-Cilag, Amgen
Marie-Lorraine Chretien
Consulting or Advisory Role: Celgene
Bruno Royer
Consulting or Advisory Role: Janssen-Cilag
Travel, Accommodations, Expenses: Amgen
Olivier Decaux
Honoraria: Janssen-Cilag, Celgene, Roche
Consulting or Advisory Role: Janssen-Cilag, Celgene, Takeda Pharmaceuticals
Research Funding: The Binding Site, Sebia, Siemens Healthcare Diagnostics
Travel, Accommodations, Expenses: Celgene, Janssen-Cilag, Roche
Arnaud Jaccard
Honoraria: Janssen Pharmaceuticals, Pfizer, Prothena, Sanofi
Research Funding: Janssen Pharmaceuticals (Inst), Celgene (Inst)
Travel, Accommodations, Expenses: Janssen Pharmaceuticals, AbbVie, Celgene
Karim Belhadj
Honoraria: Celgene, Takeda Pharmaceuticals, Amgen, Janssen Pharmaceuticals
Consulting or Advisory Role: Celgene
Travel, Accommodations, Expenses: Celgene, Amgen, Takeda Pharmaceuticals
Frédérique Orsini-Piocelle
Consulting or Advisory Role: Celgene
Mohamad Mohty
Honoraria: Celgene, Amgen, Bristol-Myers Squibb, Janssen Pharmaceuticals, Takeda Pharmaceuticals, Pfizer
Consulting or Advisory Role: Jazz Pharmaceuticals, Sanofi, MaaT Pharma, MolMed, Novartis, Atara Biotherapeutics, Xenikos, Daiichi Sankyo
Speakers’ Bureau: Janssen Pharmaceuticals, Sanofi, Jazz Pharmaceuticals
Research Funding: Sanofi (Inst), Roche (Inst), Jazz Pharmaceuticals (Inst)
Expert Testimony: Sanofi
Margaret Macro
Consulting or Advisory Role: Celgene, Amgen
Research Funding: Takeda Pharmaceuticals (Inst), Janssen Medical Affairs (Inst)
Travel, Accommodations, Expenses: Celgene, Takeda Pharmaceuticals, Janssen Pharmaceuticals, Amgen
Philippe Moreau
Honoraria: Celgene, Takeda Pharmaceuticals, Novartis, Janssen-Cilag, Amgen
Consulting or Advisory Role: Celgene, Takeda Pharmaceuticals, Janssen Pharmaceuticals, Novartis, Amgen
Xavier Leleu
Honoraria: Janssen-Cilag, Celgene, Amgen, Novartis, Bristol-Myers Squibb, Takeda Pharmaceuticals, Pierre Fabre, Sanofi, AbbVie, Merck, Roche, Mundipharma, Karyopharm Therapeutics, Carsgen Therapeutics, Oncopeptides
Consulting or Advisory Role: Janssen-Cilag, Celgene, Amgen, Takeda Pharmaceuticals, Bristol-Myers Squibb, Novartis, Merck, Gilead Sciences, AbbVie, Roche, Karyopharm Therapeutics, Mundipharma, Oncopeptides, Carsgen Therapeutics
Travel, Accommodations, Expenses: Takeda
Thierry Facon
Consulting or Advisory Role: Celgene, Janssen Pharmaceuticals, Takeda Pharmaceuticals, Amgen, Karyopharm Therapeutics, Sanofi, Oncopeptides
Speakers’ Bureau: Celgene, Janssen Pharmaceuticals, Takeda Pharmaceuticals
No other potential conflicts of interest were reported.
REFERENCES
- 1.Chng WJ, Dispenzieri A, Chim CS, et al. IMWG consensus on risk stratification in multiple myeloma. Leukemia. 2014;28:269–277. doi: 10.1038/leu.2013.247. [DOI] [PubMed] [Google Scholar]
- 2.Palumbo A, Avet-Loiseau H, Oliva S, et al. Revised International Staging System for Multiple Myeloma: A report from International Myeloma Working Group. J Clin Oncol. 2015;33:2863–2869. doi: 10.1200/JCO.2015.61.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang H, Sloan S, Li D, et al. The t(4;14) is associated with poor prognosis in myeloma patients undergoing autologous stem cell transplant. Br J Haematol. 2004;125:64–68. doi: 10.1111/j.1365-2141.2004.04867.x. [DOI] [PubMed] [Google Scholar]
- 4.Gutiérrez NC, Castellanos MV, Martín ML, et al. Prognostic and biological implications of genetic abnormalities in multiple myeloma undergoing autologous stem cell transplantation: t(4;14) is the most relevant adverse prognostic factor, whereas RB deletion as a unique abnormality is not associated with adverse prognosis. Leukemia. 2007;21:143–150. doi: 10.1038/sj.leu.2404413. [DOI] [PubMed] [Google Scholar]
- 5.Gertz MA, Lacy MQ, Dispenzieri A, et al. Clinical implications of t(11;14)(q13;q32), t(4;14)(p16.3;q32), and -17p13 in myeloma patients treated with high-dose therapy. Blood. 2005;106:2837–2840. doi: 10.1182/blood-2005-04-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moreau P, Attal M, Garban F, et al. Heterogeneity of t(4;14) in multiple myeloma. Long-term follow-up of 100 cases treated with tandem transplantation in IFM99 trials. Leukemia. 2007;21:2020–2024. doi: 10.1038/sj.leu.2404832. [DOI] [PubMed] [Google Scholar]
- 7.Fonseca R, Blood E, Rue M, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101:4569–4575. doi: 10.1182/blood-2002-10-3017. [DOI] [PubMed] [Google Scholar]
- 8.Fonseca R, Bergsagel PL, Drach J, et al. International Myeloma Working Group molecular classification of multiple myeloma: Spotlight review. Leukemia. 2009;23:2210–2221. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chng WJ, Santana-Dávila R, Van Wier SA, et al. Prognostic factors for hyperdiploid-myeloma: Effects of chromosome 13 deletions and IgH translocations. Leukemia. 2006;20:807–813. doi: 10.1038/sj.leu.2404172. [DOI] [PubMed] [Google Scholar]
- 10.Chng WJ, Kumar S, Vanwier S, et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 2007;67:2982–2989. doi: 10.1158/0008-5472.CAN-06-4046. [DOI] [PubMed] [Google Scholar]
- 11.Kumar S, Fonseca R, Ketterling RP, et al. Trisomies in multiple myeloma: Impact on survival in patients with high-risk cytogenetics. Blood. 2012;119:2100–2105. doi: 10.1182/blood-2011-11-390658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chretien ML, Corre J, Lauwers-Cances V, et al. Understanding the role of hyperdiploidy in myeloma prognosis: Which trisomies really matter? Blood. 2015;126:2713–2719. doi: 10.1182/blood-2015-06-650242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hebraud B, Leleu X, Lauwers-Cances V, et al. Deletion of the 1p32 region is a major independent prognostic factor in young patients with myeloma: The IFM experience on 1195 patients. Leukemia. 2014;28:675–679. doi: 10.1038/leu.2013.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hebraud B, Magrangeas F, Cleynen A, et al. Role of additional chromosomal changes in the prognostic value of t(4;14) and del(17p) in multiple myeloma: The IFM experience. Blood. 2015;125:2095–2100. doi: 10.1182/blood-2014-07-587964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fonseca R, Van Wier SA, Chng WJ, et al. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia. 2006;20:2034–2040. doi: 10.1038/sj.leu.2404403. [DOI] [PubMed] [Google Scholar]
- 16.Fonseca R, Barlogie B, Bataille R, et al. Genetics and cytogenetics of multiple myeloma: A workshop report. Cancer Res. 2004;64:1546–1558. doi: 10.1158/0008-5472.can-03-2876. [DOI] [PubMed] [Google Scholar]
- 17.Boyd KD, Ross FM, Chiecchio L, et al. A novel prognostic model in myeloma based on co-segregating adverse FISH lesions and the ISS: Analysis of patients treated in the MRC Myeloma IX trial. Leukemia. 2012;26:349–355. doi: 10.1038/leu.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. doi: 10.1038/leu.2017.179. Shah V, Sherborne AL, Walker BA, et al: Prediction of outcome in newly diagnosed myeloma: A meta-analysis of the molecular profiles of 1905 trial patients. Leukemia 32:102-110, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smol T, Dufour A, Tricot S, et al. Combination of t(4;14), del(17p13), del(1p32) and 1q21 gain FISH probes identifies clonal heterogeneity and enhances the detection of adverse cytogenetic profiles in 233 newly diagnosed multiple myeloma. Mol Cytogenet. 2017;10:26. doi: 10.1186/s13039-017-0327-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harousseau JL, Avet-Loiseau H, Attal M, et al. Achievement of at least very good partial response is a simple and robust prognostic factor in patients with multiple myeloma treated with high-dose therapy: Long-term analysis of the IFM 99-02 and 99-04 Trials. J Clin Oncol. 2009;27:5720–5726. doi: 10.1200/JCO.2008.21.1060. [DOI] [PubMed] [Google Scholar]
- 21.Facon T, Mary JY, Hulin C, et al. Melphalan and prednisone plus thalidomide versus melphalan and prednisone alone or reduced-intensity autologous stem cell transplantation in elderly patients with multiple myeloma (IFM 99-06): A randomised trial. Lancet. 2007;370:1209–1218. doi: 10.1016/S0140-6736(07)61537-2. [DOI] [PubMed] [Google Scholar]
- 22.Harousseau JL, Attal M, Avet-Loiseau H, et al. Bortezomib plus dexamethasone is superior to vincristine plus doxorubicin plus dexamethasone as induction treatment prior to autologous stem-cell transplantation in newly diagnosed multiple myeloma: Results of the IFM 2005-01 phase III trial. J Clin Oncol. 2010;28:4621–4629. doi: 10.1200/JCO.2009.27.9158. [DOI] [PubMed] [Google Scholar]
- 23.Attal M, Lauwers-Cances V, Hulin C, et al. Lenalidomide, bortezomib, and dexamethasone with transplantation for myeloma. N Engl J Med. 2017;376:1311–1320. doi: 10.1056/NEJMoa1611750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avet-Loiseau H, Attal M, Moreau P, et al. Genetic abnormalities and survival in multiple myeloma: The experience of the Intergroupe Francophone du Myélome. Blood. 2007;109:3489–3495. doi: 10.1182/blood-2006-08-040410. [DOI] [PubMed] [Google Scholar]
- 25.Thakurta A, Ortiz M, Blecua P, et al. High subclonal fraction of 17p deletion is associated with poor prognosis in multiple myeloma. Blood. 2019;133:1217–1221. doi: 10.1182/blood-2018-10-880831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker BA, Boyle EM, Wardell CP, et al. Mutational spectrum, copy number changes, and outcome: Results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015;33:3911–3920. doi: 10.1200/JCO.2014.59.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. doi: 10.1038/s41375-018-0037-9. Bolli N, Biancon G, Moarii M, et al: Analysis of the genomic landscape of multiple myeloma highlights novel prognostic markers and disease subgroups. Leukemia 10.1038/leu.2017.344 [Epub ahead of print on December 6, 2017] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsson T, Lenhoff S, Turesson I, et al. Cytogenetic features of multiple myeloma: Impact of gender, age, disease phase, culture time, and cytokine stimulation. Eur J Haematol. 2002;68:345–353. doi: 10.1034/j.1600-0609.2002.00724.x. [DOI] [PubMed] [Google Scholar]
- 29.Greenberg AJ, Philip S, Paner A, et al. Racial differences in primary cytogenetic abnormalities in multiple myeloma: A multi-center study. Blood Cancer J. 2015;5:e279. doi: 10.1038/bcj.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chretien ML, Hebraud B, Cances-Lauwers V, et al. Age is a prognostic factor even among patients with multiple myeloma younger than 66 years treated with high-dose melphalan: The IFM experience on 2316 patients. Haematologica. 2014;99:1236–1238. doi: 10.3324/haematol.2013.098608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Avet-Loiseau H, Malard F, Campion L, et al. Translocation t(14;16) and multiple myeloma: Is it really an independent prognostic factor? Blood. 2011;117:2009–2011. doi: 10.1182/blood-2010-07-295105. [DOI] [PubMed] [Google Scholar]