Abstract
The formation of flowers in higher plants is controlled by complex gene regulatory networks. The study of floral development in Arabidopsis is promoted and maintained by transposon-tagged mutant lines. In this study, we report a CRISPR/Cas9 genome-editing system based on RNA endoribonuclease Csy4 processing to induce high-efficiency and inheritable targeted deletion of transcription factors involved in floral development in Arabidopsis. Using AP1, SVP, and TFL1 as the target genes, multisite and multiple-gene mutations were achieved with a tandemly arrayed Csy4-sgRNA architecture to express multiplexed sgRNAs from a single transcript driven by the Pol II promoter in transgenic lines. Targeted deletions of chromosomal fragments between the first exon and second exon in either one or three genes were generated by using a single binary vector. Interestingly, the efficiency of site-targeted deletion was comparable to that of indel mutation with the multiplexed sgRNAs. DNA sequencing analysis of RT-PCR products showed that targeted deletions of AP1 and TFL1 could lead to frameshift mutations and introduce premature stop codons to disrupt the open-reading frames of the target genes. In addition, no RT-PCR amplified product was acquired after SVP-targeted deletion. Furthermore, the targeted deletions resulted in abnormal floral development in the mutant lines compared to that of wild-type plants. AP1 and SVP mutations increased plant branching significantly, while TFL1 mutant plants displayed a change from indeterminate to determinate inflorescences. Thus, our results demonstrate that CRISPR/Cas9 with the RNA endoribonuclease Csy4 processing system is an efficient tool to study floral development and improve floral traits rapidly and simply.
Subject terms: Plant molecular biology, Molecular engineering in plants, Molecular engineering in plants
Introduction
The CRISPR/Cas9 (clustered, regularly interspersed palindromic repeats-associated endonuclease 9) system has recently been developed into a powerful genome-editing technology with a role in revealing plant gene function and improving crops1–5. CRISPR/Cas9 with a single guide RNA (sgRNA) targeted to specific DNA sequences generates a DNA double-strand break (DSB) at a position three base pairs upstream of the protospacer adjacent motif (PAM) sequence. The resulting DSBs are repaired primarily by either nonhomologous end joining (NHEJ) or homology-directed repair (HDR) in vivo6–8. NHEJ occurs most frequently and can introduce indel mutations, such as random nucleotide deletions or insertions at the break site that induce a frameshift mutation in the target gene6. The targeted mutagenesis derived from genome editing by CRISPR/Cas9 is valuable for understanding the functions of specific genes, and it is becoming one of the most powerful tools to knock out an individual gene, as well as to target the insertion of a specific gene and/or control gene transcription9–13. However, the traditional CRISPR/Cas9 system is dependent on the RNA polymerase III (Pol III) promoter driving one sgRNA to generate one DSB at a target site in the genome6,8,14–17.
Previous studies have shown that mutations introduced by CRISPR/Cas9 using one sgRNA are difficult to identify by PCR analysis, and techniques are limited to homology analysis by DNA sequencing9,12,18–21. It was also found that the mutant exons generated by CRISPR/Cas9 with one sgRNA can lead to exon skipping19. Multiplex gene knockout has been performed by CRISPR/Cas9 using a single sgRNA targeting the conserved regions of multiple genes in a family22 or using multiplexed sgRNAs that were expressed from a single transcript and could generate individual functional sgRNAs after processing by exogenous ribozymes23, Csy424,25, or the plant endogenous transfer RNA (tRNA)-processing system26. Based on these technologies, the CRISPR/Cas9 toolkits enable genome editing to achieve deletion of chromosomal fragments in a target site with a pair of sgRNAs.
Flowering is an important developmental phase switch in the life cycle of higher plants. In flowering plants, APETALA1 (AP1), SHORT VEGETATIVE PHASE (SVP), and TERMINAL FLOWER1 (TFL1) are essential network genes that control the transition from a vegetative state to a reproductive state and the formation of flowers27,28. AP1 is a MADS-box transcriptional regulator that initiates flower development, which is one of the key regulatory steps of plant flowering activated by flowering locus T27,29. Meanwhile, SVP is a key conserved flowering repressor that determines inflorescence architecture by acting to suppress TFL1 in the emerging floral meristem both redundantly and directly, and causes high levels of inflorescence branching in flowering plants30. TFL1 acts as a repressor of floral initiation, regulates indeterminate conversion to a determinate architecture, controls inflorescence development and maintains the inflorescence meristem in a vegetative state via the repression of AP1 invasion of the central part of the shoot apex31.
In this study, we arrayed sgRNA sequences with a 20 bp Csy4 hairpin sequence and produced multiplexed sgRNAs processed by the RNA endoribonuclease Csy4 from Pseudomonas aeruginosa in combination with CRISPR/Cas9 to knock out transcription factors involved in floral development in Arabidopsis. AP1, SVP, and TFL1 were selected as the target genes to produce targeted deletions of chromosomal fragments between two DSBs separately or simultaneously. We showed that CRISPR/Cas9 using the RNA endoribonuclease Csy4-processing system successfully knocked out the target genes and generated mutant lines rapidly and simply. In addition, the frameshift mutations caused by the targeted deletions could be stably propagated. Our results demonstrate that the CRISPR/Cas9 system with RNA endoribonuclease Csy4 processing is a powerful tool for studying floral development with a variety of genes.
Results
Target site selection for AP1, SVP, and TFL1
To design the protospacer at the target site of AP1, SVP, and TFL1, we determined the genomic sequences of these genes through PCR using gene-specific primer pairs (AP-1/AP-2, SVP-1/SVP-2, and TFL-1/TFL-2 for AP1, SVP, and TFL1, respectively) and analysis of the nucleotide sequences by molecular cloning and DNA sequencing. The corresponding primers are provided in Table S1. The DNA sequences were compared to the corresponding loci in the TAIR database (https://www.arabidopsis.org/) as listed below: AP1 for AT1G69120.1, located on chromosome 1; SVP for AT2G22540, located on chromosome 2; and TFL1 for AT5G03840, located on chromosome 5. Finally, target sites were selected in the first and second exons of the AP1, SVP, and TFL1 genomic sequences (Table 1).
Table 1.
Summary of targeted deletions with the CRISPR/Cas9 system
| # sgRNAs | Sequence 5′–3′ | PAM 5′–3′ | GC content (%) | Target loci | Deletion mutants/# of Hyg+T1 plants |
|---|---|---|---|---|---|
| sgRNA1 | GGGGTAGGGTTCAATTGAAG | AGG | 50 | AP1 exon 1 | 8/23 |
| sgRNA2 | AGATACTTGAACGCTATGAG | AGG | 40 | AP1 exon 2 | |
| sgRNA3 | GACATCGGCGTCGCAGAGAA | CGG | 60 | SVP exon 1 | 5/17 |
| sgRNA4 | ACTGCAAGTTATGCCTCTCT | AGG | 45 | SVP exon 2 | |
| sgRNA5 | CTTCTGTTTCCTCCAAGCCT | AGG | 50 | TFL1 exon 1 | 10/30 |
| sgRNA6 | ATGATAGACCCAGATGTTCC | AGG | 45 | TFL1 exon 2 |
Assembly of CRISPR/Cas9 vectors
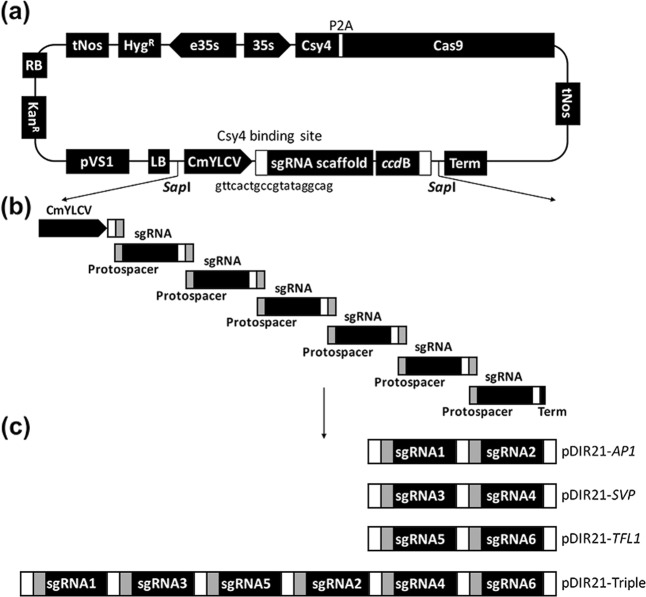
In an attempt to construct genome-editing vectors, we used the backbone of pDIRECT-21 (Fig. 1a), which contained Arabidopsis codon-optimized Cas9 driven by the 35s promoter (35s) and the Cestrum yellow leaf curling virus (CmYLCV) promoter for multiplexed sgRNA transcription. To facilitate positive/negative selection of transgenic plants, a selection marker, hygromycin B phosphotransferase, which confers tolerance to hygromycin, driven by an enhanced 35s promoter was used. The CmYLCV promoter produces a single transcript with sgRNA cassettes separated by the 20-bp Csy4 hairpins for multiplexed sgRNA release24. Fragments containing sgRNAs that targeted AP1, SVP, and TFL1 separately or simultaneously were assembled into pDIRECT-21 by the Gibson assembly method (Fig. 1b and Table S1). The assembled CRISPR/Cas9 vectors with the Csy4-processing system for target gene deletions are shown in Fig. 1c, and the sequences of the multiple sgRNAs are shown in Supplementary Information 1.
Fig. 1. A schematic diagram of the CRISPR/Cas9 system with the multiple-sgRNA expression cassette.
a The structure of the direct cloning binary vector pDIR21 was designed to speed up the cloning process of sgRNA and the transformation elements with CRISPR/Cas9. tNOS, CaMV poly(A) signal; Hyg hygromycin B phosphotransferase, e35s enhanced CaMV 35S promoter, 35s CaMV 35S promoter, Csy4 RNA endoribonuclease Csy4 from Pseudomonas aeruginosa, P2A self-splicing 2A Peptide derived from Porcine teschovirus-1, Cas9 Cas9 endonuclease, CmYLCV Cestrum yellow leaf curling virus promoter, sgRNA single guide RNA targeting specific sequence of genome locus, Csy4-binding site, a 20 bp sequence of Csy4 recognized hairpin structure, ccdB, lethal gene in E. coli. b Illustration of cloning of multiplexed sgRNA cassettes into the CRISPR/Cas9 binary vector by the Gibson assembly method. The 20nt protospacer sequences are overlapping regions for assembly, shown as gray boxes. c Overall structure of the multiplexed sgRNAs in intermediate vectors for targeted deletion of AP1, SVP, and TFL1 in Arabidopsis
Effective multiplexed CRISPR/Cas9-mediated targeted deletion
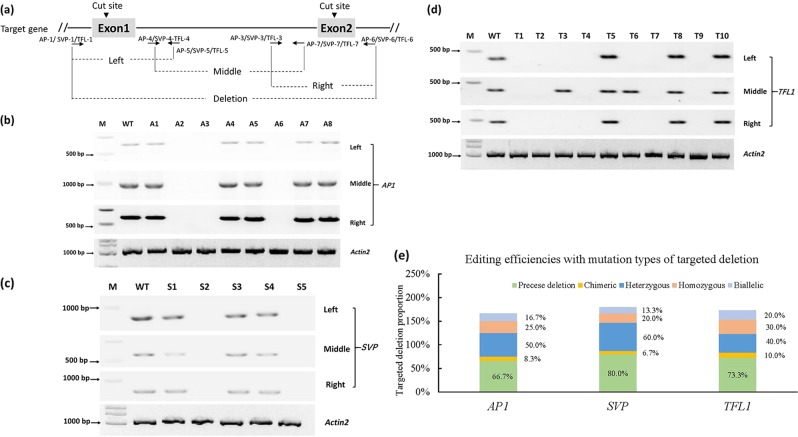
To demonstrate the efficiency of the CRISPR/Cas9-generated deletions, primers were designed upstream and downstream of various target sites, including the loci edited by the vectors pDIR21-AP1, pDIR21-SVP, and pDIR21-TFL1. The amplified fragments covered the targeted deletions in each gene (Fig. 2a). The corresponding primers are provided in Table S1. We detected genome editing in these T1 plants by PCR amplification and sequenced three clones of each PCR product cloned into a T-vector. In the AP1 lines, we obtained eight T1 lines that had the targeted deletion from 23 hygromycin-tolerant plants using a pair of AP-1/AP-3 primers (Fig. 2b). The analysis showed four types of genomic deletions at the target sites between gRNA1 and gRNA2 by DNA sequence analysis (Figs. 2c and S1). In SVP lines, five T1 lines that had the targeted deletion were obtained from 17 hygromycin-tolerant plants (Fig. 2b), and the analysis showed three types of genomic deletions at the target sites between gRNA3 and gRNA4 by DNA sequence analysis (Figs. 2c and S2). In TFL1 lines, 10 T1 lines that had the targeted deletion were obtained from 30 hygromycin-tolerant plants (Fig. 2b), and the analysis showed four types of genomic deletions at the target sites between gRNA5 and gRNA6 according to DNA sequence analysis (Figs. 2c and S3).
Fig. 2. PCR detection revealed targeted deletions of AP1, SVP, and TFL1 in T1 plants.
a Map of three floral genes targeted for deletion using a pair of sgRNAs each. sgRNA sites are shown as black lines. Arrowheads represent the primers used to detect the deletions. b Lengths of PCR products from wild-type loci and the targeted deletions of AP1, SVP, and TFL1. c DNA sequences of representative deletion products. The sequence of the unmodified locus for each of the three genes is shown on the top. sgRNA target sites are underlined, and PAM sequences are in bold. Cleavage sites are shown as black arrowheads. Deleted sequences are shown with dots
We next tested the CRISPR/Cas9 genome-targeted deletion approach with the six multiplexed sgRNAs for AP1, SVP, and TFL1 in pDIR21-Triple. From 48 hygromycin-tolerant plants, we obtained two lines with the three gene-targeted deletions; eight additional mutant lines were double mutants, and 15 had mutations in a single gene (Figs. S4–6). A summary of the targeted deletion efficiency by these four vectors in Arabidopsis T1 plants is shown in Table 1.
Mutagenesis of target sites with multiplexed sgRNAs
We explored the effectiveness of the Csy4-processing system in mutagenesis of a single targeted site of AP1, SVP, and TFL1 from pDIR21-Triple in 48 lines. We detected these T1 plants by PCR with the primers (Table S1) used to amplify the genomic regions containing the corresponding target sites. The PCR products from the six sites of the three genes were cloned and sequenced to investigate the mutation types. Overall, 109 valid sequences were obtained. A summary of the results is shown in Table 2.
Table 2.
Targeted mutagenesis of a single target site with CRISPR/Cas9 using multiplexed sgRNAs
| Target gene | Indel at sgRNA site #1 only | Indel at sgRNA site #2 only | Indel at both sgRNA sites | Inversion between sgRNA sites | Deletion between sgRNA sites | Total mutant plants |
|---|---|---|---|---|---|---|
| AP1 | 3 | 2 | 5 | 1 | 13 | 24 |
| SVP | 0 | 1 | 1 | 0 | 9 | 11 |
| TFL1 | 2 | 1 | 15 | 2 | 15 | 35 |
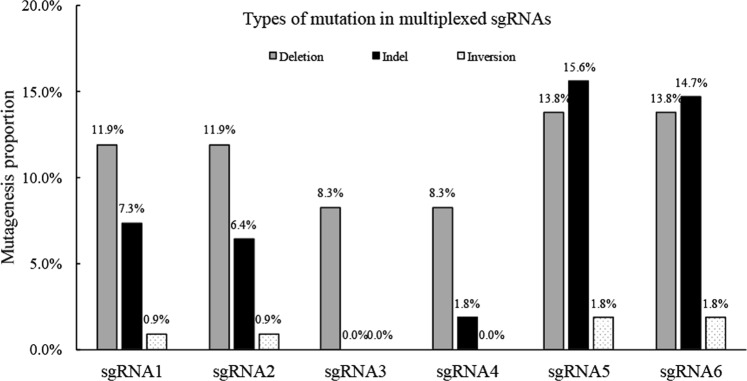
There are three types of mutagenesis: indel mutations, including insertion or deletion of nucleotides, deletions of a fragment between two sites and inversions of a fragment between two sites. The proportion of each type of mutation at the six target sites was calculated among the valid sequences, including single-target-site edits and deletions (Figs. 3 and S7–11). Thus, the results showed that the CRISPR/Cas9-processing system using the RNA endoribonuclease Csy4 provides efficient multiplexed, targeted gene deletion in Arabidopsis with multiplexed sgRNAs.
Fig. 3. Genome-editing evaluation of six sgRNAs in the pDIR21-Triple vector.
The editing efficiencies of different mutation types were determined by DNA sequencing in T1 plants
Identification of homozygous targeted deletions in mutant lines
To further confirm the homozygous mutant status of the targeted genes in the selected plants, we investigated the mutations at each disrupted gene locus by using three pairs of primers covering the two cut sites and the regions between them (Fig. 4a). The corresponding primers are provided in Table S1. In the PCR detection results, the absence of all three fragments was indicative of homozygous deletions in AP1 (Fig. 4b), SVP (Fig. 4c), and TFL1 (Fig. 4d). The middle fragment for the T3 and T6 lines of the TFL1 targeted deletion was amplified by the PCR primer pair TFL-4/TFL-7, showing that chromosomal translocations occurred between the two cut sites.
Fig. 4. Verification of homozygous deletion mutant lines.
a Primer sets were used to amplify the editing regions between two DSBs in AP1, SVP, and TFL1 separately. b–d Agarose gel electrophoresis of the fragments of targeted genes. The absence of all three fragments indicated plants with homozygous deletions. WT wild type. e Efficiencies of targeted deletion with different mutation types in T1 plants
We analyzed the targeted deletion edit types in Arabidopsis T1 plants derived from the pDIR21-AP1, pDIR21-SVP, and pDIR21-TFL1 constructs by DNA sequencing of three clones of each PCR fragment (Fig. 2b). Of the 207 obtained sequences, 16/24 (66.7%) of AP1 mutation lines, 12/15 (80%) of SVP mutation lines, and 22/30 (73.3%) of TFL1 mutation lines showed precise deletions in each gene. Meanwhile, each targeted gene contained heterozygous, homozygous, chimeric and biallelic mutations at each site (Fig. 4e and Figs. S12,13). Finally, we identified precise, targeted, and homozygous deletions in A2 of AP1, S2 of SVP and T1 of TFL1 lines.
Targeted deletion for loss-of-function mutagenesis
To understand the impact of targeted deletion of the DNA fragments between exon 1 and exon 2 of AP1, SVP, and TFL1 by CRISPR/Cas9 using two sgRNAs at the mRNA level in the homozygous mutation lines A2, S2, and T1, we screened the lines directly. We amplified the cDNA of the three genes by RT-PCR using oligo-d(T)18, which primed reverse transcription of mRNA from mutant T1 plants, and specific primer pairs binding the 5′ and 3′ UTR sequences of these genes were used separately. The corresponding primers are provided in Table S1. The results showed that the cDNA fragments of AP1 and TFL1 were amplified, but amplification failed for SVP (Fig. 5a). In the Sanger sequencing results, we identified the transcripts that corresponded to the known transcripts with partial sequences of exon 1 and exon 2 of AP1 and TFL1 separately (Figs. 5b and S14). Moreover, the stop codon TGA was introduced in the open-reading frame (ORF) of the truncated exon 2 of AP1, and TAG was introduced in the ORF of the truncated exon 2 of TFL1 (Figs. 5b and S15). This result showed that loss of function in AP1, SVP, and TFL1 induced by frameshift mutations using the CRISPR/Cas9 system using two sgRNAs was successful in T1 plants.
Fig. 5. Targeted deletion led to frameshift mutations in AP1 and TFL1.

a Agarose gel electrophoresis of the targeted deletion gene mRNA transcripts from T1 events by RT-PCR. We used Actin as an internal control. b Schematic representation of the transcripts produced by AP1 and TFL1 deletion
Floral characterization of mutant plants
To determine the functions of AP1, SVP, and TFL1 in floral development, we analyzed the phenotype of the wild-type, AP1-A2, SVP-S2, TFL1-T1, and triple mutant Tripled-26 plants.
AP1 encodes a MADS-domain transcription factor and activates floral organ identity genes to promote floral meristem formation32. Targeted deletion of AP1 resulted in floral meristem development abnormally with an increased number of petals (Fig. 6d) and three degenerated carpels united with sepals and petals (Fig. 6e). It also resulted in a significant increase in plant branching, usually with an additional secondary branch that maintained the indeterminate inflorescence phenotype in the mutant line (Fig. 6f). SVP encodes a MADS-domain transcription factor and is a flowering time regulator. However, it acts as a repressor of flowering33. The flowering of SVP mutants showed early floral meristem formation during the vegetative phase, the inflorescence and flower morphologies were aborted and failed to form pistils and stamens (Fig. 6g), and the flower buds were abnormal (Fig. 6h). There was usually an additional fourth branch growing from the secondary branch (Fig. 6i). TFL1 is a negative regulator controlling flowering time and inflorescence architecture34. TFL1 mutants showed a partial transformation of the stem meristem into a floral meristem (Fig. 6j), increased the petal number (Fig. 6k) and displayed a change from indeterminate to determinate inflorescence type (Fig. 6k). This result indicates that TFL1 can not only maintain unlimited inflorescence growth in plants with indeterminate inflorescences but also affect floral development in Arabidopsis.
Fig. 6. Partial floral development phenotypes of targeted deletion plants.
a–c Wild-type Arabidopsis. d–f AP1 mutants with strong branching characteristics and doubled or tripled carpels fused together. g–i SVP mutants with increased branching and obvious fourth-order branching showed decreased fruit number and floral abortion. j–l TFL1 mutants with dwarf determinate inflorescences, doubled carpels and increased petal numbers. m–o The floral development of the triple-gene mutant with aborted determinate inflorescences. All plants were grown for 40 days
In addition, the absence of AP1, SVP, and TFL1 in triple mutant plants caused the continuous production of inflorescence meristem in place of flowers but with the cauliflower phenotype (Fig. 6m). Moreover, some floral meristems were eventually formed and ultimately withered with the entire branch (Fig. 6n). This phenotype is very similar to that of the SVP mutant. However, the determinate inflorescence that was formed in the triple mutant was similar to that of the TFL1 mutant (Fig. 6o). Even if new shoots continued to be produced at the base to maintain the vegetative growth stage, the growth order of Arabidopsis thaliana would eventually be broken, resulting in wilting and abortion due to the disorder of the flowering gene network. This result suggests that AP1, SVP, and TFL1 together play a role in the regulation of flower formation.
Discussion
The CRISPR/Cas9 system has great potential for promoting functional studies in plants, as it can be easily adapted to generate loss-of-function mutations for single genes or multiple-gene clusters of unknown function and is becoming one of the most powerful tools for creating functional gene knockouts2,19,35–37. However, a major limitation for the use of CRISPR/Cas9 in functional genomic studies in plants is the difficulty of rapidly generating and detecting stable homozygous mutations with high efficiency, as well as the inability to simultaneously mutate multiple target genes23. Cas9 most frequently creates a blunt-ended DSB at a position three base pairs upstream of the PAM sequence7. Strikingly, we observed a very high frequency of precise ligation in our experiment: on average, 73% of all reads with long deletions were targeted deletions of 1528 bp in AP1, 995 bp in SVP, or 278 bp in TFL1. These results suggest that if a single sgRNA was used, most instances of NHEJ repair would not result in mutation. Therefore, creating deletions with two or more sgRNAs should be a more efficient approach to achieve targeted mutagenesis. Our work demonstrates a suitable approach for targeted loss-of-function deletions by CRISPR/Cas9 in plants.
Because the DSBs generated by two sgRNAs are likely to be accurately repaired, we favor the use of two sgRNAs to induce loss-of-function mutations. Selection of two sgRNAs can be used advantageously to make specific mutations. There are no concerns about creating functional gene isoforms through non-frameshift indels or alternative splicing19, and the targeted deletions can be detected by PCR, making screening for mutants significantly easier than when using one sgRNA15. In our study, the targeted deletion lines generated by two sgRNAs targeting each gene were detected by PCR analysis. The targeted deletion frequency by the pair of sgRNAs used to create the mutant lines was 30%. In our study, the target sites of AP1 and TFL1 showed higher editing efficiencies than the target sites of SVP. In contrast to the targeted deletions, gRNA5 and gRNA6 showed very high rates of single-target DSBs in most transgenic-positive lines. Additionally, we found that the GC contents of these sgRNAs ranged from 40% to 60%, which is appropriate for sgRNAs (Table 1). The editing efficiency is limited by the efficiency of the selected target locus, guide RNA structure, sufficient expression of Cas9, and multigenerational analysis of transgenic lines for more multiplexed sgRNA-based editing in plants.
There are different strategies for multiple guide RNA generation. Such polycistronic mRNAs are processed posttranscriptionally into individual sgRNAs by RNA-cleaving enzymes38. These enzymes include the CRISPR-associated RNA endoribonuclease Csy4 from P. aeruginosa, the tRNA processing enzymes naturally present in the host cells and ribozymes23,26. A significant challenge in plant genome engineering is achieving high-frequency genome editing by multiple sgRNAs derived from a polycistronic transcript. Our pDIR21-Triple construct contained six sgRNAs to target the deletion of three genes at the same time with a 4% frequency; it also generated single-gene and double-gene deletions with these multiplexed sgRNAs. We favored the Csy4 system because it consistently gave us higher deletion frequencies in Arabidopsis, and the multiple sgRNA transcripts are shorter and less repetitive than those used in the tRNA processing system26. Moreover, the polycistronic transcript sgRNAs do not have the limitation of A or G initial nucleotides in sgRNA for the Pol III promoter. There is a report that the CRISPR/Cas9-linked viral silencing suppressor p19 can generate 70% editing efficiency in Arabidopsis, but p19 induced a strong leaf developmental phenotype39. However, the Csy4 ribonuclease has no observable phenotypic consequences for transgenic plants24.
Due to the sequence alterations in the coding region of the target gene induced by the cellular DNA DSB repair mechanism, causing frameshift mutations, the CRISPR/Cas9 system is widely used as an efficient tool for specific gene knockout production. In a recent study, it was shown that indels introduced by CRISPR/Cas9 using one sgRNA can lead to random splicing as opposed to mRNA degradation or protein truncation40,41. In our demonstrated method, the cDNA sequence confirmed loss of function in AP1 and TFL1 because a stop codon was introduced by a frameshift mutation generated from the targeted deletion of exon 1 and exon 2. Furthermore, the cDNA of SVP in the mutant line was not acquired. This result indicated that the targeted deletion between exon 1 and exon 2 of SVP significantly decreased mRNA transcription, and thus, this sequence may include promoter fragments and cis-regulatory motifs that change the expression patterns of SVP. This result revealed novel insights into the functional mechanisms of divergent flowering-related gene regulation in plants42. The exon splicing mechanism reveals the existence of complex gene repair and expression43,44. Moreover, there is a report that fertility seemed normal in an SVP mutant created by transposon-tagging mutagenesis45; however, for SVP mutants with either single-gene mutation or triple-gene mutation, deletion of the chromosomal fragment of SVP exon 1 and exon 2 by CRISPR/Cas9 using gRNA3/gRNA4 of pDIR21-SVP or pDIR21-Triple directly affected the fruiting of the resultant T1 plants. Detailed studies on the differences in these parameters will require further experimentation to fully test.
Materials and methods
Plant material
Arabidopsis thaliana Col-0 plants were grown in a growth chamber that was set at 16/8 h (light/dark) and 22 °C. T1 seeds derived from plants transformed with binary vectors were selected on B5 media lacking sucrose and containing 30 mg/L hygromycin B.
Vector construction
The plasmid pDIRECT-21 purchased from ATCG (http://www.addgene.org/91130) was used as a backbone for the construction of the Cas9-sgRNA-expressing vector. Briefly, the Cas9 gene sequence, including the nuclear localization signals, was designed with codon optimization for dicot plants, and the Csy4 RNA nuclease was fused into the N terminus of Cas9 splitting by P2A. The 35s promoter was used to express the Csy4-Cas9 module, and the CmYLCV promoter drove expression of the sgRNA processed by Csy4 to release the multiplexed sgRNAs. Efficient protospacer sequences targeting the coding exons of AP1, SVP, and TFL1 were selected using the Zhang Laboratory CRISPR Design website (http://crispr.mit.edu). Oligonucleotides corresponding to the top and bottom strands of the sgRNAs were synthesized, annealed, and cloned into pDIRECT-21 as previously described24 to generate four different binary vectors encoding the Cas9 nuclease and sgRNAs. These vectors were used for Agrobacterium-mediated Arabidopsis transformation after DNA sequencing confirmation.
The genome-editing vector set uses Gibson assembly cloning to create vectors for a diverse range of genome-editing applications. There are three sets of modular cloning fragments. Fragment A contains the CmYLCV promoter and was amplified by the primers CmYLCV-F/CmYLCV-RA. Fragment B contains multiple sgRNAs targeting AP1 sites and was produced by annealing the oligos gRNA1-F/gRNA1-R or gRNA2-F/gRNA-R separately. Fragment C contains the backbone of pDIRECT-21 digested by SapI restriction enzyme. These fragments were assembled using Gibson assembly cloning kits (NEB, USA) to construct pDIR21-AP1. Using the same method, we acquired pDIR21-SVP by CmYLCV-F/ CmYLCV-RS gRNA3-F/gRNA3-R and gRNA4-F/gRNA-R and pDIR21-TFL1 by CmYLCV-F/ CmYLCV-RT, gRNA5-F/gRNA5-R, and gRNA6-F/gRNA-R separately. For pDIR21-Triple, six sgRNAs were assembled by gRNA1-F/gRNA-triple-R1, gRNA3-F/gRNA-triple-R2, gRNA5-F/gRNA-triple-R3, gRNA2-F/gRNA-triple-R4, gRNA4-F/gRNA-triple-R5, and gRNA6-F/gRNA-R separately.
Agrobacterium-mediated Arabidopsis transformation
Genetic transformation of Arabidopsis was conducted using the common floral dip method46. Wild-type (WT) Col-0 plants with flower buds were infected with activated Agrobacterium tumefaciens EHA105. The seeds from the infected plants were collected, dried, surface-disinfected with 70% ethanol for 10 min, and rinsed with 90% ethanol once and autoclaved ddH2O three times. The seeds were then cultured on agar-solidified half-strength Murashige and Skoog medium47 containing 30 mg/L hygromycin. Seed vernalization was performed at 4 °C for 3 days in the dark. Germination was performed under a photoperiod of 16/8 h (light/dark) at 22 °C for 1 day, followed by 3 days in the dark and then 3 days with a photoperiod of 16/8 (light/dark) at 22 °C. Hygromycin B-resistant seedlings screened from the dishes were transplanted onto soil for continuous growth to allow molecular analysis, phenotype observation, and seed harvesting.
Genomic DNA extraction and PCR analysis
Genomic DNA was extracted from Arabidopsis leaves using the CTAB method following the published protocol48. To detect mutagenesis at the desired sites, the target regions were amplified with specific primers (see supplementary information for primer sequences) using Premix Taq DNA Polymerase (Takara, Japan) with the following protocol: 94 °C for 10 min (94 °C for 30 s, 60–48 °C for 30 s, 72 °C for 2 min) for 13 cycles with touchdown −1 °C in each cycle (94 °C for 30 s, 52 °C for 30 s, 72 °C for 2 min) for 25 cycles, 72 °C for 10 min, 4 °C to hold. The PCR product was separated in a 1% agarose gel and stained with GelStain (TransGen Biotech, China) to detect chromosomal fragment deletions. Selected PCR products were cloned into the pGEM-T Easy Vector (Promega, USA) for Sanger DNA sequencing.
RNA extraction and RT-PCR
Total RNA was extracted using the NucleoSpin RNA Plant Kit (Takara), treated with DNase before use as the template for RT-PCR, analyzed in a 1.2% agarose gel and stained with GelStain (TransGen Biotech, China) to assess the extracted total RNA concentration and integrity. Additionally, no degradation was found in the RNA extracts, as 18S:28S was equal to 1:2 for all samples. cDNA was then generated by reverse transcription from 1 µg of total RNA using 25 U of AMV reverse transcriptase, 100 mM dNTPs, 25 U of RNase inhibitor and 100 µm Oligo-d(T)18 Primers (AMV Reverse Transcriptase Kit, Promega, USA) in a 25 µl reaction volume. The reverse transcription reaction was carried out in three steps: 60 min at 42 °C, 30 min at 55 °C and 15 min at 70 °C. PCRs were performed with 1 µl of cDNA using Premix Taq DNA Polymerase (Takara, Japan). The PCR products were separated in a 1% agarose gel and stained with GelStain (TransGen Biotech, China) to detect the cDNA of the target gene. Selected PCR products were cloned into pGEM-T Easy Vector (Promega, USA) for DNA sequencing.
Gene accession numbers in GenBank
The genes and their GenBank RefSeq accession numbers are as follows: AP1, NC_003070.9; SVP, NC_003071.7; TFL1, NC_003076.8.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31770736).
Author contributions
Y.K.G. and Y.Z.L. designed and conceived the experiment. Y.Z.L. performed vector construction, transformation, and molecular characterization of mutants. Y.Z.L., Y.K.G., and Y.H.G. analyzed the data. Y.Z.L., Y.K.G., Y.H.G., and Q.X.Z. wrote the paper. All authors have read and approved the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-019-0179-6).
References
- 1.Liu D, Mewalal R, Hu R, Tuskan GA, Yang X. New technologies accelerate the exploration of non-coding RNAs in horticultural plants. Hortic. Res. 2017;4:17031–17031. doi: 10.1038/hortres.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meng X, et al. Construction of a Genome-Wide Mutant Library in Rice using CRISPR/Cas9. Mol. Plant. 2017;10:1238–1241. doi: 10.1016/j.molp.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Samanta MK, Dey A, Gayen S. CRISPR/Cas9: an advanced tool for editing plant genomes. J. Transgenic Res. 2016;25:561–573. doi: 10.1007/s11248-016-9953-5. [DOI] [PubMed] [Google Scholar]
- 4.Voytas DF, Gao C. Precision genome engineering and agriculture: opportunities and regulatory challenges. PLOS Biol. 2014;12:e1001877. doi: 10.1371/journal.pbio.1001877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin K, Gao C, Qiu J-L. Progress and prospects in plant genome editing. Nat. Plants. 2017;3:17107. doi: 10.1038/nplants.2017.107. [DOI] [PubMed] [Google Scholar]
- 6.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jinek M, et al. A programmable Dual-RNA-guided DNA. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durr J, Papareddy R, Nakajima K, Gutierrez-Marcos J. Highly efficient heritable targeted deletions of gene clusters and non-coding regulatory regions in Arabidopsis using CRISPR/Cas9. Sci. Rep. 2018;8:4443. doi: 10.1038/s41598-018-22667-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Z, et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl Acad. Sci. USA. 2014;111:4632–4637. doi: 10.1073/pnas.1400822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao X, Chen J, Dai X, Zhang D, Zhao Y. An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol. 2016;171:1794–1800. doi: 10.1104/pp.16.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma X, Zhu Q, Chen Y, Liu Y-G. CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Mol. Plant. 2016;9:961–974. doi: 10.1016/j.molp.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Osakabe, Y. & Osakabe, K. in Progress in Molecular Biology and Translational Science, Vol. 149 (eds Weeks, Donald P. & Yang, Bing) 99–109 (Academic Press, 2017).
- 14.Chen L, et al. A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hortic. Res. 2018;5:13–13. doi: 10.1038/s41438-018-0023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J-F, et al. Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013;31:688. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma X, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Mao Y, et al. Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol. J. 2016;14:519–532. doi: 10.1111/pbi.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao J, et al. CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. J. Plant Mol. Biol. 2015;87:99–110. doi: 10.1007/s11103-014-0263-0. [DOI] [PubMed] [Google Scholar]
- 19.Lalonde S, et al. Frameshift indels introduced by genome editing can lead to in-frame exon skipping. PLoS ONE. 2017;12:e0178700. doi: 10.1371/journal.pone.0178700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishitani C, et al. Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 2016;6:31481. doi: 10.1038/srep31481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum. Mol. Genet. 2014;23:R40–R46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- 22.Yu Zhiming, Chen Qiyuan, Chen Weiwei, Zhang Xian, Mei Fengling, Zhang Pengcheng, Zhao Mei, Wang Xiaohong, Shi Nongnong, Jackson Stephen, Hong Yiguo. Multigene editing via CRISPR/Cas9 guided by a single-sgRNA seed in Arabidopsis. Journal of Integrative Plant Biology. 2018;60(5):376–381. doi: 10.1111/jipb.12622. [DOI] [PubMed] [Google Scholar]
- 23.Gao Y, Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 2014;56:343–349. doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- 24.Čermák T, et al. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell. 2017;29:1196–1217. doi: 10.1105/tpc.16.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014;32:569. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl Acad. Sci. USA. 2015;112:3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blázquez MA. The right time and place for making flowers. Science. 2005;309:1024–1025. doi: 10.1126/science.1117203. [DOI] [PubMed] [Google Scholar]
- 28.Jaeger KE, Pullen N, Lamzin S, Morris RJ, Wigge PA. Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. J. Plant Cell. 2013;25:820–833. doi: 10.1105/tpc.113.109355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, et al. MIKC(C)-type MADS-box genes in Rosa chinensis: the remarkable expansion of ABCDE model genes and their roles in floral organogenesis. Hortic. Res. 2018;5:25–25. doi: 10.1038/s41438-018-0031-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C, et al. A conserved genetic pathway determines inflorescence architecture in Arabidopsis and Rice. Dev. Cell. 2013;24:612–622. doi: 10.1016/j.devcel.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Baumann K, et al. Changing the spatial pattern of TFL1 expression reveals its key role in the shoot meristem in controlling Arabidopsis flowering architecture. J. Exp. Bot. 2015;66:4769–4780. doi: 10.1093/jxb/erv247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kempin S, Savidge B, Yanofsky M. Molecular basis of the cauliflower phenotype in Arabidopsis. Science. 1995;267:522–525. doi: 10.1126/science.7824951. [DOI] [PubMed] [Google Scholar]
- 33.Gregis V, et al. Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol. 2013;14:R56. doi: 10.1186/gb-2013-14-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanano S, Goto K. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. J. Plant Cell. 2011;23:3172–3184. doi: 10.1105/tpc.111.088641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal A, et al. Insights into maize genome editing via CRISPR/Cas9. Physiol. Mol. Biol. Plants. 2018;24:175–183. doi: 10.1007/s12298-017-0502-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demirci Y, Zhang B, Unver T. CRISPR/Cas9: an RNA-guided highly precise synthetic tool for plant genome editing. J. Cell Physiol. 2018;233:1844–1859. doi: 10.1002/jcp.25970. [DOI] [PubMed] [Google Scholar]
- 37.Hussain B, Lucas SJ, Budak H. CRISPR/Cas9 in plants: at play in the genome and at work for crop improvement. Brief. Funct. Genom. 2018;17:319–328. doi: 10.1093/bfgp/ely016. [DOI] [PubMed] [Google Scholar]
- 38.He, Y., Wang, R., Dai, X. & Zhao, Y. in Progress in Molecular Biology and Translational Science Vol. 149 (eds Weeks, Donald P. & Yang, Bing) 151-166 (Academic Press, 2017).
- 39.Mao Y, et al. Manipulating plant RNA-silencing pathways to improve the gene editing efficiency of CRISPR/Cas9 systems. J. Genome Biol. 2018;19:149. doi: 10.1186/s13059-018-1529-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen D, et al. CRISPR/Cas9-mediated genome editing induces exon skipping by complete or stochastic altering splicing in the migratory locust. BMC Biotechnol. 2018;18:60. doi: 10.1186/s12896-018-0465-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapahnke M, Banning A, Tikkanen R. Random splicing of several exons caused by a single base change in the target exon of CRISPR/Cas9 mediated gene knockout. Cells. 2016;5:45. doi: 10.3390/cells5040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhadi SR, Xu Z, Shaik R, Driscoll K, Ramakrishna W. Differential regulation of genes by retrotransposons in rice promoters. J. Plant Mol. Biol. 2015;87:603–613. doi: 10.1007/s11103-015-0300-7. [DOI] [PubMed] [Google Scholar]
- 43.Mao Y, et al. Functional analysis of alternative splicing of the FLOWERING LOCUS T orthologous gene in Chrysanthemum morifolium. Hortic. Res. 2016;3:16058–16058. doi: 10.1038/hortres.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue Chenxiao, Zhang Huawei, Lin Qiupeng, Fan Rong, Gao Caixia. Manipulating mRNA splicing by base editing in plants. Science China Life Sciences. 2018;61(11):1293–1300. doi: 10.1007/s11427-018-9392-7. [DOI] [PubMed] [Google Scholar]
- 45.Gregis V, Sessa A, Colombo L, Kater MM. AGAMOUS-LIKE24 and SHORT VEGETATIVE PHASE determine floral meristem identity in Arabidopsis. Plant J. 2008;56:891–902. doi: 10.1111/j.1365-313X.2008.03648.x. [DOI] [PubMed] [Google Scholar]
- 46.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 47.Murashige T, Skoog R. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 48.Healey A, Furtado A, Cooper T, Henry RJ. Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods. 2014;10:21–21. doi: 10.1186/1746-4811-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.