Abstract
Heat stress is a serious and widespread threat to the quality and yield of many crop species, including grape (Vitis vinifera L.), which is cultivated worldwide. Here, we conducted phosphoproteomic and acetylproteomic analyses of leaves of grape plants cultivated under four distinct temperature regimes. The phosphorylation or acetylation of a total of 1011 phosphoproteins with 1828 phosphosites and 96 acetyl proteins with 148 acetyl sites changed when plants were grown at 35 °C, 40 °C, and 45 °C in comparison with the proteome profiles of plants grown at 25 °C. The greatest number of changes was observed at the relatively high temperatures. Functional classification and enrichment analysis indicated that phosphorylation, rather than acetylation, of serine/arginine-rich splicing factors was involved in the response to high temperatures. This finding is congruent with previous observations by which alternative splicing events occurred more frequently in grapevine under high temperature. Changes in acetylation patterns were more common than changes in phosphorylation patterns in photosynthesis-related proteins at high temperatures, while heat-shock proteins were associated more with modifications involving phosphorylation than with those involving acetylation. Nineteen proteins were identified with changes associated with both phosphorylation and acetylation, which is consistent with crosstalk between these posttranslational modification types.
Subject terms: Abiotic, Protein sequencing
Introduction
Heat stress is a major factor in plant growth and productivity, and high temperature conditions are predicted to become more frequent worldwide1. Plants have evolved complex mechanisms to survive heat stress, including physiological, biochemical, cellular, and molecular processes2,3. Many studies have focused on better understanding the effects of heat stress on plants via transcriptomic and proteomic analyses4–6, and numerous genes and proteins associated with stress responses have been identified. In addition, a range of posttranslational modifications (PTMs) have been linked to plant stress responses7–9. Of these modification, perhaps the most studied is protein phosphorylation, which influences metabolism, transcription, translation, proteolysis, homeostasis, and signaling10, as exemplified by phosphoproteomic studies in plant species, such as Arabidopsis thaliana, maize (Zea mays), rice (Oryza sativa), soybean (Glycine max), and wheat (Triticum aestivum)11–18. Lys acetylation is another dynamic and reversible PTM that occurs on either the α-amino group at the N-terminus or the ε-amino group on the side chain of Lys residues19. Previous studies have indicated that protein acetylation regulates a wide range of cellular processes and phenomena, including transcription20, enzymatic activity21, protein interactions22, and protein stability23, as well as metabolic pathways24. Many Lys-acetylated proteins have been identified in both microbes and mammals, revealing roles in many cellular functions; however, the plant acetylproteome has been studied relatively little25.
Grape (Vitis vinifera L.) is an economically important crop species worldwide; however, its quality and yield are often constrained by heat stress26,27. Several studies have related changes in grape PTMs to stress responses. One example described a global comparative proteomic analysis of steady-state protein expression, as well as changes in phosphorylation and Lys acetylation of proteins from the mesocarp and exocarp of grape in response to infection by Lobesia botrana7. This analysis revealed 3059 proteins, 1135 phosphosites, and 138 Lys acetyl sites. In another study, hundreds of phosphoproteins were quantified in Cabernet Sauvignon grape leaves after abscisic acid (ABA) treatment28, and quantitative changes in the proteome and phosphoproteome of Barbera grapevine leaves after phytoplasma infection and recovery have been described. However, to our knowledge, the phosphoproteome and acetylproteome of grape plants subjected to abiotic stresses have not yet been described.
In a previous study, we analyzed transcriptomic and proteomic changes in leaves of grape plants grown under four different temperature regimes (25 °C, 35 °C, 40 °C, and 45 °C); uncovered cellular responses to sudden temperature changes; and characterized differentially expressed proteins, genes, and pathways6. Based on these results, in this study, we conducted phosphoproteomic and acetylproteomic analyses of the same material to reveal heat response PTMs in grape leaves.
Results
Protein phosphorylation and acetylation in grape leaves in response to high temperature
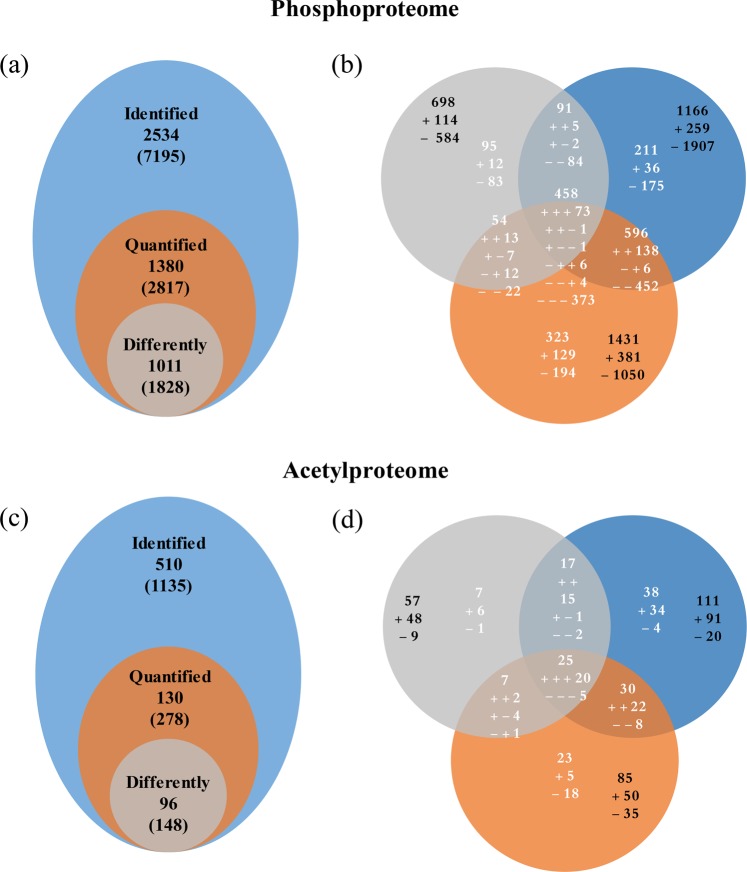
To gain insight into changes in protein phosphorylation and acetylation patterns under high temperature conditions, leaves from grape plants grown under four different temperature regimes (25 °C, 35 °C, 40 °C, and 45 °C) were collected, as we previously described6. The workflow is shown in Fig. S1. Peptides containing phosphorylation and Lys acetylation were quantified: a total of 2534 proteins with 7195 phosphosites were identified, and 1380 proteins with 2817 phosphosites were quantified. A total of 510 proteins with 1135 acetyl sites were ultimately identified, while 130 proteins with 278 acetyl sites were ultimately quantified (Fig. 1a, c).
Fig. 1. Overview of phosphoproteome and acetylproteome.
The number of identified and quantified phosphoproteins and phosphosites (a) and acetyl proteins and acetyl sites (c) in grape leaves following exposure to temperatures of 35 °C, 40 °C, and 45 °C as well as differently changed protein phosphosites (b) and acetyl sites (d) at 35 °C, 40 °C, and 45 °C. The numbers outside of parentheses indicate phosphoproteins or acetyl proteins, and the numbers inside parenthesis indicate phosphosites or acetyl sites. The “ + ” and “−” indicate upregulated and downregulated sites, respectively, and the black numbers indicate the total number of upregulated or downregulated sites at 35 °C, 40 °C, and 45 °C
Differentially changed phosphosites (DCPSs) were then determined according to the following criteria: (1) phosphosites detected at all four temperatures; (2) the quantification ratio (35 °C/25 °C, 40 °C/25 °C or 45 °C/25 °C) > 1.3 or < 0.77; and (3) the phosphosite localization probability > 0.75. As a result, 1011 differentially changed proteins with 1828 DCPSs were identified (Fig. 1a). Among these DCPSs, 114, 259, and 381 phosphosites were found to be upregulated at 35 °C, 40 °C, and 45 °C, respectively, and 73 were coupregulated. There were five common upregulated phosphosites in the 35 °C and 40 °C samples, 13 in the 35 °C and 45 °C samples, and 138 in the 40 °C and 45 °C samples. At 35 °C, 40 °C, and 45 °C, 584, 1907, and 1050 phosphosites were downregulated, respectively, and 373 were codownregulated. In addition, there were 84 commonly downregulated phosphosites in the 35 °C and 40 °C samples, 22 in the 35 °C and 45 °C samples, and 452 in the 40 °C and 45 °C samples (Fig. 1b). We identified 637 proteins with 1 phosphosite, 199 proteins with 2 phosphosites, 74 proteins with 3 phosphosites, and 53 proteins with 4 phosphosites (Fig. 2a). The dominant phosphorylated amino acid was serine (~89%), followed by threonine (~10%). Only ~0.9% of the sites detected involved tyrosine (Fig. 2b). Moreover, we found that upregulated phosphosites involving serine (pSs) increased, while downregulated pSs decreased between the 35 °C and 45 °C samples; however, upregulated phosphosites involving threonine (pTs) decreased, and downregulated pTs increased between the 35 °C and 45 °C samples (Fig. 2c).
Fig. 2. Summary of the phosphosites and acetyl sites in phosphoproteome and acetylproteome.
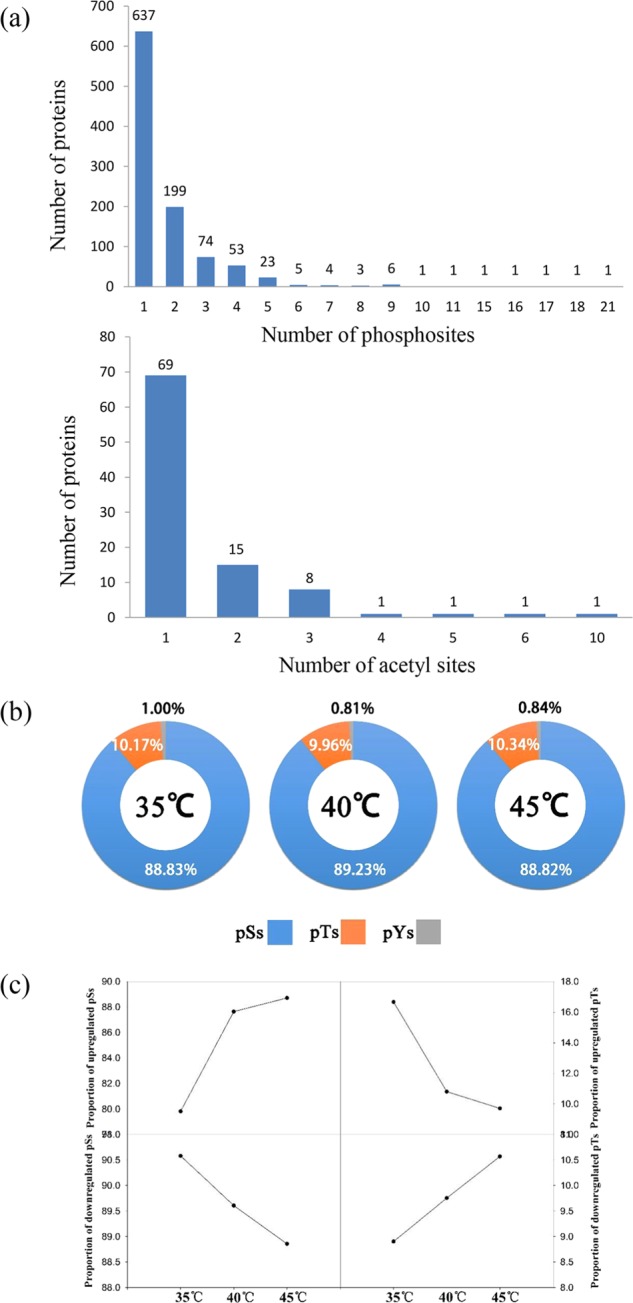
The number of phosphoproteins according to the number of identified phosphosites and acetyl sites (a). Distribution of differently changed phosphosites at 35 °C, 40 °C, and 45 °C in grape leaves (b, c). pS phosphorylated serine, pT phosphorylated threonine, pY phosphorylated tyrosine
Using similar criteria to the above to identify differentially changed acetyl sites (DCASs), we identified a total of 96 proteins with 148 DCASs (Fig. 1c). In these DCASs, 48, 91, and 50 were upregulated at 35 °C, 40 °C, and 45 °C, respectively, compared with the control, while 20 were upregulated at all three temperatures. There were 15 commonly upregulated DCASs in the 35 °C and 40 °C samples, two in the 35 °C and 45 °C samples, and 22 in the 40 °C and 45 °C samples. In the 35 °C, 40 °C, and 45 °C treatments, 9, 20, and 35 DCASs were downregulated, respectively, and five were downregulated in all three samples. In addition, there were two downregulated DCASs in the 35 °C and 40 °C samples and eight downregulated DCASs in the 40 °C and 45 °C samples (Fig. 1d). Acetylation of these proteins was modified at Lys sites. There were 69 acetyl proteins with one acetyl site, 15 proteins with two acetyl sites, and 8 proteins with three acetyl sites (Fig. 2a).
Sequence properties of phosphoproteins and acetyl proteins
To evaluate the sequence conservation around the identified phosphosites and acetyl sites, we used Motif-X software (http://motif-x.med.harvard.edu/) to compare the position-specific frequencies of the six flanking amino acid residues upstream and downstream from the sites. We identified 21 significantly enriched pS motifs from 1632 pSs and two pT motifs from 181 pTs (Fig. 3a, c). Due to limited pY (phosphorylated tyrosine) and Kac (acetylated Lys) sites, no motifs were identified for these positions. The frequency of different amino acids around the phosphosites was then determined, revealing the preferred residue for pSs to be P at + 1, R at -3, and S at -2. TP is known to be by far the most common pT motif29, and this was confirmed in our study (Fig. 3b, d, e). Moreover, we found that alanine (A), leucine (L), and glutamate (E) predominate at positions −2 to + 2 relative to Lys, which indicates that those amino acids may be conserved in grape protein acetylation sites.
Fig. 3. Motif-X analysis of the pSs, pTs and Kacs.
(a, c): Graphical representation of the amino acid environment of the identified motifs. pS phosphorylated serine, pT phosphorylated threonine, Kac acetylated Lys. Upper scales of panels (b), (d) and (e): the position of the amino acids around the phosphosites (pSs, pTs) and acetyl sites (Kacs). Lateral scales of panels (b), (d) and (e): abbreviations of the twenty amino acids
Functional classification and enrichment analysis of DCPSs and DCASs following exposure to high temperatures
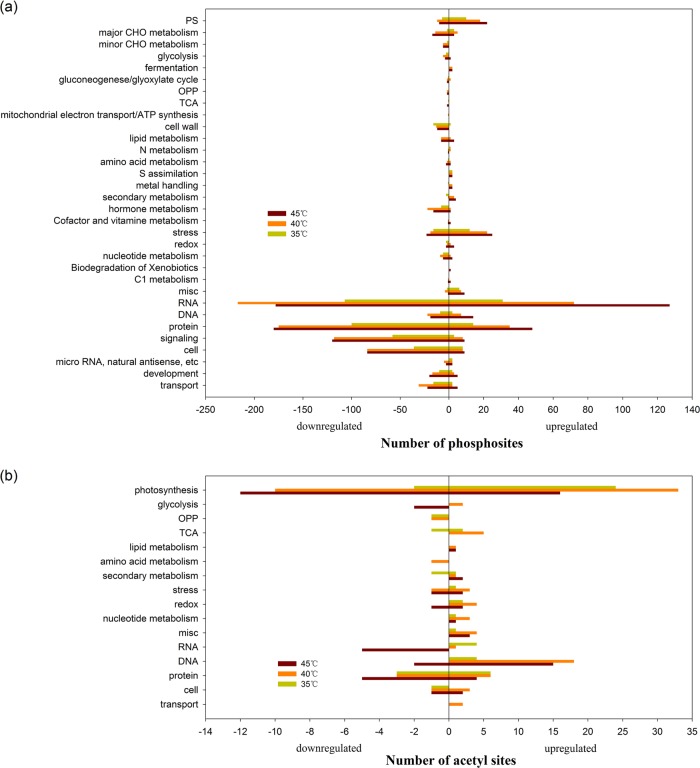
To functionally annotate the identified proteins, BLASTP (http://www.ncbi.nlm.nih.gov/BLAST/) was used to identify homologous proteins in TAIR database (http://www.arabidopsis.org/). Functional classification was then conducted using MapMan software (version 3.6.0RC1)30. The DCPSs were found to be mainly related to RNA regulation, protein synthesis, degradation, PTM, and signaling (Fig. 4a). Based on these analyses, we further analyzed proteins with up- or downregulated phosphosites in all 35 °C, 40 °C, and 45 °C samples. For 73 of these, 8 phosphosites were related to proteins involved in photosynthesis, including fructose-bisphosphate aldolase (FBA), ribulose bisphosphate carboxylase small chain 1a (RBCS1A), light-harvesting complex PSII subunit 6 (LHCB6), and photosystem II subunit R (PsbR). Twenty-three phosphosites were related to RNA, with 13 associated with RNA splicing factors. Eleven phosphosites were related to heat stress, such as heat-shock protein 23.6 (HSP23.6), HSP17.6, and CaM-binding protein 1 (Fig. S2). The 373 phosphosites that were downregulated in all three high-temperature samples were mainly associated with cells; PTM; synthesis and degradation of proteins; RNA transcriptional regulation; and signaling involving light, calcium and G-proteins (Fig. S3). For the 138 phosphosites unchanged at 35 °C but upregulated at 40 °C and 45 °C, seven were related to photosynthesis (including photorespiration), such as FBA, RuBisCO activase (RCA), peroxisomal (S)-2-hydroxy-acid oxidase (GLO1), and aminomethyltransferase (AMT). Forty-three phosphosites were associated with RNA processing, RNA regulation of transcription, and RNA binding, while 14 were SR-rich splicing factors, such as RS2Z33, RS41, and SCL30A. Ten phosphosites were related to heat stress, including HSP81.1, HSP81.2, HSP17.6, HSP23.6, HSP70.1, and CaM-binding protein 1 (Fig. S4). There were 129 phosphosites unchanged at 35 °C and 40 °C but upregulated at 45 °C, and these were mainly involved in RNA splicing and protein synthesis, degradation, and modification. There were only 13 phosphosites unchanged at 40 °C but upregulated at 35 °C and 45 °C, and they were mainly involved in RNA splicing (Fig. S5).
Fig. 4. Functional categorization in phosphoproteome and acetylproteome.
Main biological processes associated with proteins with differentially changed phosphosites (a) and acetyl sites (b) at 35 °C, 40 °C, and 45 °C in grape leaves. The x-axis indicates the number of upregulated (right) and downregulated (left) phosphosites
The DCASs were mainly related to photosynthesis, DNA synthesis, protein synthesis, degradation, and posttranslational modification (Fig. 4b). The 20 acetyl sites that were upregulated at all three relatively high temperatures were mainly associated with photosynthesis, such as PsbO2, RBCS1A, RBCL, phosphoglycerate kinase (PGK), and glyceraldehyde-3-phosphate dehydrogenase (GAPB) (Fig. S6). The five acetyl sites that were downregulated at all three relatively high temperatures were mainly associated with protein synthesis and degradation. Of the 22 acetylated proteins that were unchanged at 35 °C but upregulated at 40 °C and 45 °C, ten were related to DNA synthesis, and three were related to photosynthesis, including ribose 5-phosphate isomerase (Rib5P), RBCS1A, and PsbQ2 (Fig. S7). Five acetyl sites related to DNA synthesis, such as HTA11, were unchanged at 35 °C and 40 °C but upregulated at 45 °C, while 14 mainly related to photosynthesis, such as RCA, GAPB, glycolate oxidase, RBCS1A, and ribulose-phosphate 3-epimerase (RPE), were unchanged at 45 °C but upregulated at 35 °C and 40 °C (Fig. S8). The 33 acetyl sites upregulated only at 40 °C were mainly related to photosynthesis, respiration and DNA synthesis, such as triose phosphate isomerase (TIM), FBA, PGK, RCA, RBCS1A, PsaH2, PsbP1, PsbQ2, carbonate dehydratase, and HTB11 (Fig. S9).
Integration of the phosphoproteomic and acetylproteomic data
Out of 1011 phosphoproteins and 96 acetyl proteins exhibiting changes in response to high temperatures, there were 19 with changes in both modifications (Table S1). Eight of these proteins were related to photosynthesis or photorespiration. LHCB6 phosphorylation (T180) and acetylation (K219 and K327) levels were upregulated at 35 °C, but showed no differential change or were downregulated at 40 °C and 45 °C. LHCB6 phosphorylation (T188) was upregulated at all three high temperatures. The acetylation (K175 and K177) of RBCL was upregulated at 35 °C, 40 °C, and 45 °C, while phosphorylation (Y190 and S228) was upregulated only at 45 °C. The acetylation levels (five sites: K183, K201, K227, K236, and K314) of RBCS1A were upregulated at 35 °C, 40 °C, and 45 °C; however, the phosphorylation levels of this protein (four sites: T151, Y153, S117, and S180) were upregulated only at 45 °C. RCA acetylation increased significantly in response to all three high temperatures. Histone H2A.6 (HTA1) and H2A.4 are associated with DNA synthesis and chromatin structure. The acetylation level of H2A.6 was upregulated at 35 °C, while the phosphorylation level was downregulated at all three temperatures. The acetylation level of H2A.4 was downregulated at 45 °C, while the phosphorylation level was upregulated at 35 °C and 45 °C (Table S1).
Discussion
Phosphorylation, rather than acetylation, of SR-rich splicing factors is involved in the upregulation of alternative splicing (AS) genes in response to high temperatures in grape leaves
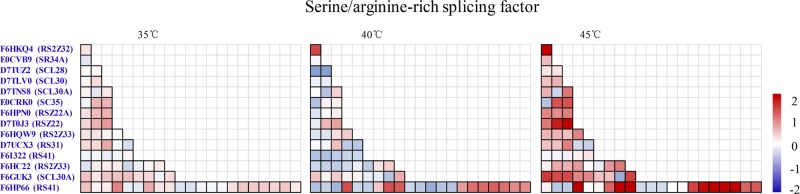
Alternative splicing involves the formation of two or more different transcripts from the same pre-mRNA molecule. The identity of the splice sites is largely determined by RNA-binding proteins, most notably SR proteins and heterogeneous nuclear ribonucleoproteins, which promote and inhibit binding of the spliceosome, respectively31. The precise formation of mRNA splice variants is determined by splicing factors that define the exon–intron boundaries between. SR proteins modulate alternative splicing via concentration-dependent and phosphorylation-dependent splice site selection, while the activity and localization of SR proteins are regulated by phosphorylation32. In plants, notable changes in alternative splicing are triggered by a wide variety of abiotic stresses33–35. A subset of human SR proteins comprise nucleocytoplasmic shuttling proteins with diverse roles in aspects of postsplicing, such as mRNA export, stability, and mRNA translation36. In our previous study, we observed a rapid change in pre-mRNA splicing in grape leaves at 35 °C, as 206 differential splicing events were detected, whereas 1075 and 748 were detected at 40 °C and 45 °C, respectively6. Moreover, we found that the abundance of SR45, SR30, and SR34 in grape leaves increased gradually as ambient temperatures increased6. These results are consistent with additional alternative splicing events that occur in response to high temperatures in grape leaves. We also found that the phosphorylation levels of 11 SR-rich splicing factors were mainly upregulated at 35 °C, 40 °C or 45 °C, which included RS2Z32, SR34A, SCL30, SCL30A, SC35, RSZ22A, RSZ22, RS31, and RS41. However, the downregulated phosphosites at each high temperature were associated only with SCL28. These results indicated that phosphorylation upregulation of SR-rich splicing factors is important for promoting alternative splicing during the response to high temperatures in grape leaves (Fig. 5; Supplementary Table S2). Some studies have also demonstrated that SCL30A, SR34, RS2Z33, and RS41 are involved in plant responses to salinity or high/low temperatures37–39. The increase in the phosphorylation level of these SR-rich splicing factors would strengthen their own activities and promote alternative splicing, which further explains why increased numbers of alternative splicing events occurred in grape leaves in response to high temperature. However, SR proteins did not appear to be acetylated during the grape leaf response to high temperature. Therefore, we conclude that phosphorylation, rather than acetylation, of SR-rich splicing factors is involved in the upregulation of alternative splicing during the grape leaf response to high temperatures.
Fig. 5. Phosphorylation level change of SR-rich splicing factors at 35 °C, 40 °C and 45 °C in grape leaves.
Each block represents a phosphosite, and the different colors of each block represent up- or downregulation of the phosphorylation level
Acetylation modulates more photosynthesis proteins and is more sensitive to high temperatures than is phosphorylation during responses to high temperatures in grape leaves
Photosynthesis involves highly heat-sensitive enzymes, and previous studies have shown that phosphorylation of photosynthesis proteins plays an important role in regulating photosynthesis40,41. The photosynthetic light reactions are catalyzed by thylakoid-embedded pigment–protein complexes (PSII, Cytb6f, and PSI), while the photooxidation of water during the light reactions of photosynthesis involves proteins of the oxygen-evolving complex (OEC), which are termed PsbO, PsbP, and PsbQ. In addition, PsbR has an auxiliary role in PsbP binding to PSII. In this study, we found that the phosphorylation levels of PsbR (at phosphosites Y52, S67, and Y77) at 35 °C, 40 °C, or 45 °C were upregulated (Fig. 6a; Table S3a). Furthermore, the acetylation levels of all oxygen-evolving complex components (PsbQ2, PsbP1, and PsbO2) were upregulated at 35 °C, 40 °C, or 45 °C, while their phosphorylation levels did not change (Fig. 6c). These results indicated that the oxygen-evolving complex may utilize acetylation and not phosphorylation to coordinate the response to high temperatures in grape leaves.
Fig. 6. Schematic representation of phosphoprotein or acetylproteins involved in photosynthesis.
Changes in phosphorylation levels (a) and acetylation levels (c) of proteins related to the light reactions of photosynthesis at 35 °C, 40 °C, and 45 °C in grape leaves. Changes in phosphorylation levels (b) and acetylation levels (d) of proteins related to the Calvin cycle of photosynthesis at 35 °C, 40 °C, and 45 °C
The photosystem II (PSII) core proteins D1 and D2, as well as the inner antenna protein CP43 and the minor PSII subunit PsbH, are phosphorylated, mostly by the kinase STN8, at Thr residues42–44. In this study, we found that the phosphorylation levels of PsbH (phosphosites T3 and T5) in grape leaves at 35 °C, 40 °C, or 45 °C were downregulated (Fig. 6a), and that the acetylation levels of PsbD2 at 35 °C were downregulated (Fig. 6c). The phosphorylation levels of PsaF and the acetylation levels of PsaH2 at the three high temperatures were upregulated (Fig. 6a, c). These results constitute the first report indicating that the acetylation of PSI proteins represents a response to high temperature in plants.
The light-harvesting proteins LHCB1, LHCB2, and LHCB4 are phosphorylated by the STN7 kinase42,45. In our study, we found that the phosphorylation levels of LHCB1 and LHCB6 were upregulated, but the phosphorylation levels of LHCB4 were downregulated in grape leaves in response to high temperatures (Fig. 6a). In addition to phosphorylation, LHC proteins undergo other PTMs such as Lys acetylation46,47. Acetylation can have a range of biochemical and biological effects, including the determination of enzyme activity, alteration of protein stability and modification of protein–protein interactions48–50. As an example, acetylation of LHCB1 and LHCB2 appears to influence LHC attachment to PSII complexes: the peripheral LHC antenna that is more loosely bound to PSII shows more extensive Lys acetylation than do PSII–LHCII supercomplexes47. In our study, the acetylation level of LHCB6 at high temperatures was downregulated or unchanged in grape leaves (Fig. 6c; Supplementary Table S3b). In addition, the phosphorylation level of LHCA3 and the acetylation level of the LHCA1 protein at high temperatures were downregulated in grape leaves (Fig. 6a, c; Supplementary Table S3a, b). These results indicated that the light-harvesting proteins of PSII and PSI responded to the phosphorylation or acetylation levels in the grape leaves in response to high temperatures.
RuBisCO is the first enzyme in carbon fixation, and it exists as a holocomplex of eight small subunits (RBCS) and eight large subunits (RBCL) that contain multiple phosphosites51,52 and are reversibly phosphorylated in many plant species52,53. RCA is an ancillary photosynthesis protein essential for RuBisCO activity. It has been reported that phosphorylation of RuBisCO results in a substantial increase in its activity, while desphosphorylation results in a major decrease in its activity54. However, the phosphorylation of RCA may result in decreased activity of RuBisCO40,41. In our study, the phosphorylation levels and phosphosites of RBCL and RBCS1A increased at high temperatures (Fig. 6b; Supplementary Table S3a), and the RCA showed the same trend as did RuBisCO. Based on these data, we concluded that the phosphorylation of RuBisCO may compensate for its decreased activity due to the phosphorylation of RCA. In addition, RuBisCO subunits undergo Lys acetylation in response to different light conditions55. The RuBisCO holocomplex has multiple Lys acetyl sites, which are located either in the RuBisCO catalytic center46,56, at the interface between the two RBCL subunits46,57, or at a location that is important in defining the RuBisCO tertiary structure57. Lys acetylation is thought to affect RuBisCO activity, as well as interactions between subunits and other molecules, and recent studies have shown that Lys acetylation suppresses RuBisCO activity46,55. In this study, almost all of the identified acetyl sites of RuBisCO (RBCL and RBCSA1) and RCA were upregulated at different high temperatures compared with the control (Fig. 6d; Supplementary Table S3b). Thus, acetylation and phosphorylation of RuBisCO and RCA may jointly help coordinate the light reactions and carbon assimilation in response to the carbon status of the cell under different high temperatures. Moreover, acetylation may have a larger effect on function rather than phosphorylation with respect to RuBisCO modification. In the green alga Chlamydomonas reinhardtii, RCA is phosphorylated at S53 by a thylakoid-localized kinase58. RCA is mainly localized in the stroma, but a small portion of the enzyme is associated with the thylakoid membrane59. It has been proposed that phosphorylation of RCA promotes its attachment to the membrane, thereby protecting Stt7 from proteolysis58,60. The relocation may also reduce the activity of RuBisCO under specific environmental conditions58. In Arabidopsis, RCA is phosphorylated at two sites, T78 and S17241, and in the dark, the proportion of phosphorylated T78 sites increases61,62. In this study, the phosphorylation levels of six sites of three RCA proteins (F6HNV2, D7SKB2, and D7THJ7) and the acetylation levels of three sites of one RCA protein (D7SKB2) were upregulated compared with the control in grape leaves under high temperature (Fig. 6b, d; Supplementary Table S3).
Other enzymes of the Calvin cycle have bene identified as being phosphorylated. Phosphoglycerate kinase (PGK) is known to be phosphorylated in Arabidopsis, rice, and maize;53,61,63,64 while the second two species have the same phosphosite, Arabidopsis PGK is phosphorylated in a domain near the N-terminus. GAPDH has several phosphorylation sites, but they differ considerably between different organisms; thus, it has been suggested that phosphorylation may not be a key factor in regulating GAPDH activity in chloroplasts64. It has been noted that PGK and GAPDH are also targets of Lys acetylation46,65, and the activities of both enzymes increase upon deacetylation. Thus, Calvin cycle enzymes are subject to complex regulation by PTMs, but the enzymes involved in such modifications have yet to be identified64,66. In our study, in addition to those of RuBisCO and RCA, the phosphorylation levels of FBA and GAPB increased at high temperatures (Fig. 6b; Supplementary Table S3a). Moreover, the acetylation levels of PGK, FBA, GAPB, RPE, and TIM increased under the heat treatments (Fig. 6d). However, the acetylation level of three sites of transketolase (TKT) decreased (Fig. 6d; Supplementary Table S3b). Therefore, both phosphorylation and acetylation are associated with the modulation of key enzymes of carbon assimilation in response to high temperatures (Fig. 6b, d). Moreover, it appears that the Calvin cycle may be affected more by acetylation than by phosphorylation. In addition, for all the investigated photosynthesis proteins in grape leaves, acetylation modification occurred largely at 35 °C, and phosphorylation modification was not prevalent (Fig. 6), which suggests that acetylation of grape leaf photosynthesis proteins is more influenced than phosphorylation is by high temperatures. We conclude that photosynthesis is fundamentally affected by high temperatures, and can be regulated by both phosphorylation and acetylation. Moreover, the acetylation of photosynthesis proteins was more sensitive to high temperatures than was phosphorylation.
HSPs exhibit more extensive phosphorylation than acetylation in response to heat exposure in grape leaves
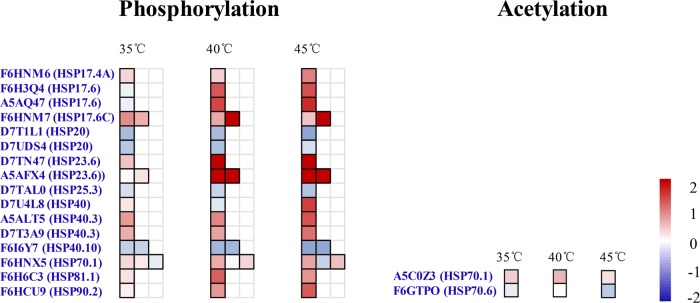
Many studies have characterized large-scale responses to heat-shock responses in a variety of cells and organisms by the use of approaches, such as transcriptional profiling, differential displays, and proteomic analyses. HSPs act as molecular chaperones, reducing the aggregation and resolubilization of denatured proteins, promoting the folding of nascent polypeptides, and facilitating the refolding of denatured proteins67–69. In plants, HSPs can be classified into five groups based on molecular mass: small HSP (sHsp) proteins, chaperonins (GroEL and Hsp60), Hsp70 (DnaK) proteins, Hsp90 proteins, and members of the Hsp100 (Clp) family. Many studies have reported that high temperatures lead to increases in HSP levels in plants70. However, there are few studies on the acetylation and phosphorylation of HSPs in plants under heat or other stresses71. Characterized the phosphoproteome of different tissues of wheat cultivars that exhibit different degrees of drought tolerance following simulated drought and recovery. Notably, the phosphorylation levels of HSP60 and HSP90 were found to be upregulated in response to drought in the drought-tolerant cultivar. In another study72, reported the effects of a short-term and moderate increase in temperature on the wheat leaf and spikelet phosphoproteome, which involved a substantial increase in the abundance of S224 of HSP90 and S577 of HSP60-3A. The acetylation of HSP70 has been described as a regulatory mechanism that temporally balances protein refolding/degradation in response to stress73,74, while HSP20 phosphorylation in mammalian systems has been implicated in a variety of pathophysiological processes but most prominently in cardiovascular disease75. In this study, we found that the phosphorylation or acetylation levels of a number of HSPs were upregulated in grape leaves in response to heat treatment. Phosphorylation levels increased substantially at two sites (S10 and S154) of HSP17.6C (F6HNM7), one site (S12) at HSP23.6 (D7TN47), and two sites (S27 and S56) of HSP17.6 (A5AQ47) at three high temperatures (Fig. 7). The phosphorylation levels of other HSPs, including HSP17.4A, HSP17.6 (F6H3Q4), HSP17.6 (A5AQ47), putative HSP40, HSP40.3 (A5ALT5), HSP40.3 (D7T3A9), HSP81.1 (F6H6C3), and HSP81.2 (F6HCU9), were regulated at high temperatures. However, phosphorylation levels declined at one site of HSP20 (D7UDS4), one site of HSP20 (D7T1L1), and two sites of HSP40.10 (Fig. 7; Supplementary Table S4). The acetylation level increased at one site (K191) in HSP70.1 (A5C0Z3) and declined at another site (K613) in HSP70.6 (F6GTP0) (Fig. 7; Supplementary Table S4). In our previous study, 11 HSPs were coupregulated at both the transcriptional and translational levels in grape leaves upon exposure to the 40 °C and 45 °C treatments compared with the control, but not by low-temperature treatments; these proteins included HSP17.4, HSP17.6, HSP23.6, HSP40, HSP25.3, HSP70.1, and HSP81.16. The results from this study indicate that these HSPs not only function in response to high temperatures by upregulating transcript and protein levels but also are involved in the heat tolerance of grape leaves via phosphorylation more so than acetylation. This study therefore underlines the importance of HSPs in grape thermotolerance.
Fig. 7.
Phosphorylation and acetylation changes in heat shock protein (HSPs) at 35 °C, 40 °C, and 45 °C in grape leaves
Crosstalk between protein phosphorylation and acetylation in response to high temperatures in grape leaves
Some proteins can undergo multiple modifications, and PTM crosstalk has recently been reported76–80. Xiong et al. found that an acetyl site (K324) on an enolase protein from rice was located next to a phosphosite (S325)81. In addition, the acetyl sites and phosphosites in two spliced isoforms of a gamma-interferon-inducible lysosomal thiol reductase precursor from rice were found to be in close proximity. Ahn et al. provided evidence for acetylation and phosphorylation crosstalk between two neighboring histone residues (S10 and K11) in vivo and in vitro in H2B (HIB4) in yeast82. Protein phosphorylation and acetylation have been associated with many cellular functions, as recently reported in Mycoplasma pneumonia83. In this study, 19 proteins exhibiting possible crosstalk between phosphorylation and acetylation were associated with the photosynthetic light reactions and the Calvin cycle, glycolysis, the TCA cycle, DNA and protein synthesis, cell division, and the cell cycle (Table S1). These results indicated that key proteins of some biological processes, especially photosynthesis, are regulated by concurrent phosphorylation and acetylation in grape leaves in response to high temperature.
Emerging evidence suggests that protein PTMs that are adjacent or in close proximity within a single protein often lead to regulation of protein function or are important in signaling84. Carlomagno et al. demonstrated that acetylation of Lys321 (contained within a KCGS motif) inhibited phosphorylation of Ser-324 in tau proteins85. Cook et al. also noted a similar competitive relationship between phosphorylation and acetylation of KIGS motifs86. Previous studies have shown that mutagenesis of the S113 and K230 sites of isocitrate dehydrogenase (Idh) results in reduced enzyme activity and substrate affinity, underlining the significance of both phosphorylation and acetylation in regulating Idh in E. coli87,88. Direct evidence of crosstalk between acetylation and phosphorylation was recently demonstrated in M. pneumonia83; however, there are no reports of phosphorylation and acetylation of the same protein in plants under stress. In this study, acetylation levels in a number of sites of RBCL increased in the grape leaves in response to 35 °C, 40 °C, and 45 °C, while the phosphorylation levels of a few sites increased only at 45 °C (Table S1). Acetylation levels at five sites and phosphorylation levels at two sites of RBCS1A increased in response to heat treatment (Table S1). These results indicated that RBCL may undergo different PTMs than does RBCS1A under high temperature in grape leaves and that their coordination modulates RuBisCO activity. Recent studies have indicated negative regulation of RuBisCO activity by Lys acetylation46,55, and we previously reported that RuBisCO activity significantly decreased in grape leaves under 43 °C high temperature24. In this study, RCA appeared to exhibit more acetylation than phosphorylation (Table S1). Other studies have indicated that the activity of RCA decreases in many plant species under high temperature89. Therefore, upregulation of acetylation levels may be a factor in the decrease in RuBisCO and RCA activities in the grape leaf response to high temperatures. In addition, at different high temperatures, the phosphorylation and acetylation levels of proteins differed. For example, the LHCB6 phosphorylation (T180) and acetylation (K219 and K327) levels were upregulated at 35 °C, but did not change or were downregulated at 40 °C and 45 °C. Grape protein modification by acetylation appears to be as common as phosphorylation, and acetylation may provide a balance to phosphorylation in the regulation of protein activity. Congruent with previous studies in humans and microbes83,87,88, an upregulated acetyl site (K183) and an upregulated phosphosite (S180) are near one another in RBCS1A, and an upregulated acetyl site (K151) and a downregulated phosphosite (S154) are very close in a SAP domain-containing protein (E0CRG0). The regulatory mechanism of protein activity under high temperature is clearly very complex.
In summary, phosphoproteomic and acetylproteomic analyses were conducted on leaves of grape plants subjected to four different temperature regimes (25 °C, 35 °C, 40 °C, and 45 °C). The results revealed significant changes in phosphosites and acetyl sites, mainly following exposure to 40 °C and 45 °C. Phosphorylation, rather than acetylation, of SR-rich splicing factors was involved in the increase in AS events. Moreover, compared with phosphorylation modification, acetylation modification modulated more photosynthesis-related proteins and was more sensitive to high temperatures. Conversely, we conclude that modifications of HSPs during heat tolerance responses in grape leaves involve phosphorylation more than acetylation. We identified 19 proteins with significantly changed phosphorylation and acetylation levels, which is indicative of crosstalk between these PTMs. Acetylation may balance phosphorylation with regard to protein activity. Additional studies are needed to determine how key phosphorylation and acetylation proteins influence grape heat tolerance.
Materials and methods
Plant materials and heat treatments
The materials and treatments were identical to those of our previous study6. One-year-old “Jingxiangyu” grapevine (Vitis vinifera L.) cuttings were planted in pots and then grown in a greenhouse under 70–80% relative humidity and at 18 °C to 25 °C. When the sixth leaf (from the base to the apex) of each grapevine became mature, all the grapevines were divided into four groups and acclimated for 2 days in a controlled-environment chamber (70% average relative humidity, 25 °C/18 °C [12-h/12-h] day/night cycle). On day 3, the grapevines were subjected to the following treatments: (1) the plants of the control group were maintained at the optimal day/night temperature (25 °C/18 °C) in the abovementioned growth chamber; (2) the plants of the treatment groups were exposed to 35 °C, 40 °C, or 45 °C from 11:30 a.m. to 1:30 p.m. (the conditions were the same as those of the control, except for the temperature). The fourth to sixth leaves (from the base to the apex) of each plant were detached at 1:30 p.m. (the end of the heat stress treatment). Each biological replicate included three plants, and two replicates were used for the three treatments and for the control. The detached leaves were frozen in liquid nitrogen immediately and then stored at −80 °C for further analysis.
Protein extraction
Proteins were extracted using the cold-acetone method and digested as described previously90, and tryptic peptides were incubated with a 4-plex iTRAQ labeling kit (114 for 25 °C, 115 for 35 °C, 116 for 40 °C, 117 for 45 °C)90.
High-pH fractionation and the enrichment of phosphopeptides
High-pH reversed-phase high-pressure liquid chromatography (HPLC) was used for peptide fractionation on a Gilson 300 series system. A total of 4 mg of each desalted iTRAQ-labeled sample corresponding to each of the four temperature treatments was solubilized individually in 200 µL of 0.02% NH4OH (pH 10) and injected onto an XBridge column (Waters, C18 3.5 µm 2.1 × 150 mm) using a linear gradient of buffer B from 2-45% for 45 min (buffer A: 0.02% NH4OH, pH 10; buffer B: 90% acetonitrile, 0.02% NH4OH, pH 10). The fractions were collected for 1 min, after which 5% of each fraction was dried under vacuum and preserved at −80 °C for total proteome analysis after desalting with a StageTip + C1891. The remaining 95% of each fraction was combined, dried under vacuum and preserved at −80 °C for phosphoproteome analysis.
The metal affinity chromatography (IMAC) enrichment of phosphopeptides was adapted from the methods of Mertins et al.91. Ion-chelated IMAC beads were prepared from Ni-NTA Superflow agarose beads (Qiagen, MA). Nickel ions were removed with 50 mM EDTA, and the iron was chelated by passing the beads through an aqueous solution of 200 mM FeCl3, followed by three water washes and one wash with binding buffer (40% acetonitrile, 1% formic acid). iTRAQ-labeled reversed-phase (RP) fractions were solubilized in binding buffer and incubated with IMAC beads for 1 h. After three washes with binding buffer, the phosphopeptides were eluted with a 2x bead volume of 500 mM potassium hydrogen phosphate (pH 7.0), and the eluate was neutralized with 10% formic acid. The enriched phosphopeptides were then desalted using an Empore 3 M C18 (2215) StageTip prior to nanoLC-MS/MS analysis92.
Enrichment of acetylpeptides
A mixture of iTRAQ-labeled samples corresponding to the four temperature treatments was desalted using a Sep-Pak tC18 cartridge (Varian, CA). Acetylpeptides were enriched using a PTMScan Acetyl-Lys Motif (Ac-K) antibody (#13416, Cell Signaling Technology, MA) according to the manufacturer’s instructions. The eluted peptides were desalted with an Empore 3 M C18 (2215) StageTip, dried and solubilized in 0.1% TFA/5% acetonitrile prior to nanoLC-MS/MS.
Mass spectrometry identification
The enriched phosphopeptides or acetylated peptides were analyzed by nanoLC-MS/MS with an RSLC system connected to a Q Exactive mass spectrometer (ThermoFisher, San Jose, CA, USA). The samples were loaded onto a self-packed 100 µm × 2 cm trap packed with Magic C18AQ, 5 µm 200 A (Michrom Bioresources, Inc., Auburn, CA, USA) and washed with buffer A (0.2% formic acid) for 5 min at a flow rate of 10 µL min-1. The trap was switched in line with a custom-made analytical column (Magic C18AQ, 3 µm 200 A, 75 µm × 50 cm), and the peptides were fractionated at 300 nL min-1 with a multistep gradient (proteome: 4–15% buffer B (0.16% formic acid 80% acetonitrile) for 30 min, 15–25% B for 40 min, and 25–50% B for 45 min; acetylome: 4–25% buffer B for 35 min followed by 25–50% B for 65 min). Mass spectrometry data were acquired using Xcalibur (ThermoFisher, San Jose, CA, USA) in data-dependent acquisition mode with a cyclic series of a full scan acquired at a resolution of 120,000 followed by MS/MS scans (30% collision energy in the HCD cell) at a resolution of 30,000 of the 20 most intense ions and with a dynamic exclusion duration of 30 s.
Phosphoproteomic and acetylproteomic data analyses
All the data were analyzed with MaxQuant proteomics data analysis workflow (version 1.6.0.1.) using the Andromeda search engine93,94. The type of LC-MS run was set to “Reporter ion MS2” with “iTRAQ4plex” as isobaric labels, and the reporter ion mass tolerance value was defined as 0.003 Da. The false discovery rate was set to 1% for protein, peptide spectrum match, and site decoy fraction levels. Peptides were required to have a minimum length of seven amino acids and a maximum mass of 4600 Da. MaxQuant was used to score fragmentation scans for identification based on a search with an allowed mass deviation of the precursor ion of up to 4.5 ppm. The allowed fragment mass deviation was 20 ppm. MS2 spectra were searched against the UniProt Vitis vinifera cultivar Pinot Noir/PN40024 database (downloaded on 01/16/2018) (https://www.uniprot.org/), which contained 29,907 entries combined with 262 common contaminants. Enzyme specificity was set as trypsin/P, which allowed three missed cleavages for phosphorylation data and five missed cleavages for acetylation data. Carbamidomethylation of cysteine was set as a fixed modification, and N-terminal protein acetylation and oxidation at methionine were set as variable modifications. The phospho (STY) and acetyl (K) parameters were set as variable modifications for the phosphorylation- and acetylation-enriched samples, respectively. The data are available via ProteomeXchange with identifier PXD014090.
The summed reporter ion intensity of each channel before enrichment was used as the normalization factor for phosphoproteomic or acetylproteomic quantitation. After filtering potential contaminants, a reverse localization score ≥0.75 for the phosphoproteome and acetylproteome was included in the data.
Bioinformatic analysis
The differentially expressed proteins were functionally categorized based on MapMan ontology30. MapMan ontology analysis was carried out based on the BLASTP results using the A. thaliana predicted proteome as a reference. Functional categories were manually verified by literature searches, and proteins that were not associated with any biological process category were assigned to MapMan (Bin35). Motif-X was used for motif analysis, and the setting parameters for searching motifs included 20 occurrences and a significance of 0.000001. Functional interaction network analysis was performed using STRING software95, and the results were visualized by Cytoscape96.
Supplementary information
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFD1000300) and the National Natural Science Foundation of China (Grant no. 31672120). We thank Baichen Wang (Institute of Botany, Chinese Academy of Sciences) and Douglas D. Archbold (Department of Horticulture, University of Kentucky) for critical reading and valuable suggestions. The authors thank PlantScribe (www.plantscribe.com) for editing this paper.
Author contributions
L.W., S.L. and C.L. and conceived and designed the experiments; G.L. and J.J. (Jianfu Jiang) performed the experiments; X.L., J.J. (Jinzhu Jiang), S.L., W.D., R.L., Y.W. and D.L. provided technical assistance to G.L. and J.J. (Jianfu Jian); and L.W. and G.L. analyzed the data and wrote the article.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-019-0183-x).
References
- 1.Fedoroff NV, et al. Radically rethinking agriculture for the 21st century. Science. 2010;327:833–834. doi: 10.1126/science.1186834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartels D, Sunkar R. Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 2005;24:23–58. [Google Scholar]
- 3.Zhuang J, Zhang J, Hou X-L, Wang F, Xiong A-S. Transcriptomic, proteomic, metabolomic and functional genomic approaches for the study of abiotic stress in vegetable crops. Crit. Rev. Plant Sci. 2014;33:225–237. [Google Scholar]
- 4.Kosová K, Vítámvás P, Prášil IT, Renaut J. Plant proteome changes under abiotic stress-contribution of proteomics studies to understanding plant stress response. J. Proteom. 2011;74:1301–1322. doi: 10.1016/j.jprot.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Grover A, Mittal D, Negi M, Lavania D. Generating high temperature tolerant transgenic plants: achievements and challenges. Plant Sci.: Int. J. Exp. plant Biol. 2013;205-206:38–47. doi: 10.1016/j.plantsci.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Jiang J-F, et al. Integrating omics and alternative splicing reveals insights into grape response to high temperature. Plant Physiol. 2017;173:1502–1518. doi: 10.1104/pp.16.01305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melo-Braga MN, et al. Modulation of protein phosphorylation, N-glycosylation and Lys-acetylation in grape(Vitis vinifera) mesocarp and exocarp owing to lobesia botrana. Infect. Mol. Cell. Proteom.: MCP. 2012;11:945–956. doi: 10.1074/mcp.M112.020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanou G, et al. Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. Plant J.: Cell Mol. Biol. 2012;72:585–599. doi: 10.1111/j.1365-313X.2012.05100.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhou LJ, et al. The small ubiquitin-like modifier E3 ligase MdSIZ1 promotes anthocyanin accumulation by sumoylating MdMYB1 under low-temperature conditions in apple. Plant, Cell Environ. 2017;40:2068–2080. doi: 10.1111/pce.12978. [DOI] [PubMed] [Google Scholar]
- 10.Thingholm TE, Jensen ON, Larsen MR. Analytical strategies for phosphoproteomics. Proteomics. 2009;9:1451–1468. doi: 10.1002/pmic.200800454. [DOI] [PubMed] [Google Scholar]
- 11.Khan M, Takasaki H, Komatsu S. Comprehensive phosphoproteome analysis in rice and identification of phosphoproteins responsive to different hormones/stresses. J. Proteome Res. 2005;4:1592–1599. doi: 10.1021/pr0501160. [DOI] [PubMed] [Google Scholar]
- 12.Kline KG, Barrett-Wilt GA, Sussman MR. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. PNAS. 2010;107:15986–15991. doi: 10.1073/pnas.1007879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, et al. Phosphoproteins regulated by heat stress in rice leaves. Proteome Sci. 2011;9:37. doi: 10.1186/1477-5956-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen THN, et al. Quantitative phosphoproteomic analysis of soybean root hairs inoculated with Bradyrhizobium japonicum. Mol. Cell. Proteom.: MCP. 2012;11:1140–1155. doi: 10.1074/mcp.M112.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonhomme L, Valot B, Tardieu Fo, Zivy M. Phosphoproteome dynamics upon changes in plant water status reveal early events associated with rapid growth adjustment in maize leaves. Mol. Cell. Proteom. 2012;11:957–72. doi: 10.1074/mcp.M111.015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lv D-W, et al. Proteome and phosphoproteome characterization reveals new response and defense mechanisms of Brachypodium distachyon leaves under salt stress. Mol. Cell Proteom. 2013;13:632–652. doi: 10.1074/mcp.M113.030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M, et al. Phosphoproteome analysis reveals new drought response and defense mechanisms of seedling leaves in bread wheat (Triticum aestivum L.) J. Proteom. 2014;109:290–308. doi: 10.1016/j.jprot.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Rotival M, et al. Integrating phosphoproteome and transcriptome reveals new determinants of macrophage multinucleation. Mol. Cell. Proteom.: MCP. 2015;14:484–498. doi: 10.1074/mcp.M114.043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mischerikow N, Heck AJR. Targeted large-scale analysis of protein acetylation. Proteomics. 2011;11:571–589. doi: 10.1002/pmic.201000397. [DOI] [PubMed] [Google Scholar]
- 20.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 21.Nambi S, et al. Cyclic AMP-dependent protein lysine acylation in mycobacteria regulates fatty acid and propionate metabolism. J. Biol. Chem. 2013;288:14114–14124. doi: 10.1074/jbc.M113.463992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 23.Liang W-X, Malhotra A, Deutscher MP. Acetylation regulates the stability of a bacterial protein: growth stage-dependent modification of RNase R. Mol. Cell. 2011;44:160–166. doi: 10.1016/j.molcel.2011.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q-J, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao, R. S. P., Thelen, J. J. & Miernyk, J. A. In silico analysis of protein Lys-Nε-acetylation in plants. Front. Plant Sci.5, 10.3389/fpls.2014.00381 (2014). [DOI] [PMC free article] [PubMed]
- 26.Parra CS, Aguirreolea J, Sánchez-Díaz M, Irigoyen JJ, Morales F. Effects of climate change scenarios on Tempranillo grapevine (Vitis vinifera L.) ripening: response to a combination of elevated CO2 and temperature, and moderate drought. Plant Soil. 2010;337:179–191. [Google Scholar]
- 27.Pillet Jrm, et al. VvGOLS1 and VvHsfA2 are involved in the heat stress responses in grapevine berries. Plant Cell Physiol. 2012;53:1776–1792. doi: 10.1093/pcp/pcs121. [DOI] [PubMed] [Google Scholar]
- 28.Rattanakan, S., George, I., Haynes, P. A. & Cramer, G. R. Relative quantification of phosphoproteomic changes in grapevine (Vitis vinifera L.) leaves in response to abscisic acid. Hortic. Res.3, 10.1038/hortres.2016.29 (2016). [DOI] [PMC free article] [PubMed]
- 29.Zhao Q, et al. Proteomics and phosphoproteomics of heat stress-responsive mechanisms in spinach. Front. Plant Sci. 2018;9:800. doi: 10.3389/fpls.2018.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thimm O, et al. MAPMAN: a user-driver tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 31.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Z-H, Fu X-D. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma. 2013;122:191–207. doi: 10.1007/s00412-013-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staiger D, Brown JWS. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell. 2013;25:3640–3656. doi: 10.1105/tpc.113.113803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laloum Tom, Martín Guiomar, Duque Paula. Alternative Splicing Control of Abiotic Stress Responses. Trends in Plant Science. 2018;23(2):140–150. doi: 10.1016/j.tplants.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Ling Y, et al. Pre-mRNA splicing repression triggers abiotic stress signaling in plants. Plant J.: Cell Mol. Biol. 2017;89:291–309. doi: 10.1111/tpj.13383. [DOI] [PubMed] [Google Scholar]
- 36.Stankovic N, et al. Dynamic distribution and interaction of the Arabidopsis SRSF1 subfamily splicing factors. Plant Physiol. 2016;170:1000–1013. doi: 10.1104/pp.15.01338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palusa SG, Ali GS, Reddy AS. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J.: Cell Mol. Biol. 2007;49:1091–1107. doi: 10.1111/j.1365-313X.2006.03020.x. [DOI] [PubMed] [Google Scholar]
- 38.Cruz T, Carvalho R, Richardson D, Duque P. Abscisic acid (ABA) regulation of Arabidopsis SR protein gene expression. Int. J. Mol. Sci. 2014;15:17541–17564. doi: 10.3390/ijms151017541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding F, et al. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genom. 2014;14:431. doi: 10.1186/1471-2164-15-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan D, et al. Phosphoproteomics unveils stable energy supply as key to flooding tolerance in Kandelia candel. J. Proteom. 2018;176:1–12. doi: 10.1016/j.jprot.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 41.Boex-Fontvieille E, et al. Phosphorylation pattern of Rubisco activase in Arabidopsis leaves. Plant Biol. 2014;16:550–557. doi: 10.1111/plb.12100. [DOI] [PubMed] [Google Scholar]
- 42.Bellafiore Sp, Barneche Fd, Peltier G, Rochaix J-D. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature. 2005;433:892–895. doi: 10.1038/nature03286. [DOI] [PubMed] [Google Scholar]
- 43.Bonardi V, et al. Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature. 2005;437:1179–1182. doi: 10.1038/nature04016. [DOI] [PubMed] [Google Scholar]
- 44.Fristedt R, Vener AV. High light induced disassembly of photosystem II supercomplexes in Arabidopsis requires STN7-dependent phosphorylation of CP29. PLoS ONE. 2011;6:e24565. doi: 10.1371/journal.pone.0024565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Depege N, Bellafiore S, Rochaix J. Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science. 2003;299:1572–1575. doi: 10.1126/science.1081397. [DOI] [PubMed] [Google Scholar]
- 46.Finkemeier I, Laxa M, Miguet L, Howden AJ, Sweetlove LJ. Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol. 2011;155:1779–1790. doi: 10.1104/pp.110.171595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X, et al. Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol. 2011;155:1769–1778. doi: 10.1104/pp.110.165852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bienvenut WV, et al. Dynamics of post-translational modifications and protein stability in the stroma of Chlamydomonas reinhardtii chloroplasts. Proteomics. 2011;11:1734–1750. doi: 10.1002/pmic.201000634. [DOI] [PubMed] [Google Scholar]
- 50.Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao X, et al. Differential expression and modification of proteins during ontogenesis in Malus domestica. Proteomics. 2011;11:4688–4701. doi: 10.1002/pmic.201100132. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, et al. Protein phosphorylation differs significantly among ontogenetic phases in Malus seedlings. Proteome Sci. 2014;12:31. doi: 10.1186/1477-5956-12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roitinger E, et al. Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and Rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol. Cell. Proteom. 2015;14:556–571. doi: 10.1074/mcp.M114.040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jr. ARP, Parry MAJ. Discoveries in Rubisco (Ribulose 1,5-bisphosphate carboxylase/oxygenase): a historical perspective. Photosynth Res. 2007;94:121–143. doi: 10.1007/s11120-007-9225-6. [DOI] [PubMed] [Google Scholar]
- 55.Gao X, et al. Downregulation of rubisco activity by non-enzymatic acetylation of RbcL. Mol. Plant. 2016;9:1018–1027. doi: 10.1016/j.molp.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 56.Cleland W, Andrews T, Gutteridge S, Hartman F, Lorimer G. Mechanism of Rubisco: the carbamate as general base. Chem. Rev. 1998;98:549–561. doi: 10.1021/cr970010r. [DOI] [PubMed] [Google Scholar]
- 57.S K, I A, CI B. Crystallographic analysis of ribulose 1,5-bisphosphate carboxylase from spinach at 2.4 A resolution. Subunit Interact. Act. Site. J. Mol. Biol. 1990;215:113–160. doi: 10.1016/S0022-2836(05)80100-7. [DOI] [PubMed] [Google Scholar]
- 58.Lemeille S, Turkina MV, Vener AV, Rochaix J-D. Stt7-dependent phosphorylation during state transitions in the green alga Chlamydomonas reinhardtii. Mol. Cell. Proteom. 2010;9:1281–1295. doi: 10.1074/mcp.M000020-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin SH, Hong J, Li XQ, Jiang DA. Antisense inhibition of Rubisco activase increases Rubisco content and alters the proportion of Rubisco activase in stroma and thylakoids in chloroplasts of rice leaves. Ann. Bot. 2006;97:739–744. doi: 10.1093/aob/mcl025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lemeille S, et al. Analysis of the chloroplast protein kinase Stt7 during state transitions. PLoS Biol. 2009;7:e45. doi: 10.1371/journal.pbio.1000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reiland S, et al. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 2009;150:889–903. doi: 10.1104/pp.109.138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim SY, et al. The plastid casein kinase 2 phosphorylates Rubisco Activase at the Thr-78 site but is not essential for regulation of Rubisco activation state. Front. Plant Sci. 2016;7:404. doi: 10.3389/fpls.2016.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Facette MR, Shen Z, Bjornsdottir FR, Briggs SP, Smith LG. Parallel proteomic and phosphoproteomic analyses of successive stages of maize leaf development. Plant Cell. 2013;25:2798–2812. doi: 10.1105/tpc.113.112227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baginsky S. Protein phosphorylation in chloroplasts—a survey of phosphorylation targets. J. Exp. Bot. 2016;67:3873–3882. doi: 10.1093/jxb/erw098. [DOI] [PubMed] [Google Scholar]
- 65.Shen Y, Wei W, Zhou DX. Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci. 2015;20:614–621. doi: 10.1016/j.tplants.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 66.Friso G, van Wijk KJ. Posttranslational protein modifications in plant metabolism. Plant Physiol. 2015;169:1469–1487. doi: 10.1104/pp.15.01378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parsell D, Kowal A, SINGER MSM, S L. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 68.Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 69.Timperio AM, Egidi MG, Zolla L. Proteomics applied on plant abiotic stresses: role of heat shock proteins (HSP) J. Proteom. 2008;71:391–411. doi: 10.1016/j.jprot.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 70.Driedonks N, Xu J, Peters JL, Park S, Rieu I. Multi-level interactions between heat shock factors, heat shock proteins, and the redox system regulate acclimation to heat. Front. plant Sci. 2015;6:999. doi: 10.3389/fpls.2015.00999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hao Pengchao, Zhu Jiantang, Gu Aiqin, Lv Dongwen, Ge Pei, Chen Guanxing, Li Xiaohui, Yan Yueming. An integrative proteome analysis of different seedling organs in tolerant and sensitive wheat cultivars under drought stress and recovery. PROTEOMICS. 2015;15(9):1544–1563. doi: 10.1002/pmic.201400179. [DOI] [PubMed] [Google Scholar]
- 72.Vu Lam Dai, Zhu Tingting, Verstraeten Inge, van de Cotte Brigitte, Gevaert Kris, De Smet Ive. Temperature-induced changes in the wheat phosphoproteome reveal temperature-regulated interconversion of phosphoforms. Journal of Experimental Botany. 2018;69(19):4609–4624. doi: 10.1093/jxb/ery204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seo, J. H. et al. ARD1-mediated Hsp70 acetylation balances stress-induced protein refolding and degradation. Nat. Commun.7, 10.1038/ncomms12882 (2016). [DOI] [PMC free article] [PubMed]
- 74.Park YH, Seo JH, Park J-H, Lee HS, Kim K-W. Hsp70 acetylation prevents caspase-dependent/independent apoptosis and autophagic cell death in cancer cells. Int. J. Oncol. 2017;51:573–578. doi: 10.3892/ijo.2017.4039. [DOI] [PubMed] [Google Scholar]
- 75.Martin TP, Currie S, Baillie dGS. The cardioprotective role of small heat-shock protein 20. Biochem. Soc. Trans. 2014;42:270–273. doi: 10.1042/BST20130272. [DOI] [PubMed] [Google Scholar]
- 76.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trinidad JC, et al. Global identification and characterization of both O-GlcNAcylation and Phosphorylation at the murine synapse. Mol. Cell. Proteom.: MCP. 2012;11:215–229. doi: 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gray VE, Liu L, Nirankari R, Hornbeck PV, Kumar S. Signatures of natural selection on mutations of residues with multiple posttranslational modifications. Mol. Biol. Evol. 2014;31:1641–1645. doi: 10.1093/molbev/msu137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pan, Z.-C. et al. Systematic analysis of the in situ crosstalk of tyrosine modifications reveals no additional natural selection on multiply modified residues. Sci. Rep.4, 10.1038/srep07331 (2014). [DOI] [PMC free article] [PubMed]
- 80.Wu Q, et al. Suberoylanilide hydroxamic acid treatment reveals crosstalks among proteome, ubiquitylome and acetylome in non-small cell lung cancer A549 cell line. Sci. Rep. 2015;5:1–11. doi: 10.1038/srep09520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiong Y-H, Peng X-J, Cheng Z-Y, Liu W-D, Wang G-L. A comprehensive catalog of the lysine-acetylation targets in rice (Oryza sativa) based on proteomic analyses. J. Proteom. 2016;138:20–29. doi: 10.1016/j.jprot.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 82.Ahn S-H, Diaz RL, Grunstein M, Allis CD. Histone H2B deacetylation at lysine 11 is required for yeast apoptosis induced by phosphorylation of H2B at serine 10. Mol. Cell. 2006;24:211–220. doi: 10.1016/j.molcel.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 83.van Noort Vera, Seebacher Jan, Bader Samuel, Mohammed Shabaz, Vonkova Ivana, Betts Matthew J, Kühner Sebastian, Kumar Runjun, Maier Tobias, O'Flaherty Martina, Rybin Vladimir, Schmeisky Arne, Yus Eva, Stülke Jörg, Serrano Luis, Russell Robert B, Heck Albert JR, Bork Peer, Gavin Anne‐Claude. Cross‐talk between phosphorylation and lysine acetylation in a genome‐reduced bacterium. Molecular Systems Biology. 2012;8(1):571. doi: 10.1038/msb.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fischle W, Wang Y-M, Allis CD. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 85.Carlomagno Y, et al. An acetylation-phosphorylation switch that regulates tau aggregation propensity and function. J. Biol. Chem. 2017;292:15277–15286. doi: 10.1074/jbc.M117.794602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cook C, et al. Acetylation of the KXGS motifs in tau is a critical determinant in modulation of tau aggregation and clearance. Hum. Mol. Genet. 2014;23:104–116. doi: 10.1093/hmg/ddt402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang J-M, et al. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteom.: MCP. 2008;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee ME, et al. Mutational analysis of the catalytic residues lysine 230 and tyrosine 160 in the NADP+-dependent isocitrate dehydrogenase from Escherichia coli+ Biochemistry. 1995;34:378–384. doi: 10.1021/bi00001a046. [DOI] [PubMed] [Google Scholar]
- 89.Scafaro AP, et al. Heat tolerance in a wild Oryza species is attributed to maintenance of Rubisco activation by a thermally stable Rubisco activase ortholog. New Phytol. 2016;211:899–911. doi: 10.1111/nph.13963. [DOI] [PubMed] [Google Scholar]
- 90.Liu GT, et al. Differential proteomic analysis of grapevine leaves by iTRAQ reveals responses to heat stress and subsequent recovery. BMC Plant Biol. 2014;14:110. doi: 10.1186/1471-2229-14-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mertins P, et al. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods. 2013;10:634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wiśniewski JR, Mann AZM. Combination of FASP and stagetip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 2009;8:5674–5678. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- 93.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 94.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 95.Franceschini A, et al. STRINGv9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.