Abstract
Protein arginine methyltransferases (PRMTs) play diverse biological roles and are specifically involved in immune cell development and inflammation. However, their role in antiviral innate immunity has not been elucidated. Viral infection triggers the TBK1–IRF3 signaling pathway to stimulate the production of type-I interferon, which mediates antiviral immunity. We performed a functional screen of the nine mammalian PRMTs for regulators of IFN-β expression and found that PRMT6 inhibits the antiviral innate immune response. Viral infection also upregulated PRMT6 protein levels. We generated PRMT6-deficient mice and found that they exhibited enhanced antiviral innate immunity. PRMT6 deficiency promoted the TBK1–IRF3 interaction and subsequently enhanced IRF3 activation and type-I interferon production. Mechanistically, viral infection enhanced the binding of PRMT6 to IRF3 and inhibited the interaction between IRF3 and TBK1; this mechanism was independent of PRMT6 methyltransferase activity. Thus, PRMT6 inhibits antiviral innate immunity by sequestering IRF3, thereby blocking TBK1-IRF3 signaling. Our work demonstrates a methyltransferase-independent role for PRMTs. It also identifies a negative regulator of the antiviral immune response, which may protect the host from the damaging effects of an overactive immune system and/or be exploited by viruses to escape immune detection.
Keywords: PRMT6, TBK1, IRF3, IFN, antiviral innate immunity
Introduction
The innate immune system protects hosts from viral infection by eliciting a robust antiviral response through the recognition of pathogen-associated molecular patterns (PAMPs).1, 2 Diverse pattern-recognition receptors (PRRs) recognize viral nucleic acids and trigger an antiviral immune response via the induction of type-I interferon (IFN-I).3–5 Toll-like receptors (TLRs) recognize viral double-stranded RNA (dsRNA) or single-stranded RNA (ssRNA) to initiate IFN-I production.3 RIG-I-like receptors (RLRs) detect viral dsRNA or ssRNA in the cytosol to facilitate IFN-I expression through the adaptor protein MAVS,6–9 while DNA sensors detect cytosolic viral DNA to induce IFN-I production via the adaptor protein STING.10–12 Both MAVS-dependent and STING-dependent signaling cascades result in the recruitment and activation of TBK1 and the subsequent activation of the transcription factors IRF3/7 and NF-κB, leading to the production of IFN-I and proinflammatory cytokines and the induction of subsequent adaptive immune responses.1 In the host, the antiviral innate immune response should be of an appropriate duration and magnitude to efficiently eliminate the invading viruses while avoiding undesirable damage.13
The TBK1–IRF3 signaling cascade, which integrates both the RNA-sensing and DNA-sensing pathways during viral infection, is essential for the production of type-I interferon and is tightly regulated. It has been reported that multiple post-translational modifications (PTMs) are involved in modulating TBK1–IRF3 signaling.14 For example, TBK1 activity is regulated by phosphorylation, ubiquitination, and acetylation.14–18 We previously showed that Nrdp1 promotes IFN-I production in response to TLR ligands by positively regulating the K63-linked ubiquitination of TBK1.19 TBK1 can undergo K48-linked ubiquitination by NLRP4/DTX4 and TRIP, leading to its proteasomal degradation.20, 21 Recently, we found that Dnmt3a upregulates HDAC9 to modify TBK1 acetylation in macrophages during viral infection.22 The transcription factor IRF3 is also tightly regulated: Pin1 negatively regulates IRF3 activity through polyubiquitination-mediated proteasome-dependent degradation,23 while PTEN, a tumor suppressor, controls IRF3 nuclear import by dephosphorylating Ser97.24 However, many mechanistic details regarding the activation and regulation of IRF3 remain unclear.
Three types of arginine methylation are catalyzed by nine protein arginine methyltransferases (PRMTs) in mammals.25 PRMTs are ubiquitously expressed and govern important and diverse cellular processes, including gene expression and pre-mRNA splicing.25–27 Accumulating evidence has shown that dysregulation of PRMTs drives the initiation and progression of several types of cancers.28 Arginine methylation also plays critical roles in the establishment and maintenance of the lymphoid and myeloid lineages and in inflammatory responses.25, 29–31 The differentiation and expansion of hematopoietic stem cells or progenitors are significantly affected by deficiencies in CARM1/PRMT4 and PRMT5.25, 30 PRMT5 is also required for lymphocyte development.30 In addition, PRMT1 and PRMT7 are required for B-cell development.32, 33 Recent reports have demonstrated a correlation between PRMTs and the inflammatory response, mainly with respect to the regulation of NF-κB.31 Although accumulating evidence has linked PRMTs with diverse cellular functions and diseases, the role of PRMTs in innate immunity, especially antiviral immunity, needs to be fully investigated.
PRMT6 catalyzes the methylation of H3R2 and H2AR29 to suppress gene expression.34–36 Recent studies have revealed that PRMT6 plays an important role in cancers28, 37 and acts as a coactivator of NF-κB to facilitate transcription.38 However, the role of PRMT6 in virus-induced innate immune signaling and type-I interferon production remains unknown. Here, we show that PRMT6 is an inhibitor of the TBK1–IRF3 signaling cascade and plays a role in attenuating the antiviral immune response.
Materials and methods
Mice
Prmt6-deficient mice were generated using the CRISPR–Cas9 gene-editing system. Two gRNAs (5′- ACG AAT CCC AGC AGG CCC CG-3′ and 5′-GAG ATC GCC TAT GCA AGT TG-3′) were designed to target the single Prmt6-encoding exon (chromosome 3). Cas9 mRNA and gRNAs were microinjected into the fertilized eggs of C57BL/6J mice to obtain F0 generation mice, and then the sequence-validated F0 generation mice mated with C57BL/6J mice to obtain positive F1 generation heterozygous mice. Heterozygous mice were bred and viable homozygotes were obtained. Genomic DNA was isolated from tails and analyzed by PCR amplification using the following primers: 5′-TTA ATC CCA GGA AAC TTG AGA CA-3′ and 5′-GAT TAG CTC GTG GAC CAA TTA T-3′. All mice were bred in pathogen-free conditions. All animal experiments were performed according to the National Institute of Health Guide for the Care and Use of Laboratory Animals with approval from the Scientific Investigation Board of Second Military Medical University, Shanghai.
Cell culture
Bone marrow-derived macrophages (BMDMs) were prepared as described previously.39 HEK293T, THP-1, A549, and Raw264.7 cell lines were obtained from the American Type Culture Collection. All cells were cultured in either DMEM or RPMI-1640 supplemented with 10% fetal bovine serum (Gibco) in a 5% CO2 atmosphere at 37 °C.
Antibodies
Antibodies against RIG-I, TBK1, p65, IRF3, JNK, ERK, p38, lamin A/C, HA-tag, Myc-tag (HRP-conjugated), Flag-tag (HRP-conjugated), and phosphor-specific antibodies against IRF3(Ser396), p65(Ser536), p38(Thr180/Tyr182), ERK(Thr202/Tyr204), and JNK(Thr183/Tyr185) were purchased from Cell Signaling Technology. Antibodies against PRMT5 and PRMT6 were from Abcam and Novus, respectively. The V5-tag antibody was from Santa Cruz. The agarose beads used for immunoprecipitation were from Sigma-Aldrich. Fluorescent anti-mouse CD11b PerCP–Cy5.5 (clone: M1/70) and anti-mouse Cd19 APC-eFluor 780 (clone: eBio1D3) antibodies were from eBioscience. Fluorescent BV605 anti-mouse CD45 (clone: 30-F11), FITC anti-mouse Ly-6G (clone: 1A8), AF700 anti-mouse Ly-6C (clone: HK1.4), and PE/Cy7 anti-mouse CD3ε (clone: 145-2C11) antibodies were from BioLegend.
Plasmid constructs and transfection
cDNA fragments encoding mouse PRMT1–9, IRF3, and TBK1 were amplified from Raw264.7 cells and cloned into separate pcDNA3.1 eukaryotic expression vectors. Deleted, truncated, and point-mutated PRMT6 constructs were generated by PCR-based amplification using the wild-type PRMT6 protein-coding construct as the template. All constructs were confirmed by sequencing. Plasmids were transiently transfected into HEK293T cells using jetPEI reagents (Polyplus Transfection) according to the manufacturer’s instructions.
Luciferase reporter assay
HEK293T cells were seeded into 24-well plates at a density of 2 × 105 cells/well. After 24 h, cells were transfected with the IFN-β reporter and expression plasmids, as well as the Renilla luciferase plasmid as an internal control. After 24 h of transfection, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
RNA quantification
Total RNA was extracted with Trizol reagent (Transgen) and reverse-transcribed using the Reverse Transcription System (Promega). Quantitative real-time RT-PCR analysis was performed on a LightCycler (Roche) with the SYBR RT-PCR kit (Takara) as described previously.40, 41 Data were normalized to Gapdh expression. The primers used for real-time RT-PCR were as follows: Gapdh (forward: 5′-AGG TCG GTG TGA ACG GAT TTG-3′, reverse: 5′-TGT AGA CCA TGT AGT TGA GGT CA-3′), Prmt6 (forward: 5′-GAT GGG CTA CGG ACT TCT GC -3′, reverse: 5′-GCA TCT GGT CGC TAA TCG GG-3′), Ifna (forward: 5′-TAC TCA GCA GAC CTT GAA CCT-3′, reverse: 5′-CAG TCT TGG CAG CAA GTT GAC-3′), Ifnb (forward: 5′-ATG AGT GGT GGT TGC AGG C-3′, reverse: 5′-TGA CCT TTC AAA TGC AGT AGA TTC A-3′), Il6 (forward: 5′-TAG TCC TTC CTA CCC CAA TTT CC-3′, reverse: 5′-TTG GTC CTT AGC CAC TCC TTC-3′), and VSV (forward: 5′-ACG GCG TAC TTC CAG ATG G-3′, reverse: 5′-CTC GGT TCA AGA TCC AGG T-3′).
ELISA
Secreted cytokines in cell culture supernatants or sera from virus-infected mice were analyzed using mouse IFN-α and IFN-β (PBL Biomedical Laboratories) and mouse TNF and IL-6 (R&D Systems) ELISA kits according to the manufacturer’s instructions.
Immunoprecipitation and immunoblotting
Total protein was extracted with cell lysis buffer (Cell Signaling Technology) containing a protease inhibitor “cocktail” (Calbiochem) and 1 mM phenylmethylsulfonyl fluoride (PMSF). Protein concentration was measured using a BCA assay (Pierce). Equivalent protein amounts were used for western blotting or immunoprecipitation as previously described.40, 41
In vitro kinase assay
The TBK1–Myc construct was cotransfected into HEK293T cells with PRMT6 expression vectors. Then TBK1–Myc was immunoprecipitated for a subsequent kinase assay as described previously.22
Nuclear and cytoplasmic extraction
Nuclear and cytoplasmic extraction was conducted with NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) according to the manufacturer’s instructions. The extracted fractions were used for immunoblotting analysis.
In vivo experiments and tissue staining
Age- and sex-matched groups of littermate mice were intraperitoneally infected with VSV (1.5 × 108 plaque-forming units per gram of body weight). Hematoxylin-and-eosin staining was performed as described previously.22
Flow cytometry
The phenotypes and proportions of neutrophils, monocytes, T cells, and B cells in the blood and spleen of Prmt6+/+ and Prmt6–/– mice were determined by flow cytometry.
Statistical analysis
Data are presented as the mean ± SEM. Statistical tests were performed using Student’s t test, one-way ANOVA, and the generalized Wilcoxon test. p < 0.05 was considered significant and denoted *p < 0.05, **p < 0.01, and ***p < 0.001.
Results
Identification of PRMT6 as a negative regulator of type-I interferon expression
To identify a potential role for PRMTs in antiviral innate immunity, we conducted a functional screen using the nine PRMT members. We individually overexpressed each of the nine PRMTs in HEK293T cells along with an IFN-β luciferase reporter, as well as key signaling proteins that activate interferon (IFN) transcription. We found that each PRMT had a different effect on type-I interferon expression (Fig. 1a). Notably, PRMT6 most potently inhibited the expression of the IFN-β luciferase reporter induced by RIG-I-2CARD, MAVS, TBK1, and IRF3–5D (Fig. 1a). We next assessed the function of PRMT6 in Raw264.7 cells. In Raw264.7 cells stably overexpressing PRMT6, we found that IFN-I mRNA levels, including IFN-α and IFN-β, were markedly reduced upon VSV infection compared with control cells (Fig. 1b). Immunoblotting analysis showed that PRMT6 overexpression inhibited the phosphorylation of IRF3 upon VSV infection (Fig. 1c). In addition, we transiently overexpressed PRMT6 in NIH3T3 mouse embryonic fibroblast cells and A549 human lung adenocarcinoma cells and then stimulated these cells with VSV; these experiments again demonstrated that PRMT6 inhibited VSV-induced IRF3 phosphorylation (Fig. S1a, b). Collectively, these results suggest that PRMT6 negatively regulates type-I interferon expression during antiviral innate immune responses.
Fig. 1.
Identification of PRMT6 as a negative regulator of type-I interferon expression. a Luciferase activity in HEK293T cells transfected with IFN-β luciferase reporter, the indicated signal proteins, and PRMT1–9. The results are expressed relative to Renilla luciferase activity. b Real-time PCR analysis of Ifna and Ifnb mRNA in control Raw264.7 cells and Raw264.7 cells stably overexpressing PRMT6 infected with VSV for the indicated lengths of time. The results are normalized to Gapdh expression. c Immunoblot analysis of total and phosphorylated IRF3 in control Raw264.7 cells and PRMT6-overexpressing Raw264.7 cells infected with VSV for the indicated lengths of time. d–h Immunoblot analysis of PRMT6 expression in mouse peritoneal macrophages (d, e), THP-1 cells (f, g), and A549 cells (h) infected with the virus for the indicated lengths of time. i Immunoblot analysis of PRMT6 expression in HBV-transfected HepG2.2.15 cells. Data in a and b are presented as the mean ± SEM of three independent experiments. Data in c–i are representative of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
Viral infections usually result in altered expression of target genes, especially those regulating antiviral innate immunity. Immunoblot analysis of virus-infected cells demonstrated that VSV and HSV-1 infection markedly increased PRMT6 expression in mouse primary peritoneal macrophages and human THP-1 cells (Fig. 1d–g). VSV infection also greatly increased PRMT6 expression in human A549 cells (Fig. 1h). We further analyzed the expression of PRMT6 in HBV-transfected HepG2.2.15 cells and found increased PRMT6 expression in these cells compared to the parental HepG2 cells (Fig. 1i). However, PRMT6 mRNA was not significantly altered upon VSV and HSV-1 infection in macrophages (Fig. S2). Altogether, these data indicate that inducible expression of PRMT6 negatively regulates antiviral innate immunity.
PRMT6 deficiency enhances antiviral innate immunity in vivo
To investigate the functional significance of PRMT6 in the host antiviral response in vivo, we generated PRMT6-deficient (Prmt6–/–) mice using the CRISPR–Cas9 gene-editing system (Fig. S3a–S3d). Prmt6–/– mice were comparable to littermate (Prmt6+/+) mice in terms of body weight and spleen weight and did not exhibit any obvious differences in immune cell populations (Fig. S3e–S3g). We challenged the mice with VSV (1.5 × 108 plaque-forming units [PFU]/g) and found that Prmt6–/– mice were significantly more resistant to VSV infection (as measured by overall survival) than Prmt6+/+ mice (Fig. 2a). The VSV loads in the liver, spleen, and lung homogenate supernatants from Prmt6–/– mice were significantly lower than those from Prmt6+/+ mice 18 h post infection (Fig. 2b). Q-PCR analysis revealed decreased VSV RNA levels in the infected organs of Prmt6–/– mice (Fig. 2c). In addition, we observed less infiltration of inflammatory cells into the lungs of Prmt6–/– mice than in the lungs of Prmt6+/+ mice following infection (Fig. 2d). Furthermore, Prmt6–/– mice produced significantly higher levels of IFN-α, IFN-β, and IL-6 than Prmt6+/+ mice in response to VSV infection (Fig. 2e). Altogether, these results reveal that PRMT6 deficiency enhances the antiviral innate immune response in vivo.
Fig. 2.
PRMT6 deficiency enhances antiviral innate immunity in vivo. a Survival of Prmt6+/+ and Prmt6–/– mice after intraperitoneal injection of VSV (1.5 × 108 plaque-forming units per gram of body weight) (n = 7). b Determination of VSV load in the liver, spleen, and lungs of Prmt6+/+ and Prmt6–/– mice 18 h after infection with VSV, as measured by TCID50 assay. c Real-time PCR analysis of VSV RNA levels in liver, spleen, and lungs of Prmt6+/+ and Prmt6–/– mice 18 h after intraperitoneal injection with a medium or VSV. d Hematoxylin-and-eosin staining of lung sections from mice in C. Scale bar, 50 μm. e ELISA demonstrating serum cytokine levels in Prmt6+/+ and Prmt6–/– mice after intraperitoneal injection of VSV for the indicated lengths of time. Data in a and d are representative of three independent experiments. Data in b, c, and e are presented as the mean ± SEM of three independent experiments. *p < 0.05, **p < 0.01
PRMT6 deficiency promotes IRF3 activation and increases type-I interferon production
To further determine the function of PRMT6 in primary cells, we prepared bone marrow-derived macrophages (BMDMs) from Prmt6+/+ and Prmt6–/– mice. Upon VSV or HSV-1 infection, Prmt6–/– BMDMs produced significantly more IFN-α, IFN-β, and IL-6 than Prmt6+/+ BMDMs at both the mRNA and protein levels (Fig. 3a–d). These data indicate that PRMT6 inhibits the antiviral response in mouse BMDMs upon both RNA and DNA virus infection. We then analyzed the signaling pathways responsible for the induction of IFN-I and found that PRMT6 deficiency increased the activation of IRF3 (Fig. 3e), consistent with our earlier results in PRMT6-overexpressing Raw264.7 cells. The activation of mitogen-activated protein kinases (MAPKs) and NF-κB signaling was not affected by PRMT6 deficiency in VSV-infected mouse BMDMs. Interestingly, PRMT6 deficiency had no observable impact on the phosphorylation of TBK1, the binding partner of IRF3. Thus, PRMT6 deficiency promotes antiviral innate immunity by increasing IRF3 activation and type-I interferon production upon viral infection.
Fig. 3.
PRMT6 deficiency promotes IRF3 activation and increases IFN-I production. a, b Real-time PCR analysis of Ifna, Ifnb, and Il6 mRNA in Prmt6+/+ and Prmt6–/– BMDMs infected with VSV (a) and HSV-1(b) for the indicated lengths of time. c ELISA demonstrating cytokine levels in supernatants from Prmt6+/+ and Prmt6–/– BMDMs infected with VSV for the indicated lengths of time. d ELISA demonstrating cytokine levels in supernatants from Prmt6+/+ and Prmt6–/– BMDMs infected with HSV-1. e Immunoblot analysis of phosphorylation of the indicated molecules in Prmt6+/+ and Prmt6–/– BMDMs infected with VSV for the indicated lengths of time. Data in a–d are presented as the mean ± SEM of three independent experiments. Data in e are representative of three independent experiments. **p < 0.01
PRMT6 inhibits IFN-I expression in a methyltransferase-independent manner
To define the mechanism by which PRMT6 regulates antiviral innate immunity, we performed a luciferase reporter assay in which we overexpressed key signaling proteins that activate IFN transcription and increasing amounts of PRMT6 vectors in HEK293T cells. We found that PRMT6 inhibited luciferase reporter expression induced by the indicated signaling proteins in a dose-dependent manner (Fig. 4a).
Fig. 4.
PRMT6 inhibits IFN-I expression in a methyltransferase-independent manner. a Luciferase activity in HEK293T cells transfected with the IFN-β luciferase reporter, the indicated adaptor proteins, and increasing amounts of PRMT6. The results are expressed relative to Renilla luciferase activity. b Luciferase activity in HEK293T cells transfected with the IFN-β luciferase reporter, the indicated adaptor proteins, and the empty vector (EV), PRMT6, or the catalytically inactive PRMT6 (PRMT6(dead)). The results are expressed relative to Renilla luciferase activity. Data in a and b are presented as the mean ± SEM of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
Methyltransferase activity is important for many physiological functions of PRMT family members. Many histone and non-histone proteins have recently been identified as substrates of PRMT6.28, 37 Thus, we investigated whether the function of PRMT6 in antiviral immunity depends on its methyltransferase activity. Several studies have reported a catalytically inactive form of PRMT6 (PRMT6(dead)), which carries the mutation V86-L-D88 to K–L–A.38, 42 Therefore, we overexpressed key signaling proteins, either catalytically active or inactive PRMT6, and the IFN-β luciferase reporter in HEK293T cells. Unexpectedly, we found that both PRMT6 and PRMT6(dead) overexpression significantly inhibited luciferase reporter activity induced by all of the signaling proteins tested (Fig. 4b). These data indicate that the inhibitory effect of PRMT6 on IFN-I expression does not depend on its methyltransferase activity.
PRMT6 targets the TBK1–IRF3 signaling complex
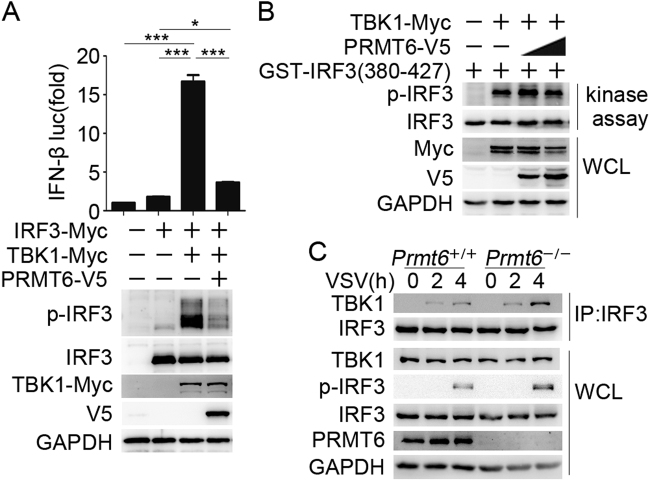
The TBK1–IRF3 signaling cascade is a common pathway that is activated by the adaptor proteins TRIF, MAVS, and STING. Our data demonstrating that PRMT6 inhibits TRIF-induced, MAVS-induced, and STING-induced IFN transcription suggested that PRMT6 may target the TBK1 and IRF3 signaling complex. To test this, we co-overexpressed TBK1, IRF3, and PRMT6 together with the IFN-β luciferase reporter in HEK293T cells. As expected, expression of the luciferase reporter was potently upregulated upon co-overexpression of TBK1 and IRF3. However, luciferase reporter expression was significantly inhibited in the presence of PRMT6 (Fig. 5a). Consistent with the decrease in luciferase activity, PRMT6 overexpression also decreased IRF3 phosphorylation (Fig. 5a). These results indicate that PRMT6 inhibits antiviral innate immunity by targeting the TBK1–IRF3 signaling complex.
Fig. 5.
PRMT6 targets the TBK1–IRF3 signaling complex. a Luciferase activity (top) and immunoblot analysis (below) of HEK293T cells transfected with IFN-β luciferase reporter and the indicated vectors. b In vitro kinase assay demonstrating the levels of phosphor-IRF3 (Ser396) and total IRF3 in HEK293T cells transfected with GST–IRF3(380–427) as the substrate and the indicated combinations of TBK1–Myc and PRMT6–V5. Immunoblot analysis was used to assess proteins immunoprecipitated with an anti-Myc antibody or GST-fused IRF3 peptide (top) as well as whole-cell lysates (below). c Co-immunoprecipitation and immunoblot analysis of Prmt6+/+ and Prmt6–/– BMDMs infected with VSV for the indicated lengths of time. Data in a–c are representative of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
The above data showed that TBK1 activation was unchanged, while IRF3 phosphorylation was enhanced in VSV-infected PRMT6-deficient BMDMs (Fig. 3e). Furthermore, PRMT6 functioned in a methyltransferase-independent manner (Fig. 4b). These data implied that PRMT6 instead influences signal transduction from TBK1 to IRF3. To explore this, we first tested whether PRMT6 affects the kinase activity of TBK1. We transfected HEK293T cells with TBK1 and increasing amounts of PRMT6 expression vectors and then immunoprecipitated TBK1. A glutathione S-transferase (GST)-fused IRF3 peptide (IRF3 amino acids 380–427) was used as a substrate for the immunoprecipitated TBK1 in a kinase assay. Immunoblotting analysis showed that IRF3 phosphorylation by TBK1 was not affected in the presence of PRMT6 (Fig. 5b), indicating that PRMT6 does not affect the kinase activity of TBK1. Thus, we further investigated the interaction between TBK1 and IRF3 in Prmt6–/– BMDMs upon VSV infection. We found that the TBK1–IRF3 interaction was enhanced and that consequently, IRF3 phosphorylation was increased in Prmt6–/– BMDMs compared to Prmt6+/+ BMDMs upon VSV infection (Fig. 5c). Taken together, these results indicate that PRMT6 negatively regulates the antiviral innate immune response by targeting the assembly of the TBK1–IRF3 signaling complex without affecting the kinase activity of TBK1.
PRMT6 disrupts the assembly of the TBK1–IRF3 signaling complex by binding and sequestering IRF3
To determine how PRMT6 modulates the TBK1–IRF3 interaction, we analyzed the molecular targets of PRMT6. We overexpressed PRMT6 with TBK1, IKKε, IRF3, or IRF7 in HEK293T cells. Co-immunoprecipitation followed by immunoblotting revealed that PRMT6 interacts with IRF3 but not with TBK1, IKKε, or IRF7 (Fig. 6a, b). To examine the interaction between endogenous PRMT6 and IRF3 under physiological conditions, we first determined the cellular localization of PRMT6. By analyzing nuclear and cytoplasmic cellular fractions, we found that PRMT6 was mainly localized to the cytoplasm in both human THP-1 cells and mouse BMDMs (Fig. S4a, b). In addition, microscopic analysis showed that overexpressed PRMT6 was also mainly localized to the cytoplasm of HEK293T cells (Fig. S4c). Co-immunoprecipitation followed by immunoblotting revealed that PRMT6 constitutively interacted with IRF3 in unstimulated BMDMs (Fig. 6c). Following VSV infection, the interaction between PRMT6 and IRF3 was enhanced, and the bound IRF3 was weakly phosphorylated (Fig. 6c).
Fig. 6.
PRMT6 binds and sequesters IRF3 to disrupt assembly of the TBK1–IRF3 signaling complex. a Co-immunoprecipitation and immunoblot analysis of HEK293T cells transfected with PRMT6–V5 and Myc-tagged TBK1, IKKε, IRF3, or IRF7. b Co-immunoprecipitation and immunoblot analysis of HEK293T cells transfected with PRMT6–V5 and IRF3-Flag. c Co-immunoprecipitation and immunoblot analysis of BMDMs infected with VSV for the indicated lengths of time. d Co-immunoprecipitation and immunoblot analysis of HEK293T cells transfected with IRF3-Flag, TBK1–Myc, and PRMT6 or PRMT6(dead). e Co-immunoprecipitation and immunoblot analysis of BMDMs infected with VSV for the indicated lengths of time. Data in a–e are representative of three independent experiments
We then investigated whether the binding of PRMT6 to IRF3 affected the interaction between TBK1 and IRF3. We overexpressed TBK1, IRF3, and PRMT6 in HEK293T cells. Co-immunoprecipitation followed by immunoblotting revealed that TBK1 strongly interacts with IRF3 to promote IRF3 activation. This interaction and the activation of IRF3 were markedly reduced by PRMT6 overexpression (Fig. 6d). Interestingly, PRMT6(dead) had the same effect as wild-type PRMT6 (Fig. 6d), which was consistent with the methyltransferase-independent role of PRMT6 in the inhibition of innate immune signaling and IFN-I expression. We further analyzed the dynamic interactions between IRF3, TBK1, and PRMT6 under physiological conditions at various time points after VSV infection. In resting BMDMs, TBK1 was not associated with IRF3, but a small amount of PRMT6 was constitutively associated with IRF3 (Fig. 6e). After infection with VSV, the TBK1–IRF3 signaling complex was assembled, triggering the antiviral innate response; TBK1–IRF3 association peaked at 4 h post infection and then weakened and returned to resting levels (Fig. 6e). The interaction of PRMT6 with IRF3 was maintained during the first 4 h of VSV infection, was enhanced beginning at 6 h, peaked at 8 h post infection, and then returned to resting levels (Fig. 6e). Thus, the enhanced interaction of PRMT6 and IRF3 can inhibit the TBK1–IRF3 interaction. This dynamic interaction between IRF3, TBK1, and PRMT6 allowed the antiviral innate immune response to be finely triggered and tightly regulated. Altogether, these results suggest that PRMT6 binds and sequesters IRF3, blocking the TBK1–IRF3 interaction and inhibiting the activation of IRF3 and the subsequent production of IFN-I upon viral infection.
The AA189–318 domain of PRMT6 is critical for blocking the TBK1–IRF3 interaction
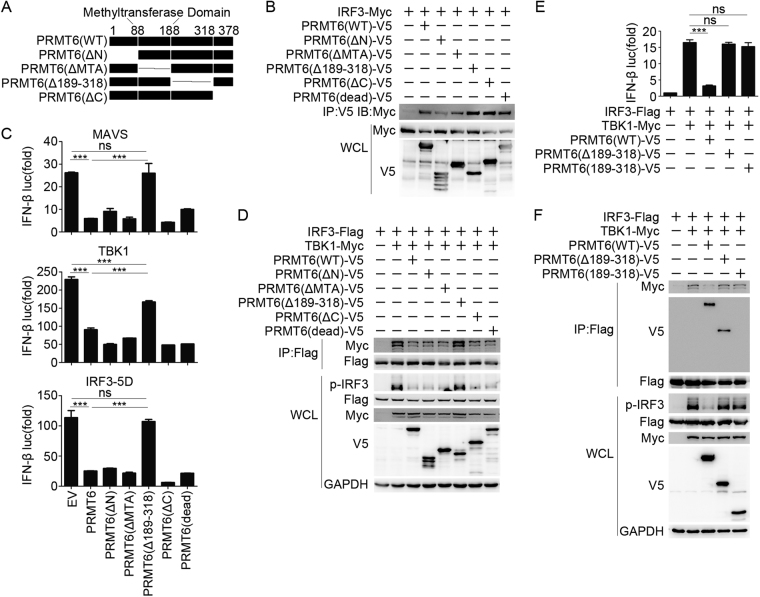
To better identify the specific domain of PRMT6 responsible for mediating the inhibition of TBK1–IRF3 signaling, we constructed PRMT6 deletion mutants according to its domain structure (Fig. 7a). We first tried to determine which domain of PRMT6 is responsible for the interaction with IRF3 and found that the N-terminal domain of PRMT6 was sufficient for the interaction with IRF3 (Fig. 7b). We also found that the domain deletion mutants of PRMT6 exhibited altered dimerization (Fig. S5); however, it seemed that the binding of PRMT6 to IRF3 was not affected by the ability of PRMT6 to dimerize (Fig. 7b). Additionally, we found that the domain deletion mutants of IRF3 failed to interact with PRMT6 (Fig. S6), indicating that the complete conformational structure of IRF3 is critical for PRMT6 binding.
Fig. 7.
PRMT6 blocks TBK1–IRF3 signaling in a manner dependent on its 189–318 amino acid domain. a Domain structure and domain deletion mutants of PRMT6. b Co-immunoprecipitation and immunoblot analysis of HEK293T cells transfected with IRF3–Myc and PRMT6 mutants. c Luciferase activity in HEK293T cells transfected with IFN-β luciferase reporter, the indicated adaptor proteins, and empty vector (EV), PRMT6, or PRMT6 mutants. The results are expressed relative to Renilla luciferase activity. d Co-immunoprecipitation and immunoblot analysis of HEK293T cells transfected with IRF3-Flag, TBK1–Myc, and PRMT6 or PRMT6 mutants. e Luciferase activity in HEK293T cells transfected with IFN-β luciferase reporter, IRF3-Flag, TBK1–Myc, and PRMT6(WT), PRMT6(Δ189–318), or PRMT6(189–318). The results are expressed relative to Renilla luciferase activity. f Co-immunoprecipitation and immunoblot analysis of HEK293T cells transfected with IRF3-Flag, TBK1–Myc, and PRMT6 or PRMT6 mutants. Data in c and e are presented as the mean ± SEM of three independent experiments. Data in b, d, and f are representative of three independent experiments. ***p < 0.001
We then overexpressed PRMT6 mutants together with key signaling proteins and the IFN-β luciferase reporter in HEK293T cells. We found that PRMT6(Δ189–318) rescued the expression of the luciferase reporter induced by MAVS, TBK1, and IRF3–5D overexpression (Fig. 7c). PRMT mutants, including one with deletion of the methyltransferase domain (MTA), inhibited luciferase activity to the same level as wild-type PRMT6 (Fig. 7c). Additionally, co-immunoprecipitation followed by immunoblotting showed that PRMT6 blocked the TBK1–IRF3 interaction and subsequent IRF3 phosphorylation; PRMT6(Δ189–318) rescued these effects, while other mutants of PRMT6 failed to do so (Fig. 7d). These data indicate that the 189–318 amino acids of PRMT6 are critical for blocking the TBK1–IRF3 interaction and inhibiting type-I interferon production, indicating a unique mechanism of TBK1–IRF3 signaling modulation.
To better understand how PRMT6 blocks TBK1–IRF3 signaling via its AA189–318 domain, we examined the conservation of this sequence. We found that these amino acids are highly conserved between mouse and human PRMT6 and that this region is quite different from that found in other PRMTs (Fig. S7). Thus, we cloned the 189–318 amino acids of PRMT6 (PRMT6(189–318)) and co-overexpressed TBK1, IRF3, and PRMT6 mutants together with the IFN-β luciferase reporter in HEK293T cells. Unexpectedly, PRMT6(189–318) failed to inhibit the expression of the luciferase reporter induced by TBK1 and IRF3 co-overexpression (Fig. 7e). Further co-immunoprecipitation and immunoblotting showed that PRMT6 bound IRF3 and blocked the TBK1–IRF3 interaction and subsequent IRF3 activation. Although PRMT6(Δ189–318) and WT PRMT6 bound equally well to IRF3, the mutant failed to inhibit the TBK1–IRF3 interaction and IRF3 activation. PRMT6(189–318) neither failed to bind IRF3 nor blocked the TBK1–IRF3 interaction and IRF3 activation (Fig. 7f). Collectively, these data indicate that both the N-terminal domain and the AA189–318 domain of PRMT6 are required to block the TBK1–IRF3 interaction and IRF3 activation: the N-terminal domain of PRMT6 binds to IRF3, and the AA189–318 domain may act to block the interaction between TBK1 and IRF3.
IRF3–5D is constitutively active and has been widely used for IFN gene induction. Our earlier data showed that PRMT6 could inhibit the expression of the IFN-β luciferase reporter induced by IRF3–5D (Figs. 1a, 4, and 7c). To determine the possible mechanism of this inhibition, we co-overexpressed IRF3–5D and PRMT6 in HEK293T cells and found that PRMT6 could bind to IRF3–5D (Fig. S8a). Furthermore, PRMT6 overexpression decreased the nuclear translocation of IRF3–5D and increased IRF3–5D retention in the cytoplasm (Fig. S8b). These results suggest that PRMT6 can inhibit the nuclear translocation of IRF3–5D.
Discussion
In this report, we identified PRMT6 as a virally induced protein that negatively regulates antiviral innate immunity. We found that PRMT6 deficiency enhanced the antiviral immune response by promoting the activation of IRF3 and the production of type-I interferon. Mechanistically, PRMT6 acted as an inhibitor of TBK1–IRF3 signaling to prevent IRF3 activation. Therefore, our results provide new insight into the regulation of TBK1–IRF3 signaling and present a PRMT molecule as a key regulator of antiviral innate immunity.
The function of PRMT6 in gene expression has been largely validated. PRMT6-mediated methylation of H3R2 and H2AR29 induces repression of gene transcription. Furthermore, PRMT6 plays oncogenic roles in several types of cancers, typically through the regulation of p53/p21 gene expression or the modulation of p21 localization by direct arginine methylation.42–44 However, we have demonstrated a methyltransferase-independent role for PRMT6 in the regulation of antiviral innate immunity, which is dependent on its N-terminal domain and AA189–318 domain. The N-terminal domain of PRMT6 binds to IRF3, and its AA189–318 domain may act as an inhibitor of the TBK1–IRF3 interaction, thus displaying a unique method of TBK1–IRF3 signaling modulation. These findings also provide a new understanding of the function of PRMTs in different cellular processes.
The TBK1–IRF3 signaling cascade is the main pathway responsible for the induction of type-I interferon production. The activities of TBK1 and IRF3 have been reported to be under precise control through distinct mechanisms.14 PTMs, including phosphorylation, ubiquitination, and acetylation, stand out as the critical mechanism, by which the activation of TBK1 and IRF3 is regulated during viral infection. Other studies have also revealed distinct mechanisms by which TBK1–IRF3 signaling can be modulated: MSX1 modulates RLR-mediated activation of IRF3 by facilitating the assembly of a TBK1-associated complex;45 S6K enhances antiviral immunity by facilitating the recruitment of IRF3 to STING in response to DNA virus infection in BMDCs;46 and cFLIPL inhibits IFN-I production by interrupting the interaction between IRF3, CBP, and target DNA.47 In contrast, our study found that PRMT6 acts to block TBK1–IRF3 signaling: the binding of PRMT6 to IRF3 significantly inhibited the viral infection-induced TBK1–IRF3 interaction. These findings reveal a new strategy to attenuate the antiviral immune response, which may be an important mechanism to check the potential destructive effects of an overactive immune system on the host. Our findings also lead us to question whether viruses have evolved this strategy to escape the host immune response. Although this needs further investigation, there are some preliminary data supporting this model. According to published gene-profiling data, PRMT6 expression is upregulated in whole blood cells of dengue hemorrhagic fever and dengue fever patients compared with healthy control volunteers, though PRMT6 expression returns to normal levels in convalescent patients (Fig. S9a; GEO accession no. GDS5093). In addition, PRMT6 is upregulated in Nef-expressing CD4+ T cells (Nef is a critical factor involved in simian immunodeficiency virus replication) (Fig. S9b; GEO accession no. GDS2164). Moreover, HPV-positive head and neck squamous cell carcinoma tumors also show a slightly upregulated PRMT6 expression (Fig. S9c; GEO accession no. GDS1667). These data indicate that PRMT6 is upregulated during viral infection, possibly helping the virus to inhibit the immune response and escape immune system detection. The elevated PRMT6 expression returns to normal when effective treatments are used and invading viruses are eliminated. These data, together with our results, suggest an important potential connection between PRMT6 and human infectious diseases and indicate that the targeting of PRMT6 has possible clinical applications in the treatment of viral infectious diseases.
Arginine methylation governs important cellular processes that affect many aspects of cellular functions. Using hydrophilic interaction liquid chromatography (HILIC) coupled to heavy methyl-stable isotope labeling by amino acids in cell culture (SILAC)-based mass spectrometry, researchers have found that the arginine-methylated proteins were mainly implicated in endosomal trafficking, chromatin remodeling, transcription, RNA processing, and translation in T cells.48 The arginine-methylated proteins in mouse brains and embryos showed a diverse distribution of protein functions, as revealed by immunoaffinity purification coupled with liquid chromatography–tandem mass spectrometry (IAP–LC–MS/MS).49 Although PRMT6 was the most powerful negative regulator of all of the PRMTs in our functional screening experiment, other PRMT members may be able to modulate innate immunity to some extent. On the other hand, arginine methylation of key adaptors in the innate immune response has not been identified. Thus, it is of great value to define the role of PRMTs and arginine methylation in antiviral innate immunity.
In addition to their important roles in antiviral innate immunity, TBK1 and IRF3 are also involved in many other cellular processes. TBK1 and IRF3 are ubiquitously expressed in many cell types, including cancer cells. Given that PRMT6 also plays a role in cancer, our findings regarding the mechanisms of PRMT6 may also be relevant in cancer, though this will require further investigation.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the National Key R&D program of China (2018YFA0507401), National Natural Science Foundation of China (31390431, 31522019, 81471568, 80178104, and 31770945), and the CAMS Innovation Fund for Medical Sciences (2016-12M-1-003). We thank Ms. Xiaofei Li for technical assistance and Life Science Editors for editing assistance.
Author contributions
X.C. designed and supervised the study. H.Z., C.H., T.L., and N.L. performed the experiments. H.Z., C.H., and X.C. analyzed the data and wrote the paper.
Competing interests
The authors declare no competing financial interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hua Zhang, Chaofeng Han.
Change history
11/27/2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Electronic supplementary material
The online version of this article (10.1038/s41423-018-0057-4) contains supplementary material.
References
- 1.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 2.Cao X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 2016;16:35–50. doi: 10.1038/nri.2015.8. [DOI] [PubMed] [Google Scholar]
- 3.Gurtler C, Bowie AG. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013;21:413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loo YM, Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goubau D, et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5’-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol. Cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 11.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen K, Liu J, Cao X. Regulation of type I interferon signaling in immunity and inflammation: a comprehensive review. J. Autoimmun. 2017;83:1–11. doi: 10.1016/j.jaut.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Qian C, Cao X. Post-translational modification control of innate immunity. Immunity. 2016;45:15–30. doi: 10.1016/j.immuni.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 15.Li X., et al. The tyrosine kinase Src promotes phosphorylation of the kinase TBK1 to facilitate type I interferon production after viral infection. Sci Signal10, eaae0435 (2017). [DOI] [PubMed]
- 16.Gabhann JN, et al. Absence of SHIP-1 results in constitutive phosphorylation of tank-binding kinase 1 and enhanced TLR3-dependent IFN-beta production. J. Immunol. 2010;184:2314–2320. doi: 10.4049/jimmunol.0902589. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, et al. PPM1B negatively regulates antiviral response via dephosphorylating TBK1. Cell. Signal. 2012;24:2197–2204. doi: 10.1016/j.cellsig.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCoy CE, Carpenter S, Palsson-McDermott EM, Gearing LJ, O’Neill LA. Glucocorticoids inhibit IRF3 phosphorylation in response to Toll-like receptor-3 and -4 by targeting TBK1 activation. J. Biol. Chem. 2008;283:14277–14285. doi: 10.1074/jbc.M709731200. [DOI] [PubMed] [Google Scholar]
- 19.Wang C, et al. The E3 ubiquitin ligase Nrdp1 ‘preferentially’ promotes TLR-mediated production of type I interferon. Nat. Immunol. 2009;10:744–752. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- 20.Cui J, et al. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat. Immunol. 2012;13:387–395. doi: 10.1038/ni.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, et al. TRAF-interacting protein (TRIP) negatively regulates IFN-beta production and antiviral response by promoting proteasomal degradation of TANK-binding kinase 1. J. Exp. Med. 2012;209:1703–1711. doi: 10.1084/jem.20120024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, et al. Methyltransferase Dnmt3a upregulates HDAC9 to deacetylate the kinase TBK1 for activation of antiviral innate immunity. Nat. Immunol. 2016;17:806–815. doi: 10.1038/ni.3464. [DOI] [PubMed] [Google Scholar]
- 23.Saitoh T, et al. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 2006;7:598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- 24.Li S, et al. The tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat. Immunol. 2015;17:241–249. doi: 10.1038/ni.3311. [DOI] [PubMed] [Google Scholar]
- 25.Blanc RS, Richard S. Arginine methylation: the coming of age. Mol. Cell. 2017;65:8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Biggar KK, Li SS. Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 2015;16:5–17. doi: 10.1038/nrm3915. [DOI] [PubMed] [Google Scholar]
- 27.Wei H, Mundade R, Lange KC, Lu T. Protein arginine methylation of non-histone proteins and its role in diseases. Cell Cycle. 2014;13:32–41. doi: 10.4161/cc.27353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer. 2012;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 29.Greenblatt SM, Liu F, Nimer SD. Arginine methyltransferases in normal and malignant hematopoiesis. Exp. Hematol. 2016;44:435–441. doi: 10.1016/j.exphem.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Liu F, et al. Arginine methyltransferase PRMT5 is essential for sustaining normal adult hematopoiesis. J. Clin. Invest. 2015;125:3532–3544. doi: 10.1172/JCI81749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JH, et al. The role of protein arginine methyltransferases in inflammatory responses. Mediat. Inflamm. 2016;2016:4028353. doi: 10.1155/2016/4028353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Infantino S, et al. Arginine methylation of the B cell antigen receptor promotes differentiation. J. Exp. Med. 2010;207:711–719. doi: 10.1084/jem.20091303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ying Z, et al. Histone arginine methylation by PRMT7 controls germinal center formation via regulating Bcl6 transcription. J. Immunol. 2015;195:1538–1547. doi: 10.4049/jimmunol.1500224. [DOI] [PubMed] [Google Scholar]
- 34.Guccione E, et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 35.Hyllus D, et al. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waldmann T, et al. Methylation of H2AR29 is a novel repressive PRMT6 target. Epigenetics Chromatin. 2011;4:11. doi: 10.1186/1756-8935-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poulard C, Corbo L, Le Romancer M. Protein arginine methylation/demethylation and cancer. Oncotarget. 2016;7:67532–67550. doi: 10.18632/oncotarget.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Lorenzo A, Yang Y, Macaluso M, Bedford MT. A gain-of-function mouse model identifies PRMT6 as a NF-kappaB coactivator. Nucleic Acids Res. 2014;42:8297–8309. doi: 10.1093/nar/gku530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, et al. Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining activation of the kinase Btk. Nat. Immunol. 2011;12:416–424. doi: 10.1038/ni.2015. [DOI] [PubMed] [Google Scholar]
- 40.Han C, et al. Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat. Immunol. 2010;11:734–742. doi: 10.1038/ni.1908. [DOI] [PubMed] [Google Scholar]
- 41.Chen W, et al. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell. 2013;152:467–478. doi: 10.1016/j.cell.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 42.Neault M, Mallette FA, Vogel G, Michaud-Levesque J, Richard S. Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Res. 2012;40:9513–9521. doi: 10.1093/nar/gks764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phalke S, et al. p53-Independent regulation of p21Waf1/Cip1 expression and senescence by PRMT6. Nucleic Acids Res. 2012;40:9534–9542. doi: 10.1093/nar/gks858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakakido M, et al. PRMT6 increases cytoplasmic localization of p21CDKN1A in cancer cells through arginine methylation and makes more resistant to cytotoxic agents. Oncotarget. 2015;6:30957–30967. doi: 10.18632/oncotarget.5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen LT, Hu MM, Xu ZS, Liu Y, Shu HB. MSX1 modulates RLR-mediated innate antiviral signaling by facilitating assembly of TBK1-associated complexes. J. Immunol. 2016;197:199–207. doi: 10.4049/jimmunol.1600039. [DOI] [PubMed] [Google Scholar]
- 46.Wang F, et al. S6K-STING interaction regulates cytosolic DNA-mediated activation of the transcription factor IRF3. Nat. Immunol. 2016;17:514–522. doi: 10.1038/ni.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gates LT, Shisler JL. cFLIPL Interrupts IRF3-CBP-DNA Interactions To Inhibit IRF3-Driven Transcription. J. Immunol. 2016;197:923–933. doi: 10.4049/jimmunol.1502611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uhlmann T, et al. A method for large-scale identification of protein arginine methylation. Mol. Cell. Proteomics. 2012;11:1489–1499. doi: 10.1074/mcp.M112.020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo A, et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteomics. 2014;13:372–387. doi: 10.1074/mcp.O113.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.