Abstract
Microbial endosymbiosis is widespread in animals, with major ecological and evolutionary implications. Successful symbiosis relies on efficient vertical transmission through host generations. However, when symbionts negatively affect host fitness, hosts are expected to evolve suppression of symbiont effects or transmission. Here, we show that sex chromosomes control vertical transmission of feminizing Wolbachia endosymbionts in the isopod Armadillidium nasatum. Theory predicts that the invasion of an XY/XX species by cytoplasmic sex ratio distorters is unlikely because it leads to fixation of the unusual (and often lethal or infertile) YY genotype. We demonstrate that A. nasatum X and Y sex chromosomes are genetically highly similar and that YY individuals are viable and fertile, thereby enabling Wolbachia spread in this XY-XX species. Nevertheless, we show that Wolbachia cannot drive fixation of YY individuals, because infected YY females do not transmit Wolbachia to their offspring, unlike XX and XY females. The genetic basis fits the model of a Y-linked recessive allele (associated with an X-linked dominant allele), in which the homozygous state suppresses Wolbachia transmission. Moreover, production of all-male progenies by infected YY females restores a balanced sex ratio at the host population level. This suggests that blocking of Wolbachia transmission by YY females may have evolved to suppress feminization, thereby offering a whole new perspective on the evolutionary interplay between microbial symbionts and host sex chromosomes.
Microbial endosymbiosis is widespread in animals and relies on efficient vertical transmission through host generations. This study shows that sex chromosomes control vertical transmission of feminizing Wolbachia endosymbionts in an isopod, offering a new perspective on the evolutionary interplay between microbial symbionts and host sex chromosomes.
Introduction
Microbial endosymbiosis is widespread in animals, with major effects on host ecology and evolution [1,2]. Successful symbiosis relies on efficient symbiont transmission through host generations. Vertical transmission occurs when symbionts are transferred from parent to offspring, usually through the maternal germ line [3,4]. From a symbiont perspective, faithful maternal inheritance can be achieved by conferring benefits to hosts, such as nutritional provisioning [5], defense against natural enemies [6], and pathogen resistance [7,8]. This transmission strategy leads to convergence of symbiont and host fitness. Conversely, symbionts may follow a selfish strategy consisting of favouring their transmission at the expense of host fitness. This is achieved through manipulation of host reproduction, which can result in highly distorted sex ratios [9,10]. Because balanced sex ratios are optimal for most nuclear genes owing to biparental inheritance, hosts are predicted to evolve suppression mechanisms to control symbiont effects or transmission [10,11]. Genetic conflicts between sex ratio distorters, such as feminizing symbionts, and the rest of the genome are increasingly recognized as drivers of the evolution of host sex determination systems [10–16]. Here, we take the reverse perspective and investigate whether host sex chromosome systems can influence symbiont transmission and dynamics.
In many animals, sex is determined by a locus located on sex chromosomes that is heterozygous in one sex (the heterogametic sex) and homozygous in the other sex (the homogametic sex). Two major types of sex chromosomes exist: male heterogamety (XY males and XX females) and female heterogamety (ZZ males and ZW females) [13,15]. Sex chromosomes evolve from autosomes that acquired a sex-determining locus characterized by sex-specific inheritance. Subsequently, genomic regions around sex-determining loci often stop recombining, leading to gradual accumulation of nucleotide and structural variation and repetitive DNA [13,15,17–19]. This so-called degeneration process also causes the formation of pseudogenes and gene loss, resulting in increasing differentiation of sex chromosomes over time. This is well illustrated by the human X and Y sex chromosomes, which dramatically differ in size and gene content [19].
Arthropods host a diverse array of intracellular symbionts, including bacteria (such as Wolbachia, Cardinium, Rickettsia, Spiroplasma, and others) and unicellular eukaryotes (microsporidia), which are able to distort host sex ratios toward females [9,10]. The evolutionary impact of cytoplasmic sex ratio distorters on host sex determination systems has been particularly investigated in terrestrial isopods (crustaceans). This is because both female and male heterogametic systems of sex chromosomes are found in this speciose taxonomic group [20], and many species are naturally infected with Wolbachia bacteria [21,22]. Wolbachia are intracellular, maternally inherited endosymbionts of arthropods, often acting as reproductive parasites that manipulate host reproduction to favour infected females in host populations [23]. In terrestrial isopods, Wolbachia is best known as a sex ratio distorter because of its ability to feminize genetic males into phenotypic females [10,24–26]. For example, the presence of feminizing Wolbachia in Armadillidium vulgare leads to highly female-biased progenies [27,28] because symbionts override the ZZ/ZW system of sex chromosomes [29,30]. As a result, genetic ZZ male embryos develop as phenotypic ZZ females when infected by Wolbachia. Theoretical models predict the extinction of the W sex chromosome in A. vulgare lines infected by Wolbachia [31,32], and empirical evidence verified this prediction [33,34]. Thus, all individuals of Wolbachia-infected lineages end up with ZZ sex chromosomes at equilibrium; those inheriting Wolbachia develop as females and those lacking Wolbachia develop as males. This phenomenon is known as cytoplasmic sex determination [10,24–26]. In sum, invasion of a ZZ/ZW species by feminizing Wolbachia is not problematic, because it leads to the fixation of the ZZ genotype, which is the natural male genotype.
In sharp contrast with ZZ/ZW heterogamety, the conditions allowing feminizing Wolbachia to spread in an XY/XX heterogametic system are much more stringent. Indeed, theoretical work predicts that infection of an XY/XX system by a cytoplasmic sex ratio distorter should drive the loss of the X chromosome and concomitantly lead to fixation of the YY genotype at equilibrium [31]. In this case, YY individuals inheriting Wolbachia should develop as females, and those lacking Wolbachia should develop as males. However, YY is not a standard genotype, and it may often be lethal or infertile owing to Y chromosome degeneration causing the loss of essential genes [13,15,17–19]. Therefore, XY/XX heterogamety is expected to be incompatible with invasion of feminizing Wolbachia symbionts unless the X and Y chromosomes are not substantially divergent—i.e., they both carry all vital loci and are genetically very similar. To the best of our knowledge, this prediction has never been explored empirically.
Here, we investigated the interplay between sex chromosomes and Wolbachia symbionts in A. nasatum, a terrestrial isopod species related to A. vulgare [20]. Analogous to A. vulgare, a feminizing Wolbachia strain (wNas) is naturally present in A. nasatum [21,35,36]. However, unlike A. vulgare, chromosomal sex determination follows XY/XX heterogamety in A. nasatum [20,35]. This result was established using an original strategy consisting of experimentally reversing young genetic females into phenotypic males, crossing them with their sisters, and analyzing sex ratios of the resulting progenies [20,35]. Here, based on whole-genome sequencing, pedigree analyses, and simulations, we show that Wolbachia endosymbionts can spread in A. nasatum populations because A. nasatum sex chromosomes are genetically highly similar, and YY males and females are both viable and fertile. Nevertheless, Wolbachia cannot drive the loss of the X chromosome because infected YY females do not transmit Wolbachia to their offspring, unlike XX and XY females. As infected YY females produce all-male progenies, a balanced sex ratio is maintained at the host population level despite the presence of feminizing Wolbachia, suggesting that blocking of Wolbachia transmission by YY females may have evolved to suppress feminization.
Results
De novo assembly and annotation of male (XY) A. nasatum genome
We sequenced the male (XY) genome of an A. nasatum line derived from wild animals sampled in Thuré, France, in 2008. Males and females from this line have been consistently producing progenies with balanced sex ratios in our laboratory. In addition, sex reversal and crossing experiments demonstrated that sex determination in this line follows male heterogamety [20]. Absence of Wolbachia endosymbionts in the individuals selected for sequencing was confirmed by PCR.
We assembled the A. nasatum genome using a hybrid approach combining short paired-end Illumina reads and long PacBio reads (S1 Table). The initial assembly was processed through polishing, decontamination, and scaffolding by a transcriptome assembly. The final assembly had a total length of 1,223,175,971 bp. It was composed of 25,196 contigs and scaffolds (hereafter collectively referred to as “contigs”), with an N50 (minimum contig length to cover 50% of the genome) of 86,284 bp and containing barely no (0.01%) undetermined nucleotides (Table 1). Genome completeness assessment using Benchmarking Universal Single-Copy Orthologs (BUSCO 3.0.1) [37] revealed that 1,001 of 1,066 (94%) conserved specific arthropod genes were present in the assembly (Table 1). Furthermore, transcriptome assembly alignment on the genome assembly yielded 99.0% of transcripts longer than 1 kb aligned. Thus, we have obtained a reliable assembly of the XY male genome from A. nasatum.
Table 1. Summary information of A. nasatum genome assembly.
| Genome assembly features | Assembly figures |
|---|---|
| Assembly statistics | |
| Number of contigs and scaffolds | 25,196 |
| Total size | 1,223,175,971 bp |
| Longest contig/scaffold | 877,941 bp |
| Number of contigs and scaffolds >1 kb | 25,166 |
| N50 contig size | 75,116 bp |
| N50 scaffold size | 86,284 bp |
| Undetermined nucleotides | 0.01% |
| G + C nucleotide content | 25.77% |
| Analysis of BUSCO genes | |
| Complete genes | 974/1,066 (91.4%) |
| Complete and single-copy genes | 932/1,066 (87.4%) |
| Complete and duplicated genes | 42/1,066 (3.9%) |
| Fragmented genes | 27/1,066 (2.5%) |
| Missing genes | 65/1,066 (6.1%) |
Abbreviation: BUSCO, Benchmarking Universal Single-Copy Orthologs
The annotation of the A. nasatum assembly included 14,636 predicted gene models, with an average length of 9,422 bp and representing 11.3% of the total assembly length (S2 Table). Among these predicted genes, 9,281 (63.4%) had Blastp hits to the UniProt-SwissProt database (release September 2016), 9,459 (64.6%) had InterProScan hits to Pfam domains (version 30.0), and 6,137 (41.9%) had Gene Ontology terms. Repeats accounted for 64.6% (or 790 Mb) of the A. nasatum assembly (S3 Table). Specifically, transposable elements and simple tandem repeats accounted for 40.6% (or 496 Mb) and 23.1% (or 283 Mb) of the genome. The annotated genome sequence of A. nasatum is available in DNA DataBank of Japan/European Nucleotide Archive/GenBank under accession number SEYY00000000. The version described in this article is version SEYY01000000.
High genetic similarity of the X and Y chromosomes
To investigate the extent of genomic differentiation between sex chromosomes, we searched the A. nasatum assembly for contigs containing Y-specific sequences. We mapped short paired-end Illumina reads generated for XY males and XX females (S1 Table) onto the A. nasatum assembly and performed a Chromosome Quotient (CQ) analysis [38]. This analysis consists of comparing the ratios of female-to-male sequencing depths for each contig, with the expectations that (1) Y-specific contigs should be mapped by male reads only (CQ approximately 0), (2) X-specific contigs should be mapped by female reads at twice the sequencing depth of male reads (CQ approximately 2), and (3) autosomal contigs should be mapped at similar sequencing depths by female and male reads (CQ approximately 1). The resulting frequency distribution of CQ scores was unimodal and centered at CQ approximately 1 (mean: 1.03, median: 1.04), with no peak at CQ scores of approximately 0 and 2 (Fig 1A). In addition, we used the mapping-free Y chromosome Genome Scan (YGS) method [39], which computes the proportion of single-copy k-mers for each contig of the genome assembly of the heterogametic (male) sex that are unmatched to sequencing reads of the homogametic (female) sex. Y-specific contigs are expected to be unmatched by female reads (YGS approximately 100%), whereas autosomal and X-linked contigs are expected to be matched entirely (YGS approximately 0%). The resulting frequency distribution of YGS scores indicated that most contigs have very low YGS scores (mean: 14.5%, median: 11.2%), and very few contigs had high YGS scores (i.e., only 43 contigs with YGS ≥ 80% and just 2 with YGS ≥ 90%) (Fig 1B). Thus, the CQ and YGS analyses consistently indicated that the A. nasatum assembly mostly contains autosomal contigs and very few contigs containing X- and Y-specific sequences. Intersecting the results of the CQ and YGS analyses identified only 78 out of the 25,196 contigs of the assembly as containing putative Y-specific sequences, despite the use of permissive thresholds—i.e., CQ ≤ 0.35 and YGS ≥ 35% (Fig 1C). The 78 contigs comprised a total length of 1,327 kb, corresponding to just 0.1% of the A. nasatum genome assembly (S4 Table). Although most contigs lacked any gene (65/78, or 83%), a total of 20 genes (out of 14,636 genes in the assembly) were annotated from 13 contigs (S4 Table). They constitute candidate master genes for sex determination in A. nasatum. Thus, the Y-specific region of the A. nasatum genome is extremely small (around 1 Mb) and contains very few genes, leading to the conclusion that the X and Y chromosomes are poorly differentiated at the genomic and genic levels.
Fig 1. Identification of sex-specific contigs in the A. nasatum genome assembly.
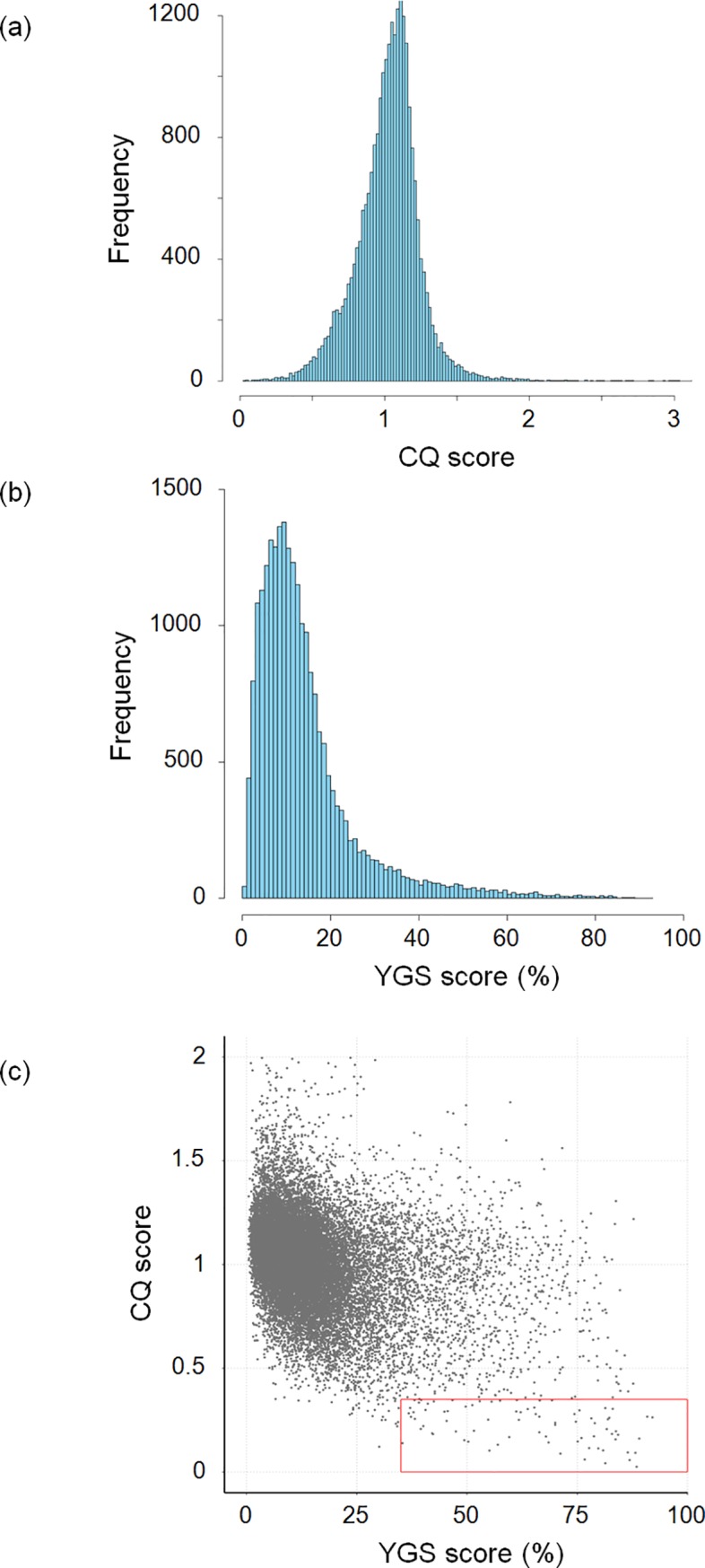
Frequency distribution of CQ (a) and YGS (b) scores calculated for each contig and scaffold of the assembly. (c) Comparison of CQ and YGS scores. Each dot corresponds to one contig or scaffold (those with CQ > 2 are not represented). The red box contains 78 contigs with CQ ≤ 0.35 and YGS ≥ 35%. CQ, Chromosome Quotient; YGS, Y chromosome Genome Scan.
To investigate sex chromosome differentiation at the molecular level, we analyzed the patterns of sequence divergence and repeat content in the 78 contigs containing putative Y-specific sequences relative to the other contigs of the A. nasatum genome. First, we analyzed the density of single nucleotide polymorphisms (SNPs) in the 78 contigs (after removal of hemizygous regions, to focus on regions with orthologs on the X chromosome) versus the other contigs of the assembly. We calculated a median SNP density of 2.61 SNPs/kb between allelic X/Y regions of the 78 contigs, compared with 2.55 SNPs/kb across all other contigs of the assembly (S1 Fig). Thus, we observed a slight but nonsignificant (Mann-Whitney two-sided U test, U = 969,520, p = 0.88) excess of SNP density in allelic X/Y regions relative to other regions of the genome. In addition, the repeat content in the 78 contigs was slightly higher than (median 69.6%), but not significantly different (Mann-Whitney bilateral U test, U = 1,048,600, p = 0.28) from, the repeat content of all other contigs (median 67.8%) (S1 Fig). Furthermore, even when focusing on 25 of the 78 contigs that were independently validated as Y-linked by PCR (see below), there was no significant difference with all other contigs of the assembly, both in terms of SNP density (median 3.91 SNPs/kb, Mann-Whitney two-sided U test, U = 371,560, p = 0.11) and repeat content (median 65.3%, Mann-Whitney two-sided U test, U = 301,380, p = 0.73) (S1 Fig). Thus, the potential presence of (autosomal) false positives among the 78 contigs containing putative Y-specific sequences has not affected our results. In sum, our results indicated that A. nasatum sex chromosomes present patterns of molecular evolution that are quite similar to those of other genomic regions, with a slight elevation of SNP density and repeat content in contigs containing putative Y-specific sequences that is consistent with expectations for sex-specific sequences of the genome [40]. We conclude that the X and Y chromosomes of A. nasatum are highly similar not only at the genomic and genic levels but also at the molecular level.
YY individuals are viable and fertile
To test for the existence and, if so, viability and fertility of YY individuals, we tracked Y chromosome inheritance in a Wolbachia-infected A. nasatum pedigree. First, we established robust Y-specific molecular markers using the putative Y-specific contigs identified previously. Reliable PCR assays were successfully designed for 42 of the 78 contigs containing putative Y-specific sequences, most of which (25/42) exhibited male-specific amplification, with DNA samples from males and females closely related to those used for genome sequencing (S4 Table). However, only nine of the 25 confirmed markers displayed male-specific amplification when tested in more distantly related individuals from the same population and in individuals from two other populations. Lack of sex specificity of a subset of markers in some populations indicated that recombination has occurred between some of the tested contigs and the sex-determining locus. This result provided further evidence that the Y-specific region of the genome is extremely small in A. nasatum. Concomitantly, the universal male specificity of nine markers across all tested populations demonstrated that these A. nasatum populations possess a homologous Y chromosome. Hence, we used a subset of these robust markers to track the Y chromosome in an A. nasatum pedigree spanning three generations.
The pedigree was initiated by crossing a male with a female naturally infected by the feminizing Wolbachia strain wNas (F0), which produced an F1 progeny comprising 17 sisters (Fig 2). Two F1 sisters were then crossed with genetic males (XY) to produce two F2 progenies: I-F2-1 comprising 24 females and I-F2-2 comprising 32 males. Wolbachia infection was tested in the 62 individuals of the pedigree (except the F0 father), indicating that all females carried Wolbachia and all males lacked Wolbachia infection, as expected. Next, we used previously designed Y-specific markers to assess the presence of the Y chromosome in the 62 individuals (except the F0 father). In the I-F2-1 progeny, half of the females showed amplification of the Y markers (indicating they are XY or YY), and half did not (indicating they are XX). The F2-1 mother carried XX chromosomes, as she did not amplify the Y markers. As all I-F2-1 offspring necessarily carry one X chromosome from their mother, the 11 I-F2-1 females amplifying the Y markers must be XY. In the I-F2-2 progeny, all individuals amplified the Y markers (indicating they are XY or YY), as did their mother. Given that the F2-2 father is XY, if the F2-2 mother was XY, we would have expected 25% XX individuals in the I-F2-2 progeny, but there was none. Thus, the I-F2-2 mother must be YY, which is the only genotype that can explain that the entire I-F2-2 progeny carries at least one Y chromosome. Given the parental genotypes, we predicted that the I-F2-2 progeny is composed of half XY males and half YY males. Finally, with the two F1 sisters being XX and YY, it followed that the F0 father and mother are XY. To independently assess these predictions, we devised a quantitative PCR assay measuring Y chromosome dose relative to autosomes (S2 Fig). This assay confirmed the predicted genotypes of the F0 and F1 individuals we tested and the composition of the I-F2-2 progeny (15 XY and 17 YY individuals) (Fig 2, S5 Table). In sum, the resolution of sex chromosome genotypes in the pedigree demonstrated that the YY genotype is viable both as male and as female in A. nasatum, as is the XY genotype. Moreover, the YY and XY genotypes are both fertile as females. Finally, we show below that the YY genotype is also fertile as male.
Fig 2. A. nasatum pedigree used to track inheritance of the Y chromosome and Wolbachia.
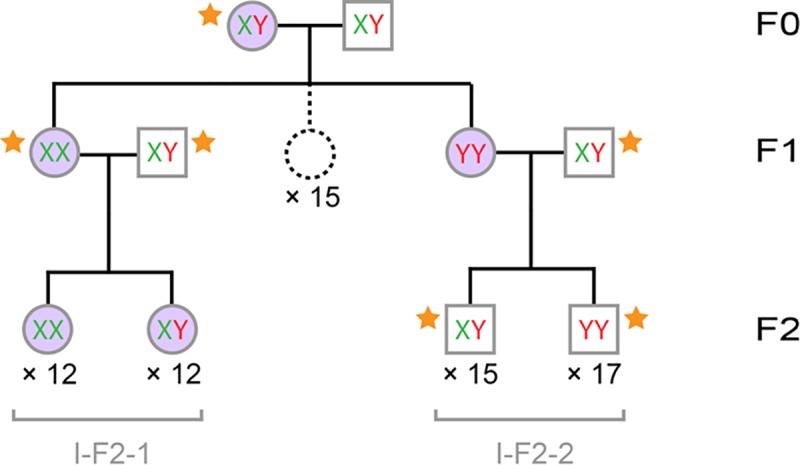
The pedigree spans three generations (F0–F2) and comprises 62 individuals (35 males and 27 females) for which sex chromosome genotype (XX, XY, or YY) was identified and 15 F1 females (not included in the molecular analyses; dotted circle). Males are shown as squares, and females are shown as circles. Individuals carrying Wolbachia are shown in purple. Progeny identifiers are shown in gray. Sex chromosome genotype of individuals marked with an orange star (34 males and two females) was also assessed with a quantitative PCR assay.
Blocking of Wolbachia transmission by YY females prevents X chromosome loss
Surprisingly, the Wolbachia-infected YY mother in the previous pedigree analysis produced an all-male progeny (I-F2-2) entirely lacking Wolbachia endosymbionts (Fig 2). This is unexpected because infection by feminizing Wolbachia endosymbionts is usually associated with highly female-biased progenies because of the maternal transmission of Wolbachia to usually >80% of the offspring [10,24–26]. To test whether lack of Wolbachia transmission from mother to I-F2-2 progeny was a random event or was due to the unusual YY maternal genotype, we extended our previous pedigree analysis to span five generations (S3 Fig) and analyzed a second, independent pedigree spanning four generations (S4 Fig). In total, 20 families (defined as father, mother, and progeny) were included in the two pedigrees, representing 799 individuals (252 males and 547 females). We tested 464 individuals for the presence of Wolbachia (Table 2). As expected, most tested females were infected by Wolbachia (180/214), whereas no tested male was (0/250). Based on the patterns of Y chromosome amplification and following a reasoning similar to that described in the previous section, we were able to infer sex chromosome genotypes for most analyzed individuals. In addition, we independently verified sex chromosome genotypes of the parents of the 20 families using the aforementioned quantitative PCR assay (S5 Table). The sex chromosome genotypes of the 20 mothers included 11 XX, 5 XY, and 4 YY females (S3 and S4 Figs, Table 2). All tested fathers were XY males, except the father of the II-F2-1 progeny, which was a YY male. Interestingly, this individual demonstrated that the YY genotype is viable and fertile as male.
Table 2. Composition and frequency of Wolbachia infection in 20 families of A. nasatum.
Wolbachia frequency in progenies was calculated as the Wolbachia frequency observed among tested females weighted by female proportion in the progenies (as all males lack Wolbachia).
| Progeny identifier | Parental genotypes | Progeny size | Female Nb. | Female proportion | Wolbachia presence in males | Wolbachia presence in females | Wolbachia frequency | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Mother | Father | Nb. tested | Nb. infected | Nb. tested | Nb. infected | |||||
| I-F1-1 | XY | XY | 17 | 17 | 100% | 0 | 0 | 3 | 3 | 100% |
| I-F2-1 | XX | XY | 24 | 24 | 100% | 0 | 0 | 24 | 24 | 100% |
| I-F2-2 | YY | XY | 32 | 0 | 0% | 32 | 0 | 0 | 0 | 0% |
| I-F2-3 | XX | XY | 52 | 52 | 100% | 0 | 0 | 2 | 2 | 100% |
| I-F3-1 | XY | XY | 68 | 68 | 100% | 0 | 0 | 12 | 12 | 100% |
| I-F3-2 | XX | XY | 33 | 33 | 100% | 0 | 0 | 10 | 8 | 80% |
| I-F3-3 | XX | XY | 59 | 59 | 100% | 0 | 0 | 12 | 12 | 100% |
| I-F3-4 | XX | XY | 40 | 40 | 100% | 0 | 0 | 10 | 9 | 90% |
| I-F4-1 | XX | XY | 50 | 39 | 78% | 11 | 0 | 15 | 12 | 62% |
| I-F4-2 | XX | XY | 41 | 24 | 59% | 17 | 0 | 13 | 0 | 0% |
| I-F4-3 | XX | XY | 32 | 10 | 31% | 22 | 0 | 10 | 2 | 6% |
| I-F4-4 | XX | XY | 16 | 16 | 100% | 0 | 0 | 10 | 9 | 90% |
| I-F4-5 | XX | XY | 51 | 51 | 100% | 0 | 0 | 15 | 15 | 100% |
| I-F4-6 | XY | XY | 42 | 19 | 45% | 23 | 0 | 13 | 7 | 24% |
| II-F1-1 | XX | Y? | 13 | 13 | 100% | 0 | 0 | 2 | 2 | 100% |
| II-F2-1 | XY | YY | 60 | 60 | 100% | 0 | 0 | 60 | 60 | 100% |
| II-F2-2 | XY | XY | 20 | 20 | 100% | 0 | 0 | 1 | 1 | 100% |
| II-F3-1 | YY | XY | 28 | 0 | 0% | 28 | 0 | 0 | 0 | 0% |
| II-F3-2 | YY | XY | 42 | 0 | 0% | 42 | 0 | 0 | 0 | 0% |
| II-F3-3 | YY | XY | 57 | 0 | 0% | 57 | 0 | 0 | 0 | 0% |
Abbreviation: Nb., number
Wolbachia transmission rate from mother to offspring differed significantly between XX, XY, and YY mothers, considering all 20 families (Kruskal-Wallis test, χ2 = 8.84, p = 0.01) or only the 16 families in which Wolbachia presence was tested in ≥10 individuals (Kruskal-Wallis test, χ2 = 7.91, p = 0.02). Wolbachia transmission rate was high (generally ≥80%) and did not differ significantly between XX and XY mothers (Dunn's post hoc tests, p = 0.73 for 20 families and p = 0.56 for 16 families) (Fig 3). By contrast, Wolbachia infected none of the 159 offspring of the 4 YY mothers. This result cannot be ascribed to Wolbachia transmission to offspring and subsequent selective death of embryos carrying Wolbachia infection, because progeny size did not differ significantly between XX, XY, and YY mothers (Kruskal-Wallis tests, χ2 = 0.18, p = 0.91 for 20 families and χ2 = 3.32, p = 0.19 for 16 families) (Table 2). Instead, the YY genotype is associated with a lack of Wolbachia transmission from mother to offspring. As a result, Wolbachia transmission rate differed significantly between YY mothers and both XX mothers (Dunn's post hoc tests: p = 0.006 for 20 families and p = 0.01 for 16 families) and XY mothers (Dunn's post hoc tests: p = 0.008 for 20 families and p = 0.01 for 16 families) (Fig 3). Consequently, as all offspring inherited a Y chromosome from their YY mother but no Wolbachia endosymbiont, they all developed as males (Table 2). Importantly, YY mothers originated from the two independent pedigrees we analyzed (S3 and S4 Figs), indicating that the observed pattern is robust to A. nasatum genetic background.
Fig 3. Boxplot of Wolbachia transmission rate from mother to offspring (measured as the frequency of Wolbachia-carrying individuals in each progeny) according to mother’s sex chromosome genotype.
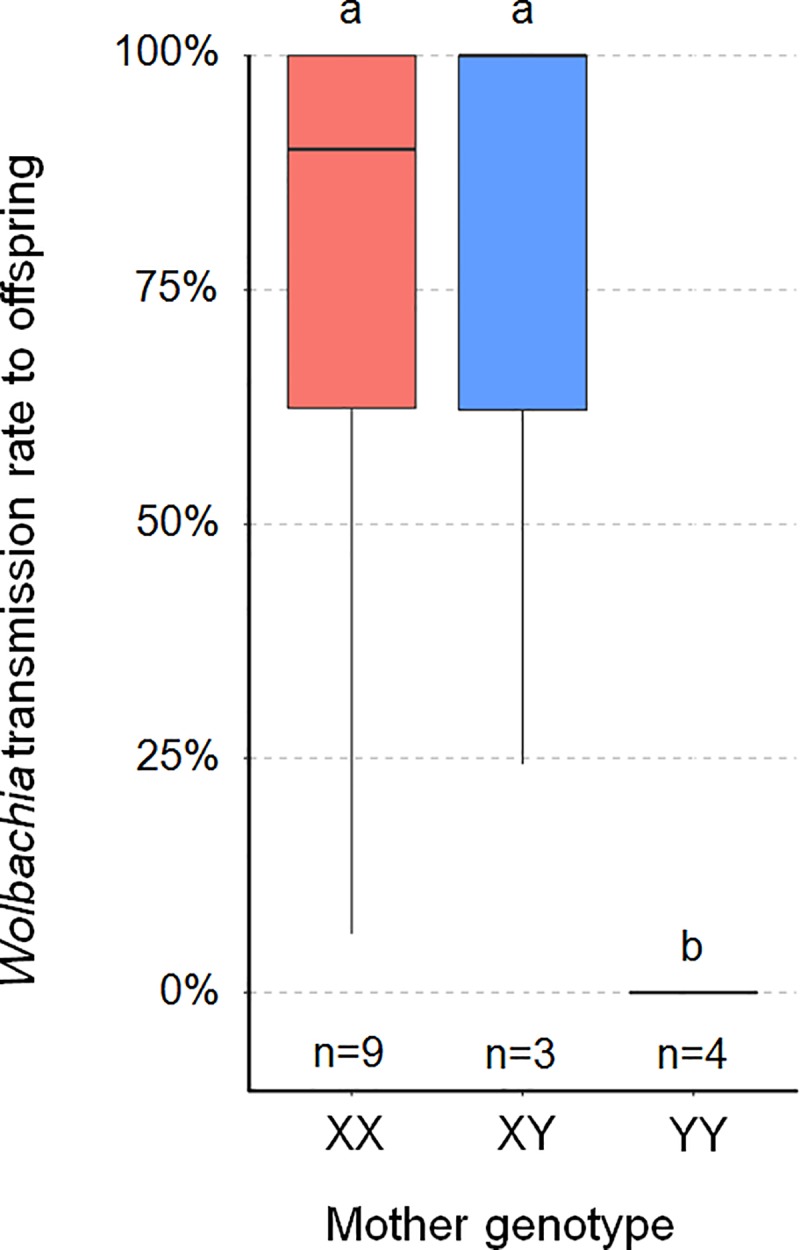
The analysis is based on 16 progenies (n), in which Wolbachia presence was tested in ≥10 individuals (the underlying data for this figure can be found in Table 2). Thick lines and boxes depict median and interquartile range, respectively. Whiskers are bounded to the most extreme data point within the 1.5 interquartile range. Plots marked with the same letter (a, b) are not statistically different from each other (Kruskal-Wallis test followed by pairwise comparison Dunn test).
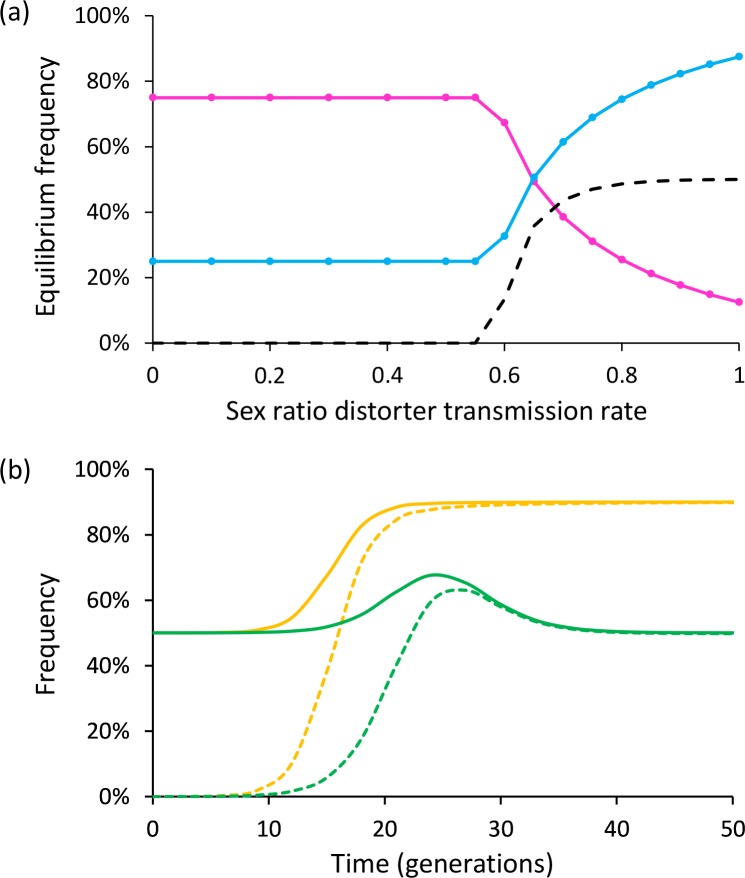
As YY females exclusively produce males, only XX and XY females can produce female offspring in Wolbachia-infected lines of A. nasatum. Thus, the X chromosome is necessarily transmitted to at least a subset of each progeny of these Wolbachia-infected females. To evaluate consequences on sex chromosome frequencies in the long term, we extended Taylor's theoretical work [31] to enable the transmission rate of a cytoplasmic sex ratio distorter to vary depending on female sex chromosome genotype. Specifically, we simulated the equilibrium frequencies of X and Y chromosomes in a population of a diploid genetic model containing a distorter with transmission rates of α, α, and 0 in XX, XY, and YY females (with α varying from 0 to 1), respectively, to reflect our empirical results. Under these conditions, equilibrium frequencies are 25% and 75% for the X and Y chromosomes for α < 0.55 (Fig 4A). In such cases, the distorter is lost from the population at equilibrium, and the population comprises XY males and XX females in equal proportions. By contrast, for higher α values, equilibrium frequencies of the X and Y chromosomes vary in opposite directions, to the extent that the Y chromosome becomes more frequent than the X chromosome, but X equilibrium frequency is always ≥12.5%, whatever the value of α (Fig 4A). Thus, a consequence of the lack of transmission of the distorter by YY females is that the X chromosome cannot be lost from the population. Another consequence evidenced by our simulations is that the equilibrium frequency of the distorter never exceeds 50%, whatever the value of α (Fig 4A). Remarkably, a balanced sex ratio is maintained in the population at equilibrium, despite infected females individually producing progenies highly biased toward either females (XX and XY mothers) or males (YY mothers) (Fig 4B). This is in sharp contrast with the highly female-biased sex ratios at equilibrium expected for α > 0.5 when the distorter is transmitted by all three types of females [31] (Fig 4B).
Fig 4. Evolutionary consequences of a cytoplasmic sex ratio distorter at the population level.
(a) Equilibrium frequencies of X chromosome (pink line), Y chromosome (blue line), and distorter (dashed line), according to distorter transmission rate in XX and XY females (YY females do not transmit the distorter). (b) Evolution of the frequencies of females (solid lines) and individuals carrying the distorter (dashed lines) through time, with a distorter transmission rate of 0.9. Green: YY females do not transmit the distorter (only XX and XY females do); orange: all females transmit the distorter.
Discussion
We sequenced the genome of the terrestrial isopod A. nasatum. With an N50 of 86 kb, high completeness (BUSCO = 94%), and near absence of unidentified nucleotides (0.01%), our approximately 1.2-Gb assembly is among the best assembled of all large-genome crustaceans sequenced to date [30]. Our results establish that A. nasatum X and Y chromosomes are genetically highly similar. The Y-specific region of the genome may be as small as 1 Mb, and it contains very few genes. Even at nucleotide resolution, sequence divergence and repeat content indicate very limited sex chromosome differentiation. This result explains why the unusual YY genotype is viable in this species. The underlying causes of the very high similarity of A. nasatum sex chromosomes include young evolutionary age, ongoing recombination, or both. In any event, this high similarity was likely instrumental to the establishment of feminizing Wolbachia infection in the male heterogametic isopod A. nasatum.
An important outcome of the infection of A. nasatum by feminizing Wolbachia is the unusual production of YY individuals and XY females. This situation challenges the classical model of sex chromosome evolution by affecting sex chromosome effective population size (Ne) and recombination patterns. Classically, recombination arrest occurs between the X and Y chromosomes, leading to a drastic reduction of Y chromosome Ne to one-third of X chromosome Ne and one-fourth of autosome Ne (because there is one Y chromosome dose for three X chromosome doses and four autosome doses per mating pair). Lowered Ne results in enhanced genetic drift and concomitant reduction of selection efficiency, leading the Y chromosome to accumulate deleterious mutations and degenerate [13,15,17–19]. However, the existence of YY individuals and XY females implies that the standard expectation of sex chromosome Ne does not apply to A. nasatum in the presence of Wolbachia. Indeed, our simulations indicate that X chromosome Ne should stabilize at about one-fourth of Y chromosome Ne and about one-fifth of autosome Ne at observed Wolbachia transmission rates (Fig 4A). Thus, X chromosome Ne, not Y chromosome Ne, is predicted to be dramatically reduced in A. nasatum genetic backgrounds infected by feminizing Wolbachia. Furthermore, the occurrence of YY individuals opens the possibility of Y-Y recombination and more efficient selection, which may prevent the accumulation of deleterious mutations on the Y chromosome. By contrast, XX individuals are rare according to our simulations (<2%), thereby reducing the opportunity of X-X recombination. This suggests that the X chromosome, rather than the Y chromosome, may accumulate deleterious mutations and degenerate in the long term. Another possibility to consider is that recombination patterns may differ between sexes (heterochiasmy) in some species so that the X and Y chromosomes may not recombine simply because males do not recombine (whereas females do) [15]. If so, the existence of XY females in A. nasatum as a consequence of Wolbachia infection would open the possibility of X-Y recombination in phenotypic females, as previously reported in frogs [41]. Unfortunately, male and female patterns of recombination are unknown in terrestrial isopods. Their investigation therefore represents a perspective for future research. In any event, there appears to be ample scope for microbial symbionts to drive the molecular evolution of their host sex chromosomes.
Remarkably, infected YY females did not transmit Wolbachia to any offspring, as confirmed in two independent pedigrees. An evolutionary consequence is that the YY genotype cannot become fixed (nor the X chromosome lost) in infected A. nasatum lines. Lack of Wolbachia transmission by YY females also raises the question of the underlying mechanism controlling vertical transmission of the symbionts. In sharp contrast with the YY genotype, the alternative XX and XY genotypes were associated with similarly high rates of Wolbachia transmission in both pedigrees. Thus, the genetic basis of Wolbachia transmission control fits the model of a recessive allele linked to the Y chromosome (associated with an X-linked dominant allele), in which the homozygous state blocks Wolbachia transmission. It has been shown previously that within-host microbial interactions can enhance or reduce symbiont vertical transmission [42]. This applies to Wolbachia, whose vertical transmission is prevented by Asaia bacteria in Anopheles mosquitoes [43]. Host nuclear genotype is also an important factor that can affect symbiont vertical inheritance. For example, Wolbachia maternal transmission is strongly reduced by actin in Drosophila [44] and the wds gene in Nasonia [45]. Our results show that sex chromosome genotype represents yet another case of host nuclear control over symbiont transmission. Wolbachia symbionts were previously found to prevent sex chromosome transmission in Eurema mandarina butterflies [46]. Here, we show that sex chromosomes can prevent Wolbachia transmission in A. nasatum, thereby offering a whole new perspective on the molecular interplay between feminizing symbionts and host sex chromosomes.
The mutation causing the suppression of Wolbachia transmission in A. nasatum may have existed in the standing pool of sex chromosome variants prior to Wolbachia infection, having evolved either neutrally or under natural selection. Alternatively, the mutation may have evolved as an adaptation subsequent to Wolbachia infection. An excellent empirical example supporting the latter hypothesis is offered by the butterfly Hypolimnas bolina, in which suppression of Wolbachia-mediated male killing was found to have evolved in Southeast Asia secondarily to Wolbachia spread in the Indo-Pacific region [47]. Whereas infection by feminizing symbionts normally leads to highly female-biased sex ratios, the ability of infected YY females to impede Wolbachia transmission in A. nasatum results in the production of all-male progenies because of the systematic inheritance of a maternal Y chromosome. Interestingly, this situation restores balanced sex ratios at the population level, as indicated by our simulations (Fig 4) and consistent with empirical evidence [35,48]. This opens the possibility that blocking of Wolbachia transmission by YY females may have evolved to suppress feminization and ensuing sex ratio biases, as predicted by sex ratio selection [10–12,15]. Indeed, strong sex ratio biases toward females imposed by feminizing symbionts induce nucleocytoplasmic conflicts with most nuclear genes, which are biparentally inherited and optimally benefit from balanced sex ratios. In principle, conflict resolution may be achieved by restoring balanced sex ratios at the parental level through selection of females producing balanced sex ratios [49]. Alternatively, resolution may occur at the population level through selection of females producing male-biased sex ratios to compensate for the female-biased progenies of infected females [49]. YY females in A. nasatum may represent an original example of conflict resolution at the population level. Several cases of nuclear suppression preventing the action or transmission of sex ratio–distorting symbionts have been reported [11], including in the isopod A. vulgare [50,51]. A distinguishing feature of symbiont suppression in A. nasatum is its connection with sex chromosomes.
Materials and methods
Genome sequencing and assembly
All A. nasatum individuals used for sequencing were from our inbred laboratory line ANa, which is originally derived from wild animals caught in Thuré, France, in 2008. Specifically, we used XY genetic males and XX genetic females descended from a single pair of grandparents of the family ANa2 (according to the crossing scheme shown in S5 Fig) to minimize heterozygosity. Total genomic DNA was extracted using the QiagenDNeasy Blood and Tissue Kit, according to the protocol for animal tissues (3 h of incubation in proteinase K at 56°C and 30 min of RNase treatment at 37°C). Absence of Wolbachia endosymbionts in all samples was confirmed by PCR using the ftsZ and wsp markers [21,36]. Short paired-end libraries with approximately 200-bp insert sizes were sequenced with the Illumina HiSeq2000 technology (S1 Table). In addition, PacBio RS II sequencing (P6C4 chemistry) was performed to obtain long sequencing reads (S1 Table). Accession numbers for Illumina and PacBio sequence datasets are provided in S1 Table.
Sequencing reads were subjected to quality control and filtering as described previously [30]. Male Illumina and PacBio sequencing reads were used in a hybrid strategy to assemble the male (XY) genome of A. nasatum, as described previously [30]. A summary of the workflow we used (including assembly, polishing, contaminant removal, and scaffolding) is shown in S6 Fig. Genome assembly completeness was evaluated using BUSCO, version 3.0.1 [37], with the arthropod profile library (-l arthropoda) composed of 1,066 arthropod core genes. Repeat identification and gene annotation were performed as described previously [30].
Identification and analyses of contigs containing putative Y-specific sequences
Contigs containing putative Y-specific sequences in the A. nasatum assembly were identified using the CQ [38] and YGS [39] methods, as described previously [30]. The maximum CQ score was set to 0.35 to retain contigs as Y-specific candidates, as recommended by CQ authors [38]. The minimum YGS score was set to 35% to retain contigs as Y-specific candidates. This threshold was selected to account for the high repetitive nature of the A. nasatum genome. To identify heterozygous SNPs in the contigs of the male genome assembly, we applied the Genome Analysis ToolKit (GATK) pipeline (version 3.8-0-ge9d806836) [52], as described previously [30].
PCR assays were designed on the candidate contigs as follows (S4 Table). Primers were designed with Primer-BLAST [53] in unique regions of the contigs, and primer specificity was checked using Blastn (version 2.2.30+) [54] by aligning primers to the unmasked A. nasatum assembly. PCR reactions were carried out in 25 μL with 5 μL of buffer 5×, 0.5 μL of dNTPs (2.15 mM), 0.7 μL of each primer (10 μM), 0.25 μL of Taq polymerase 5 u/μL, and 1 μL of DNA. PCRs were conducted using the following temperature cycle: 3 min at 94°C for initial denaturation, followed by 35 cycles of 30 s at 94°C, 30 s at 48°C/50°C/52°C (depending on primer annealing temperature), and 1 min at 72°C. The final elongation step was 10 min at 72°C. PCR tests were then conducted in three successive steps: (1) test on one male and three pools of two females of the ANa2 family (used for genome sequencing), (2) test on six males and 12 females from other families of our ANa laboratory line, and (3) test on two males and a pool of three females from the Beauvoir-sur-Niort population (France) and on two males and a pool of three females from the Piriápolis population (Uruguay). After each step, loci amplifying in all males and no females were retained for the next step. PCR tests targeting the autosomal 18S rRNA [30] and mitochondrial COI [55] genes were used as positive controls in all samples. Absence of Wolbachia endosymbionts was also confirmed in all samples by PCR using the ftsZ and wsp markers [21,36].
Pedigree construction and analyses
We generated two independent A. nasatum pedigrees spanning five (pedigree I, S3 Fig) and four (pedigree II, S4 Fig) generations (A. nasatum has a generation time of 1 y). We started in 2013 with two F0 female founders isolated from a laboratory cage population (NASw) initiated in 2008 from wild animals (caught in Thuré, France) infected by the feminizing Wolbachia strain wNas [36]. Each F0 female produced an F1 progeny (resulting from mating with unknown F0 males from the cage population). Each year, two to six females were selected from the previous generation and crossed with XY genetic males from our ANa laboratory line (except the father of the II-F2-1 progeny, which was selected from the NASw cage population and could carry XY or YY sex chromosomes). Pedigrees I and II were composed of 14 and six families, respectively.
At each generation, total genomic DNA was extracted from the progenitors (except the F0 male founders and individuals that died prematurely) and from males and females of their progenies. Presence or absence of Wolbachia was tested by PCR using the ftsZ and wsp markers [21,36]. We also used two of the previously designed Y-specific markers (contig12740 and contig18908) to assess the presence of the Y chromosome in the individuals, interpreting PCR results as follows: amplification indicating the individual is XY or YY and lack of amplification indicating that the individual is XX. PCR tests targeting the autosomal 18S rRNA [30] and mitochondrial COI [55] genes were used as positive controls in all samples. Based on PCR amplification patterns and pedigree structures, we were able to infer sex chromosome genotypes (XX, XY, or YY) for most analyzed individuals.
To independently assess sex chromosome genotypes (XX, XY, or YY), we developed a quantitative PCR assay measuring Y chromosome abundance relative to autosomes. Y-specific primers were designed within the Y-specific PCR amplicon previously validated in contig10349 (primer sequences: 5’-CCCTACACAGCATACTTGACAG and 5’-CAGGTGCTCCTTCAGAGAAAC, product size: 129 bp). Contig10349 abundance was normalized against EF2 gene abundance [56]. EF2 is a single-copy gene in the A. nasatum assembly located in autosomal contig8976 (YGS = 8.8% and CQ = 1.09). PCR reactions were run in duplicates in a 480 LightCycler (Roche) using SYBR Green I assays under the following conditions: 10 min at 95°C and 45 cycles of the following: 10 sec at 95°C, 10 sec at 60°C, and 20 sec at 72°C. A melting curve (65°C to 97°C) was recorded at the end of each reaction to check PCR product uniqueness. Reaction mixture consisted of 0.5 μL of each primer (10 μM), 5 μL of Fast SYBR Green Master Mix (Roche), 3 μL of bidistilled water, and 1 μL of extracted DNA. Fluorescence crossing points (Ct) were calculated with the second derivative maximum method using the LightCycler 1.5 software. PCR amplification efficiency was determined with a calibration curve for each primer pair. Only PCR reactions producing a single product and with Ct ≤ 35 cycles were considered. Relative abundance of the Y-specific marker relative to autosomal marker was calculated as 2−ΔCt, where ΔCt = Ct(Y chromosome)−Ct(autosome). XX, XY, and YY genotypes corresponded to 2−ΔCt values of approximately 0, 0.5, and 1, respectively.
Simulation of sex chromosome frequencies
To evaluate the evolutionary impact of cytoplasmic sex ratio distorters on sex chromosome frequencies, we extended Taylor's theoretical work [31] to enable the transmission rate of the distorter to vary according to female sex chromosome genotypes. Specifically, we simulated the frequencies of the X chromosome, the Y chromosome, the distorter, and females in a population of a diploid genetic model with a distorter whose transmission rate can vary according to the sex chromosome genotype of females (i.e., XX, XY, and YY). We used the following equations:
- Genotypes
- Males: G1 = XY; G3 = YY; FM = males’ frequency
- Females: G2 = XX; G4 = XX.Wo+; G6 = XY.Wo+; G8 = YY.Wo+; FF = females’ frequency
- Gametes
- Males: P1 = Y; P3 = X
- Females: P2 = X; P4 = Y; P6 = X.Wo+; P8 = Y.Wo+
- Wolbachia transmission rates by females
- tX by XX.Wo+; tXY by XY.Wo+; tY by YY.Wo+
- Gametes production formula
- Male gametes:
Female gametes:
- Next-generation genotypes production formula
- Males:
Females:
with and at equilibrium.
Supporting information
Box plots of SNP density (a) and repeat proportion (b) in the 78 computationally inferred, putative Y-linked contigs, a subset of 25 of the 78 contigs that were independently validated as Y-linked by PCR, and all the other contigs of the assembly. Thick lines and boxes depict median and interquartile range, respectively. Whiskers are bounded to the most extreme data point within the 1.5 interquartile range. Open circles represent outliers. The underlying data for this figure can be found in S6 Table. SNP, single nucleotide polymorphism.
(TIF)
Y chromosome to autosome ratios were calculated for 60 individuals and compared with expected ratios: 1 for YY individuals (corresponding to 18 males and three females), 0.5 for XY individuals (28 males and five females), and 0 for XX individuals (six females). Thick lines and boxes depict median and interquartile range, respectively. Whiskers are bounded to the most extreme data point within the 1.5 interquartile range.
(TIF)
The pedigree spans five generations (F0–F4) and comprises 572 individuals (119 males and 453 females), 269 of which were included in molecular analyses (individuals not included in the molecular analyses are shown in dotted circles). Males are shown as squares, and females are shown as circles. Individuals carrying Wolbachia are shown in purple. Progeny identifiers are shown in gray. Sex chromosome genotype of individuals marked with an orange star was also assessed with a quantitative PCR assay.
(TIF)
The pedigree spans four generations (F0–F3) and comprises 226 individuals (132 males and 94 females), 196 of which were included in molecular analyses (individuals not included in the molecular analyses are shown in dotted circles). Males are shown as squares, and females are shown as circles. Individuals carrying Wolbachia are shown in purple. Progeny identifiers are shown in gray. Sex chromosome genotype of individuals marked with an orange star was also assessed with a quantitative PCR assay.
(TIF)
Males are shown as squares, and females are shown as circles.
(TIF)
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
nt, not tested because the male individual originated from the Wolbachia-free line and, hence, is necessarily XY (except II-F1-1 father from Wolbachia-infected line, hence XY or YY). Y chromosome-to-autosome ratio was calculated as 2−ΔCt.
(XLSX)
SNP, single nucleotide polymorphism.
(XLSX)
Abbreviations
- BUSCO
Benchmarking Universal Single-Copy Orthologs
- CQ
Chromosome Quotient
- Ne
effective population size
- SNP
single nucleotide polymorphism
- YGS
Y chromosome Genome Scan
Data Availability
The annotated genome sequence of Armadillidium nasatum is available in DDBJ/ENA/GenBank under accession number SEYY00000000. Accession numbers for Illumina and PacBio sequence datasets are SRR8759099-SRR8759101. All other relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by European Research Council Starting grant 260729 (EndoSexDet) and Agence Nationale de la Recherche grant ANR-15-CE32-0006-01 (CytoSexDet) to RC, the 2015-2020 State-Region Planning Contracts (CPER) and European Regional Development Fund (FEDER), and intramural funds from the Centre National de la Recherche Scientifique and the University of Poitiers. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci USA. 2007;104: 8627 10.1073/pnas.0611659104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110: 3229 10.1073/pnas.1218525110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bright M, Bulgheresi S. A complex journey: transmission of microbial symbionts. Nature Reviews Microbiology. 2010;8: 218 10.1038/nrmicro2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funkhouser LJ, Bordenstein SR. Mom Knows Best: The Universality of Maternal Microbial Transmission. PLoS Biol. 2013;11: e1001631 10.1371/journal.pbio.1001631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumann P. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005;59: 155–189. 10.1146/annurev.micro.59.030804.121041 [DOI] [PubMed] [Google Scholar]
- 6.Oliver KM, Degnan PH, Hunter MS, Moran NA. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science. 2009;325: 992–994. 10.1126/science.1174463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedges LM, Brownlie JC, O’Neill SL, Johnson KN. Wolbachia and Virus Protection in Insects. Science. 2008;322: 702–702. 10.1126/science.1162418 [DOI] [PubMed] [Google Scholar]
- 8.Teixeira L, Ferreira Á, Ashburner M. The Bacterial Symbiont Wolbachia Induces Resistance to RNA Viral Infections in Drosophila melanogaster. PLoS Biol. 2008;6: e1000002 10.1371/journal.pbio.1000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelstädter J, Hurst GDD. The Ecology and Evolution of Microbes that Manipulate Host Reproduction. Annual Review of Ecology, Evolution, and Systematics. 2009;40: 127–149. 10.1146/annurev.ecolsys.110308.120206 [DOI] [Google Scholar]
- 10.Cordaux R, Bouchon D, Grève P. The impact of endosymbionts on the evolution of host sex-determination mechanisms. Trends Genet. 2011;27: 332–341. 10.1016/j.tig.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 11.Hurst GDD, Frost CL. Reproductive parasitism: maternally inherited symbionts in a biparental world. Cold Spring Harbor perspectives in biology. 2015;7: a017699 10.1101/cshperspect.a017699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Werren JH, Beukeboom LW. Sex Determination, Sex Ratios, and Genetic Conflict. Annual Review of Ecology and Systematics. 1998;29: 233–261. 10.1146/annurev.ecolsys.29.1.233 [DOI] [Google Scholar]
- 13.Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman T-L, et al. Sex determination: why so many ways of doing it? PLoS Biol. 2014;12: e1001899 10.1371/journal.pbio.1001899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mank JE, Hosken DJ, Wedell N. Conflict on the Sex Chromosomes: Cause, Effect, and Complexity. Cold Spring Harb Perspect Biol. 2014;6: a017715 10.1101/cshperspect.a017715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beukeboom LW, Perrin N. The Evolution of Sex Determination. Oxford University Press; 2014. [Google Scholar]
- 16.Leclercq S, Thézé J, Chebbi MA, Giraud I, Moumen B, Ernenwein L, et al. Birth of a W sex chromosome by horizontal transfer of Wolbachia bacterial symbiont genome. PNAS. 2016; 15036–15041. 10.1073/pnas.1608979113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends in Ecology & Evolution. 2009;24: 94–102. 10.1016/j.tree.2008.09.010 [DOI] [PubMed] [Google Scholar]
- 18.Ming R, Bendahmane A, Renner SS. Sex Chromosomes in Land Plants. Annu Rev Plant Biol. 2011;62: 485–514. 10.1146/annurev-arplant-042110-103914 [DOI] [PubMed] [Google Scholar]
- 19.Bachtrog D. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 2013;14: 113–124. 10.1038/nrg3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becking T, Giraud I, Raimond M, Moumen B, Chandler C, Cordaux R, et al. Diversity and evolution of sex determination systems in terrestrial isopods. Scientific Reports. 2017;7: 1084 10.1038/s41598-017-01195-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouchon D, Rigaud T, Juchault P. Evidence for widespread Wolbachia infection in isopod crustaceans: molecular identification and host feminization. Proceedings of the Royal Society of London B: Biological Sciences. 1998;265: 1081–1090. 10.1098/rspb.1998.0402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cordaux R, Pichon S, Hatira HBA, Doublet V, Grève P, Marcadé I, et al. Widespread Wolbachia infection in terrestrial isopods and other crustaceans. Zookeys. 2012; 123–131. 10.3897/zookeys.176.2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Micro. 2008;6: 741–751. 10.1038/nrmicro1969 [DOI] [PubMed] [Google Scholar]
- 24.Rigaud T, Juchault P, Mocquard J-P. The evolution of sex determination in isopod crustaceans. Bioessays. 1997;19: 409–416. 10.1002/bies.950190508 [DOI] [Google Scholar]
- 25.Bouchon D, Cordaux R, Grève P. Feminizing Wolbachia and the evolution of sex determination in isopods Insect Symbiosis, Volume 3 Boca Raton, FL: CRC Press; 2008. pp. 273–294. [Google Scholar]
- 26.Cordaux R, Gilbert C. Evolutionary Significance of Wolbachia-to-Animal Horizontal Gene Transfer: Female Sex Determination and the f Element in the Isopod Armadillidium vulgare. Genes. 2017;8: 186 10.3390/genes8070186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin G, Juchault P, Legrand JJ. Mise en evidence d’un micro-organisme intracytoplasmique symbiote de l’oniscoide Armadillidium vulgare Latr. dont la presence accompagne l’intersexualite ou la feminisation totale des males genetiques de la lignee thelygene. C R Acad Sci Paris. 1973;276: 2213–2216. [Google Scholar]
- 28.Cordaux R, Michel-Salzat A, Frelon-Raimond M, Rigaud T, Bouchon D. Evidence for a new feminizing Wolbachia strain in the isopod Armadillidium vulgare: evolutionary implications. Heredity. 2004;93: 78–84. 10.1038/sj.hdy.6800482 [DOI] [PubMed] [Google Scholar]
- 29.Juchault P, Legrand J-J. Croisements de néo-mâles expérimentaux chez Armadillidium vulgare Latr. (Crustacé Isopode Oniscoïde). Mise en évidence d’une hétérogamétie femelle. C R Acad Sc Paris. 1972;1387–1389: 274–276. [Google Scholar]
- 30.Chebbi MA, Becking T, Moumen B, Giraud I, Gilbert C, Peccoud J, et al. The genome of Armadillidium vulgare (Crustacea, Isopoda) provides insights into sex chromosome evolution in the context of cytoplasmic sex determination. Molecular Biology and Evolution. 2019;36: 727–741. 10.1093/molbev/msz010 [DOI] [PubMed] [Google Scholar]
- 31.Taylor DR. Evolutionary consequences of cytoplasmic sex ratio distorters. Evol Ecol. 1990;4: 235–248. 10.1007/BF02214332 [DOI] [Google Scholar]
- 32.Ferdy J-B, Liu N, Sicard M. Transmission modes and the evolution of feminizing symbionts. J Evol Biol. 2016;29: 2395–2409. 10.1111/jeb.12963 [DOI] [PubMed] [Google Scholar]
- 33.Juchault P, Legrand J-J, Mocquard J-P. Contribution à l’étude qualitative et quantitative des facteurs contrôlant le sexe dans les populations du Crustacé Isopode terrestre Armadillidium vulgare Latr. I. La population de Niort (Deux-Sèvres). Arch Zool Exp Gén. 1980;121: 3–27. [Google Scholar]
- 34.Juchault P, Rigaud T, Mocquard JP. Evolution of sex determination and sex ratio variability in wild populations of Armadillidium vulgare (Latr.)(Crustacea, Isopoda): a case study in conflict resolution. Acta Oecologica. 1993;14: 547–562. [Google Scholar]
- 35.Juchault P, Legrand J-J. Analyse génétique et physiologique de la détermination du sexe dans une population du Crustacé Isopode Oniscoïde Armadillidium nasatum. Arch Zool exp gén. 1979;120: 25–43. [Google Scholar]
- 36.Cordaux R, Michel-Salzat A, Bouchon D. Wolbachia infection in crustaceans: novel hosts and potential routes for horizontal transmission. Journal of Evolutionary Biology. 2001;14: 237–243. 10.1046/j.1420-9101.2001.00279.x [DOI] [Google Scholar]
- 37.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- 38.Hall AB, Qi Y, Timoshevskiy V, Sharakhova MV, Sharakhov IV, Tu Z. Six novel Y chromosome genes in Anopheles mosquitoes discovered by independently sequencing males and females. BMC Genomics. 2013;14: 273 10.1186/1471-2164-14-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carvalho AB, Clark AG. Efficient identification of Y chromosome sequences in the human and Drosophila genomes. Genome Res. 2013;23: 1894–1907. 10.1101/gr.156034.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95: 118–128. 10.1038/sj.hdy.6800697 [DOI] [PubMed] [Google Scholar]
- 41.Rodrigues N, Studer T, Dufresnes C, Perrin N. Sex-Chromosome Recombination in Common Frogs Brings Water to the Fountain-of-Youth. Mol Biol Evol. 2018;35: 942–948. 10.1093/molbev/msy008 [DOI] [PubMed] [Google Scholar]
- 42.Rock DI, Smith AH, Joffe J, Albertus A, Wong N, O’Connor M, et al. Context-dependent vertical transmission shapes strong endosymbiont community structure in the pea aphid, Acyrthosiphon pisum. Mol Ecol. 2018;27: 2039–2056. 10.1111/mec.14449 [DOI] [PubMed] [Google Scholar]
- 43.Hughes GL, Dodson BL, Johnson RM, Murdock CC, Tsujimoto H, Suzuki Y, et al. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc Natl Acad Sci USA. 2014;111: 12498–12503. 10.1073/pnas.1408888111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newton ILG, Savytskyy O, Sheehan KB. Wolbachia utilize host actin for efficient maternal transmission in Drosophila melanogaster. PLoS Pathog. 2015;11: e1004798 10.1371/journal.ppat.1004798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Funkhouser-Jones LJ, van Opstal EJ, Sharma A, Bordenstein SR. The Maternal Effect Gene Wds Controls Wolbachia Titer in Nasonia. Curr Biol. 2018;28: 1692–1702.e6. 10.1016/j.cub.2018.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kageyama D, Ohno M, Sasaki T, Yoshido A, Konagaya T, Jouraku A, et al. Feminizing Wolbachia endosymbiont disrupts maternal sex chromosome inheritance in a butterfly species. Evol Lett. 2017;1: 232–244. 10.1002/evl3.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hornett EA, Charlat S, Duplouy AMR, Davies N, Roderick GK, Wedell N, et al. Evolution of Male-Killer Suppression in a Natural Population. PLoS Biol. 2006;4: e283 10.1371/journal.pbio.0040283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Juchault P, Legrand JJ. Sex determination and monogeny in terrestrial isopods Armadillidium vulgare (Latreille, 1804) and Armadillidium nasatum Budde-Lund, 1885. Monografia Monitore zoologico italiano. 1989;4: 359–375. [Google Scholar]
- 49.Werren JH. The coevolution of autosomal and cytoplasmic sex ratio factors. Journal of Theoretical Biology. 1987;124: 317–334. 10.1016/S0022-5193(87)80119-4 [DOI] [Google Scholar]
- 50.Rigaud T, Juchault P. Genetic control of the vertical transmission of a cytoplasmic sex factor in Armadillidium vulgare Latr.(Crustacea, Oniscidea). Heredity. 1992;68: 47–52. [Google Scholar]
- 51.Caubet Y, Hatcher MJ, Mocquard JP, Rigaud T. Genetic conflict and changes in heterogametic mechanisms of sex determination. Journal of Evolutionary Biology. 2000;13: 766–777. 10.1046/j.1420-9101.2000.00225.x [DOI] [Google Scholar]
- 52.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline: The Genome Analysis Toolkit Best Practices Pipeline In: Bateman A, Pearson WR, Stein LD, Stormo GD, Yates JR, editors. Current Protocols in Bioinformatics. Hoboken, NJ: John Wiley & Sons; 2013. pp. 11.10.1–11.10.33. 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13: 134 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Marine Biol Biotechnol. 1994;3: 294–299. [PubMed] [Google Scholar]
- 56.Chevalier F, Herbinière-Gaboreau J, Charif D, Mitta G, Gavory F, Wincker P, et al. Feminizing Wolbachia: a transcriptomics approach with insights on the immune response genes in Armadillidium vulgare. BMC Microbiology. 2012;12: S1 10.1186/1471-2180-12-S1-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Box plots of SNP density (a) and repeat proportion (b) in the 78 computationally inferred, putative Y-linked contigs, a subset of 25 of the 78 contigs that were independently validated as Y-linked by PCR, and all the other contigs of the assembly. Thick lines and boxes depict median and interquartile range, respectively. Whiskers are bounded to the most extreme data point within the 1.5 interquartile range. Open circles represent outliers. The underlying data for this figure can be found in S6 Table. SNP, single nucleotide polymorphism.
(TIF)
Y chromosome to autosome ratios were calculated for 60 individuals and compared with expected ratios: 1 for YY individuals (corresponding to 18 males and three females), 0.5 for XY individuals (28 males and five females), and 0 for XX individuals (six females). Thick lines and boxes depict median and interquartile range, respectively. Whiskers are bounded to the most extreme data point within the 1.5 interquartile range.
(TIF)
The pedigree spans five generations (F0–F4) and comprises 572 individuals (119 males and 453 females), 269 of which were included in molecular analyses (individuals not included in the molecular analyses are shown in dotted circles). Males are shown as squares, and females are shown as circles. Individuals carrying Wolbachia are shown in purple. Progeny identifiers are shown in gray. Sex chromosome genotype of individuals marked with an orange star was also assessed with a quantitative PCR assay.
(TIF)
The pedigree spans four generations (F0–F3) and comprises 226 individuals (132 males and 94 females), 196 of which were included in molecular analyses (individuals not included in the molecular analyses are shown in dotted circles). Males are shown as squares, and females are shown as circles. Individuals carrying Wolbachia are shown in purple. Progeny identifiers are shown in gray. Sex chromosome genotype of individuals marked with an orange star was also assessed with a quantitative PCR assay.
(TIF)
Males are shown as squares, and females are shown as circles.
(TIF)
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
nt, not tested because the male individual originated from the Wolbachia-free line and, hence, is necessarily XY (except II-F1-1 father from Wolbachia-infected line, hence XY or YY). Y chromosome-to-autosome ratio was calculated as 2−ΔCt.
(XLSX)
SNP, single nucleotide polymorphism.
(XLSX)
Data Availability Statement
The annotated genome sequence of Armadillidium nasatum is available in DDBJ/ENA/GenBank under accession number SEYY00000000. Accession numbers for Illumina and PacBio sequence datasets are SRR8759099-SRR8759101. All other relevant data are within the paper and its Supporting Information files.