Clostridium difficile represents today a real danger for human and animal health. It is the leading cause of diarrhea associated with health care in adults in industrialized countries. The incidence of these infections continues to increase, and this trend is accentuated by the general aging of the population. Many questions about the mechanisms contributing to C. difficile's success inside the host remain unanswered. The set of genetic tools available for this pathogen is limited, and new developments are badly needed. C. difficile has developed efficient defense systems that are directed against foreign DNA and that could contribute to its survival in phage-rich gut communities. We show how one such defense system, named CRISPR-Cas, can be hijacked for C. difficile genome editing. Our results also show a great potential for the use of the CRISPR-Cas system for the development of new therapeutic strategies against C. difficile infections.
KEYWORDS: Clostridium difficile, CRISPR, endogenous subtype I-B CRISPR-Cas system, genome editing
ABSTRACT
The human enteropathogen Clostridium difficile constitutes a key public health issue in industrialized countries. Many aspects of C. difficile pathophysiology and adaptation inside the host remain poorly understood. We have recently reported that this bacterium possesses an active CRISPR-Cas system of subtype I-B for defense against phages and other mobile genetic elements that could contribute to its success during infection. In this paper, we demonstrate that redirecting this endogenous CRISPR-Cas system toward autoimmunity allows efficient genome editing in C. difficile. We provide a detailed description of this newly developed approach and show, as a proof of principle, its efficient application for deletion of a specific gene in reference strain 630Δerm and in epidemic C. difficile strain R20291. The new method expands the arsenal of the currently limiting set of gene engineering tools available for investigation of C. difficile and may serve as the basis for new strategies to control C. difficile infections.
IMPORTANCE Clostridium difficile represents today a real danger for human and animal health. It is the leading cause of diarrhea associated with health care in adults in industrialized countries. The incidence of these infections continues to increase, and this trend is accentuated by the general aging of the population. Many questions about the mechanisms contributing to C. difficile's success inside the host remain unanswered. The set of genetic tools available for this pathogen is limited, and new developments are badly needed. C. difficile has developed efficient defense systems that are directed against foreign DNA and that could contribute to its survival in phage-rich gut communities. We show how one such defense system, named CRISPR-Cas, can be hijacked for C. difficile genome editing. Our results also show a great potential for the use of the CRISPR-Cas system for the development of new therapeutic strategies against C. difficile infections.
INTRODUCTION
The strictly anaerobic spore-forming bacterium Clostridium difficile (novel name, Clostridioides difficile [1]) is one of the major nosocomial pathogenic clostridia. This enteropathogen causes the majority of cases of antibiotic therapy-associated diarrhea and can lead to pseudomembranous colitis, a potentially lethal disease (2, 3). Over the last few decades, C. difficile infections have become one of the most important public health problems due to the emergence of hypervirulent strains (such as the PCR ribotype 027 R20291 strain) (4) and the increased incidence of C. difficile antibiotic resistance (5). The disruption of the colonic microflora caused by antibiotic therapy allows C. difficile to colonize the intestinal tract after the germination of preexisting or acquired spores (2, 6). Following gut colonization, C. difficile produces one or both of the large toxins TcdA and TcdB. These toxins trigger alterations in the intestinal cell cytoskeleton, resulting in cell lysis and inflammation (3, 7). Many aspects of C. difficile pathogenesis, including the molecular mechanisms of the infection cycle, remain poorly understood. Therefore, it is important to develop new genome editing approaches for further investigations of this emerging human pathogen.
CRISPR (clustered regularly interspaced short palindromic repeat)-Cas (CRISPR-associated) systems protect bacteria and archaea from phages and other mobile genetic elements (8). These adaptive immunity systems are highly diverse (9) and have been discovered in half of the sequenced bacterial genomes and in almost all archaeal genomes (10). CRISPR-Cas systems comprise CRISPR arrays and cas gene operons. CRISPR arrays are arranged into short direct repeats (20 to 40 bp) separated by variable spacers. Some spacers are complementary to protospacers, which are sequences within phage and other mobile genetic element genomes (11). CRISPR arrays are transcribed from promoters localized in leader regions into long pre-CRISPR RNAs (pre-crRNAs). Pre-crRNAs are processed into small protective CRISPR RNAs (crRNAs). In complex with Cas proteins, crRNAs serve as guides to recognize and direct the cleavage of foreign genetic elements by Cas nucleases in a process known as “interference” (12). New spacers are acquired into CRISPR arrays from foreign genomes during the adaptation process (13).
For many CRISPR-Cas systems, an important component of immunity mechanism is a protospacer-adjacent motif (PAM). PAMs are short sequences located on the 3′ or 5′ end of the protospacer. PAMs are necessary for protospacer recognition, and they are absent in CRISPR arrays; this allows avoidance of autoimmunity (8).
According to a recent classification based on the cas genes involved in interference, the CRISPR-Cas systems are divided into two classes and are further subdivided into six types and 33 subtypes (9). Class 1 includes type I, III, and IV CRISPR-Cas systems, which are characterized by multisubunit effector complexes, while class 2 includes type II, V, and VI CRISPR-Cas systems, which carry single-protein effectors. Recent studies showed that C. difficile strains possess an active subtype I-B CRISPR-Cas system (14–17). The C. difficile CRISPR-Cas system is characterized by an unusually high number of CRISPR arrays (on average, 8.5 CRISPR arrays per genome, with some arrays being localized in prophages) (16) and the presence of two or three cas operons belonging to the same subtype (15, 17). In our previous studies, we demonstrated active expression of all CRISPR arrays for the C. difficile 630 and R20291 strains, as well as the ability of C. difficile 630 to mount robust CRISPR interference (14, 15). We also bioinformatically predicted and experimentally validated C. difficile CRISPR-Cas PAMs (15).
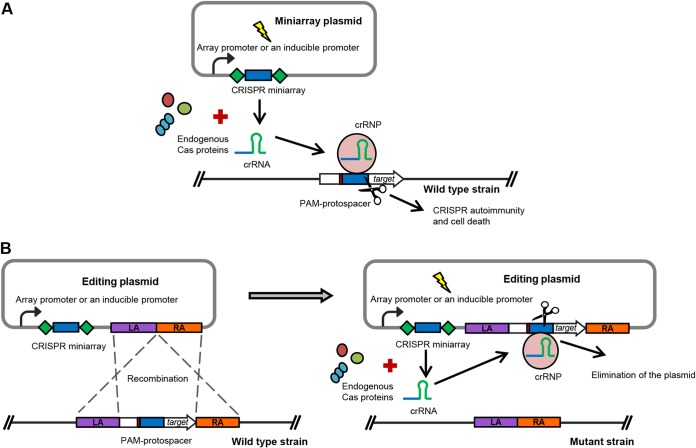
During the last few years, substantial efforts have been concentrated on the development of various CRISPR-based biotechnological tools (18). In particular, the type II Cas9- and type V Cpf1 (Cas12a)-based technologies are widely used for genome editing in different organisms (19, 20). Nevertheless, the application of other types of CRISPR-Cas systems has also attracted the attention of the scientific community. Harnessing of endogenous CRISPR-Cas systems for genome editing in bacteria and archaea appears to be a particularly attractive strategy (18, 21). This approach is based on the use of plasmid vectors containing artificial CRISPR miniarrays with spacers targeting a chromosomal gene (21). crRNAs expressed from a plasmid-borne miniarray utilize the endogenous Cas machinery to form an effector complex which recognizes the protospacer of choice, leading to its cleavage. Destruction of chromosomal DNA leads to the killing of wild-type cells (Fig. 1A). An editing plasmid with sequences homologous to sequences flanking the protospacer triggers homologous recombination and allelic exchange with the targeted chromosomal region (Fig. 1B). This results in elimination of the wild-type allele and preservation of chromosomal mutants since they no longer possess the targeted protospacer (Fig. 1B). The endogenous CRISPR-based method is often easier to set up for editing in prokaryotes than the CRISPR-Cas9 and CRISPR-Cpf1 (Cas12a) technologies. Another advantage of this approach is that there is no need to heterologously express potentially toxic Cas proteins inside bacterial or archaeal cells. The genome editing approach based on an endogenous CRISPR-Cas system was successfully applied in several prokaryotic organisms using CRISPR-Cas subtype I-A and III-B or subtype I-B in archaea, Sulfolobus islandicus (21) and Haloarcula hispanica (22), respectively, and subtype I-B in several clostridial species, C. pasteurianum (23), C. tyrobutyricum (24), and C. saccharoperbutylacetonicum (25).
FIG 1.
General scheme of using endogenous CRISPR-Cas systems for genome editing in bacteria and archaea. (A) The crRNA is expressed from a vector-borne CRISPR miniarray under the control of native or inducible promoters. The crRNA forms a ribonucleoprotein (crRNP) complex with endogenous Cas proteins, which recognizes and directs the cleavage of the PAM-associated protospacer, localized at the target chromosome region. This leads to chromosome disruption and cell death. (B) An editing plasmid, additionally carrying homologous arms (the left arm [LA] and the right arm [RA]), allows recombination between the plasmid and the chromosome to occur before CRISPR interference. The crRNP targets the PAM-protospacer on the plasmid, which leads to the elimination of the plasmid and preservation of the chromosomal mutants.
In C. difficile, various genetic tools for genome manipulation have been established. One of the most widely used methods is the ClosTron technology, based on mobile altered type II introns and the utilization of retrotransposable activated markers (RAM) (26, 27). Though this genome editing technique allows targeting of almost any chromosomal region and RAM enable one to easily identify potential mutants, the method has some disadvantages. Most importantly, ClosTron generates insertion mutations that may cause polar effects on downstream genes. An additional limitation comes from difficulties in finding an efficient insertion site within genes of a small size.
Another popular C. difficile genome editing approach is the allele-coupled exchange technique, based on a semisuicidal plasmid vector carrying the Escherichia coli cytosine deaminase gene (codA) or the C. difficile orotate phosphoribosyltransferase gene (pyrE) as a counterselection marker (28, 29). This method includes a two-step recombination event between the editing plasmid and the genome and the selection of double-crossing-over clones that lost the plasmid on nutrient-poor medium supplemented with 5-fluorocytosine (for codA-based plasmids) or fluoroorotic acid (for the pyrE allelic exchange system). The counterselection procedure is based on the generation of highly toxic compounds from these substrates. Despite the fact that this approach allows the creation of C. difficile mutants carrying point mutations, deletions, and insertions, it can be difficult to apply in some cases. First, mutations that result in a growth deficiency phenotype or the inactivation of metabolic genes may affect growth on nutrient-poor medium. Second, there are some difficulties with losing the editing plasmids in mutant strains after editing, which could lead to the spontaneous creation of revertant strains.
Recently, a method based on the DNA double-strand breaks in C. difficile has been reported (30). This technology uses site-specific cleavage by the Saccharomyces cerevisiae yeast homing endonuclease I-SceI, whose recognition site is introduced in the editing plasmid vector. After the integration of the editing vector into the chromosome, another vector containing the I-SceI endonuclease gene under the control of a constitutive promoter is transferred to the single-crossing-over integrants to induce double-strand breaks and genome editing via homologous recombination. The advantage of this method is the possibility to create markerless deletions and the fast loss of the vector. Nevertheless, this method includes time-consuming two-step conjugations and the expression of I-SceI endonuclease, which could induce side effects.
During the past few years, the successful application of CRISPR-Cas9 and CRISPR-Cpf1 (Cas12a) for genome editing in C. difficile has been reported (31–34). These approaches have enhanced the possibilities of genetic manipulation in C. difficile and have proven to be efficient. However, the Cas9 and Cpf1 technologies require the design of plasmids harboring specific single guide RNAs (sgRNAs), and the editing plasmid is not automatically cured after the editing is complete. The use of an endogenous CRISPR-Cas system can enhance the possibilities of the genetic manipulation of C. difficile. The present work describes the utilization of a native C. difficile subtype I-B CRISPR-Cas system to generate deletion mutants in the 630Δerm and R20291 strains.
To evaluate the possibility of using an endogenous C. difficile CRISPR-Cas system for the targeting of specific sequences on the bacterial chromosome, we have chosen the hfq gene. Hfq is a bacterial RNA-binding protein that plays major roles in RNA metabolism and the global posttranscriptional network, in particular, in Gram-negative bacteria (35). The study of Hfq depletion in C. difficile 630Δerm (36) suggested a pleiotropic role of this protein in C. difficile physiology, with the most pronounced effect being on sporulation. The availability of an hfq deletion mutant would open new perspectives for further characterization of its role in RNA-based regulation in C. difficile. Our previous attempts to inactivate the hfq gene using a ClosTron gene knockout system were unsuccessful (36). Additionally, we have tried to delete hfq using the codA allelic exchange approach (28, 29), but also without success (data not shown).
RESULTS
Construction of targeting miniarray plasmids and verification of their functionality.
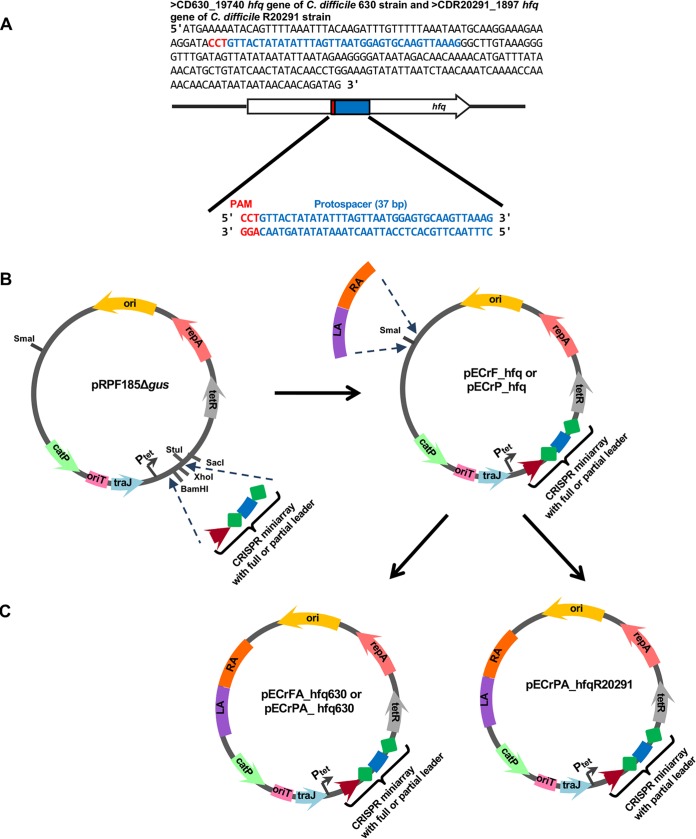
The general strategy for the construction of functional editing plasmids pECrFA_hfq630 and pECrPA_hfqR20291 for use in the 630Δerm and R20291 strains, respectively, is shown in Fig. 2. We first constructed two CRISPR miniarray plasmids targeting the hfq gene (pECrF_hfq and pECrP_hfq). The miniarray was based on the C. difficile 630Δerm CRISPR 16 array, which is highly expressed and capable of interference (15). Two variants of the leader sequence upstream of the miniarray were used (see Fig. S1A and B in the supplemental material): the full leader (a 403-bp sequence upstream of the first direct repeat of the CRISPR 16 array) containing all native promoters, which should allow autonomous expression of the miniarray (pECrF_hfq), and a partial leader (a 154-bp region upstream of the first direct repeat of the CRISPR 16 array), which lacked native promoters but which should allow the inducible expression of the miniarray from a vector-borne anhydrotetracycline (ATc)-inducible promoter (Ptet) (pECrP_hfq). The repeat-spacer-repeat motif of the synthetic miniarray was also based on 29-bp repeat sequences of the C. difficile 630Δerm CRISPR 16 array (Fig. 2A and Fig. S1A and B). For successful recognition of protospacers by the C. difficile CRISPR-Cas system, a functional PAM-flanking protospacer at the 5′ end is necessary (15). Two functional trinucleotide PAMs of the C. difficile CRISPR-Cas system, 5′ CCA and CCT, have been experimentally validated, and additional alternative motifs, such as CCC, CCG, and TCA, have been predicted (15). The coding region of the hfq gene possesses at least three functional CCW motifs and two alternative TCA motifs. A 37-bp sequence inside the hfq gene sequence associated with the 5′ CCT PAM was chosen (Fig. 2A; the mean length of the C. difficile spacers is 37 bp). The pECrF_hfq and pECrP_hfq plasmids (Fig. 2B) were conjugated to C. difficile 630Δerm cells using the heat shock method to ensure the highest conjugation efficiency (37). No transconjugants were obtained after conjugation of the pECrF_hfq plasmid in C. difficile 630Δerm, suggesting CRISPR autoimmunity due to self-targeting (Fig. 3A). The conjugation efficiency of 380 transconjugants/ml was observed after conjugation with pECrP_hfq (approximately 1.9 × 10−6 transconjugants/donor or recipient cell). A control conjugation with the pRPF185Δgus vector resulted in 5,480 transconjugants/ml (approximately 27.4 × 10−6 transconjugants/donor or recipient cell). The smaller number of transconjugants in the pECrF_hfq conjugation reaction could be due to Ptet promoter leakage leading to autoimmunity caused by self-cleavage in some transconjugants. To check for the efficiency of self-targeting by crRNA expressed from the pECrP_hfq plasmid, eight transconjugant colonies were restreaked on brain heart infusion (BHI) agar plates supplemented with 500 ng/ml ATc to fully induce the expression of the miniarray. No growth was observed on these plates, indicating highly efficient self-targeting by the induced miniarray (Fig. 3B). The same effects were observed after conjugation of the pECrF_hfq plasmid in C. difficile R20291 cells, suggesting that the synthetic array based on the C. difficile 630Δerm CRISPR 16 leader and repeat sequences mimics well native subtype I-B CRISPR arrays in C. difficile for at least the 630 and R20291 strains. Therefore, the C. difficile endogenous CRISPR-Cas system can recognize and target protospacers on the bacterial chromosome using crRNAs expressed from a plasmid-borne artificial miniarray, and this feature can be utilized for genome editing.
FIG 2.
Strategy for the design of the editing plasmids to delete the hfq gene in the C. difficile 630Δerm and R20291 strains. (A) The coding sequences of the C. difficile 630 and R20291 hfq genes and a 37-bp sequence associated with the 5′ CCT PAM, selected as a protospacer for the miniarray. (B) Construction of the pECrF_hfq and pECrP_hfq miniarray plasmids on the basis of the pRPF185Δgus vector. The miniarray sequences were cloned into the BamHI and XhoI restriction sites. (C) Construction of the pECrFA_hfq630, pECrPA_hfq630, and pECrPA_hfqR20291 editing plasmids on the basis of pECrF_hfq and pECrP_hfq. The homologous arms (LA and RA) were cloned into the SmaI restriction site. The F in the plasmid names represents the full-length leader region for autonomous expression of the miniarray under the control of native promoters, while the P points out the presence of a partial leader region without native promoters for miniarray expression under the control of an inducible Ptet promoter. The presence of homologous arms for recombination within the 630Δerm or R20291 strain is indicated by A and the strain name. The pECrFA_hfq630 plasmid carrying the miniarray with the full-length leader region was not efficient for gene deletion in the 630Δerm strain; in contrast, pECrPA_hfq630 and pECrPA_hfqR20291 were efficiently used for hfq gene deletion in the 630Δerm and R20291 strains, respectively.
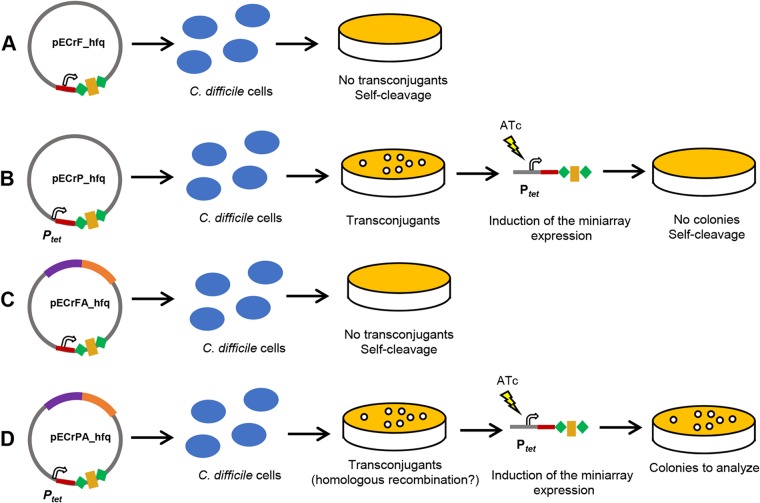
FIG 3.
Different effects of the conjugation of the miniarray and editing plasmids into C. difficile cells. (A) CRISPR self-cleavage induced by immediate expression of the miniarray from the plasmid pECrF_hfq after conjugation. (B) CRISPR self-cleavage resulted from the ATc-induced expression of the miniarray from the plasmid pECrP_hfq after second plating of the transconjugants. (C) CRISPR self-cleavage induced by the immediate expression of the miniarray from the pECrFA_hfq plasmid after conjugation. (D) Homologous recombination between the chromosome and the pECrPA_hfq plasmid and cleavage of the plasmid resulted from the ATc-induced expression of the miniarray from the plasmid after the second plating of the transconjugants. The effects were tested in the 630Δerm strain (B and C), and the effects were tested and gene deletion was performed in both the 630Δerm and R20291 strains (A and D).
Construction of the genome editing plasmid and deletion of the hfq gene of C. difficile 630Δerm and R20291.
We first assessed which miniarray plasmid, pECrF_hfq or pECrP_hfq, is best for C. difficile genome manipulation. Approximately 1,200-bp-long regions flanking the hfq gene of the 630Δerm strain (Fig. S1C) were amplified by PCR and introduced into the SmaI restriction sites of pECrF_hfq or pECrP_hfq using the Gibson assembly reaction (Fig. 2C). No transconjugants were obtained after conjugation of C. difficile 630Δerm with pECrFA_hfq630, carrying the miniarray with the full-length leader region (Fig. 3C). This may mean that the CRISPR-induced autoimmune degradation of DNA around the targeted protospacer is more efficient than homologous recombination between the chromosome and the homologous region of pECrFA_hfq630. Whatever the reason, the plasmid with the full-length CRISPR array leader sequence is clearly not suitable for genome editing. After conjugation with the pECrPA_hfq630 plasmid carrying the miniarray under the control of the inducible Ptet promoter, about 460 transconjugants/ml (approximately 2.3 × 10−6 transconjugants/donor or recipient cell) were obtained. To induce expression of the hfq-targeting miniarray, 10 transconjugants were restreaked on BHI agar supplemented with 500 ng/ml ATc. We observed the growth of each transconjugant tested, suggesting that homologous recombination between the chromosome and plasmid had occurred (Fig. 3D) or that CRISPR interference was not efficient. One clone from each plate was then restreaked on BHI plates with or without thiamphenicol (Tm) to check for plasmid loss. Three out of 10 clones lost the plasmid. When analyzed by PCR, these clones turned out to be Δhfq mutants (Fig. 4A). The experiment was independently repeated at least three times. In all cases, when testing 6 to 10 clones, the mutant strains could be reproducibly obtained with an overall efficiency varying from 30% to 100%. Thus, a plasmid containing an inducibly transcribed CRISPR miniarray and arms for homologous recombination at the targeted protospacer allows efficient genome editing in C. difficile.
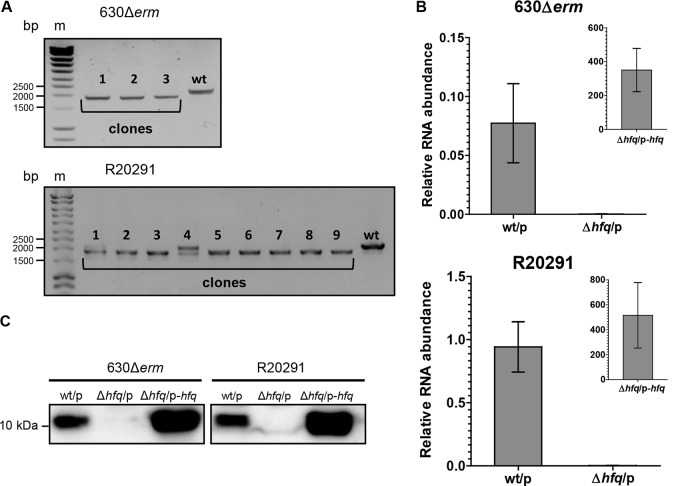
FIG 4.
Validation of hfq deletion mutants. (A) PCR analysis of the C. difficile clones which lost the plasmid after genome editing. The 2,151-bp PCR bands correspond to the wild-type genotype; the 1,893-bp PCR bands correspond to the mutant genotype. For the R20291 strain, both the wild-type and mutant copies were detected with clone 4 (lane 4); this clone was discarded from further analysis. Lanes m, molecular mass markers. (B) qRT-PCR analysis of the wild-type and Δhfq mutant strains carrying an empty pRPF185Δgus (the wt/p and Δhfq/p strains, respectively) and the complemented Δhfq C. difficile strain (the Δhfq/p-hfq strain). mRNA levels are relative to those of 16S rRNA. (C) Western blot analysis of the wt/p, Δhfq-p, and Δhfq/p-hfq C. difficile strains. As loading controls, InstantBlue dye-stained protein gels were used (see Fig. S2 in the supplemental material).
The coding region of the hfq gene of the C. difficile R20291 strain is identical to that of the 630Δerm strain, but the flanking sequences are different. Therefore, to delete the R20291 hfq gene, we constructed the pECrPA_hfqR20291 plasmid on the basis of the pECrP_hfq miniarray plasmid with homologous arms of R20291 hfq flanking sequences (Fig. 2C and Fig. S1D). Nine out of 10 selected transconjugants had lost the plasmid, and PCR analysis showed that seven out of nine clones without the plasmid were Δhfq mutants (Fig. 4A).
Validation and complementation of hfq deletion strains.
To validate the hfq deletion, we assessed hfq mRNA expression in the wild-type and Δhfq mutant strains carrying an empty pRPF185Δgus vector (the wt/p and Δhfq/p strains, respectively) as well as in complemented C. difficile Δhfq strain Δhfq/p-hfq expressing plasmid-borne hfq. Quantitative reverse transcription-PCR (qRT-PCR) analysis confirmed the absence of hfq expression in the C. difficile 630Δerm Δhfq and R20291 Δhfq strains and the presence of the transcript in the wild-type strains (Fig. 4B). A high 400- to 500-fold increase in hfq mRNA abundance compared to that in the wild type was detected in complemented strains due to strong Ptet induction in the presence of ATc (Fig. 4B). Western blotting with polyclonal anti-Hfq antibodies confirmed the lack of the Hfq protein in the Δhfq/p strains (Fig. 4C; InstantBlue dye-stained protein gels, used as loading controls, are shown in Fig. S2).
Sporulation assay of C. difficile 630Δerm Δhfq mutants.
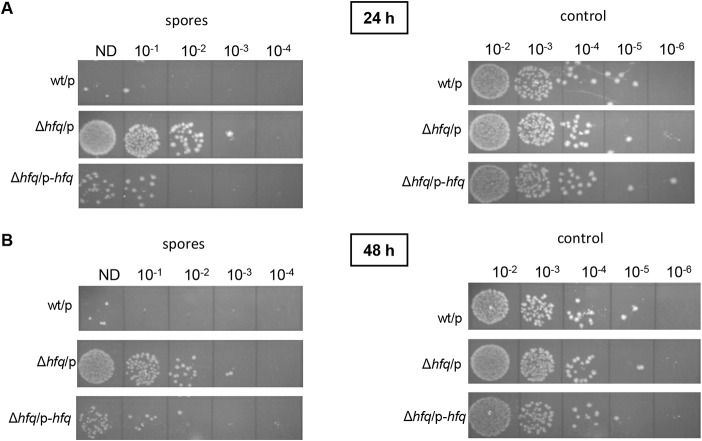
Sporulation represents one of the crucial features of C. difficile as a successful pathogen. In our previous work, we revealed that the Hfq protein likely controls the sporulation rates in C. difficile 630Δerm-derived strains (36). The Hfq-depleted strain demonstrated higher levels of sporulation than the control strain. To analyze the effect of the hfq gene deletion on this phenotype, we compared the sporulation rates in the 630Δerm wt/p, Δhfq/p, and Δhfq/p-hfq strains. After 24 h and 48 h in BHIS medium supplemented with Tm and ATc, the mutant strain (the Δhfq/p strain) demonstrated a higher level of sporulation than the wild-type strain (the wt/p strain) (Fig. 5). In addition, the complemented strain (the Δhfq/p-hfq strain) showed a reversion of sporulation efficiency to a level close to that seen in the wild type (Fig. 5). Thus, these results are consistent with previously obtained data and confirm the potential involvement of the Hfq protein in the control of sporulation in C. difficile (36).
FIG 5.
Sporulation levels in the C. difficile 630Δerm wt/p, Δhfq/p, and Δhfq/p-hfq strains (as numbers of spores) and the total amount of bacteria (as numbers of CFU) (control) after 24 h (A) and 48 h (B) of growth in BHIS supplemented with Tm and ATc. The serial dilutions of the cultures spotted on BHI plates supplemented with taurocholate are indicated (ND, not diluted). Spore samples were heated to kill all cells other than spores, while the control samples were not heated to estimate the total amount of bacteria.
DISCUSSION
Over the last decade, the rapid development of various biotechnological tools based on prokaryotic adaptive immune CRISPR-Cas systems has occurred (18). In addition to the most popular CRISPR tools, based on class 2 Cas9 and Cpf1 (Cas12a) proteins (19, 20), other CRISPR-Cas systems are also being actively explored for genetic manipulation purposes. One of the most promising applications is the use of endogenous CRISPR-Cas systems for genome editing and engineering of bacteria and archaea (18, 21).
In the present work, we utilized the endogenous CRISPR-Cas system for genome editing of enteropathogenic C. difficile. Although other techniques for genome manipulation in this bacterium are available (26–34), they present some limitations in their applications. Harnessing the native subtype I-B CRISPR-Cas system for genome editing in C. difficile allowed us to create deletion mutants of the hfq gene, encoding the RNA chaperone Hfq. Attempts to inactivate this gene using other approaches, including the ClosTron technology (36) and codA allelic exchange, were not successful (data not shown). Though a strain depleted of Hfq by expression of antisense RNA is available, the construction of the hfq deletion mutant opens up interesting possibilities for future studies of the regulatory role of Hfq and its RNA network in C. difficile.
The general work flow for the application of native CRISPR-Cas genome editing method in C. difficile is presented in Fig. 6. To repurpose the endogenous CRISPR-Cas system for deletion of the hfq gene, we designed plasmid vectors carrying targeting miniarray and editing plasmids carrying, in addition, homologous arms for recombination (Fig. 2B and C). The C. difficile 630Δerm CRISPR 16 array was chosen as a basis for synthetic miniarray construction, since it is functional for interference (15). The repeat-spacer-repeat motif for the artificial miniarray was composed of 29-bp repeat sequences and a 37-bp spacer sequence associated with a functional 5′ CCT PAM inside the hfq gene coding region. To facilitate the genome editing procedure, we used the pECrPA_hfq630 plasmid containing the miniarray under the control of the inducible Ptet promoter. This strategy allowed us to successfully generate hfq deletion mutants in both C. difficile 630Δerm and epidemic R20291 strains. The CRISPR repeats in the 630Δerm and R20291 strains have similar consensus sequences (15). Moreover, both strains possess homologous complete and partial subtype I-B cas operons also present in the majority of sequenced C. difficile strains (15). The Cas machineries of the R20291 strain (and, by extension, those of other C. difficile isolates) can successfully recognize and utilize crRNAs expressed from a 630Δerm-based miniarray. Thus, the artificial miniarray designed from the C. difficile 630Δerm CRISPR 16 leader and repeat sequences is suitable for targeting specific chromosomal protospacer sequences and can be used for genome editing in at least two C. difficile strains. The general conservation of subtype I-B cas operons in C. difficile makes it likely that the same targeting arrays will be suitable for the majority of C. difficile strains, though this conjecture remains to be experimentally verified.
FIG 6.
General work flow for application of endogenous CRISPR-Cas-based genome editing method in C. difficile.
Repurposing of native CRISPR-Cas systems for genome editing in C. difficile has considerable advantages over other techniques applied to this bacterium. First of all, this method does not require the expression of heterologous proteins inside C. difficile cells, which may have toxic or other unpredictable effects. A miniarray localized on an editing plasmid mimics the natural C. difficile CRISPR array and should not have an undesirable impact during genome manipulation. Second, this approach includes only one conjugation round and fewer plating steps, giving significant time savings (Fig. 6). For example, the codA allelic exchange method requires at least three more colony plating steps than the method with the miniarray editing plasmid, increasing the time needed to complete the editing experiment by at least 3 days. Finally, the miniarray editing plasmid is readily lost after the editing process, preventing the spontaneous emergence of revertants.
Among the possible challenges for the application of the method could be the choice of the best protospacer on the target genome region. The presence of a functional PAM upstream of the protospacer is imperative for successful targeting. For this reason, the choice of the genome sequence for editing should be guided by the availability of PAMs. For the moment, only two PAMs (CCA and CCT) have been experimentally confirmed for C. difficile CRISPR-Cas target recognition (15). At the same time, general in silico analysis of CRISPR spacer homology to phage protospacers revealed a rather unconstrained PAM consensus CCN/TCN for the C. difficile CRISPR-Cas system (15). These data increase the possibilities of target sequence selection. In addition, type I CRISPR-Cas systems can recognize protospacers on both strands of the target DNA, which expands the opportunities to find functional PAMs in the target region (21).
The applications of endogenous CRISPR-Cas system for genome editing in C. difficile could be potentially larger than those for the generation of deletion mutants. This technique could be readily applied for introducing other types of mutations, i.e., point mutations and insertions (21). For a point mutation, the homologous arms on the editing plasmid could be designed to introduce changes in the functional PAMs at the editing region to a nonfunctional motif. Alternatively, substitutions could be introduced into a seed region, the first 8 nucleotides of the protospacer, crucial for CRISPR targeting (38). As a priority choice, a point mutation design could be achieved by introducing changes at the first or second position of PAMs. Combining the changes within PAMs and the seed region could even increase the efficiency of editing, as reported for other endogenous CRISPR editing tools (21, 25). We have previously shown that a nonfunctional PAM and mutation in the first position of protospacer within the seed region abolished or considerably impaired CRISPR interference (15). Genome insertions designed to make a break in the integrity of the chosen protospacer or/and PAM of the targeted genome sequence (21) or to insert a mutation to knock out the PAM (25) could be introduced by the homologous arms.
The role of essential genes cannot be easily investigated since no deletion mutant can be generated. Therefore, the CRISPRi method (utilizing CRISPR interference), which allows repression of the expression of target genes, has recently been developed (39). This technology is primarily based on CRISPR-Cas9 systems with a mutated catalytic site of the Cas9 protein (catalytically dead Cas9 [dCas9]) (40). The dCas9-based method has already been used in C. difficile (41). In addition, it was shown that an E. coli native subtype I-E CRISPR-Cas system lacking cas3 could be repurposed for programmable transcriptional repression (42). Furthermore, a recent study showed that the subtype I-B CRISPR-Cas system of Haloferax volcanii lacking the cas3 and cas6 genes could be used for gene repression in this archaeon (43). Altogether, these data suggest that the C. difficile native CRISPR-Cas system may be used for this goal, too, in a particular context. However, about 90% of the sequenced C. difficile strains possess two subtype I-B cas operons, each carrying the cas3 nuclease gene. An additional partial cas operon with the cas3 gene is present in the majority of the multilocus sequence type 3 group of C. difficile strains, including the PCR ribotype 027 strains (15). Thus, depending on the strain, the creation of a double- or triple-cas3-mutant background would be necessary to consider application of this CRISPRi method.
CRISPR self-targeting could lead to bacterial cell death. This feature of CRISPR-Cas systems can be applied for the development of new antimicrobial agents (44). Among the suggested strategies reside the use of phage particles and phagemids as vectors to deliver all the necessary autotargeting CRISPR-Cas components inside the cell of the targeted pathogen (44). In the present study, we showed the active killing of C. difficile cells by CRISPR self-targeting via expression of the miniarray from a plasmid vector. Therefore, in perspective, this approach could be promising for the future development of alternative strategies for the treatment of C. difficile infections.
In conclusion, the repurposing of the endogenous CRISPR-Cas system for genome editing in C. difficile extends the range of biotechnological techniques available for this enteropathogenic bacterium and could be valuable in further studies.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All the plasmids and bacterial strains used in this study are listed in Table 1. C. difficile strains were grown in brain heart infusion (BHI; Difco) or tryptone, yeast extract (TY) (45) medium at 37°C under anaerobic conditions (5% H2, 5% CO2, 90% N2) in an anaerobic chamber (Jacomex). BHI medium supplemented with yeast extract (5 mg/ml) and l-cysteine (0.1%) (BHIS) was used in the sporulation experiments. When needed, thiamphenicol (Tm) at a final concentration of 15 μg/ml was added to the C. difficile cultures. The E. coli strains (Table 1) were grown in LB medium (46) supplemented with ampicillin (100 μg/ml) and chloramphenicol (15 μg/ml) when it was suitable. The nonantibiotic analog anhydrotetracycline (ATc) was used for induction of the Ptet promoter of pRPF185 vector derivatives in C. difficile (47).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| NEB-10 beta | Δ(ara-leu)7697 araD139 fhuA ΔlacX74 galK16 galE15 e14 mutant ϕ80dlacZΔM15 recA1 relA1 endA1 nupG rpsL (Strr) rph spoT1 Δ(mrr hsdRMS-mcrBC) | New England Biolabs |
| HB101(RP4) | supE44 aa14 galK2 lacY1 Δ(gpt-proA)62 rpsL20 (Strr) xyl-5 mtl-1 recA13 Δ(mcrC-mrr) hsdSB(rB− mB−) RP4 (Tra+ IncP Apr Kmr Tcr) | Laboratory stock |
| C. difficile | ||
| 630Δerm | Sequenced reference strain, ΔermB | Laboratory stock (52) |
| R20291 | PCR ribotype 027 epidemic strain | Laboratory stock |
| wt/p | 630Δerm or R20291 carrying the pRPFΔgus plasmid | This work |
| Δhfq/p | 630Δerm Δhfq or R20291 Δhfq carrying the pRPFΔgus plasmid | This work |
| Δhfq/p-hfq | 630Δerm Δhfq or R20291 Δhfq carrying the p-hfq plasmid | This work |
| Plasmid | ||
| pRPF185Δgus | pRPF185Δgus vector derivative | 14, 47 |
| pECrF_hfq | pRPF185Δgus carrying the hfq gene targeting the CRISPR miniarray with the full leader sequence | This work |
| pECrP_hfq | pRPF185Δgus carrying the hfq gene targeting the CRISPR miniarray with the partial leader sequence under the control of the Ptet promoter | This work |
| pECrFA_hfq630 | pECrF_hfq carrying arms for recombination in the 630Δerm strain | This work |
| pECrPA_hfq630 | pECrP_hfq carrying arms for recombination in the 630Δerm strain | This work |
| pECrPA_hfqR20291 | pECrP_hfq carrying arms for recombination in the R20291 strain | This work |
| p-hfq | pRPF185Δgus carrying the hfq gene under the control of the Ptet promoter | This work |
Plasmid construction and conjugation into C. difficile.
All the oligonucleotides used in this work are listed in Table 2. To create artificial CRISPR miniarrays targeting the C. difficile hfq gene, the full leader sequence (positions −403 to −1 relative to the first nucleotide of the first repeat in the array) and a partial leader sequence (positions −154 to −1 relative to the first nucleotide of the first repeat in the array) of the C. difficile 630Δerm CRISPR 16 array were amplified by PCR of genomic DNA (see Fig. S1A and B in the supplemental material). The artificial repeat-spacer-repeat motif was amplified by PCR from synthetic oligonucleotides to generate the double-stranded fragment. The full or partial leader sequence and the repeat-spacer-repeat motif were assembled and cloned into the BamHI and XhoI sites of the pRPF185Δgus plasmid vector (14) using the Gibson assembly reaction (48), giving the pECrF_hfq and pECrP_hfq miniarray plasmids (Fig. 2B).
TABLE 2.
Oligonucleotides used in this study
| Primer purpose and name | Sequence (5′–3′)a | Descriptionb |
|---|---|---|
| Construction of CRISPR miniarray plasmid | ||
| AM81 | TAACAGATCTGAGCTCCAGGCCTTCAATTATATGATAGGTTTTTTATTAAGCATACTAGCTGGTGTTATATC | Full leader CR16 F |
| AM82 | GTTAATCTAAAACCCCAAAATAAACTTAGTATTTCCAATATCTACACATACAC | Leader CR16-R |
| AM83 | TAACAGATCTGAGCTCCAGGCCTTCTGAGCAATATTTGCGATAAATTGAAGTTTAACAATTG | Partial leader CR16-F |
| AM91 | GTTTTAGATTAACTATATGGAATGTAAATGTTACTATATATTTAGTTAATGGAGTGCAAGTTAAAGGTTTTAGATTAACTATATGGAATGTAAAT | hfq repeat-spacer-repeat motif |
| AM92 | AGTTTATTTTGGGGTTTTAGATTAACTATATGGAATGTAAATGTTACTATATATTTAGTTAATGGAGTG | Repeat-spacer-repeat F |
| AM93 | TTTAAAGTTTTATTAAAACTTATAGATTTACATTCCATATAGTTAATCTAAAACCTTTAACTTGCAC | Repeat-spacer-repeat R |
| Construction of editing plasmids | ||
| AM158 | GAACACTTGCCGAAAAAGAAAAACTGCCGGGTACGTACCCCGATATTGAAATAAAAAGTTTATTG | Left arm 630 and R20291 F |
| AM159 | TCTTAAATTAAATTAATTATTAGATTTGTACCCTCCCAAG | Left arm 630 and R20291 R |
| AM160 | CTTGGGAGGGTACAAATCTAATAATTAATTTAATTTAAGATGATTGAG | Right arm 630 and R20291 F |
| AM161 | GAGCGAGGAAGCGGAAGAGCGCTCGGCGGGGATCGATCCCGGAACAGGTTTTACATAAGAATC | Right arm 630 and R20291 R |
| Δhfq mutant detection | ||
| AM106 | ACTAAAAGGGTCATAAGAGC | Δhfq F |
| AM169 | TATAAGGAGGTCTTATTGGAGC | Δhfq R |
| Construction of plasmids for complementation | ||
| HFQ1 | GAAGGCCTGGTAGGAATATTTTAGAAGT | 5′ hfq StuI |
| HFQ2 | GGGGATCCCATTAAGCATTTTATCACCTGTC | 3′ hfq BamHI |
| qRT-PCR | ||
| QRTBD37 | GGGAGACTTGAGTGCAGGAG | 16S RNA F |
| QRTBD38 | GTGCCTCAGCGTCAGTTACA | 16S RNA R |
| IMV447 | AGGGCTTGTAAAGGGGTTTG | qRT-PCR hfq F |
| IMV448 | TTGTTGTTTTGGTTTTGATTTGTT | qRT-PCR hfq R |
Overlapping regions are indicated in boldface, and underlined sequences represent those of the restriction endonucleases.
CR16, CRISPR 16 array; F, forward; R, reverse.
To construct editing plasmids, approximately 1,200-bp-long regions flanking the hfq gene of the 630Δerm and R20291 strains (Fig. S1C and D) were amplified by PCR and introduced into the SmaI restriction site of pECrF_hfq or pECrP_hfq using the Gibson assembly reaction, resulting in the pECrFA_hfq630, pECrPA_hfq630, and pECrPA_hfqR20291 plasmids (Fig. 2C).
To construct a plasmid for complementation of the hfq deletion, the hfq gene sequence, including the ribosome-binding site (positions −50 to +397 relative to the translational start site), was amplified by PCR and cloned into the StuI and BamHI sites of pRPF185Δgus under the control of the ATc-inducible Ptet promoter, giving the p-hfq plasmid.
DNA sequencing was performed to verify the plasmid constructs. pRPF185Δgus is a shuttle vector that replicates both in E. coli (ColE1 origin) and in C. difficile. All resulting plasmids were transformed into the E. coli HB101(RP4) strain and further transferred into C. difficile cells by conjugation. The heat shock method with incubation for 15 min at 50°C was used to get the highest conjugation efficiency (37). C. difficile transconjugants were selected on BHI agar containing Tm (15 μg/ml), d-cycloserine (25 μg/ml), and cefoxitin (8 μg/ml).
Deletion of the hfq gene and validation of Δhfq mutants.
To induce the expression of the CRISPR miniarrays under the control of the Ptet promoter, C. difficile transconjugants containing the pECrP_hfq, pECrPA_hfq630, or pECrPA_hfqR20291 plasmid were subsequently restreaked onto BHI agar supplemented with ATc (500 ng/ml). The resulting C. difficile colonies were then restreaked in parallel onto BHI agar supplemented or not with Tm (15 μg/ml) to check for plasmid loss. Subsequently, selected clones without plasmids were analyzed by PCR to detect the chromosomal deletion of the hfq gene. The resulting PCR fragments were sequenced to confirm the gene deletion.
RNA extraction and qRT-PCR.
For total RNA extraction, C. difficile 630Δerm- and R20291-derived pRPF185Δgus- and p-hfq-carrying strains were grown for 6 h or 8 h in TY medium supplemented with Tm (7.5 μg/ml) and ATc (250 ng/ml). Total RNA isolation was performed as previously described (49). cDNA synthesis by reverse transcription and quantitative reverse transcription PCR (qRT-PCR) was performed as previously described (50) using a Bio-Rad CFX Connect real-time system. The expression level of the hfq gene relative to that of the 16S RNA gene was calculated (51).
Protein extract preparation and Western blotting.
To extract total proteins, C. difficile 630Δerm- and R20291-derived pRPF185Δgus- and p-hfq-carrying strains were grown for 6 h or 16 h in TY medium supplemented with Tm (7.5 μg/ml) and ATc (250 ng/ml). Cell lysis and protein extraction were performed as previously described (36).
For each sample, 30 μg of protein extract was loaded onto two 15% SDS polyacrylamide gels in parallel. After the electrophoresis, proteins from the 1st gel were transferred to a polyvinylidene fluoride membrane. Membrane hybridization with primary and secondary antibodies was then performed as described before (36). The bioluminescent signal from the secondary antibodies was detected using the SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) and a Fusion FX (Vilber Lourmat) digital camera. The 2nd gel was stained with InstantBlue dye (Expedeon) and used as a loading control (Fig. S2).
Sporulation assay.
C. difficile strains harboring the pRPF185Δgus and p-hfq plasmids were grown overnight in TY medium containing Tm (15 μg/ml). Overnight cultures were used to inoculate the strain at an optical density at 600 nm (OD600) of 0.1 in fresh TY medium supplemented with taurocholate (0.1%), d-fructose (0.5%), Tm (7.5 μg/ml), and ATc (10 ng/ml) to get only vegetative cells. When the cultures had reached an OD600 of 1.0 to 1.5, they were diluted to an OD600 of 0.01 in BHIS medium containing Tm (7.5 μg/ml) and ATc (10 ng/ml) and grown at 37°C. After 24 h and 48 h of growth, 1 ml of each culture was divided into two samples. To determine the total amount of bacteria (in number of CFU), the first sample was serially diluted and spotted (10 μl per spot) onto BHI agar containing 0.1% taurocholate. The second sample was incubated at 65°C for 30 min to eliminate vegetative cells. Subsequently, the sample was serially diluted and spotted (10 μl per spot) onto BHI agar containing 0.1% taurocholate to estimate the number of spores.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Agence Nationale de la Recherche (CloSTARn; grant ANR-13-JSV3-0005-01 to O.S.), the Institut Universitaire de France (to O.S.), the University Paris-Sud, the Institute for Integrative Biology of the Cell, the DIM-1HEALTH regional Ile de France program (LSP grant no. 164466), the CNRS-RFBR PRC 2019 (grant no. 288426) to O.S. and K.S., a Vernadski fellowship to A.M., and a Skoltech Biomedical Initiative grant (grant no. SBI RF-0000000136) to K.S.
We are grateful to P. Boudry, E. Semenova, and J. Peltier for helpful discussions during the preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01416-19.
REFERENCES
- 1.Oren A, Rupnik M. 2018. Clostridium difficile and Clostridioides difficile: two validly published and correct names. Anaerobe 52:125–126. doi: 10.1016/j.anaerobe.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 3.Abt MC, McKenney PT, Pamer EG. 2016. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol 14:609–620. doi: 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 5.Banawas SS. 2018. Clostridium difficile infections: a global overview of drug sensitivity and resistance mechanisms. Biomed Res Int 2018:8414257. doi: 10.1155/2018/8414257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seekatz AM, Young VB. 2014. Clostridium difficile and the microbiota. J Clin Invest 124:4182–4189. doi: 10.1172/JCI72336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Just I, Selzer J, Wilm M, von Eichel-Streiber C, Mann M, Aktories K. 1995. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375:500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- 8.Sorek R, Lawrence CM, Wiedenheft B. 2013. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem 82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 9.Koonin EV, Makarova KS, Zhang F. 2017. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grissa I, Vergnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shmakov SA, Sitnik V, Makarova KS, Wolf YI, Severinov KV, Koonin EV. 2017. The CRISPR spacer space is dominated by sequences from species-specific mobilomes. mBio 8:e01397-17. doi: 10.1128/mBio.01397-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garneau JE, Dupuis M-V, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 13.Marraffini LA. 2015. CRISPR-Cas immunity in prokaryotes. Nature 526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 14.Soutourina O, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, Semenova E, Severinov K, Le Bouguenec C, Coppée JY, Dupuy B, Martin-Verstraete I. 2013. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet 9:e1003493. doi: 10.1371/journal.pgen.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boudry P, Semenova E, Monot M, Datsenko KA, Lopatina A, Sekulovic O, Ospina-Bedoya M, Fortier LC, Severinov K, Dupuy B, Soutourina O. 2015. Function of the CRISPR-Cas system of the human pathogen Clostridium difficile. mBio 6:e01112-15. doi: 10.1128/mBio.01112-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen JM, Shoup M, Robinson C, Britton R, Olsen KEP, Barrangou R. 2016. CRISPR diversity and microevolution in Clostridium difficile. Genome Biol Evol 8:2841–2855. doi: 10.1093/gbe/evw203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hargreaves KR, Flores CO, Lawley TD, Clokie RJ, Trevor D, Hargreaves KR, Flores CO, Lawley TD, Clokie RJ. 2014. Abundant and diverse clustered regularly interspaced short palindromic repeat spacers in Clostridium difficile strains and prophages target multiple phage types within this pathogen. mBio 5:e01045-13. doi: 10.1128/mBio.01045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrangou R, Horvath P. 2017. A decade of discovery: CRISPR functions and applications. Nat Microbiol 2:17092. doi: 10.1038/nmicrobiol.2017.92. [DOI] [PubMed] [Google Scholar]
- 19.Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Safari F, Zare K, Negahdaripour M, Barekati-Mowahed M, Ghasemi Y. 2019. CRISPR Cpf1 proteins: structure, function and implications for genome editing. Cell Biosci 9:36. doi: 10.1186/s13578-019-0298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Pan S, Zhang Y, Ren M, Feng M, Peng N, Chen L, Liang YX, She Q. 2016. Harnessing type I and type III CRISPR-Cas systems for genome editing. Nucleic Acids Res 44:e34. doi: 10.1093/nar/gkv1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng F, Gong L, Zhao D, Yang H, Zhou J, Li M, Xiang H. 2017. Harnessing the native type I-B CRISPR-Cas for genome editing in a polyploid archaeon. J Genet Genomics 44:541–548. doi: 10.1016/j.jgg.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Pyne ME, Bruder MR, Moo-Young M, Chung DA, Chou CP. 2016. Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium. Sci Rep 6:25666. doi: 10.1038/srep25666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Zong W, Hong W, Zhang Z-T, Wang Y. 2018. Exploiting endogenous CRISPR-Cas system for multiplex genome editing in Clostridium tyrobutyricum and engineer the strain for high-level butanol production. Metab Eng 47:49–59. doi: 10.1016/j.ymben.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Atmadjaja AN, Holby V, Harding AJ, Krabben P, Smith HK, Jenkinson ER. 2019. CRISPR-Cas, a highly effective tool for genome editing in Clostridium saccharoperbutylacetonicum N1-4(HMT). FEMS Microbiol Lett 366:fnz059. doi: 10.1093/femsle/fnz059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, Minton NP. 2010. The ClosTron: mutagenesis in Clostridium refined and streamlined. J Microbiol Methods 80:49–55. doi: 10.1016/j.mimet.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 27.Kuehne SA, Heap JT, Cooksley CM, Cartman ST, Minton NP. 2011. ClosTron-mediated engineering of Clostridium. Methods Mol Biol 765:389–407. doi: 10.1007/978-1-61779-197-0_23. [DOI] [PubMed] [Google Scholar]
- 28.Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP. 2012. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl Environ Microbiol 78:4683–4690. doi: 10.1128/AEM.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng YK, Ehsaan M, Philip S, Collery MM, Janoir C, Collignon A, Cartman ST, Minton NP. 2013. Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PLoS One 8:e56051. doi: 10.1371/journal.pone.0056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Theophilou ES, Vohra P, Gallagher MP, Poxton IR, Blakely GW. 2018. Generation of markerless deletions in the nosocomial pathogen Clostridium difficile by induction of DNA double-strand breaks. Appl Environ Microbiol 85:e02055-18. doi: 10.1128/AEM.02055-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McAllister KN, Bouillaut L, Kahn JN, Self WT, Sorg JA. 2017. Using CRISPR-Cas9-mediated genome editing to generate C. difficile mutants defective in selenoproteins synthesis. Sci Rep 7:14672. doi: 10.1038/s41598-017-15236-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingle P, Groothuis D, Rowe P, Huang H, Cockayne A, Kuehne SA, Jiang W, Gu Y, Humphreys CM, Minton NP. 2019. Generation of a fully erythromycin-sensitive strain of Clostridioides difficile using a novel CRISPR-Cas9 genome editing system. Sci Rep 9:8123. doi: 10.1038/s41598-019-44458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inés I, Cañ C, Groothuis D, Zygouropoulou M, Rodrigues R, Minton NP, 2019. RiboCas: a universal CRISPR-based editing tool for Clostridium. ACS Synth Biol 8:1379–1390. doi: 10.1021/acssynbio.9b00075. [DOI] [PubMed] [Google Scholar]
- 34.Hong W, Zhang J, Cui G, Wang L, Wang Y. 2018. Multiplexed CRISPR-Cpf1-mediated genome editing in Clostridium difficile toward the understanding of pathogenesis of C. difficile infection. ACS Synth Biol 7:1588–1600. doi: 10.1021/acssynbio.8b00087. [DOI] [PubMed] [Google Scholar]
- 35.Sobrero P, Valverde C. 2012. The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit Rev Microbiol 38:276–299. doi: 10.3109/1040841X.2012.664540. [DOI] [PubMed] [Google Scholar]
- 36.Boudry P, Gracia C, Monot M, Caillet J, Saujet L, Hajnsdorf E, Dupuy B, Martin-Verstraete I, Soutourina O. 2014. Pleiotropic role of the RNA chaperone protein Hfq in the human pathogen Clostridium difficile. J Bacteriol 196:3234–3248. doi: 10.1128/JB.01923-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirk JA, Fagan RP. 2016. Heat shock increases conjugation efficiency in Clostridium difficile. Anaerobe 42:1–5. doi: 10.1016/j.anaerobe.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJJ, Severinov K. 2011. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A 108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CHS, Koo BM, Marta E, Shiver AL, Whitehead EH, Weissman JS, Brown ED, Qi LS, Huang KC, Gross C. 2016. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Müh U, Pannullo AG, Weiss DS, Ellermeier CD. 2019. A xylose-inducible expression system and a CRISPRi-plasmid for targeted knock-down of gene expression in Clostridioides difficile. J Bacteriol 201:e00711-18. doi: 10.1128/JB.00711-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo ML, Mullis AS, Leenay RT, Beisel CL. 2015. Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res 43:674–681. doi: 10.1093/nar/gku971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stachler A-E, Marchfelder A. 2016. Gene repression in Haloarchaea using the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas I-B system. J Biol Chem 291:15226–15242. doi: 10.1074/jbc.M116.724062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bikard D, Barrangou R. 2017. Using CRISPR-Cas systems as antimicrobials. Curr Opin Microbiol 37:155–160. doi: 10.1016/j.mib.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 45.Dupuy B, Sonenshein AL. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol 27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- 46.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fagan RP, Fairweather NF. 2011. Clostridium difficile has two parallel and essential Sec secretion systems. J Biol Chem 286:27483–27493. doi: 10.1074/jbc.M111.263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 49.André G, Even S, Putzer H, Burguière P, Croux C, Danchin A, Martin-Verstraete I, Soutourina O. 2008. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res 36:5955–5969. doi: 10.1093/nar/gkn601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. 2011. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J Bacteriol 193:3186–3196. doi: 10.1128/JB.00272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Metcalf D, Sharif S, Weese JS. 2010. Evaluation of candidate reference genes in Clostridium difficile for gene expression normalization. Anaerobe 16:439–443. doi: 10.1016/j.anaerobe.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 52.Hussain HA, Roberts AP, Mullany P. 2005. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Δerm) and demonstration that the conjugative transposon Tn916ΔE enters the genome of this strain at multiple sites. J Med Microbiol 54:137–141. doi: 10.1099/jmm.0.45790-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.