The ubiquitous protozoan Toxoplasma gondii is the subject of renewed interest due to the spread of oocysts in water and food causing endemic and epidemic outbreaks of toxoplasmosis in humans and animals worldwide. Displaying a sensitivity close to animal models, cell culture represents a real alternative to assess the infectivity of oocysts in water and in biological sentinel mussels. This method opens interesting perspectives for evaluating human exposure to infectious T. gondii oocysts in the environment, where oocyst amounts are considered to be very small.
KEYWORDS: Toxoplasma gondii, biomonitoring, in vitro cell culture, infectivity, mussels, oocysts, qPCR, sporocysts, waterborne pathogens
ABSTRACT
Toxoplasma gondii is a ubiquitous foodborne protozoan that can infect humans at low dose and displays different prevalences among countries in the world. Ingestion of food or water contaminated with small amounts of T. gondii oocysts may result in human infection. However, there are no regulations for monitoring oocysts in food, mainly because of a lack of standardized methods to detect them. The objectives of this study were (i) to develop a reliable method, applicable in biomonitoring, for the rapid detection of infectious oocysts by cell culture of their sporocysts combined with quantitative PCR (sporocyst-CC-qPCR) and (ii) to adapt this method to blue and zebra mussels experimentally contaminated by oocysts with the objective to use these organisms as sentinels of aquatic environments. Combining mechanical treatment and bead beating leads to the release of 84% ± 14% of free sporocysts. The sporocyst-CC-qPCR detected fewer than ten infectious oocysts in water within 4 days (1 day of contact and 3 days of cell culture) compared to detection after 4 weeks by mouse bioassay. For both mussel matrices, oocysts were prepurified using a 30% Percoll gradient and treated with sodium hypochlorite before cell culture of their sporocysts. This assay was able to detect as few as ten infective oocysts. This sporocyst-based CC-qPCR appears to be a good alternative to mouse bioassay for monitoring infectious T. gondii oocysts directly in water and also using biological sentinel mussel species. This method offers a new perspective to assess the environmental risk for human health associated with this parasite.
IMPORTANCE The ubiquitous protozoan Toxoplasma gondii is the subject of renewed interest due to the spread of oocysts in water and food causing endemic and epidemic outbreaks of toxoplasmosis in humans and animals worldwide. Displaying a sensitivity close to animal models, cell culture represents a real alternative to assess the infectivity of oocysts in water and in biological sentinel mussels. This method opens interesting perspectives for evaluating human exposure to infectious T. gondii oocysts in the environment, where oocyst amounts are considered to be very small.
INTRODUCTION
The apicomplexan Toxoplasma gondii, an obligate intracellular parasite, can infect humans and a wide range of warm-blooded vertebrates, leading to toxoplasmosis. This generally benign infection can cause severe life-threatening disease, particularly in immunocompromised patients and congenitally infected children (1).
There are two major infective stages of the parasite that can infect humans: tissue cysts (bradyzoites), found only in meat (2, 3), and oocysts (sporozoites), which are shed exclusively by infected felids and contaminate soil, water, or food (fruits, vegetables, and mollusks) (4). One infected cat can excrete up to 100 million oocysts, which become infective following sporulation. Oocysts are known to resist environmental conditions, most physical and/or chemical treatments (5), and some industrial processes applied to foods, such as high hydrostatic pressure (6).
Toxoplasma gondii oocysts were responsible for 2% of parasitic protozoan outbreaks between January 2004 and December 2010 (4, 7). Oocysts transmitted by water were associated with 21% of waterborne outbreaks between 1976 and 2009 (7–9). Several studies reported the detection of the parasite in surface and drinking waters (10, 11) and in fruits, vegetables, and mollusks exposed to contaminated waters (12, 13).
Monitoring approaches of water quality are based upon punctual sampling (time and location), and methods used to detect T. gondii oocysts in water require the filtration of large volumes of water (up to 1,000 liters) to concentrate parasites before their detection. Moreover, in aquatic habitats, oocysts are subjected to dilution events, and water characteristics such as salinity, organic matter content, and temperature can affect oocyst transport dynamics as well as their spatial and temporal distributions (14). Using water for monitoring oocysts in water bodies can thus lead to variable results depending on physicochemical and meteorological parameters, which are particularly important in the present context of global climate change. To circumvent the main drawbacks of water analyses, new alternative approaches to water analyses have recently emerged in water quality surveys using host-associated microorganisms as natural biosamplers (15). Special attention has been paid to bivalves, because their intense filtering activity leads to a high accumulation of pathogens (15, 16). Hence, studying bivalves can highlight pathogen contamination while water analysis results are negative (17). Laboratory studies have shown that marine and freshwater bivalves can concentrate waterborne protozoan parasites (18–20). Consistent with this, some studies have reported the detection of T. gondii oocysts in different marine (12, 21–23) or continental (24) bivalves, allowing the study of a large spatial scale (freshwater-seawater continuum).
Experiments have demonstrated that T. gondii oocysts can sporulate in seawater, be concentrated by mussels, and remain infectious for laboratory mice (25–27). Usually, DNA-based methods are applied to detect protozoa in mollusks (23, 28, 29). However, DNA can persist for a long time in dead cells (30), thus preventing a distinction between viable and dead parasites. As only viable parasites are potentially infectious and can lead to illness, viability is a major feature for assessing the health risk. Methods to measure the viability and infectivity of protozoa, including T. gondii, were recently reviewed (31). Among the molecular techniques, propidium monoazide based-PCR assays appeared to not be relevant to measure the viability of T. gondii oocysts (32), and RNA-based methods overestimated the exposure of humans to viable oocysts because of the persistence of RNA in dead parasites (33, 34). Considering that all viable parasites are not necessarily infectious, i.e., able to replicate within host cells, the methods allowing the characterization of infectivity remain the most reliable ones.
Many authors have used animal models, the gold standard, to evaluate the infectiosity of T. gondii oocysts spiked on raspberries or blueberries (6, 35) or in naturally contaminated mussels and oysters (12, 25) or in water (11). However, bioassays are time-consuming, labor intensive, and expensive and raise ethical concerns. Moreover, bioassays only provide a qualitative assessment of oocyst viability. It is therefore essential to develop complementary methods, easier to implement in the laboratory and for stakeholders, to provide tools to quantify viable infective parasites in the environment.
In vitro cell culture assays are alternative approaches to bioassays. They have been widely applied associated with quantitative PCR (qPCR) or reverse transcriptase quantitative PCR (RT-qPCR) methods to detect infectious viruses (36, 37) and Cryptosporidium parvum (38–43) and T. gondii oocysts (44, 45) following sporozoite excystation. In these latter studies, excystation relied on a mechanical treatment to obtain free sporocysts, followed by their incubation with bile salts to release the sporozoites. However, this method is long and nonreproducible and results in a poor sporozoite yield mainly because of parasite loss due to mechanical treatment and bile salt exposure. The objective of this study was therefore to propose a rapid, accessible, and sensitive method to detect infectious T. gondii oocysts for water quality monitoring based on the use of bivalves as indicators of water contamination. An approach based on the infection of cell cultures using free sporocysts, combined with qPCR, was first developed in a simple matrix (water) and characterized in terms of limit of detection and correlation with mouse bioassays. Then, the developed method was adapted to the detection of oocysts by using the continental zebra mussel (Dreissena polymorpha) and the coastal blue mussel (Mytilus edulis) as reliable indicators of water quality assessment in the freshwater-seawater continuum.
RESULTS
Selection of the protocol for the release of sporocysts from T. gondii oocysts.
As sporozoite excystation protocols usually lead to low sporozoite yield, we chose to test different protocols to break the oocyst wall while keeping the sporocysts intact for cell culture. For this, viable oocysts were exposed to different mechanical disruption protocols to optimize the release of intact sporocysts. The assessed parameters were types (ceramic, glass, or a mix [Lysing Matrix E from MP Biomedicals]) and sizes (0.4 to 2 mm) of beads, suspension solution (0.05% SDS, Iscove's modified Dulbecco's growth medium [IMDM]), time of agitation, and instrument for agitation (vortex, TissueLyser). However, considering the high variability observed using the vortex mixer (data not shown), the TissueLyser was selected for further optimization (see Fig. S1 in the supplemental material).
Overall, irrespective of the time of TissueLyser agitation and of the suspension solution (0.05% SDS and IMDM), less than 40% of sporocysts were released with ceramic and glass beads. A higher percentage was obtained following agitation with small glass beads (425 to 600 μm) for 30 s in 0.05% SDS and with larger glass beads (2 mm) for 3 min in 0.05% SDS. However, under both conditions, the interassay variability was high (11%). Mechanical disruption with a TissueLyser associated with a Lysing Matrix E tube led to the release of more than 25% of sporocysts irrespective of the agitation time. The highest percentage rate was obtained after 30 s in IMDM, with 84% ± 14% of released sporocysts. Hence, a 30-s agitation of oocysts in the Lysing Matrix E tube in IMDM was the condition used for further sporocyst-cell culture (CC)-qPCR experiments.
CC-qPCR based on the infection of cells challenged with T. gondii sporocysts.
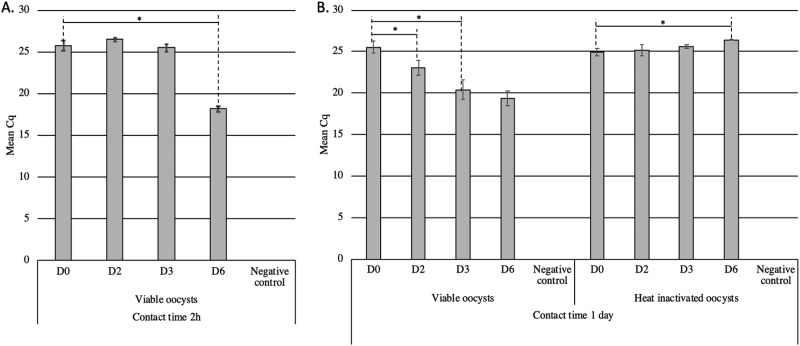
The optimal contact time required for the sporocysts to naturally excyst and release sporozoites and for the sporozoites to penetrate the cells was determined in vitro. To that aim, Vero cells and sporocysts were left in contact for 2 h or 1 day before washing of the cells (i.e., D0) (Fig. 1A and B). Irrespective of the contact time, T. gondii DNA was systematically detected by qPCR in total DNA extracted from the cell pellet. These results suggest that the two tested contact times were sufficient to allow some sporozoites to penetrate the cells and/or that some sporocysts remained stuck to the surfaces of Vero cells, even after washing. Then, the ability of the sporozoites to differentiate into replicative tachyzoites in Vero cells was assessed by qPCR following 2 to 6 days (D2 to D6) of culture. When cell culture was stopped at D2 or D3 following a contact time of 2 h (Fig. 1A), the quantification cycle (Cq) values did not vary significantly (P value > 0.05), suggesting that no or few tachyzoites multiplied within the cells. But after 6 days of cell culture, the Cq significantly decreased compared to that at D0 (Cq D0 − Cq D6 = 7.6; P value < 0.05), demonstrating the presence of replicative tachyzoites (Fig. 1A).
FIG 1.
Kinetics of cell infection by T. gondii sporocysts in simple matrix assessed by sporocyst-CC-qPCR assay. Following 2 h (A) or 1 day (B) of contact between the sporocysts and Vero cells (D0), the cells were cultivated for 2 (D2), 3 (D3), or 6 days (D6). Sporocysts were obtained after TissueLyser agitation (30 s, 33 Hz) from viable and heat-inactivated oocysts (5 min at 99°C). Error bars indicate standard deviation (2 < n < 5 independent experiments). Negative control corresponds to Vero cells without sporocysts. *, P < 0.05.
When the contact between cells and sporocysts was lengthened to 1 day, the qPCR signal decreased from 25.51 ± 0.70 at D0 to 23.03 ± 0.95 at D2 and 20.40 ± 1.17 at D3 of culture. Although increasing the time of culture resulted in a larger decrease in Cq values (Cq D0 − Cq D6 = 6.2; P value < 0.05), we selected the protocol allowing the detection of infectious oocysts within 4 days (1 day contact time plus 3 days cell culture) as an optimum. This assay was able to specifically detect infective oocysts as demonstrated by the absence of qPCR signal reduction following contact of cells with heat-inactivated oocysts (Fig. 1B).
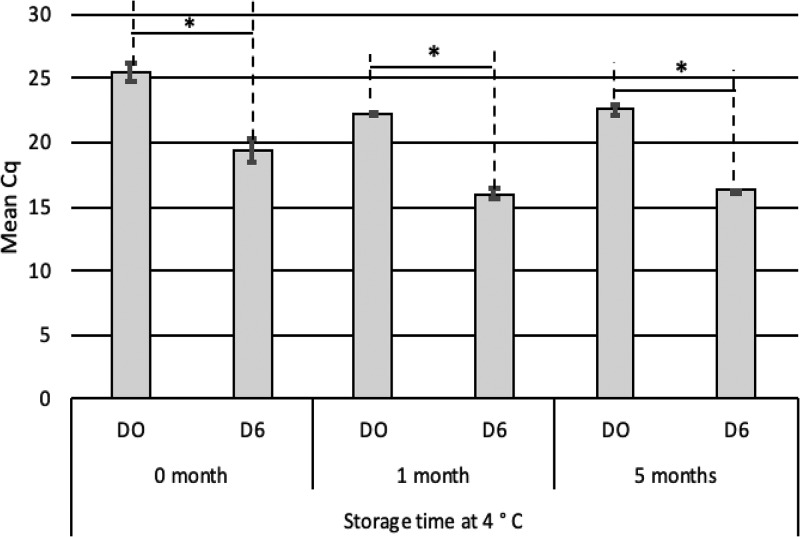
To assess the effect of the TissueLyser method on the infective potential of sporocysts, free sporocysts were stored for 1 or 5 months in phosphate-buffered saline (PBS) at 4°C (Fig. 2). The cell culture was stopped at D6 to be under the most favorable condition to observe parasite infectivity. Irrespective of storage time, the Cq decreases between D0 and D6 (Cq D0 − Cq D6 = 6) were similar (Fig. 2). Thus, mechanical agitation by the TissueLyser did not alter the infectivity of sporocysts stored up to 5 months, demonstrating the robustness of the sporocyst stage compared to sporozoites, which are very fragile.
FIG 2.
Infectivity of T. gondii sporocysts after storage at 4°C assessed by sporocyst-CC-qPCR. After 1 day of contact between the sporocysts and Vero cells (D0), the cells were cultivated for 6 days (D6). Sporocysts were obtained after TissueLyser agitation (30 s, 33 Hz) from viable oocysts and stored 0, 1, or 5 months at 4°C. Error bars indicate standard deviation (2 < n < 9 independent experiments). *, P < 0.05.
Limit of detection of the T. gondii sporocyst-based CC-qPCR assay in a simple matrix.
The protocol allowing the detection of infective oocysts within the shortest time, i.e., 4 days (1 day contact time plus 3 days cell culture), was selected for further characterization. A serial dilution ranging from 101 to 104 oocysts was used to prepare sporocysts for Vero cell infection using the protocol selected above. Ten infective oocysts were consistently detected (3/3 positive replicates at day 3) (Table 1), indicating that the limit of detection of the method was <10 oocysts. The linear correlation between the initial number of infectious parasites and the Cq values from 10,000 to 100 (R2 > 0.99) suggested that this model was quantitative in this range and that the limit of quantification was between 10 and 100 oocysts.
TABLE 1.
Limit of detection of the T. gondii sporocyst-CC-qPCR assay in simple matrixa
| Oocyst quantity | D0 |
D3 |
||
|---|---|---|---|---|
| Cq (mean ± SD) | No. positive exptsb/total no. | Cq (mean ± SD) | No. positive exptsb/total no. | |
| 10,000 | 25.36 ± 0.59 | 3/3 | 20.40 ± 1.17c | 3/3 |
| 1,000 | 29.37 ± 0.27 | 3/3 | 24.06 ± 0.09c | 3/3 |
| 100 | 32.71 ± 0.41 | 3/3 | 27.38 ± 0.75c | 3/3 |
| 10 | 33.33 ± 1.08 | 3/3 | 28.01 ± 2.07c | 3/3 |
A serial dilution ranging from 10 to 10,000 T. gondii oocysts was used to obtain sporocysts resuspended in culture medium and then deposited on Vero cells (80% confluence) incubated at 37°C. qPCR specific to T. gondii was performed from the cell pellet DNA extract after a contact time between sporozoites and cells of 1 day (D0) and after culture of the cells for 3 days (D3) (n = 3 independent experiments).
Positive signal by qPCR in well culture.
Significantly different from D0 (P < 0.05).
Assessing infectivity of heat-treated T. gondii oocysts by CC-qPCR and bioassay.
To be reliable, the measure of infectivity by sporocyst-based CC-qPCR needs to correlate with that of the bioassay, which is the reference method to detect infective T. gondii oocysts. To address this, oocysts were inactivated by heating for 5 min at 99°C, 2 min at 80°C, or 2 min at 60°C. The two latter treatments were selected because they were shown to lead to the absence of correlation between animal bioassays and the viability method based on reverse transcriptase qPCR (46). However, the developed sporocyst-CC-qPCR showed a valid correlation with mouse bioassays irrespective of the applied heat treatment (see Table S1). Indeed, untreated oocysts were infectious using both methods, while no tachyzoites were detected by sporocyst-CC-qPCR and mice remained seronegative with heat-treated T. gondii. Hence, the proposed sporocyst-based CC-qPCR assay showed good agreement with the bioassay to determine the infectivity of oocysts exposed to heat treatments and was able to specifically detect infective oocysts.
Potential of the CC-qPCR assay to determine the infectivity of T. gondii oocysts in spiked mussels (M. edulis and D. polymorpha).
To determine the infectivity of oocysts in mussel matrices, some adjustments of the sporocyst-based CC-qPCR assay were required (see details in Materials and Methods). First, the oocysts recovered from M. edulis and D. polymorpha were purified using a Percoll gradient (30%) combined with a sodium hypochlorite treatment, prior to sporocyst release. Second, in order to avoid cell culture contamination due to mussel tissues, cell infection was performed in 24-well plates to dilute mussel tissue homogenates, and the contact between sporocysts and cells was reduced from 1 day to 3 h.
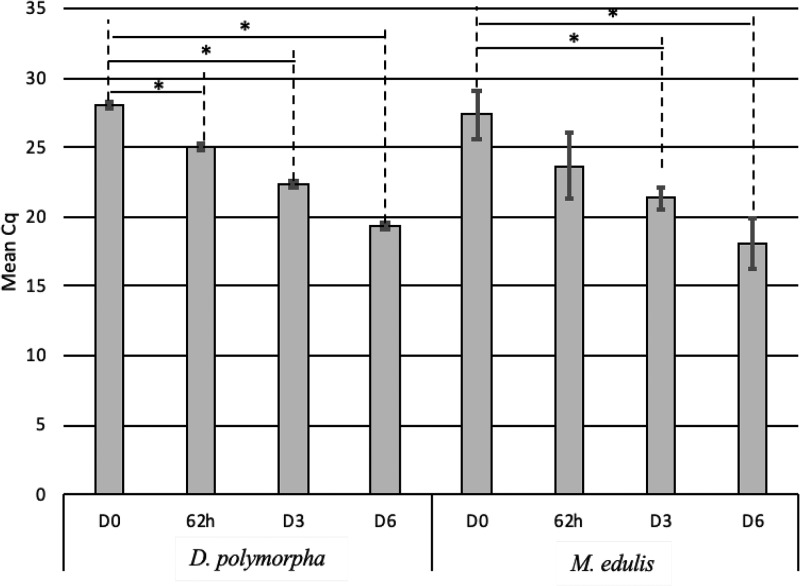
The kinetics of Vero cell infection with sporocysts isolated from M. edulis and D. polymorpha homogenates are presented in Fig. 3. For D. polymorpha, a significant decrease of Cq was observed from 62 h (P value < 0.05) and was maximal after 6 days (Cq D0 − Cq D6 = 8.52). For M. edulis, the decrease was remarkable after 3 days of culture (D3) and was maximal after 6 days (ΔCq = 9.3). To detect infectious oocysts in mussel matrix as quickly as possible, a culture time of D3 was used for both mussel matrices. Then, the limit of detection of the sporocyst-based assays to detect infective parasites in both mussel species was determined (Table 2). To that aim, mussel tissues were spiked with a number of oocysts ranging from 10 to 104 and then were processed as described in Materials and Methods to recover oocysts and to release sporocysts before infecting cell cultures. For both mussels, as few as 10 infective oocysts initially spiked to tissues led to parasite multiplication and cell infection within 3 days (Cq = 24.1 for M. edulis and Cq = 19.44 for D. polymorpha) (Table 2). These results indicated that the limit of detection of this sporocyst-based CC-qPCR was below 10 oocysts. However, the Cq values obtained at D3 were not correlated to the initial amount of oocysts spiked on tissues (r2 ≤ 0.81) (Table 2), suggesting this in vitro model is not quantitative.
FIG 3.
Kinetics of cell infection by T. gondii sporocysts isolated from D. polymorpha and M. edulis assessed by sporocyst-CC-qPCR. Sporocysts were released from oocysts isolated from mussels, and following 3 h of contact between the sporocysts and Vero cells (D0), the cells were cultivated for 62 h or 3 (D3) or 6 (D6) days. Error bars indicate standard deviation (n = 3 independent experiments). *, P < 0.05.
TABLE 2.
Limit of detection of the sporocyst-CC-qPCR assay of the sporocyst-CC-qPCR assay in D. polymorpha and M. edulisa
| Parameter | Oocyst quantity |
Cq (mean ± SD) |
|
|---|---|---|---|
| D. polymorpha | M. edulis | ||
| D0 | 10,000 | 25.73 ± 0.42 | 27.34 ± 1.73 |
| D3 | 50,000 | 20.25 ± 0.36 | 18.13 ± 0.21 |
| 10,000 | 21.83 ± 0.54 | 18.82 ± 0.41 | |
| 5,000 | 22.43 ± 0.55 | 19 ± 0.34 | |
| 1,000 | 23.85 ± 0.13 | 19.49 ± 0.64 | |
| 500 | 23.77 ± 0.44 | 20.15 ± 0.16 | |
| 100 | 24.42 ± 0.17 | 19.94 ± 0.69 | |
| 50 | 25.08 ± 1.23 | 19.72 ± 0.14 | |
| 10 | 24.1 ± 0.20 | 19.44 ± 0.36 | |
| Equation | y = −1.14x + 26.47 | y = −0.39x + 20.47 | |
| r² | 0.81 | 0.57 | |
Oocysts (10 to 104) were spiked to tissues from one mussel. Then, the tissues were processed to recover oocysts and to release sporocysts before infecting cells (see Materials and Methods). qPCR was performed at D0 (i.e., after 3 h of contact between sporocysts and cells) and at D3 (i.e., after 3 days of cell culture) (n = 3 independent experiments).
DISCUSSION
The waterborne transmission route of T. gondii to humans via the dissemination of oocysts through surface and drinking waters and its epidemiological impact are now more significant than previously believed (4, 7, 47, 48). Through 2010, T. gondii had been identified as the etiologic agent in seven reported waterborne outbreaks in Brazil, Panama, Canada, French Guiana, and India (7, 9). The parasite has been detected in surface water bodies used for recreation, in commercially harvested shellfish generally consumed raw or undercooked (see references 40 and 49 for reviews), and in drinking water that could infect humans. Moreover, in contrast to running water, using filter-feeding attached organisms makes the contamination measurement representative of the sample or exposure site. Measurements in biological matrices (mussels) could (i) limit the variability (temporal integration) of measurements compared to those taken in water, and (ii) provide more reliable data on the degree of contamination of water bodies, thus facilitating their comparison. The continuous improvement of methods to detect T. gondii oocysts could help to determine the prevalence of the parasite in different environmental samples and to study a large spatial scale (freshwater-seawater continuum); marine and seawater invertebrates could be tested at the same time.
To assess the human health risk linked to T. gondii oocysts, information is required on parasite viability. Viability assays postulate that only viable oocysts can lead to host infection. However, viable parasites are not necessarily all infectious, and consequently, such assays can lead to an overestimation of the risk. This is particularly the case for nonsporulated T. gondii oocysts that are noninfectious while being viable. The current method used to assess oocyst infectivity is bioassay, which is considered the gold standard. However, as well as being expensive, there are ethical concerns related to the use of animal models. Cell culture assays have been described as promising alternative methods in terms of cost and response delay. However, although cell culture combined with qPCR has been largely described for C. parvum oocysts (38, 39, 42, 43, 50), only a few studies have described cell culture from T. gondii oocysts. They are based on sporozoite-host cell coculture and the subsequent detection of proliferating parasites by qPCR (44, 45). Moreover, no study has reported CC-qPCR assays with the objective of detecting small amounts of T. gondii oocysts under conditions similar to those of environmental samples with natural contamination (31). Here we describe a cell culture assay based on T. gondii sporocysts which is suitable for environmental applications in terms of performance and implementation.
Several protocols have been described for the excystation of T. gondii oocysts. The effectiveness of the sporozoite-releasing methods represents a critical step. Given the robust nature of the oocyst wall (51), protocols usually rely on mechanical breaking of the wall to allow release of the sporocysts. Then, sporozoite excystation is achieved by incubating sporocysts in a biliary salt suspension. We tested different protocols to break the oocyst walls while keeping the sporocyst walls intact (44, 45, 52). Here, a TissueLyser combined with Lysing Matrix E gave the best results.
To obtain the best conditions for opening the sporocyst wall, several studies used bile salt or sodium taurocholate treatment associated or not with Na2CO3 and medium saturated with CO2 (53, 54). However, we observed significant sporozoite loss, huge mortality, and nonreproducible protocols because of sporozoite fragility (data not shown), which has already been described for Isospora suis (55) but not corroborated with the release of 80.6% sporozoites in the study by Villegas et al. (45). As no concluding results were obtained for the sporozoite release step, we tested the ability of sporocysts to naturally excyst in cell culture at 37°C. Only mechanical grinding with a TissueLyser (30 s) associated with a Lysing Matrix E tube in IMDM achieved the release of 84% ± 14% of sporocysts. However, this percentage of released sporocysts is probably underestimated, because 10% to 15% of oocysts never sporulate (32). This method can be easily implemented in laboratories and can be applied to oocysts isolated from environmental or food samples, such as mussels.
To determine the optimal conditions for measuring infectivity, two parameters were studied: (i) the contact time between the sporocysts and cells that is required for the release of sporozoites and cell infection and (ii) the time of cell culture that is required to measure parasite multiplication by qPCR.
In water, infective oocysts were detected following 1 day of contact between sporocysts and cells and 2 days of cell culture, demonstrating that sporocysts were able to excyst, to release sporozoites that could eventually invade cells, and to differentiate into replicative tachyzoites in 3 days. However, the optimal sporocyst-based CC-qPCR condition was established at 3 days of cell culture following 1 day of contact, allowing a large and significant parasite multiplication in the shortest time (i.e., 4 days). Our results also demonstrated that mechanical grinding with a TissueLyser combined with Lysing Matrix E did not damage fresh sporocysts and also stored sporocysts (up to 5 months at 4°C), which were shown to still be able to induce cell infection. Using this 4-day sporocyst-based CC-qPCR assay, the time to response was divided by 2.5, relative to the 10 days usually described for cell culture based on sporozoite infection (44, 45). Obviously, this sporocyst-based CC-qPCR assay also represented a significant improvement compared to animal models (3 weeks). Reducing the time of contact between the sporocysts and the cells to 2 h led to an increase of the time to response, with the requirement of 6 days of cell culture to significantly detect parasite multiplication. The limit of detection of the 4-day sporocyst-CC-qPCR assay was <10 oocysts in water, which makes it suitable for environmental applications where the oocyst level of contamination can be low. Moreover, a linear correlation between the initial number of infective oocysts (101 to 104) and the Cq values obtained after 3 days of cell culture was observed, suggesting that this approach could be used to assess treatment efficacy in water.
To obtain a reliable assessment of the exposure of humans to infective oocysts, the proposed approach needs to correlate with an infectivity measure using the reference method, i.e., animal models. Heat treatments that showed correlation between bioassays and a viability method based on reverse transcriptase qPCR for 5 min at 99°C, but not for 2 min at 80°C or 2 min at 60°C (46), were applied to oocysts. Irrespective of the treatment, no parasite multiplication was detected using the sporocyst-CC-qPCR assay, indicating that this approach was able to specifically detect infective oocysts. Moreover, results from the sporocyst-based CC-qPCR assay were in agreement with those of the bioassay, indicating that this approach is as reliable as animal models, to assess the exposure of humans or mammals to infective oocysts.
The sporocyst-based CC-qPCR developed in water matrix was adapted to mussel matrices (M. edulis and D. polymorpha). Percoll gradient purification has been used on clams but also on other types of matrices, such as meat, cheese, eggs, and fish (56). We tested a 30% Percoll gradient purification step for eliminating most of the mussel matrix contaminants and sodium hypochlorite for removing bacteria, and we observed the infectivity of T. gondii oocysts in mussel matrices. Percoll did not alter T. gondii oocyst infectivity, as shown for C. parvum oocysts (57), and seems therefore compatible with sporocyst-CC-qPCR on a mollusk matrix.
To avoid fungal growth during cell culture, a 24-well plate format assay was used to dilute the mussel matrices still present after Percoll separation and sodium hypochlorite treatment, and the contact time between sporocysts and cells was reduced to 3 h (versus 1 day for oocysts in water). Longer culture times enhanced the detection of infectious parasites in mussel matrices. These results are consistent with previous observations for C. parvum oocysts that highlighted that increasing the culture time to 72 h led to higher levels of infection (58). With the objective of using the sporocyst-based CC-qPCR assay in a monitoring application, the assay allowing the significant detection of infective oocysts at a high level within 3 days (3 h of contact between sporocysts and cells plus 3 days of cell culture) was selected. This method was able to detect fewer than 10 infective T. gondii oocysts in both M. edulis and D. polymorpha, with a result in only 3 days. In comparison, in vivo, the limit of detection on the Mytella guyanensis mussel was between 10 and 100 T. gondii oocysts, with a result obtained after 8 weeks (12). Nevertheless, the sporocyst-based CC-qPCR appeared to not be quantitative in mussels, indicating that it could not be used to study the efficiency of industrial processes in these matrices. Considering that a single oocyst is sufficient to cause infection/disease (in some hosts), rather than quantification, a low level of detection is crucial in the context of risk assessment. Thus, the sporocyst-based CC-qPCR method represents a promising alternative to bioassays to detect infective T. gondii oocysts in M. edulis and D. polymorpha in environmental monitoring.
To conclude, we have developed a laboratory-accessible method based on Vero cell infection using sporocyst cell culture combined with qPCR detection, allowing the specific characterization of fewer than 10 infective oocysts in water (4 days) and in mussels (3 days). This new approach, which correlates with bioassays, is relevant to assess the exposure to infective T. gondii oocysts in water, by direct analyses of water samples, but also in mussels as biological sentinels for a more representative characterization of the risk. Overall, this tool appears to be of benefit to the food/water industries to help them assess the health risk associated with the presence of infective oocysts.
MATERIALS AND METHODS
T. gondii oocysts.
Oocysts of type III strain VEG were produced as described previously (59), containing 87.2% of sporulated oocysts, provided in H2SO4 aqueous solution (2%), and stored at 4°C until use. Before experiments, oocysts were washed three times in sterile distilled water (dH2O) to remove sulfuric acid. The concentrations of parasite suspensions used for experiments were calibrated by counting oocysts in sodium dodecyl sulfate (SDS) aqueous solution (0.5%) on a Kova Glasstic Slide 10 (with 10 wells; Kova International) using a phase-contrast microscope (Axioskop 40; Zeiss). Oocysts used throughout this study were aged less than 14 months, and their viability was controlled before use by RT-qPCR targeting the sporo-SAG gene as previously described (31). Oocysts were inactivated by heating in a dry heating block as follows: 5 min at 99°C, 2 min at 80°C, and 2 min at 60°C. To minimize experimental variability, each treatment condition was tested the same day for CC-qPCR and the mouse bioassay. Three aliquots were dedicated to bioassays and six to CC-qPCR analyses.
Release of sporocysts.
To optimize the release of sporocysts, 104 oocysts in IMDM (see below) or 0.05% SDS (sufficient quantity for 350 μl) were subjected to mechanical agitation using a vortex mixer (33 rpm) and a TissueLyser (Qiagen) at 33 Hz. Different sizes and types of beads were tested (2 mm or 425 to 600 μm for glass beads, 1.2 mm for ceramic beads, or a Lysing Matrix E tube from MP Biomedicals that contains 1.4-mm ceramic spheres, 0.1-mm silica spheres, and one 4-mm glass bead), as well as different beating times (0 s, 30 s, 1 min, 2 min, 3 min, or 5 min). The free sporocysts and oocysts were counted in a Kova slide cell (with 10 wells; Kova International). To calculate the percentage of released sporocysts (number of sporocysts observed/number of theoretical sporocysts), we considered that 104 oocysts corresponded to 2 × 104 sporocysts, assuming that 100% oocysts were sporulated and each contained two sporocysts. Each experiment was performed singly or in duplicates.
Sporocyst-based cell culture infection assay for T. gondii oocysts in a simple matrix.
A Vero cell line (ATCC, CCL-81) was used to support the development of T. gondii parasites from sporocysts. Cells were maintained at 37°C in IMDM containing glutaMAX (Thermo Fisher, USA) supplemented with 5% (vol/vol) heat-inactivated fetal bovine serum (Eurobio, France), 100 μg/ml of streptomycin (Gibco), and 100 μg/ml of penicillin (Gibco). Cells were grown to 80% confluence in a 75-cm2 culture flask (VWR, Canada) in a 5% CO2 atmosphere at 37°C. Once confluent, cells were washed with pH 7.4 PBS and trypsinized to remove the cell monolayer from the flask. Approximately 2 × 105 cells were seeded into each well of 96-well culture plates and grown in medium for 24 h as indicated above. The sporocysts obtained from 104 oocysts in cell culture medium were deposited on the confluent monolayer, allowing the sporozoites to be spontaneously released and to invade cells for 2 h or 1 day. After the contact time, cells were washed to remove the parasites that had not penetrated the cells and were considered “noninfective” (D0). Then, cells were cultivated for 2 (D2), 3 (D3), or 6 days (D6) at 37°C, allowing conversion from sporozoites to tachyzoites and their detection by qPCR. A control culture was incubated alone without sporocysts. A serial dilution of oocysts, ranging from 10 to 104, in cell culture medium was used to estimate the limit of detection of the sporocyst-based CC-qPCR assay in a simple matrix. For each dilution, three independent experiments were performed. The limit of detection was defined as the lowest quantity of oocysts that led to a qPCR signal in all the replicates.
Sporocyst-based cell culture infection assay for T. gondii oocysts recovered from mussels.
The seawater blue mussels (4- to 5-cm shell length), Mytilus edulis, were collected on the intertidal rocky shore of Yport (Seine-Maritime, France). The freshwater zebra mussels, Dreissena polymorpha, were collected at an approximately 5-m depth in November at Lake Der-Chantecoq (Marne, France). Whole mussels were frozen before being sliced with a scalpel. Then, tissues from one mussel were introduced into stomacher bags (Bagpage R400; Interscience, Saint-Nom-la-Bretèche, France) and then spiked with 104 oocysts for infection kinetics characterization or with different amounts of oocysts (ranging from 10 to 5 × 104) to estimate the limit of detection of the sporocyst-based CC-qPCR assay. Each spiking was performed in triplicates. A negative control was performed without oocysts. Each stomacher bag was incubated for 2 h at room temperature and then overnight at 4°C. Mussel tissue digestion was performed using 1× trypsin (Thermo Fisher Scientific; Villebon-sur-Yvette, France) for 1 h 30 min at 37°C at 90-rpm agitation. Filtrates were collected, and 25 ml of 0.9% NaCl was added to wash the stomacher bag. Samples were washed twice at 2,500 × g at 10°C for 10 min. The pellets were suspended with 1.5 ml of 30% Percoll (density, 1.04). After 5 min at 12,000 × g, 1 ml of supernatant was discarded and 1 ml of 0.9% NaCl was added to break the Percoll gradient. Samples were then exposed to household bleach containing 1.6% sodium hypochlorite for 10 min at 4°C to prevent bacterial contamination. The oocysts were then washed twice at 5,000 × g for 5 min in IMDM to remove bleach before sporocyst release using a Lysing Matrix E tube and agitation for 30 s with a TissueLyser. The pellet was resuspended in culture medium and inoculated into wells of a 24-well culture plate, each containing a monolayer of 1 × 106 Vero cells established as indicated above. After 3 h of contact between sporocysts and cells, cells were washed (D0) and then cultured for 62 h or 3 (D3) or 6 (D6) days. The limit of detection was defined as the lowest quantity of oocysts that led to qPCR signal in all the replicates.
DNA extraction and real-time qPCR.
Using the QIAamp DNA minikit (Qiagen), the supernatant was discarded from each well and cells were suspended in 180 μl of the ATL buffer and 20 μl of proteinase K. The suspension was transferred into a 1.5-ml Eppendorf tube, and the sample was processed further for DNA extraction according to the manufacturer-recommended procedures.
The 529-bp repeat element was targeted using previously described primers and probe (58). qPCR was performed using a SimpliAmp thermal cycler (Thermo Fisher Scientific Inc, Villebon-sur-Yvette, France) in a final volume of 25 μl containing 12.5 μl of iQ Supermix (Bio-Rad, Marnes-la-Coquette, France), 1 μl of bovine serum albumin (BSA; 10 mg/ml) (Sigma, France), 1 μl of each 20 μM primer (forward, 5′-AGAGACACCGGAATGCGATCT-3′; reverse, 5′-CCCTCTTCTCCACTCTTCAATTCT-3′), 0.5 μl of 10 μM probe (5′-6-carboxyfluorescein [FAM]-ACGCTTTCCTCGTGGTGATGGCG-3′-black hole quencher 1 [BHQ1]), 5 μl of DNA template, and 4 μl of DNase-RNase-free water. Each DNA extract was tested in duplicates. The cycling parameters included a denaturation step at 95°C for 3 min, followed by 40 cycles at 95°C for 15 s and 65°C for 1 min. A decrease in Cq (quantification cycle) values between D0 and the end of culture (water matrix: D2, D3, and D6; mussel matrices: 62 h, D2, D3, or D6) highlighted the infectivity of the parasites, and a signal difference (Cq D0 − Cq end culture) was calculated.
Mouse bioassay for infectivity.
As described in a previous study (11), 103 T. gondii oocysts were inoculated in outbred female Swiss Webster mice (Charles River Laboratory; Neuilly-sur-Seine, France) weighing 20 to 30 g. Each mouse was intraperitoneally inoculated, as this procedure allows control of the dose of inoculation. Mice were housed in cages, provided granules and water ad libitum, and tested for T. gondii seroconversion with the modified agglutination test (MAT) 4 weeks postinoculation (11).
Statistical analysis.
The Cq values obtained in qPCR were compared using the nonparametric Kruskal-Wallis test. If the null hypothesis H0 (the tested conditions have no effect on the measured value) was rejected, then the post hoc Wilcoxon-Mann-Whitney test was applied for two independent samples. All statistical tests were performed using StatXact7. Statistical difference was considered a P value of <0.05.
Supplementary Material
ACKNOWLEDGMENTS
A. Rousseau and this work were supported by the UMT ACTIA PROTORISK and the Universities of Reims Champagne-Ardenne and Rouen Normandie, in the framework of a PhD with a CIFRE agreement between ACTALIA and the French Ministry of Higher Education, Research and Innovation. This study was also supported by the French national Research Agency (grant ANR-15-CE34-0005), the Normandie Region and the European Union through the European Regional Development Fund (FEDER). A. Dumètre is supported by the Institut Hospitalo-Universitaire (IHU), the National Research Agency (grants 10-IAHU-03 and 17-CE21-0005-07), the Région Provence Alpes Côte d’Azur, and European funding FEDER PRIMI.
We thank HYDREOS, a French organization that promotes the water sector in northeast France, for its backing. We also thank Nikki Sabourin-Gibbs, Rouen University Hospital, for editing the manuscript.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01189-19.
REFERENCES
- 1.AFSSA. 2005. Toxoplasmose: état des connaissances et évaluation du risque lié à l’alimentation. Rapport du groupe de travail “Toxoplasma gondii.” Sauter à la Recherche Agence Française de Sécurité Sanitaire des Aliments, Val-de-Marne, France. [Google Scholar]
- 2.Belluco S, Mancin M, Conficoni D, Simonato G, Pietrobelli M, Ricci A. 2016. Investigating the determinants of Toxoplasma gondii prevalence in meat: a systematic review and meta-regression. PLoS One 11:e0153856. doi: 10.1371/journal.pone.0153856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Condoleo R, Rinaldi L, Sette S, Mezher Z. 2018. Risk assessment of human toxoplasmosis associated with the consumption of pork meat in Italy. Risk Anal 38:1202–1222. doi: 10.1111/risa.12934. [DOI] [PubMed] [Google Scholar]
- 4.Jones JL, Dubey JP. 2010. Waterborne toxoplasmosis—recent developments. Exp Parasitol 124:10–25. doi: 10.1016/j.exppara.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Dumètre A, Dardé M-L. 2003. How to detect Toxoplasma gondii oocysts in environmental samples? FEMS Microbiol Rev 27:651–661. doi: 10.1016/S0168-6445(03)00071-8. [DOI] [PubMed] [Google Scholar]
- 6.Lindsay DS, Holliman D, Flick GJ, Goodwin DG, Mitchell SM, Dubey JP. 2008. Effects of high pressure processing on Toxoplasma gondii oocysts on raspberries. J Parasitol 94:757–758. doi: 10.1645/GE-1471.1. [DOI] [PubMed] [Google Scholar]
- 7.Baldursson S, Karanis P. 2011. Waterborne transmission of protozoan parasites: review of worldwide outbreaks—an update 2004–2010. Water Res 45:6603–6614. doi: 10.1016/j.watres.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Meireles LR, Ekman CC, Andrade HF, Luna EJ. 2015. Human toxoplasmosis outbreaks and the agent infecting form finding from a systematic review. Rev Inst Med Trop Sao Paulo 57:369–376. doi: 10.1590/S0036-46652015000500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karanis P, Kourenti C, Smith H. 2007. Waterborne transmission of protozoan parasites: a worldwide review of outbreaks and lessons learnt. J Water Health 5:1–38. doi: 10.2166/wh.2006.002. [DOI] [PubMed] [Google Scholar]
- 10.Plutzer J, Karanis P. 2016. Neglected waterborne parasitic protozoa and their detection in water. Water Res 101:318–332. doi: 10.1016/j.watres.2016.05.085. [DOI] [PubMed] [Google Scholar]
- 11.Villena I, Aubert D, Gomis P, Ferté H, Inglard JC, Denis-Bisiaux H, Dondon JM, Pisano E, Ortis N, Pinon JM. 2004. Evaluation of a strategy for Toxoplasma gondii oocyst detection in water. Appl Environ Microbiol 70:4035–4039. doi: 10.1128/AEM.70.7.4035-4039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esmerini PO, Gennari SM, Pena HF. 2010. Analysis of marine bivalve shellfish from the fish market in Santos city, São Paulo state, Brazil, for Toxoplasma gondii. Vet Parasitol 170:8–13. doi: 10.1016/j.vetpar.2010.01.036. [DOI] [PubMed] [Google Scholar]
- 13.Lass A, Pietkiewicz H, Szostakowska B, Myjak P. 2012. The first detection of Toxoplasma gondii DNA in environmental fruits and vegetables samples. Eur J Clin Microbiol Infect Dis 6:1101–1108. doi: 10.1007/s10096-011-1414-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniels ME, Hogan J, Smith WA, Oates SC, Miller MA, Hardin D, Shapiro K, Huertos ML, Conrad PA, Dominik C, Watson FGR. 2014. Estimating environmental conditions affecting protozoal pathogen removal in surface water wetland systems using a multi-scale, model-based approach. Sci Total Environ 493:1036–1046. doi: 10.1016/j.scitotenv.2014.06.053. [DOI] [PubMed] [Google Scholar]
- 15.Roslev P, Bukh AS, Iversen L, Sonderbo H, Iversen N. 2010. Application of mussels as biosamples for characterization of faecal pollution in coastal recreational waters. Water Sci Technol 62:586–593. doi: 10.2166/wst.2010.910. [DOI] [PubMed] [Google Scholar]
- 16.Grodzki M, Schaeffer J, Piquet J-C, Le Saux J-C, Chevé J, Ollivier J, Le Pendu J, Le Guyader FS. 2014. Bioaccumulation efficiency, tissue distribution, and environmental occurrence of hepatitis E virus in bivalve shellfish from France. Appl Environ Microbiol 80:4269–4276. doi: 10.1128/AEM.00978-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayres PA, Burton HW, Cullum ML. 1978. Sewage pollution and shellfish, p 51–62. In Lovelock DM, Davies R (ed), Techniques for the study of mixed populations. Society for Applied Bacteriology technical series, number 11. Academic Press, London, United Kingdom. [Google Scholar]
- 18.Graczyk TK, Conn DB, Marcogliese DJ, Graczyk H, de Lafontaine Y. 2003. Accumulation of human parasites by zebra mussels (Dreissena polymorpha) and Asian freshwater clams (Corbicula fluminea). Parasitol Res 89:107–112. doi: 10.1007/s00436-002-0729-x. [DOI] [PubMed] [Google Scholar]
- 19.Robertson LJ, Gjerde B. 2008. Development and use of a pepsin digestion method for analysis of shellfish for Cryptosporidium oocysts and Giardia cysts. J Food Prot 71:959–966. doi: 10.4315/0362-028X-71.5.959. [DOI] [PubMed] [Google Scholar]
- 20.Palos Ladeiro M, Aubert D, Villena I, Geffard A, Bigot A. 2014. Bioaccumulation of human waterborne protozoa by zebra mussel (Dreisseina polymorpha): interest for water biomonitoring. Water Res 48:148–155. doi: 10.1016/j.watres.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Aksoy U, Marangi M, Papini R, Ozkoc S, Bayram Delibas S, Giangaspero A. 2014. Detection of Toxoplasma gondii and Cyclospora cayetanensis in Mytilus galloprovincialis from Izmir Province coast (Turkey) by real time PCR/high-resolution melting analysis (HRM). Food Microbiol 2:128–135. doi: 10.1016/j.fm.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 22.Shapiro K, Van Wormer E, Aguilar B, Conrad PA. 2015. Surveillance for Toxoplasma gondii in California mussels (Mytilus californianus) reveals transmission of atypical genotypes from land to sea. Environ Microbiol 17:4177–4188. doi: 10.1111/1462-2920.12685. [DOI] [PubMed] [Google Scholar]
- 23.Staggs SE, Keely SP, Ware MW, Schable N, See MJ, Gregorio D, Zou X, Su C, Dubey JP, Villegas E. 2015. The development and implementation of a method using blue mussels (Mytilus sp.) as biosentinels of Cryptosporidium spp. and Toxoplasma gondii contamination in marine aquatic environments. Parasitol Res 114:4655–4667. doi: 10.1007/s00436-015-4711-9. [DOI] [PubMed] [Google Scholar]
- 24.Kerambrun E, Palos Ladeiro M, Bigot-Clivot A, Dedourge-Geffard O, Dupuis E, Villena I, Aubert D, Geffard A. 2016. Zebra mussel as a new tool to show evidence of freshwater contamination by waterborne Toxoplasma gondii. J Appl Microbiol 120:498–508. doi: 10.1111/jam.12999. [DOI] [PubMed] [Google Scholar]
- 25.Lindsay DS, Phelps KK, Smith SA, Flick G, Sumner SS, Dubey JP. 2001. Removal of Toxoplasma gondii oocysts from sea water by eastern oysters (Crassostrea virginica). J Eukaryot Microbiol 48:197S–198S. doi: 10.1111/j.1550-7408.2001.tb00517.x. [DOI] [PubMed] [Google Scholar]
- 26.Arkush KD, Miller MA, Leutenegger CM, Gardner IA, Packham AE, Heckeroth AR, Tenter AM, Barr BC, Conrad PA. 2003. Molecular and bioassay-based detection of Toxoplasma gondii oocyst uptake by mussels (Mytilus galloprovincialis). Int J Parasitol 33:1087–1097. doi: 10.1016/S0020-7519(03)00181-4. [DOI] [PubMed] [Google Scholar]
- 27.Lindsay DS, Dubey JP. 2009. Long-term survival of Toxoplasma gondii sporulated oocysts in seawater. J Parasitol 95:1019–1020. doi: 10.1645/GE-1919.1. [DOI] [PubMed] [Google Scholar]
- 28.Giangaspero A, Cirillo R, Lacasella V, Lonigro A, Marangi M, Cavallo P, Berrilli F, Di Cave D, Brandonisio O. 2009. Giardia and Cryptosporidium in inflowing water and harvested shellfish in a lagoon in southern Italy. Parasitol Int 58:12–17. doi: 10.1016/j.parint.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Tedde T, Marangi M, Papini R, Salza S, Normanno G, Virgilio S, Giangaspero A. 2019. Toxoplasma gondii and other zoonotic protozoans in Mediterranean mussel (Mytilus galloprovincialis) and blue mussel (Mytilus edulis): a food safety concern? J Food Prot 27:535–542. doi: 10.4315/0362-028X.JFP-18-157. [DOI] [PubMed] [Google Scholar]
- 30.Garcés G, Effenberger M, Najdrowski M, Wackwitz C, Gronauer A, Wilderer PA, Lebuhn M. 2006. Quantification of Cryptosporidium parvum in anaerobic digesters treating manure by (reverse-transcription) quantitative real-time PCR, infectivity and excystation tests. Water Sci Technol 53:195–202. doi: 10.2166/wst.2006.250. [DOI] [PubMed] [Google Scholar]
- 31.Rousseau A, La Carbona S, Dumètre A, Robertson LJ, Gargala G, Escotte-Binet S, Favennec L, Villena I, Gérard C, Aubert D. 2018. Assessing viability and infectivity of foodborne and waterborne stages (cysts/oocysts) of Giardia duodenalis, Cryptosporidium spp., and Toxoplasma gondii: a review of methods. Parasite 25:14. doi: 10.1051/parasite/2018009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rousseau A, Villena I, Dumètre A, Escotte-Binet S, Favennec L, Dubey JP, Aubert D, La Carbona S. 2019. Evaluation of propidium monoazide-based qPCR to detect viable oocysts of Toxoplasma gondii. Parasitol Res 118:999–1010. doi: 10.1007/s00436-019-06220-1. [DOI] [PubMed] [Google Scholar]
- 33.Smith JJ, Gunasekera TS, Barardi CRM, Veal D, Vesey G. 2004. Determination of Cryptosporidium parvum oocyst viability by fluorescence in situ hybridization using a ribosomal RNA-directed probe. J Appl Microbiol 96:409–417. doi: 10.1046/j.1365-2672.2004.02150.x. [DOI] [PubMed] [Google Scholar]
- 34.Habtewold T, Groom Z, Duchateau L, Christophides GK. 2015. Detection of viable plasmodium ookinetes in the midguts of Anopheles coluzzi using PMA-qrtPCR. Parasit Vectors 8:455. doi: 10.1186/s13071-015-1087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kniel KE, Lindsay DS, Sumner SS, Hackney CR, Pierson MD, Dubey JP. 2002. Examination of attachment and survival of Toxoplasma gondii oocysts on raspberries and blueberries. J Parasitol 88:790–793. doi: 10.1645/0022-3395(2002)088[0790:EOAASO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 36.Grabow WO, Taylor MB, de Villiers JC. 2001. New methods for the detection of viruses: call for review of drinking water quality guidelines. Water Sci Technol 43:1–8. doi: 10.2166/wst.2001.0703. [DOI] [PubMed] [Google Scholar]
- 37.Lee HK, Jeong YS. 2004. Comparison of total culturable virus assay and multiplex integrated cell culture-PCR for reliability of waterborne virus detection. Appl Environ Microbiol 70:3632–3636. doi: 10.1128/AEM.70.6.3632-3636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rochelle PA, Marshall MM, Mead JR, Johnson AM, Korish DG, Rosen JS, De Leon R. 2002. Comparison of in vitro cell culture and a mouse assay for measuring infectivity of Cryptosporidium parvum. Appl Environ Microbiol 68:3809–3817. doi: 10.1128/AEM.68.8.3809-3817.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jenkins M, Trout J, Higgins J, Dorsch M, Veal D, Fayer R. 2003. Comparison of tests for viable and infectious Cryptosporidium parvum oocysts. Parasitol Res 89:1–5. doi: 10.1007/s00436-002-0720-6. [DOI] [PubMed] [Google Scholar]
- 40.Palos Ladeiro M, Bigot A, Aubert D, Hohweyer J, Favennec F, Villena I, Geffard A. 2013. Protozoa interaction with aquatic invertebrate: interest for watercourses biomonitoring. Environ Sci Pollut Res 20:778–789. doi: 10.1007/s11356-012-1189-1. [DOI] [PubMed] [Google Scholar]
- 41.Parr JB, Sevilleja JE, Samie A, Amidou S, Alcantara C, Stroup SE, Kohli A, Fayer R, Lima AAM, Houpt ER, Guerrant RL. 2007. Detection and quantification of Cryptosporidium in HCT-8 cells and human fecal specimens using real-time polymerase chain reaction. Am J Trop Med Hyg 76:938–942. doi: 10.4269/ajtmh.2007.76.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shahiduzzaman M, Dyachenko V, Keidel J, Schmäschke R, Daugschies A. 2010. Combination of cell culture and quantitative PCR (cc-qPCR) to assess disinfectants efficacy on Cryptosporidium oocysts under standardized conditions. Vet Parasitol 167:43–49. doi: 10.1016/j.vetpar.2009.09.042. [DOI] [PubMed] [Google Scholar]
- 43.Johnson AM, Di Giovanni GD, Rochelle PA. 2012. Comparison of assays for sensitive and reproducible detection of cell culture-infectious Cryptosporidium parvum and Cryptosporidium hominis in drinking water. Appl Environ Microbiol 78:156–162. doi: 10.1128/AEM.06444-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ware MW, Augustine SAJ, Erisman DO, See MJ, Wymer L, Hayes SL, Dubey JP, Villegas EN. 2010. Determining UV inactivation of Toxoplasma gondii oocysts by using cell culture and a mouse bioassay. Appl Environ Microbiol 76:5140–5147. doi: 10.1128/AEM.00153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villegas EN, Augustine SAJ, Villegas LF, Ware MW, See MJ, Lindquist HDA, Schaefer FW III, Dubey JP. 2010. Using quantitative reverse transcriptase PCR and cell culture plaque assays to determine resistance of Toxoplasma gondii oocysts to chemical sanitizers. J Microbiol Methods 81:219–225. doi: 10.1016/j.mimet.2010.03.023. [DOI] [PubMed] [Google Scholar]
- 46.Travaillé E, La Carbona S, Gargala G, Aubert D, Guyot K, Dumètre A, Villena I, Houssin M. 2016. Development of a qRT-PCR method to assess the viability of Giardia intestinalis cysts, Cryptosporidium spp. and Toxoplasma gondii oocysts. Food Control 59:359–365. doi: 10.1016/j.foodcont.2015.06.007. [DOI] [Google Scholar]
- 47.Karanis K, Aldeyarbi HM, Mirhashemi EM, Khalil KM. 2013. The impact of the waterborne transmission of Toxoplasma gondii and analysis efforts for water detection: an overview and update. Environ Sci Pollut Res 20:86–99. doi: 10.1007/s11356-012-1177-5. [DOI] [PubMed] [Google Scholar]
- 48.Efstratiou A, Ongerth JE, Karanis P. 2017. Waterborne transmission of protozoan parasites: review of worldwide outbreaks—an update 2011–2016. Water Res 144:14–22. doi: 10.1016/j.watres.2017.01.036. [DOI] [PubMed] [Google Scholar]
- 49.Hohweyer J, Dumètre A, Aubert D, Azas N, Villena I. 2013. Tools and methods for detecting and characterizing Giardia, Cryptosporidium, and Toxoplasma parasites in marine mollusks. J Food Prot 76:1649–1657. doi: 10.4315/0362-028X.JFP-13-002. [DOI] [PubMed] [Google Scholar]
- 50.Najdrowski M, Joachim A, Daugschies A. 2007. An improved in vitro infection model for viability testing of Cryptosporidium parvum oocysts. Vet Parasitol 150:150–154. doi: 10.1016/j.vetpar.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 51.Dumètre A, Dubey JP, Ferguson DJ, Bongrand P, Azas N, Puech PH. 2013. Mechanics of the Toxoplasma gondii oocyst wall. Proc Natl Acad Sci U S A 110:11535–11540. doi: 10.1073/pnas.1308425110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Everson WV, Ware MW, Dubey JP, Lindquist HD. 2002. Isolation of purified oocyst walls and sporocysts from Toxoplasma gondii. J Eukaryot Microbiol 49:344–349. doi: 10.1111/j.1550-7408.2002.tb00381.x. [DOI] [PubMed] [Google Scholar]
- 53.Kasper LH, Bradley MS, Pfefferkorn ER. 1984. Identification of stage-specific sporozoite antigens of Toxoplasma gondii by monoclonal antibodies. J Immunol 132:443–449. [PubMed] [Google Scholar]
- 54.Freyre A, Falcon J. 2004. Massive excystation of Toxoplasma gondii sporozoites. Exp Parasitol 107:72–77. doi: 10.1016/j.exppara.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 55.Lindsay DS, Current WL, Ernst JV. 1983. Excystation of Isospora suis Biester, 1934 of swine. Z Parasitenkd 69:27–34. [DOI] [PubMed] [Google Scholar]
- 56.Fukushima H, Katsube K, Hata Y, Kishi R, Fujiwara S. 2007. Rapid separation and concentration of food-borne pathogens in food samples prior to quantification by viable-cell counting and real-time PCR. Appl Environ Microbiol 73:92–100. doi: 10.1128/AEM.01772-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suresh P, Rehg JE. 1996. Comparative evaluation of several techniques for purification of Cryptosporidium parvum oocysts from rat feces. J Clin Microbiol 34:38–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lélu M, Villena I, Dardé ML, Aubert D, Geers R, Dupuis E, Marnef F, Poulle ML, Gotteland C, Dumètre A, Gilot-Fromont E. 2012. Quantitative estimation of the viability of Toxoplasma gondii oocysts in soil. Appl Environ Microbiol 78:5127–5132. doi: 10.1128/AEM.00246-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dubey JP, Lunney JK, Shen SK, Kwok OCH, Ashford DA. 1996. Infectivity of low numbers of Toxoplasma gondii oocysts to pigs. J Parasitol 82:428–443. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.