Candida-secreted quorum-sensing molecules (i.e., farnesol and tyrosol) are key regulators in fungal physiology, which induce phenotypic adaptations, including morphological changes, altered biofilm formation, and synchronized expression of virulence factors. Moreover, they have a remarkable antifungal activity at supraphysiological concentrations. Limited data are available concerning the tyrosol-induced molecular and physiological effects on non-albicans Candida species such as C. parapsilosis. In addition, the background of the previously observed antifungal effect caused by tyrosol remains unknown. This study reveals that tyrosol exposure enhanced the oxidative stress response and the expression of efflux pump genes, while it inhibited growth and ribosome biogenesis as well as several virulence-related genes. Metabolism was changed toward glycolysis and ethanol fermentation. Furthermore, the initial adherence was not influenced significantly in the presence of tyrosol. Our results provide several potential explanations for the previously observed antifungal effect.
KEYWORDS: Candida parapsilosis, RNA-Seq, oxidative stress, quorum sensing, tyrosol
ABSTRACT
Tyrosol plays a key role in fungal morphogenesis and biofilm development. Also, it has a remarkable antifungal effect at supraphysiological concentrations. However, the background of the antifungal effect remains unknown, especially in the case of non-albicans Candida species such as Candida parapsilosis. We examined the effect of tyrosol on growth, adhesion, redox homeostasis, virulence, as well as fluconazole susceptibility. To gain further insights into the physiological consequences of tyrosol treatment, we also determined genome-wide gene expression changes using transcriptome sequencing (RNA-Seq). A concentration of 15 mM tyrosol caused significant growth inhibition within 2 h of the addition of tyrosol, while the adhesion of yeast cells was not affected. Tyrosol increased the production of reactive oxygen species remarkably, as revealed by a dichlorofluorescein test, and it was associated with elevated superoxide dismutase, glutathione peroxidase, and catalase activities. The interaction between fluconazole and tyrosol was antagonistic. Tyrosol exposure resulted in 261 and 181 differentially expressed genes with at least a 1.5-fold increase or decrease in expression, respectively, which were selected for further study. Genes involved in ribosome biogenesis showed downregulation, while genes related to the oxidative stress response and ethanol fermentation were upregulated. In addition, tyrosol treatment upregulated the expression of efflux pump genes, including MDR1 and CDR1, and downregulated the expression of the FAD2 and FAD3 virulence genes involved in desaturated fatty acid formation. Our data demonstrate that exogenous tyrosol significantly affects the physiology and gene expression of C. parapsilosis, which could contribute to the development of treatments targeting quorum sensing in the future.
IMPORTANCE Candida-secreted quorum-sensing molecules (i.e., farnesol and tyrosol) are key regulators in fungal physiology, which induce phenotypic adaptations, including morphological changes, altered biofilm formation, and synchronized expression of virulence factors. Moreover, they have a remarkable antifungal activity at supraphysiological concentrations. Limited data are available concerning the tyrosol-induced molecular and physiological effects on non-albicans Candida species such as C. parapsilosis. In addition, the background of the previously observed antifungal effect caused by tyrosol remains unknown. This study reveals that tyrosol exposure enhanced the oxidative stress response and the expression of efflux pump genes, while it inhibited growth and ribosome biogenesis as well as several virulence-related genes. Metabolism was changed toward glycolysis and ethanol fermentation. Furthermore, the initial adherence was not influenced significantly in the presence of tyrosol. Our results provide several potential explanations for the previously observed antifungal effect.
INTRODUCTION
The proliferation and virulence of Candida cells are under strict cell density-based control mediated by various quorum-sensing molecules, including farnesol and tyrosol (1, 2). While farnesol induces the hypha-to-yeast transition in Candida albicans, tyrosol has an opposite effect. Tyrosol is a tyrosine-derived molecule, which is released into the growth medium continuously during the exponential growth phase and is capable of decreasing the duration of the lag phase before cells begin germination (3). Previous studies have reported that the accumulation of tyrosol in the culture medium is directly related to the increase in fungal cell density (3, 4). Moreover, 20 μM tyrosol could stimulate germ tube and hypha formation in the early and intermediate stages of biofilm development (4). Based on the gene expression profile of C. albicans, tyrosol affects cell cycle regulation, DNA replication, and chromosome segregation (3). Besides its role in fungal physiology, tyrosol also possesses remarkable antifungal activity at supraphysiological concentrations (1 to 50 mM) against both planktonic and sessile Candida populations; therefore, its potential role in certain aspects of antifungal therapy has been postulated, e.g., in catheter lock therapy (5–7).
In recent years, alternative treatments targeting quorum sensing against Candida species have become an intensively researched area; however, tyrosol remains a mysterious molecule, and the exact background of its antifungal mechanism is still poorly understood (1, 2, 8–10). In the case of C. albicans, both the transcriptional and physiological responses exerted by tyrosol have been addressed previously (3, 4), aiding in the understanding of the observed antifungal effect against C. albicans. However, the tyrosol-induced physiological and molecular events, especially in non-albicans Candida species, remained unknown. Here, we report the effects of tyrosol at supraphysiological concentrations on cell growth, adhesion, oxidative stress-related enzyme production, virulence, fluconazole susceptibility, as well as gene expression, which may help in understanding the potential physiological and molecular backgrounds of its antifungal effect on Candida parapsilosis.
RESULTS
Effects of tyrosol on growth, morphology, extracellular phospholipase and proteinase production, biofilm formation, as well as adhesion to Caco-2 cells.
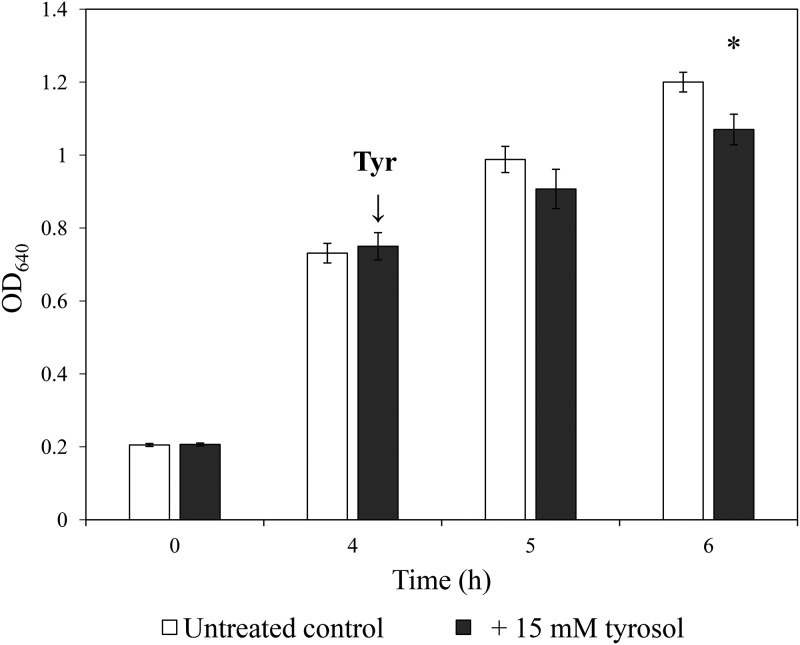
Growth of C. parapsilosis was examined following 15 mM tyrosol exposure in yeast extract-peptone-dextrose (YPD). The addition of tyrosol to preincubated cells resulted in significant inhibition starting at 6 h postinoculation, which was confirmed by both absorbance (A) measurements and CFU determination. Growth was significantly inhibited within 2 h of the addition of tyrosol in terms of both CFU changes (8.25 × 107 ± 0.8 × 107 and 4.5 × 107 ± 0.4 × 107 CFU/ml for untreated control and tyrosol-exposed cells, respectively) (P = 0.002) and observed absorbance values (optical density at 640 nm [OD640]) (P = 0.002) (Fig. 1). The ratios of yeast cells to pseudohyphae were comparable between control cells (69.3% ± 2.1% and 28.7% ± 3.5% for yeast and pseudohyphae, respectively) and treated cells (66.0% ± 1.7% and 37.7% ± 1.5% for yeast and pseudohyphae, respectively) with a 2-h stress exposure time (P > 0.05). Tyrosol treatment did not influence significantly the extracellular proteinase activity (precipitation zone [Pz] values were 0.82 ± 0.04 and 0.80 ± 0.05 for untreated control and tyrosol-exposed cells, respectively; P > 0.05). There was no remarkable phospholipase activity for untreated control and tyrosol-exposed cells (the Pz value was 1.0). Biofilm-forming abilities were comparable with and without 15 mM tyrosol (4.9 × 105 ± 1.1 × 105 and 6.4 × 105 ± 1.9 × 105 living fungal cells at 24 h for untreated control and tyrosol-exposed cells, respectively) (P > 0.05); however, metabolic activity was significantly increased in the tyrosol treatment group (P = 0.001). Adhesion of C. parapsilosis to Caco-2 cells in the presence of tyrosol (4.2% ± 1.1% adherent cells) did not differ significantly from that observed for untreated control cells (3.1% ± 1.1% adherent cells) (P > 0.05).
FIG 1.
Effect of tyrosol on growth of C. parapsilosis. Growth of C. parapsilosis CLIB 214 in YPD medium was monitored by measurement of the absorbance (OD640). Tyrosol was added at a 4-h incubation time at a 15 mM final concentration. Data represent mean values ± SD calculated from 10 independent experiments. The asterisk indicates a significant difference between control and tyrosol-treated cultures calculated by paired Student’s t test.
Tyrosol-induced oxidative stress and stress response in C. parapsilosis.
Tyrosol caused significantly higher reactive species production than in untreated control cells (P < 0.001), as presented in Table 1. This tyrosol-related higher reactive species level was associated with elevated superoxide dismutase (P = 0.021), glutathione peroxidase (P = 0.012), and catalase (P = 0.013) activities in tyrosol-exposed cells (Table 1). In contrast, the measured glutathione reductase values were statistically comparable (P > 0.05) in tyrosol-treated and untreated Candida cells (Table 1).
TABLE 1.
Tyrosol-induced oxidative stress response in Candida parapsilosisa
| Oxidative stress-related parameter | Mean value ± SD |
|
|---|---|---|
| Untreated cultures | Tyrosol-treated cultures | |
| Catalase [kat (kg protein)−1] | 1.4 ± 0.24 | 2.2 ± 0.4* |
| GR [mkat (kg protein)−1] | 8.9 ± 0.4 | 9.6 ± 0.5 |
| GPx [mkat (kg protein)−1] | 1.8 ± 0.3 | 2.5 ± 0.4* |
| SOD [mU (mg protein)−1] | 72.5 ± 4.5 | 129.4 ± 10.9* |
| DCF [nmol DCF (OD640)−1] | 11.7 ± 2 | 19.3 ± 2*** |
Mean values ± standard deviations calculated from three independent experiments are presented. * and *** indicate significant differences at P values of <0.05 and 0.001, respectively, calculated by paired Student’s t test, comparing untreated control and tyrosol-treated cultures. GR, glutathione reductase; GPx, glutathione peroxidase; SOD, superoxide dismutase.
Susceptibility of planktonic and sessile C. parapsilosis cells to fluconazole and tyrosol.
The median planktonic MICs of fluconazole and tyrosol were 0.5 mg/liter and 30 mM, respectively. Functional inhibitory concentration index (FICI) determination showed a clear antagonistic interaction; the median FICI value from three independent experiments was 4.125. In the case of biofilms, the median MIC values were 4 mg/liter and 30 mM for fluconazole and tyrosol, respectively, while the interaction was also antagonistic (FICI of 4.5), similar to that of planktonic cells.
In vivo experiments.
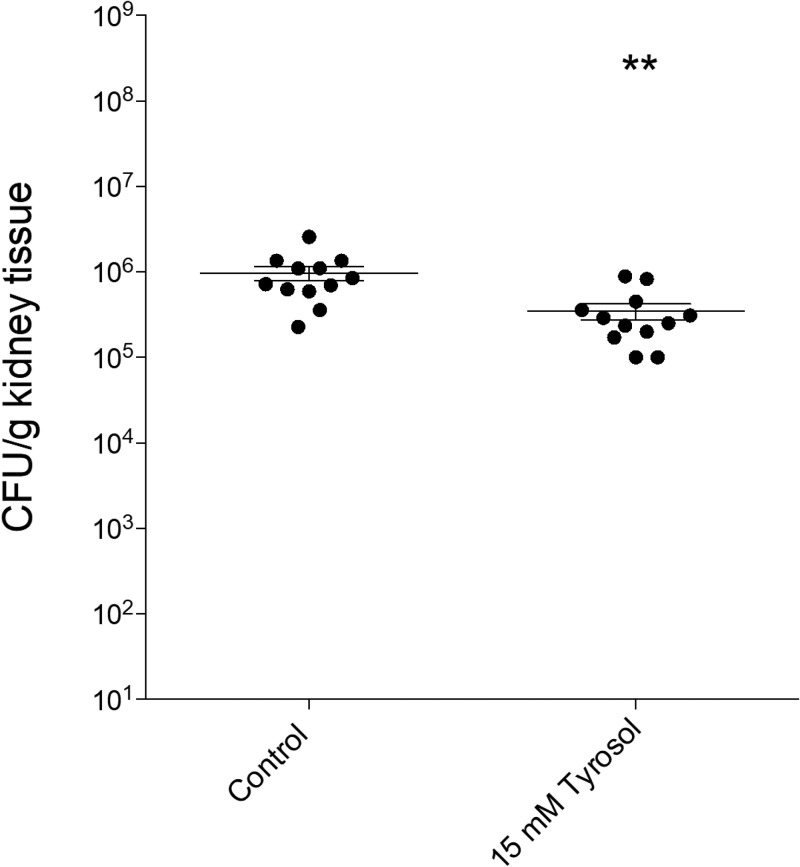
Daily treatment with 15 mM tyrosol decreased the fungal tissue burden in the kidneys by at least half a log degree, which corresponded to a significantly lower tissue burden (P = 0.004) than that seen in the untreated controls (Fig. 2).
FIG 2.
Results of in vivo experiments. Shown are kidney tissue burdens of permanently neutropenic BALB/c mice infected intravenously with C. parapsilosis strain CLIB 214. Daily intraperitoneal tyrosol (15 mM) treatment was started at 24 h postinoculation. Fungal kidney tissue burden was determined at the end of the experiments on day 6. Bars represent means ± SD. The level of statistical significance compared with the untreated control group on day 6 is indicated (**, P < 0.01).
Transcriptional profiling and RNA-Seq data validation.
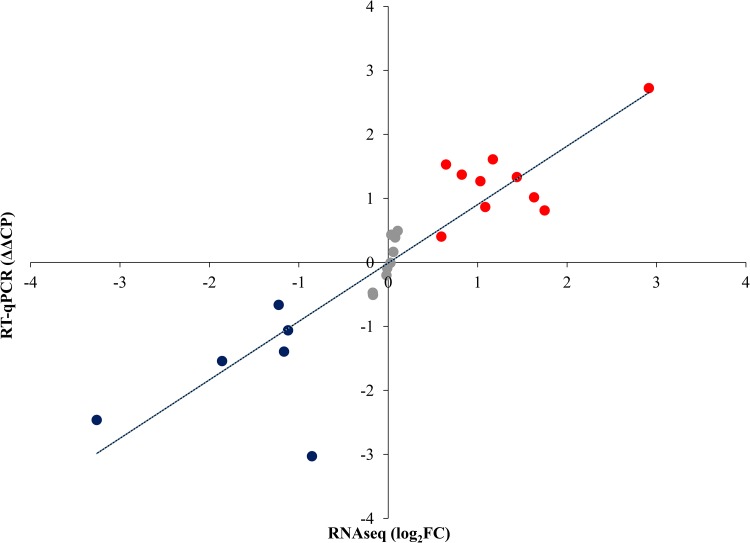
Comparison of the tyrosol-treated C. parapsilosis cell gene expression profile with that of untreated cells revealed 1,462 differentially expressed genes (see Fig. S1 in the supplemental material). The reverse transcriptase quantitative PCR (qRT-PCR) data for the selected 25 genes showed a strong correlation with the obtained transcriptome sequencing (RNA-Seq) data (Fig. 3 and Table S2).
FIG 3.
Correlation between RT-qPCR and transcriptome data. RNA-Seq data are presented as log2(FC) values, where FC is “fold change.” Relative transcription levels were quantified as ΔΔCP = ΔCPcontrol − ΔCPtreated, where ΔCPtreated = CPtested gene − CPreference gene, measured from treated cultures, and ΔCPcontrol = CPtested gene − CPreference gene, measured from control cultures. CP values represent the qRT-PCR cycle numbers of crossing points. The ACT1 gene was used as a reference gene. ΔΔCP values significantly (P < 0.05 by Student’s t test; n = 3) higher or lower than zero (up- or downregulated genes) are marked in red and blue, respectively. Pearson’s correlation coefficient between the RT-qPCR and RNA-seq values was 0.88.
Tyrosol-responsive genes were defined as differentially expressed genes with a log2(FC) greater than 0.585 (upregulated genes) or a log2(FC) less than −0.585 (downregulated genes), where FC is the fold change in fragments per kilobase per million mapped reads (FPKM) values (tyrosol-treated versus untreated cultures). Results of the gene set enrichment analysis of the 261 upregulated and 181 downregulated genes are presented in Table 2 and in Tables S3 and S4 and are summarized below.
TABLE 2.
Selected significant shared gene ontology terms
| Parameter for tyrosol-treated group relative to untreated group | Upregulated genesb | Downregulated genesb |
|---|---|---|
| No. of genes | 261 | 181 |
| Significant shared GO terms (no. of genes belonging to GO term)a | Cell surface (16), cell wall (18), hyphal cell wall (13), extracellular matrix (14), peroxisomal matrix (5), biofilm matrix (14), carbohydrate metabolic process (32), glycolytic process (11), glycolytic fermentation (4), purine nucleotide biosynthetic process (12), response to oxidative stress (18), oxidoreductase activity (38), peroxidase activity (5), cofactor binding (29), drug catabolic process (8), positive regulation of defense response (6) | Transmembrane transport (42), rRNA transport (6), amino acid transport (11), anion transmembrane transporter activity (14), cell wall (14), hyphal cell wall (8), cytosol (28), cytosolic ribosome (26), translation (34), cytoplasmic translation (12), ribosome biogenesis (25), large ribosomal subunit (14), small ribosomal subunit (12) |
Significant shared GO terms (P < 0.05) were determined with the Candida Genome Database Gene Ontology Term Finder (http://www.candidagenome.org/cgi-bin/GO/goTermFinder). Shown are selected significant shared terms only. The whole data set is available in Table S3 in the supplemental material. Numbers in parentheses represent the number of upregulated or downregulated genes belonging to the appropriate GO term.
Up- and downregulated genes were defined as differentially expressed genes (corrected P value of <0.05), with a log2(FC) higher than 0.585 or a log2(FC) lower than −0.585, respectively, where FC is the fold change in the FPKM value.
Evaluation of tyrosol-responsive genes.
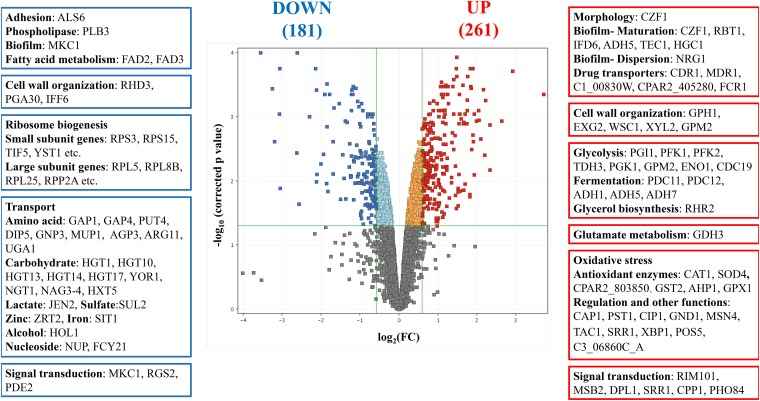
(i) Virulence-related genes. Selected genes involved in the genetic control of C. parapsilosis virulence were determined according to methods described previously by Tóth et al. (11). Virulence-related genes were significantly enriched within the tyrosol-responsive upregulated gene group according to Fisher’s exact test (Table S4). Most of these 16 putative genes are involved in biofilm production (e.g., CZF1, RBT1, IFD6, TEC1, HGC1, and NRG1) (Fig. 4, Table 2, and Tables S3 and S4). Three downregulated virulence-related genes, FAD2 and FAD3 (involved in saturated fatty acid formation) and MKC1 (a putative regulator of biofilm formation), are notable (Fig. 4, Table 2, and Tables S3 and S4). It is noteworthy that tyrosol may also enhance its production through increasing the gene expression level of ADH5, which is involved in tyrosine-tyrosol conversion in C. albicans and in Saccharomyces cerevisiae (12).
FIG 4.
Genome-wide transcriptional changes induced by tyrosol in C. parapsilosis. Upregulated (red) and downregulated (dark blue) genes were defined as differentially expressed genes (corrected P value of <0.05), where the log2(FC) was greater than 0.585 or less than −0.585, respectively, and FC is the fold change of FPKM values (tyrosol treated versus untreated). On the sides of the volcano plot are representative genes upregulated or downregulated by tyrosol treatment.
(ii) Cell wall-related genes. Regarding cell wall assembly, the expression levels of the putative genes PDC11, TDH3, and ADH1 were upregulated by tyrosol treatment (Fig. 4, Table 2, and Tables S3 and S4).
(iii) Antifungal drug transport-related genes. Significant upregulation of putative genes encoding antifungal drug transport proteins was observed (e.g., MDR1, CDR1, and FCR1) (Fig. 4, Table 2, and Tables S3 and S4).
(iv) Metabolic pathway-related genes. Selected genes involved in glucose catabolism were determined using the Candida Genome Database (http://www.candidagenome.org). Tyrosol exposure resulted in increased expression levels of several genes related to the “carbohydrate metabolic process” gene ontology (GO) term, including glycolysis (PGI1, PFK1, PFK2, TDH3, PGK1, GPM2, ENO1, and CDC19) and fermentation (PDC11, PDC12, ADH1, ADH5, and ADH7) genes but not tricarboxylic acid cycle genes (Fig. 4, Table 2, and Tables S3 and S4). In contrast, tyrosol treatment led to reduced expression of several genes (42 genes altogether) involved in transmembrane transport, including 10 putative carbohydrate transport genes and 12 putative amino acid transport genes (Fig. 4, Table 2, and Tables S3 and S4). Downregulation of ribosome biogenesis genes (36 genes altogether) was also notable (Fig. 4, Table 2, and Tables S3 and S4).
(v) Oxidative-stress-related genes. Genes belonging to the “response to oxidative stress” GO term were enriched in the tyrosol-responsive upregulated gene group (Fig. 4, Table 2, and Tables S3 and S4). Altogether, 18 genes were upregulated after tyrosol treatment, including CAT1, SOD4, CPAR2_803850, GPX1, GST2, AHP1, CAP1, PST1, CIP1, TAC1, and MSN4 (Fig. 4, Table 2, and Tables S3 and S4).
DISCUSSION
Alternative treatments targeting fungal quorum sensing have become an intensely researched area in recent years (1, 2, 5–8, 10). However, the majority of studies focused on farnesol-related antifungal effects, especially in the case of C. albicans (1, 10). Therefore, there are limited data about tyrosol-induced changes, particularly among non-albicans Candida species such as C. parapsilosis, which is the second most frequently isolated Candida species from blood in Asia, Latin America, and several Mediterranean European countries (13).
Previous studies reported that tyrosol has a potential antifungal effect similar to that of farnesol (5–7, 14–16); moreover, it enhances the activity of caspofungin and micafungin against C. parapsilosis (6). Nevertheless, the exact physiological and transcriptional background of the antifungal activity exerted by tyrosol remains to be elucidated. In the present study, tyrosol exposure was associated with physiological alterations as well as genome-wide transcriptional changes in C. parapsilosis.
Concerning our growth-based experiments, a significant inhibitory effect was induced by the tyrosol concentration used, which is in line with previous investigations concerned with C. albicans, C. tropicalis, and C. krusei (5, 7, 14). The observed significant downregulation of ribosome biogenesis genes is concordant with this marked growth inhibition. Another explanation for the antifungal effect of tyrosol is the induction of oxidative stress suggested by the increased level of reactive species in the presence of tyrosol. These reactive species can alter the ratio of saturated to unsaturated fatty acids in the cell membrane; furthermore, hydrogen peroxide and hydroxyl radicals cause irreversible damage to proteins, lipids, and nucleic acids, resulting in impaired viability (15). A well-known response of fungal species to reactive oxygen species is the rapid induction of oxidative stress detoxification at the mRNA level (15). In this study, several putative oxidative stress-responsive genes, CAT1, CPAR2_803850 (both coding for catalase activity), GPX1 (encoding glutathione peroxidase), and SOD4 (encoding superoxide dismutase), were upregulated following exposure to tyrosol, which was associated with increased catalase, glutathione peroxidase, and superoxide dismutase activities. Furthermore, CAP1, the major transcription factor in oxidative stress elimination, was also overexpressed following tyrosol exposure (15).
The elevated levels of reactive species may be explained by the disruption of polyunsaturated fatty acid (PUFA) metabolism. The synthesis of PUFAs is significantly downregulated in tyrosol-treated cells, as tyrosol decreased the expression of the FAD2 and FAD3 genes encoding delta12-fatty acid desaturase and omega-3 fatty acid desaturase enzymes, respectively. These are orthologs of genes participating in PUFA synthesis in C. albicans (16, 17). PUFAs (stearidonic acid, eicosapentaenoic acid, and docosapentaenoic acid) are major components of the cell membrane of C. parapsilosis, accounting for approximately 30% of the fatty acid content, and have a remarkable antioxidant effect (17). Tyrosol-induced inhibition of the expression of FAD2 and FAD3 found in this study therefore decreases the antioxidant level in cell membranes, which may contribute to the antifungal effect of tyrosol against C. albicans or C. parapsilosis (16–18). Although tyrosol was reported to exert an antioxidant effect, this was measured only at micromolar concentrations (19), while the concentration used in this study was 15 mM.
Tyrosol is a well-known regulator molecule in the biofilm formation of C. albicans (3, 4); however, its exact role in the development of C. parapsilosis biofilms remained unknown. In this study, the ortholog of the C. albicans CZF1 gene, which is one of the key transcription factors of biofilm development in C. parapsilosis, was upregulated following tyrosol exposure (20). CZF1 promotes the yeast-to-pseudohypha transition (20); CZF1 mutants showed reduced colony wrinkling, and the biofilms produced contained mainly yeast cells (20). Nonetheless, we did not observe higher rates of adherence and biofilm-forming ability in the presence of tyrosol. Monteiro et al. observed that tyrosol did not induce increased adhesion in C. albicans and C. glabrata, which is in line with the gene expression pattern observed and the biofilm formation experiments in our study with C. parapsilosis (21). Tyrosol had no effect on putative morphology-related genes (CPH2, EFG1, UME6, OCH1, SPT3, and CWH41), which is in line with our microscopic observations.
Regarding antifungal susceptibility, a clear antagonistic interaction was observed between fluconazole and tyrosol against both planktonic cells and biofilms. Tyrosol increased the expression of MDR1 and CDR1 orthologs as well as the FCR1, C1_00830W, and CPAR2_405280 drug transporter genes in C. parapsilosis, which may explain the observed antagonistic interactions. In contrast, in previous studies, farnesol exposure decreased the expression levels of MDR1, CDR1, as well as ERG genes, which was associated with the observed reversion of azole resistance in C. albicans (22, 23). In C. parapsilosis, neither MDR1 nor CDR1 genes were influenced by farnesol (24). Furthermore, ergosterol biosynthesis genes were not affected by tyrosol; therefore, the observed antagonistic interaction was probably not linked to the ergosterol pathway but rather was due to the overexpression of the above-mentioned efflux pumps.
Based on our in vivo experiments, daily intraperitoneal treatment with 15 mM tyrosol for 5 days significantly reduced the fungal kidney burden in systemic infection with CLIB 214, which was in parallel with the results of the gene expression pattern. Transcriptional profiling showed that genes involved in adhesion (ALS6) had reduced expression in response to tyrosol. In addition, genes encoding secreted aspartyl proteinases (SAPP1 and SAPP3) (25) were not upregulated significantly, which may explain the above-mentioned decrease in in vivo virulence. Furthermore, downregulation of the expression of FAD2 and FAD3, coding for proteins involved in PUFA synthesis, may also contribute to lower virulence by decreasing the tolerance to oxidative stress.
Our study analyzes changes in gene expression in C. parapsilosis following tyrosol exposure using RNA-Seq (11), providing important insights into the mechanism of antifungal action of tyrosol and the response of C. parapsilosis, which may aid in better understanding tyrosol-related antifungal activity in non-albicans Candida species. In summary, tyrosol exposure enhanced the oxidative stress response and upregulated efflux pumps while inhibiting growth, ribosome biogenesis, as well as virulence. Metabolism was modulated toward glycolysis and ethanol fermentation. Initial adherence was not influenced by the presence of tyrosol. Our findings suggest that tyrosol may be a potential locally active and/or adjuvant agent in the development of alternative treatments targeting quorum sensing against C. parapsilosis in the future.
MATERIALS AND METHODS
Fungal strain, culture medium, and epithelial cell line.
The CLIB 214 C. parapsilosis (sensu stricto) reference strain was used in all experiments. The strain was maintained and cultured on yeast extract-peptone-dextrose (YPD) agar (1% yeast extract, 2% mycological peptone, 2% glucose, and 2% agar [pH 5.6]). The Caco-2 epithelial cell line was cultured as described previously by Nemes et al. (26). The Caco-2 cell line was obtained from the European Collection of Cell Cultures (ECACC) (Salisbury, United Kingdom). Cells were grown in plastic cell culture flasks in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 3.7 g/liter NaHCO3, 10% (vol/vol) heat-inactivated fetal bovine serum, a 1% (vol/vol) nonessential amino acid solution, 0.584 g/liter l-glutamine, 4.5 g/liter d-glucose, 100 IU/ml penicillin, and 100 mg/liter streptomycin at 37°C in the presence of 5% CO2. The cells were routinely maintained by regular passaging, and glutamine was supplemented with GlutaMAX. The cells used for adhesion and toxicity experiments were between passages 20 and 40 (26).
Tyrosol [2-(4-hydroxyphenyl) ethanol; Sigma, Budapest, Hungary] was prepared as a 0.1 M stock solution in YPD or sterile physiological saline for physiology experiments and susceptibility testing or in vivo experiments, respectively. The stock solution of fluconazole was prepared in sterile distilled water and preserved according to the manufacturer’s instructions. Susceptibility testing of C. parapsilosis against tyrosol and fluconazole was performed using RPMI 1640 medium [with l-glutamine and without bicarbonate (pH 7.0), with 3-(N-morpholino)propanesulfonic acid (MOPS); Sigma, Budapest, Hungary].
Toxicity experiments.
In preliminary experiments, 100 μM, 1 mM, 10 mM, and 15 mM tyrosol were evaluated in terms of toxicity to the Caco-2 cell line using a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay (Sigma, Budapest, Hungary) (26, 27) and xCELLigence real-time cell analysis (ACEA Biosciences, Inc., San Diego, CA, USA) (28), where none of them caused relevant toxicity. As 100 μM, 1 mM, and 10 mM tyrosol concentrations caused physiological and transcriptional changes that were less marked in C. parapsilosis, we focused exclusively on the effect of 15 mM tyrosol in further experiments in order to reveal the potential mechanisms of the antifungal effect.
Growth conditions.
Precultures were grown in YPD medium at 30°C at a 3.7-Hz shaking frequency for 18 h, diluted to an OD640 of 0.2 (corresponding to 2.9 × 106 ± 0.5 × 106 CFU/ml) in 20 ml of YPD, and incubated at 37°C with a 2.3-Hz shaking frequency (29). Following a 4-h incubation time, we added 15 mM tyrosol to the YPD cultures, and growth was then examined at 1-h intervals by determination of cell density both by means of measuring the absorbance (at 640 nm) and by counting living cells (CFU) as described previously (30). Morphological alterations were monitored using phase-contrast microscopy (Euromex Holland) with the MicroQ-W Pro camera, which was used to evaluate the ratio of yeast cells to pseudohyphae based on 100 cells per sample. Statistical comparison of the growth-based data was performed by paired Student’s t test using GraphPad Prism 6.05 software. The differences between values for treated and control cells were considered significant if the P value was <0.05.
For reactive species production, antioxidant enzyme activities, and RNA extraction, the yeast precultures were diluted to an OD640 of 0.2 in YPD broth, and cultures were then grown for 4 h at 37°C. Next, YPD medium was supplemented with a final concentration of 15 mM tyrosol, and fungal cells were collected 2 h following tyrosol exposure by centrifugation (5 min at a relative centrifugal force [RCF] of 4,000 × g at 4°C). The cells were washed three times with phosphate-buffered saline and stored at −70°C until use (31).
Reactive species production and antioxidant enzyme activities.
Reactive species were measured in the presence or absence of tyrosol by a technique that converts 2′,7′-dichlorofluorescin diacetate to 2′,7′-dichlorofluorescein (DCF) (Sigma, Budapest, Hungary). The amount of DCF produced is directly proportional to the number of reactive species (31).
Glutathione reductase, glutathione peroxidase, catalase, and superoxide dismutase activities were determined as described previously by Jakab et al. (31). Reactive species and enzyme activities were measured in three independent experiments and are presented as means ± standard deviations (SD). Statistical comparisons of reactive species and enzyme production data were performed by paired Student’s t test using GraphPad Prism 6.05 software. The differences between values for treated and control cells were considered significant if the P value was <0.05.
Adhesion experiments.
The Caco-2 epithelial cell line was inoculated with 1 × 105 CFU/ml C. parapsilosis in order to evaluate the adhesion of fungal cells as described previously (32, 33). In the adhesion assay, Caco-2 and C. parapsilosis cells were coincubated for 1 h in DMEM at 37°C in the presence of 5% CO2 with and without 15 mM tyrosol. Afterwards, nonadherent fungal cells were removed by rinsing with phosphate-buffered saline, and the epithelial cells were fixed with 4% formaldehyde. Adherent C. parapsilosis cells were stained with calcofluor white (Sigma, Budapest, Hungary), and stained fungal cells were examined with a Zeiss AxioScope A1 fluorescence microscope with a Zeiss AxioCam ICm1 camera (32, 33). Adhesion (percent) was calculated using the following formula: [(average cell count in the field × area of the well in square micrometers)/(area of the field in square micrometers × inoculated fungal cells in each well)] × 100. Adhesion was evaluated in three independent experiments and is presented as the mean ± standard deviation. Statistical comparison of adhesion-related data was performed by paired Student’s t test using GraphPad Prism 6.05 software. The differences between values for treated and control cells were considered significant if the P value was <0.05.
Evaluation of extracellular phospholipase and aspartic proteinase activities.
Extracellular phospholipase production by tyrosol-treated and untreated C. parapsilosis cells was examined on egg yolk medium (5.85% [wt/vol] NaCl, 0.05% [wt/vol] CaCl2, and 10% [vol/vol] sterile egg yolk [Sigma, Budapest, Hungary] in YPD medium). Aspartic proteinase activity was evaluated on solid medium supplemented with bovine serum albumin (Sigma, Budapest, Hungary).
According to methods described previously by Kantarcioglu and Yücel, 5-μl suspensions of 1 × 107 cells/ml were inoculated onto the surface of the agar plates (34). Colony diameters and the colony and precipitation zones were measured after 7 days of incubation at 37°C (35).
Biofilm formation.
One-day-old C. parapsilosis biofilms were prepared as described previously (6, 36). Briefly, isolates were suspended in RPMI 1640 broth at a concentration of 1 × 106 cells/ml, and aliquots of 100 μl were inoculated onto flat-bottom 96-well sterile microtiter plates (TPP, Trasadingen, Switzerland) in the presence or absence of 15 mM tyrosol and then incubated statically at 37°C for 24 h (6, 36). Biofilm formation was determined by quantitative CFU determination of adhered cells and metabolic activity measurement using the XTT [2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide salt] assay (6, 36). Statistical comparison of CFU data and metabolic activity changes between tyrosol-treated and untreated cells was performed by the Mann-Whitney test using GraphPad Prism 6.05 software. The results between treated and control values were considered significant if the P value was <0.05.
Susceptibility of planktonic cells and biofilms to fluconazole and tyrosol.
Planktonic MIC determination was performed in line with the guidelines in Clinical and Laboratory Standards Institute document M27-A3 (37). MICs of fluconazole and tyrosol were determined in RPMI 1640. The tested drug concentrations ranged from 0.03 to 2 mg/liter for fluconazole, while tyrosol concentrations ranged from 0.5 to 60 mM. MICs were determined as the lowest drug concentration that produces at least a 50% growth reduction compared to the growth control (37). MICs represent data from three independent experiments per isolate and are expressed as medians.
The examined concentrations for sessile MIC (sMIC) determination ranged from 0.125 to 8 mg/liter and from 0.5 to 60 mM for fluconazole and tyrosol, respectively. The preformed biofilms were washed three times with sterile physiological saline. Different drug concentrations in RPMI 1640 were added to biofilms, and the plates were then incubated for 24 h at 37°C. Afterwards, sMIC determination was performed using the XTT assay as described previously (6, 36). The percent change in metabolic activity was calculated on the basis of the absorbance (A) at 492 nm as 100% × (Awell − Abackground)/(Adrug-free well − Abackground). sMICs for the biofilms were defined as the lowest drug concentration resulting in at least a 50% metabolic activity reduction compared to growth control cells (6, 36). sMICs represent data from three independent experiments per isolate and are expressed as median values.
Evaluation of interactions of fluconazole and tyrosol by fractional concentration index.
The interaction between fluconazole and tyrosol was evaluated by a two-dimensional broth microdilution chequerboard assay against planktonic and sessile cells. Afterwards, the nature of the interaction was analyzed by FICI determination. The tested concentration ranges were the same as those described above for the MIC determination. FICIs were calculated using the formula ΣFIC = FICA + FICB = MICAcomb/MICAalone + MICBcomb/MICBalone, where MICAalone and MICBalone are MICs of drugs A and B when used alone and MICAcomb and MICBcomb represent the MIC values of drugs A and B at isoeffective combinations, respectively (36, 38). FICIs were determined as the lowest ΣFIC. FICI values of ≤0.5 were defined as synergistic, values between >0.5 and 4 were defined as indifferent, and values of >4 were defined as antagonistic. FICIs were determined in three independent experiments, and median values are presented (36, 38).
In vivo experiments.
Groups of 12 female BALB/c mice (19 to 22 g) were maintained in accordance with guidelines for the care and use of laboratory animals; experiments were approved by the Animal Care Committee of the University of Debrecen (permission no. 12/2014 DEMÁB). Mice were immunosuppressed with 4 doses of intraperitoneal cyclophosphamide, i.e., 4 days prior to infection (150 mg/kg of body weight), 1 day prior to infection (100 mg/kg), 2 days postinfection (100 mg/kg), and 5 days postinfection (100 mg/kg) (30). Mice were inoculated intravenously through the lateral tail vein with an infectious dose of 7 × 106 CFU/mouse. The inoculum density was confirmed by plating serial dilutions onto Sabouraud dextrose agar plates. A 5-day intraperitoneal treatment with 15 mM tyrosol daily was started at 24 h postinoculation. On day 6 after infection, all mice were sacrificed, and kidneys were removed, weighed, and homogenized aseptically. Homogenates were diluted 10-fold; aliquots of 0.1 ml of the undiluted and diluted homogenates were plated onto Sabouraud dextrose agar plates and incubated at 37°C for 48 h. The lower limit of detection was 50 CFU/g of tissue. Statistical analysis of the kidney tissue burden was performed using an unpaired t test (30). The kidney burden was analyzed using a Kruskal-Wallis test with Dunn’s posttest (GraphPad Prism 6.05). Significance was defined as a P value of <0.05.
RNA sequencing.
Total RNA was isolated from untreated control fungal cells and 15 mM tyrosol-treated cultures in three replicates. Freeze-dried cells were processed using Trizol reagent (Invitrogen, Austria) according to methods described previously by Chomczynski (39).
To obtain global transcriptome data, high-throughput mRNA sequencing was performed on an Illumina NextSeq sequencing platform. Total RNA sample quality was checked on an Agilent Bioanalyzer using the eukaryotic total RNA nano kit (Agilent Technologies, Inc., Santa Clara, CA, USA) according to the manufacturer’s protocol. Samples with an RNA integrity number (RIN) value of >7 were accepted for the library preparation process. RNA-Seq libraries were prepared from total RNA using the TruSeq RNA sample preparation kit (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. Sequencing libraries were normalized to the same molar concentration and pooled. The library pool was sequenced on a NextSeq500 instrument (Illumina, San Diego, CA, USA), generating single-read 75-bp-long sequencing reads. Fastq files were generated automatically after the sequencing run by Illumina BaseSpace. The library preparations and the sequencing run were performed by the Genomic Medicine and Bioinformatics Core Facility of the Department of Biochemistry and Molecular Biology, Faculty of Medicine, University of Debrecen, Hungary.
Quality control of the sequencing data was performed using the FastQC package (http://www.bioinformatics.babraham.ac.uk/projects/fastqc), and the STAR RNA-Seq aligner was then used to map the sequenced reads to the reference genome (C_parapsilosis_CDC317_version_s01m03r27_features_with_chromosome_sequences.gff.gz; http://www.candidagenome.org/download/gff/C_parapsilosis_CDC317/archive/). The DESeq algorithm (StrandNGS software) was used to obtain normalized gene expression (FPKM [fragments per kilobase per million mapped fragments]) values. Gene expression differences between treated and control groups were compared by a moderated t test; the Benjamini-Hochberg false discovery rate was used for multiple-testing correction, and a corrected P value of <0.05 was considered significant (differentially expressed genes). Up- and downregulated genes were defined as differentially expressed genes with >1.5-fold change (FC) values. The FC ratios were calculated from the FPKM values.
Quantitative reverse transcriptase PCR assays.
To confirm the RNA-Seq results, 10 upregulated and 6 downregulated genes as well as 9 genes without a significant change in expression were selected for RT-qPCR analysis.
RT-qPCRs with the Xceed qPCR SG 1-step 2× Mix Lo-ROX kit (Institute of Applied Biotechnologies, Czech Republic) were performed according to the manufacturer’s protocol, using 500 ng of DNase (Sigma, Budapest, Hungary)-treated total RNA per reaction.
Primer pairs (see Table S1 in the supplemental material) were designed using OligoExplorer (version 1.1.) and Oligo Analyser (version 1.0.2) software and were purchased from Integrated DNA Technologies. Three parallel measurements were performed with each sample in a LightCycler 480 II real-time PCR instrument (Roche, Switzerland) (40). Relative transcription levels were quantified with the ΔΔCP method using the formula ΔΔCP = ΔCPcontrol − ΔCPtreated, where ΔCPtreated = CPtested gene − CPreference gene, measured from treated cultures, and ΔCPcontrol = CPtested gene − CPreference gene, measured from control cultures. CP values represent qRT-PCR cycle numbers of crossing points. Three reference genes (ACT1, TUB2, and TUB4) were tested. All of them showed stable transcription in our experiments. Only data calculated with the ACT1 (CPAR2_201570) transcription values are presented. ΔΔCP values are expressed as means ± SD calculated from three independent measurements, and ΔΔCP values significantly (P < 0.05) higher or lower than zero were determined using Student’s t test (40).
Gene set enrichment analysis.
Significant shared GO (gene ontology) terms were determined with the Candida Genome Database Gene Ontology Term Finder (http://www.candidagenome.org/cgi-bin/GO/goTermFinder). Only hits with an adjusted P value of <0.05 were taken into consideration during the evaluation process.
Data availability.
The data discussed have been deposited in the NCBI Gene Expression Omnibus (GEO) (41) (https://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession no. GSE129372.
Supplementary Material
ACKNOWLEDGMENTS
Renátó Kovács was supported by the EFOP-3.6.3-VEKOP-16-2017-00009 program. Ágnes Jakab was supported by the NTP-NFTÖ-18 scholarship program and EFOP-3.6.1-16-2016-00022. Zoltán Tóth and Fruzsina Nagy were supported by the ÚNKP-18-3 New National Excellence Program of the Ministry of Human Capacities. Research was financed by the European Union and the European Social Fund through project EFOP-3.6.1-16-2016-00022 and by the Higher Education Institutional Excellence Program of the Ministry of Human Capacities in Hungary, within the framework of the Biotechnology Thematic Program of the University of Debrecen.
László Majoros received conference travel grants from MSD, Astellas, and Pfizer. All other authors declare no conflicts of interest.
We thank Eszter Csoma for valuable suggestions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01388-19.
REFERENCES
- 1.Polke M, Leonhardt I, Kurzai O, Jacobsen ID. 2018. Farnesol signalling in Candida albicans—more than just communication. Crit Rev Microbiol 44:230–243. doi: 10.1080/1040841X.2017.1337711. [DOI] [PubMed] [Google Scholar]
- 2.Wongsuk T, Pumeesat P, Luplertlop N. 2016. Fungal quorum sensing molecules: role in fungal morphogenesis and pathogenicity. J Basic Microbiol 56:440–447. doi: 10.1002/jobm.201500759. [DOI] [PubMed] [Google Scholar]
- 3.Chen H, Fujita M, Feng Q, Clardy J, Fink GR. 2004. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc Natl Acad Sci U S A 101:5048–5052. doi: 10.1073/pnas.0401416101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alem MA, Oteef MD, Flowers TH, Douglas LJ. 2006. Production of tyrosol by Candida albicans biofilms and its role in quorum sensing and biofilm development. Eukaryot Cell 5:1770–1779. doi: 10.1128/EC.00219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monteiro DR, Arias LS, Fernandes RA, Deszo da Silva LF, de Castilho MOVF, da Rosa TO, Vieira APM, Straioto FG, Barbosa DB, Delbem ACB. 2017. Antifungal activity of tyrosol and farnesol used in combination against Candida species in the planktonic state or forming biofilms. J Appl Microbiol 123:392–400. doi: 10.1111/jam.13513. [DOI] [PubMed] [Google Scholar]
- 6.Kovács R, Tóth Z, Nagy F, Daróczi L, Bozó A, Majoros L. 2017. Activity of exogenous tyrosol in combination with caspofungin and micafungin against Candida parapsilosis sessile cells. J Appl Microbiol 122:1529–1536. doi: 10.1111/jam.13452. [DOI] [PubMed] [Google Scholar]
- 7.Shanmughapriya S, Sornakumari H, Lency A, Kavitha S, Natarajaseenivasan K. 2014. Synergistic effect of amphotericin B and tyrosol on biofilm formed by Candida krusei and Candida tropicalis from intrauterine device users. Med Mycol 52:853–861. doi: 10.1093/mmy/myu046. [DOI] [PubMed] [Google Scholar]
- 8.Padder SA, Prasad R, Shah AH. 2018. Quorum sensing: a less known mode of communication among fungi. Microbiol Res 210:51–58. doi: 10.1016/j.micres.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Grainha TRR, Jorge PADS, Pérez-Pérez M, Pérez Rodríguez G, Pereira MOBO, Lourenço AMG. 2018. Exploring anti-quorum sensing and anti-virulence based strategies to fight Candida albicans infections: an in silico approach. FEMS Yeast Res 18:foy022. doi: 10.1093/femsyr/foy022. [DOI] [PubMed] [Google Scholar]
- 10.Polke M, Jacobsen ID. 2017. Quorum sensing by farnesol revisited. Curr Genet 63:791–797. doi: 10.1007/s00294-017-0683-x. [DOI] [PubMed] [Google Scholar]
- 11.Tóth R, Nosek J, Mora-Montes HM, Gabaldon T, Bliss JM, Nosanchuk JD, Turner SA, Butler G, Vágvölgyi C, Gácser A. 2019. Candida parapsilosis: from genes to the bedside. Clin Microbiol Rev 32:e00111-18. doi: 10.1128/CMR.00111-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh S, Kebaara BW, Atkin AL, Nickerson KW. 2008. Regulation of aromatic alcohol production in Candida albicans. Appl Environ Microbiol 74:7211–7218. doi: 10.1128/AEM.01614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cattana ME, Dudiuk C, Fernández M, Rojas F, Alegre L, Córdoba S, Garcia-Effron G, Giusiano G. 2017. Identification of Candida parapsilosis sensu lato in pediatric patients and antifungal susceptibility testing. Antimicrob Agents Chemother 61:e02754-16. doi: 10.1128/AAC.02754-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cordeiro RDA, Teixeira CE, Brilhante RS, Castelo-Branco DS, Alencar LP, de Oliveira JS, Monteiro AJ, Bandeira TJ, Sidrim JJ, Moreira JL, Rocha MF. 2015. Exogenous tyrosol inhibits planktonic cells and biofilms of Candida species and enhances their susceptibility to antifungals. FEMS Yeast Res 15:fov012. doi: 10.1093/femsyr/fov012. [DOI] [PubMed] [Google Scholar]
- 15.Dantas ADS, Day A, Ikeh M, Kos I, Achan B, Quinn J. 2015. Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules 5:142–165. doi: 10.3390/biom5010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thibane VS, Ells R, Hugo A, Albertyn J, van Rensburg WJ, Van Wyk PW, Kock JL, Pohl CH. 2012. Polyunsaturated fatty acids cause apoptosis in C. albicans and C. dubliniensis biofilms. Biochim Biophys Acta 1820:1463–1468. doi: 10.1016/j.bbagen.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Bucek A, Matouskova P, Sychrova H, Pichova I, Hruskova-Heidingsfeldova O. 2014. Delta12-fatty acid desaturase from Candida parapsilosis is a multifunctional desaturase producing a range of polyunsaturated and hydroxylated fatty acids. PLoS One 9:e93322. doi: 10.1371/journal.pone.0093322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrisic L, Collinson EJ, Tehlivets O, Perak E, Zarkovic T, Dawes IW, Zarkovic N, Cipak Gasparovic A. 2015. Transcriptional and antioxidative responses to endogenous polyunsaturated fatty acid accumulation in yeast. Mol Cell Biochem 399:27–37. doi: 10.1007/s11010-014-2229-6. [DOI] [PubMed] [Google Scholar]
- 19.Cremer J, Vatou V, Braveny I. 1999. 2,4-(Hydroxyphenyl)-ethanol, an antioxidative agent produced by Candida spp., impairs neutrophilic yeast killing in vitro. FEMS Microbiol Lett 170:319–325. doi: 10.1111/j.1574-6968.1999.tb13390.x. [DOI] [PubMed] [Google Scholar]
- 20.Holland LM, Schroder MS, Turner SA, Taff H, Andes D, Grozer Z, Gacser A, Ames L, Haynes K, Higgins DG, Butler G. 2014. Comparative phenotypic analysis of the major fungal pathogens Candida parapsilosis and Candida albicans. PLoS Pathog 10:e1004365. doi: 10.1371/journal.ppat.1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monteiro DR, Feresin LP, Arias LS, Barão VA, Barbosa DB, Delbem AC. 2015. Effect of tyrosol on adhesion of Candida albicans and Candida glabrata to acrylic surfaces. Med Mycol 53:656–665. doi: 10.1093/mmy/myv052. [DOI] [PubMed] [Google Scholar]
- 22.Jabra-Rizk MA, Shirtliff M, James C, Meiller T. 2006. Effect of farnesol on Candida dubliniensis biofilm formation and fluconazole resistance. FEMS Yeast Res 6:1063–1073. doi: 10.1111/j.1567-1364.2006.00121.x. [DOI] [PubMed] [Google Scholar]
- 23.Sharma M, Prasad R. 2011. The quorum-sensing molecule farnesol is a modulator of drug efflux mediated by ABC multidrug transporters and synergizes with drugs in Candida albicans. Antimicrob Agents Chemother 55:4834–4843. doi: 10.1128/AAC.00344-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossignol T, Logue ME, Reynolds K, Grenon M, Lowndes NF, Butler G. 2007. Transcriptional response of Candida parapsilosis following exposure to farnesol. Antimicrob Agents Chemother 51:2304–2312. doi: 10.1128/AAC.01438.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horvath P, Nosanchuk JD, Hamari Z, Vagvolgyi C, Gacser A. 2012. The identification of gene duplication and the role of secreted aspartyl proteinase 1 in Candida parapsilosis virulence. J Infect Dis 205:923–933. doi: 10.1093/infdis/jir873. [DOI] [PubMed] [Google Scholar]
- 26.Nemes D, Kovács R, Nagy F, Mező M, Poczok N, Ujhelyi Z, Pető Á, Fehér P, Fenyvesi F, Váradi J, Vecsernyés M, Bácskay I. 2018. Interaction between different pharmaceutical excipients in liquid dosage forms—assessment of cytotoxicity and antimicrobial activity. Molecules 23:E1827. doi: 10.3390/molecules23071827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berridge MV, Herst PM, Tan AS. 2005. Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnol Annu Rev 11:127–152. doi: 10.1016/S1387-2656(05)11004-7. [DOI] [PubMed] [Google Scholar]
- 28.Atienza JM, Zhu J, Wang X, Xu X, Abassi Y. 2005. Dynamic monitoring of cell adhesion and spreading on microelectronic sensor arrays. J Biomol Screen 10:795–805. doi: 10.1177/1087057105279635. [DOI] [PubMed] [Google Scholar]
- 29.Tóth R, Alonso MF, Bain JM, Vágvölgyi C, Erwig LP, Gácser A. 2015. Different Candida parapsilosis clinical isolates and lipase deficient strain trigger an altered cellular immune response. Front Microbiol 6:1102. doi: 10.3389/fmicb.2015.01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovács R, Gesztelyi R, Berényi R, Domán M, Kardos G, Juhász B, Majoros L. 2014. Killing rates exerted by caspofungin in 50% serum and its correlation with in vivo efficacy in a neutropenic murine model against Candida krusei and Candida inconspicua. J Med Microbiol 63:186–194. doi: 10.1099/jmm.0.066381-0. [DOI] [PubMed] [Google Scholar]
- 31.Jakab Á, Emri T, Sipos L, Kiss Á, Kovács R, Dombrádi V, Kemény-Beke Á, Balla J, Majoros L, Pócsi I. 2015. Betamethasone augments the antifungal effect of menadione—towards a novel anti-Candida albicans combination therapy. J Basic Microbiol 55:973–981. doi: 10.1002/jobm.201400903. [DOI] [PubMed] [Google Scholar]
- 32.Dalle F, Wächtler B, L’Ollivier C, Holland G, Bannert N, Wilson D, Labruère C, Bonnin A, Hube B. 2010. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell Microbiol 12:248–271. doi: 10.1111/j.1462-5822.2009.01394.x. [DOI] [PubMed] [Google Scholar]
- 33.Jakab Á, Mogavero S, Förster TM, Pekmezovic M, Jablonowski N, Dombrádi V, Pócsi I, Hube B. 2016. Effects of the glucocorticoid betamethasone on the interaction of Candida albicans with human epithelial cells. Microbiology 162:2116–2125. doi: 10.1099/mic.0.000383. [DOI] [PubMed] [Google Scholar]
- 34.Kantarcioglu AS, Yücel A. 2002. Phospholipase and protease activities in clinical Candida isolates with reference to the sources of strains. Mycoses 45:160–165. doi: 10.1046/j.1439-0507.2002.00727.x. [DOI] [PubMed] [Google Scholar]
- 35.Price MF, Wilkinson ID, Gentry LO. 1982. Plate method for detection of phospholipase activity in Candida albicans. Sabouraudia 20:7–14. doi: 10.1080/00362178285380031. [DOI] [PubMed] [Google Scholar]
- 36.Kovács R, Bozó A, Gesztelyi R, Domán M, Kardos G, Nagy F, Tóth Z, Majoros L. 2016. Effect of caspofungin and micafungin in combination with farnesol against Candida parapsilosis biofilms. Int J Antimicrob Agents 47:304–310. doi: 10.1016/j.ijantimicag.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 37.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard, 3rd ed Document M27-A3 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 38.Meletiadis J, Verweij PE, Te Dorsthorst DT, Meis JF, Mouton JW. 2005. Assessing in vitro combinations of antifungal drugs against yeasts and filamentous fungi: comparison of different drug interaction models. Med Mycol 43:133–152. doi: 10.1080/13693780410001731547. [DOI] [PubMed] [Google Scholar]
- 39.Chomczynski P. 1993. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 15:532–534. [PubMed] [Google Scholar]
- 40.Kurucz V, Kiss B, Szigeti ZM, Nagy G, Orosz E, Hargitai Z, Harangi S, Wiebenga A, de Vries RP, Pócsi I, Emri T. 2018. Physiological background of the remarkably high Cd2+ tolerance of the Aspergillus fumigatus Af293 strain. J Basic Microbiol 58:957–967. doi: 10.1002/jobm.201800200. [DOI] [PubMed] [Google Scholar]
- 41.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed have been deposited in the NCBI Gene Expression Omnibus (GEO) (41) (https://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession no. GSE129372.