Bacterial leaf spot of tomato and pepper is an endemic plant disease with a global distribution. In this study, we investigated the evolutionary processes leading to the emergence of novel X. perforans lineages identified in Alabama. While one lineage was isolated from symptomatic tomato and pepper plants, confirming the host range expansion of X. perforans, the other lineage was isolated from tomato and acquired a novel transcription activation-like effector, here designated PthXp1. Functional analysis of PthXp1 indicated that it does not induce Bs4-mediated resistance in tomato and contributes to virulence, providing an adaptive advantage to strains on tomato. Our findings also show that different phylogenetic groups of the pathogen have experienced independent recombination events originating from multiple Xanthomonas species. This suggests a continuous gene flux between related xanthomonads associated with diverse plant hosts that results in the emergence of novel pathogen lineages and associated phenotypes, including host range.
KEYWORDS: TAL effector, Xanthomonas, emerging, host specificity, novel lineages, recombination
ABSTRACT
Xanthomonas perforans is the predominant pathogen responsible for bacterial leaf spot of tomato and X. euvesicatoria for that of pepper in the southeast United States. Previous studies have indicated significant changes in the X. perforans population collected from Florida tomato fields over the span of 2 decades, including a shift in race and diversification into three phylogenetic groups driven by genome-wide homologous-recombination events derived from X. euvesicatoria. In our sampling of Xanthomonas strains associated with bacterial spot disease in Alabama, we were readily able to isolate X. perforans from symptomatic pepper plants grown in several Alabama counties, indicating a recent shift in the host range of the pathogen. To investigate the diversity of these pepper-pathogenic strains and their relation to populations associated with tomatoes grown in the southeast United States, we sequenced the genomes of eight X. perforans strains isolated from tomatoes and peppers grown in Alabama and compared them with previously published genome data available from GenBank. Surprisingly, reconstruction of the X. perforans core genome revealed the presence of two novel genetic groups in Alabama that each harbored a different transcription activation-like effector (TALE). While one TALE, AvrHah1, was associated with an emergent lineage pathogenic to both tomato and pepper, the other was identified as a new class within the AvrBs3 family, here designated PthXp1, and was associated with enhanced symptom development on tomato. Examination of patterns of homologous recombination across the larger X. euvesicatoria species complex revealed a dynamic pattern of gene flow, with multiple donors of Xanthomonas spp. associated with diverse hosts of isolation.
IMPORTANCE Bacterial leaf spot of tomato and pepper is an endemic plant disease with a global distribution. In this study, we investigated the evolutionary processes leading to the emergence of novel X. perforans lineages identified in Alabama. While one lineage was isolated from symptomatic tomato and pepper plants, confirming the host range expansion of X. perforans, the other lineage was isolated from tomato and acquired a novel transcription activation-like effector, here designated PthXp1. Functional analysis of PthXp1 indicated that it does not induce Bs4-mediated resistance in tomato and contributes to virulence, providing an adaptive advantage to strains on tomato. Our findings also show that different phylogenetic groups of the pathogen have experienced independent recombination events originating from multiple Xanthomonas species. This suggests a continuous gene flux between related xanthomonads associated with diverse plant hosts that results in the emergence of novel pathogen lineages and associated phenotypes, including host range.
INTRODUCTION
Knowledge of the importance of genetic exchange in bacterial evolution can be traced back as early as the 1960s, when plasmid-mediated transfer of penicillin resistance was documented among members of the Enterobacteriaceae (1). With the increasing availability of bacterial genomes, it has since become clear that the exchange of mobile genetic elements, including plasmids, bacteriophage, genomic islands, and other mechanisms of horizontal gene transfer, is commonplace among bacterial populations. Such genetic exchanges can confer traits imparting phenotypic and genotypic plasticity to a bacterial species in response to changes in the environment (2), thus facilitating adaptive evolution. In addition to horizontal gene transfer, many bacteria undergo homologous recombination. This is a process similar to meiotic recombination, in which segments of a bacterial genome are replaced by closely related sequences from a donor organism, resulting in a mosaic pattern of loci with distinct evolutionary histories (3).
In the absence of barriers such as adaptive incompatibility, related bacterial lineages are expected to display evidence of admixture in their evolutionary histories when they inhabit overlapping environmental niches (4). Therefore, patterns of recombination are hypothesized to reflect a corresponding microbial ecology and to maintain the cohesion of various bacterial species as monophyletic groups (5). In contrast to many genera of plant-pathogenic bacteria, including Pseudomonas, Ralstonia, and Burkholderia, that are abundant in multiple environments, including water and soil, the life histories of Xanthomonas spp. have traditionally been considered to be restricted to plants (6). Most xanthomonads exhibit a high degree of host specificity, with the individual species (or the pathovars found within them) each forming a genetically monomorphic cluster of lineages (7). This evolutionary trend suggests that the divergence of Xanthomonas species/pathovars is primarily driven by ecological isolation associated with adaptation to a particular host (7–9).
Among the numerous plant diseases caused by xanthomonads, bacterial leaf spot of tomato (Solanum lycopersicum) and pepper (Capsicum annuum) is atypical, as four distinct species, Xanthomonas cynarae pv. gardneri, Xanthomonas euvesicatoria, Xanthomonas perforans, and Xanthomonas vesicatoria, have converged on these hosts to cause the same disease (10, 11). These species may differ in geographic distribution, as well as in the molecular mechanisms employed in pathogenesis (12–14). Likewise, temporal shifts in the species and pathogen races responsible for the disease in specific production regions have been documented (15). In the southeastern United States, X. euvesicatoria and X. perforans are currently the dominant pathogens of pepper and tomato, respectively, while X. cynarae pv. gardneri and X. vesicatoria have not been observed and are more distantly related (13, 14). Earlier studies classified X. euvesicatoria and X. perforans into distinct bacterial species based on DNA-DNA hybridization values, biochemical profiling, and different host specificities (10). However, whole-genome sequence analysis of diverse strains from the two groups indicated that their average nucleotide identity (ANI) of >98% falls well within the recommended threshold of 95% for species delimitation of prokaryotes (16–21). These findings have led to the recent proposals of consideration of X. euvesicatoria and X. perforans as pathovars of X. euvesicatoria rather than separate species (16, 17). The two species form sister lineages within the larger X. euvesicatoria species complex, which is composed of strains that infect diverse monocot and dicotyledonous plant families and generally display ANI values of ≥98%. The X. euvesicatoria species complex also includes strains classified as X. alfalfae, X. axonopodis, and X. euvesicatoria, which may be further classified at the infrasubspecific level into numerous pathovars based on host range (17, 20, 21). The phenotypic heterogeneity, genetic variation, and large collective host range observed within this phylogenetic group present a challenge that is often faced in defining microbial species. Among the important issues associated with this challenge are their widespread geographical distribution, their unexplored ecological niches, and the extraordinary ability to transfer genes across species boundaries.
The studies investigating the extent of gene flow within the X. euvesicatoria species complex have been focused mostly on bacterial spot xanthomonads, mainly X. perforans and X. euvesicatoria. Our previous population genomic studies with X. perforans strains collected in Florida revealed homologous recombination to be a primary factor driving the diversification of the pathogen into three different phylogenetic groups, with X. euvesicatoria identified as the primary donor of recombinant sequences (18). Similar observations were recorded among X. perforans strains collected in Nigeria, where X. euvesicatoria was also identified as the likely donor in recent recombination (19). Given the close evolutionary relationship of strains found within the X. euvesicatoria species complex and their underexplored ecology, the effective population size of X. euvesicatoria and related pathogens and the extent of gene flux may be considerably larger than previously recognized.
Until the isolation of a single X. perforans strain (Xp2010) from an infected pepper sample during the 2010 season in Florida (22), the host range of the pathogen was thought to be restricted to tomato. Most X. perforans strains carry a type 3 secreted effector (T3SE) (either AvrBsT or AvrXv3) that, when injected into the host cell, elicits effector-triggered immunity in susceptible pepper varieties (23). Mutational analysis of AvrBsT and AvrXv3 in different genetic backgrounds of X. perforans revealed different abilities among strains to multiply and cause disease when infiltrated into pepper leaves (22). Intriguingly, the lineages determined to be pathogenic to pepper based on the artificial-inoculation experiments displayed evidence of extensive recent recombination originating from X. euvesicatoria, whereas similar signatures were reported to be minimal among the tomato-limited X. perforans lineages (14, 18).
In a recent survey of the Xanthomonas population responsible for the bacterial spot disease in Alabama, we were readily able to isolate X. perforans strains from pepper plants grown in several Alabama counties (N. Potnis and R. R. Bhandari, unpublished data). These findings were surprising, as they indicated a recent shift in the host range of X. perforans and an emerging threat to pepper production. Our initial survey with small-scale and commercial growers indicated differences in the production practices, variety choice, and scale of production compared to neighboring tomato and pepper production regions. These observations led us to hypothesize that X. perforans exhibits additional genome-wide diversity beyond that which has been documented among the strains sampled in Florida. In order to investigate the diversity of these pepper-pathogenic X. perforans strains and their relation to populations associated with tomatoes grown in the southeast United States, we sequenced the genomes of eight X. perforans strains isolated from tomatoes and peppers grown in Alabama and compared them with previously published genome data available from GenBank. Using a Bayesian statistical approach, we refined the population structure of X. perforans with the identification of two novel phylogenetic groups. Our findings indicate that the recent emergence of these lineages was associated with the acquisition of novel transcription activator-like effectors and independent recombination events originating from members of the X. euvesicatoria species complex associated with diverse hosts of isolation.
RESULTS
Reconstruction of the X. perforans core genome reveals the presence of two novel genetic clusters composed of X. perforans strains collected in Alabama.
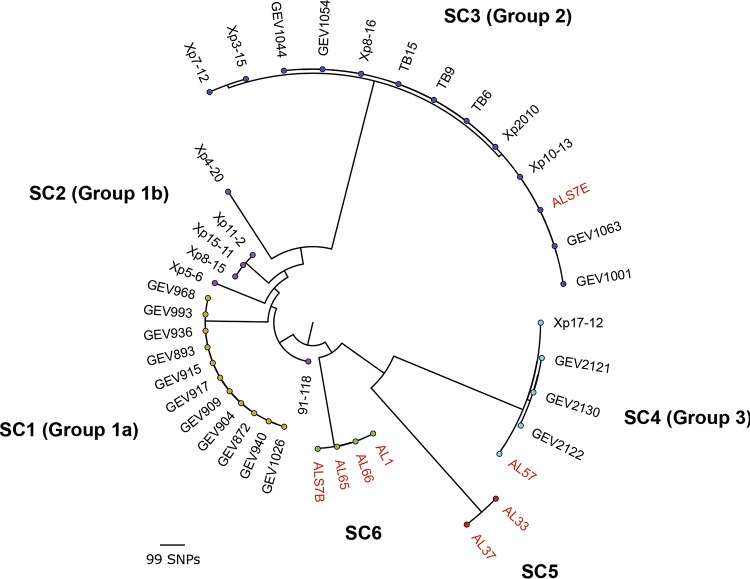
In order to investigate the diversity of the strains sequenced in this study in the context of the larger X. perforans population structure previously described in Florida (18, 22), a subset of 33 Florida X. perforans strains were reexamined. Five of the strains were isolated from symptomatic pepper plants (four from Alabama and one from Florida), while the remaining strains were isolated from tomato. A maximum-likelihood phylogeny constructed from a concatenated alignment of 16,501 high-quality core genome single-nucleotide polymorphisms (SNPs), coupled with a Bayesian analysis of population structure (24), revealed the presence of six distinct genetic clusters within X. perforans (Fig. 1). Sequence cluster 1 (SC1) through SC4 corresponded to the previously described population structure of X. perforans strains collected in Florida (18, 22), while SC5 and SC6 were composed exclusively of Alabama strains sequenced in this study. Each of the genetic clusters inferred in this analysis were primarily clonal within the lineage, except for SC2 (referred to as group 1b by Schwartz et al. [22]), which displayed the greatest amount of variation. SC3 (group 2) contained the Florida strain originally isolated from pepper (Xp2010), as well as other strains demonstrated to be pathogenic to this host under artificial-inoculation conditions (22). A single Alabama strain (ALS7E) isolated from pepper clustered within the group, while the remaining pepper strains (ALS7B, AL65, and AL66) and one tomato strain (AL1) formed a novel phylogenetic lineage, here designated SC6. The Alabama strain AL57 grouped with other X. perforans strains collected in Florida within SC4 (group 3). This genetic cluster branched from SC5, which was composed of two Alabama strains (AL33 and AL37) isolated from tomato plants.
FIG 1.
Midpoint-rooted maximum-likelihood phylogeny of 41 X. perforans strains isolated from tomato and pepper plants grown in Florida and Alabama based on a concatenated alignment of 16,501 core genome SNPs. The tips are color coded according to the SCs identified in the first level of the HierBAPS hierarchy (24). The phylogroup designations of strains described previously (18, 22) are shown in parentheses, and strains sequenced in this study are highlighted in red. The scale bar indicates the number of substitutions per site.
Analysis of T3SE repertoires provides evidence for the plasmid-mediated acquisition of several accessory T3SEs, including the transcription activation-like effector (TALE) AvrHah1, among X. perforans strains collected in Alabama.
A total of 24 T3SE genes were conserved among the X. perforans strains sequenced in this study (Table 1). These results were largely concordant with conserved effectors identified across the X. perforans population characterized in Florida (22), except for xopE2, which was carried by only three strains (AL33, AL37, and ALS7E) sequenced here and was present in contigs that displayed 100% identity to plasmid pLH3.2 (NZ_CP018476.1) from X. perforans strain LH3. These contigs also contained genes encoding copper resistance, including copA, copB, and copF. Strains AL33, AL37, and AL57 carried an intact copy of the avirulence gene avrXv3, which was disrupted by an insertion sequence in the other Alabama strains. The inability of the latter strains to induce hypersensitive resistance (HR) in the pepper cultivar Early Cal Wonder (ECW) confirmed the nonfunctionality of avrXv3, while the former stains with the intact allele elicited HR in ECW (Table 1).
TABLE 1.
Pathogenicity phenotyping and distribution of type 3 secreted effectors among X. perforans strains isolated from tomato and pepper in Alabama
| T3SE | Presencea or pathogenicityb |
|||||||
|---|---|---|---|---|---|---|---|---|
| ALS7E | AL33 | AL37 | AL57 | ALS7B | AL1 | AL65 | AL66 | |
| ECW | C | HR | HR | HR | C | C | C | C |
| ECW30R | C | HR | HR | HR | C | HR | HR | HR |
| avrXv3 | IS | + | + | + | IS | IS | IS | IS |
| xopAQc | − | − | + | + | − | − | − | − |
| pthXp1 | − | − | + | + | − | − | − | − |
| avrBsT homologuec | − | − | + | + | − | − | − | − |
| xopE3c | − | − | IS | IS | − | − | − | − |
| xopE2c | + | + | + | − | − | − | − | − |
| avrHah1c | − | − | − | − | − | + | + | + |
| xopAOc | − | − | − | − | − | + | + | + |
| avrXv4 | + | + | + | + | + | + | + | + |
| xopA | + | + | + | + | + | + | + | + |
| xopAD | + | + | + | + | + | + | + | + |
| xopAE | + | + | + | + | + | + | + | + |
| xopAK | + | + | + | + | + | + | + | + |
| avrBs2 | + | + | + | + | + | + | + | + |
| xopAP | + | + | + | + | + | + | + | + |
| xopAR | + | + | + | + | + | + | + | + |
| xopC2 | + | + | + | + | + | + | + | + |
| xopD | + (Ctg) | + | + | + | + | + | + | + |
| xopE1 | + | + | + | + | + | + | + | + |
| xopF1 | + | + | + | + | + | + | + | + |
| xopF2 | + | + | + | + | + | + | + | + |
| xopI | + | + | + | + | + | + | + | + |
| xopK | + | + | + | + | + | + | + | + |
| xopL | + | + | + | + | + | + | + | + |
| xopN | + | + | + | + | + | + | + | + |
| xopP1 | + | + | + | + | + | + | + | + |
| xopP2 | + | + | + | + | + | + | + | + |
| xopQ | + | + | + | + | + | + | + | + |
| xopR | + | + | + | + | + | + | + | + |
| xopV | + | + | + | + | + | + | + | + |
| xopX | + | + | + | + | + | + | + | + |
| xopZ | + | + | + | + | + | + | + | + |
+, the gene was present with an intact coding sequence; −, the gene was absent; IS, the gene was disrupted by an insertion sequence; Ctg, the gene was present at the end of a contig break.
Pathogenicity phenotyping on pepper cultivars ECW and ECW30R. A compatible reaction is indicated by C and hypersensitive resistance by HR.
The T3SE was present in contigs assembled by plasmidSPAdes in at least one strain.
Several other variable T3SE genes were identified among the Alabama strains, along with evidence of their presence in putative plasmids. The T3SE genes xopAQ and xopE3 were present in strains AL37 and AL57 while they were absent from the genome assemblies of other Alabama strains. The latter effector gene was disrupted by a transposon insertion in both strains. Both effectors were present in the same contig assembled by plasmidSPAdes software in strain AL57, which displayed significant similarity (99% nucleotide identity and 94% query coverage) to an unnamed plasmid found in X. campestris pv. campestris strain CN18 (CP017322.1). Likewise, a putative T3SE with similarity to AvrBsT (71% amino acid identity) was also carried on contigs assembled by plasmidSPAdes in the same two strains. The top BLAST hit for these contigs (94% nucleotide identity; over 87% query coverage) was plasmid pXCARECAE29 (NZ_CP034654.1) from X. campestris pv. arecae strain NCPPB 2649.
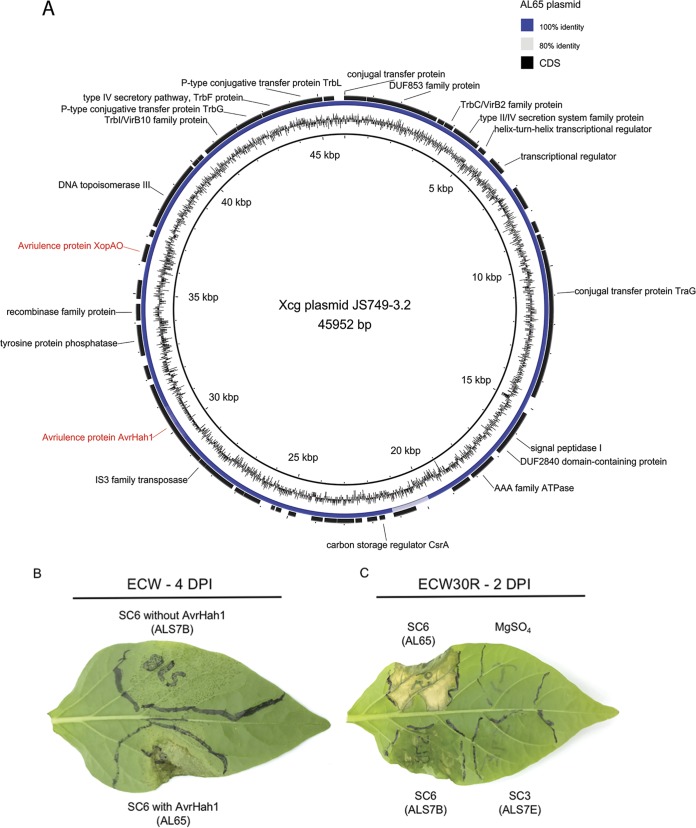
Analysis with the plasmidSPAdes software also revealed the presence of several contigs in all but one of the strains (ALS7B) located in genetic cluster SC6, which displayed 100% nucleotide identity and over 99% query coverage to the complete sequence of plasmid pJS749-3.2 (NZ_CP018730.1) from X. cynarae pv. gardneri strain JS749-3 (Fig. 2A). The sequences corresponding to this putative plasmid were divided between two and five contigs per strain and totaled from 43 to 45 kb, which was consistent with the size of pJS749-3.2 (46 kb). Analysis of these contigs with tBLASTn produced significant hits for the TALE AvrHah1 and the T3SE XopAO. The activity of AvrHah1 among these strains was supported through inoculations in the pepper cultivar ECW, which produced the profuse water-soaking phenotype associated with this TALE (Fig. 2B). Further evidence was obtained through inoculations into the pepper cultivar ECW30R, which resulted in hypersensitive resistance (Fig. 2C). A summary of the contigs assembled by plasmidSPAdes and BLAST search results is presented in Table S2 in the supplemental material.
FIG 2.
X. perforans strains collected in Alabama carry a plasmid nearly identical to one found in X. cynarae pv. gardneri, which harbors the transcription activation-like effector AvrHah1, and are pathogenic to pepper. (A) Alignment of contigs assembled with plasmidSPAdes software from X. perforans strain AL65 and the X. cynarae pv. gardneri (Xcg) plasmid JS749-3.2. The inner ring shows the GC content of the plasmid, and the annotations of coding sequences (CDS) (shown in black) are provided where available. (B) Bacterial spot symptoms produced by two strains from SC6, with and without AvrHah1, shown 4 days postinoculation (DPI). Leaves of the susceptible pepper cultivar ECW were infiltrated with a needleless syringe using a bacterial suspension adjusted to 104 CFU ml−1. The individual strains used for inoculation are shown in parentheses. (C) Compatible and incompatible interactions on pepper cultivar ECW30R with Bs3 resistance, shown 2 days postinoculation with a bacterial suspension raised to 108 CFU ml−1. Hypersensitive resistance mediated by the recognition of AvrHah1 in strain AL65 is shown on the top left portion of the leaf. Pepper strains ALS7B and ALS7E, from SC6 and SC3, are shown on the bottom left and bottom right portions of the leaf, respectively.
Identification of a novel class of TALE within the AvrBs3 family among X. perforans strains with different genetic backgrounds.
Screening of the genome assemblies for T3SEs with tBLASTn indicated the presence of a putative TALE with similarity to AvrBs3 in strains AL57 and AL37 of SC4 and SC5, respectively. Phusion PCR utilizing primers designed to anneal to conserved loci within the N- and C-terminal domains of the AvrBs3 effector family produced an ∼2.9-kb amplicon (see Fig. S1 in the supplemental material). Sanger sequencing of the amplicon utilizing internal primers flanking the repeat region of avrBs3 revealed that the TALE was comprised of 15.5 tandem repeats, each 102 bp in length. A BLAST search of the NCBI nonredundant protein database using the N- and C-terminal domains of the protein indicated that it displayed 99 and 100% identity, respectively, to the avrBs3 allele (X16130.1) found in X. vesicatoria.
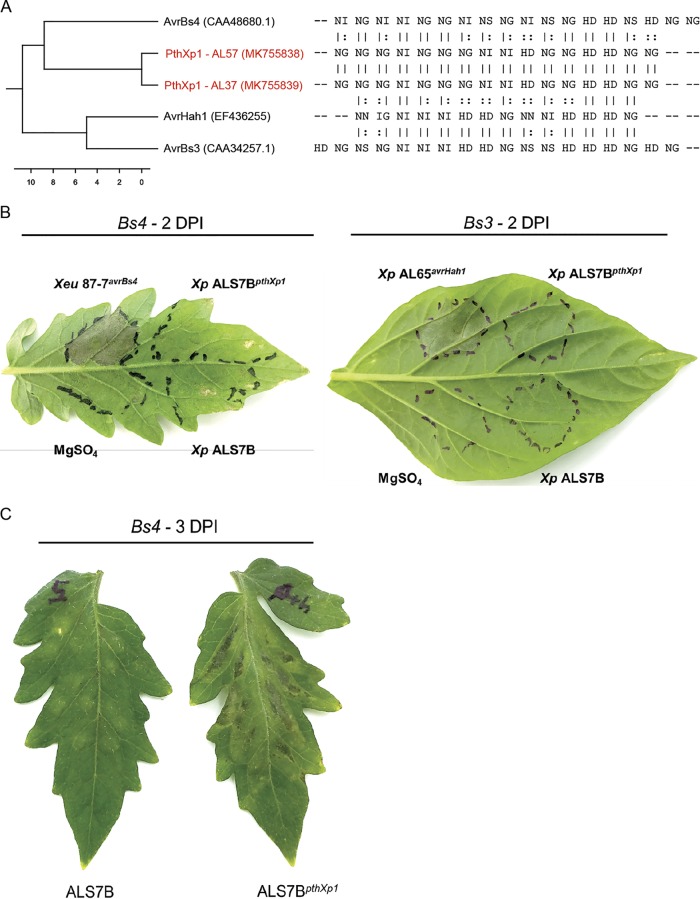
Despite this similarity to AvrBs3, analysis of the repeat variable diresidue (RVD) sequences with AnnoTALE software indicated that the gene could not be placed into the same class as any TALE for which sequence data were available based on the recommended cutoff of ≤5.0 pairwise difference and alignment probability of ≤0.01 (data not shown). Alignment of the RVDs with other TALEs found in bacterial spot xanthomonads showed the effector was most closely related to AvrBs4 and shared two blocks of homology with the effector; however, it displayed a pairwise difference in RVDs of 8.8 (Fig. 3A). In order to investigate the function and host recognition of this novel TALE, we cloned the gene into a broad-host-range cosmid and conjugated it into an X. perforans strain (ALS7B) pathogenic to both tomato and pepper. Inoculations on the tomato cultivar MoneyMaker with Bs4 resistance and the pepper cultivar ECW30R with Bs3 resistance resulted in compatible interactions, indicating host recognition distinct from those of previously described TALEs in bacterial spot pathogens (Fig. 3B). When infiltrated into tomato leaves, the TALE was associated with enhanced chlorosis and lesion development relative to the nearly isogenic wild-type strain 72 h postinoculation (Fig. 3C). No TALE-associated phenotype was observed when strains were infiltrated into pepper leaves (data not shown).
FIG 3.
Identification of a new class of transcription activation-like effector, PthXp1, in X. perforans. (A) Dendrogram (left) constructed from an alignment of repeat variable diresidue sequences (right) found among the previously described transcription activation-like effectors carried by bacterial spot pathogens and PthXp1 from X. perforans strains AL37 and AL57 (shown in red). The GenBank accession numbers of the sequences are shown in parentheses, and the scale bar indicates the number of substitutions per site. (B) Differential reactions in tomato cultivar MoneyMaker with Bs4 resistance and pepper cultivar ECW30R with Bs3 resistance. Leaves were infiltrated with a bacterial suspension adjusted to 108 CFU ml−1 using X. perforans strain ALS7B, pathogenic to tomato and pepper, and the nearly isogenic strain ALS7BpthXp1 carrying the pthXp1 gene on a broad-host-range cosmid. Hypersensitive resistance is indicated by the appearance of collapsed/necrotic leaf tissue. X. euvesicatoria strain 87-7 with avrBs4 and X. perforans strain AL65 with avrHah1 were included as positive controls for Bs4 and Bs3 recognition, respectively. (C) Symptom development associated with PthXp1 on tomato cultivar MoneyMaker leaves infiltrated with a bacterial suspension raised to 108 CFU ml−1, shown 3 days postinoculation (DPI).
Novel lineages within X. perforans have emerged through recent recombination derived from outside the bacterial spot species complex.
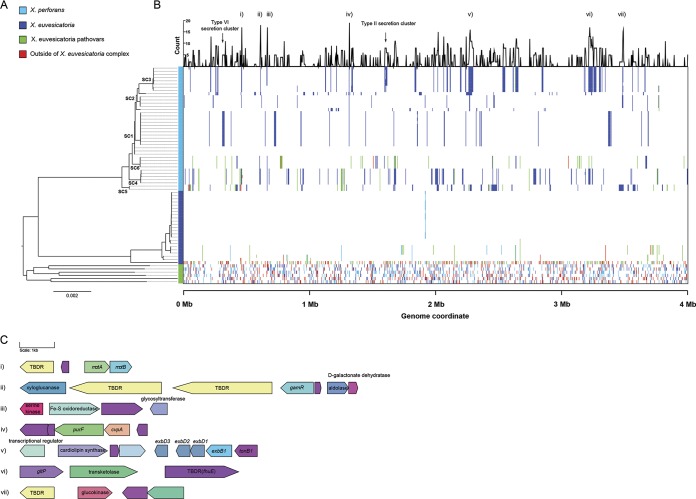
To examine patterns of homologous recombination between X. perforans, X. euvesicatoria, and other closely related species within the X. euvesicatoria species complex, a second core genome alignment was constructed utilizing the strains sequenced in this study and genome assemblies available from GenBank (n = 68). A maximum-likelihood phylogeny generated from the resulting 3.98-Mb alignment was largely congruent to the population structure inferred by previous studies (19, 22) and showed X. perforans and X. euvesicatoria branching from each other into two distinct phylogenetic groups, while other related X. euvesicatoria pathovars displayed considerably longer branch lengths and clustered into a paraphyletic group of strains. Analysis with the FastGear software indicated the presence of three distinct lineages, which were defined as a group of strains that shared a common ancestry in at least half of the alignment (3) and that corresponded to the three phylogenetic groups described above (Fig. 4A).
FIG 4.
Landscape of homologous recombination within the X. euvesicatoria species complex. (A) Phylogeny of 67 X. perforans, X. euvesicatoria, and closely related X. euvesicatoria pathovars based on a core genome alignment of 3.98 Mb. The branches corresponding to the sequence clusters inferred within X. perforans (Fig. 1) are labeled at the nodes, and lineages predicted by the FastGear software are color coded at the right of the tree. (B) Patterns of recent recombination across the core genome of the X. euvesicatoria species complex. The colors indicate the donor lineages of the recent recombination events, and the line graph above shows the frequency of recombination within X. perforans at a particular site in the alignment. Recombination events originating from outside the sampled Xanthomonas population are shown in red. (C) Gene contents of core genome loci with elevated recombination frequencies (numbered i to vii) within X. perforans. Genes are color coded according to their annotation, with hypothetical proteins shown in purple. Gene names/functional annotations are shown where available. TBDR, putative TonB-dependent receptor.
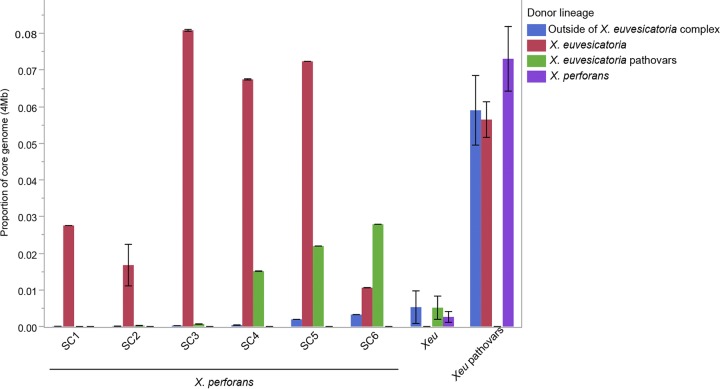
A total of 3,174 recent recombination events were identified across the alignment (Fig. 4B). Consistent with the long branch lengths, the lineage composed of various X. euvesicatoria pathovars displayed evidence of extensive recent recombination, with an average (±standard deviation) 19.00% ± 0.88% of the core genome predicted to be recombinant among the individual strains. A considerable amount of recent recombination found within this group originated from both the X. perforans and X. euvesicatoria lineages (7.31% and 5.90% of the alignment, respectively); however, the proportion of gene flux from each donor lineage was somewhat variable at the strain level (standard deviations of ±2.16% and ±1.19% for X. perforans and X. euvesicatoria, respectively). Interestingly, approximately a third of the recombinant sequences detected in this lineage (5.90% ± 2.33% of the alignment) were predicted to have originated outside the sampled Xanthomonas population (Fig. 5).
FIG 5.
Summary of the proportions and origins of recent recombinations among lineages within the X. euvesicatoria (Xeu) species complex and X. perforans SCs as predicted by FastGear. “Outside of X. euvesicatoria complex” indicates the proportion of recombinant sequences donated from outside the sampled Xanthomonas population. The error bars indicate standard deviations of the mean.
Evidence of recent recombination was also apparent within the X. perforans lineage. Nearly all of the recent recombination events detected in the Florida X. perforans strains collected prior to 2015 (SC1 through SC3) were predicted to have originated from X. euvesicatoria, whereas recombination events derived from both X. euvesicatoria and the lineage composed of various X. euvesicatoria pathovars were detected in the recently described group 3 X. perforans strains (here designated SC4), as well as the Alabama strains located in SC5 and SC6. The average proportions of core genome donated by various species within the X. euvesicatoria species complex ranged from 1.51% to 2.79% among these groups, and they were the primary donors in recent recombination (66.7% of total recent recombinant sequences) to the pepper-pathogenic strains of SC6. Variability in the overall proportions of recent recombination was also observed among the X. perforans sequence clusters, ranging from 1.41% ± 1.70% in SC1 to 9.64% ± 0.00% in SC5 (Fig. 5).
Recombination hot spots are characterized by pathways involved in the acquisition/metabolism of plant-derived nutrients and motility.
We investigated seven recombination hot spots within X. perforans based on the frequency of recombination events at a particular site in the core genome alignment (Fig. 4C). Hot spots ii, vi, and vii mapped to loci with genes involved in carbohydrate and amino acid metabolism, including a xyloglucanase (lysR)-type regulator of galactose metabolism (gamR), transketolase, a symporter of protons/glutamate (gltP), and a glucokinase. All of these genes were adjacent to at least one TonB-dependent receptor (TBDR), while a fourth TBDR locus, hot spot i, contained the flagellar motor components motA and motB. It was therefore interesting to find the entire TonB transduction system (tonB-exbB-exbD1-exbD2) at recombination hot spot v. Other genes in recombination hot spots included an iron-sulfur oxidoreductase (iii) and a gene with homology to a colicin V production protein, cvpA (iv). Finally, while not necessarily present in hot spots, we noted that most of the genes that encode the Xps type II and type VI secretion systems were recombinant among strains from SC3 and SC1, respectively.
DISCUSSION
Recent emergence of a novel X. perforans lineage pathogenic to tomato and pepper.
Maximum-likelihood reconstruction of the X. perforans core genome revealed the presence of diversity among the strains sequenced in this study, with the identification of two additional phylogenetic groups, here designated SC5 and SC6 (Fig. 1). To our surprise, the majority of pepper strains did not group with others that were previously reported to be pathogenic to pepper within SC3 but comprised the newly emerged SC6. Three of the four strains within this phylogenetic group carried a plasmid nearly identical to one often found in X. cynarae pv. gardneri that harbored two different type 3 secreted effectors, XopAO and the TALE AvrHah1 (Table 1 and Fig. 2A). The latter effector is commonly associated with the enhanced virulence of strains in tomato and pepper plants through hijacking the host expression of pectate lyase genes. This results in profuse water soaking of leaf tissue and is hypothesized to increase the uptake of bacterial cells into the apoplast, facilitating ingress and/or dissemination (25, 26). We noted that one pepper strain located in SC6 (ALS7B) lacked the plasmid associated with the horizontal transfer of AvrHah1 yet was still capable of producing water-soaked lesions when infiltrated into pepper leaves (Fig. 2B). This indicated that while AvrHah1 serves as a virulence/fitness factor for X. perforans in a fashion similar to what has been reported for X. cynarae pv. gardneri, it is not required for pathogenicity among strains of this genetic background.
Significant changes in the international Xanthomonas population were documented over the course of the 1990s, where X. perforans displaced X. euvesicatoria as the predominant pathogen of tomato in several countries simultaneously while X. euvesicatoria maintained its dominance in pepper fields (27–30). Recently, Burlakoti and colleagues (31) isolated four X. perforans strains from naturally infected pepper samples in Taiwan, providing further evidence for the host as a niche for X. perforans. However, when these strains were inoculated back into their original host of isolation, they produced a hypersensitive response and were avirulent. The majority of X. perforans strains located in SC3 carry the avirulence gene avrBsT on a self-transmissible plasmid (15, 23). While the loss of avrBsT may result in a host range expansion to pepper (22), it is also associated with a fitness penalty to the pathogen (32). The same is not true of the pepper and tomato strains located in SC6, indicating that the recent emergence of this lineage may pose a high risk to both tomato and pepper production. This highlights the importance of continued surveillance of bacterial spot pathogens, as rapidly changing populations have important implications for resistance breeding and disease management strategies.
Identification of PthXp1, a new class of TALE in X. perforans.
Another diverse X. perforans lineage, referred to here as SC5, was identified in our analysis and consisted of two Alabama strains that were isolated from tomato plants collected from commercial production fields. This phylogenetic group branched from SC4 (Fig. 1), which was recently found to be prevalent in Florida tomato fields (18). Screening of the genome assemblies with tBLASTn revealed the presence of sequences nearly identical to the N- and C-terminal domains of AvrBs3/AvrBs4 in the Alabama strains AL57 and AL37 of SC4 and SC5, respectively. This was surprising to find in strains isolated from tomato, as AvrBs4 induces a hypersensitive response in plants with the cognate Bs4 resistance gene. While AvrBs3 does not elicit a hypersensitive response, it is likely a negative factor and has been reported only in Xanthomonas spp. isolated from pepper plants (33–35). We therefore obtained the complete TALE sequence using a Phusion PCR approach, which revealed an unusual effector gene composed of 15.5 tandem repeats. Analysis of the RVD sequence showed that while the effector was most closely related to AvrBs4 (Fig. 3A), it represented a new class of TALE in bacterial spot pathogens based on the suggested cutoff of 5.0 pairwise difference in RVD sequence (36). Therefore, we propose the name PthXp1.
Because the strains carrying this TALE also harbored the avirulence gene avrXv3 (Table 1), we cloned pthXp1 into a broad-host-range cosmid and conjugated it into an X. perforans strain (ALS7B) pathogenic to both tomato and pepper. As predicted by the RVD sequence, PthXp1 was not recognized by the tomato Bs4 or pepper Bs3 resistance gene (Fig. 3B). Pathogenicity analysis showed that expression of the effector in tomato leaves was associated with enhanced chlorosis and lesion development relative to the nearly isogenic wild-type strain (Fig. 3C), indicating that PthXp1 plays a clear role in promoting bacterial virulence. Given its similarity in RVD sequence to AvrBs4, it is tempting to speculate that PthXp1 has evolved to overcome Bs4 recognition in tomato. However, we did not find evidence of PthXp1 in contigs assembled with the plasmidSPAdes software that would suggest a common mechanism of horizontal transfer/descent. Further work is needed to elucidate the origin and host target of this novel TALE. Interestingly, a BLAST search of NCBI GenBank revealed the N- and C-terminal sequences of PthXp1 (which can be differentiated from AvrBs3 by two amino acid substitutions) in the recently published draft genomes of SC4 strains collected in Florida (data not shown). Therefore, the acquisition of PthXp1 may be a significant factor contributing to the recent emergence of SC4 and SC5 in the southeast United States.
Analysis of interlineage recombination within the X. euvesicatoria species complex provides insights into pathogen emergence and ecologically significant loci.
Investigation of X. perforans in the context of the larger X. euvesicatoria species complex revealed a dynamic pattern of gene flow between X. perforans, X. euvesicatoria, and a third lineage that was composed of related X. euvesicatoria pathovars (Fig. 4). The last lineage exhibited evidence of extensive admixture, with ∼19% of the core genome affected by recent recombination events derived from both X. perforans, X. euvesicatoria, and an unidentified donor outside the sampled Xanthomonas population (Fig. 5). As these strains were isolated from numerous hosts, including pepper, alfalfa, citrus, onion, anthurium, and commiphora (see Table S1 in the supplemental material), this could be an indication of a cryptic ecology within the X. euvesicatoria species complex. Host range studies of X. euvesicatoria and related pathogens have shown that strains often have the capacity to colonize and/or infect several plant species beyond their original host of isolation (21, 37–39), thus supporting this hypothesis. However, because the individual branches found within this apparently host generalist lineage were as divergent as those of the X. perforans and X. euvesicatoria lineages (Fig. 4A), it is possible that these contrasting evolutionary patterns may reflect adaptation to diverse hosts.
Consistent with previous observations (18), X. euvesicatoria was likely the primary donor of recent recombination to X. perforans (Fig. 4B). Each of the SCs inferred within this lineage was affected by recent recombination to varying degrees (Fig. 5), which correlated with the branch lengths observed in the core genome phylogeny (Fig. 1). Interestingly, the recent emergence of SC4, SC5, and SC6 was associated in part with the acquisition of recombinant sequences from the lineage composed of various X. euvesicatoria pathovars, which were absent from the X. perforans strains found to be predominant in Florida prior to 2015 (SC1 through SC3) (Fig. 5). Surprisingly, this lineage was suggested to be the primary donor of recent recombination to the pepper-pathogenic strains of SC6, for which only a minor proportion of the core genome (4.18%) carried signals of homologous recombination. This contrasted with the pepper-pathogenic strains of SC3, which acquired ∼8.10% of the core genome from X. euvesicatoria (Fig. 5), and suggests that the evolution of pathogenicity to pepper among strains from these two phylogenetic groups has likely occurred independently.
Examination of recombination hot spots within X. perforans revealed that several loci implicated in the plant-pathogen interaction were frequently recombining (Fig. 4C). Most of these recombination tracts contained genes associated with nutrient acquisition/metabolism (gamR, gltP, fhuE, and transketolase and glucokinase genes) and motility (motA and motB) and were often adjacent to at least one TonB-dependent receptor. This class of outer membrane receptors binds with high specificity to a variety of macromolecules, including plant-derived carbohydrates and iron, and facilitates the active transport of substrates into the periplasm via the TonB transduction system (40). It was therefore curious that the core components of this system (tonB1, exbB1, exbD1, exbD2, and exbD3) were also present in a separate recombination hot spot. One gene, exbD2, distinguished the TonB operon found in Xanthomonas spp. from those of most other bacterial genera and is essential for pathogenicity and induction of bacterial pectate lyase activity (41, 42). Interestingly, we found that, in addition to the TonB system, the majority of the Xps type II secretion cluster was recombinant among the pepper-pathogenic strains of SC3. The importance of the Xps type II gene cluster in the secretion of numerous plant cell wall-degrading enzymes and virulence has been well established in X. euvesicatoria (43–45). Therefore, it is possible that the recombination-mediated remodeling of the TonB and Xps type II systems may affect the secretion of plant cell wall-degrading enzymes and the regulation of downstream pathogenesis processes, leading to adaptation within the pepper niche.
Overall, the diversity of the X. perforans population observed in Alabama was striking in relation to that reported in neighboring tomato and pepper production regions in the United States and was indicative of adaptive evolution. Based on the evolutionary dynamics observed in this study, both X. perforans and X. euvesicatoria more closely resemble a clonal complex rather than a bona fide bacterial species. In the background of a high degree of genetic conservation, X. perforans presents an apt example of adaptive evolution mediated by homologous recombination and horizontal gene transfer originating from multiple donors (Fig. 6). The independent acquisition of novel virulence factors, such as at least two TALE genes in X. perforans strains, warrants our attention to this important family of effectors, particularly from the aspect of identifying corresponding host resistance or susceptibility genes. It was curious that the recent acquisition of a nearly identical plasmid harboring AvrHah1 in X. perforans was not inconsistent with the reemergence of X. cynarae pv. gardneri as a global pathogen of both tomato and pepper (14). This highlights the adaptive potential of a bacterial spot species complex. We also observed that the genomic flux in diverse X. perforans lineages is not limited to the tomato/pepper-pathogenic strains but can extend to other closely related Xanthomonas species/pathovars within the X. euvesicatoria species complex that can infect a wide range of hosts. This finding indicates that genetic heterogeneity in X. perforans makes it an ideal system to address the gap in our knowledge of the ecological niche of the X. euvesicatoria species complex.
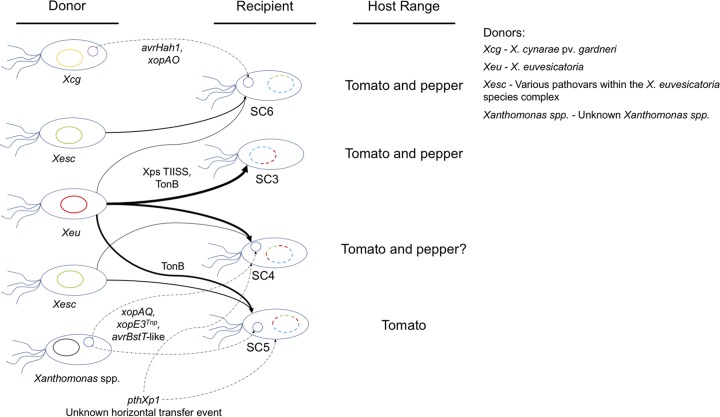
FIG 6.
Model illustrating the putative origins of recent recombination events (solid arrows) and horizontal gene transfers (dotted arrows) leading to the emergence of X. perforans SC3 to SC6, along with the corresponding host range. The thickness of the arrows showing recent recombination is proportional to the overall amount of sequence donated to each X. perforans lineage. The specific genes or functional pathways associated with recombination/horizontal-transfer events are labeled where available; Xps TIISS, Xps type II secretion system; TonB, TonB transduction system; xopE3Tnp, xopE3 disrupted by a transposon insertion; avrBsT-like, putative type 3 secreted effector gene with 71% amino acid identity to AvrBsT. The chromosomes (ovals) and plasmids (smaller circles) of donor lineages are color coded accordingly. We currently do not know if the pthXp1 gene was acquired via plasmid or Tn3-like transposase-mediated horizontal transfer or the origin of this novel transcription activation-like effector. A question mark was placed after the host range of SC4, as pathogenicity to pepper has been documented experimentally for only one strain from the group (22).
MATERIALS AND METHODS
Bacterial strain collection, sequencing, and assembly.
Eight bacterial strains, isolated from symptomatic tomato and pepper plants grown in three different Alabama counties during the summer of 2017 (Table 2), were selected at random for draft genome sequencing. Prior to DNA extraction, the bacterial strains were cultured on nutrient agar (NA) for 48 h at 28°C. Single colonies were selected from plates, cultured overnight, and then suspended in sterile deionized water. Genomic DNA was isolated using the cetyltrimethylammonium bromide (CTAB)-NaCl method, as described previously (46), and submitted to the Georgia Genomics and Bioinformatics Core, University of Georgia, for library preparation and sequencing. Paired-end reads were generated by multiplexing 12 libraries in a single lane on an Illumina MiSeq Micro (PE150), and de novo assemblies were constructed using the A5-miseq pipeline v.20160825 under the default settings (47). Briefly, adapter and quality trimming were performed with Trimmomatic (48), followed by error correction with the k-mer-based string graph assembler algorithm (49) and contig assembly using the iterative de Bruijn graph assembler (50). The genome assemblies were annotated using the NCBI Prokaryotic Genome Annotation Pipeline (51).
TABLE 2.
Collection information, assembly statistics, and pathogenicity phenotyping of different pepper varieties for the X. perforans strains sequenced in this study
| Strain | Host | County | No. of contigs | N50a (kb) | Genome size (Mb) | GenBank accession no. |
|---|---|---|---|---|---|---|
| AL1 | Tomato | Lee | 86 | 1.78 | 5.02 | SMVJ00000000 |
| AL65 | Pepper | Lee | 58 | 1.91 | 5.02 | SMVI00000000 |
| AL66 | Pepper | Lee | 52 | 2.49 | 5.02 | SMVH00000000 |
| AL57 | Tomato | Lee | 55 | 2.68 | 4.97 | SMVC00000000 |
| AL33 | Tomato | Tuscaloosa | 60 | 2.27 | 5.22 | SMVE00000000 |
| AL37 | Tomato | Tuscaloosa | 98 | 1.31 | 5.28 | SMVD00000000 |
| ALS7B | Pepper | Tuscaloosa | 65 | 1.87 | 5.06 | SMVG00000000 |
| ALS7E | Pepper | Tuscaloosa | 88 | 1.39 | 5.15 | SMVF00000000 |
N50, minimum contig length needed to cover 50% of the genome.
Reconstruction of the X. perforans core genome.
To test for a nested population structure within X. perforans, a read-mapping approach was taken using the Snippy pipeline (v.4.3.5) (52). The sequencing reads for 33 X. perforans strains described by Schwartz et al. (22) and Timilsina et al. (18) were downloaded from the Sequence Read Archive database (PRJNA526741) and subjected to adapter and quality trimming with Scythe (v.0.991) (https://github.com/vsbuffalo/scythe) and SolexaQA (v.3.1.7.1) (53), respectively. These data were combined with the quality-trimmed reads generated in this study and were individually aligned against the completed genome of X. perforans strain 91-118 using the Burrows-Wheeler alignment tool (v.0.7.17-r1188) (54). Only reads with a mapping quality of 60 (i.e., uniquely mapped reads) and a Phred quality score of 20 (at least 99% consensus) were included in the analysis, while soft-clipped reads were filtered from the data sets with SAMtools (v.1.9) (55). Variants were called from the whole-genome alignments using FreeBayes (v.1.2.0) (56), and SNPs common to all the genomes were extracted to generate a concatenated set of high-quality core genome SNPs. This concatenated SNP alignment was used to infer the population structure of X. perforans using HierBAPS software with four hierarchy levels and an upper cluster limit of 20 (24). Additionally, a maximum-likelihood phylogeny was inferred from the concatenated SNP alignment using iQTree (v.1.6.4) (57). The best-fitting model (TVM plus ASC plus G4) was chosen using Model Finder (58), and branch support was assessed with the ultrafast bootstrap method using 1,000 replicates (59). The phylogenetic tree was visualized and annotated using FigTree (v.1.4.2) (http://tree.bio.ed.ac.uk/software/figtree/ ).
Interlineage recombination.
To examine patterns of gene flow across the X. euvesicatoria species complex as described by Parkinson et al. (20), a core genome alignment was generated with the program Parsnp (60) using the genome assemblies of 39 X. perforans strains, 23 X. euvesicatoria strains, and 6 related Xanthomonas sp. strains available from GenBank (see Table S1). Highly fragmented genome assemblies (≥400 contigs) were excluded from this analysis, as they were found to significantly reduce the alignment coverage of the reference genome. The xmfa alignment file produced by Parsnp was converted to a multi-fasta format with the perl script xmfa2fasta.pl (https://github.com/kjolley/seq_scripts/blob/master/xmfa2fasta.pl) and used as input for the FastGear algorithm (3). The software was run under default settings, with the statistical significance of recombination predictions tested using a Bayes factor (BF) of >1 for recent recombination events and a BF of >10 for ancestral recombination events. The resulting output was visualized using the Phandango Web server (61), and recombination hot spots within X. perforans were visually assessed based on the frequency of recombination events at a particular site in the alignment after correcting for oversampling of clonal populations. Gene neighborhoods located in recombination hot spots were extracted for the 91-118 reference chromosome and visualized using Gene Graphics (62). Finally, the core genome alignment was used to construct a maximum-likelihood phylogeny using RAxML (v8.2.10) (63) with the gamma time-reversible model of nucleotide substitution and 1,000 bootstrap replicates.
Prediction of T3SEs and plasmids.
A database of T3SEs was compiled using the Xanthomonas Web resource (http://www.xanthomonas.org/t3e.html) and used to query the genome assemblies generated in this study using tBLASTn (E value ≤ 10−5). An effector was considered to be present if it displayed ≥80% amino acid identity over 80% of the query length. Nucleotide sequences of putative effector genes were subsequently extracted from the genomes and examined for frameshift mutations and other potential disruptions of the coding sequence with BLASTx. Plasmids were computationally predicted using plasmidSPAdes software (64). Contigs assembled by the program were subsequently screened against the NCBI nucleotide collection (nr/nt) database to assess for the presence of putative plasmid sequences. These contigs were also screened for T3SEs as described above.
Sequencing and analysis of pthXp1 from X. perforans strains AL37 and AL57.
After noting evidence for an avrBs3-like effector in the genome assemblies of strains AL37 and AL57, forward (5′-ATGAGGTGCAATCGGGTCTG-3′) and reverse (5′-GTCCTCATCTTGTTCCCGCA-3′) primers were designed to anneal to conserved loci within the N- and C-terminal domains of avrBs3. Phusion high-fidelity PCR was used to amplify the gene with a T100 thermal cycler (Bio-Rad, Hercules, CA). Each PCR mixture contained a final volume of 50 μl with 1× Phusion Master Mix containing HF buffer (Thermo Scientific, Waltham, MA), 0.5 μM each primer, and 1 μl of cells treated at 95°C for 10 min. The cycling conditions consisted of initial denaturation at 98°C for 30 s, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 63°C for 30 s, extension at 72°C for 90 s, and a final extension of 72°C for 10 min. The resulting amplicons were purified and submitted to Eurofins Genomics (Louisville, KY) for Sanger sequencing using the Power Read Service optimized for tandem-repeat stretches with the internal sequencing primers 5′-AAGATTGCAAAACGTGGCGG-3′ and 5′-CCGGATCAGGGCGAGATAAC-3′. Classification of the completed transcription activation-like effector sequences and alignment of the repeat variable diresidues were conducted using AnnoTALE (v.1.4) software utilizing the default alignment parameters (36).
Cloning of PthXp1.
PthXp1 with an endogenous promoter region of 1,000 bp was amplified as described above. The purified PCR product was cloned in the pGEMT-Easy vector (Promega) and then transferred into a broad-host-range cosmid vector, pLAFR3, using a previously described method (65). PthXp1-pLAFR3 and empty-vector constructs were conjugated into X. perforans strain ALS7B via triparental mating using the pRK2073 helper plasmid to increase transformation efficiency.
Plant inoculations.
Three- to 4-week-old pepper cultivars ECW and ECW30R with Bs3 resistance and tomato cultivar MoneyMaker with Bs4 resistance were used in all pathogenicity assays. Seedlings were grown under standard greenhouse conditions (average temperature, 28 ± 5°C, and 70% to 90% relative humidity) in individual pots with commercial potting soil amended with Osmocote (The Scotts Company, Marysville, OH). Prior to pathogenicity assays, plants were transferred to a growth chamber and maintained at 25°C with a 12-h photoperiod. The bacterial strains sequenced in this study were assessed for the capacity to induce a hypersensitive response by infiltrating individual leaves with a needleless syringe using a bacterial suspension raised to 108 CFU ml−1 (optical density at 600 nm [OD600] = 0.3) in a sterile MgSO4·7H2O solution. The presence of necrotic collapsed tissue 24 to 48 h after inoculation was scored as a positive result. The pathogenicity of strains was further assessed using bacterial suspensions prepared as described above and diluted to a concentration of 104 CFU ml−1 using 10-fold serial dilutions. A strain was considered to be pathogenic based on the development of water-soaked lesions 4 to 5 days postinoculation. A sterile MgSO4·7H2O solution was used as a negative control in the experiments described above, and each strain was tested at least twice.
Accession number(s).
The completed pthXp1 sequences were submitted NCBI GenBank with the accession numbers MK755838 and MK755839. The genome assemblies were deposited, along with the raw sequencing reads, in GenBank under BioProject accession number PRJNA526717.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the USDA National Institute of Food and Agriculture, Hatch project 1012760, and the Alabama Agricultural Experiment Station.
We thank four anonymous reviewers for their helpful suggestions and critical review of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00885-19.
REFERENCES
- 1.Datta N, Kontomichalou P. 1965. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 208:239. doi: 10.1038/208239a0. [DOI] [PubMed] [Google Scholar]
- 2.Wu X, Monchy S, Taghavi S, Zhu W, Ramos J, van der Lelie D. 2011. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol Rev 35:299–323. doi: 10.1111/j.1574-6976.2010.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mostowy R, Croucher NJ, Andam CP, Corander J, Hanage WP, Marttinen P. 2017. Efficient inference of recent and ancestral recombination within bacterial populations. Mol Biol Evol 34:1167–1182. doi: 10.1093/molbev/msx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiedenbeck J, Cohan FM. 2011. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol Rev 35:957–976. doi: 10.1111/j.1574-6976.2011.00292.x. [DOI] [PubMed] [Google Scholar]
- 5.Dillon MM, Thakur S, Almeida RND, Wang PW, Weir BS, Guttman DS. 2019. Recombination of ecologically and evolutionarily significant loci maintains genetic cohesion in the Pseudomonas syringae species complex. Genome Biol 20:3. doi: 10.1186/s13059-018-1606-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacques M-A, Arlat M, Boulanger A, Boureau T, Carrère S, Cesbron S, Chen NWG, Cociancich S, Darrasse A, Denancé N, Fischer-Le Saux M, Gagnevin L, Koebnik R, Lauber E, Noël LD, Pieretti I, Portier P, Pruvost O, Rieux A, Robène I, Royer M, Szurek B, Verdier V, Vernière C. 2016. Using ecology, physiology, and genomics to understand host specificity in Xanthomonas. Annu Rev Phytopathol 54:163–187. doi: 10.1146/annurev-phyto-080615-100147. [DOI] [PubMed] [Google Scholar]
- 7.Mhedbi-Hajri N, Hajri A, Boureau T, Darrasse A, Durand K, Brin C, Saux M-L, Manceau C, Poussier S, Pruvost O, Lemaire C, Jacques M-A. 2013. Evolutionary history of the plant pathogenic bacterium Xanthomonas axonopodis. PLoS One 8:e58474. doi: 10.1371/journal.pone.0058474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamza AA, Robene-Soustrade I, Jouen E, Lefeuvre P, Chiroleu F, Fisher-Le Saux M, Gagnevin L, Pruvost O. 2012. Multilocus sequence analysis- and amplified fragment length polymorphism-based characterization of xanthomonads associated with bacterial spot of tomato and pepper and their relatedness to Xanthomonas species. Syst Appl Microbiol 35:183–190. doi: 10.1016/j.syapm.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Huang C-L, Pu P-H, Huang H-J, Sung H-M, Liaw H-J, Chen Y-M, Chen C-M, Huang M-B, Osada N, Gojobori T, Pai T-W, Chen Y-T, Hwang C-C, Chiang T-Y. 2015. Ecological genomics in Xanthomonas: the nature of genetic adaptation with homologous recombination and host shifts. BMC Genomics 16:188. doi: 10.1186/s12864-015-1369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones JB, Lacy GH, Bouzar H, Stall RE, Schaad NW. 2004. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst Appl Microbiol 27:755–762. doi: 10.1078/0723202042369884. [DOI] [PubMed] [Google Scholar]
- 11.Timilsina S, Kara S, Jacques MA, Potnis N, Minsavage GV, Vallad GE, Jones JB, Fischer-Le Saux M. 2019. Reclassification of Xanthomonas gardneri (ex Šutič 1957) Jones et al. 2006 as a later heterotypic synonym of Xanthomonas cynarae Trébaol et al. 2000 and description of X. cynarae pv. cynarae and X. cynarae pv. gardneri based on whole genome analyses. Int J Syst Evol Microbiol 69:343–349. doi: 10.1099/ijsem.0.003104. [DOI] [PubMed] [Google Scholar]
- 12.Potnis N, Krasileva K, Chow V, Almeida NF, Patil PB, Ryan RP, Sharlach M, Behlau F, Dow JM, Momol M, White FF, Preston JF, Vinatzer BA, Koebnik R, Setubal JC, Norman DJ, Staskawicz BJ, Jones JB. 2011. Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genomics 12:146. doi: 10.1186/1471-2164-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Potnis N, Timilsina S, Strayer A, Shantharaj D, Barak JD, Paret ML, Vallad GE, Jones JB. 2015. Bacterial spot of tomato and pepper: diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol Plant Pathol 16:907–920. doi: 10.1111/mpp.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Timilsina S, Jibrin MO, Potnis N, Minsavage GV, Kebede M, Schwartz A, Bart R, Staskawicz B, Boyer C, Vallad GE, Pruvost O, Jones JB, Goss EM. 2015. Multilocus sequence analysis of xanthomonads causing bacterial spot of tomato and pepper plants reveals strains generated by recombination among species and recent global spread of Xanthomonas gardneri. Appl Environ Microbiol 81:1520–1529. doi: 10.1128/AEM.03000-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timilsina S, Abrahamian P, Potnis N, Minsavage GV, White FF, Staskawicz BJ, Jones JB, Vallad GE, Goss EM. 2016. Analysis of sequenced genomes of Xanthomonas perforans identifies candidate targets for resistance breeding in tomato. Phytopathology 106:1097–1104. doi: 10.1094/PHYTO-03-16-0119-FI. [DOI] [PubMed] [Google Scholar]
- 16.Barak JD, Vancheva T, Lefeuvre P, Jones JB, Timilsina S, Minsavage GV, Vallad GE, Koebnik R. 2016. Whole-genome sequences of Xanthomonas euvesicatoria strains clarify taxonomy and reveal a stepwise erosion of type 3 effectors. Front Plant Sci 7:1805. doi: 10.3389/fpls.2016.01805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Constantin EC, Cleenwerck I, Maes M, Baeyen S, Malderghem CV, Vos PD, Cottyn B. 2016. Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathol 65:792–806. doi: 10.1111/ppa.12461. [DOI] [Google Scholar]
- 18.Timilsina S, Pereira-Martin JA, Minsavage GV, Iruegas Bocardo F, Abrahamian P, Potnis N, Kolaczkowski B, Vallad GE, Goss EM, Jones J. 2019. Multiple recombination events drive the current genetic structure of Xanthomonas perforans in Florida. Front Microbiol 10:448. doi: 10.3389/fmicb.2019.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jibrin MO, Potnis N, Timilsina S, Minsavage GV, Vallad GE, Roberts PD, Jones JB, Goss EM. 2018. Genomic inference of recombination-mediated evolution in Xanthomonas euvesicatoria and X. perforans. Appl Environ Microbiol 84:e00136-18. doi: 10.1128/AEM.00136-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkinson N, Cowie C, Heeney J, Stead D. 2009. Phylogenetic structure of Xanthomonas determined by comparison of gyrB sequences. Int J Syst Evol Microbiol 59:264–274. doi: 10.1099/ijs.0.65825-0. [DOI] [PubMed] [Google Scholar]
- 21.Bansal K, Kumar S, Patil PB. 2018. Complete genome sequence reveals evolutionary dynamics of an emerging and variant pathovar of Xanthomonas euvesicatoria. Genome Biol Evol 10:3104–3109. doi: 10.1093/gbe/evy238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz AR, Potnis N, Timilsina S, Wilson M, Patané J, Martins JJ, Minsavage GV, Dahlbeck D, Akhunova A, Almeida N, Vallad GE, Barak JD, White FF, Miller SA, Ritchie D, Goss E, Bart RS, Setubal JC, Jones JB, Staskawicz BJ. 2015. Phylogenomics of Xanthomonas field strains infecting pepper and tomato reveals diversity in effector repertoires and identifies determinants of host specificity. Front Microbiol 6:535. doi: 10.3389/fmicb.2015.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minsavage GV, Dahlbeck D, Whalen MC, Kearney B, Bonas U, Staskawicz BJ, Stall RE. 1990. Gene-for-gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria-pepper interactions. Mol Plant Microbe Interact 3:41–47. doi: 10.1094/MPMI-3-041. [DOI] [Google Scholar]
- 24.Cheng L, Connor TR, Sirén J, Aanensen DM, Corander J. 2013. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol 30:1224–1228. doi: 10.1093/molbev/mst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schornack S, Minsavage GV, Stall RE, Jones JB, Lahaye T. 2008. Characterization of AvrHah1, a novel AvrBs3-like effector from Xanthomonas gardneri with virulence and avirulence activity. New Phytol 179:546–556. doi: 10.1111/j.1469-8137.2008.02487.x. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz AR, Morbitzer R, Lahaye T, Staskawicz BJ. 2017. TALE-induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proc Natl Acad Sci 114:E897–E903. doi: 10.1073/pnas.1620407114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horvath DM, Stall RE, Jones JB, Pauly MH, Vallad GE, Dahlbeck D, Staskawicz BJ, Scott JW. 2012. Transgenic resistance confers effective field level control of bacterial spot disease in tomato. PLoS One 7:e42036. doi: 10.1371/journal.pone.0042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abbasi PA, Khabbaz SE, Weselowski B, Zhang L. 2015. Occurrence of copper-resistant strains and a shift in Xanthomonas spp. causing tomato bacterial spot in Ontario. Can J Microbiol 61:753–761. doi: 10.1139/cjm-2015-0228. [DOI] [PubMed] [Google Scholar]
- 29.Araújo ER, Costa JR, Ferreira M, Quezado‐Duval AM. 2017. Widespread distribution of Xanthomonas perforans and limited presence of X. gardneri in Brazil. Plant Pathol 66:159–168. doi: 10.1111/ppa.12543. [DOI] [Google Scholar]
- 30.Kebede M, Timilsina S, Ayalew A, Admassu B, Potnis N, Minsavage GV, Goss EM, Hong JC, Strayer A, Paret M, Jones JB, Vallad GE. 2014. Molecular characterization of Xanthomonas strains responsible for bacterial spot of tomato in Ethiopia. Eur J Plant Pathol 140:677–688. doi: 10.1007/s10658-014-0497-3. [DOI] [Google Scholar]
- 31.Burlakoti RR, Hsu C, Chen J, Wang J. 2018. Population dynamics of xanthomonads associated with bacterial spot of tomato and pepper during 27 years across Taiwan. Plant Dis 102:1348–1356. doi: 10.1094/PDIS-04-17-0465-RE. [DOI] [PubMed] [Google Scholar]
- 32.Abrahamian P, Timilsina S, Minsavage GV, Kc S, Goss EM, Jones JB, Vallad GE. 2018. The type III effector AvrBsT enhances Xanthomonas perforans fitness in field-grown Tomato. Phytopathology 108:1355–1362. doi: 10.1094/PHYTO-02-18-0052-R. [DOI] [PubMed] [Google Scholar]
- 33.Schornack S, Ballvora A, Gürlebeck D, Peart J, Baulcombe D, Ganal M, Baker B, Bonas U, Lahaye T. 2004. The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J 37:46–60. doi: 10.1046/j.1365-313X.2003.01937.x. [DOI] [PubMed] [Google Scholar]
- 34.Schornack S, Peter K, Bonas U, Lahaye T. 2005. Expression levels of avrBs3-like genes affect recognition specificity in tomato Bs4- but not in pepper Bs3-mediated perception. Mol Plant Microbe Interact 18:1215–1225. doi: 10.1094/MPMI-18-1215. [DOI] [PubMed] [Google Scholar]
- 35.Boch J, Bonas U. 2010. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol 48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 36.Grau J, Reschke M, Erkes A, Streubel J, Morgan RD, Wilson GG, Koebnik R, Boch J. 2016. AnnoTALE: bioinformatics tools for identification, annotation, and nomenclature of TALEs from Xanthomonas genomic sequences. Sci Rep 6:21077. doi: 10.1038/srep21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang C-H, Vallad GE, Adkison H, Summers C, Margenthaler E, Schneider C, Hong J, Jones JB, Ong K, Norman DJ. 2013. A novel Xanthomonas sp. causes bacterial spot of rose (Rosa spp.). Plant Dis 97:1301–1307. doi: 10.1094/PDIS-09-12-0851-RE. [DOI] [PubMed] [Google Scholar]
- 38.Osdaghi E, Taghavi SM, Hamzehzarghani H, Lamichhane JR. 2016. Occurrence and characterization of the bacterial spot pathogen Xanthomonas euvesicatoria on pepper in Iran. J Phytopathol 164:722–734. doi: 10.1111/jph.12493. [DOI] [Google Scholar]
- 39.Yaripour Z, Mohsen Taghavi S, Osdaghi E, Lamichhane JR. 2018. Host range and phylogenetic analysis of Xanthomonas alfalfae causing bacterial leaf spot of alfalfa in Iran. Eur J Plant Pathol 150:267–274. doi: 10.1007/s10658-017-1271-0. [DOI] [Google Scholar]
- 40.Schauer K, Rodionov DA, de Reuse H. 2008. New substrates for TonB-dependent transport: do we only see the “tip of the iceberg”? Trends Biochem Sci 33:330–338. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 41.Wiggerich H-G, Pühler A. 2000. The exbD2 gene as well as the iron-uptake genes tonB, exbB and exbD1 of Xanthomonas campestris pv. campestris are essential for the induction of a hypersensitive response on pepper (Capsicum annuum). Microbiology 146:1053–1060. doi: 10.1099/00221287-146-5-1053. [DOI] [PubMed] [Google Scholar]
- 42.Vorhölter F-J, Wiggerich H-G, Scheidle H, Sidhu VK, Mrozek K, Küster H, Pühler A, Niehaus K. 2012. Involvement of bacterial TonB-dependent signaling in the generation of an oligogalacturonide damage-associated molecular pattern from plant cell walls exposed to Xanthomonas campestris pv. campestris pectate lyases. BMC Microbiol 12:239. doi: 10.1186/1471-2180-12-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baptista JC, Machado MA, Homem RA, Torres PS, Vojnov AA, do Amaral AM. 2010. Mutation in the xpsD gene of Xanthomonas axonopodis pv. citri affects cellulose degradation and virulence. Genet Mol Biol 33:146–153. doi: 10.1590/S1415-47572009005000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solé M, Scheibner F, Hoffmeister A-K, Hartmann N, Hause G, Rother A, Jordan M, Lautier M, Arlat M, Büttner D. 2015. Xanthomonas campestris pv. vesicatoria secretes proteases and xylanases via the Xps type II secretion system and outer membrane vesicles. J Bacteriol 197:2879–2893. doi: 10.1128/JB.00322-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szczesny R, Jordan M, Schramm C, Schulz S, Cogez V, Bonas U, Büttner D. 2010. Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv vesicatoria. New Phytol 187:983–1002. doi: 10.1111/j.1469-8137.2010.03312.x. [DOI] [PubMed] [Google Scholar]
- 46.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1994. Current protocols in molecular biology. John Wiley and Sons, New York, NY. [Google Scholar]
- 47.Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 48.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simpson JT, Durbin R. 2012. Efficient de novo assembly of large genomes using compressed data structures. Genome Res 22:549–556. doi: 10.1101/gr.126953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng Y, Leung HCM, Yiu SM, Chin FYL. 2010. IDBA—a practical iterative de Bruijn graph de novo assembler, p 426–440. In Berger B. (ed), Research in computational molecular biology. Springer, Berlin, Germany. [Google Scholar]
- 51.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seemann T. 2015. Snippy: fast bacterial variant calling from NGS reads. https://github.com/tseemann/snippy.
- 53.Cox MP, Peterson DA, Biggs PJ. 2010. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv 1207.3907v2 [q-bio.GN] https://arxiv.org/abs/1207.3907.
- 57.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, Harris SR. 2018. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34:292–293. doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harrison KJ, de Crécy-Lagard V, Zallot R. 2018. Gene Graphics: a genomic neighborhood data visualization Web application. Bioinformatics 34:1406–1408. doi: 10.1093/bioinformatics/btx793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. 2016. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32:3380–3387. doi: 10.1093/bioinformatics/btw493. [DOI] [PubMed] [Google Scholar]
- 65.Sambrook J, Fritsch E, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.