The catabolism of complex carbohydrates that otherwise escape the endogenous digestive enzymes of humans and other animals drives the composition and function of the gut microbiota. Thus, detailed molecular characterization of dietary glycan utilization systems is essential both to understand the ecology of these complex communities and to manipulate their compositions, e.g., to benefit human health. Our research reveals new insight into how ubiquitous members of the human gut microbiota have evolved a set of microheterogeneous gene clusters to efficiently respond to the structural variations of plant xyloglucans. The data here will enable refined functional prediction of xyloglucan utilization among diverse environmental taxa in animal guts and beyond.
KEYWORDS: human gut microbiota, microbiome, Bacteroides, carbohydrate-active enzymes, carbohydrate-binding proteins, Bacteroidetes, CAZymes, carbohydrate-binding modules, environmental bacteria
ABSTRACT
Genome sequencing has revealed substantial variation in the predicted abilities of individual species within animal gut microbiota to metabolize the complex carbohydrates comprising dietary fiber. At the same time, a currently limited body of functional studies precludes a richer understanding of how dietary glycan structures affect the gut microbiota composition and community dynamics. Here, using biochemical and biophysical techniques, we identified and characterized differences among recombinant proteins from syntenic xyloglucan utilization loci (XyGUL) of three Bacteroides and one Dysgonomonas species from the human gut, which drive substrate specificity and access to distinct polysaccharide side chains. Enzymology of four syntenic glycoside hydrolase family 5 subfamily 4 (GH5_4) endo-xyloglucanases revealed surprising differences in xyloglucan (XyG) backbone cleavage specificity, including the ability of some homologs to hydrolyze congested branched positions. Further, differences in the complement of GH43 alpha-l-arabinofuranosidases and GH95 alpha-l-fucosidases among syntenic XyGUL confer distinct abilities to fully saccharify plant species-specific arabinogalactoxyloglucan and/or fucogalactoxyloglucan. Finally, characterization of highly sequence-divergent cell surface glycan-binding proteins (SGBPs) across syntenic XyGUL revealed a novel group of XyG oligosaccharide-specific SGBPs encoded within select Bacteroides.
IMPORTANCE The catabolism of complex carbohydrates that otherwise escape the endogenous digestive enzymes of humans and other animals drives the composition and function of the gut microbiota. Thus, detailed molecular characterization of dietary glycan utilization systems is essential both to understand the ecology of these complex communities and to manipulate their compositions, e.g., to benefit human health. Our research reveals new insight into how ubiquitous members of the human gut microbiota have evolved a set of microheterogeneous gene clusters to efficiently respond to the structural variations of plant xyloglucans. The data here will enable refined functional prediction of xyloglucan utilization among diverse environmental taxa in animal guts and beyond.
INTRODUCTION
Humans have coevolved with a complex assemblage of microbes (1), among which the human gut microbiota (HGM) is a key driver of our metabolism and systemic health (2). The populous HGM of the lower gastrointestinal (GI) tract is fueled primarily by complex carbohydrates that are otherwise not accessed by our own limited cohort of digestive enzymes (3–5). Specifically, the metabolism of plant glycans that comprise “dietary fiber” significantly contributes to energy uptake (6) and strongly influences HGM composition (4, 7–10). Deleterious shifts in the composition of the HGM have been associated with serious metabolic, inflammatory, neurological, and oncological diseases (11). Hence, there is significant, sustained interest in the maintenance of an individual’s “healthy microbiota” (12) using prebiotic, probiotic (including HGM transplants), and synbiotic approaches (13).
Revealing the molecular basis of diet-HGM interactions is essential to understand the ecological and evolutionary dynamics that govern this complex ecosystem (14). Within the HGM, the evolution of numerous complex polysaccharide utilization loci (PUL) has positioned members of the phylum Bacteroidetes (e.g., Bacteroides and Prevotella) as prodigious colonizers of the lower GI tract (15). PUL encode coregulated cohorts of glycan-binding proteins, carbohydrate-active enzymes (CAZymes), transporters, and sensors/regulators that work in concert to saccharify individual cognate glycans (16–20). As HGM studies transition into a postgenomic era (15, 17, 21, 22), a growing body of integrated studies has revealed the combined genetic, biochemical, and structural basis of complex glycan utilization by human gut Bacteroides species (23–35). Indeed, these studies, which are underpinned by precise biochemical characterization, demonstrate that the enzyme cohort of PUL-encoded systems is exquisitely tuned to address source-specific polysaccharide substructures (9, 19, 36).
The xyloglucans (XyGs) constitute a family of complex plant polysaccharides found in the cell walls of all terrestrial plants, including vegetables and fruits in the human diet (37). XyGs are united by a β(1→4)-glucan backbone that is ramified by regular α(1→6)-xylopyranosyl residues in two general patterns, XXXG type and XXGG type {where G refers to an unbranched β(1→4)-Glcp residue and X refers to a branched [α(1→6)-Xylp]-β(1→4)-Glcp disaccharide unit} (37). Further substitution of the xylosyl branches is species, tissue, and/or developmental stage specific. XXXG-type xyloglucans are typically found in dicots (e.g., most vegetables and fruits) as fucogalactoxyloglucan (Fig. 1A), while XXGG-type XyGs are found in the Poales (e.g., cereals) as galactoxyloglucan and in Solanales (e.g., tomato and potato) as arabinogalactoxyloglucan (Fig. 1B) (37).
FIG 1.
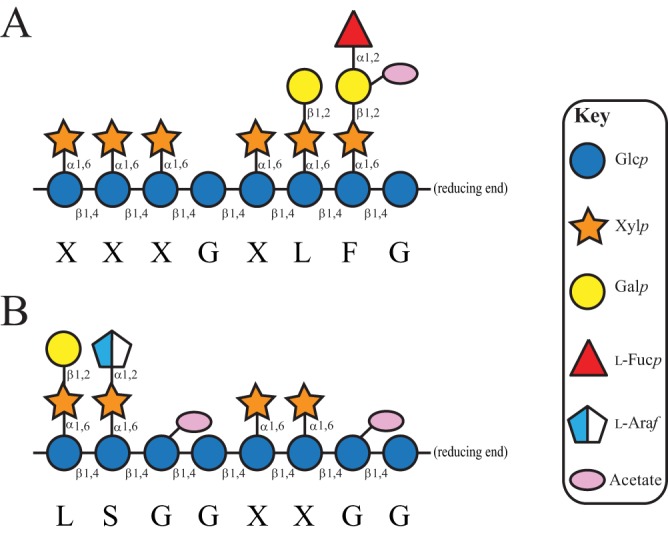
Representative xyloglucan structures (37). (A) XXXG-type fucogalactoxyloglucan, typical of dicots. (B) XXGG-type arabinogalactoxyloglucan, typical of Poales and Solanales. For both, two exemplar repeating units are shown. Monosaccharides are represented by Consortium for Functional Glycomics notation (https://www.ncbi.nlm.nih.gov/glycans/snfg.html).
Using reverse genetics, biochemistry, and structural biology, we recently characterized a xyloglucan utilization locus (XyGUL) in the human gut symbiont Bacteroides ovatus, syntenic loci of which serve as molecular markers of xyloglucan metabolism in the HGM (24, 25, 38). By analogy with the archetypal starch utilization system (Sus) (39), the B. ovatus XyGUL encodes all necessary XyG-binding, -hydrolyzing, and -sensing proteins, including a canonical SusC/SusD homolog pair (a TonB-dependent transporter [TBDT] and associated cell surface glycan-binding protein [SGBP-A]) (40) (Fig. 2). An additional “SusE-positioned” (39) SGBP-B assists with substrate capture at the outer membrane (38), while the glycoside hydrolase (GH) complement comprises all cell surface endo-hydrolase and periplasmic exo-hydrolase activities required for the complete saccharification of solanaceous arabinogalactoxyloglucan (24, 25) (Fig. 2A; cf. Fig. 1B). Specifically, the B. ovatus XyGUL encodes two glycoside hydrolase family 43 (GH43) α-l-arabinofuranosidases but notably lacks a fucosidase (e.g., a GH29 or GH95 member [41, 42]) required to hydrolyze terminal side chain residues in dicot XyG (Fig. 2A; cf. Fig. 1A).
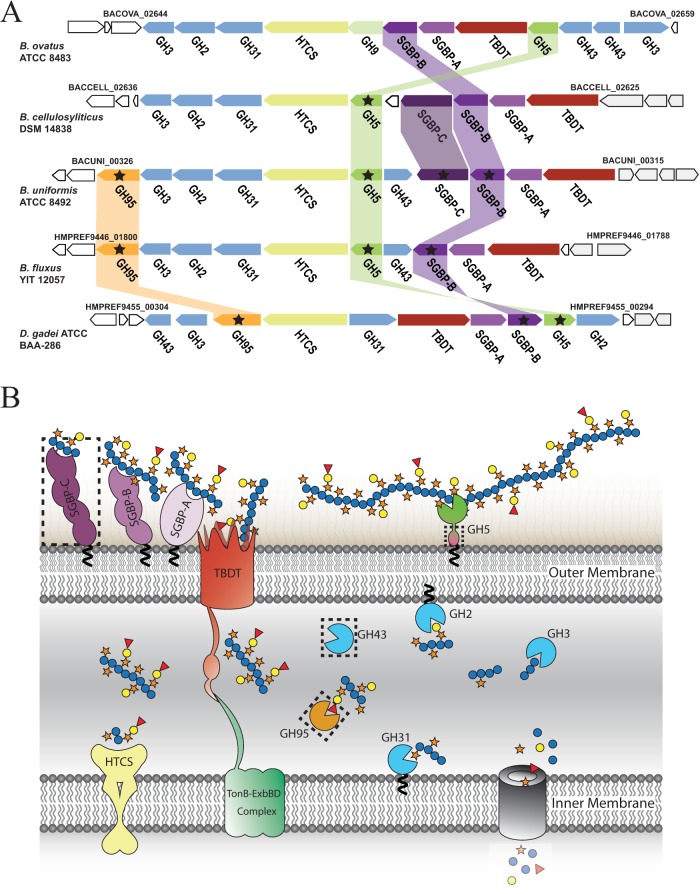
FIG 2.
XyGUL in Bacteroidetes. (A) Individual genes are represented as arrows indicating orientation, with genomic locus tags given for bounding genes. The encoded protein family is indicated below each gene: GHn, glycoside hydrolase family n member; SGBP, cell surface glycan-binding protein (SGBP-As are SusD homologs); HTCS, hybrid two-component system; TBDT, TonB-dependent transporter (SusC homologs). Homologous gene products studied in this work are connected by colored bars and are marked with stars. For clarity, homology between other genes/gene products (24) is not indicated. (B) Model of XyG saccharification in the cell envelope of Bacteroidetes. Gene products are colored as in panel A. SGBPs initiate polysaccharide recognition at the cell surface (38), where the GH5 (subfamily 4) endo-xyloglucanase cleaves the substrate (24) for active transport into the periplasm via the TBDT/SGBP-A complex (40). In the periplasm, exo-acting GHs work in a concerted fashion to liberate monosaccharides (24, 25) for cytosolic import and metabolism. The dashed rectangles indicate nonconserved protein products of the XyGUL. The model shows dicot fucogalactoxyloglucan utilization, which does not contain α-l-arabinofuranosyl residues that are the substrates of the GH43 members (24).
We observed that XyGUL synteny is not strictly conserved among closely related Bacteroides species or the more distant Bacteroidetes member Dysgonomonas gadei (24) (Fig. 2A). Regarding the B. ovatus XyGUL, it is particularly notable that some species lack one or both GH43 members (predicted α-l-arabinofuranosidases), while some possess a GH95 member (predicted α-l-fucopyranosidase) and/or an additional “SusF-positioned” gene (39) (predicted here to encode an additional SGBP). This may suggest ongoing microevolution of XyGUL in autochthonous Bacteroidales (24, 43), possibly balancing functional redundancy and nutrient niche specialization (44).
Here, we explored the possibility that the assortment of GHs and SGBPs might be responsible for xyloglucan subtype specificity among four members of the phylum Bacteroidetes, using biochemical and biophysical approaches. We first defined general XyG specificity through analysis of the vanguard GH5 subfamily 4 (GH5_4) endo-xyloglucanases, which surprisingly revealed distinct backbone cleavage specificities. Where present, GH95 members provided complementary α-l-fucosidase activity to address dicot fucogalactoxyloglucan. Lastly, characterization of SusE- and SusF-positioned SGBP gene products revealed conservation of XyG-binding function among these proteins despite very high sequence diversity. Together, these data on a diverse cohort of GHs and SGBPs provide refined insight into molecular markers of XyG metabolism in the HGM.
RESULTS
The XyGUL is not completely syntenic among Bacteroidetes.
We showed previously that B. cellulosilyticus DSM 14838, B. uniformis ATCC 8492, B. fluxus YIT 12057, and D. gadei ATCC BAA-286 all used XyG as a sole carbon source and that each of the homologous XyGUL was induced under these conditions (24). All of the XyGUL contained GH5_4 members (predicted endo-xyloglucanases) and GH31 members (predicted α-xylosidases), concordant with XyG specificity (24, 45–47). Likewise, the presence of GH2 (predicted β-galactosidase [24]) and GH3 (predicted β-glucosidase [24]) members was ubiquitously conserved. However, comparison with the well-characterized B. ovatus XyGUL (24) revealed notable differences in overall locus arrangement and encoded GH and SGBP content (Fig. 2).
For example, the GH5_4 members present different modular architectures comprising either one or two PFAM PF13004 domains following a signal peptidase II lipidation motif and have poor sequence conservation within their catalytic modules (see Fig. S1 and Table S1 in the supplemental material). Poor sequence conservation was also observed for the TBDTs and SGBP-As, with the SGBP-Bs exhibiting particularly low identity and similarity values (<19% and 32%, respectively) (see Tables S1 to S4 in the supplemental material).
Most notably, the remaining complements of predicted exo-glycosidases were significantly different among XyGUL. As introduced above, an α-l-fucosidase would be required to remove the terminal side chain of dicot XyG, whereas an α-l-arabinofuranosidase is required to cleave the terminal side chain of solanaceous XyG (Fig. 1). The B. ovatus XyGUL lacks a predicted α-l-fucosidase, as does the B. cellulosilyticus XyGUL, while the B. uniformis, B. fluxus, and D. gadei XyGUL each possess a GH95 member (42). On the other hand, only the B. cellulosilyticus XyGUL lacks both a predicted α-l-fucosidase (GH95 member) and an α-l-arabinofuranosidase (GH43 member); the B. cellulosilyticus and B. uniformis XyGUL are otherwise syntenic. Of particular note, the B. cellulosilyticus and B. uniformis XyGUL both contain an additional gene encoding a protein of previously unknown function (24) (annotated here as SGBP-C) (see below).
XyGUL GH5_4 endo-xyloglucanases generate distinct product profiles.
The presence of conserved signal peptidase II extracellular secretion signal peptides (48) and PFAM PF13004 domains is consistent with cell surface outer membrane localization of all GH5_4 homologs through N-terminal lipidation (24). As anticipated from their subfamily membership (46), initial screening of the recombinant, full-length GH5_4 members indicated high specificity for tamarind seed XyG (tamXyG) (a galactoxyloglucan), with no detectable activity on barley mixed-linkage β-glucan or konjac glucomannan (GM). pH rate profiles on tamXyG indicated optima of pH 6.0 to 6.5, which is consistent with activity in the human lower GI tract (see Fig. S2 in the supplemental material). Subsequent Michaelis-Menten kinetic analysis at the pH optima and 37°C underscored the high XyG specificity of the four enzymes, with low Km and high kcat values (Table 1; see Fig. S3 in the supplemental material).
TABLE 1.
Michaelis-Menten kinetic constants of BoGH5A, BuGH5, BcGH5, BfGH5, and DgGH5 against tamXyG
| Bacterial strain | Enzyme | Substrate | kcat (s−1) | Km (mg · ml−1) | kcat/Km (s−1 · mg−1 · ml) | Assay |
|---|---|---|---|---|---|---|
| B. ovatus ATCC 8483 | BoGH5 | tamXyG | 8.9 ± 0.2a | 0.046 ± 0.006 | 190 | BCA |
| B. cellulosilyticus DSM 14838 | BcGH5 | tamXyG | 31.0 ± 1.0 | 0.036 ± 0.003 | 862 | BCA |
| B. uniformis ATCC 8492 | BuGH5 | tamXyG | 19.6 ± 0.8 | 0.043 ± 0.007 | 456 | BCA |
| B. fluxus YIT 12057 | BfGH5 | tamXyG | 50.1 ± 3.0 | 0.17 ± 0.02 | 295 | BCA |
| D. gadei ATCC DAA-286 | DgGH5 | tamXyG | 19.0 ± 1.0 | 0.039 ± 0.009 | 487 | BCA |
All error values correspond to errors in fitting the Michaelis-Menten equation to the data.
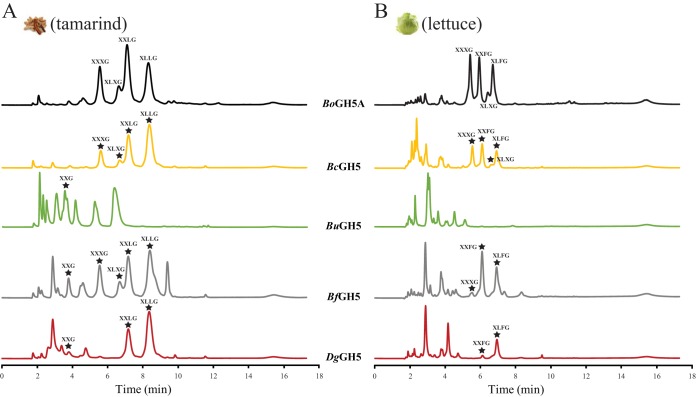
Despite having apparently similar kinetic properties, each of the four GH5_4 members generated distinct limit digestion (terminal hydrolysis) products on tamXyG and lettuce XyG (lettXyG) (a fucogalactoxyloglucan) versus the previously characterized B. ovatus GH5 (BoGH5) endo-xyloglucanase. Consistent with our earlier study (24), BoGH5 cleaved both types of XyG at unbranched glucosyl residues to generate typical Glc4-based xyloglucan oligosaccharides (XyGOs) (49) (XXXG, XLXG, XXLG, and XLLG from tamXyG and XXXG, XLXG, XXFG, and XLFG from lettXyG) (Fig. 3, Fig. S4; see Fig. 1 for XyGO nomenclature). B. cellulosilyticus GH5 (BcGH5) similarly followed this typical cleavage pattern, especially on tamXyG, although a very limited ability to generate shorter XyGOs from lettXyG was suggested by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis (Fig. 3; see Fig. S5 in the supplemental material).
FIG 3.
HPAEC-PAD analysis of XyG hydrolysis products of GH5 endo-xyloglucanases. Two micromoles of the recombinant form of the GH5 enzymes investigated in this study was incubated with tamXyG (A) or lettXyG (B) at 37°C for 22 h. Peaks corresponding to known XyGOs are indicated (compare with Fig. 1).
In contrast, B. uniformis GH5 (BuGH5), B. fluxus GH5 (BfGH5), and D. gadei GH5 (DgGH5) all convincingly demonstrated the ability to generate a range of shorter and longer XyGOs by cleavage both at unbranched G units and at branched positions in tamXyG and lettXyG (especially X) (Fig. 3; see Fig. S6 to S8 in the supplemental material). For example, MALDI-TOF MS analysis of the BfGH5 limit-digest of lettXyG revealed the presence of XyGOs with masses corresponding to XX, LG/XGG, XXX, XXXG, XLXG/XXLG, XXFG, XLFG, GXLFG, and XFGXX (see Fig. S7). BuGH5 notably produced the unique pentasaccharide FG and other atypical fucosylated XyGOs in high abundance (see Fig. S6). Apparently rapid cleavage of XXXG resulted in the total absence of this product in the limit-digests of DgGH5 (Fig. 3; compare Fig. S8 to Fig. S4).
Corroborating these observations on XyG polysaccharides, BcGH5, BuGH5, BfGH5, and DgGH5 were each able to hydrolyze the heptasaccharide XXXG by cleavage between two X units (see Fig. S9 in the supplemental material). BuGH5 and DfGH5 were especially efficient and were able to completely hydrolyze the starting material at equal enzyme loads. Both of the enzymes were additionally able to hydrolyze the galactosylated congener XXLG, with BuGH5 especially competent (see Fig. S9). The ability to bind an X unit in the −1 subsite, as a prerequisite to cleaving an X-X backbone bond in XyG, is known in some GH44 and GH74 endo-(xylo)glucanases (reviewed in reference 46). However, among GH5 members, this previously had been demonstrated only in a single endo-xyloglucanase, XEG5B, from a rumen metagenome (50). Tertiary-structure analysis indicated that XEG5B has a large −1′ pocket to accommodate a xylosyl residue, which is absent in the G-unit-specific rumen endo-xyloglucanase XEG5A (50). The presence or absence of this pocket is not readily defined by amino acid sequence alignment, which unfortunately precludes correlation with BcGH5, BuGH5, BfGH5, and DgGH5 here, in the absence of three-dimensional structures of these enzymes.
GH95 members are effective XyG fucosidases.
BuGH95, BfGH95, and DgGH95, which do not have homologs in the B. ovatus XyGUL, were produced recombinantly in Escherichia coli to assess the abilities of these putative α-fucosidases to remove the terminal residue from the F units of dicot XyGOs. BuGH95 and BfGH95 share high amino acid sequence identity (81%), while DgGH95 is more divergent (45% identity versus either Bacteroides GH95 member). The GH95 catalytic carboxylic residues are conserved within the sequences of these targets, despite an overall low degree of sequence similarity with known structural representatives (26, 51) (see Table S5 in the supplemental material). As GH95 is known to contain α-l-galactosidases and α-l-fucosidases with diverse linkage specificities (42), detailed enzymatic characterization was warranted.
Initial tests indicated that BuGH95, BfGH95, and DgGH95 were active on 2-chloro-4-nitrophenyl α-l-fucoside (Fuc-α-CNP). Using this substrate, classical bell-shaped pH rate profiles were obtained with pH optima of ca. 6.0 (see Fig. S10 in the supplemental material). The observed kcat/Km values for the three GH95 members (Table 2) are similar to that of the GH95 α-fucosidase from Bifidobacterium longum subsp. infantis, encoded by locus Blon_2335 (0.12 mM−1 s−1) (52). However, compared to CjAfc95 from the soil saprophyte Cellvibrio japonicus, kcat/Km values for the Bacteroidetes GH95 members were approximately 10- to 25-fold lower (53). When tested on readily available commercial substrates, all three Bacteroidetes GH95 members were able to hydrolyze 2′-fucosyllactose and 3′-fucosyllactose, while only DgGH95 showed limited activity on fucosyl-α(1→6)-N-acetylglucosamine (see Fig. S11 in the supplemental material).
TABLE 2.
Michaelis-Menten kinetic constants of BuGH95, BfGH95, and DgGH95 against 2-chloro-4-nitrophenyl α-l-fucopyranoside
| Bacterial strain | Enzyme | Substrate | kcat (s−1) | Km (mg · ml−1) | kcat/Km (s−1 · mg−1 · ml) | Assay |
|---|---|---|---|---|---|---|
| B. uniformis ATCC 8492 | BuGH95 | l-Fuc-α-CNP | 2.0 ± 0.1a | 1.7 ± 0.1 | 1.2 | CNP |
| B. fluxus YIT 12057 | BfGH95 | l-Fuc-α-CNP | 1.9 ± 0.1 | 3.8 ± 0.3 | 0.49 | CNP |
| D. gadei ATCC DAA-286 | DgGH95 | l-Fuc-α-CNP | 3.4 ± 0.2 | 4.7 ± 0.4 | 0.73 | CNP |
All error values correspond to errors in fitting the Michaelis-Menten equation to the data.
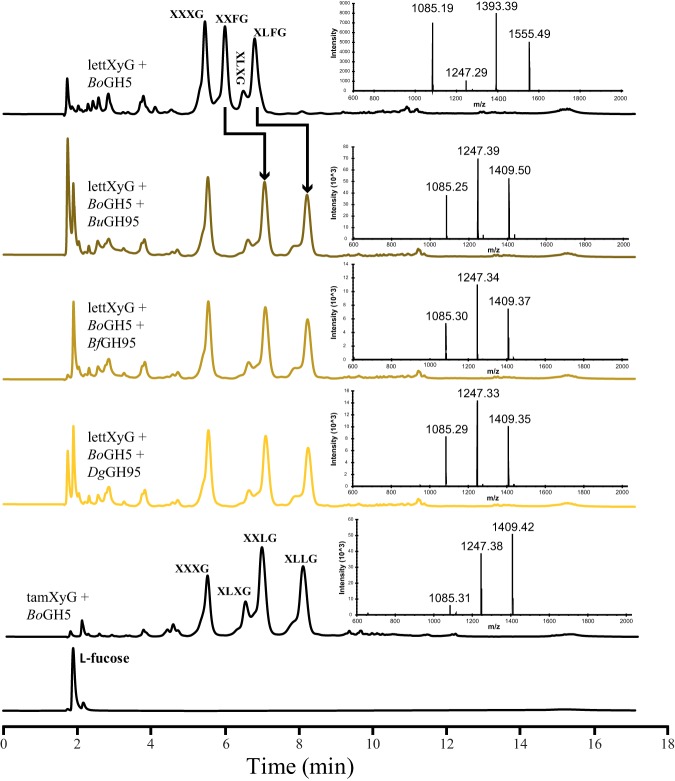
Consistent with their ability to hydrolyze the Fucα(1→2)-Gal linkage of 2′-fucosyllactose, the fucosylated XyGOs produced from lettXyG by BoGH5 were competent substrates for all three Bacteroidetes GH95 members. Thus, XXFG and XLFG in the mixture comprising XXXG, XXFG, XLXG, and XLFG are hydrolyzed to XXLG and XLLG by BuGH95, BfGH95, and DgGH95 (Fig. 4). Similarly, we examined the actions of these GH95 members on the fucosylated XyGOs produced by their cognate GH5s, which again indicated that fucose was completely released (see Fig. S5 to S8 and S12 in the supplemental material). This activity is concordant with that of characterized GH95 α-l-fucosidases from the bacterium C. japonicus (53), the fungus Aspergillus niger (54), and the plants Arabidopsis thaliana and Lilium longiflorum (55, 56), as well as the GH29 α-l-fucosidase FgFCO1 from the plant-pathogenic fungus Fusarium graminearum (57, 58). Site-directed mutation of the predicted general acid residues (Glu) to Ala in both BuGH95 and BfGH95 abolished activity (see Fig. S13 in the supplemental material), consistent with their essential roles in catalysis (51).
FIG 4.
HPAEC-PAD and MALDI-TOF MS analyses of GH95 hydrolysis products from fucosylated XyGOs. A mixture of fucosylated XyGOs (XXXG, XXFG, XLXG, and XLFG) was produced by the action of BoGH5A (24) on lettXyG, followed by incubation with 2 μM recombinant BuGH95, BfGH95, or DgGH95 at 37°C for 16 h in 50 mM sodium citrate buffer, pH 6.0. In each case, XLFG and XXFG were converted into XLLG and XXLG, as confirmed by comparison with the XyGOs produced by BoGH5 on tamXyG. The observed molecular masses of the peaks in MALDI-TOF MS correspond to the [M + Na]+ ions of XXXG (calculated: 1,085.9), XLXG/XXLG (calculated: 1,248.05), XXFG (calculated: 1,394.21), XLLG (calculated: 1,410.19), and XLFG (calculated: 1,556.35).
Characterization of diverse in-tandem gene products reveals new SGBPs.
Binding of bacteria in the HGM to plant cell wall material constitutes the first step in complex carbohydrate utilization (59). We previously characterized the XyG specificities and tertiary structures of the SGBP-A and SGBP-B proteins of the B. ovatus XyGUL (24, 38). BoSGBP-A displays a canonical SusD-like protein fold with an extended substrate binding platform of aromatic amino acid side chains (38). Given the predictable structural conservation (39) among syntenic SGBP-As in the homologous XyGUL (Fig. 2; see Table S3), we focused our attention here on characterizing the substrate specificities of the highly sequence-divergent SusE-positioned SGBP-Bs (Fig. 2; see Table S4).
All syntenic XyGUL SGBP-Bs contain a predictive N-terminal signal peptidase II sequence for lipidation and outer membrane targeting (48), consistent with a role in substrate adherence. Affinity PAGE analysis of full-length recombinant BuSGBP-B, BfSGBP-B, and DgSGBP-B revealed that they all bind to tamXyG and have no affinity for mixed-linkage β(1-3)/β(1-4)-glucan (MLG) and GM (Fig. 5) or the artificial derivative hydroxyethylcellulose (data not shown). BcSGBP-B has high sequence similarity (70%) and identity (56%) to BuSGBP-B (see Table S4) and thus was not produced and characterized.
FIG 5.
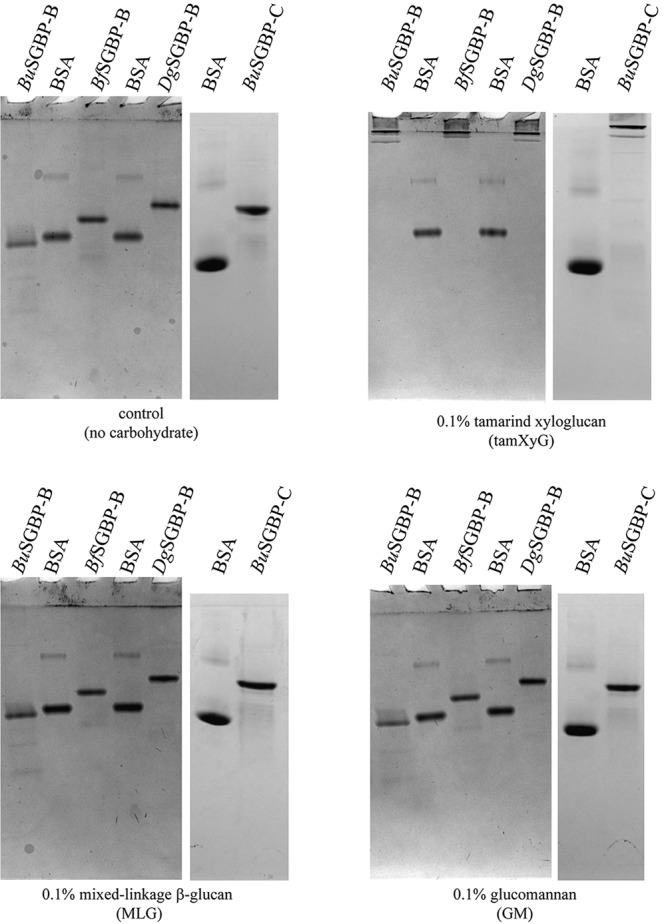
Affinity PAGE analysis of SGBP-B and SGBP-C proteins. BSA was used as a nonbinding control protein.
Isothermal titration calorimetry (ITC) confirmed that the three SGBP-Bs bind to tamXyG, with affinities up to 2 orders of magnitude higher than for the syntenic BoSGBP-B (38). Each SGBP-B bound variably galactosylated tamXyGOs comprising Glc8 or Glc4 backbones, with association constant (Ka) values 1 and 2 orders of magnitude lower than for tamXyG, respectively (tamXyGO2 and tamXyGO1) (Table 3; see Fig. S14 and S15 in the supplemental material). The presence of fucosylation in a lettXyGO1 mixture had either limited impact or slightly reduced binding (Table 3; see Fig. S16 in the supplemental material). We were unable to test binding on lettXyG polysaccharide due to inherent difficulties in producing a sufficiently pure sample from the natural source. Nonetheless, these data suggest that BuSGBP-B, BfSGBP-B, and DgSGBP-B all present an extended polysaccharide-binding platform and that binding is primarily driven by backbone Glc and side chain Xyl units, similar to BoSGBP-B, for which the three-dimensional structure is known (38).
TABLE 3.
Thermodynamic parameters for BuSGBP-B, BfSGBP-B, DgSGBP-B, and BuSGBP-C obtained by isothermal titration calorimetry at 25°C in 50 mM sodium phosphate buffer (pH 7.0)
| Carbohydrate |
Ka (M−1) |
ΔG (kcal · mol−1) |
ΔH (kcal · mol−1) |
TΔS (kcal · mol−1) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BuSGBP-B | BfSGBP-B | DgSGBP-B | BuSGBP-C | BuSGBP-B | BfSGBP-B | DgSGBP-B | BuSGBP-C | BuSGBP-B | BfSGBP-B | DgSGBP-B | BuSGBP-C | BuSGBP-B | BfSGBP-B | DgSGBP-B | BuSGBP-C | |
| XyGa | 3.0 (±0.5) × 106 | 3.7 (±0.5) × 105 | 1.3 (±0.2) × 106 | Weakb | −8.3 | −7.6 | −8.3 | Weakb | −31.2 ± 0.5 | −33.8 ± 0.8 | −44.4 ± 0.7 | Weakb | −22.9 | −26.2 | −36.1 | Weak |
| tamXyGO2 | 1.6 (±0.1) × 105 | 2.1 (±0.1) × 105 | 1.5 (±0.2) × 105 | 6.8 (±0.1) × 104 | −7 | −7.2 | −7.2 | −6.6 | −31.1 ± 0.7 | −25.3 ± 0.2 | −35.7 ± 0.2 | −21.8 ± 0.2 | −24.1 | −18.1 | −28.5 | −15.2 |
| tamXyGO1 | 6.4 (±1.0) × 104 | 6.0 (±0.1) × 103 | 1.8 (±0.1) × 104 | 8.6 (±0.6) × 103 | −7 | −5.1 | −5.8 | −5.3 | −27.7 ± 2.9 | −35.8 ± 0.5 | −29.2 ± 0.3 | −26.5 ± 3.8 | −20.7 | −30.7 | −23.4 | −21.2 |
| lettXGO1 cognate GH5 | 0.67 (±0.04) × 103 | 2.6 (±0.2) × 103 | 1.0 (±0.1) × 103 | Weakb | −3.9 | −4.6 | −4.2 | Weakb | −70.4 ± 3.9 | −26.7 ± 1.2 | −118 | Weakb | −66.5 | −22.1 | −113.8 | Weak |
Binding thermodynamics for XyG were based on the concentration of the binding unit, tamXyGO2. All error values correspond to errors from model fitting to ITC data.
Weak binding represents a Ka of 500 M−1.
In addition to SGBP-Bs, the B. uniformis and B. cellulosilyticus XyGUL notably also contain larger, divergent downstream genes (39) whose functions are not predicable based on homology (Fig. 2). The proteins encoded by these genes are significantly different in size than the correspondingly positioned GH9 endo-(xylo)glucanase of the BoXyGUL (24, 60), and reducing-sugar assays of recombinant BuSGBP-C failed to reveal any polysaccharide hydrolase activity (data not shown). Rather, affinity PAGE revealed significant binding of BuSGBP-C to tamXyG, but not to mixed-linkage glucan or glucomannan (Fig. 5). BuSGBP-C and BcSGBP-C share 65% amino acid sequence identity, suggesting conserved function, as for the other syntenic gene products of the B. uniformis and B. cellulosilyticus XyGUL.
Despite apparently specific binding observed by affinity PAGE, ITC indicated that BuSGBP-C in fact had a low affinity for tamXyG vis-à-vis the SGBP-Bs; indeed, binding was too low to be quantified (Table 3; see Fig. S14). On the other hand, quantitation of binding to tamXyGO2 and tamXyGO1 indicated that BuSGBP-C bound to these substrates with Ka values similar to those of the three SGBP-Bs (Table 3; see Fig. S14 and S15). Binding to fucosylated lettXyGO1 was again too low to be quantified (Table 3; see Fig. S16). Taken together, the affinity PAGE and ITC data indicate that BuSGBP-C constitutes a new XyG-binding protein that is, moreover, unique in the broader context of XyGUL and PUL in general. However, in the absence of a tertiary structure of this SGBP, the reasons for the specificity for XyGOs over the polysaccharide remain opaque.
DISCUSSION
Postgenomic functional characterization of the proteins and enzymes in the HGM is essential to define and refine models of complex carbohydrate utilization by the HGM (23–35). Such models both advance understanding of HGM metabolism in the context of human health and bring insight into the molecular mechanisms that connect diet to microbiome evolution. In the present study, we have explored the molecular diversity of syntenic XyGUL from the HGM (Fig. 2), which has revealed (i) novel differences in specificity among homologous GH5_4 endo-xyloglucanases, (ii) complementary α-fucosidase activity among select XyGUL to address dicot fucogalactoxyloglucan, and (iii) convergent XyG specificity among sequence-diverse SGBPs, including (iv) a new nonsyntenic group of proteins (XyGUL SGBP-C).
Surprisingly, the five GH5 subfamily 4 endo-xyloglucanases from the XyGUL generate different products from tamXyG and lettXyG, due to differences in backbone cleavage regiospecificities (Fig. 3; see Fig. S4 to S9). Across all GH families, most endo-xyloglucanases specifically cleave dicot XyG at unbranched glucosyl units (positioned in the −1 subsite) (46), which produces the typical “XXXG-type” XyGOs (49). BoGH5A exemplifies this mode of action (Fig. 3). In contrast, alternative cleavage patterns have been reported for some endo-(xylo)glucanases from GH44 (61, 62) and GH74 (63–66), indicating the ability of these enzymes to accommodate xylosyl-substituted residues (X) in the −1 subsite. Within GH5, this ability has been reported for only a single GH5_4 endo-xyloglucanase, XEG5B from a ruminant gut metagenome (50). Thus, BuGH5, BfGH5, and DgGH5 comprise additional examples of atypical backbone regiospecificity in GH5_4, which could not have been predicted by protein sequence analysis. Notably, the limited ability of these GH5_4 members to hydrolyze XXLG, with the exception of BuGH5, indicates that the extension of the side chain in subsite +2 with a galactosyl residue inhibits cleavage efficiency (see Fig. S9). Future tertiary-structural analysis will be required to fully illuminate the molecular basis of XyG recognition and hydrolysis by these GH5 enzymes.
These observations are reminiscent of analyses on endo-xylanases from the ruminant Bacteroidetes Prevotella bryantii, in which two homologous GH5 proteins with similar domain organization released different oligosaccharides from xylan (67). Current data indicate that the majority of characterized endo-xyloglucanases hydrolyze the polysaccharide backbone in a regular manner at G units (46), perhaps because of steric or ring-conformational factors (68). Regardless, such specificity is unlikely to be the result of any biological selection pressure, because it is assumed that large saccharide chain fragments are imported from the cell surface to the periplasm for subsequent saccharification (16, 39). In such cases, the point of chain cleavage should not matter.
Comparative analysis of periplasmic exo-glycosidases further revealed how syntenic XyGUL vary in their biochemical capacity to address the distinct side chains observed in XyGs from disparate dietary sources. Whereas GH43 α-l-ararabinofuranosidases, as found, for example, in the BoXyGUL, are required to completely saccharify solanaceous arabinogalactoxyloglucan, GH95 α-l-fucosidases are necessary to remove the terminal side chain residue of F units in dicot fucogalactoxyloglucan (Fig. 1). Of the XyGUL studied here, only those from B. uniformis, B. fluxus, and D. gadei encode this capacity. Indeed, the GH95 members all actively release fucose from fucosylated XyGOs produced by the action of their cognate endo-xyloglucanase. Even those Bacteroidetes that lack XyGUL-encoded α-l-fucosidases may still access a significant proportion of the monosaccharides in fucogalactoxyloglucan through the concerted action of α-xylosidases, β-galactosidases, and β-glucosidases acting from the nonreducing end of XyGOs (24) (Fig. 1). Further, the extent to which other fucosidases encoded elsewhere in individual Bacteroidetes genomes might complement deficiencies in XyGUL GH cohorts is currently unknown.
In the broader context of GH95, the α-l-fucosidase specificity observed here is concordant with recent structure-function studies. Of the nine bacterial and eukaryotic GH95 members characterized to date (41), the majority (seven) have been described as α-l-fucosidases (52–56, 69, 70), while two Bacteroides members have been shown to be specific α-l-galactosidases (26, 33) (l-fucose is the 6-deoxy congener of α-l-galactose). Tertiary-structural analysis of the B. ovatus α-l-galactosidase vis-à-vis an exemplar GH95 α-l-fucosidase from Bifidobacterium bifidum (51, 69) indicated that these related specificities likely result from a single amino acid substitution in the active-site pocket: The α-l-galactosidase presents a threonine side chain that makes polar interactions with O-6 of l-Gal, while the α-l-fucosidase has a bulkier histidine side chain that is sterically compatible with the deoxysugar (26). Sequence alignment indicates that BuGH95, BfGH95, and DgGH95 each contain a histidine residue at this position (see Fig. S17), consistent with the α-l-fucosidase activity observed here.
Analogous to the archetypal Sus system (39, 59), substrate binding is mediated by the noncatalytic SGBPs of the XyGUL. Interestingly, pairwise sequence similarity among the SGBP-Bs is significantly lower (ca. 30%) than among the SGBP-As (ca. 40%), GHs (51 to 56%), and TBDTs (ca. 55%) of homologous XyGUL (see Tables S1 to S4). The evolutionary reason for this enhanced sequence diversity is unclear, but it prevents straightforward construction of a sequence-related carbohydrate-binding module family (71). Nonetheless, all the syntenic SGBP-B gene products had homologous functions in the context of the XyGUL, viz., high binding specificity for the cognate polysaccharide.
Most striking in our SGBP analysis was the revelation of a third, sequence-diverse group of SGBPs encoded downstream from the SGBP-Bs in the B. uniformis and B. cellulosilyticus XyGUL. The exemplar BuSGBP-C failed to bind the XyG polysaccharide with any appreciable affinity yet bound XyGOs, as well as the various SGBP-Bs. The biological significance of this distinct specificity is currently unclear in the absence of detailed reverse-genetics studies and tertiary-structural analysis (29, 38, 59, 60). It is notable that the SGBP-C proteins are significantly larger (BuSGBP-C, 95 kDa; BcSGBP-C, 93 kDa) than the adjacent SGBP-B proteins (BuSGBP-B, 71 kDa; BcSGBP-B, 74 kDa), which suggests the presence of additional, or unique, domain structures (for comparison, BoSGBP-B is 53 kDa, BfSGBP-B is 66 kDa, and DgSGBP-B is 66 kDa). Variable, multidomain, extended architectures are typical of SGBP-Bs. For example, B. thetaiotaomicron SusE and SusF have three and four domains, respectively (59), the B. ovatus XyGUL and MLGUL SGBP-Bs both have four domains (29, 38), and the SGBP-B from the B. thetaiotaomicron heparin/heparin-sulfate PUL has six domains (30).
Conclusions.
Recent studies have demonstrated the importance of structural differences among plant xylans in differentially impacting the growth of closely related human gut Bacteroides species (26, 72, 73). Such observations align with the concept of “discrete (carbohydrate) structures” that correspond directly to individual gene products in microbiomes (9, 19, 36). Here, we have illustrated this concept as it pertains to discrete XyG side chain structures and their concordance with the gene complements of individual XyGUL addressing plant arabinogalactoxyloglucan and fucogalactoxyloglucan.
Moreover, precise growth analyses are currently revealing that individual species of the HGM differentially prioritize the utilization of specific complex carbohydrates from mixtures (74–78). Despite their ubiquity in the human diet from vegetables and fruits, xyloglucans appear to be secondary in this utilization hierarchy. Nonetheless, it has recently been shown that fucogalactoxyloglucan elicits larger shifts than the higher-priority pectins or fructooligossaccharides (FOS) in human fecal samples in vitro (79). The observation of a strong, specific enrichment of B. uniformis in this analysis is particularly striking in light of the XyGUL described here. As such, our present study provides further insight into how individual Bacteroidetes members “navigate the gut buffet” through refined specificity of PUL-encoded systems (20).
MATERIALS AND METHODS
Carbohydrate sources.
Tamarind seed XyG, barley mixed-linkage beta-glucan, and konjac glucomannan were purchased from Megazyme International (Bray, Ireland). Hydroxyethyl cellulose was purchased from Amresco (Solon, OH, USA), and 2-fucosyllactose, 3-fucosyllactose, 2-acetamido-2-deoxy-6-O-(α-l-fucopyranosyl)-d-glucopyranose, and 2-chloro-4-nitrophenyl α-l-fucopyranoside were purchased from Carbosynth (Compton, United Kingdom).
Fucosylated XyGOs were isolated from lettuce using a modified and scaled-up version of a previously described method (80). Thus, 190 g of plant material (iceberg lettuce) was homogenized in 0.5 liter of 70% aqueous ethanol using a blender and filtered onto Miracloth (Millipore). The residue was then transferred to a ceramic mortar and pestle and ground to a fine powder using liquid nitrogen. The powder was incubated in 1.5 liters of 70% aqueous ethanol at 60°C for 6 h to remove alcohol-soluble polysaccharides and smaller sugars. The residue was subsequently washed and filtered two times with 1 liter of 70% aqueous ethanol and 1 liter of acetone, respectively. Alcohol-insoluble residues were extracted for 16 h at 37°C with 6 M NaOH (100 ml) containing 1% NaBH4 to prevent alkaline peeling. The suspension was filtered onto Miracloth, cooled on ice for 20 min, and neutralized by adding acetic acid. Polysaccharides were extracted by precipitation by adding ethanol to the solution to a final concentration of 70%. The solution was then centrifuged at 20,000 × g for 15 min, and the pellet was washed two times with 70% ethanol. The washed pellet was dissolved in water, to which was added 2 mg endo-xyloglucanase GH5 (37°C overnight; produced in house [this study and reference 24]). After lyophilization, the XGOs were dissolved in water.
Cloning, expression, and purification of recombinant proteins.
B. uniformis ATCC 8492 and B. cellulosilyticus DSM 14838 were grown in tryptone-yeast extract-glucose (TYG) medium at 37°C under anaerobic conditions. Total genomic DNA (gDNA) was extracted using the Genomic-tip 20/G (Qiagen), following the manufacturer’s instructions. The gDNAs from B. fluxus YIT 12057 and D. gadei ATCC BAA-286 were a kind gift from Eric Martens (University of Michigan, Ann Arbor, MI, USA).
The open reading frames corresponding to BACUNI_00317, BACUNI_00318, BACUNI_00321, and BACUNI_00326 (encoding BuSGBP-B, BuSGBP-C, BuGH5, and BuGH95); HMPREF9446_01791, HMPREF9446_01793, and HMPREF9446_01800 (encoding BfSGBP-B, BfGH5, and BfGH95); HMPREF9455_00295, HMPREF9455_00296, and HMPREF9455_00301 (encoding DgGH5, DgSGBP-B, and DgGH95); and BACCELL_02631 (encoding BcGH5) were PCR amplified from genomic DNA using Q5 high-fidelity polymerase (NEB) with appropriate primers designed to remove signal peptides and lipidation cysteines (81, 82) (Table 4). PCR products of BACUNI_00318, HMPREF9455_00301, and BACCELL_02631 containing appropriate pMCSG complementary sequences were generated for subsequent ligation-independent cloning into the pMCSG53 plasmid, providing an N-terminal His6 tag (83). The PCR products of BACUNI_00317, HMPREF9446_01791, and HMPREF9455_00296 were ligated into a modified version of pET28a (EMD Biosciences) containing a recombinant tobacco etch virus (rTEV) protease recognition site (pET28a-TEV), as previously described (38). The rest of the PCR products introduced NdeI/XhoI or NheI/XhoI (NEB) restriction sites into the flanks of the target genes, and the amplified DNA was cloned into expression vector pET28a so that the encoded recombinant proteins contained an N-terminal His6 tag. Successful cloning was confirmed by colony PCR (GoTaq polymerase; Promega) and sequencing (Genewiz). BuGH95 E531A and BfGH95 E543A were generated using pET28a::BuGH95 and pET28a::BfGH95 as template DNAs (Table 5).
TABLE 4.
Cloning primers and E. coli expression vectors used in this study
| Gene | Vector | Forward primer (5′ → 3′)a | Reverse primer (5′ → 3′)a |
|---|---|---|---|
| BACOVA_02653 | pET21a | CTTTTATATCATGTAGCCATATGGATGAAAAAGGAGG | GGAGCTCTCGAGATTTAATCCTTTCATAATCGC |
| BACCELL_02631 | pMCSG53 | TACTTCCAATCCAATGCCATGGATCCTGTAACATCGGAAATAA | TTATCCACTTCCAATGTTATTATTGCGGTGCATTGTCATAGACG |
| BACUNI_00321 | pET28a | GGAGCTCATATGTGTTCCGGCAATGATGATAC | GGAGCTCTCGAGTCAATAAGGGTATTTGCCGGC |
| HMPREF9446_01793 | pET28a | GGAGCTCATATGTCGGATGACAATTCCGGTAC | GGAGCTCTCGAGTCAGTAAGGATACTTTCCGTCTC |
| HMPREF9455_00295 | pET28a | GGAGCTCATATGGGCGACGATGATGATCCGATA | GGAGCTCTCGAGTTAATTTCCTGCATCTATAATAGCACTGATAG |
| BACUNI_00326 | pET28a | GGAGCTCATATGAATCATCAGCTATACTATACATCG | GGAGCTCTCGAGCTATAAGAAATGTATGGTCACCGCCTC |
| HMPREF9446_01800 | pET28a | GGAGCTCATATGAACCACCGGCTTTATTATACCA | GGAGCTCTCGAGTCAAAAAGAAAAGAAAGTGA |
| HMPREF9455_00301 | pMCSG53 | TACTTCCAATCCAATGCCATGAAATGGCAATATTATTTCAACGA | TTATCCACTTCCAATGTTATTATAGAAATCTTGATATTCTGATT |
| BACUNI_00317 | pET28a-TEV | AAAAACATATGGGGGCAAAAACCTATGTTTCCG | AAAAACTCGAGTTATTCAATGGTCATCGAAATTATCTC |
| HMPREF9446_01791 | pET28a-TEV | AAAAACATATGGACAAGGGGCTGACCACTGATGAC | AAAAACTCGAGTTACAATACCTTCAGACTATCTATCG |
| HMPREF9455_00296 | pET28a-TEV | AAAAAGCTAGCGACAAATACGAGATGGGTGCAAGC | AAAAACTCGAGTTATTTTAGAATATAATGATAAATAG |
| BACUNI_00318 | pMCSG53 | TACTTCCAATCCAATGCCATGCACACCACCCCGATTCTGAACGAATC | TTATCCACTTCCAATGTTATTACCATTCAGGAACAGAGAAAAGAG |
Restriction sites are underlined, and pMCSG LIC vector complementary sequences are in boldface.
TABLE 5.
Primers used for site-directed mutagenesis
| Gene | Residue change | Forward primer (5′ → 3′)a | Reverse primer (5′ → 3′)a |
|---|---|---|---|
| BACUNI_00326 | E531A | ACTGCCCCCACTTCTTCTCCGGCGAACAGCTTCTATGTGCCGGGT | ACCCGGCACATAGAAGCTGTTCGCCGGAGAAGAAGTGGGGGCAGT |
| HMPREF9446_01800 | E543A | ACCGCTCCCACTTCTTCACCGGCTAACAGTTTCTATGTGCCGGGC | GCCCGGCACATAGAAACTGTTAGCCGGTGAAGAAGTGGGAGCGGT |
Changed bases are underlined.
Recombinant proteins were produced in E. coli BL21(DE3) cells cultured in Terrific broth containing ampicillin (50 μg · ml−1) or kanamycin (50 μg · ml−1) at 37°C (200 rpm). The cells were grown to mid-exponential phase (optical density at 600 nm [OD600] ≈ 0.4 to 0.6). Overexpression was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM, and the cultures were grown further at 16°C (200 rpm) for 18 h. The cells were harvested by centrifugation, and sonicated, and His6-tagged recombinant proteins were purified using a HisTrap IMAC FF nickel-nitrilotriacetic acid column (GE Healthcare) with a gradient elution up to 100% elution buffer containing 20 mM sodium phosphate, pH 7.4, 500 mM NaCl, and 500 mM imidazole in a BioLogic fast protein liquid chromatography (FPLC) system (Bio-Rad). The purity of the recombinant proteins was determined by SDS-PAGE, and their concentrations were determined from their calculated molar extinction coefficients at 280 nm using an Epoch microplate spectrophotometer (BioTek).
Carbohydrate analytical methods.
High-pH anion-exchange chromatography–pulsed amperometric detection (HPAEC-PAD) carbohydrate analysis was performed as previously described (63). Solvent A was ultrapure water, solvent B was 1 M sodium hydroxide, and solvent C was 1 M sodium acetate prepared from anhydrous BioUltra-grade solid (Sigma). Depending on the sample, the following gradients were used for gradient A (0 to 5 min, 10% B and 3.5% C; 5 to 12 min, 10% B, linear gradient from 3.5 to 30% C; 12.0 to 12.1 min, 50% B, 50% C; 12.1 to 13.0 min, exponential gradient [curve setting 9] of B and C back to initial conditions; 13 to 17 min, initial conditions) and for gradient B (0 to 5 min, 10% B; 5 to 10 min, 10% B, linear gradient from 0 to 30% C; 10.1 to 11 min, 50% B, 50% C; 11 to 11.9 min, linear gradient of B and C back to initial conditions; 11.9 to 17 min, initial conditions).
MALDI-TOF MS analysis was performed on oligosaccharide samples as described previously (63). A standard mix of XXXG, XXLG/XLXG, and XLLG was used for an external calibration.
Enzyme kinetic analysis.
Polysaccharide hydrolysis was quantified using a bicinchoninic acid (BCA) reducing-sugar assay (84). The reactions were conducted in a final volume of 100 μl at the optimum pH at 37°C for 10 min. Reactions were terminated by the addition of an equal volume (100 μl) of BCA reagent. Color was developed by heating to 80°C for 20 min before reading the absorbance at 563 nm. A glucose series (25 to 150 μM) was performed with each assay to quantify the reducing ends released.
The pH optimum of each enzyme used in this work was initially determined using the same enzyme reaction assay to quantify reducing ends over 10 min of incubation with 1.0 mg/ml XyG in different buffers at 50 mM: sodium citrate (pH 3.0 to 6.5), sodium phosphate (pH 6.5 to 8.0), and glycine (pH 9.0 to 10.5).
To determine Michaelis-Menten parameters, nine different concentrations of XyG solutions were used over the range 0.025 to 2 mg · ml−1 with the appropriate concentration of enzyme for 10 min, and the reducing ends released were quantified as described above.
The release of 2-chloro-4-nitrophenolate from 2-chloro-4-nitrophenyl α-l-fucopyranoside was monitored continuously by following the A410 with a 1-cm-path-length quartz cuvette using a Cary 60 UV-visible (Vis) spectrophotometer (Agilent Technologies). Reactions, carried out in 250-μl volumes in the optimum pH buffer at 37°C, were assayed with nine different substrate concentrations, and the rate was calculated using an extinction coefficient determined according to the buffer used.
Enzyme product analysis.
To determine limit-digestion products, 2 μM of the recombinant enzyme was incubated with 0.25 mg · ml−1 polysaccharide or 0.5 mM oligosaccharide for 18 h (37°C) in a 500-μl reaction mixture containing 50 mM citrate buffer (pH 6.0). The reaction mixture was then diluted two times for the polysaccharide reactions and five times for the oligosaccharide reactions prior to product analysis by HPAEC-PAD.
Affinity gel electrophoresis.
Qualitative analysis of binding to tamarind xyloglucan, barley β-glucan, konjac glucomannan, and artificial hydroxyethyl cellulose was performed exactly as described previously (38). SGBPs (5 μg) were loaded, with bovine serum albumin (BSA) used as a noninteracting negative control.
Isothermal titration calorimetry.
ITC of glycan binding by the SGBPs was performed using a MicroCal VP-ITC titration calorimeter equilibrated to 25°C. The proteins (20 to 40 μM) were placed in the sample cell, and the syringe was loaded with 2.5 mg/ml of XyG polysaccharide or 1 mM oligosaccharide (XyGO2, tamXyGO1, and lettXyGO1). Following an initial injection of 2 μl, 25 subsequent injections of 10 μl were performed with stirring at 280 rpm, and the resulting heat of reaction was recorded. Integrated heats were fitted to a single-site model using MicroCal Origin v7.0 to derive n, Ka, and ΔH values.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eric Martens (University of Michigan) for providing Bacteroidetes strains and gDNA. We thank Nicole Koropatkin (University of Michigan) for providing the pET28a-TEV plasmid. We thank Charles Haynes (Michael Smith Laboratories, University of British Columbia) for access to ITC equipment.
Work at the University of British Columbia was generously supported by operating grants from the Canadian Institutes for Health Research (MOP-137134 and MOP-142472) and infrastructure support from the Canadian Foundation for Innovation (project no. 30663) and the British Columbia Knowledge Development Fund.
G.D. cloned, produced, and performed final biochemical analyses of GH and SGBP targets and cowrote the article. A.S.T. performed initial cloning, recombinant production, and preliminary characterization of GH and SGBP targets, with the assistance of S.W.B. A.L.C. assisted with ITC data analysis. H.B. devised the overall study, supervised research, and cowrote the article with input from all of us. We all read and approved the final manuscript.
We declare that we have no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01491-19.
REFERENCES
- 1.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 3.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. 2013. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 4.Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porter NT, Martens EC. 2017. The critical roles of polysaccharides in gut microbial ecology and physiology. Annu Rev Microbiol 71:349–369. doi: 10.1146/annurev-micro-102215-095316. [DOI] [PubMed] [Google Scholar]
- 6.McNeil NI. 1984. The contribution of the large-intestine to energy supplies in man. Am J Clin Nutr 39:338–342. doi: 10.1093/ajcn/39.2.338. [DOI] [PubMed] [Google Scholar]
- 7.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujimura KE, Slusher NA, Cabana MD, Lynch SV. 2010. Role of the gut microbiota in defining human health. Expert Rev Anti Infect Ther 8:435–454. doi: 10.1586/eri.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamaker BR, Tuncil YE. 2014. A perspective on the complexity of dietary fiber structures and their potential effect on the gut microbiota. J Mol Biol 426:3838–3850. doi: 10.1016/j.jmb.2014.07.028. [DOI] [PubMed] [Google Scholar]
- 10.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lynch SV, Pedersen O. 2016. The human intestinal microbiome in health and disease. N Engl J Med 375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 12.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS, Giglio MG, Hallsworth-Pepin K, Lobos EA, Madupu R, Magrini V, Martin JC, Mitreva M, Muzny DM, Sodergren EJ, Versalovic J, Wollam AM, Worley KC, Wortman JR, Young SK, Zeng QD, Aagaard KM, Abolude OO, Allen-Vercoe E, Alm EJ, Alvarado L, Andersen GL, Anderson S, Appelbaum E, Arachchi HM, Armitage G, Arze CA, Ayvaz T, Baker CC, Begg L, Belachew T, Bhonagiri V, Bihan M, Blaser MJ, Bloom T, Bonazzi V, Brooks JP, Buck GA, Buhay CJ, Busam DA, Campbell JL. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elzinga J, van der Oost J, de Vos WM, Smidt H. 2019. The use of defined microbial communities to model host-microbe interactions in the human gut. Microbiol Mol Biol Rev 83:e00054-18. doi: 10.1128/MMBR.00054-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terrapon N, Lombard V, Drula E, Lapebie P, Al-Masaudi S, Gilbert HJ, Henrissat B. 2018. PULDB: the expanded database of polysaccharide utilization loci. Nucleic Acids Res 46:D677–D683. doi: 10.1093/nar/gkx1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grondin JM, Tamura K, Dejean G, Abbott DW, Brumer H. 2017. Polysaccharide utilization loci: fueling microbial communities. J Bacteriol 199:e00860-16. doi: 10.1128/JB.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. 2011. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol 9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hemsworth GR, Dejean G, Davies GJ, Brumer H. 2016. Learning from microbial strategies for polysaccharide degradation. Biochem Soc Trans 44:94–108. doi: 10.1042/BST20150180. [DOI] [PubMed] [Google Scholar]
- 19.Martens EC, Kelly AG, Tauzin AS, Brumer H. 2014. The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol 426:3851–3865. doi: 10.1016/j.jmb.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwalm ND, Groisman EA. 2017. Navigating the gut buffet: control of polysaccharide utilization in Bacteroides spp. Trends Microbiol 25:1005–1015. doi: 10.1016/j.tim.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 22.Waldor MK, Tyson G, Borenstein E, Ochman H, Moeller A, Finlay BB, Kong HH, Gordon JI, Nelson KE, Dabbagh K, Smith H. 2015. Where next for microbiome research? PLoS Biol 13:e1002050. doi: 10.1371/journal.pbio.1002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. 2010. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, Klinter S, Pudlo NA, Urs K, Koropatkin NM, Creagh AL, Haynes CA, Kelly AG, Cederholm SN, Davies GJ, Martens EC, Brumer H. 2014. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506:498–502. doi: 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemsworth GR, Thompson AJ, Stepper J, Sobala LF, Coyle T, Larsbrink J, Spadiut O, Goddard-Borger ED, Stubbs KA, Brumer H, Davies GJ. 2016. Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut. Open Biol 6:160142. doi: 10.1098/rsob.160142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC, Basle A, Morland C, Day AM, Zheng H, Rogers TE, Thompson P, Hawkins AR, Yadav MP, Henrissat B, Martens EC, Dupree P, Gilbert HJ, Bolam DN. 2015. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun 6:7481. doi: 10.1038/ncomms8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, Rogowski A, Hamilton BS, Chen R, Tolbert TJ, Piens K, Bracke D, Vervecken W, Hakki Z, Speciale G, Munōz-Munōz JL, Day A, Peña MJ, McLean R, Suits MD, Boraston AB, Atherly T, Ziemer CJ, Williams SJ, Davies GJ, Abbott DW, Martens EC, Gilbert HJ. 2015. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Hemsworth GR, DeJean G, Rogers TE, Pudlo NA, Urs K, Jain N, Davies GJ, Martens EC, Brumer H. 2017. Molecular mechanism by which prominent human gut Bacteroidetes utilize mixed-linkage beta-glucans, major health-promoting cereal polysaccharides. Cell Rep 21:417–430. doi: 10.1016/j.celrep.2017.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K, Foley MH, Gardill BR, Dejean G, Schnizlein M, Bahr CME, Louise Creagh A, van Petegem F, Koropatkin NM, Brumer H. 2019. Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus. Cell Mol Life Sci doi: 10.1007/s00018-019-03115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cartmell A, Lowe EC, Basle A, Firbank SJ, Ndeh DA, Murray H, Terrapon N, Lombard V, Henrissat B, Turnbull JE, Czjzek M, Gilbert HJ, Bolam DN. 2017. How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans. Proc Natl Acad Sci U S A 114:7037–7042. doi: 10.1073/pnas.1704367114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bagenholm V, Reddy SK, Bouraoui H, Morrill J, Kulcinskaja E, Bahr CM, Aurelius O, Rogers T, Xiao Y, Logan DT, Martens EC, Koropatkin NM, Stalbrand H. 2017. Galactomannan catabolism conferred by a polysaccharide utilization locus of Bacteroides ovatus: enzyme synergy and crystal structure of a beta-mannanase. J Biol Chem 292:229–243. doi: 10.1074/jbc.M116.746438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Temple MJ, Cuskin F, Basle A, Hickey N, Speciale G, Williams SJ, Gilbert HJ, Lowe EC. 2017. A Bacteroidetes locus dedicated to fungal 1,6-beta-glucan degradation: unique substrate conformation drives specificity of the key endo-1,6-beta-glucanase. J Biol Chem 292:10639–10650. doi: 10.1074/jbc.M117.787606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ndeh D, Rogowski A, Cartmell A, Luis AS, Basle A, Gray J, Venditto I, Briggs J, Zhang X, Labourel A, Terrapon N, Buffetto F, Nepogodiev S, Xiao Y, Field RA, Zhu Y, O’Neill MA, Urbanowicz BR, York WS, Davies GJ, Abbott DW, Ralet MC, Martens EC, Henrissat B, Gilbert HJ. 2017. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544:65–70. doi: 10.1038/nature21725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cartmell A, Munoz-Munoz J, Briggs JA, Ndeh DA, Lowe EC, Basle A, Terrapon N, Stott K, Heunis T, Gray J, Yu L, Dupree P, Fernandes PZ, Shah S, Williams SJ, Labourel A, Trost M, Henrissat B, Gilbert H. 2018. A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation. Nat Microbiol 3:1314–1326. doi: 10.1038/s41564-018-0258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luis AS, Briggs J, Zhang XY, Farnell B, Ndeh D, Labourel A, Basle A, Cartmell A, Terrapon N, Stott K, Lowe EC, McLean R, Shearer K, Schuckel J, Venditto I, Ralet MC, Henrissat B, Martens EC, Mosimann SC, Abbott DW, Gilbert HJ. 2018. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol 3:210–219. doi: 10.1038/s41564-017-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lapebie P, Lombard V, Drula E, Terrapon N, Henrissat B. 2019. Bacteroidetes use thousands of enzyme combinations to break down glycans. Nat Commun 10:7. doi: 10.1038/s41467-019-10068-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pauly M, Keegstra K. 2016. Biosynthesis of the plant cell wall matrix polysaccharide xyloglucan. Annu Rev Plant Biol 67:235–259. doi: 10.1146/annurev-arplant-043015-112222. [DOI] [PubMed] [Google Scholar]
- 38.Tauzin AS, Kwiatkowski KJ, Orlovsky NI, Smith CJ, Creagh AL, Haynes CA, Wawrzak Z, Brumer H, Koropatkin NM. 2016. Molecular dissection of xyloglucan recognition in a prominent human gut symbiont. mBio 7:e02134-15. doi: 10.1128/mBio.02134-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foley MH, Cockburn DW, Koropatkin NM. 2016. The Sus operon: a model system for starch uptake by the human gut Bacteroidetes. Cell Mol Life Sci 73:2603–2617. doi: 10.1007/s00018-016-2242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glenwright AJ, Pothula KR, Bhamidimarri SP, Chorev DS, Baslé A, Firbank SJ, Zheng H, Robinson CV, Winterhalter M, Kleinekathöfer U, Bolam DN, van den Berg B. 2017. Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature 541:407–411. doi: 10.1038/nature20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.CAZypedia Consortium. 2018. Ten years of CAZypedia: a living encyclopedia of carbohydrate-active enzymes. Glycobiology 28:3–8. doi: 10.1093/glycob/cwx089. [DOI] [PubMed] [Google Scholar]
- 43.Zitomersky NL, Atkinson BJ, Franklin SW, Mitchell PD, Snapper SB, Comstock LE, Bousvaros A. 2013. Characterization of adherent Bacteroidales from intestinal biopsies of children and young adults with inflammatory bowel disease. PLoS One 8:e63686. doi: 10.1371/journal.pone.0063686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ley RE, Peterson DA, Gordon JI. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 45.Aspeborg H, Coutinho PM, Wang Y, Brumer H, Henrissat B. 2012. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol Biol 12:186. doi: 10.1186/1471-2148-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Attia MA, Brumer H. 2016. Recent structural insights into the enzymology of the ubiquitous plant cell wall glycan xyloglucan. Curr Opin Struct Biol 40:43–53. doi: 10.1016/j.sbi.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 47.Attia MA, Nelson CE, Offen WA, Jain N, Davies GJ, Gardner JG, Brumer H. 2018. In vitro and in vivo characterization of three Cellvibrio japonicus glycoside hydrolase family 5 members reveals potent xyloglucan backbone-cleaving functions. Biotechnol Biofuels 11:16. doi: 10.1186/s13068-018-1039-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paetzel M, Karla A, Strynadka NCJ, Dalbey RE. 2002. Signal peptidases. Chem Rev 102:4549–4579. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 49.Hoffman M, Jia ZH, Pena MJ, Cash M, Harper A, Blackburn AR, Darvill A, York WS. 2005. Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr Res 340:1826–1840. doi: 10.1016/j.carres.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 50.dos Santos CR, Cordeiro RL, Wong DWS, Murakami MT. 2015. Structural basis for xyloglucan specificity and alpha-d-Xylp(1 → 6)-d-Glcp recognition at the −1 subsite within the GH5 family. Biochemistry 54:1930–1942. doi: 10.1021/acs.biochem.5b00011. [DOI] [PubMed] [Google Scholar]
- 51.Nagae M, Tsuchiya A, Katayama T, Yamamoto K, Wakatsuki S, Kato R. 2007. Structural basis of the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase from Bifidobacterium bifidum. J Biol Chem 282:18497–18509. doi: 10.1074/jbc.M702246200. [DOI] [PubMed] [Google Scholar]
- 52.Sela DA, Garrido D, Lerno L, Wu S, Tan K, Eom HJ, Joachimiak A, Lebrilla CB, Mills DA. 2012. Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol 78:795–803. doi: 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larsbrink J, Thompson AJ, Lundqvist M, Gardner JG, Davies GJ, Brumer H. 2014. A complex gene locus enables xyloglucan utilization in the model saprophyte Cellvibrio japonicus. Mol Microbiol 94:418–433. doi: 10.1111/mmi.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bauer S, Vasu P, Persson S, Mort AJ, Somerville CR. 2006. Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc Natl Acad Sci U S A 103:11417–11422. doi: 10.1073/pnas.0604632103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ishimizu T, Hashimoto C, Takeda R, Fujii K, Hase S. 2007. A novel α1,2-l-fucosidase acting on xyloglucan oligosaccharides is associated with endo-β-mannosidase. J Biochem 142:721–729. doi: 10.1093/jb/mvm186. [DOI] [PubMed] [Google Scholar]
- 56.Leonard R, Pabst M, Bondili JS, Chambat G, Veit C, Strasser R, Altmann F. 2008. Identification of an Arabidopsis gene encoding a GH95 α1,2-fucosidase active on xyloglucan oligo- and polysaccharides. Phytochemistry 69:1983–1988. doi: 10.1016/j.phytochem.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 57.Cao H, Walton JD, Brumm P, Phillips GN Jr.. 2014. Structure and substrate specificity of a eukaryotic fucosidase from Fusarium graminearum. J Biol Chem 289:25624–25638. doi: 10.1074/jbc.M114.583286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paper JM, Scott-Craig JS, Cavalier D, Faik A, Wiemels RE, Borrusch MS, Bongers M, Walton JD. 2013. Alpha-fucosidases with different substrate specificities from two species of Fusarium. Appl Microbiol Biotechnol 97:5371–5380. doi: 10.1007/s00253-012-4423-3. [DOI] [PubMed] [Google Scholar]
- 59.Cameron EA, Kwiatkowski KJ, Lee B-H, Hamaker BR, Koropatkin NM, Martens EC. 2014. Multifunctional nutrient-binding proteins adapt human symbiotic bacteria for glycan competition in the gut by separately promoting enhanced sensing and catalysis. mBio 5:e01441-14. doi: 10.1128/mBio.01441-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Foley MH, Dejean G, Hemsworth GR, Davies GJ, Brumer H, Koropatkin NM. 2019. A cell-surface GH9 endo-glucanase coordinates with surface glycan-binding proteins to mediate xyloglucan uptake in the gut symbiont Bacteroides ovatus. J Mol Biol 431:981–995. doi: 10.1016/j.jmb.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ravachol J, de Philip P, Borne R, Mansuelle P, Mate MJ, Perret S, Fierobe HP. 2016. Mechanisms involved in xyloglucan catabolism by the cellulosome-producing bacterium Ruminiclostridium cellulolyticum. Sci Rep 6:22770. doi: 10.1038/srep22770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ariza A, Eklof JM, Spadiut O, Offen WA, Roberts SM, Besenmatter W, Friis EP, Skjot M, Wilson KS, Brumer H, Davies G. 2011. Structure and activity of Paenibacillus polymyxa xyloglucanase from glycoside hydrolase family 44. J Biol Chem 286:33890–33900. doi: 10.1074/jbc.M111.262345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arnal G, Stogios PJ, Asohan J, Skarina T, Savchenko A, Brumer H. 2018. Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74 endo-xyloglucanase. Biochem J 475:3963–3978. doi: 10.1042/BCJ20180763. [DOI] [PubMed] [Google Scholar]
- 64.Feng T, Yan KP, Mikkelsen MD, Meyer AS, Schols HA, Westereng B, Mikkelsen JD. 2014. Characterisation of a novel endo-xyloglucanase (XcXGHA) from Xanthomonas that accommodates a xylosyl-substituted glucose at subsite −1. Appl Microbiol Biotechnol 98:9667–9679. doi: 10.1007/s00253-014-5825-1. [DOI] [PubMed] [Google Scholar]
- 65.Ichinose H, Araki Y, Michikawa M, Harazono K, Yaoi K, Karita S, Kaneko S. 2012. Characterization of an endo-processive-type xyloglucanase having a beta-1,4-glucan-binding module and an endo-type xyloglucanase from Streptomyces avermitilis. Appl Environ Microbiol 78:7939–7945. doi: 10.1128/AEM.01762-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yaoi K, Mitsuishi Y. 2002. Purification, characterization, cloning, and expression of a novel xyloglucan-specific glycosidase, oligoxyloglucan reducing end-specific cellobiohydrolase. J Biol Chem 277:48276–48281. doi: 10.1074/jbc.M208443200. [DOI] [PubMed] [Google Scholar]
- 67.Dodd D, Moon Y-H, Swaminathan K, Mackie RI, Cann I. 2010. Transcriptomic analyses of xylan degradation by Prevotella bryantii and insights into energy acquisition by xylanolytic Bacteroidetes. J Biol Chem 285:30261–30273. doi: 10.1074/jbc.M110.141788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Speciale G, Thompson AJ, Davies GJ, Williams SJ. 2014. Dissecting conformational contributions to glycosidase catalysis and inhibition. Curr Opin Struct Biol 28:1–13. doi: 10.1016/j.sbi.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katayama T, Sakuma A, Kimura T, Makimura Y, Hiratake J, Sakata K, Yamanoi T, Kumagai H, Yamamoto K. 2004. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-alpha-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J Bacteriol 186:4885–4893. doi: 10.1128/JB.186.15.4885-4893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fan S, Zhang H, Chen X, Lu L, Xu L, Xiao M. 2016. Cloning, characterization, and production of three α-l-fucosidases from Clostridium perfringens ATCC 13124. J Basic Microbiol 56:347–357. doi: 10.1002/jobm.201500582. [DOI] [PubMed] [Google Scholar]
- 71.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Centanni M, Hutchison JC, Carnachan SM, Daines AM, Kelly WJ, Tannock GW, Sims IM. 2017. Differential growth of bowel commensal Bacteroides species on plant xylans of differing structural complexity. Carbohydr Polym 157:1374–1382. doi: 10.1016/j.carbpol.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 73.Mendis M, Martens EC, Simsek S. 2018. How fine structural differences of xylooligosaccharides and arabinoxylooligosaccharides regulate differential growth of Bacteroides species. J Agric Food Chem 66:8398–8405. doi: 10.1021/acs.jafc.8b01263. [DOI] [PubMed] [Google Scholar]
- 74.Rogers TE, Pudlo NA, Koropatkin NM, Bell JSK, Balasch MM, Jasker K, Martens EC. 2013. Dynamic responses of Bacteroides thetaiotaomicron during growth on glycan mixtures. Mol Microbiol 88:876–890. doi: 10.1111/mmi.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pudlo NA, Urs K, Kumar SS, German JB, Mills DA, Martens EC. 2015. Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. mBio 6:e01282-15. doi: 10.1128/mBio.01282-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tuncil YE, Nakatsu CH, Kazem AE, Arioglu-Tuncil S, Reuhs B, Martens EC, Hamaker BR. 2017. Delayed utilization of some fast-fermenting soluble dietary fibers by human gut microbiota when presented in a mixture. J Funct Foods 32:347–357. doi: 10.1016/j.jff.2017.03.001. [DOI] [Google Scholar]
- 77.Tuncil YE, Xiao Y, Porter NT, Reuhs BL, Martens EC, Hamaker BR. 2017. Reciprocal prioritization to dietary glycans by gut bacteria in a competitive environment promotes stable coexistence. mBio 8:e01068-17. doi: 10.1128/mBio.01068-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwalm ND, Townsend GE, Groisman EA. 2017. Prioritization of polysaccharide utilization and control of regulator activation in Bacteroides thetaiotaomicron. Mol Microbiol 104:32–45. doi: 10.1111/mmi.13609. [DOI] [PubMed] [Google Scholar]
- 79.Cantu-Jungles TM, do Nascimento GE, Zhang XW, Iacomini M, Cordeiro LMC, Hamaker BR. 2019. Soluble xyloglucan generates bigger bacterial community shifts than pectic polymers during in vitro fecal fermentation. Carbohydr Polym 206:389–395. doi: 10.1016/j.carbpol.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 80.Hsieh YS, Harris PJ. 2009. Xyloglucans of monocotyledons have diverse structures. Mol Plant 2:943–965. doi: 10.1093/mp/ssp061. [DOI] [PubMed] [Google Scholar]
- 81.Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nielsen H. 2017. Predicting secretory proteins with SignalP. Methods Mol Biol 1611:59–73. doi: 10.1007/978-1-4939-7015-5_6. [DOI] [PubMed] [Google Scholar]
- 83.Eschenfeldt WH, Makowska-Grzyska M, Stols L, Donnelly MI, Jedrzejczak R, Joachimiak A. 2013. New LIC vectors for production of proteins from genes containing rare codons. J Struct Funct Genomics 14:135–144. doi: 10.1007/s10969-013-9163-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arnal G, Attia MA, Asohan J, Brumer H. 2017. A low-volume, parallel copper-bicinchoninic acid (BCA) assay for glycoside hydrolases. Methods Mol Biol 1588:3–14. doi: 10.1007/978-1-4939-6899-2_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.