Abstract
Background
This study aimed to investigate the plasma long noncoding RNA metastasis‐associated lung adenocarcinoma transcript 1 (lncRNA MALAT1) expression with risk, severity, inflammation level, and prognosis in sepsis.
Methods
One hundred and ninety sepsis patients and 190 health controls (HCs) were consecutively recruited. Blood samples within 24 hours after admission of sepsis patients and those on enrollment of HCs were collected, and then, plasma was separated for lncRNA MALAT1 and miR‐125b expressions detections by RT‐qPCR. In sepsis patients, clinical data and 28‐day mortality were recorded, and plasma inflammatory cytokines expressions were detected by ELISA.
Results
Plasma lncRNA MALAT1 expression was elevated in sepsis patients than HCs (P < 0.001), and ROC curve disclosed that it had a good value in predicting sepsis risk with an area under curve (AUC) of 0.823 (95% CI: 0.783‐0.864). Additionally, lncRNA MALAT1 expression was positively correlated with Scr (P = 0.005), WBC (P = 0.017), CRP (P < 0.001), PCT (P < 0.001), TNF‐α (P < 0.001), IL‐8 (P < 0.001), IL‐17 (P = 0.001), APACHE II score (P < 0.001), and SOFA score (P < 0.001). LncRNA MALA1 expression was elevated in deaths compared with survivors (P < 0.001) and could predict the risk of 28‐day mortality with an AUC of 0.755 (95% CI: 0.682‐0.828). Accumulating survival was worse in patients with lncRNA MALAT1 high expression compared with patients who had lncRNA MALAT1 low expression (P < 0.001). Moreover, lncRNA MALAT1 expression was negatively correlated with miR‐125b level in both sepsis patients (P < 0.001) and HCs (P < 0.001).
Conclusion
LncRNA MALAT1 could be developed as a potential biomarker for facilitating diagnosis and management in sepsis patients.
Keywords: lncRNA MALAT1, miR‐125b, prognosis, risk, sepsis, severity
1. INTRODUCTION
Sepsis, a systemic inflammatory response syndrome (SIRS) triggered by imbalanced and intricate host response to infection, attacks over 19 million persons annually and is responsible for approximately 30% of mortality in the intensive care unit (ICU).1, 2 Multiple risk factors for sepsis have been identified through years of investigations, such as very old age, immunosuppressive diseases, and cancers, which assist in the prevention of sepsis to some extent, and along with the reduced time delay to initial intervention, the survival of sepsis patients has been improved over the past decades.3, 4, 5 However, despite a decrease in in‐hospital mortality, the sepsis survivors still experience disability, chronic health problems, and readmission to hospital.6, 7, 8, 9 Thus, the prevention, diagnosis, and personalized management of sepsis remain to be optimized.
Long noncoding RNAs (lncRNAs) are a category of promising noncoding RNAs that have been found to interplay with multiple factors in various diseases, which are characterized by a length more than 200 nt and almost no protein‐coding function.10, 11, 12 However, although lncRNAs are not involved in protein coding, these molecules are able to mediate gene levels at different aspects, including transcription, post‐transcription processing, and so on.13 LncRNA metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1) locates on chromosome 11 has been found to be essential in the regulation of tumorigenesis and inflammation, and for instance, it is reported that lncRNA MALAT1 promotes inflammation‐related hepatocellular carcinoma by binding to the chromatin‐remodeling subunit BRG1.14 Although it seems that lncRNA MALAT1 plays a dual role in inflammation according to the previous findings, recent studies report that lncRNA MALAT1 might be a pro‐inflammatory gene in sepsis.15, 16, 17 Therefore, we presumed that lncRNA MALAT1 may have the potential to serve as a biomarker for risk prediction of sepsis and prognosis in patients with sepsis. Nonetheless, the diagnostic and prognostic role of lncRNA MALAT1 in sepsis patients remains largely unknown.
Thus, the aim of this study was to investigate the plasma lncRNA MALAT1 expression with risk, severity, inflammation level, and prognosis in sepsis.
2. MATERIALS AND METHODS
2.1. Participants
This study consecutively recruited 190 sepsis patients who were admitted to intensive care units (ICUs) of The Central Hospital of Wuhan Hospital from January 2015 to December 2018. The patients included in this study were diagnosed as sepsis according to the Surviving Sepsis Campaign: International Guidelines for Management of Severe Sepsis and Septic Shock, 2012.4 Patients were excluded if they were less than 18 years old, accompanied with primary malignancies, received immunosuppressant before enrollment, were pregnant, lactating, human immunodeficiency virus (HIV)‐positive, or in end‐of‐life conditions. In addition, 190 healthy subjects who underwent physical examination in The Central Hospital of Wuhan were enrolled as healthy controls (HCs), and all of them had no history of severe infection or malignancies, or other obvious abnormalities by physical examination. The study protocol was approved by Ethics Committee of The Central Hospital of Wuhan. All participants or their guardians signed informed consents.
2.2. Data collection
Patients’ clinical data were collected, which consisted of age, gender, body mass index (BMI) smoke, history of chronic obstructive pulmonary disease (COPD), history of cardiomyopathy, history of chronic kidney failure, history of cirrhosis, serum creatinine (Scr), albumin, white blood cell (WBC), C‐reactive protein (CRP), procalcitonin (PCT), acute physiology and chronic health evaluation (APACHE) II score, and sequential organ failure assessment (SOFA) score. The APACHE II score and SOFA score were assessed within 24 hours after ICU admission. After hospital admission, all enrolled patients were followed up to 28 days, and 28‐day mortality was calculated, and according to that, the accumulating survival was calculated as well.
2.3. Samples collection and measurements
Blood samples of sepsis patients were collected in anticoagulant tube within 24 hours after ICU admission, and blood samples of HCs were collected in anticoagulant tube on the enrollment. All the blood samples were processed immediately after collection for the separation of plasma. The conditions were as follows: The whole blood samples were centrifuged at 1600 g for 10 minutes at 4°C, and supernatant was transferred to the Eppendorf (EP) tube, then were centrifuged again at 2500 g for 10 minutes at 4°C. After centrifugation, the plasma was collected and stored at −80°C until determination. LncRNA MALAT1 and miR‐125b relative expressions in plasma were determined using real‐time quantitative polymerase chain reaction (RT‐qPCR). The levels of inflammatory cytokines including tumor necrosis factor‐α (TNF‐α), interleukin‐1β (IL‐1β), IL‐6, and IL‐17 in plasma were determined by enzyme‐linked immunosorbent assay (ELISA) using commercial human ELISA Kits (eBioscience) following the manufacturer's protocol.
2.4. RT‐qPCR
First, the total RNA was elicited from plasma by using the TRIzol reagent (Invitrogen) and then was assessed by spectrometer for its purity. Then, the total RNA was reversely transcribed to complementary DNAs by PrimeScript RT reagent Kit (Takara). Subsequently, qPCR was performed by using TB Green Fast qPCR Mix (Takara), and the relative expressions of lncRNA and miRNA were calculated according to the 2−ΔΔt formula using GAPDH as internal reference for lncRNA MALAT1 and U6 as internal reference for miR‐125b. In addition, the primers used in the present study were as follows: lncRNA MALAT1, forward: TCCTAAGGTCAAGAGAAGTGTCAG, reverse: GTGGCGATGTGGCAGAGAA; GAPDH, forward: GAGTCCACTGGCGTCTTCAC, reverse: ATCTTGAGGCTGTTGTCATACTTCT; U6, forward: CTCGCTTCGGCAGCACA, reverse: AACGCTTCACGAATTTGCGT; miR‐125b, forward: ACACTCCAGCTGGGTCCCTGAGACCCTAACTT, reverse: TGTCGTGGAGTCGGCAATTC.
2.5. Statistical analysis
Statistical analysis was performed using SPSS 24.0 (IBM), figure was plotted using GraphPad Prism 7.00 (GraphPad Software). Data were presented as mean ± standard deviation (SD), median (interquartile range, IQR), or count (percentage). Comparisons were determined by Wilcoxon rank sum test. Correlations were determined by Spearman's rank correlation test. Receiver operating characteristic (ROC) curves were plotted, and the area under curve (AUC) with 95% confidence interval (CI) was calculated to assess the discrimination ability of LncRNA MALAT1 and miR‐125b relative expressions between sepsis patients and HCs or between deaths and survivors. Kaplan‐Meier curve was used to illustrate accumulating survival, and accumulating survival difference between groups was determined by log‐rank test. All tests were 2‐sided, and P value < 0.05 was considered as significant.
3. RESULTS
3.1. Clinical characteristics of sepsis patients
There were 127 males and 63 females in sepsis patients with a mean age of 58.5 ± 13.8 years and a mean BMI of 23.1 ± 5.5 kg/m2 (Table 1). The numbers of patients who had history of COPD, cardiomyopathy, chronic kidney failure, and cirrhosis were, respectively, 33 (17.4%), 70 (36.8%), 14 (7.4%), and 35 (18.4%). And the mean values of APACHE II score and SOFA score were 12.9 ± 5.7 and 5.1 ± 2.8, respectively. In addition, the medians of TNF‐α, IL‐6, IL‐8, and IL‐17 were 174.1 (103.7‐272.0) pg/mL, 63.1 (35.8‐129.9) pg/mL, 52.7 (23.8‐131.8) pg/mL, and 123.5 (61.6‐183.5) pg/mL.
Table 1.
Clinical characteristics of sepsis patients
| Items | Sepsis patients (N = 190) |
|---|---|
| Age (y), mean ± SD | 58.5 ± 13.8 |
| Gender (male/female) | 127/63 |
| BMI (kg/m2), mean ± SD | 23.1 ± 5.5 |
| Smoke, No. (%) | 58 (30.5) |
| History of COPD, No. (%) | 33 (17.4) |
| History of cardiomyopathy, No. (%) | 70 (36.8) |
| History of chronic kidney failure, No. (%) | 14 (7.4) |
| History of cirrhosis, No. (%) | 35 (18.4) |
| Scr (mg/dL), median (IQR) | 1.6 (1.1‐2.4) |
| Albumin (g/L), median (IQR) | 25.6 (21.3‐33.0) |
| WBC (×109/L), median (IQR) | 18.6 (3.2‐28.2) |
| CRP (mg/L), median (IQR) | 98.7 (52.3‐150.0) |
| PCT (ng/mL), median (IQR) | 12.6 (7.5‐24.4) |
| APACHE II score, mean ± SD | 12.9 ± 5.7 |
| SOFA score, mean ± SD | 5.1 ± 2.8 |
| TNF‐α (pg/mL), median (IQR) | 174.1 (103.7‐272.0) |
| IL‐6 (pg/mL), median (IQR) | 63.1 (35.8‐129.9) |
| IL‐8 (pg/mL), median (IQR) | 52.7 (23.8‐131.8) |
| IL‐17 (pg/mL), median (IQR) | 123.5 (61.6‐183.5) |
Abbreviations: APACHE, acute physiology and chronic health evaluation; BMI, body mass index; COPD, chronic obstructive pulmonary disease; CRP, C‐reactive protein; IL, interleukin; IQR, interquartile range; PCT, procalcitonin; Scr, serum creatinine; SD, standard deviation; SOFA, sequential organ failure assessment; TNF, tumor necrosis factor; WBC, white blood cell.
3.2. Predictive value of plasma lncRNA MALAT1 for sepsis risk
Plasma lncRNA MALAT1 expression was elevated in sepsis patients than that in HCs (P < 0.001; Figure 1A), and ROC curve analysis disclosed that lncRNA MALAT1 expression had a good value in predicting sepsis risk with an AUC of 0.823 (95%CI: 0.783‐0.864; Figure 1B). This result indicated that lncRNA MALAT1 expression in plasma may have potential in predicting sepsis risk.
Figure 1.
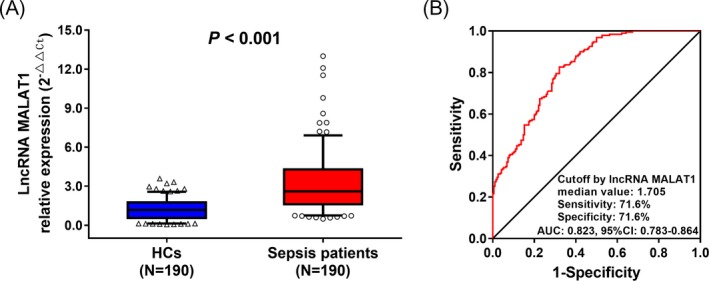
Expression of lncRNA MALAT1 in predicting sepsis risk. The plasma expression of lncRNA MALAT1 in sepsis patients and HCs (A), and its predictive value for sepsis risk by ROC curve analysis (B). Comparisons were determined by Wilcoxon rank sum test. ROC curves were plotted, and AUC with 95% confidence interval was calculated to assess the discrimination ability of LncRNA MALAT1 for sepsis risk. P value < 0.05 was considered significant. MALAT1, metastasis‐associated lung adenocarcinoma transcript 1; HCs, healthy controls; ROC, receiver operating characteristic; AUC, area under the curve
3.3. Correlation of plasma lncRNA MALAT1 with clinical characteristics in sepsis patients
Regarding all the continuous variables of clinical characteristics in sepsis patients, lncRNA MALAT1 expression in plasma was positively correlated with Scr (P = 0.005), WBC (P = 0.017), CRP (P < 0.001), PCT (P < 0.001), TNF‐α (P < 0.001), IL‐8 (P < 0.001), and IL‐17 (P = 0.001) levels as well as the APACHE II score (P < 0.001) and SOFA score (P < 0.001; Table 2). However, lncRNA MALAT1 expression was not correlated with age (P = 0.643), BMI (P = 0.569), albumin (P = 0.099), or IL‐6 (P = 0.064) levels. As to the categorical variables of clinical characteristics, lncRNA MALAT1 level was not associated with gender (P = 0.519), smoke (P = 0.289), history of COPD (P = 0.739), history of cardiomyopathy (P = 0.402), history of chronic kidney failure (P = 0.127), or history of cirrhosis (P = 0.261; Table 3). These results suggested that lncRNA MALAT1 in plasma positively associated with severity and inflammation level in sepsis patients.
Table 2.
Correlations of lncRNA MALAT1 relative expression with clinical characteristics (continuous variables)
| Items | LncRNA MALAT1 | |
|---|---|---|
| P value | Correlation coefficient (r) | |
| Age | 0.643 | −0.034 |
| BMI | 0.569 | 0.042 |
| Scr | 0.005 | 0.202 |
| Albumin | 0.099 | −0.120 |
| WBC | 0.017 | 0.173 |
| CRP | <0.001 | 0.296 |
| PCT | <0.001 | 0.280 |
| APACHE II score | <0.001 | 0.430 |
| SOFA score | <0.001 | 0.475 |
| TNF‐α | <0.001 | 0.354 |
| IL‐6 | 0.064 | 0.135 |
| IL‐8 | <0.001 | 0.331 |
| IL‐17 | 0.001 | 0.236 |
Correlation analyses were determined by Spearman's rank correlation test.
Abbreviations: APACHE, acute physiology and chronic health evaluation; BMI, body mass index; CRP, C‐reactive protein; IL, interleukin; LncRNA, long noncoding RNA; PCT, procalcitonin; Scr, serum creatinine; SOFA, sequential organ failure assessment; TNF, tumor necrosis factor; WBC, white blood cell.
Table 3.
Correlations of lncRNA MALAT1 relative expression with clinical characteristics (categorical variables)
| Items | LncRNA MALAT1, median (IQR) | P value |
|---|---|---|
| Gender | ||
| Male | 2.550 (1.618‐4.232) | 0.519 |
| Female | 2.800 (1.606‐4.352) | |
| Smoke | ||
| Yes | 2.382 (1.686‐4.367) | 0.289 |
| No | 2.709 (1.347‐4.185) | |
| History of COPD | ||
| Yes | 2.143 (1.649‐4.803) | 0.739 |
| No | 2.693 (1.590‐4.227) | |
| History of cardiomyopathy | ||
| Yes | 2.450 (1.617‐4.152) | 0.402 |
| No | 2.676 (1.585‐4.628) | |
| History of chronic kidney failure | ||
| Yes | 4.155 (1.889‐5.915) | 0.127 |
| No | 2.554 (1.600‐4.230) | |
| History of cirrhosis | ||
| Yes | 1.852 (1.050‐4.412) | 0.261 |
| No | 2.672 (1.663‐4.280) | |
Correlation analyses were determined by Wilcoxon rank sum test.
Abbreviations: COPD, chronic obstructive pulmonary disease; LncRNA, long noncoding RNA.
3.4. Predictive value of plasma lncRNA MALAT1 for 28‐day mortality in sepsis patients
The 28‐day mortality rate was 30.5%, and lncRNA MALA1 expression in plasma was elevated in deaths compared with survivors (P < 0.001) (Figure 2A), and then, the ROC curve analysis illuminated that lncRNA MALAT1 expression could predict the higher risk of 28‐day mortality with an AUC of 0.755 (95% CI: 0.682‐0.828; Figure 2B). Furthermore, according to median value of lncRNA MALAT1 expression in sepsis patients, patients were divided into those with lncRNA MALAT1 high expression and those with lncRNA MALAT1 low expression, and the accumulating survival was worse in patients with lncRNA MALAT1 high expression compared with that in patients who had lncRNA MALAT1 low expression (P < 0.001; Figure 2C). These data indicated that plasma lncRNA MALAT1 expression may serve as a prognostic biomarker in sepsis.
Figure 2.
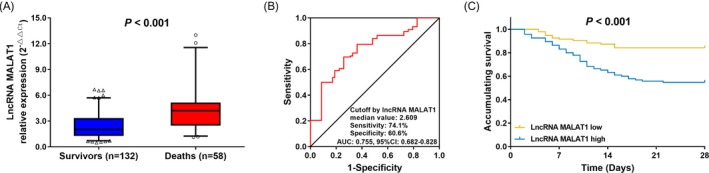
Expression of lncRNA MALAT1 in predicting 28‐day mortality in sepsis patients. The plasma expression of lncRNA MALAT1 in deaths and survivors (A), the ROC curve of lncRNA MALAT1 in predicting 28‐day mortality (B) and Kaplan‐Meier curves of patients with high and low lncRNA MALAT1 expressions (C). Comparisons were determined by Wilcoxon rank sum test. ROC curves were plotted, and AUC with 95% confidence interval was calculated to assess the discrimination ability of LncRNA MALAT1 for 28‐day mortality. Kaplan‐Meier curve was used to illustrate accumulating survival, and difference between two groups was determined by log‐rank test. P value < 0.05 was considered significant. MALAT1, metastasis‐associated lung adenocarcinoma transcript 1; ROC, receiver operating characteristic; AUC, area under the curve
3.5. Correlation of plasma lncRNA MALAT1 with miR‐125b and implication of miR‐125b in sepsis
Moreover, we evaluated the correlation between lncRNA MALAT1 expression and miR‐125b expression in plasma, and found that lncRNA MALAT1 expression was negatively correlated with the miR‐125b expression in both sepsis patients (P < 0.001; Figure 3A) and HCs (P < 0.001; Figure 3B). Furthermore, miR‐125b expression was downregulated in sepsis patients compared with HCs (P < 0.001; Figure 3C), and ROC curve analysis showed that miR‐125b expression had a good value in differentiating sepsis patients from HCs with an AUC of 0.859 (95% CI: 0.824‐0.895; Figure 3D). In addition, miR‐125b level was decreased in deaths compared with survivors (P < 0.001; Figure 3E), and ROC curve analysis revealed that miR‐125b had a good predictive value for 28‐day mortality with an AUC of 0.802 (95% CI: 0.728‐0.876; Figure 3F). Moreover, patients with miR‐125b low expression presented with worse accumulating survival compared with patients who had miR‐125b high expression (P < 0.001; Figure 3G). In addition, with respect to all the continuous variables of clinical characteristics, miR‐125b expression was negatively correlated with SCr (P < 0.001), WBC (P = 0.007), CRP (P < 0.001), PCT (P < 0.001), TNF‐α (P < 0.001), IL‐6 (P < 0.001), IL‐8 (P < 0.001) and IL‐17 (P < 0.001) levels, and the APACHE II score (P < 0.001) as well as SOFA score (P < 0.001), while was not associated with age (P = 0.641), BMI (P = 0.659), or albumin level (P = 0.171) in sepsis patients (Table 4). As for the categorical variables, the miR‐125b expression was negatively correlated with history of cirrhosis (P < 0.001), but it was not correlated with gender (P = 0.506), smoke (P = 0.706), history of COPD (P = 0.133), history of cardiomyopathy (P = 0.174), or history of chronic kidney failure (P = 0.170) in sepsis patients (Table 5).
Figure 3.
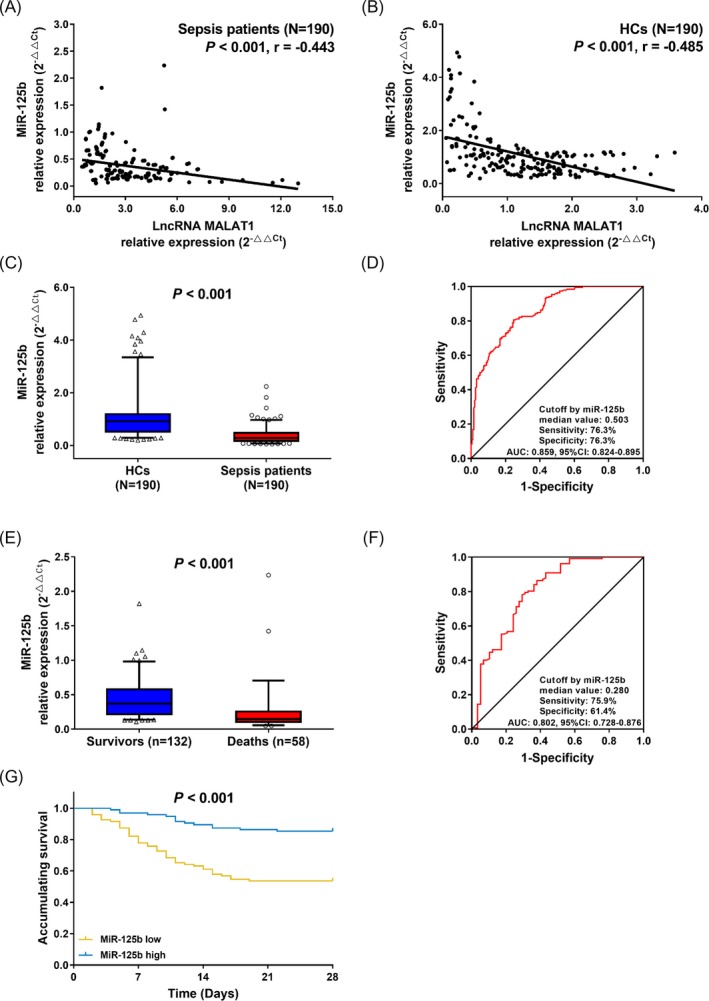
Correlation of lnRNA MALAT1 expression with miR‐125b. The correlation of plasma lncRNA MALAT1 expression with miR‐125b level in sepsis patients (A) and HCs (B), the expression of miR‐125b in sepsis patients and HCs (C), ROC curve of miR‐125b expression in predicting sepsis risk (D), miR‐125b expression in deaths and survivors (E), ROC curve of miR‐125b expression in predicting 28‐day mortality, (F) and the Kaplan‐Meier curves of patients with high and low miR‐125b expressions (G). Comparisons were determined by Wilcoxon rank sum test. ROC curves were plotted, and AUC with 95% confidence interval was calculated to assess the discrimination ability of miR‐125b for sepsis risk and 28‐day mortality. Kaplan‐Meier curve was used to illustrate accumulating survival, and difference between two groups was determined by log‐rank test. P value < 0.05 was considered significant. MALAT1, metastasis‐associated lung adenocarcinoma transcript 1; HCs, healthy controls; ROC, receiver operating characteristic; AUC, area under the curve
Table 4.
Correlations of miR‐125b relative expression with clinical characteristics (continuous variables)
| Items | MiR‐125b | |
|---|---|---|
| P value | Correlation coefficient (r) | |
| Age | 0.641 | −0.034 |
| BMI | 0.659 | −0.032 |
| Scr | <0.001 | −0.265 |
| Albumin | 0.171 | 0.100 |
| WBC | 0.007 | −0.194 |
| CRP | <0.001 | −0.374 |
| PCT | <0.001 | −0.260 |
| APACHE II score | <0.001 | −0.511 |
| SOFA score | <0.001 | −0.399 |
| TNF‐α | <0.001 | −0.418 |
| IL‐6 | <0.001 | −0.339 |
| IL‐8 | <0.001 | −0.324 |
| IL‐17 | <0.001 | −0.418 |
Correlation analyses were determined by Spearman's rank correlation test.
Abbreviations: APACHE, acute physiology and chronic health evaluation; BMI, body mass index; CRP, C‐reactive protein; IL, interleukin; PCT,, procalcitonin; Scr, serum creatinine; SOFA, sequential organ failure assessment; TNF, tumor necrosis factor; WBC, white blood cell.
Table 5.
Correlations of miR‐125b relative expression with clinical characteristics (categorical variables)
| Items | MiR‐125b, median (IQR) | P value |
|---|---|---|
| Gender | ||
| Male | 0.283 (0.171‐0.471) | 0.506 |
| Female | 0.272 (0.146‐0.482) | |
| Smoke | ||
| Yes | 0.288 (0.128‐0.578) | 0.706 |
| No | 0.273 (0.176‐0.434) | |
| History of COPD | ||
| Yes | 0.504 (0.188‐0.658) | 0.133 |
| No | 0.272 (0.165‐0.427) | |
| History of cardiomyopathy | ||
| Yes | 0.239 (0.171‐0.619) | 0.174 |
| No | 0.286 (0.144‐0.422) | |
| History of chronic kidney failure | ||
| Yes | 0.183 (0.063‐0.861) | 0.170 |
| No | 0.286 (0.168‐0.471) | |
| History of cirrhosis | ||
| Yes | 0.471 (0.413‐0.640) | <0.001 |
| No | 0.240 (0.160‐0.394) | |
Correlation analyses were determined by Wilcoxon rank sum test.
Abbreviation: COPD, chronic obstructive pulmonary disease.
4. DISCUSSION
Sepsis, a relatively common and critically fatal SIRS in ICU, requires for immediate diagnosis and intervention for the enhancement of satisfactory clinical outcomes. However, the present biomarkers in clinical practice are not sensitive or specific enough for enhancing the timely intervention of sepsis, such as CRP, PCT, and IL‐6.18, 19, 20 Therefore, the effort for exploring biomarkers assisting in diagnosis and prognosis is continuing even at the time of writing, and increasingly profound genetic mechanisms of the interactions that take place in sepsis have been discovered, in which the roles of ncRNAs have been implied in the regulation of sepsis etiology.21 To be specific, lncRNAs have been revealed to play essential roles in the regulations of immune responses and inflammation in sepsis, indicating that they may hold missing drivers of the sepsis pathogenesis.14, 22 Herein, we investigated the correlation of a lncRNA that has a promising role in inflammation, lncRNA MALAT1, with the sepsis risk, and severity, inflammation level, and prognosis in sepsis patients, and found that: (a) plasma lncRNA MALAT1 was upregulated in sepsis patients and had good value in predicting sepsis risk; (b) lncRNA MALAT1 expression was positively correlated with severity and inflammation level in sepsis patients; (c) high lncRNA MALAT1 had good value in predicting increased 28‐day mortality and was associated with worse accumulating survival in sepsis patients; and (d) lncRNA MALAT1 expression was negatively correlated with miR‐125b expression in both sepsis patients and HCs.
LncRNA MALAT1 has been reported to participate in several inflammation‐related diseases in regulating the inflammatory or immune responses. An in vitro experiment shows that lncRNA MALAT1, mainly located in nuclear speckles, was upregulated in endothelial cells incubated with high glucose concentration and is correlated with higher expressions of inflammatory cytokines, and the diabetic‐induced inflammatory cytokines levels elevations are reduced in lncRNA MALAT1 knockdown diabetic rat models.23 Another in vitro experiment reports that lncRNA MALAT1 might play a pro‐inflammatory role in lung injury by regulating macrophage polarization via mediating the IL‐4 expression.24 And an in vitro and in vivo experiments elucidate that lncRNA MALAT1 advocates the inflammation‐mediated progression of hepatocellular carcinoma via binding to chromatin‐remodeling subunit BRG1.14 Recently, a study investigates the effect of lncRNA MALAT1 on septic lung injury and finds that lncRNA MALAT1 knockdown markedly reduced lung injury induced by sepsis possibly via the repression of p38 MAPK/p65 NF‐κB signaling pathway.25 Another experiment illuminates that lncMALAT1 knockdown helps with the suppression of inflammatory response via elevating the miR‐146a in lipopolysaccharide‐induced acute lung damage.26 These previous studies indicate that lncRNA MALAT1 acts as a pro‐inflammatory gene and also promotes organ injury, such as the heart and lung injuries. In addition, there are reports illuminating that lncRNA MALAT1 reduces inflammation in several diseases as well. For instance, a recent experiment reveals that lncRNA MALAT1 represses proliferation and inflammation by inhibiting Wnt signaling pathway in fibroblast‐like synoviocytes.15 And another study elucidates that lncRNA MALAT1 expression is elevated in ischemia‐reperfusion injury and represses hypoxia‐induced inflammation via the NF‐κB pathway.27 And these findings indicate a dual role of this lncRNA in the regulation of inflammation probably because of that in lncRNA MALAT1 might function as a gene that keep balance in inflammatory responses, while in sepsis the main pathological feature is an inflammation cascade, which results in a pro‐inflammatory role of lncRNA MALAT1 in the complex interactions of sepsis. In this study, we discovered that upregulated plasma lncRNA MALAT1 expression correlated with higher sepsis risk, and worse disease condition, increased inflammation level, and poorer prognosis in sepsis patients, which could be explained by that lncRNA MALAT1 might promote the development and progression of sepsis through (a) enhancing inflammation via mediating multiple signaling pathways or proteins, such as regulating the IL‐4 expression and (b) advocating the organ dysfunction by regulating various pathways, for instance the p38 MAPK/p65 NF‐κB signaling pathway.14, 23, 24, 25
In addition, we evaluated the expression of lncRNA MALAT1 in plasma in sepsis patients, and the possible sources of lncRNA MALAT1 in plasma might be exosomes or cell debris from inflammatory cells or immune cells such as lymphocytes and macrophages due to that most lncRNAs in plasma are abundantly expressed in exosomes or cell debris or exist as nucleic acid, and sepsis is a type of systemic inflammatory response syndrome closely related to inflammation and immune disorders.28, 29
MiR‐125b is an inflammatory‐related miRNA and has been reported to play an anti‐inflammatory role in various diseases. For example, a cells experiment reveals that miR‐125b suppresses lipopolysaccharide‐induced inflammatory damage in chondrogenic cells ATDC5 by targeting macrophage inflammatory protein‐1 alpha (MIP‐1α).30 And another study elucidates that miR‐125b promotes the recovery and regeneration of spinal cord injury via decreasing cell apoptosis and inflammatory responses in neurons obtained from mouse models of cervical spinal cord contusion.31 In addition, there are several previous experiments illustrating that lncRNA MALAT1 targets miR‐125b to regulated inflammation or other pathogenesis progresses in several diseases. For instance, an experiment elucidates that lncRNA MALAT1 promotes neovascularization via mediating the miR‐125b/VE‐cadherin axis in endothelial cells models of diabetic retinopathy.32 More importantly, a study illuminates that lncRNA MALAT1 promotes cardiac inflammation and dysfunction in sepsis mice by mediating miR‐125b and p38 MAPK/NFκB.17 In our study, the plasma expression of lncRNA MALAT1 was negatively associated with miR‐125b expression in both sepsis patients and HCs, and further analysis showed that miR‐125b level was downregulated in sepsis patients and nonsurvivors, and its downregulation was correlated with increased severity, inflammation level, and worse survival in sepsis patients, indicating that miR‐125b is a target of lncRNA MALAT1 in sepsis and plays an anti‐inflammatory role in sepsis.
There were several limitations in our study. First, our study was conducted in a single center, which caused a selection bias due to that the patients were mostly from the same region in China. Second, the molecular mechanisms of lncRNA MALAT1 in promoting the development or progression of sepsis was not evaluated in our study. Third, long‐term clinical outcomes were not assessed in this study. Thus, multicenter studies with longer follow‐up time, and in vivo as well as in vitro experiments, should be performed in the future.
In conclusion, lncRNA MALAT1 could be developed as a biomarker for facilitating diagnosis and management in sepsis patients.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
ACKNOWLEDGMENTS
None.
Geng F, Liu W, Yu L. Potential role of circulating long noncoding RNA MALAT1 in predicting disease risk, severity, and patients' survival in sepsis. J Clin Lab Anal. 2019;33:e22968 10.1002/jcla.22968
Feng Geng and Wei Liu are contributed equally to this work.
REFERENCES
- 1. Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (sepsis‐3). JAMA. 2016;315(8):801‐810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fleischmann C, Scherag A, Adhikari N, et al. Assessment of global incidence and mortality of hospital‐treated sepsis. Current estimates and limitations. Am J Respir Crit Care Med. 2016;193(3):259‐272. [DOI] [PubMed] [Google Scholar]
- 3. Vincent J‐L, Sakr Y, Sprung CL, et al. Sepsis in European intensive care units: results of the SOAP study. Crit Care Med. 2006;34(2):344‐353. [DOI] [PubMed] [Google Scholar]
- 4. Dellinger RP, Levy MM, Rhodes A, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 2013;39(2):165‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vincent JL, Bihari DJ, Suter PM, et al. The prevalence of nosocomial infection in intensive care units in Europe. Results of the European Prevalence of Infection in Intensive Care (EPIC) Study. EPIC international advisory committee. JAMA. 1995;274(8):639‐644. [PubMed] [Google Scholar]
- 6. Iwashyna TJ, Ely EW, Smith DM, Langa KM. Long‐term cognitive impairment and functional disability among survivors of severe sepsis. JAMA. 2010;304(16):1787‐1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shah FA, Pike F, Alvarez K, et al. Bidirectional relationship between cognitive function and pneumonia. Am J Respir Crit Care Med. 2013;188(5):586‐592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yende S, Linde‐Zwirble W, Mayr F, Weissfeld LA, Reis S, Angus DC. Risk of cardiovascular events in survivors of severe sepsis. Am J Respir Crit Care Med. 2014;189(9):1065‐1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mayr FB, Talisa VB, Balakumar V, Chang C‐C, Fine M, Yende S. Proportion and cost of unplanned 30‐day readmissions after sepsis compared with other medical conditions. JAMA. 2017;317(5):530‐531. [DOI] [PubMed] [Google Scholar]
- 10. Mercer TR, Dinger ME, Mattick JS. Long non‐coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155‐159. [DOI] [PubMed] [Google Scholar]
- 11. Degirmenci U, Li J, Lim YC, et al. Silencing an insulin‐induced lncRNA, LncASIR, impairs the transcriptional response to insulin signalling in adipocytes. Sci Rep. 2019;9(1):5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li H, Han S, Sun Q, et al. Long non‐coding RNA CDKN2B‐AS1 reduces inflammatory response and promotes cholesterol efflux in atherosclerosis by inhibiting ADAM10 expression. Aging (Albany NY). 2019;11(6):1695‐1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun Y, Zhong L, He X, et al. LncRNA H19 promotes vascular inflammation and abdominal aortic aneurysm formation by functioning as a competing endogenous RNA. J Mol Cell Cardiol. 2019;131:66‐81. [DOI] [PubMed] [Google Scholar]
- 14. Huang M, Wang H, Hu X, Cao X. lncRNA MALAT1 binds chromatin remodeling subunit BRG1 to epigenetically promote inflammation‐related hepatocellular carcinoma progression. Oncoimmunology. 2019;8(1):e1518628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li G‐Q, Fang Y‐X, Liu Y, et al. MALAT1‐driven inhibition of Wnt signal impedes proliferation and inflammation in fibroblast‐like synoviocytes through CTNNB1 promoter methylation in rheumatoid arthritis. Hum Gene Ther. 2019. 10.1089/hum.2018.212. [DOI] [PubMed] [Google Scholar]
- 16. Yu Z, Rayile A, Zhang X, Li Y, Zhao Q. Ulinastatin protects against lipopolysaccharide‐induced cardiac microvascular endothelial cell dysfunction via downregulation of lncRNA MALAT1 and EZH2 in sepsis. Int J Mol Med. 2017;39(5):1269‐1276. [DOI] [PubMed] [Google Scholar]
- 17. Chen H, Wang X, Yan X, et al. LncRNA MALAT1 regulates sepsis‐induced cardiac inflammation and dysfunction via interaction with miR‐125b and p38 MAPK/NFκB. Int Immunopharmacol. 2018;55:69‐76. [DOI] [PubMed] [Google Scholar]
- 18. Bloos F, Reinhart K. Rapid diagnosis of sepsis. Virulence. 2014;5(1):154‐160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pierrakos C, Vincent JL. Sepsis biomarkers: a review. Crit Care. 2010;14(1):R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sankar V, Webster NR. Clinical application of sepsis biomarkers. J Anesth. 2013;27(2):269‐283. [DOI] [PubMed] [Google Scholar]
- 21. Zhang T‐N, Li DA, Xia J, et al. Non‐coding RNA: a potential biomarker and therapeutic target for sepsis. Oncotarget. 2017;8(53):91765‐91778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu X, Hong C, Wu S, et al. Downregulation of lncRNA TUG1 contributes to the development of sepsis‐associated acute kidney injury via regulating miR‐142‐3p/sirtuin 1 axis and modulating NF‐kappaB pathway. J Cell Biochem. 2019; 120(7). [DOI] [PubMed] [Google Scholar]
- 23. Gordon AD, Biswas S, Feng B, Chakrabarti S. MALAT1: a regulator of inflammatory cytokines in diabetic complications. Endocrinol Diabetes Metab. 2018;1(2):e00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cui H, Banerjee S, Guo S, et al. Long noncoding RNA Malat1 regulates differential activation of macrophages and response to lung injury. JCI Insight. 2019;4(4):124522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin LP, Niu GH, Zhang XQ. Influence of lncRNA MALAT1 on septic lung injury in mice through p38 MAPK/p65 NF‐kappaB pathway. Eur Rev Med Pharmacol Sci. 2019;23(3):1296‐1304. [DOI] [PubMed] [Google Scholar]
- 26. Dai L, Zhang G, Cheng Z, et al. Knockdown of LncRNA MALAT1 contributes to the suppression of inflammatory responses by up‐regulating miR‐146a in LPS‐induced acute lung injury. Connect Tissue Res. 2018;59(6):581‐592. [DOI] [PubMed] [Google Scholar]
- 27. Tian H, Wu M, Zhou P, Huang C, Ye C, Wang LI. The long non‐coding RNA MALAT1 is increased in renal ischemia‐reperfusion injury and inhibits hypoxia‐induced inflammation. Ren Fail. 2018;40(1):527‐533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hutchinson JN, Ensminger AW, Clemson CM, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007;8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dahariya S, Paddibhatla I, Kumar S, et al. Long non‐coding RNA: classification, biogenesis and functions in blood cells. Mol Immunol. 2019;112:82‐92 [DOI] [PubMed] [Google Scholar]
- 30. Jia J, Wang J, Zhang J, et al. MiR‐125b Inhibits LPS‐Induced inflammatory injury via targeting MIP‐1alpha in chondrogenic cell ATDC5. Cell Physiol Biochem. 2018;45(6):2305‐2316. [DOI] [PubMed] [Google Scholar]
- 31. Dai J, Xu LJ, Han GD, et al. MicroRNA‐125b promotes the regeneration and repair of spinal cord injury through regulation of JAK/STAT pathway. Eur Rev Med Pharmacol Sci. 2018;22(3):582‐589. [DOI] [PubMed] [Google Scholar]
- 32. Liu P, Jia S‐B, Shi J‐M, et al. LncRNA‐MALAT1 promotes neovascularization in diabetic retinopathy through regulating miR‐125b/VE‐cadherin axis. Biosci Rep. 2019;39(5):BSR20181469. [DOI] [PMC free article] [PubMed] [Google Scholar]


