Abstract
Background
To study the expression of microribonucleic acid (miR)‐205 in breast cancer and its effects on the proliferation and apoptosis of breast cancer cells.
Methods
Breast cancer cell line MCF‐7 cells with stable expression of miR‐205‐3p were constructed. Cell proliferation, invasion, and apoptosis were detected via MTT assay, transwell assay, and flow cytometry, respectively. The expressions of Ezrin, LaminA/C, cleaved caspase‐3, Bcl‐2, and Bax were detected via Western blotting. The expressions of miR‐205‐3p in breast cancer tissues and para‐carcinoma tissues were detected via quantitative PCR (qPCR).
Results
In transfection group, cell proliferation and invasion capacities were increased significantly (P < 0.01), but apoptotic cells were significantly reduced (P < 0.01). In addition, the expressions of Ezrin, LaminA/C, and cleaved caspase‐3 in the transfection group were significantly decreased (P < 0.01), but the Bcl‐2/Bax ratio was significantly increased (P < 0.01). The miR‐205‐3p expression in tumor tissues of breast cancer patients was significantly higher than that in para‐carcinoma tissue, but Ezrin, LaminA/C, and cleaved caspase‐3 expressions in tumor tissues were remarkably declined (P < 0.01), while the Bcl‐2/Bax ratio was remarkably increased (P < 0.01). Moreover, the 5‐year survival of patients with high expression of miR‐205‐3p was significantly shorter than patients with normal or low expression (P < 0.01).
Conclusion
Highly expressed miR‐205‐3p can promote the proliferation and invasion and reduce the apoptosis of breast cancer cells, and the high expression of miR‐205‐3p can significantly reduce the survival time of patients.
Keywords: apoptosis, breast cancer, miR‐205‐3p, prognosis, proliferation
1. INTRODUCTION
Both morbidity and mortality rates of breast cancer, as one of the common malignant tumors in women, are ranked 1st in a variety of female tumors, seriously threatening the female health1 The number of breast cancer patients diagnosed and treated each year is not large in China compared with that in Western developed countries, but the incidence rate of breast cancer has displayed an increasing trend in the last 2 decades. In particular, breast cancer has become the malignant tumor with the highest incidence rate in women in big cities2 A variety of stress responses in cells will be produced when various external stimuli (various carcinogenic factors) act on breast epithelial cells, resulting in cell proliferation, thus leading to breast cancer3 At present, surgical operation is dominated in the clinical treatment approaches of breast cancer, but the range of surgical resection in clinic is wide, the trauma is large, and a lot of related complications often occur in patients after clinical surgical treatment, causing serious physiological and psychological damage to patients4.
A large number of studies in recent years have revealed that various specific gene mutations were identified in tumor patients and several microribonucleic acids (miRNAs) are closely related to the tumor metastasis and invasion5, 6 Osipov et al7 found high expression of miR‐205 in epithelial ovarian cancer cells, indicating that miR‐205 is possibly associated with the incidence of ovarian cancer. There is a great amount of research evidence proving that the miR‐205 expressions in a variety of tumors are lower than that in normal tissues8, 9, 10, 11 miRNAs are therefore applied in the diagnosis of cancer12, 13 For instance, miR‐205 presents as the potential biomarkers for early prognosis or diagnosis while its level is proposed in predicting lymph node metastasis in triple‐negative breast cancer patients14, 15 Moreover, it has been demonstrated that miR‐205/ASPP2 axis promoted cell migration and also increased cell proliferation, which may be potential diagnostic and therapeutic biomarkers in cervical and lung cancers16 miR‐205 enhances chemosensitivity of breast cancer cells to TAC chemotherapy by suppressing both VEGFA and FGF2, leading to evasion of apoptosis17 However, the exact role of miR‐205‐3p in the pathogenesis and development of breast cancer remains poorly understood. The present study aims to investigate the role and possible mechanism of miR‐205‐3p in breast cancer, in order to provide a theoretical basis for further elaborating the effect of miR‐205‐3p in the course of breast cancer.
2. PATIENTS AND METHODS
2.1. Patients and grouping
A total of 58 patients who were pathologically diagnosed with breast cancer, treated, and underwent mastectomy in Guangdong Provincial People's Hospital from June 2011 to June 2013 were collected. None of the patients received chemotherapy and radiotherapy before operation. The para‐carcinoma tissues of breast cancer (more than 5 cm away from the breast cancer tissues) were taken during operation as the control. All enrolled patients were females aged 42‐69 years with a median age of 56.2 years, and they all suffered from invasive ductal carcinoma. According to the tumor‐node‐metastasis (TNM) staging criteria issued by the International Union Against Cancer, patients were staged, and there were 8 cases in stage I, 41 cases in stage II, 6 cases in stage III, and 3 cases in stage IV. Inclusion criteria were as follows: (1) patients without severe hepatic and renal function impairment, (2) patients who agreed to be followed up, (3) patients without a history of serious cardiovascular and immune system diseases, and (4) patients without chronic or acute infectious diseases. All enrolled patients personally signed the informed consent and had complete clinical and pathological data and complete therapeutic regimens. The experimental scheme was reviewed and approved ethically by our hospital.
2.2. Detection of miR‐205‐3p expression level in breast cancer tissues
After breast cancer tissues and para‐carcinoma tissues were obtained, they were added into the tube (100 mg/mL) together with TRIzol, and smashed using an ultrasonic disruptor followed by being incubated on ice for 10 minutes, followed by centrifugation at 12 000 g and 4°C for 10 minutes. 700 μL supernatant was taken, added with chloroform, and manually vibrated for 10 times, followed by incubation on ice for 10 minutes and centrifugation at 12 000 g and 4°C for 10 minutes. After the supernatant was discarded, 1 mL freshly prepared 75% ethanol was added into each tube, followed by centrifugation at 12 000 g and 4°C for 15 minutes. The ethanol was discarded, the cap of the centrifuge tube was removed for natural air‐drying, and 30 μL diethyl pyrocarbonate (DEPC)‐treated water was added to dissolve the RNA. The optical density (OD) value and purity of RNA were measured. The reaction system was prepared strictly according to instructions of the reverse transcription kit (Invitrogen): 5 μg total RNA, 1 μL Oligo(dT)20, 1 μL 10 mmol/L dNTP, and 3 μL 0.1mol/L DTT were added, and DEPC‐treated water was also added until the total volume was 20 μL. After reaction at 60°C for 15 minutes and incubation at 37°C for 2 minutes, the reverse transcription was performed. After RT‐qPCR system was prepared, the amplification was performed on the RT‐qPCR instrument. Amplification conditions are as follows: pre‐denaturation at 92°C for 2 minutes, denaturation at 95°C for 20 seconds, annealing at 60°C for 40 seconds, extension at 72°C for 2 minutes, and a total of 35 cycles. Primers were synthesized by Invitrogen, and RNU6B was used as the internal control. The primer sequence was as follows: 5′‐CGG GAT TTC AGT GGA GTG AAG TTC‐3′ (miR‐205‐3p); 5′‐CTC GCT TCG GCA GCA CA‐3′ (RNU6B, sense) and 5′‐AAC GCT TCA CGA ATT TGC GT‐3′ (RNU6B, antisense). The expression of miR‐205‐3p was quantified as a relative expression to RNU6B using 2−△△CT method. All RT‐qPCR experiments were conducted according to the MIQE (minimum information for publication of quantitative real‐time PCR experiments) guidelines18 Each amplification reaction was performed in triplicate, and mean value of the three‐cycle threshold was used for further analysis. Based on the median expression of miR‐205‐3p, patients were further divided into high and low expression group.
2.3. Survival curve analysis
The miR‐205‐3p expression in cancer tissues of each patient with breast cancer was detected and compared with that in para‐carcinoma tissue, and divided into normal or low expression group and high expression group. Data of patients were recorded in detail, and patients were followed up for 5 years. The 5‐year survival rate of patients was recorded, and the survival curve was drawn. Influencing factors of patient's prognosis in both groups were analyzed via Cox multivariate regression analysis.
2.4. Cell culture and construction of cell lines with miR‐205‐3p overexpression
Human breast cancer MCF‐7 cell lines were purchased from the Kunming Cell Bank of the Chinese Academy of Sciences. After resuscitation, cells were cultured using Dulbecco's modified Eagle medium (DMEM) (Gibco) containing 10% fetal bovine serum (FBS) in an incubator with 5% CO2 at 37°C till the logarithmic growth phase. Then, cells were digested with trypsin and continued to be cultured, followed by passage till the 3rd generation used for the experiment. The gene sequence (5′‐GAUUUCAGUGGAGUGAAGUUC‐3′) of hsa‐miR‐205‐3p was found in the Sanger microRNA database, and primers were synthesized by Invitrogen: forward primer: 5′‐GAGGATCCCCGGGTACCGGTAGGCCTTT‐3′ and reverse primer: 5′‐CACACATTCCACAGGCTGCTACGGTGGTGGCGGCGGGCGGT‐3′. After amplification and electrophoretic separation using agarose gel, the target band was recycled. The GV369 vector (Shanghai GeneChem Co., Ltd.) was cut via double digestion, and the target fragment (5′‐GAUUUCAGUGGAGUGAAGUUC‐3′) was connected to the vector using T4 ligase (Takara), followed by plate clone using LB solid medium at 37°C and sequencing identification using positive colonies. The miR‐205‐3p lentivirus was packaged with the plasmid (TransGen), the titer was detected, and the miR‐205‐3p lentivirus was used to infect MCF‐7 cells after meeting requirements: After digestion, MCF‐7 cells were inoculated into a 24‐well plate at a density of 5 × 104/well. After culturing for 24 hours, 440 μL DMEM, 10 μL hsa‐miR‐205‐3p, and 50 μL polybrene were added into the centrifuge tube, mixed evenly, and inoculated into the plate. After culturing for 12 hours, the original medium was replaced with DMEM containing 10% FBS for culture for another 72 hours. Then, the fluorescence intensity of cells was observed under a microscope. After cells were screened using puromycin (Sigma) and cloned, the MCF‐7 cell lines with stable infection were constructed. Overexpression of miR‐205‐3p was then confirmed by real‐time quantitative polymerase chain reaction (RT‐qPCR) using RNU6B as an internal control.
2.5. Detection of cell proliferation and invasion capacities
Cell proliferation capacity was detected via methyl thiazolyl tetrazolium (MTT) assay: After cells in the logarithmic growth phase were digested, the cell density was adjusted to 1 × 104/well, and cells were inoculated into a 96‐well plate and divided into blank control group (MCF‐7 cells without any treatment), negative control group (MCF‐7cells inoculated with empty vector), and hsa‐miR‐205‐3p group (MCF‐7 cells inoculated with hsa‐miR‐205‐3p), followed by culture in an incubator for 24 hours. Then, 100 μL MTT was added to culture cells for another 4 hours, the supernatant was discarded, and 150 μL dimethyl sulfoxide (DMSO) was added into each well and vibrated on a shaking table for 1 minute. Finally, cell proliferation in each group was detected using a microplate reader.
Cell invasion capacity was detected via transwell assay: After cells in the logarithmic growth phase were digested, the cell density was adjusted to 1.5 × 106/well, and the cell suspension was prepared and divided into blank control group (MCF‐7 cells without any treatment), negative control group (MCF‐7 cells inoculated with empty vector), and hsa‐miR‐205‐3p group (MCF‐7 cells inoculated with hsa‐miR‐205‐3p). 300 μL cell suspension was added into the upper side of polycarbonate transwell filter (Corning) containing 50 μL mixture of Matrigel (BD Biosciences) and DMEM (1:4), while 700 μL DMEM containing 10% FBS was added into the lower chamber, followed by culture in the incubator for 24 hours. After being removed from the chamber, cells were fixed with methanol and stained with crystal violet. The cell invasion was observed under the microscope, and the number of cells passing through the membrane was calculated.
2.6. Detection of apoptosis
Apoptosis was detected via flow cytometry using FITC Annexin V Apoptosis Detection Kit (BD Bioscience): After cells in the logarithmic growth phase were digested, the cell density was adjusted to 2 × 106/well, and the cell suspension was prepared and divided into blank control group (MCF‐7 cells without any treatment), negative control group (MCF‐7 cells inoculated with empty vector), and hsa‐miR‐205‐3p group (MCF‐7 cells inoculated with hsa‐miR‐205‐3p). Then, the cell suspension was inoculated into a 6‐well plate and added with serum‐free DMEM, followed by culture in the incubator for 24 hours. After the culture solution was discarded, cells were washed with phosphate‐buffered saline (PBS), scraped off, centrifuged, and resuspended with PBS for 3 times strictly according to instructions of the apoptosis kit (BD Bioscience). The fluorescence maker was added and cells were incubated in a dark place at room temperature for 15 minutes, followed by sample loading and detection via flow cytometry. All samples should be detected within 1 hour to ensure the effect.
2.7. Detection of expression levels of related proteins
Cells in each group were cultured in the 6‐well plate, collected, followed by addition of radioimmunoprecipitation assay (RIPA) lysis solution (Wuhan Servicebio) and 1% protease inhibitor (Wuhan Servicebio) into each well (500 μL/well). After breast cancer tissues and para‐carcinoma tissues were obtained, they were added with RIPA lysis solution and 1% protease inhibitor (100 mg/mL) and smashed using the ultrasonic disruptor. The above cell suspension and tissue homogenate were incubated on ice, followed by centrifugation at 1000 g and 4°C for 10 minutes to obtain the total protein solution. The total protein concentration in each sample was detected using the BCA protein assay kit (PIERCE). After the protein loading buffer in an equal concentration was prepared, 10% gel was prepared and added with an equal amount of loading buffer, followed by sodium dodecyl sulfate polyacrylamide gel electrophoresis under constant pressure. Then, the protein was transferred onto a PVDF membrane (Millipore), and the target band was cut and sealed with freshly prepared 5% skim milk powder for 2 hours. Ezrin, LaminA/C, cleaved caspase‐3, Bcl‐2, Bax, and glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) antibodies (purchased from CST, USA, and prepared at a volume ratio of 1:1000) were incubated at 4°C overnight. After the band was washed with PBS with Tween‐20 (PBST) for 3 times (5 minutes per time), it was incubated with the horseradish peroxidase‐conjugated secondary antibody (1:5000, Shanghai YIHYSON Biotechnology Co., Ltd.) at room temperature for 1 hour, and then the membrane was washed with Tris‐buffered saline with Tween‐20 (TBST), followed by image development in a dark room: Enhanced chemiluminescence (ECL) solution was added, and images were obtained using the fluorescence imaging technique. The expression level of each protein was presented as Ezrin/GAPDH, LaminA/C/GAPDH, cleaved caspase‐3/GAPDH, and Bcl‐2/Bax, respectively.
2.8. Statistical analysis
In this study, three independent experiments were performed for in vitro assay. Data were presented as mean ± SD and processed using Statistical Product and Service Solutions (SPSS) 19.0 software (SPSS Inc.). Paired Student's t test was used for the comparison between two groups, chi‐square test was used for enumeration data, and analysis of variance was used for the comparison among groups. The homogeneity test of variance was performed, and Bonferroni's method was used for the pairwise comparison in case of homogeneity of variance, while Welch's method was used in case of heterogeneity of variance. The Pearson correlation analysis was adopted for correlation analysis. Cox proportional hazards model was performed for multivariate analysis of the independent factors for the survival of breast cancer patients. P < 0.05 suggested that the difference was statistically significant.
3. RESULTS
3.1. miR‐205‐3p expression in breast cancer tissues
The miR‐205‐3p expressions in tumor tissues and para‐carcinoma tissues of breast cancer patients were detected via RT‐qPCR. According to results, the miR‐205‐3p expression in tumor tissues of breast cancer patients was increased significantly compared with that in para‐carcinoma tissues (1.87 ± 0.63 vs 1, P < 0.01) (Figure 1).
Figure 1.
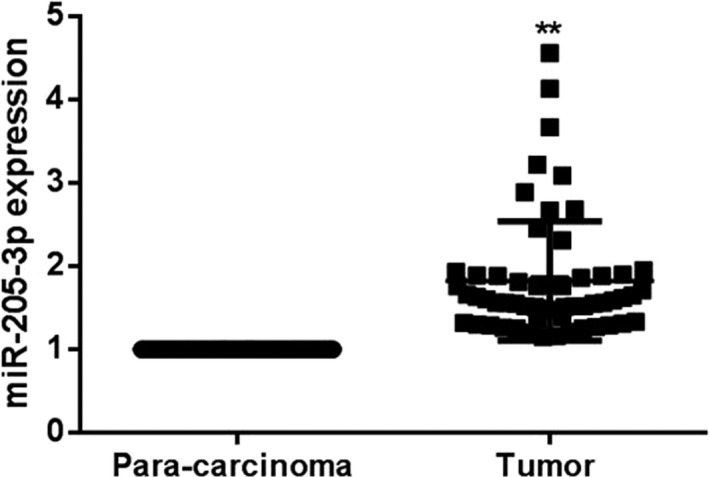
Detection of miR‐205‐3p expressions in tumor tissues and para‐carcinoma tissues in each group via RT‐qPCR. The miR‐205‐3p expression in tumor tissues of breast cancer patients is obviously higher than that in para‐carcinoma tissues, ** P < 0.01
3.2. Related protein expressions in breast cancer tissues
Ezrin, LaminA/C, cleaved caspase‐3, Bcl‐2, and Bax protein expressions in tumor tissues and para‐carcinoma tissues of breast cancer patients were detected via Western blotting. Results manifested that compared with those in para‐carcinoma tissues, Ezrin, LaminA/C, and cleaved caspase‐3 expressions were significantly declined in tumor tissues (P < 0.01), but the Bcl‐2/Bax ratio was increased significantly (P < 0.01) (Figure 2).
Figure 2.
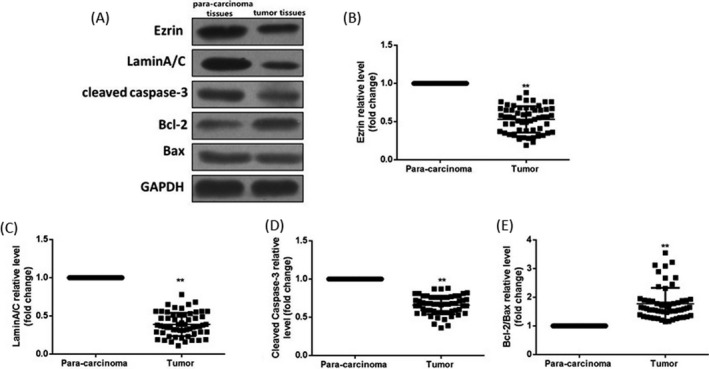
Detection of related protein expressions in breast cancer tissues and para‐carcinoma tissues via Western blotting. (A) Protein bands. (B) Statistical graph of Ezrin protein expression level. (C) Statistical graph of LaminA/C protein expression level. (D) Statistical graph of cleaved caspase‐3 protein expression level. (E) Statistical graph of Bcl‐2/Bax protein expression level. Compared with those in para‐carcinoma tissues, Ezrin, LaminA/C, and cleaved caspase‐3 expressions are remarkably lower in breast cancer tissues, but the Bcl‐2/Bax ratio is higher. Data were presented as a fold change relative to GAPDH (mean ± SD). ** P < 0.01
3.3. Correlations of miR‐205‐3p with clinicopathological features of breast cancer patients
The miR‐205‐3p level was classified as high or low in relation to the median value (cutoff value = 1.704). The above patients were accordingly divided into low or normal expression group and high expression group based on the miR‐205‐3p expression level in breast cancer tissues, with para‐carcinoma tissues as the control. The clinicopathological data of patients in each group were recorded in details, and correlations of miR‐205‐3p expression level with clinicopathological data of patients were analyzed. Results showed that miR‐205‐3p was not related to the age and menstrual condition of breast cancer patients, but statistically closely related to the size, TNM staging, lymph node metastasis, and recurrence of breast cancer (P < 0.05) (Table 1). Further univariate and multivariate analysis showed that miR‐205‐3p expression was an independent factor for the survival (recurrence‐free survival) of breast cancer patients (Table 2).
Table 1.
Clinicopathological data of breast cancer patients
| Groups | n | miR‐205‐3p expression level | χ2 | df | P value | Contingency coefficient | |
|---|---|---|---|---|---|---|---|
| Low or normal expression | High expression | ||||||
| Age | |||||||
| ≥50 y old | 46 | 25 | 21 | 0.0611 | 1 | 0.8047 | 0.2472 |
| <50 y old | 12 | 7 | 5 | ||||
| Menstrual condition | |||||||
| Premenopause | 38 | 20 | 18 | 0.2877 | 1 | 0.5917 | 0.5363 |
| Postmenopause | 20 | 12 | 8 | ||||
| Tumor size | |||||||
| ≥2 cm | 16 | 6 | 10 | 13.48 | 1 | 0.0002 | 3.672 |
| <2 cm | 42 | 36 | 6 | ||||
| TNM staging | |||||||
| Stages I and II | 49 | 32 | 17 | 9.106 | 1 | 0.0025 | 3.108 |
| Stages III and IV | 9 | 1 | 8 | ||||
| Lymph node metastasis | |||||||
| Yes | 22 | 7 | 15 | 5.625 | 1 | 0.0177 | 2.372 |
| No | 36 | 23 | 13 | ||||
| Recurrence | |||||||
| Yes | 25 | 7 | 18 | 8.507 | 1 | 0.0035 | 2.917 |
| No | 33 | 22 | 11 | ||||
Table 2.
Multivariate analysis of clinicopathological factors for overall survival (Cox proportional hazards model)
| Factors | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age (≥50 y) | 1.08 (0.43‐2.61) | 0.796 | / | / |
| Gender | 0.53 (0.21‐1.63) | 0.195 | / | / |
| Tumor size (≥2 cm) | 6.90 (2.36‐38.2) | 0.006 | 10.6 (0.51‐567) | 0.092 |
| TNM staging (III‐IV/I‐II) | 1.65 (1.35‐1.93) | 0.021 | 2.76 (0.05‐236) | 0.693 |
| Lymph node metastasis (yes/no) | 1.03 (1.01‐1.21) | 0.031 | 1.16 (0.15‐25.6) | 0.961 |
| miR‐205‐3p expression (high/low) | 26.7 (18.3‐45.7) | <0.0001 | 16.5 (2.61‐359) | 0.002 |
3.4. Correlation of miR‐205‐3p with prognosis of breast cancer patients
All breast cancer patients enrolled were followed up for 5 years, the survival rate of patients was recorded, and the survival curve was drawn. According to results, the 5‐year survival rate of breast cancer patients with low or normal expression of miR‐205‐3p was significantly higher than that in patients with high expression of miR‐205‐3p (P < 0.01) (Figure 3).
Figure 3.
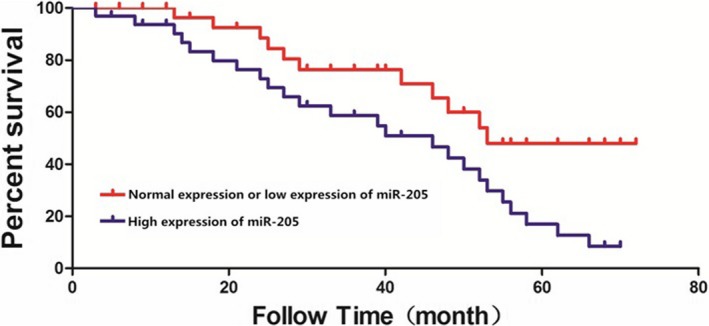
5‐year survival curves of breast cancer patients. The 5‐year survival rate of patients with low or normal expression of miR‐205‐3p is significantly higher than that in patients with high expression of miR‐205‐3p
3.5. Construction of cell lines with miR‐205‐3p overexpression
After human breast cancer MCF‐7 cells were transfected with miR‐205‐3p lentivirus, the miR‐205‐3p expression in each group was detected via RT‐qPCR. Results showed that miR‐205‐3p expression levels in cells in blank control group and negative control group were significantly lower than that in hsa‐miR‐205‐3p group (P < 0.01), indicating successful construction of MCF‐7 cell lines with miR‐205‐3p overexpression (Figure 4).
Figure 4.
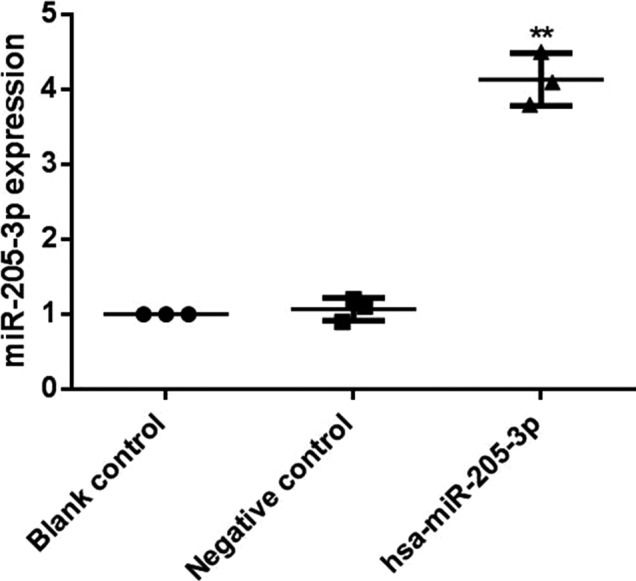
Detection of miR‐205‐3p expression after transfection by RT‐qPCR. After MCF‐7 cells were transfected with has‐miR‐205‐3p, total RNA was isolated for analysis of the expression of miR‐205‐3p by RT‐qPCR. The miR‐205‐3p expression level in cells in hsa‐miR‐205‐3p group is significantly higher than those in blank control group and negative control group, ** P < 0.01
3.6. Effects of miR‐205‐3p on proliferation, invasion, and apoptosis of breast cancer cells
Effects of miR‐205‐3p on proliferation, invasion, and apoptosis of MCF‐7 cells were detected via MTT assay, transwell assay, and flow cytometry, respectively. Results showed that both cell proliferation and invasion capacities in hsa‐miR‐205‐3p group were significantly higher than those in blank control group and negative control group (P < 0.01, P < 0.01). After transfection with miR‐205‐3p, the number of apoptotic cells in hsa‐miR‐205‐3p group was significantly decreased compared with those in the blank control group and negative control group (P < 0.01) (Figure 5).
Figure 5.
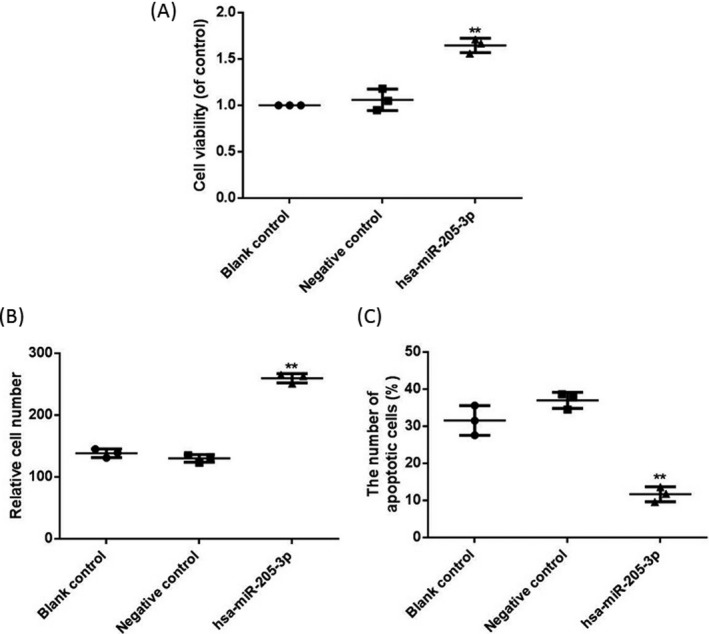
Effects of miR‐205‐3p on cell proliferation, invasion, and apoptosis. (A) Cell proliferation detected via MTT assay. (B) Cell invasion detected via transwell assay. (C) Apoptosis detected via flow cytometry. After transfection with miR‐205‐3p, both cell proliferation and invasion capacities in hsa‐miR‐205‐3p group are significantly higher than those in blank control group and negative control group, but the apoptosis level is obviously decreased, ** P < 0.01
3.7. Effects of miR‐205‐3p on related protein expressions
Ezrin, LaminA/C, cleaved caspase‐3, Bcl‐2, and Bax protein expressions in each group were detected via Western blotting. Results demonstrated that compared with those in the blank control group and negative control group, Ezrin, LaminA/C, and cleaved caspase‐3 expressions were significantly declined in hsa‐miR‐205‐3p group (P < 0.01), but the Bcl‐2/Bax ratio was increased significantly (P < 0.01) (Figure 6).
Figure 6.
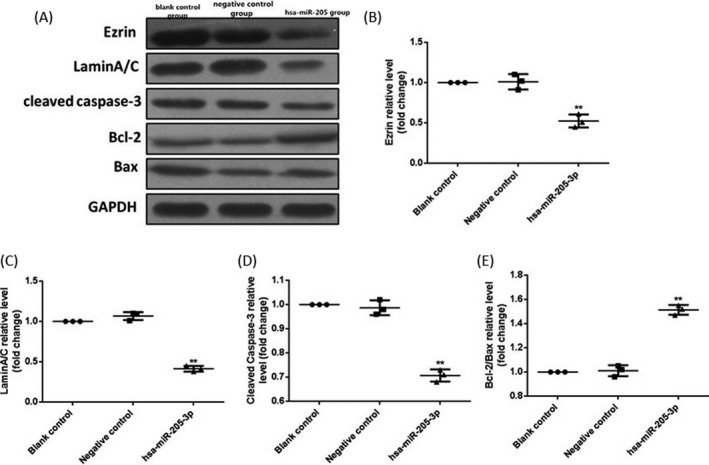
Detection of related protein expressions in MCF‐7 cells via Western blotting. (A) Protein bands. (B) Statistical graph of Ezrin protein expression level. (C) Statistical graph of LaminA/C protein expression level. (D) Statistical graph of cleaved caspase‐3 protein expression level. (E) Statistical graph of Bcl‐2/Bax protein expression level. Compared with those in the blank control group and negative control group, Ezrin, LaminA/C, and cleaved caspase‐3 expressions are remarkably lower in hsa‐miR‐205‐3p group, but the Bcl‐2/Bax ratio is higher. Data were presented as a fold change relative to GAPDH (mean ± SD). ** P < 0.01
4. DISCUSSION
Breast cancer ranks first in the common malignant tumors in women, the current incidence of which accounts for more than 10% of the systemic tumors and seriously affects the physical health of women19 Breast cancer occurs frequently in postmenopausal women mainly at the age of 45‐65 years. With the environmental deterioration and other factors, a younger trend of breast cancer occurrence has been shown in recent years20 The radical surgical resection, combined with radiotherapy and chemotherapy, marks the improvement regarding the survival time of early breast cancer patients, but its therapeutic effect is not satisfactory on advanced breast cancer patients21 miRNA, as a kind of endogenous small noncoding RNA, plays an important role in cell differentiation, proliferation, and apoptosis through regulating the signaling pathway. Accumulative evidences indicate that a variety of miRNAs are involved in the regulation process of several tumors and play similar roles to tumor suppressor genes and oncogenes22, 23 The abnormal expressions of miR‐21 and miR‐205 are possibly related to the occurrence and development of breast cancer12 Li et al24 found that the miR‐205 expression level is increased in epithelial ovarian cancer, and is closely related to the proliferation and invasion of ovarian cancer. In this study, we found that the level of miR‐205‐3p was significantly upregulated in breast cancer tissues, and further in vitro assay by overexpressing miR‐205‐3p validated its role on proliferation, invasion, and apoptosis of cancer cells, which was in line with previous findings24.
Our data showed that Ezrin and LaminA/C protein expressions in MCF‐7 cells with miR‐205‐3p overexpression were significantly decreased compared with those in normal cells, which was consistent with previous finding25 Ezrin protein is an important protein molecule in the ERM protein family, which, as a connexin between cytoskeleton and cell membrane, plays an important regulatory role in cell movement and adhesion, cytoskeletal remodeling, etc Ezrin protein also exerts important effects on the occurrence and metastasis of tumors26 It has been demonstrated that Ezrin functions as a prognostic factor and may predict potential lung metastasis in osteosarcoma and could also possibly be used as a new therapeutic target27 Ezrin is closely linked with the metastatic progression of cancer and is required in tumor‐induced angio/lymphangiogenesis in vivo, which is frequently abnormally expressed in aggressive cancer types28 Interestingly, there is some controversy in the literature regarding the role of Ezrin in cancer metastasis and invasion. It has been indicated that the overexpression of Ezrin might promote ovarian cancer cell invasion and might predict poor prognosis29 However, the study revealed that patients who had negative or low Ezrin expression had a significantly shorter survival than those whose tumors had moderate or high Ezrin expression,30 which is consistent with our finding that Ezrin expressions were significantly declined in breast cancer tissues. Also, it has been shown that radixin, as a member of the ERM (ezrin/radixin/moesin) protein family, along with moesin and ezrin, was reduced in lung adenocarcinoma, including early‐stage bronchioloalveolar carcinoma31 An intriguing implication of our finding suggests that ezrin may function as tumor suppressors in breast cancer, besides lung adenocarcinoma oncogenesis, and have a distinct function in tumor cell invasion. Analysis of caveolin‐1 expression in ovarian carcinoma cell lines demonstrated DNA methylation and histone deacetylation as a means to downregulate caveolin expression32 The similar mechanisms may reduce ezrin expression, in addition to genetic alterations associated with breast carcinogenesis.
Expression of A‐type lamins increases the risk of death from colorectal cancer because its presence gives rise to increased invasiveness but not affect cell proliferation33 By contrast, previous data indicate that low expression of lamin A/C is associated with an increased disease recurrence in stage II and III colon cancer patients34 Moreover, evidence unraveled that cultured breast cancer cell lines expressed less lamin A/C compared with noncancerous mammary gland cells, which is in line with our result that LaminA/C was significantly decreased in breast cancer tissues35 The loss of lamin A/C expression is associated with variables of poor prognosis and shorter outcome in patients with breast cancer36 The loss of A‐type lamins was shown to induce changes in nuclear distribution of telomeres and to affect telomere function, thereby inducing chromosomal instability37 The malfunction of A‐type lamins will increase chromosomal instability and stimulate carcinogenesis. Increasing genomic instability may enhance tumor progression, including generation of tumor cell clones that have the capacity to cause disease recurrence.
Mitochondrial pathway plays an important role in mediating the apoptosis process. Antiapoptotic protein Bcl‐2, proapoptotic protein Bax, and downstream cleaved caspase‐3 are involved in apoptosis cascade reaction and apoptosis of tumor cells38 Both in vitro and in vivo experiments in this study manifested that the increased miR‐205‐3p expression led to the significant decrease in the cleaved caspase‐3 expression, significant increase in the Bcl‐2/Bax ratio in cells, thus reducing the apoptosis level.
Wei et al39 studied and found that miR‐205 has close correlations with clinical progression and prognosis of ovarian cancer patients. In this study, correlations of miR‐205‐3p expression in tumor tissues with clinicopathological conditions and recurrence of breast cancer patients were analyzed. According to results, the miR‐205‐3p expression was related to the size, TNM staging, metastasis, and recurrence of tumor, and the 5‐year survival rate of patients with high expression of miR‐205‐3p was obviously lower than that of patients with normal or low expression. Moreover, miR‐205‐3p was identified as an independent factor for the survival of breast cancer patients. Our data demonstrate that correlation of miR‐205‐3p with prognosis of breast cancer patients was established in those patients that died because the breast cancer but no other causes and miR‐205‐3p may serve as an important gene for prognosis evaluation of breast cancer. The limitation in our study still exists that the controversial result regarding the changes in Ezrin, LaminA/C in tumor tissues, and the potential mechanisms remain to be further investigated. We propose that cancer cells exhibit a variety of features indicative of atypical nuclei, while the changes in these indicators may be further related to tumor's pathological classification and cancer progression.
In conclusion, significantly higher miR‐205‐3p expression is found in breast cancer tissues and associated with shorter survival of patients. Highly expressed miR‐205‐3p promotes the proliferation and invasion and reduces the apoptosis of breast cancer cells in vitro, suggesting therapeutic targeting miR‐205‐3p might be a novel approach for the treatment of breast cancer.
Qiu C, Huang F, Zhang Q, Chen W, Zhang H. miR‐205‐3p promotes proliferation and reduces apoptosis of breast cancer MCF‐7 cells and is associated with poor prognosis of breast cancer patients. J Clin Lab Anal. 2019;33:e22966 10.1002/jcla.22966
REFERENCES
- 1. Liu JC, Granieri L, Shrestha M, et al. Identification of CDC25 as a common therapeutic target for triple‐negative breast cancer. Cell Rep. 2018;23(1):112‐126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bambhroliya A, Van Wyhe RD, Kumar S, et al. Gene set analysis of post‐lactational mammary gland involution gene signatures in inflammatory and triple‐negative breast cancer. PLoS ONE. 2018;13(4):e0192689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang H, Xu HL, Wang YC, Lu ZY, Yu XF, Sui DY. 20(S)‐protopanaxadiol‐induced apoptosis in MCF‐7 breast cancer cell line through the Inhibition of PI3K/AKT/mTOR signaling pathway. Int J Mol Sci. 2018;19(4):E1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Evans DG, Howell SJ, Howell A. Personalized prevention in high risk individuals: Managing hormones and beyond. Breast. 2018;18(8):356‐372. [DOI] [PubMed] [Google Scholar]
- 5. Yunqi H, Fangrui Y, Yongyan Y, et al. MiR‐455 functions as a tumor suppressor through targeting GATA6 in colorectal cancer. Oncol Res. 2018;3(11):53‐62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen M, Xia Y, Tan Y, Jiang G, Jin H, Chen Y. Downregulation of microRNA‐370 in esophageal squamous‐cell carcinoma is associated with cancer progression and promotes cancer cell proliferation via upregulating PIN1. Gene. 2018;661(18):68‐77 [DOI] [PubMed] [Google Scholar]
- 7. Osipov ID, Zaporozhchenko IA, Bondar AA, Zaripov MM, Voytsitskiy VE, Vlassov VV. Cell‐free miRNA‐141 and miRNA‐205 as prostate cancer biomarkers. Adv Exp Med Biol. 2016;924(3):9‐12. [DOI] [PubMed] [Google Scholar]
- 8. Zhang H, Li B, Zhao H, Chang J. The expression and clinical significance of serum miR‐205‐3p for breast cancer and its role in detection of human cancers. Int J Clin Exp Med. 2015;8(2):3034‐3042. [PMC free article] [PubMed] [Google Scholar]
- 9. Mittal A, Chitkara D, Behrman SW, Mahato RI. Efficacy of gemcitabine conjugated and miRNA‐205 complexed micelles for treatment of advanced pancreatic cancer. Biomaterials. 2014;35(25):7077‐7087. [DOI] [PubMed] [Google Scholar]
- 10. Huo L, Wang Y, Gong Y, et al. MicroRNA expression profiling identifies decreased expression of miR‐205 in inflammatory breast cancer. Mod Pathol. 2016;29(4):330‐346. [DOI] [PubMed] [Google Scholar]
- 11. Asghari F, Haghnavaz N, Shanehbandi D, Khaze V, Baradaran B, Kazemi T. Differential altered expression of let‐7a and miR‐205 tumor‐suppressor miRNAs in different subtypes of breast cancer under treatment with Taxol. Adv Clin Exp Med. 2018;27(7):941‐945. [DOI] [PubMed] [Google Scholar]
- 12. Teoh SL, Das S. The role of microRNAs in diagnosis, prognosis, metastasis and resistant cases in breast cancer. Curr Pharm Des. 2017;23(12):1845‐1859. [DOI] [PubMed] [Google Scholar]
- 13. Chernyy V, Pustylnyak V, Kozlov V, Gulyaeva L. Increased expression of miR‐155 and miR‐222 is associated with lymph node positive status. J Cancer. 2018;9(1):135‐140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Adhami M, Haghdoost AA, Sadeghi B, Malekpour AR. Candidate miRNAs in human breast cancer biomarkers: a systematic review. Breast Cancer. 2018;25(2):198‐205. [DOI] [PubMed] [Google Scholar]
- 15. Berber U, Yilmaz I, Narli G, Haholu A, Kucukodaci Z, Demirel D. miR‐205 and miR‐200c: predictive micro RNAs for lymph node metastasis in triple negative breast cancer. J Breast Cancer. 2014;17(2):143‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X, Yu M, Zhao K, et al. Upregulation of MiR‐205 under hypoxia promotes epithelial‐mesenchymal transition by targeting ASPP2. Cell Death Dis. 2016;7(12):e2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu Y, Qiu Y, Yague E, Ji W, Liu J, Zhang J. miRNA‐205 targets VEGFA and FGF2 and regulates resistance to chemotherapeutics in breast cancer. Cell Death Dis. 2016;7(6):e2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum Information for publication of quantitative real‐time PCR experiments. Clin Chem. 2009;55:611‐622. [DOI] [PubMed] [Google Scholar]
- 19. Xu L, Du J, Wan C, et al. Ultrasound molecular imaging of breast cancer in MCF‐7 orthotopic mice using gold nanoshelled poly(lactic‐co‐glycolic acid) nanocapsules: a novel dual‐targeted ultrasound contrast agent. Int J Nanomedicine. 2018;21(13):1791‐1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Samanta SK, Lee J, Hahm ER, Singh SV. Peptidyl‐prolyl cis/trans isomerase Pin1 regulates withaferin A‐mediated cell cycle arrest in human breast cancer cells. Mol Carcinog. 2015;3(8):15‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee KM, Jung D, Hwang H, et al. Pre‐treatment anxiety is associated with persistent chemotherapy‐induced peripheral neuropathy in women treated with neoadjuvant chemotherapy for breast cancer. J Psychosom Res. 2018;108(9):14‐19. [DOI] [PubMed] [Google Scholar]
- 22. Jia X, Liu X, Li M, et al. Potential tumor suppressing role of microRNA‐545 in epithelial ovarian cancer. Oncol Lett. 2018;15(5):6386‐6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao JM, Huang LZ, Huang ZG, He RQ. Clinical value and potential pathways of miR‐183‐5p in bladder cancer: a study based on miRNA‐seq data and bioinformatics analysis. J Natl Cancer Inst. 2018;15(4):5056‐5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li M, Zhou Y, Xia T, et al. Circulating microRNAs from the miR‐106a‐363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res Treat. 2018;12(15):32‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li J, Li L, Li Z, et al. The role of miR‐205 in the VEGF‐mediated promotion of human ovarian cancer cell invasion. Gynecol Oncol. 2015;137(1):125‐133. [DOI] [PubMed] [Google Scholar]
- 26. Zacapala‐Gómez AE, Navarro‐Tito N, Alarcón‐Romero L, Ortuño‐Pineda C, Illades‐Aguiar B. Ezrin and E‐cadherin expression profile in cervical cytology: a prognostic marker for tumor progression in cervical cancer. BMC Cancer. 2018;18(1):349‐356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bulut G, Hong SH, Chen K, et al. Small molecule inhibitors of ezrin inhibit the invasive phenotype of osteosarcoma cells. Oncogene. 2012;31(3):269‐281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ghaffari A, Hoskin V, Szeto A, et al. A novel role for ezrin in breast cancer angio/lymphangiogenesis. Breast Cancer Res. 2014;16(5):438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kobel M, Gradhand E, Zeng K, et al. Ezrin promotes ovarian carcinoma cell invasion and its retained expression predicts poor prognosis in ovarian carcinoma. Int J Gynecol Pathol. 2006;25(2):121‐130. [DOI] [PubMed] [Google Scholar]
- 30. Moilanen J, Lassus H, Leminen A, Vaheri A, Butzow R, Carpen O. Ezrin immunoreactivity in relation to survival in serous ovarian carcinoma patients. Gynecol Oncol. 2003;90(2):273‐281. [DOI] [PubMed] [Google Scholar]
- 31. Tokunou M, Niki T, Saitoh Y, Imamura H, Sakamoto M, Hirohashi S. Altered expression of the ERM proteins in lung adenocarcinoma. Lab Invest. 2000;80(11):1643‐1650. [DOI] [PubMed] [Google Scholar]
- 32. Wiechen K, Diatchenko L, Agoulnik A, et al. Caveolin‐1 is down‐regulated in human ovarian carcinoma and acts as a candidate tumor suppressor gene. Am J Pathol. 2001;159(5):1635‐1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Willis ND, Cox TR, Rahman‐Casans SF, et al. Lamin A/C is a risk biomarker in colorectal cancer. PLoS ONE. 2008;3(8):e2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Belt EJ, Fijneman RJ, van den Berg EG, et al. Loss of lamin A/C expression in stage II and III colon cancer is associated with disease recurrence. Eur J Cancer. 2011;47(12):1837‐1845. [DOI] [PubMed] [Google Scholar]
- 35. Matsumoto A, Hieda M, Yokoyama Y, et al. Global loss of a nuclear lamina component, lamin A/C, and LINC complex components SUN1, SUN2, and nesprin‐2 in breast cancer. Cancer Med. 2015;4(10):1547‐1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alhudiri IM, Nolan CC, Ellis IO, et al. Expression of Lamin A/C in early‐stage breast cancer and its prognostic value. Breast Cancer Res Treat. 2019;174(3):661‐668. [DOI] [PubMed] [Google Scholar]
- 37. Gonzalez‐Suarez I, Redwood AB, Perkins SM, et al. Novel roles for A‐type lamins in telomere biology and the DNA damage response pathway. EMBO J. 2009;28(16):2414‐2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yin H, Zhu Q, Liu M, et al. GPER promotes tamoxifen‐resistance in ER+ breast cancer cells by reduced Bim proteins through MAPK/Erk‐TRIM2 signaling axis. Int J Oncol. 2017;51(4):1191‐1198. [DOI] [PubMed] [Google Scholar]
- 39. Wei J, Zhang L, Li J, et al. MicroRNA‐205 promotes cell invasion by repressing TCF21 in human ovarian cancer. J Ovarian Res. 2017;10(1):33‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]


