Abstract
Objective
Candida species is implicated in a wide array of clinical infections. Speciation of Candida strains is of prime importance in the epidemiological survey and laboratory diagnosis as there is an upswing of antifungal resistance and changing trends in the antifungal resistance pattern among C. albicans and non albicans Candida. Varied phenotypic methods are available for the identification of Candida species which vary in principles and cost factors. Chromogenic agar medium (HiCrome Candida differential agar) is one of the preferred phenotypic methods in limited resource laboratories. Hence, this study was aimed to assess the reliability of HiCrome Candida differential agar, M1297A (HiMedia) in the identification of Candida species compared polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP). Oral Candida isolates (n = 194) were inoculated onto HiCrome Candida differential agar and the potential of Candida differential Agar was compared with PCR–RFLP.
Results
The results were not in agreement with PCR–RFLP. Percentage of disagreement was 40.2, 50.0, 100.0 and 25.0 for Candida albicans, Candida krusei, Candida glabrata and Candida tropicalis respectively. PCR–RFLP demonstrated a very high discriminatory power in the identification of Candida species compared to agar.
Keywords: Candida, HiCrome Candida differential agar, PCR–RFLP
Introduction
Identification of Candida strains to the species level is increasingly necessary because of their variation both in their ability to cause infection and also in their susceptibility to antifungal agents. Species-level of yeast identification is mandatory for epidemiological purpose and laboratory diagnosis. A large variety of phenotypic methods for identification of Candida spp. are available which vary in principles and cost factors. As phenotypic methods demand more time and labour, a chromogenic substrate containing culture media have been used in research and clinical laboratories for the identification of Candida species [1]. The chromogenic media helps in identifying microbial colonies based on the colours produced due to chromogenic substrates that react with enzymes differentially secreted by microorganisms [1]. Molecular methods have high discriminatory power and hence more reliable for the identification of species [2]. This study aimed to evaluate the performance and reliability of HiCrome Candida differential Agar, M1297A (HiMedia, Mumbai, India) for the identification of Candida species. The potential of the chromogenic media was assessed by comparing with an economical, quick and consistent PCR–RFLP system.
Main text
Methods
A single colony of oral clinical Candida isolates (n = 194) from Sabouraud Dextrose Agar (SDA) (HiMedia, Mumbai, India) plate were inoculated onto HiCrome Candida differential agar and incubated at 37 °C aerobically for 24 h. The speciation of Candida isolates was based on the colony colour as per the manufacturer’s instructions (Table 1). Four to six isolates were inoculated per plate.
Table 1.
Disagreement in speciation of Candida isolates by HiCrome agar and PCR RFLP
| Candida species | Colour on HiCrome media as per manufacturer’s instruction/(no of isolates showing the respective colony colour) | PCR RFLP results consistent with HiCrome agar | PCR-RFLP disagreement with HiCrome agar |
|---|---|---|---|
| C. albicans | Light green (132) | 79 (59.8%) | 53 (40.2%) |
| C. krusei | Purple (38) | 19 (50%) | 19 (50.0%) |
| C. glabrata | Cream to white (6) | 0 (0%) | 6 (100%) |
| C. tropicalis | Blue to metallic blue (20) | 15 (75%) | 5 (25.0%) |
DNA was extracted from all the Candida isolates (n = 194) by boiling lysis method [3]. Briefly, a single colony from fresh culture of each Candida isolate on SDA plate was inoculated into 200 µl of sterile PCR grade water and incubated in a heat block (Rivotek, India) at 100 °C for 10 min. Following incubation the sterile PCR grade water containing the DNA was immediately cooled to − 20 °C for 10 min, then centrifuged at 10,000 rpm for 5 min. The supernatant collected was used for PCR assay. PCR targeting ITS1-5.8SrDNA-ITS2 region was performed for all the Candida isolates. The 25 µl reaction volume consisted of 10 pM of Candida-ITS-primers as described by Mohammadi et al. [4] ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′), 2.5 µl of 10× PCR buffer with MgCl2, 0.4 mM of dNTP mix, 1 unit of Taq polymerase, 2 µl of DNA template. The PCR amplification was performed in Veriti 96 Thermal Cycler (Applied Biosystems, USA) with initial denaturation at 94 °C for 3 min followed by 40 cycles at 94 °C for 20 s, 55 °C for 30 s and 72 °C for 45 s, and subsequently final extension at 72 °C for 5 min. The PCR amplicons were resolved along with DNA markers in 1% agarose with ethidium bromide (0.5 µg/ml) by gel electrophoresis for 25 min at 135 V using Mupid-exU system (Takara, Japan). The gel was analysed by BioGlow UV Transilluminators (Crystal Technology, USA). To speciate Candida isolates, 8.8 µl of each ITS PCR product was digested with 0.2 µl MspI (4U) restriction enzyme (New England Biolabs) along with 1 µl of 10× Enzyme Buffer [4]. The restriction digestion was carried out in Veriti 96 Thermal Cycler (Applied Biosystems, USA), by incubating the mix at 37 °C for 60 min followed by heat inactivation at 85 °C for 5 min. The ITS PCR–RFLP products were resolved by electrophoresis on 2% agarose gel with 0.5 µg/ml ethidium bromide and restriction patterns were documented and compared with in silico restriction pattern by pDRAW32 (V 1.1.140) using sequences from NCBI.
Results
Percent agreement was determined by the number of isolates positive by HiCrome Candida differential Agar/number of PCR–RFLP positive isolates × 100. Percent disagreement was derived by subtracting percent agreement from 100. Among the 194 candida isolates screened, 132 were identified as C. albicans, 36 as C. krusei, 6 as C. glabrata and 20 as C. tropicalis based on colour code on HiCrome agar (Fig. 1a, b). All the isolates were further identified genotypically by PCR–RFLP method. All Candida isolates identified as C. albicans based on colour (light green) by HiCrome agar were not in agreement with PCR–RFLP method as shown in Table 1. Similarly, the identification of three non albicans Candida species (C. krusei, C. glabrata and C. tropicalis) by colour code on HiCrome agar also showed a discrepancy with PCR–RFLP (Table 1).
Fig. 1.
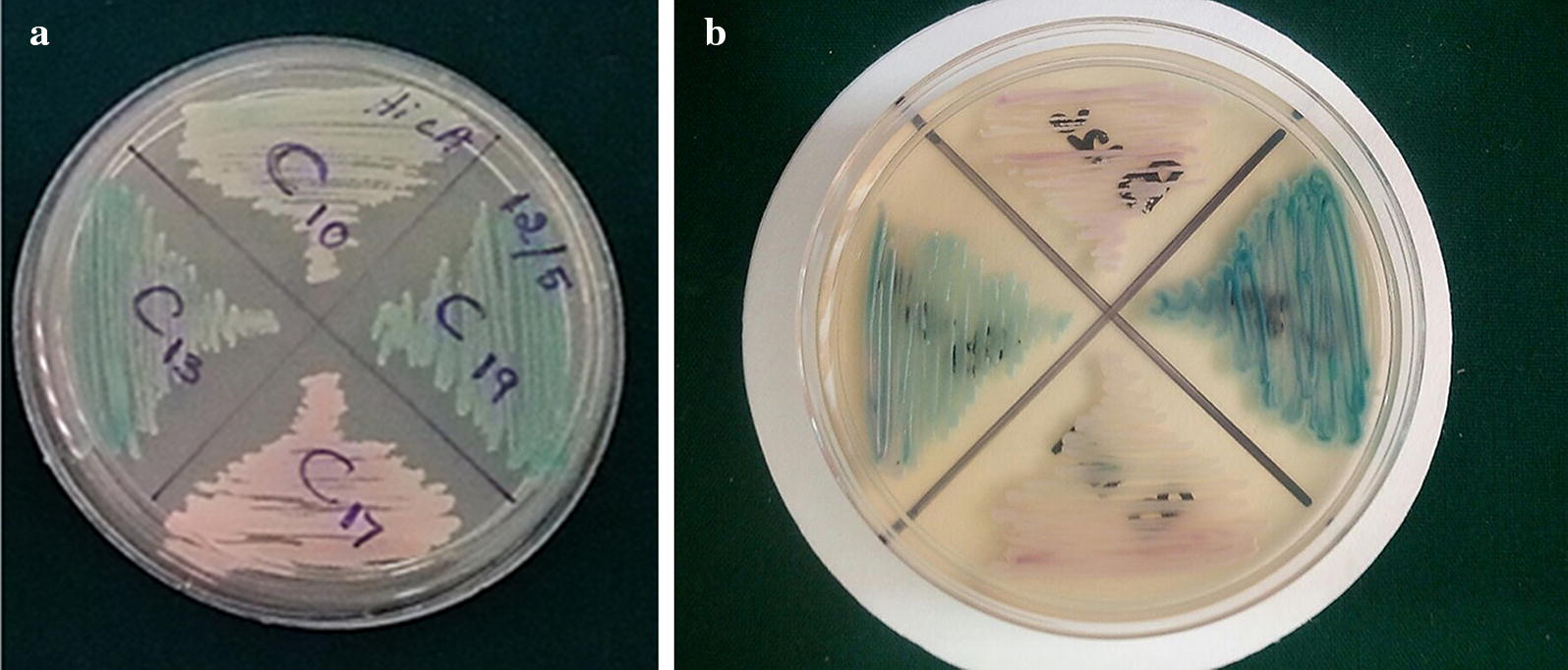
a, b HiCrome Agar plates showing different colours for identification of Candida species
Discussion
The results of the present study revealed that HiCrome Candida differential Agar method of speciation is unreliable compared to PCR–RFLP. The results of this study are not in concurrence with prior studies [5–8]. The colour codes mentioned by the manufacturers on HiCrome Candida differential agar for C. albicans, C. krusei, C. glabrata and C. tropicalis were also shown by other species. This may be due to the production of similar enzymes by different species of Candida. The enzyme–substrate reaction was not unique to each species of Candida. Similar colour was produced by more than one species and hence, chromogenic media was not able to identify the species as mentioned by the manufacturer’s instruction. Genotypic methods are potentially more sensitive and reliable means for the identification of yeasts. DNA amplification with universal fungal primers followed by detection using species-specific probes greatly enhances the sensitivity of Candida detection [9]. Time taken by PCR–RFLP is similar to routine phenotypic conventional methods [2] but then PCR–RFLP method is highly sensitive in identifying all species of Candida. The sensitivity of PCR–RFLP was found to be 100% compared to HiCrome Candida differential agar [4, 10]. To conclude, PCR–RFLP method is more reliable for identifying Candida species than HiCrome Candida differential agar even though it may be a preferred method in a resource—limited lab setting. Hence, a molecular technique with more discriminatory power and expeditious like PCR–RFLP can be strongly recommended in the identification of Candida species.
Limitations
Different types of chromogenic media were not compared in the present study.
All Candida species cannot be identified by chromogenic media.
Acknowledgements
The authors wish to acknowledge DST-FIST (Ref.No.SR/FST/College-23,2017) India, for providing research facilities.
Abbreviations
- PCR–RFLP
polymerase chain reaction-restriction fragment length polymorphism
- ITS
internal transcribed spacer
- NCBI
National Center for Biotechnology Information
Authors’ contributions
SLS and KM wrote the proposal, KM and SLS participated in data collection and analysis VN, SLS and KM participated in the analysis, KM, SLS and VN participated in drafting, writing, reviewing of the manuscript for publication. All authors read and approved the manuscript.
Funding
The work was supported by Bharath Institute of Higher Education and Research. Funding body had no role in the design of the study and collection, analysis, and interpretation of the data, or writing of the manuscript.
Availability of data and materials
The research data are available in the main document.
Ethics approval and consent to participate
Ethical approval was given by The Institutional Ethics Committee (Ethics committee DCGI Registration No. ECR/761/Inst/TN/2015), Sree Balaji Dental College and Hospital. IEC Approval No: SBDCH/IEC/03/2016/1.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sankar Leena Sankari, Email: drleena.sankari@gmail.com.
Krishnan Mahalakshmi, Email: kmagvenkat@gmail.com.
Venkatesan Naveen Kumar, Email: naveenkumar.nsd1527@gmail.com.
References
- 1.Bernal S, Mazuelos ME, Garcia M, Aller AI, Martinez MA, Gutierrez MJ. Evaluation of CHROMagar Candida medium for the isolation and presumptive identification of species of Candida of clinical importance. Diagn Microbiol Infect Dis. 1996;24(4):201–204. doi: 10.1016/0732-8893(96)00063-6. [DOI] [PubMed] [Google Scholar]
- 2.Fatima A, Bashir G, Wani T, Jan A, Kohli A, Khan MS. Molecular identification of Candida species isolated from cases of neonatal candidemia using polymerase chain reaction-restriction fragment length polymorphism in a tertiary care hospital. Indian J Pathol Microbiol. 2017;60:61–65. doi: 10.4103/0377-4929.200023. [DOI] [PubMed] [Google Scholar]
- 3.Silva GA, Bernardi TL, Schaker PD, Menegotto M, Valente P. Rapid yeast DNA extraction by boiling and freeze-thawing without using chemical reagents and DNA purification. Braz Arch Biol Technol. 2012;55(2):319–327. doi: 10.1590/S1516-89132012000200020. [DOI] [Google Scholar]
- 4.Mohammadi R, Mirhendi H, Rezaei-Matehkolaei A, Ghahri M, Shidfar MR, Jalalizand N, Makimura K. Molecular identification and distribution profile of Candida species isolated from Iranian patients. Med Mycol. 2013;51(6):657–663. doi: 10.3109/13693786.2013.770603. [DOI] [PubMed] [Google Scholar]
- 5.Juyal D, Sharma M, Pal S, Rathaur VK, Sharma N. Emergence of non albicans Candida species in neonatal candidemia. N Am J Med Sci. 2013;5:541–545. doi: 10.4103/1947-2714.118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vijayakumar R, Giri S, Kindo AJ. Molecular species identification of Candida from blood samples of intensive care unit patients by polymerase chain reaction-restricted fragment length polymorphism. J Lab Physicians. 2012;4:1–4. doi: 10.4103/0974-2727.98661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mousavi SA, Khalesi E, Bonjar GH, Aghighi S, Sharifi F, Aram F. Rapid molecular diagnosis for Candida species using PCR-RFLP. Biotechnology. 2007;6:583–587. doi: 10.3923/biotech.2007.583.587. [DOI] [Google Scholar]
- 8.Daef E, Moharram A, Eldin SS, Elsherbiny N, Mohammed M. Evaluation of chromogenic media and seminested PCR in the identification of Candida species. Braz J Microbiol. 2014;45(1):255–262. doi: 10.1590/S1517-83822014005000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wahyuningsih R, Freisleben HJ, Sonntag HG, Schnitzler P. Simple and rapid detection of Candida albicans DNA in serum by PCR for diagnosis of invasive candidiasis. J Clin Microbiol. 2000;38:3016–3021. doi: 10.1128/jcm.38.8.3016-3021.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altman DG, Bland JM. Diagnostic tests. 1: sensitivity and specificity. BMJ. 1994;308(6943):1552. doi: 10.1136/bmj.308.6943.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The research data are available in the main document.


