Abstract
Foodborne salmonellosis causes approximately 1 million illnesses annually in the United States. In the summer of 2017, we investigated four multistate outbreaks of Salmonella infections associated with Maradol papayas imported from four Mexican farms. PulseNet initially identified a cluster of Salmonella Kiambu infections in June 2017, and early interviews identified papayas as an exposure of interest. Investigators from Maryland, Virginia and Food and Drug Administration (FDA) collected papayas for testing. Several strains of Salmonella were isolated from papayas sourced from Mexican Farm A, including Salmonella Agona, Gaminara, Kiambu, Thompson and Senftenberg. Traceback from two points of service associated with illness sub-clusters in two states identified Farm A as a common source of papayas, and three voluntary recalls of Farm A papayas were issued. FDA sampling isolated four additional Salmonella strains from papayas sourced from Mexican Farms B, C and D. In total, four outbreaks were identified, resulting in 244 cases with illness onset dates from 20 December 2016 to 20 September 2017. The sampling of papayas and the collaborative work of investigative partners were instrumental in identifying the source of these outbreaks and preventing additional illnesses. Evaluating epidemiological, laboratory and traceback evidence together during investigations is critical to solving and stopping outbreaks.
Key words: Food-borne infections, outbreaks, salmonellosis
Introduction
Nontyphoidal salmonellosis is the most common bacterial cause of foodborne illness in the United States, leading to approximately 1 million domestically acquired foodborne illnesses, 19 000 hospitalisations and 380 deaths annually [1]. Most people infected with Salmonella develop diarrhoea, fever and abdominal cramps that usually lasts 4–7 days [2]. Young children, older adults and immunocompromised people are most likely to have severe infections [2].
Public health laboratories in the United States conduct serotyping and subtyping on Salmonella isolates by pulsed-field gel electrophoresis (PFGE). These PFGE patterns are uploaded to PulseNet, the national molecular subtyping network for foodborne disease surveillance, and are monitored to detect clusters of illness with indistinguishable PFGE patterns. Isolates with indistinguishable PFGE patterns are more likely to share a common source [3, 4].
The proportion of foodborne outbreaks, including Salmonella outbreaks, attributed to raw produce, such as fruits and vegetables, has been increasing in the United States since the 1970s [5–8]. Approximately half of the fresh fruit and about 20% of the fresh vegetables consumed in the United States are imported from other countries, most of which come from Mexico [9, 10]. There have been several multistate Salmonella outbreaks attributed to imported raw fresh produce over the past decade [11–18]. The implicated foods include cucumbers, mangoes, papayas, hot peppers and cantaloupe. Most of these foods were imported from Mexico, although two outbreaks were associated with produce imported from other countries in Central America [11–18].
After an outbreak of Salmonella infections in 2011 was linked to imported papayas from Mexico and a large proportion of import sampling identified Salmonella, the US Food and Drug Administration (FDA) issued a broad import alert to detain all whole, fresh papayas entering the United States from Mexico [12, 19]. To be removed from import alert status and supply Mexican papayas to the United States, firms need to provide documentation with sufficient evidence that future shipments of their papaya will not be adulterated; FDA may consider five consecutive commercial shipments over a period of time, analysed for Salmonella as prescribed by the FDA, as being adequate to be added to the ‘green list’ [19]. The 2011 import alert was still in effect when the outbreaks described below occurred; however, suppliers of Mexican papayas available in the United States at the time had been through the ‘green list’ process.
In this paper, we report the results of four multistate outbreaks of Salmonella infections linked to imported Maradol papayas from Mexico.
Methods
Outbreak detection
In June 2017, PulseNet detected a cluster of 13 Salmonella Kiambu infections with an indistinguishable PFGE XbaI restriction enzyme pattern. This pattern is the most common Salmonella Kiambu pattern in the PulseNet database. However, Salmonella Kiambu is a rare serotype, and the number of isolates uploaded to PulseNet in June 2017 was higher than expected based on data from previous years. State and local health departments and the Centers for Disease Control and Prevention (CDC) initiated a multistate investigation to determine the source of the outbreak and prevent additional illnesses.
Initial case definition and case finding
Additional illnesses were identified from isolate uploads to PulseNet. The initial outbreak case definition was an infection with the outbreak strain of Salmonella Kiambu in a person with illness onset on or after 10 April 2017.
Epidemiological investigation
Local and state investigators interviewed case-patients with routine enteric disease questionnaires to gather exposure information. Due to the increasing number of uploads to PulseNet and to gather more detailed exposure information, CDC requested that local and state health departments re-interview case-patients with the national hypothesis generating questionnaire (NHGQ), which includes over 300 questions on various food, animal and other exposures in the week prior to illness onset. Based on initial exposure information received, CDC developed a modified NHGQ to include detailed questions on Hispanic-style foods and eventually a focused questionnaire to collect details for foods of most interest in the outbreak. Food exposures were compared with expected background rates for foods reported by healthy people interviewed in the 2006–2007 FoodNet Population Survey, using the binomial test [20].
Traceback investigation
State and local regulatory agencies collected invoices and records from points of service where case-patients reported eating or purchasing food items of interest, and the FDA conducted traceback investigations to trace these items to their original source. Traceback investigations were focused on illness sub-clusters: locations where two or more unrelated case-patients reported eating or purchasing suspected food items. Traceback investigations involve a thorough review of product distribution records and supporting information to identify or rule out a common supplier or source for the food item of interest across sub-clusters.
Laboratory investigation
State public health laboratories serotyped and subtyped clinical Salmonella isolates by PFGE using standard methodology [18, 19]. State and local public health laboratories and CDC's enteric disease laboratory further characterised a sub-set of clinical isolates by whole-genome sequencing (WGS). Genomic DNA was extracted using a QIAGEN DNeasy Blood and Tissue Kit (Qiagen, Aarhus, Denmark). The DNA libraries were prepared using a Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA) and the DNA sequencing was performed on the Illumina MiSeq Sequencing System using the 2 × 250 base pair sequencing chemistry. High-quality single nucleotide polymorphism (hqSNP) analysis was performed using the Lyve-SET pipeline [21]. Depending on the serotype, either closed PacBio sequences or draft Illumina assemblies were used as references with prophages removed from the analysis. Read mapping was performed using SMALT and SNPs were called using VarScan at >20× coverage, >95% read support and clustered SNPs <5 base pair apart were filtered out.
Local and state investigators collected samples of suspected food items from grocery stores and distribution centres for Salmonella testing. FDA also collected samples of suspected foods at domestic distributors and at import as products were entering the country. State and local public health and regulatory laboratories and FDA laboratories tested these food products for Salmonella and further characterised any Salmonella isolates identified by serotype, PFGE and WGS.
Results
Epidemiological investigation
By 20 July, 18 of 30 (60%) case-patients identified themselves as Hispanic or Latino, and common foods reported from the NHGQ data received from 24 case-patients included mangoes (50%), papayas (42%), cilantro (35%), fresh hot peppers (26%) and sprouts (22%). These frequencies were compared with consumption rates reported by healthy Hispanic people in the 2006–2007 FoodNet Population Survey in the months of May and June in the week before they were interviewed or to consumption rates reported by Hispanic ill people in CDC's NHGQ database, which includes NHGQs collected for cluster cases or sporadic illnesses [20]. The reported frequencies among case-patients for mangoes (50% vs. 34%), papayas (42% vs. 16%) and sprouts (22% vs. 7%) were statistically significantly higher than the expected background rates, while the frequencies for cilantro (35% vs. 48%) and fresh hot peppers (26% vs. 17%) were not significantly higher than expected.
On 5 July, New York City notified CDC of an illness sub-cluster including two ill people who were residents of the same nursing home prior to their illness onsets. One of these ill people was a confirmed case-patient in the outbreak, and the other had reported Salmonella infection with sub-typing pending. The nursing home served papayas and mangoes regularly to the residents. However, we were unable to confirm definitively when or if papayas or mangoes were consumed by either case-patient.
On 13 July, Maryland investigators reported to CDC a second potential illness sub-cluster where five ill people reported shopping at the same international grocery store prior to their illness. Three of these ill people were confirmed case-patients in the outbreak, and two others had reported Salmonella infections with sub-typing not yet completed. These ill people had illness onset dates ranging from 31 May to 26 June. Among three ill people with purchase information, the only common food purchased from this store was papayas.
Traceback investigation
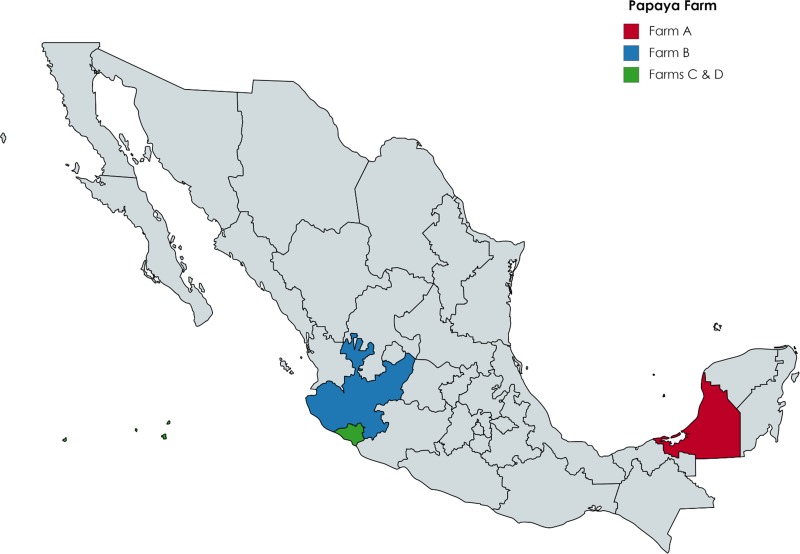
State regulatory agencies and FDA conducted traceback for the papayas purchased and served at the two sub-cluster locations in New York City and Maryland. Investigators found that both locations sold or served fresh Maradol papayas from Mexico, and both received papayas from the same Mexican farm: Farm A (Fig. 1). The investigation did not identify a single shipment of interest. Preliminary traceback documents related to mangoes reportedly purchased by case-patients were also collected; however, traceback for mangoes was not completed once papayas were confirmed as the vehicle based on multiple product samples and traceback convergence.
Fig. 1.
Farm locations of papaya samples yielding Salmonella outbreak strains, by state, Mexico, 2016–2017.
Laboratory investigations
On 12 July, Maryland investigators collected 10 single, whole papayas for Salmonella testing from the sub-cluster store location in Maryland: five yellow Maradol papayas from Mexico and five green papayas from Guatemala. The two papaya varieties were stored in separate bins at the store. Salmonella was isolated from three of the five Mexican Maradol papayas that were sampled, which originated from Farm A in Mexico; no Salmonella was identified in the five green papayas from Guatemala. Four separate Salmonella serotypes were identified: Kiambu, Thompson, Gaminara and Senftenberg. The Salmonella Kiambu strain identified differed from the clinical isolates in the outbreak by PFGE by 2–3 bands; this strain was very rare and had not been seen in the PulseNet database since 2012. The Salmonella Thompson and Salmonella Gaminara strains were both new to the PulseNet database, and the Salmonella Senftenberg strain was very rare.
On 20 and 24 of July, investigators in Virginia collected seven single, whole Maradol papayas from two Virginia grocery store locations that received papayas from the same distributor as the Maryland sub-cluster grocery store location. Salmonella was isolated from five of these papayas, which were all collected from the same store location and originated from Farm A in Mexico. Three strains were identified: the same strain of Salmonella Gaminara that had been identified previously, as well as a strain of Salmonella Agona, and a second variant strain of Salmonella Thompson.
From July through August 2017, FDA collected and tested 13 targeted samples of Maradol papayas as part of the outbreak investigation, including both domestic and import samples. The sampling targeted papaya firms based on preliminary traceback and state laboratory information. In total, there were 185 single, whole Maradol papayas that made up the 13 targeted samples, and each sample consisted of between 10 and 30 single papaya subsamples. Salmonella was isolated from seven of these samples. Five Salmonella strains were identified in four FDA samples of papayas originating from Farm A in Mexico, including the original Salmonella Kiambu outbreak strain found in clinical isolates, as well as the same Salmonella Thompson, Salmonella Agona and Salmonella Senftenberg strains isolated from papayas collected in Maryland and Virginia. FDA testing also isolated two additional strains of Salmonella Senftenberg from Maradol papayas. One strain was very rare, last seen in the PulseNet database in 2011, and the other strain was new to the PulseNet database. All papaya sampling was conducted on the surface of the fruits. Since pulp analysis was not conducted, it is unknown if the contamination migrated to the interior of the fruit.
Investigators in New Jersey and New York collected nine single Maradol papaya samples for testing; none of these yielded Salmonella. In total, Salmonella was isolated from 8 of 26 (31%) single papayas collected by state and local investigators and in seven of 13 targeted papaya samples collected by federal investigators.
From late July to early August, state investigators collected 25 samples of mangoes from retail locations where case-patients reported purchasing them in Maryland (12), New Jersey (7), New York (3) and Virginia (3). No mangoes samples tested yielded Salmonella.
Summary of final outbreak findings
As additional Salmonella strains were isolated from papaya samples, the PulseNet database was queried for recent clinical isolates that were indistinguishable from the identified patterns. Based on laboratory and epidemiological evidence, strains identified in papaya samples originating from Farm A in Mexico were added to the case definition. The final case definition was an infection with one of the identified outbreak strains in a person with illness onset on or after 1 May 2017. In total, we identified 213 outbreak-associated cases of Salmonella Kiambu (50 cases), Salmonella Thompson (142 cases), Salmonella Agona (12 cases), Salmonella Gaminara (7 cases) and Salmonella Senftenberg (2 cases) (Table 1). Case-patients were from 22 states: Connecticut (6), Delaware (4), Florida (1), Georgia (1), Iowa (2), Illinois (6), Kentucky (4), Massachusetts (10), Maryland (11), Michigan (1), Minnesota (4), North Carolina (6), New Jersey (41), New York (70), Ohio (2), Oklahoma (5), Pennsylvania (8), South Carolina (1), Tennessee (2), Texas (9), Virginia (18) and Wisconsin (1).
Table 1.
Number of outbreak-associated clinical and papaya isolates by Mexican farm and Salmonella strain, 2016–2017
| Onset date | Serotype | PFGE pattern | Number of clinical isolates | Number of papaya isolates | |
|---|---|---|---|---|---|
| Farm A | 17 May–20 September 2017 | Agona | JABX01.0258 | 12 | 17 |
| Gaminara | TDJX01.0463 | 7 | 16 | ||
| Kiambu | TENX01.0003 | 50 | 4 | ||
| TENX01.0049 | 0 | 3 | |||
| Senftenberg | JMPX01.0157 | 5 | 15 | ||
| JMPX01.0296 | 0 | 1 | |||
| JMPX01.0526 | 0 | 1 | |||
| Thompson | JP6X01.0757 | 55 | 1 | ||
| JP6X01.0838 | 87 | 6 | |||
| Farm B | 23 July–14 August 2017 | Urbana | JQGX01.0194 | 7 | 6 |
| Farm C | 19 July–7 August 2017 | Infantis | JFXX01.1235 | 1 | 1 |
| Newport | JJPX01.5863 | 3 | 2 | ||
| Farm D | 20 December, 2016–16 August 2017 | Anatum | JAGX01.0013 | 20 | 4 |
| JAGX01.0721 | 0 | 1 | |||
| Unrelated farma | N/A | Abaetetuba | JREX01.0011 | 0 | 1 |
This farm was not associated with any identified illnesses or outbreaks during the investigations described in this paper.
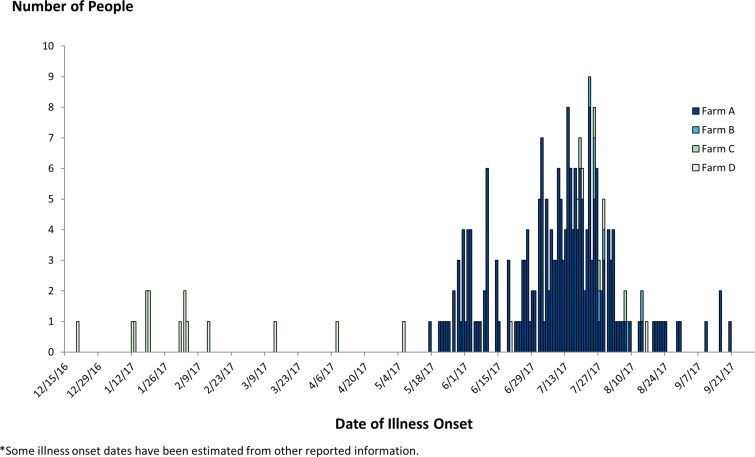
Case-patients ranged in age from <1 to 95 years (median: 40), and 62% were female. Among 169 case-patients with information, 68 (40%) were hospitalised and one died. Illness onset dates ranged from 17 May to 20 September 2017 (Fig. 2). In total, among 168 case-patients with race/ethnicity information, 113 (67%) identified as Hispanic or Latino. Exposure information was available for 143 case-patients, of whom 78 (55%) reported consuming fresh papaya in the week prior to illness. This proportion was significantly higher than results from the 2006–2007 FoodNet Population Survey, where 22% of healthy Hispanic people reported eating papayas in the summer months in the week before they were interviewed [20]. No other food items were reported by case-patients significantly more than expected compared with background consumption rates.
Fig. 2.
People infected with the outbreak strains of Salmonella (n = 244), by date of illness onset and papaya farm, United States, 2016–2017. *Some illness onset dates have been estimated from other reported information.
Identification of additional outbreaks
Among the seven targeted papaya samples collected by FDA which yielded Salmonella, three did not originate from Farm A in Mexico. Four Salmonella strains were identified in these papaya samples, all of which were new or very rare in the PulseNet database.
One rare strain of Salmonella Abaetetuba was identified, which had only been seen in PulseNet once previously from a papaya sample originating from Mexico in 2011. No clinical isolates were identified with this strain. The other three Salmonella strains identified from FDA's targeted testing led to the identification of additional papaya-associated outbreaks of salmonellosis, after recent clinical isolates with these strains were found in PulseNet. Additionally, FDA non-targeted, increased surveillance sampling identified two strains of Salmonella Anatum that were ultimately linked to a previously unsolved outbreak. In total, testing identified 15 strains of Salmonella from papaya samples associated with five different Mexican farms (Table 1).
Outbreak of Salmonella Urbana infections
FDA targeted sampling identified a strain of Salmonella Urbana in papayas from Farm B in Mexico (Fig. 1). This pattern was new to the PulseNet database and had not been seen prior to the summer of 2017. In total, we identified seven Salmonella Urbana outbreak-associated cases from three states: New Jersey (5), New York (1) and Pennsylvania (1). Case-patients ranged in age from <1 to 57 years (median: 1), and 4 (57%) were female. Illness onset dates ranged from 23 July to 14 August 2017. Five (71%) of the seven case-patients were of Hispanic ethnicity. Four (57%) reported being hospitalised and no deaths were reported. Among five case-patients with exposure information, three (60%) reported consuming fresh papaya in the week prior to their illness.
Outbreak of Salmonella Newport and Salmonella Infantis infections
FDA targeted sampling identified strains of Salmonella Newport and Infantis in papayas from Farm C in Mexico (Fig. 1). The strain of Salmonella Newport was very rare and was last seen in PulseNet in 2006, and the Salmonella Infantis strain was new to the database and had not been seen prior to the summer of 2017. In total, we identified four outbreak-associated cases of Salmonella Newport (3 cases) and Salmonella Infantis (1 case) from 4 states: Illinois (1), Massachusetts (1), Michigan (1) and New York (1). Case-patients ranged in age from 40 to 82 years (median: 63), and 2 (50%) were female. Two (50%) of the four case-patients were hospitalised and no deaths were reported. Illness onset dates ranged from 19 July to 7 August 2017. Two (50%) of the case-patients were of Hispanic ethnicity. Among three case-patients with exposure information, all three (100%) reported consuming fresh papaya in the week prior to their illness.
Outbreak of Salmonella Anatum infections
In August, Salmonella Anatum was identified by FDA in a non-targeted, increased surveillance sample from an imported papaya from Farm D in Mexico (Fig. 1); the collection of this sample was not directly related to the previously described outbreak investigation. This strain of Salmonella was indistinguishable from an unsolved Salmonella Anatum outbreak from the spring of 2017, in which papayas had been the suspected source of the outbreak at the time based on epidemiological evidence. While FDA determined that 16 different papaya suppliers in Mexico could have accounted for product purchased by several case-patients in this outbreak, the traceback investigation was limited due to the lack of sub-clusters. As the traceback findings could not identify a specific supplier due to limited papaya exposure information and limited lot code documentation throughout the distribution chain, no conclusions or actionable findings could be drawn; therefore, the investigation for this Salmonella Anatum outbreak ended in May of 2017.
After the product testing results identified the outbreak strain, the investigation was re-opened and additional related illnesses were identified. In total, we identified 20 cases of Salmonella Anatum from three states: Arizona (1), California (18) and Colorado (1). Case-patients ranged in age from <1 to 89 years (median: 43), and 85% were female. Among 11 case-patients with available information, five (45%) were hospitalised. One death was reported from California. Illness onset dates ranged from 20 December 2016 to 16 August 2017. Among 11 case-patients with information, 10 (91%) were of Hispanic ethnicity. Ten (91%) of 11 case-patients interviewed reported eating papayas in the week prior to their illness.
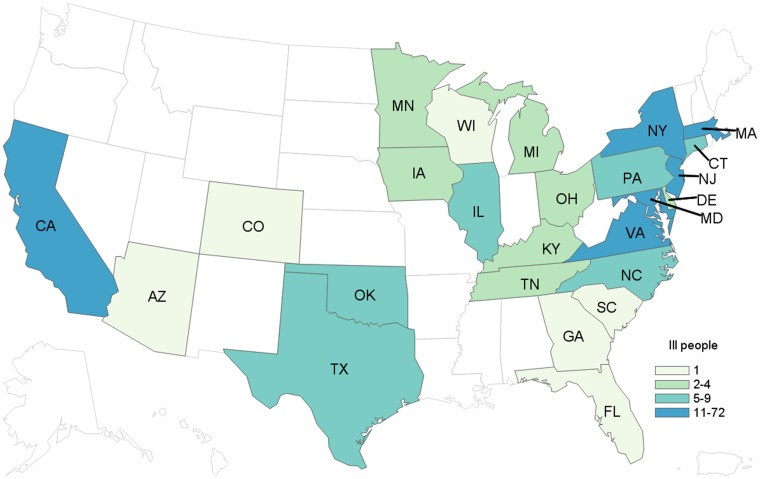
In total, we identified 244 cases from 25 states across all four outbreaks, with illness onset dates ranging from 20 December 2016 to 20 September 2017 (Fig. 3).
Fig. 3.
People infected with the outbreak strains of Salmonella (n = 244), by state of residence, United States, 2016–2017.
Whole-genome sequencing
WGS was completed on 191 clinical and 48 food isolates representing each of the identified PFGE patterns. Among each of the PFGE patterns, WGS analysis showed clinical isolates to be closely related genetically to food isolates. For most strains, there were less than four hqSNP differences between isolates; for strains serotyped as Salmonella Thompson, the difference was between 0 and 13 hqSNPs. All PFGE patterns involved in these outbreaks were either novel to the PulseNet database or identified previously only rarely (suggesting they were all very rare).
Control measures
During the outbreak, three firms issued recalls for three brands of Maradol papayas sourced from Farm A in Mexico on 26 July, 5 August and 9 August. Products from Farms B and C were not recalled because the shipments associated with Salmonella-contaminated papayas were destroyed, and the previous shipments from these firms were past shelf-life by the time sample results were available. On 10 September 2017, one firm recalled Maradol papayas from Farm D in Mexico, since contaminated papayas were still on the market.
At the time the investigation began, all firms associated with papaya samples which yielded Salmonella were on Import Alert 21-17's ‘green list,’ meaning they had previously satisfied the requirements to be removed from the import alert. During the investigation, these firms were removed from the ‘green list’. Additionally, firms implicated in these outbreaks were added to the ‘red list’ for a different import alert, updated in 2017. This alert calls for the detention of fresh produce that appeared to have been prepared, packed or held under insanitary conditions and is updated as needed [22]. Firms placed on red-list status must submit sufficient evidence of corrective actions to FDA to be removed from this list [22].
Discussion
Four concurrent multistate outbreaks of Salmonella infections were linked to imported Maradol papayas from four geographically dispersed Mexican farms in 2017. This was the first time papayas have been implicated as a multistate outbreak vehicle since the 2011 Salmonella Agona outbreak, and the first time four papaya-associated outbreaks were identified in 1 year, one of which was the largest papaya outbreak identified to date in the United States. The identification of multiple outbreaks and multiple implicated papaya farms highlights the potential for local contamination of an imported produce product like papayas to lead to widespread, disseminated illness in the United States. Collaborative work of local, state and federal public health and regulatory partners was instrumental in identifying the source of these outbreaks and preventing additional illnesses. These outbreak investigations demonstrate that industry vigilance and dedication to providing a safe product must be maintained long after issues have been identified and addressed, and that FDA import alerts and initial microbial testing alone are not always enough to prevent outbreaks from recurring. This is particularly relevant considering that there was not a sole supplier linked to all outbreaks.
Quick and targeted sampling of suspected foods during outbreak investigations, based on identifying common foods and sub-cluster locations, can provide crucial clues to identifying outbreak sources. Early in the Salmonella Kiambu investigation, interview data alone were not sufficient to identify a food item responsible for the outbreak, but a common retail sub-cluster was identified. By isolating the outbreak strain from targeted retail sampling of papayas, investigators were able to confirm papayas were the source of the illnesses. In 2017, sampling of papayas was essential for solving the originally identified cluster of Salmonella Kiambu infections and in identifying additional Salmonella strains and outbreaks. In the United States, we have a highly trained network of foodborne outbreak investigators around the country who are able to rapidly respond to foodborne outbreak investigations. Programmes such as FDA's Rapid Response Teams and CDC's OutbreakNet Enhanced programme are designed to support the ability of local and state health and regulatory agencies to rapidly respond to foodborne outbreaks [23, 24]. It is important to maintain and expand such programmes to strengthen our ability to respond to foodborne outbreaks around the country.
These outbreaks are examples of complicated investigations where multiple, cross-cutting investigation tools and strong collaboration are necessary to identify the source. Traditional epidemiological tools, such as the NHGQ, proved ineffective at identifying a source among multiple suspected food vehicles early in the investigation. During early hypothesis generation the percentage of case-patients reporting consumption of mangoes was slightly higher than the percentage reporting papaya consumption (50% vs. 42%), and both were higher than expected background rates for those foods, making it difficult to identify which food was most likely to be the source of the outbreak. Additionally, there are often higher overall consumption rates among case-patients for implicated produce items during multistate Salmonella outbreaks, in recent years ranging from 69% to 89% for outbreaks associated with cantaloupe, cucumbers and mangoes compared with 57% for the 2011 Salmonella Agona outbreak associated with papayas and 55% for the outbreak described in this paper, suggesting that papayas may be a food item that is more difficult to identify through epidemiological investigations than other produce items [10, 11, 13–15]. Smaller overall consumption rates for suspected food items and having multiple food items reported above expected background levels limit the ability of investigators to develop hypotheses based on early epidemiological data. Rather than relying solely on early epidemiological data to inform hypothesis generation, it is essential to evaluate epidemiological data along with laboratory and traceback data to determine the source of outbreaks.
Additionally, it would be beneficial to develop predictive epidemiological tools that could assist with identifying potential outbreak vehicles early in investigations. For example, a tool that could generate a list of potential vehicles based on early case-patient demographic information, such as race, ethnicity and gender, could help narrow the focus of an epidemiological investigation. One such pilot model has been developed using retrospective data for Shiga toxin-producing Escherichia coli outbreaks in the United States, which demonstrated the feasibility of using case demographic and outbreak characteristics to assist in predicting food sources [25]. However, more work is needed to develop and validate a tool that can be used with data that is readily available early in an investigation and adapt it for use with other pathogens, such as Salmonella [25]. Additionally, information on food import and distribution patterns within the United States coupled with geographical information for early outbreak case-patients could inform hypothesis generation. An analysis of historical import data for five produce-associated outbreaks demonstrated that the geographical distribution of illnesses differed between produce vehicles which were grown domestically compared with those imported into the United States from Mexico and Central America. Investigators concluded that the geospatial distribution of early illnesses might be useful in identifying suspected ports of entry for produce-associated outbreaks [26]. Targeted sampling of imported produce items from ports of entry, coupled with focused traceback investigations might accelerate the identification of outbreak vehicles during multistate outbreak investigations.
These four outbreaks highlight important steps investigators can take now and in the future to more rapidly and effectively respond to multistate foodborne outbreaks. Maintaining and expanding capacity building programmes around the country will assist investigators to more quickly solve future outbreaks. It may also be useful to increase targeted product testing during outbreak investigations to help find the source of illnesses, such as at points of services of interest identified through early epidemiological investigations and at certain ports of entry based on early case-patient locations. Novel hypothesis generation tools could also be developed to assist investigators in finding signals early in data collection, such as predictive models which generate potential food vehicles based on available early demographic and geographical data. The strengthening of current investigational tools along with the development of new tools could help in the evaluation of epidemiological, laboratory and traceback evidence together during investigations, to more quickly solve and stop outbreaks.
Acknowledgements
We thank public health, environmental health, laboratory and regulatory officials in the following jurisdictions: Arizona, California, Colorado, Connecticut, Delaware, Florida, Georgia, Iowa, Illinois, Kentucky, Los Angeles County, Massachusetts, Maryland, Michigan, Minnesota, North Carolina, New Jersey, New York City, New York, Ohio, Oklahoma, Pennsylvania, South Carolina, Tennessee, Texas, Virginia and Wisconsin.
Conflict of interest
The authors declare no conflicts of interest.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
References
- 1.Scallan E et al. (2011) Foodborne illness acquired in the United States–unspecified agents. Emerging Infectious Diseases 17, 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Salmonella: Questions and Answers. March 5, 2018 [cited June 21, 2018]; Available at https://www.cdc.gov/salmonella/general/index.html.
- 3.Centers for Disease Control and Prevention. Standard operating Procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. Available at https://www.cdc.gov/pulsenet/pdf/ecoli-shigella-salmonella-pfge-protocol-508c.pdf (Accessed 13 July 2018).
- 4.Ribot EM and Hise KB (2016) Future challenges for tracking foodborne diseases: PulseNet, a 20-year-old US surveillance system for foodborne diseases, is expanding both globally and technologically. EMBO Reports 17, 1499–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sivapalasingam S et al. (2004) Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. Journal of Food Protection 67, 2342–2353. [DOI] [PubMed] [Google Scholar]
- 6.Bennett SD et al. (2018) Produce-associated foodborne disease outbreaks, USA, 1998–2013. Epidemiology and Infection 146, 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callejón R et al. (2015) Reported foodborne outbreaks due to fresh produce in the United States and European Union: trends and causes. Foodborne Pathogens and Disease 12, 32–38. [DOI] [PubMed] [Google Scholar]
- 8.Hanning I, Nutt JD and Ricke S (2009) Salmonellosis outbreaks in the United States due to fresh produce: sources and potential intervention measures. Foodborne Pathogens and Disease 6, 635–648. [DOI] [PubMed] [Google Scholar]
- 9.United States Department of Agriculture Economic Research Service. Fruit & Tree Nuts. Available at https://www.ers.usda.gov/topics/crops/fruit-tree-nuts/ (Accessed 22 June 2018).
- 10.United States Department of Agriculture Economic Research Service. Vegetables & Pulses. Available at https://www.ers.usda.gov/topics/crops/vegetables-pulses/ (Accessed 22 June 2018).
- 11.Barton Behravesh C et al. (2011) 2008 outbreak of Salmonella Saintpaul infections associated with raw produce. The New England Journal of Medicine 364, 918–927. [DOI] [PubMed] [Google Scholar]
- 12.Mba Jonas A et al. (2018) A multistate outbreak of human Salmonella Agona infections associated with consumption of fresh, whole papayas imported from Mexico-United States, 2011. Clinical Infectious Diseases 66, 1756–1761. [DOI] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. Multistate outbreak of Salmonella Braenderup Infections Associated with Mangoes (Final Update). Available at https://www.cdc.gov/salmonella/braenderup-08-12/index.html (Accessed 22 June 2018).
- 14.Centers for Disease Control and Prevention. Multistate Outbreak of Salmonella Litchfield Infections Linked to Cantaloupes (Final Update). Available at https://www.cdc.gov/salmonella/2008/cantaloupes-4-2-2008.html (Accessed 22 June 2018).
- 15.Centers for Disease Control and Prevention. Multistate Outbreak of I Panama Infections Linked to Cantaloupe (Final Update). Available at https://www.cdc.gov/salmonella/2011/cantaloupes-6-23-2011.html (Accessed 22 June 2018).
- 16.Centers for Disease Control and Prevention. Multistate Outbreak of I Poona Infections Linked to Imported Cucumbers (Final Update). Available at https://www.cdc.gov/salmonella/poona-09-15/index.html (Accessed 22 June 2018).
- 17.Centers for Disease Control and Prevention. Multistate Outbreak of Salmonella Saintpaul Infections Linked to Imported Cucumbers (Final Update). Available at https://www.cdc.gov/salmonella/saintpaul-04-13/index.html (Accessed 22 June 2018).
- 18.Hassan R et al. (2017) Multistate outbreak of Salmonella Anatum infections linked to imported hot peppers – United States, May–July 2016. Morbidity and Mortality Weekly Report 66, 663–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.U.S. Food & Drug Administration. Import Alert 21-17. Available at https://www.accessdata.fda.gov/cms_ia/importalert_721.html (Accessed 26 June 2018).
- 20.Centers for Disease Control and Prevention. Foodborne Active Surveillance Network (FoodNet) Population Survey Atlas of Exposures. 2006–2007: Atlanta, GA.
- 21.Katz L et al. (2017) A comparative analysis of the Lyve-SET phylogenomics pipeline for genomic epidemiology of foodborne pathogens. Frontiers in Microbiology 8, 375–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.U.S. Food and Drug Administration. Import Alert 99-35. Available at https://www.accessdata.fda.gov/cms_ia/importalert_1128.html (Accessed 29 June 2018).
- 23.U.S. Food and Drug Administration. Rapid Response Teams (RRTs). Available at https://www.fda.gov/forfederalstateandlocalofficials/programsinitiatives/ucm475021.htm (Accessed 13 September 2018).
- 24.Centers for Disease Control and Prevention. OutbreakNet Enhanced. Available at https://www.cdc.gov/foodsafety/outbreaknetenhanced/index.html (Accessed 13 September 2018).
- 25.White A et al. (2016) Food source prediction of Shiga toxin-producing Escherichia coli outbreaks using demographic and outbreak characteristics, United States, 1998–2014. Foodborne Pathogens and Disease 13, 527–534. [DOI] [PubMed] [Google Scholar]
- 26.Chen Parker C et al. (2017) Geospatial mapping of early cases in multistate foodborne disease outbreaks: a strategy to expedite identification of contaminated imported produce, United States, 2006 to 2013. Journal of Food Protection 80, 1821–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]